Abstract
Purpose:
To determine the presence of herpes simplex virus and varicella zoster virus (HSV 1 and 2, VZV) in the cornea of normal subjects by multiplex real time quantitative (qPCR) assay and evaluate its utility in the diagnosis of viral keratitis.
Methods:
Corneal epithelial cells from 33 eyes of 22 patients undergoing photorefractive keratectomy surgery (controls) and 50 corneal scrapings from 50 patients with suspected HSV keratitis were analyzed for the presence of HSV1 by conventional PCR and for presence of HSV1 and 2 and/or VZV by multiplex real-time PCR. Corneal scrapings of patients were also tested for HSV1 antigen by immunofluorescence assay (IFA). The results were compared and clinical records reviewed.
Results:
HSV1 and VZV DNA were detected in 8/33 controls (mean-14.3 ± 7.96, range: 3-29.1 copies/mL) and 2/33 controls (mean-10.7 ± 10.9, range 3-18.5 copies/ml) respectively. HSV2 was not detected in any of the controls. Copy numbers above the mean + 1SD of controls were considered significant for viral load in patient samples. Significantly higher number of corneal scrapings (39/50, 78%) from patients were positive for HSV1 (1.2 × 106 copies/mL ± 3.7 × 106 copies/mL) by real time qPCR compared to IFA (11/48, 23%, P value 0.0001) and conventional PCR (20/50, 40%, P value 0.0002). Double infection with HSV-1 (1.5 × 107 copies/ml) and HSV-2 (3.57 × 104 copies/ml) in one case and VZV infection (1.03 × 102 copies/ml) in another was also detected by the multiplex real-time PCR.
Conclusion:
Multiplex real-time PCR reliably detects HSV1 and 2 and VZV DNA and is ideal for the diagnosis of HSV and VZV keratitis in an ocular microbiology laboratory.
Keywords: HSV 1, HSV 2, keratitis, real-time PCR, VZV
Herpes simplex viruses 1 and 2 (HSV-1 and -2) and varicella-zoster virus (VZV) are human double-stranded DNA viruses belonging to the Herpesviridae family[1] and infections caused by these viruses are common throughout the world, with considerable variation within population groups. Among these, only HSV-1 keratitis has been reported to be an important cause of ocular morbidity.[2] HSV-1 is reported to be involved in a number of clinical manifestations ranging from blepharitis, acute infectious epithelial keratitis to the potentially blinding chronic stromal keratitis.[3] Frequent recurrences followed by immunological responses may lead to corneal scarring, thinning, neovascularization[4,5,6] and consequently the patient may need to undergo keratoplasty.[7] At present, the diagnosis of herpes simplex virus keratitis (HSK) is primarily dependent on the clinical opinion of the ophthalmologist. A study by Rübben et al.[8] reported that VZV was misdiagnosed as HSV-1 in 8% of patients. Uncommon causes of viral keratitis also include VZV, adenovirus, enterovirus and CMV.[9,10,11] HSK can be diagnosed by multiple laboratory tests, of which the gold standard is the isolation of HSV from the cornea; however, this technique is time consuming and possesses low level of sensitivity. Another technique is detection of intranuclear inclusions and multinucleated giant cells or fluorescence based immunological detection of HSV-1 antigen,[12] which is not sensitive enough, and additional molecular tests are important to avoid inappropriate diagnosis of HSK and enable appropriate treatment. Polymerase chain reaction (PCR) is however not without its own inherent problems, large variations were observed in various studies between the rate of HSV-1 detection by PCR and the clinical diagnosis[13,14] in addition to it being laborious and prone to contamination. Thus, real-time polymerase chain reaction or quantitative PCR (qPCR), is now being developed as an alternative approach in the diagnosis of viral keratitis. Along with being more rapid and sensitive, qPCR overcomes the drawbacks of conventional PCR by reducing the risk for carry-over contamination and eliminates the time-consuming detection step.[14] The purpose of this study was to determine the efficacy and feasibility of a commercial multiplex Real-Time PCR Assay for detection and quantitation of herpes simplex virus 1 and 2 and varicella-zoster virus DNA in corneal scrapings of patients diagnosed with viral keratitis and compare the results with Immunofluorescence assay (IFA) and in-house PCR for herpes simplex virus 1.
Methods
Clinical samples
All patients diagnosed to have microbial keratitis of possible HSV etiology, seen at the Institute, over the period June 2016 – October 2016, were included in this study. It was a retrospective study and was approved by institutional review board (LEC 08-16-064) and it adhered to the tenets of the Declaration of Helsinki. Data collected included demographic details, clinical and microbiology data. The clinical features included dense infiltration of stroma with an overlying epithelial defect with or without ghost scars and deep vessels. Patients also had single or multiple grey/white creamy homogenous abscesses with edema with or without associated keratic precipitates and severe iridocyclitis with hypopyon.
