Abstract
Recent explorations of knee biomechanics have benefited from computational modeling, specifically leveraging advancements in finite element analysis and rigid body dynamics of joint and tissue mechanics. A large number of models have emerged with different levels of fidelity in anatomical and mechanical representation. Adapted modeling and simulation processes vary widely, based on justifiable choices in relation to anticipated use of the model. However, there are situations where modelers' decisions seem to be subjective, arbitrary, and difficult to rationalize. Regardless of the basis, these decisions form the “art” of modeling, which impact the conclusions of simulation-based studies on knee function. These decisions may also hinder the reproducibility of models and simulations, impeding their broader use in areas such as clinical decision making and personalized medicine. This document summarizes an ongoing project that aims to capture the modeling and simulation workflow in its entirety—operation procedures, deviations, models, by-products of modeling, simulation results, and comparative evaluations of case studies and applications. The ultimate goal of the project is to delineate the art of a cohort of knee modeling teams through a publicly accessible, transparent approach and begin to unravel the complex array of factors that may lead to a lack of reproducibility. This manuscript outlines our approach along with progress made so far. Potential implications on reproducibility, on science, engineering, and training of modeling and simulation, on modeling standards, and on regulatory affairs are also noted.
Keywords: computational modeling, reproducibility, knee biomechanics, finite element analysis, joint mechanics, tissue mechanics
1. Background and Motivation
Computational modeling has become ubiquitous to support scientific and clinical studies on knee biomechanics (Fig. 1). State-of-the-art simulation frameworks for finite element analysis [1] and multibody dynamics [2] permit high-fidelity representations of joint anatomy and mechanics, providing the opportunity to understand interactions between body level loads, joint movements, and tissue deformations. Naturally, such techniques have been leveraged to explore tibiofemoral and patellofemoral joint mechanics in health and disease, and after reconstruction [3]. Essentially, all of the passive tissue structures are focus areas of a growing number of simulation-based examinations including the cartilage, bone, meniscus, and ligaments [4]. Subsequently, computational models of the knee have penetrated all aspects of healthcare delivery and research in many ways: to support intervention design, to predict surgical outcomes, to deliver medical training, and for personalized care [3–5].
Fig. 1.
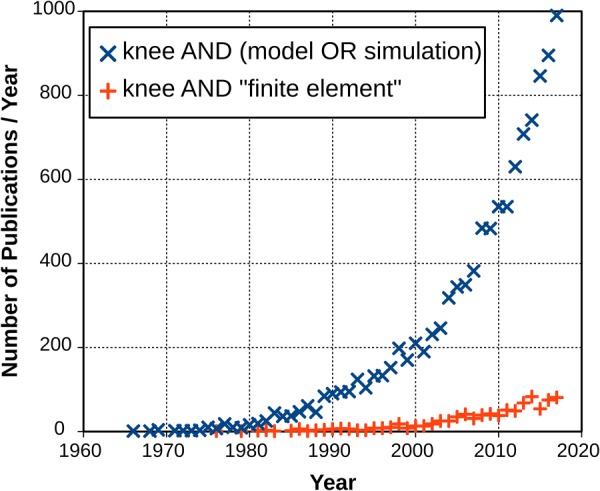
Number of publications focusing on knee modeling or simulation reaches up to 1000 per year. Total number of publications (up to year 2018) is 10,895 (data from PubMed2). With the increased fidelity of simulation software and computing hardware, use of finite element analysis in computational knee mechanics has gained traction. Annual number of studies are approaching 100 (total up to year 2018 is 882).
Fundamental abstraction of a knee model is straightforward (Fig. 2). Virtual reconstruction of anatomical properties is needed to represent geometry and arrangement of tissue structures at a desired level of specificity (population-based or subject/specimen-specific) [6]. Physiological properties, i.e., mechanical properties, in Fig. 2, of all modeled tissue components are also required [6]. In biomechanical modeling, these include properties defined at the structural or material levels [4]. Note that connectivity between tissues, e.g., bone and ligaments, as well as mechanical interactions, e.g., contact between articulating surfaces, are usually incorporated as part of the anatomical or mechanical features of the model [6]. Loading, e.g., muscular forces, external loads, prescribed kinematics, boundary conditions, etc., are imperative to conduct meaningful simulations that have scientific and/or clinical relevance [6]. The end goal of building a computational model is to perform simulations, i.e., finite element analysis, to predict the mechanical response of the joint [6]. In a majority of cases, the response that is sought is related to joint movement (kinematics–kinetics) and tissue deformations (stress–strain distribution) [6]. Despite the simplicity of this generalized description of computational models in biomechanics, its interpretation by a modeler may vary when developing and using a virtual representation of the knee [4] (Fig. 3).