Whenever there was equivocal clinical presentation, the patients were subjected to complete microbiological investigations as described earlier[15] In cases, where strong viral etiology was suspected, only two corneal scrapings were collected under topical anaesthesia with a sterile blade number 15 on Bard Parker handle for IFA and PCR. Additionally, corneal epithelial cells were collected in 0.5 mL of PBS (pH 7.2) from 33 eyes of 22 patients undergoing photorefractive keratectomy surgery in essentially normal eyes (control group) and stored at 4°C until processed for PCR.
Immunofluorescence assay
Immunofluorescence assay (IFA) was done as described earlier.[12] Rabbit anti HSV-1 polyclonal antibody (DAKO, Denmark) was used as primary antibody on fixed smears. A positive control of Vero cell line infected with HSV 1 was used for each batch of test.
Nucleic acid extraction
DNA was extracted from 200 μL of PBS containing corneal scraping/epithelial cells from both patient samples and controls using spin-column based QIAamp Mini Kit (Germany), according to the manufacturer's instructors.
In-house conventional polymerase chain reaction
The target gene for the in-house conventional PCR was HSV-1 glycoprotein D gene with primer positions being F: 19–43 and R: 218–239. The primer sequences were Forward (5′-CACGGTAGCCCGGCCGTGTGTGACA) and Reverse (5′- CATACCGGAACGCACCACACAA). The PCR conditions were as described in an earlier publication.[12] The positive control DNA was obtained from HSV-1 ATCC, VR-539, USA, grown and maintained in Vero cell line.
Real Time Quantitative PCR (qPCR)
While performing conventional PCR, a portion of the DNA (patients and controls) was stored at −20°C and tested later for qPCR. The presence of HSV 1 and 2 and/or VZV by real-time qPCR (R-gene® kit, Argene/bioMérieux) according to the manufacturer's instructions on the Applied Biosystems Real Time 7900 instrument (ABI7900HT, Applied Biosystems, CA, USA) programmed for a three-step protocol: 15 minutes at 95°C for polymerase activation for one cycle, 10 seconds at 95°C for denaturation, 40 seconds at 60°C for annealing, extension for 45 cycles and data collection. The target gene was HSV-1: US7 gene HSV-2: US2 gene VZV: gene coding for gp19 (ORF 17). The quantification standard for HSV-1, HSV-2, and VZV was linear between 50 copies/mL to 5 × 105 copies/mL and Creating standard curves wherein the limit of detection determined was HSV-1: 2 Copies/PCR HSV-2: 2 Copies/PCR VZV: 2 Copies/PCR. The results were expressed in number of copies/mL of sample. The results were validated with the internal extraction control, quantification standards and water for negative controls provided in the HSV1/HSV2 VZV R-gene® kit. Valid results were reported quantitatively (1 U/ml) as “positive” if the value obtained was between the limit of quanitation _(LOQ) and the limit of detection (LOD).
Statistical analysis
All data were analyzed by the Student's t- test or χ2 test. Results were considered statistically significant at P < 0.05.
Results
Corneal scrapings from 50 eyes of 50 consecutive patients with suspected viral keratitis and corneal epithelial cells from 33 eyes of 22 normal individuals were included in the study. The patients included 19 (38%) women and 31 (62%) men, with a median age of 42.16 (range: 1 to 82 years). In addition, 48/50 corneal scrapings were also tested for HSV-1 antigen by IFA while all 50 were tested by both conventional and qPCR [Table 1]. All 33 samples collected from normal individuals were also tested by conventional PCR for HSV 1 and for detection of HSV-1 and 2/VZV DNA by qPCR.
Table 1.
Details of results of IFA and PCR for HSV-1 and qPCR results (HSV 1&2 and VZV) of the 50 patients included in the study
| IFA | PCR | qHSV1 Copies/mL | qHSV2 Copies/mL | VZV Copies/mL |
|---|---|---|---|---|
| Pos | Pos | 2.55×106 | ||
| Neg | Neg | |||
| Neg | Neg | |||
| Neg | Neg | |||
| Neg | Neg | 76.7 | ||
| Neg | Neg | |||
| Neg | Neg | |||
| Neg | Neg | 1.38×107 | ||
| Neg | Neg | 179 | ||
| Neg | Neg | 96.0 | ||
| Neg | Neg | 103 | ||
| Neg | Pos | 2.26×103 | ||
| Neg | Neg | |||
| Neg | Neg | |||
| Neg | Neg | 120 | ||
| Neg | Neg | 6.5×105 | ||
| Pos | Pos | 1.25×107 | ||
| Pos | Pos | 240 | ||
| Pos | Pos | 189 | ||
| ND | Pos | 3.49×105 | ||
| ND | Pos | 7.57×103 | ||
| Neg | Neg | 8.00×104 | ||
| Neg | Neg | 63.1 | ||
| Neg | Pos | 208 | ||
| Neg | Pos | 256 | ||
| Neg | Neg | 182 | ||
| Neg | Neg | 3.52×103 | ||
| Neg | Neg | 8.63×103 | ||
| Neg | Neg | 131 | ||
| Pos | Pos | 50.5 | ||
| Neg | Neg | - | - | |
| Neg | Neg | 155 | ||
| Neg | Neg | 5.93×103 | ||
| Pos | Pos | 4.42×104 | ||
| Pos | Pos | 5.85×104 | ||
| Neg | Pos | 99.0 | ||
| Neg | Neg | 203 | ||
| Neg | Pos | 6.15×103 | ||
| Neg | Pos | 4.30×103 | ||
| Pos | Pos | 1.86×105 | ||
| Pos | Pos | 1.55×107 | 3.57×104 | |
| Neg | Pos | 4.29×103 | ||
| Neg | Neg | |||
| Neg | Neg | 39.7 | ||
| Neg | Neg | 9.00 x104 | ||
| Neg | Neg | 7.69 x103 | ||
| Pos | Pos | 8.81 x103 | ||
| Neg | Neg | |||
| Neg | Neg | 8.16×104 | ||
| Pos | Pos | 619 |
Legend: Pos: positive; Neg: Negative; ND not done
The quantification of DNA from corneal scrapings was performed using a standard curve of known HSV1 and 2 as well as VZV DNA concentrations provided in the kit. The data revealed a correct discrimination between HSV-1 and 2 and VZV as well as a comparable sensitivity for both virus types for the triplex real-time PCR. While all 33 samples from normal individuals were negative for HSV-1 DNA by conventional PCR, real time qPCR showed presence of HSV-1 DNA in 8/33 (mean-14.3 ± 7.96, range: 3-29.1 copies/mL) and VZV DNA was detected in 2/33 (VZV, mean-10.7 ± 10.9, range 3-18.5 copies/ml). HSV-2 however was not detected in any of the control samples. Thus we considered that copy numbers above the mean + 2 SD of controls i.e. >=31 copies/mL to be significant or indicative of HSV1 and VZV infection in patient samples.
In patient samples, 39/50 samples (78%, 95% CI 0.66-0.89) were positive for HSV-1 by qPCR and the virus load of the HSV-1 positive samples from all clinical specimens varied over a wide range, showing threshold cycles between 18 and 41 (corresponding to copy numbers 39.7-1.5 × 107 HSV copies/mL). Comparatively, only 20/50 test samples (40%, 95% CI 0.26– 0.54) showed presence of HSV-1 DNA by conventional PCR and 11/48 samples, (23%, 95% CI 0.11-0.35) showed presence of HSV-1 antigen by IFA as depicted in Fig. 1.
Figure 1.
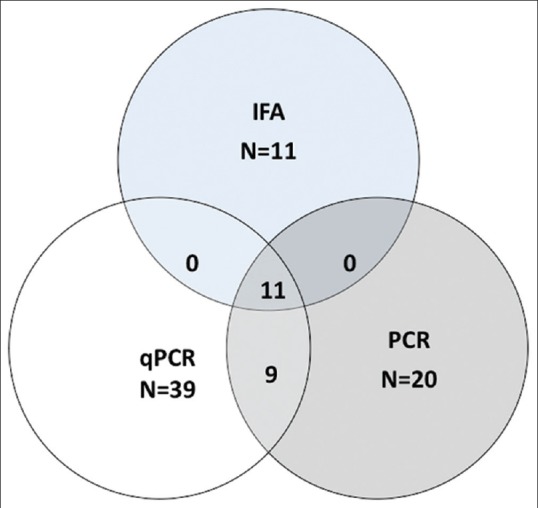
Venn diagram depicting HSV-1 keratitis diagnosis by PCR, IFA and qPCR (real time qPCR) in the 50 patients with keratitis included in the study
All DNA samples positive for HSV 1 by conventional PCR and IFA were also positive by qPCR [Fig. 1] however, the sample with the lowest viral load (39.7 copies/mL) that detected by qPCR was not diagnosed by the conventional PCR. Thus the efficacy of real time qPCR was significantly higher to conventional PCR (P = 0.0002) and antigen detection (P = 0.0001) in patients with keratitis. Comparing the different investigative methods and taking IFA as gold standard the sensitivity and specificity of PCR was 100% and 76.9% respectively and for qPCR was 100% and 28.2% respectively. Additionally, one patient sample (#11) was positive for VZV DNA (1.03 × 102/mL) [Table 1] while another sample (#41) showed a double infection with both HSV-1 (1.5 × 107/mL) and 2 (3.57 × 104/mL) by real-time qPCR assay. Table 1 shows the results of qPCR, IFA and conventional PCR for all patient samples included in the study.