Fig. 2.
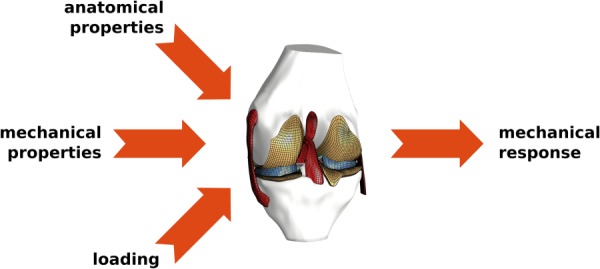
Fundamental abstraction of modeling in knee biomechanics. Required input parameters include anatomical (geometry, mesh, etc.) and mechanical representations (stiffness, material properties, etc.) of joint components (bones, cartilage, ligaments, menisci, muscles, etc.), and loading and boundary conditions (external loads, muscular forces, etc.). Simulations look for predictions of mechanical response, e.g., joint movements, tissue stresses, and strains.
Fig. 3.
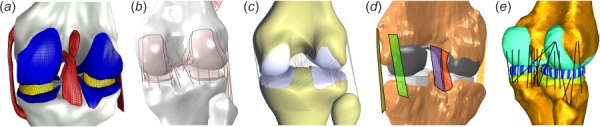
Even visually, computational models of the knee joint exhibit large variations in anatomical and mechanical representations of tissue structures. Shown are samples of work by teams collaborating in a comprehensive study to understand the art of modeling and simulation in knee biomechanics: (a) open knee(s)—generation 1 from Cleveland Clinic team (Reproduced from [7]), (b) a model from the group at University of Denver (Reproduced from [8]), (c) work by researchers at Auckland Bioengineering Institute, (d) a current model from Cleveland State University, and (e) recent modeling by the team at Hospital for Special Surgery (Reproduced from [9]).
Computational biomechanics of the knee inherits a large variety of activities of the broad modeling and simulation lifecycle [10], which are labor and resource intensive, and demand significant intellectual capacity (Fig. 4). Naturally, a computational model is built to answer a question, which is the starting point of any modeling task. Questions can range from clinical to basic research: Will a new ligament reconstruction provide mechanical stability? What is the increased risk of cartilage degeneration due to altered joint mechanics? and so on. Defining this question clearly dictates the model's context of use and motivates the required modeling fidelity and relevant simulation scenarios. Commonly, anatomical imaging (computed tomography and magnetic resonance imaging) [11], in vitro mechanical testing (joint and tissue testing) [12], in vivo data collection [13], and research and clinical literature provide the foundational information to build the model. Geometries of tissues are usually generated through image segmentation [14] and discretized to generate a mesh for finite element analysis [6] or multibody dynamics simulations [15]. Tissue testing data, if available, can provide material properties of the bone and soft tissues (cartilage, ligaments, meniscus, etc.) [4]. Noninvasive imaging may also be used to infer subject-specific material properties in certain cases, e.g., computed tomography to estimate apparent bone mineral density [16]. Yet, tissue properties are often adapted from prior reports. Loading conditions can be derived from experiments conducted on a subject's knee [17] or on a cadaver specimen [7]; or, they can be approximated from known or anticipated loads of daily activities [18], a clinical test [19], or an injury scenario [20]. A model calibration phase commonly follows model development, where properties of the model, e.g., ligament stiffness or slack lengths [8], are optimized such that differences between an experimental condition with known joint/tissue mechanics and model predictions are minimized [21]. Uncertainty quantification, verification and validation studies, and sensitivity analysis may follow [22,23]. These activities encompass model benchmarking, which aims to understand the performance of the model within the context of its intended use. With established confidence in the model, the developers return back to the question they want to answer and conduct simulations to gain insight into the mechanics of the joint and tissues. It is important to recognize the distinction between calibration, where model results have been tuned to a specific set of data, versus validation, where model predictions are assessed relative to unseen data. Some studies have performed specimen-specific tuning of constraint, followed by predictions of other activities, e.g., gait [24], or model states, e.g., a ligament or structure removed [25]. With recent emphasis on model sharing [26] and the increased interest in model reuse, e.g., for multiscale analysis [27], developers, researchers, or others may repurpose a knee model to explore joint and tissue function for different cases. For example, a comprehensive knee model, which was originally used to understand coupling of ligament function and joint movements [28] may later be used to investigate meniscectomy [29]. Model reuse is a fundamental element that we are hoping to provide and it is arguably not achieved in the field of knee biomechanics extensively. As models are repurposed during their lifetime, iterations are anticipated for additional data acquisition, model modification, and evaluation.