The qPCR data of 39 patients positive for HSV-1 DNA were analyzed against the clinical records to determine possible associations between the viral DNA load in corneas and the clinical parameters at the time of presentation. The results are shown in Table 2. It showed that the reduced vision was the most common complaint reported at presentation (100%), and the median duration of symptoms was 25 (range 3–365) days. The pre-disposing factors included injury with dust particle, stick or insect in 10 patients while four patients reported a history of previous episode of HSV keratitis while one patient had systemic herpes zoster infection. At ophthalmic examination, a clinical diagnosis of HSV epithelial keratitis was made in 7 patients, and HSV stromal disease in 19 patients, while one had both HSV related epithelial and stromal disease. All of these patients received antivirals in the form of acyclovir eye ointment or tablet 400 mg. Additionally neurotrophic keratopathy was diagnosed in two cases and in 11 cases the causative agent was not identified as HSV-1 by conventional PCR and hence the patients were not started on antivirals. The co-infections with bacteria and fungus was observed in six and four patients respectively, following which the patient was started on antibacterials and/or antifungals [Table 2].
Table 2.
Clinical Features and viral load in the corneal scraping of patients diagnosed as HSV keratitis by real time qPCR
| S. No. (Patient. #) | Pre disposing factor | Initial VA | Clinical Diagnosis | Treatment | Final VA | PCR | q HSV-1 copies/mL | Microbiology culture |
|---|---|---|---|---|---|---|---|---|
| 1 (1) | Contact lens | HM+ | HSV stro Stromal Keratitis | ACV + Mx + At | 20/200p | + | 2.55x10^6 | NG |
| 2 (5) | Injury (stick) | HM+ | NNeurotrophic keratitis | Cz + Cip + At | HM+ | 76.7 | S. epidermidis | |
| 3 (8) | Injury | 20/60 | Epithelial Keratitis | ACV + Pf | 20/40 | - | 1.38x10^7 | - |
| 4 (9) | Injury | CFCF | Infectious KERATITIS | Mx + At + Pf | 20/125p | - | 179 | NG |
| 5 (10) | injury | 20/40 | Infectious KERATITIS | Nat + At | LFU | - | 96 | S.epidermidis Microbacterium sp. |
| 6 (12) | - | 20/80 | Epithelial Keratitis | ACV | 20/20 | + | 2.26×10^3 | ND |
| 7 (15) | h/o HERPES ZOSTER infection | HM+ | Neurotrophic keratitis | Cz + ACV + Cip | HM+ | - | 1.20×10^2 | S.aureus |
| 8 (16) | - | HM+ | Epithelial Keratitis | ACV + At | HM+ | - | 6.52×10^5 | NG |
| 9(17) | - | FFL | Epithelial Keratitis | ACV | FFL | + | 1.25×10^7 | NG |
| 10 (18) | Injury (stick) | 20/800 | Infectious KERATITIS | Nat + KTZ + Ch | 20/50 | + | 2.40×10^2 | UID |
| 11 (19) | injury (dust) | 20/60p | Infectious KERATITIS | Cz + At + Cip | 20/50p | + | 189 | P.acnes |
| 12 (20) | - | 20/600 | Epithelial Keratitis | Cz + ACV + Cip + Mx | PK | + | 3.49×10^5 | ND |
| 13 (21) | Injury | HM+ | Infectious KERATITIS | Cz + Mx | PK | + | 7.57×10^3 | NG |
| 14 (22) | - | 20/400 | stromal Keratitis | ACV | CFCF | 8.00×10^4 | NG | |
| 15 (23) | HSV -reactivation | CFCF | Epithelial Keratitis | ACV + Cip + Cz | 20/25 (pinhole) | - | 63.1 | Sphingomonas paucimobilis S.epidermidis |
| 16 (24) | h/o jaundice | CF 2m | Infectious KERATITIS | Cz + Cip | 20/60p (unaided) | + | 2.08×10^2 | S.pneumoniae |
| 17 (25) | HSV -reactivation | 20/60p | stromal keratitis | ACV | 20/60p | + | 2.56×10^2 | NG |
| 18 (26) | - | HM+ | Microbial Keratitis | Cz + Cip + PHMB + Chlorhexidine | HM+ | - | 1.82×10^2 | Acanthamoeba species |
| 19 (27) | - | CF 1m | stromal keratitis | Cz + ACV + Cip | PK | - | 3.52×10^3 | Sphingomonas paucimobilis |
| 20(28) | - | CFCF | stromal keratitis | Cz + ACV | 20/80p | - | 8.63×10^3 | NG |
| 21 (29) | - | HM+ | Fungal KERATITIS | Nat | 20/20 | - | 1.31×10^2 | Fusarium solani |
| 22 (30) | HSV -reactivation | 20/125p | stromal keratitis | ACV + Ch | 20/40 | + | 50.5 | ND |
| 23 (32) | - | HM+ | MICROBIAL KERATITIS | Cz + Cip | HM+ | - | 1.55×10^2 | NG |
| 24 (33) | injury(sugarcane) | CF1m | HSV -Stromal Keratitis | ACV + Ch | 20/40p | - | 5.93×10^3 | NG |
| 25 (34) | - | 20/500 | HSV stromal keratitis | Cz + ACV + Cip | 20/400 | + | 4.42×10^4 | Sphingomonas paucimobilis |
| 26 (35) | - | 20/100 | HSV stromal keratitis | ACV + Mx | 20/80 | + | 5.85×10^4 | Sphingomonas paucimobilis |
| 27 (36) | - | CFCF | HSV -Stromal Keratitis | Cz + ACV + Cip + Ch | 20/200 | + | 99.1 | Staphylococcus haemolyticus |
| 28 (37) | - | PL+PR+ | MICROBIAL KERATITIS | Ch | PL + PR Accurate | - | 2.03×10^2 | NG |
| 29 (38) | HSV -reactivation | PL+PR + | HSV stromal keratitis | ACV + Ch | HM+ | + | 6.15×10^3 | NG |
| 30 (39) | - | CFCF | HSV stromal keratitis | ACV + Ch | PK | + | 4.30×10^3 | NG |
| 31 (40) | - | CF 1m | HSV Stromal Keratitis | Mx + ACV + Pf | 20/320 | + | 1.86×10^5 | NG |
| 32 (41) | Injury (stick) | HM+ | HSV Stromal Keratitis | Mx + ACV + Pf | 20/600 | + | 1.55×10^7 | Methylobacterium sp. |
| 33 (42) | - | CFCF | HSV Stromal Keratitis | Cz + ACV + Cip | PL+PR Accurate | + | 4.29×10^3 | NG |
| 34 (44) | - | HM+ | MICROBIAL KERATITIS | At + Ch + Pf | HM+ | - | 39.7 | NG |
| 35 (45) | post-PK | HM+ | HSV Epithelial Keratitis | Cz + At + Cip | LFU | - | 9.00×10^4 | NG |
| 36 (46) | - | PL+PR- | HSV Stromal Keratitis w/o sec fungal | Cz + ACV + Cip + KTZ + Nat | PK | - | 7.69×10^3 | Alternaria species |
| 37 (47) | - | CF 1m | Fungal KERATITIS | Nat | CF 1m | + | 8.81×10^3 | (fungal filaments on smear) NG |
| 38 (49) | - | CFCF | HSV stromal and epithelial keratitis | ACV + Ch | 20/125 | - | 8.16×10^4 | NG |
| 39 (50) | HM | HSV Stromal Keratitis | Ch + ACV + KTZ + Nat | LFU | + | 6.19×10^2 | Curvulariasp. |
Legend: HM+: Hand Movements; FFL: Following and fixing light; CFCF: Counting fingers close to face; CF: Counting fingers; PL+PR-: Accurate Projection of Light, Inaccurate projection of rays; PL+PR+: Accurate Projection of Light, accurate projection of rays, NG: No growth; ACV: Acyclovir; Ch: Chloramphenicol, Mx: Moxifloxacin; Nat: natamycin; KTZ: Ketoconazole; Cip: Ciprofloxacin; At: Atropine; Pf: Predforte; PHMB: Polyhexa methylene biguanide
In 11/39 patients that showed presence of HSV-1/2/VZV DNA, the visual outcome was 20/200 or worse. Additionally, 3/39 patients needed penetrating keratoplasty and two were lost to follow-up. The one case that showed positivity to presence of VZV DNA (patients # 11) was clinically diagnosed as Acanthamoeba keratitis and since Acanthamoeba grew in culture the patient did not receive any antivirals. This could explain why his vision improved from light perception to merely 20/200 on anti-acanthamoeba medications. Patient (#41) who presented with vision of hand movements (HM+) was suspected to have HSV-1 infection, but it also showed presence of HSV-2 DNA and his vision improved minimally to 20/600 though he was treated with antivirals.