Fig. 4.
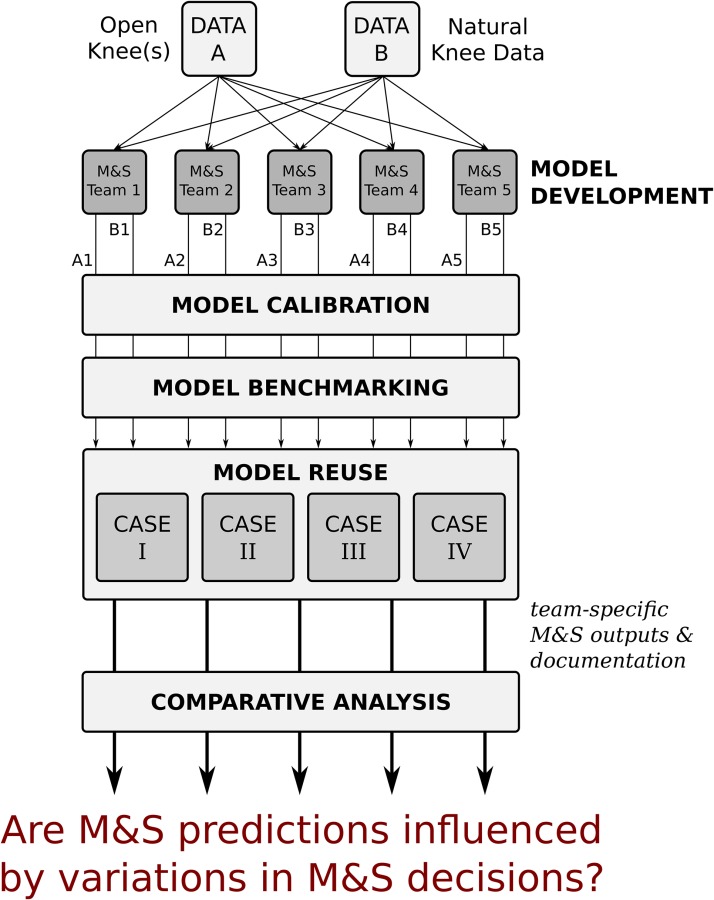
Starting with the same data sets, each modeling and simulation (M & S) team goes through a sequence of modeling and simulation phases to come up with their own flavor of models representing the knee specimens of the data sets. The overarching goal of this study is to understand if the decisions of the modeling teams influence simulation predictions, and their interpretation to reach scientific and clinically relevant conclusions. Dissemination of all modeling and simulation outcomes and documentation of the whole lifecycle of the models will provide the opportunity to understand the source of variations in modeling decisions and the motivations behind them.
A large number of modeling and simulation decisions need to be made throughout the lifecycle of models, which define the modeler's art. Modeler's choices can be biased by their preferences, which are formed through their experience and knowledge. The modeler needs to tackle many questions during the process, while accounting for the ultimate intended use of the model. While knee experimentation relies upon sample populations [30], in silico studies of the knee rely on a single or a handful of models (a few statistical shape models for population based studies are exceptions [31]) giving rise to many questions: Which knee should be modeled? What type of demographics or disease population should it represent? Should it be subject/specimen-specific or a generic representation? Are data on the specific knee available? Or, can it be collected at a desired level of detail? The fidelity of anatomical and mechanical representation can vary. The geometric representation of articular surfaces can be as simple as an average parametric representation [32] or as detailed as an explicit surface accommodating the specificity of the knee [33]. Mechanics can be approximated at a structural level or at a material level. This is demonstrated by the variety of ligament representations, which can range from springs to three-dimensional continuum models [34]. The modeler may decide to use a continuum representation for soft tissues such as cartilage, for example. Yet, many different constitutive models have been used to describe cartilage behavior, from linear elastic to nonlinear elastic, to multiphasic [11]. For a chosen constitutive model, literature provides the opportunity (or the uncertainty) to choose from a large span of material coefficients [4], i.e., for cartilage, the reported elastic modulus ranges within two orders of magnitude [4]. Preferences can be based on convenience, data limitations, perceived level of representation required to address the question, access to software and hardware, simulation software capabilities, and computational cost. Data analysis to derive model components may introduce process uncertainties, e.g., strategy to determine tissue boundaries [35]. Mesh convergence analysis [36] may indicate a denser discretization but it may challenge computational tractability. A substitute material model may be used because the simulation software may not have a readily available implementation of the desired constitutive model or to improve run time, e.g., deformable versus rigid body with a pressure-overclosure relationship [37]. Loading conditions may need to be simplified to accommodate for the uncertainties in data and to support simulation convergence. Similarly, a tissue structure may be omitted, e.g., meniscus, or additional stabilizing structures, e.g., springs and dampers, are introduced to converge to a solution [38]. These potentially subjective decisions of the modeler collectively define his/her art. Not only do they influence model development but also the means to conduct simulations and postprocess and interpret results. The spectrum of individual journeys in modeling and simulation can explain the immense variations in models of the knee [3,4].