Discussion
In this study we demonstrated that qPCR significantly increased the rate of HSV detection in corneal scrapings compared to IFA and conventional PCR and that its implementation is feasible for routine diagnostic settings. The patients enrolled in the present study, were all clinically suspected HSK and referred for microbiological analysis for confirmation. Out of the 50 cases, 39 (78%) were found to be positive for HSV-1 DNA. Our qPCR, is more specific and sensitive than conventional PCR, and can be used to detect the virus more quickly as it eliminates the need for laborious post PCR methods to help make diagnosis sooner. The possibility of the real-time PCR to quantify HSV virus load and to distinguish between different HSV types could also be important for prognosis or for monitoring treatment success. Also, viruses are usually latent in the trigeminal ganglia after primary infection. With repeated reactivation cycles, viruses can also be found in corneal epithelial scrapings, stroma, or tears.[16,17,18,19,20] Unlike HSV-1 seroprevalence, little is known about asymptomatic shedding of HSV-1 in the eye. Few studies have tested the presence of HSV-1 in tears of healthy individuals.[21,22,23] Kaye et al.[21] found no shedding of HSV-1, and Kaufman et al.[14] and Okinaga[22] found 0.8% and 0.05%, respectively, but these studies used relatively insensitive culturing techniques. In comparison to those findings, our present data showed significantly higher HSV-DNA–positive results (~15%) in corneal epithelial cells of normal individuals. Since we could not find any study describing the cut off value or viral load in normal population, we decided to first establish a normal accepted copy number of HSV virus that could be present in the population. A value greater than one standard deviation above the average number of HSV-1 copy number i.e. >=25 copies/mL was arbitrarily set as cut-off for true infection. However, this should be interpreted with caution as qPCR is a very sensitive assay that can detect very low numbers of DNA in samples, and this need not be reflective of the infectious virus. It is highly possible that remnants of viruses or replication defective viruses may persist in tissues after active infection is controlled. However, all samples included in the study were clinically suspected to have active HSV infection.[24]
But, the question arises whether this technique has been validated and why this assay can be considered to be a reliable test to be used as a decisive judge. Towards this end a sub analysis of 19 patients who were only qPCR-positive and a confirmative microbiological diagnosis of HSV-1 was missed, showed that in 10/19 cases acyclovir was started on clinical suspicion alone and 5/10 (50%) patients showed improvement in visual outcome. All the remaining 5 patients had a viral load of >1000 copies/ml. Only in 1/19 cases, the load of HSV-1 virus was >1000 copies/ml (#45) and antivirals were not started which could probably explain the unsatisfactory visual outcome. High levels of HSV DNA in patient's samples may indicate active HSV reproduction in lesions resulting in corneal disease. Although, the gold standard for determining viral load in these samples is by culturing them in various cell lines like HCE, this method is however more laborious and time consuming. Domingues and colleagues[25] reported that patients with a high HSV copy number (105/ml) had a poorer prognosis than those with lower copy numbers, whereas others did not see such a correlation in patients with HSV encephalitis.[26] Our data shows a similar trend with 11/13 cases who had a viral load >10,000 copies/ml achieving a final visual outcome of 20/200 or worse post treatment. Additionally, greater amounts of viral DNA may not be reflective of greater amounts of infectious virus, but may be also due to plaque to particle differences. Another limitation in this study is that the results of viral loads should have been expressed in viral copies/number of cells (with a q PCR of a housekeeping gene). Since the standards were given in the kit as copies/mL, the data from the standard curve is also expressed in the same way. Due to exhaustion of DNA, we could not repeat the test using an external housekeeping gene. Thus, more clinical studies using quantitative HSV DNA assays will be necessary for determining their actual value in establishing the prognosis and improving treatment results in herpes viral keratitis. Our further future study aims at that, and we would assess the time point at which a decrease in copy count is observed.
Dual infections with both HSV-1 and -2 are also well described.[27,28] An earlier report had shown that as long as the quantitative difference is within 1,000-fold, both HSV-1 and 2 viruses could be detected. And when the difference is between 1,000 and 10,000-fold, then the one with less quantity is likely to be competed out.[29] In our series too, one patient sample was dual positive and the difference in titre load was 1000 fold. There was no obvious difference between the clinical course of mixed infection and those of single HSV-1 or HSV-2 infections. The question however remains about which infection was the initial one. The coincidence of VZV and acanthamoeba in our data series remains a curiosity. This study also highlights the frequent coinfections with both bacteria (six cases) and fungus (four cases) in patients with herpetic keratitis. This is an important consideration for the medical management, in addition to acyclovir treatment, especially in tropical climates with a history of trauma. In 3/6 patients who had co-infection with bacteria, and 3/4 patients who had co-infection with fungus, there was no improvement in visual outcome. Non-responding ulcers should be immediately suspected of co-infection or super added infection.
Conclusion
Our results suggest that detecting ocular herpetic disease by quantitative PCR method is informative as it not only helps in distinguishing between HSV-1 and 2 and VZV virus, the viral load is particularly helpful in the decision for treatment.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgements
Hyderabad Eye Research Foundation.
References
- 1.Kinchington PR, Leger AJ, Guedon JM, Hendricks RL. Herpes simplex virus and varicella zoster virus, the house guests who never leave. Herpesviridae. 2012;3:5. doi: 10.1186/2042-4280-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farooq AV, Shukla D. Herpes simplex epithelial and stromal keratitis: An epidemiologic update. Surv Ophthalmol. 2012;57:448–62. doi: 10.1016/j.survophthal.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaye S, Choudhary A. Herpes simplex keratitis. Prog Retin Eye Res. 2006;25:355–80. doi: 10.1016/j.preteyeres.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Kaufman HE, Azcuy AM, Varnell ED, Sloop GD, Thompson HW, Hill JM. HSV-1 DNA in tears and saliva of normal adults. Invest Ophthalmol Vis Sci. 2005;46:241–7. doi: 10.1167/iovs.04-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toma HS, Murina AT, Areaux RG, Neumann DM, Bhattacharjee PS, Foster TP, et al. Ocular HSV-1 latency, reactivation and recurrent disease. Semin Ophthalmol. 2008;23:249–73. doi: 10.1080/08820530802111085. [DOI] [PubMed] [Google Scholar]
- 6.Suryawanshi A, Mulik S, Sharma S, Reddy PB, Sehrawat S, Rouse BT. Ocular neovascularization caused by HSV-1 infection results from breakdown of binding between VEGF-A and its soluble receptor. J Immunol. 2011;186:3653–65. doi: 10.4049/jimmunol.1003239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inoue Y. Immunological aspects of herpetic stromal keratitis. Semin Ophthalmol. 2008;23:221–7. doi: 10.1080/08820530802111390. [DOI] [PubMed] [Google Scholar]
- 8.Rübben A, Baron JM, Grussendorf-Conen EI. Routine detection of herpes simplex virus and varicella zoster virus by polymerase chain reaction reveals that initial herpes zoster is frequently misdiagnosed as herpes simplex. Br J Ophthalmol. 1997;137:259–61. doi: 10.1046/j.1365-2133.1997.18161913.x. [DOI] [PubMed] [Google Scholar]
- 9.Chodosh J, Miller D, Stroop WG, Pflugfelder SC. Adenovirus epithelial keratitis. Cornea. 1995;14:167–74. [PubMed] [Google Scholar]
- 10.Wilhelmus KR, Font RL, Lehmann RP, Cernoch PL. Cytomegalovirus keratitis in acquired immunodeficiency syndrome. Arch Ophthalmol. 1996;114:869–72. doi: 10.1001/archopht.1996.01100140083016. [DOI] [PubMed] [Google Scholar]
- 11.Marangon FB, Miller D, Alfonso E. Laboratory results in ocular viral diseases: Implications in clinical-laboratory correlation. Arq Bras Oftalmol. 2007;79:189–94. doi: 10.1590/s0004-27492007000200002. [DOI] [PubMed] [Google Scholar]
- 12.Subhan S, Jose RJ, Duggirala A, Hari R, Krishna P, Reddy S, et al. Diagnosis of herpes simplex virus-1 keratitis: Comparison of Giemsa stain, immunofluorescence assay and polymerase chain reaction. Curr Eye Res. 2004;29:209–13. doi: 10.1080/02713680490504911. [DOI] [PubMed] [Google Scholar]