Modeler's choices likely impact the simulation results and the conclusions reached by their interpretation. These decisions may diminish the reproducibility potential of a modeling and simulation workflow. Ideally, one should be able to reproduce the modeler's work, generate practically similar results, and reach the same conclusions in regard to the simulated knee's state and function. Subjectivity may cloud documentation of models, modeling and simulation activities, and related justifications. Scholarly publication platforms have promoted detailed documentation of “what a model is” [6] but may not provide an effective means to communicate how it is actually built and used at a detail to support reproducibility. The broader modeling and simulation community launched initiatives on credible practice of modeling and simulation [39]. The computational biomechanics community has primarily focused on verification and validation [40]. However, strategies to capture the modeling and simulation process in its entirety have not been widely implemented. In the specialized area of musculoskeletal movement simulations, competitions such as the grand knee challenge [41] provided important insight on the impact of simulation approaches on model predictions such as knee joint contact forces. Nonetheless, documentation of decisions and related justifications were limited to scholarly publications and models were not necessarily disseminated. Most importantly, the modeling paradigm focused on musculoskeletal movements and implant function and did not target higher fidelity models of the knee joint and its tissue structures that are more relevant to explore natural and pathological knee mechanics [4].
The goal of this manuscript is to outline a collaborative study to capture the nature of modeling and simulation processes in computational knee biomechanics. Such a study should enable curation of all aspects of the simulation workflow from start to end; including specifications, protocol deviations, source data, final, and intermediate outcomes (model components, models, and simulation results). It has the potential to uncover the art of knee joint modeling. In this way, this document, and the project described within, are aimed to raise awareness of subjectivity and reproducibility in modeling and simulation, focusing on in silico explorations of knee joint mechanics.
2. A Comprehensive Study
2.1. Overall Workflow.
Comprehension of subjective decisions in modeling and simulation and their impact on reproducibility of the workflows necessitate a controlled study. Such an investigation should minimize the variability of source data used by different teams and it should promote substantive documentation and dissemination to characterize the art of computational modeling. Recently, the co-authors of this manuscript launched a multisite collaboration to characterize the variations in modeling and simulation workflows and to document their influence on the reproducibility of joint and tissue level predictions in computational knee biomechanics [42,43]. The intent of this project is to answer a fundamental question of modeling and simulation: Do the predictions of natural knee biomechanics depend on the modeling decisions of separate development teams when the target simulation scenarios and the source data to build models remain the same? Five research teams, who are experienced in knee joint modeling (Fig. 3), will separately model two knees relying on the same data (Fig. 4). Modeling and simulation phases include model development—to deliver initial working models, model calibration—to tune model parameters, model benchmarking—to evaluate model performance, and model reuse—to understand the utility of simulation for scientific and clinical decision making. Specimen-specific data available to the teams will be limited to data earmarked for the individual phases. Nonetheless, the teams will be permitted to leverage literature and choose their own tools and strategies for computational modeling. For each phase of the modeling and simulation workflow, team-specific approaches will be documented as specifications of the phase before conducting any modeling activities. This documentation will be supported by a record of protocol deviations that describe any changes to the specifications as the phase proceeds. The outcomes of each modeling and simulation phase (models, preprocessed data, etc.) will be disseminated along with documentation. Third-party review of workflow documentation and modeling outcomes will be conducted by an independent group who will not be involved in the modeling phases.
2.2. Data.
The project leverages two existing data sets of knee anatomy and mechanics: Open Knee(s) [44] and Natural Knee Data [45]. Open Knee(s) provides data collected on eight cadaver knees from eight donors. These data include anatomical imaging based on magnetic resonance imaging modality [46]: general purpose (3D T1-weighted without fat suppression), for cartilage (3D T1-weighted with fat suppression), and for connective tissue (proton density acquired for three orthogonal planes). Spherical registration markers were attached to bones (three each) and were also imaged. Mechanical testing data include tibiofemoral joint kinematics–kinetics [47]: passive flexion (0–90 deg), joint laxity (at 0 deg, 30 deg, 60 deg, and 90 deg flexion for internal–external rotation, varus–valgus, and anterior–posterior translation), and combined loading (at 0 deg, 30 deg, 60 deg, and 90 deg flexion; permutations of internal–external rotation moments, varus–valgus moments, and anterior–posterior drawer forces). Patellofemoral joint kinematics–kinetics data are also available for quadriceps loading (0–600 N) at 0 deg, 15 deg, 30 deg, 45 deg, and 60 deg flexion, including patellofemoral contact pressures. Digitization was performed during mechanical testing providing coordinates of anatomical landmarks and spherical registration markers. Natural knee data were obtained from seven cadaver knees from five donors. Anatomical imaging of the knee included computed tomography and T2-weighted magnetic resonance scans. Experimentation consisted of digitizing landmarks on the bone, cartilage, and ligament attachments. Specific features were the femoral and tibial bone surfaces; the articular surfaces of the femur, tibia, and patella cartilage; and the attachment sites of the anterior and posterior cruciate ligaments, and medial and lateral collateral ligaments. Available mechanical data span tibiofemoral joint kinematics and kinetics [8]: passive flexion (0–120 deg), joint laxity at 0 deg, 15 deg, 30 deg, 45 deg, 60 deg, 75 deg, 90 deg, and 120 deg flexion for internal–external rotation, varus–valgus, and anterior–posterior translation, repeated passive flexion, and laxity testing after sequential resection of the cruciate ligaments. Patellofemoral mechanics data are also available that were acquired under quadriceps loading as the tibia was flexed from 0 deg to 90 deg flexion [48]. This data set also includes measurements after sequential resection of cruciate ligaments.
2.3. Modeling and Simulation.
The modeling and simulation workflow adapted for this project reflects the lifecycle of computational models in biomechanics research and innovation. The four phases are described below. Before their execution, the teams will reach a consensus agreement on the primary and secondary deliverables of the phase and the extent of data that will be earmarked for it. Following dissemination of earmarked data, individual teams will prepare and submit specifications to accomplish the modeling stage in their own way, i.e., their procedures and operational burden. Upon release of team specifications, modeling and simulation activities will start. The teams are asked to document and submit any protocol deviations. In addition, final and intermediate products of the modeling and simulation stage, including but not limited to computational models, geometric representations, etc., will be disseminated.
2.3.1. Model Development.
In the model development phase, the participating teams aim to deliver initial working models. Earmarked data include specimen-specific anatomical imaging and possibly any other anatomical information measured by other means, e.g., digitization of landmarks. If available, use of specimen-specific tissue testing data can be considered. It is anticipated that the teams will rely upon data from the literature to define a complete model. Performing a sample simulation, e.g., passive flexion, ensures models are at a “working” condition. At the end of this phase, dissemination of two knee models from each team is anticipated along with preprocessed data for representation of anatomy (image segmentation, geometry, and mesh) and tissue mechanical behavior (constitutive models, tissue stress–strain response, and tissue bulk response).
2.3.2. Model Calibration.
The goal of the model calibration phase is to obtain tuned knee models. The teams start with their initial working models (the outcome of the previous phase) and use specimen-specific joint kinematics–kinetics data to calibrate model parameters that may otherwise be missing or uncertain, e.g., ligament stiffness properties and slack lengths. Joint laxity tests, available in Open Knee(s) and Natural Knee Data, can serve for this purpose where model parameters can be updated to match experimental data [8]. It is possible that the teams may choose to update model parameters to match information available in the literature describing joint and tissue level function of the knee. The outcome of this phase is a set of calibrated specimen-specific models with documented fit errors. Updated model parameters, and loading and boundary conditions of calibration related simulations are part of phase outputs including any changes in anatomical and tissue mechanical representations.