- 13.Fukuda M, Deai T, Hibino T, Higaki S, Hayashi K, Shimomura Y. Quantitative analysis of herpes simplex virus genome in tears from patients with herpetic keratitis. Cornea. 2003;22:S55–60. doi: 10.1097/00003226-200310001-00008. [DOI] [PubMed] [Google Scholar]
- 14.Ma JX, Wang LN, Zhou RX, Yu Y, Du TX. Real-time polymerase chain reaction for the diagnosis of necrotizing herpes stromal keratitis. Int J Ophthalmol. 2016;9:682–6. doi: 10.18240/ijo.2016.05.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunimoto DY, Sharma S, Garg P, Gopinathan U, Miller D, Rao GN. Corneal ulceration in the elderly in Hyderabad, south India. Br J Ophthalmol. 2000;84:54–9. doi: 10.1136/bjo.84.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kennedy DP, Clement C, Arceneaux RL, Bhattacharjee PS, Huq TS, Hill JM. Ocular herpes simplex virus type 1: Is the cornea a reservoir for viral latency or a fast pit stop? Cornea. 2011;30:251–9. doi: 10.1097/ICO.0b013e3181ef241d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shoji J, Sakimoto T, Inada N, Kamei Y, Matsubara M, Takamura E, et al. A diagnostic method for herpes simplex keratitis by simultaneous measurement of viral DNA and virus-specific secretory IgA in tears: An evaluation. Jpn J Ophthalmol. 2016;60:294–301. doi: 10.1007/s10384-016-0448-y. [DOI] [PubMed] [Google Scholar]
- 18.Kaneko H, Higaki S, Fukuda M, Shimomura Y, Ishioka K, Fukushima E, et al. The quantitative detection of herpes simplex virus, varicella zoster virus, and cytomegalovirus DNAs in recipient corneal buttons. Cornea. 2010;29:1436–9. doi: 10.1097/ICO.0b013e3181d3d69d. [DOI] [PubMed] [Google Scholar]
- 19.Remeijer L, Duan R, van Dun JM, Wefers Bettink MA, Osterhaus AD, Verjans GM. Prevalence and clinical consequences of herpes simplex virus type 1 DNA in human cornea tissues. J Infect Dis. 2009;200:11–9. doi: 10.1086/599329. [DOI] [PubMed] [Google Scholar]
- 20.Fukuda M, Deai T, Higaki S, Hayashi K, Shimomura Y. Presence of a large amount of herpes simplex virus genome in tear fluid of herpetic stromal keratitis and persistent epithelial defect patients. Semin Ophthalmol. 2008;23:217–20. doi: 10.1080/08820530802111366. [DOI] [PubMed] [Google Scholar]
- 21.Kaye SB, Madan N, Dowd TC, Hart CA, McCarthy K, Patterson A. Ocular shedding of herpes simplex virus. Br J Ophthalmol. 1990;74:114–6. doi: 10.1136/bjo.74.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okinaga S. Shedding of herpes simplex virus type 1 into tears and saliva in healthy Japanese adults. Kurume Med J. 2000;47:273–7. doi: 10.2739/kurumemedj.47.273. [DOI] [PubMed] [Google Scholar]
- 23.Higaki S, Fukuda M, Shimomura Y. Virological and molecular biological evidence supporting herpes simplex virus type 1 corneal latency. Jpn J Ophthalmol. 2015;59:131–4. doi: 10.1007/s10384-014-0369-6. [DOI] [PubMed] [Google Scholar]
- 24.Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–8. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 25.Domingues RB, Lakeman FD, Mayo MS, Whitley RJ. Application of competitive PCR to cerebrospinal fluid samples from patients with herpes simplex encephalitis. J Clin Microbiol. 1998;36:2229–34. doi: 10.1128/jcm.36.8.2229-2234.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Revello MG, Baldanti F, Sarasini A, Zella D, Zavattoni M, Gerna G. Quantitation of herpes simplex virus DNA in cerebrospinal fluid of patients with herpes simplex encephalitis by the polymerase chain reaction. Clin Diagn Virol. 1997;7:183–191. doi: 10.1016/s0928-0197(97)00269-9. [DOI] [PubMed] [Google Scholar]
- 27.Samarai A, Shareef A, Kinghorn G, Potter C. Sequential genital infections with herpes simplex virus types 1 and 2. Genitourin Med. 1989;65:39–41. doi: 10.1136/sti.65.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.SucatoG, Wald A, Wakabayashi E, Vieira J, Corey L. Evidence of latency and reactivation of both herpes simplex virus (HSV-1) and HSV-2 in the genital region. J Infect Dis. 1998;177:1069–72. doi: 10.1086/515261. [DOI] [PubMed] [Google Scholar]
- 29.Corey L, Huang ML, Selke S, Wald A. Differentiation of herpes simplex virus types 1 and 2 in clinical samples by a real-time taqman PCR assay. J Med Virol. 2005;76:350–5. doi: 10.1002/jmv.20365. [DOI] [PubMed] [Google Scholar]