2.3.3. Model Benchmarking.
The model benchmarking phase evaluates model performance against experimental data obtained from the specimens that were modeled, which are intentionally separated from data used in previous phases [49]. Open Knee(s) data include multiplanar loading of the tibiofemoral joint and contact pressure measurements for the patellofemoral joint, which can be leveraged for this purpose. Natural Knee Data provide joint kinematics–kinetics after ligament resection, which is likely to be used for benchmarking. Each team will perform simulations using their calibrated models to quantify the performance of their models against data designated for benchmarking. The goal is to obtain and disseminate benchmarked models with documented benchmark error relative to the amount of data used for calibration, representing each team's individualized workflow. Comparisons against literature are also welcome. Secondary outcomes of the workflow are the representation of loading and boundary conditions based on each team's interpretation of the experiments conducted on each cadaver knee specimen.
2.3.4. Model Reuse.
In the model reuse phase, individual teams will use their models for in silico investigations that have scientific and clinical relevance. Four simulation cases were selected to demonstrate the utility of the knee models. Simulations of passive flexion (case I) target the ability of the models to predict knee joint kinematics–kinetics at low loads. Coupled movements of tibiofemoral joint degrees-of-freedom to flexion angle are well documented [50]. Models of natural knees should therefore reproduce such behavior, which is a property of knee mechanics due to the structural arrangement of the ligaments and the contact surface geometry of the tibiofemoral joint but may also be influenced by coordinate system definitions. Pivot shift simulations (case II) can be used to evaluate a model's performance when predicting the ability of the anterior cruciate ligament to stabilize the knee. The pivot shift exam is a common clinical test [51] where the physician flexes the knee while applying internal rotation and valgus torques. In the intact case, the exam loads the anterior cruciate ligament. If the anterior cruciate ligament is compromised, an abrupt change in anterior–posterior translation of the tibia relative to the femur should be observed during the maneuver [51]. The third simulation case will reproduce loading and boundary conditions of a weight-bearing, standing X-ray (case III). This is a common clinical exam to understand the progression of osteoarthritis by evaluating joint space [52]. In this simulation case, contact mechanics of the tibiofemoral joint (dictated by cartilage and menisci) can be evaluated in addition to the change in joint space due to loading. The final simulation scenario imitates sit-to-stand (case IV) for evaluation of functional biomechanics of the knee during a common activity of daily living. Sit-to-stand is a demanding weight-bearing activity for the young and elderly [53], requiring a large range of knee extension. With this simulation scenario, each models' ability to predict tibiofemoral and patellofemoral joint movement and loading in a functional setting can be evaluated. Specimen-specific experimental data to support these simulation cases are not necessarily available. Therefore, the teams are challenged to rely on their expertise and the literature to make modeling and simulation decisions, i.e., for the implementation of loading and boundary conditions. All teams are expected to disseminate customized models and simulation results including predictions of joint kinematics–kinetics and tissue mechanics.
2.4. Comparative Analysis.
Full dissemination of intermediate and final modeling and simulation outcomes and extensive documentation of each team's processes provide a comprehensive foundation for quantitative and qualitative comparisons. Simulation results, in particular joint kinematics–kinetics predictions and tissue mechanics metrics, can be used to evaluate predictive capacity of the knee models. For the model calibration phase, fit error provides an absolute metric to assess the performance of the model, e.g., in the form of root-mean-square error. Similarly, for the model benchmarking phase, specimen-specific mechanical testing is available to quantify performance. When an experimental benchmark is not available, simulation results of individual teams can be compared against each other using a generalized Bland-Altman analysis to provide limits of agreement among multiple methods [54]. All these strategies provide the means to rank performance of models for selected simulation cases or for all cases. Any discrepancies in simulation results are likely a function of differences in model components that span geometries, tissue representations and material properties, and implementation of loading and kinematic definitions. Variation of geometric representations among models can be calculated using surface comparison techniques, e.g., Hausdorff distance [55]. Mechanics of the same tissue component of different models can be compared at the material level—based on constitutive parameters, and at the structural level—based on the response of the tissue as a whole, e.g., total ligament force. For loading and boundary conditions, deviations in coordinate system representations can be calculated along with differences in prescribed forces and moments, and displacements and rotations.
Specifications and protocol deviations for each modeling and simulation phase will be assessed to examine the reproducibility potential of the workflow and to understand the art of modeling and simulation. Reproducibility potential is likely to be related to the completeness of information, which should state what the model is, how it is built and evaluated, and what the simulation conditions are. The documents can be compared to guidance in literature [6] to objectively evaluate the correspondence of the provided information against the minimum required to redo the work. An interested party can access the earmarked data, follow the specifications (and corresponding protocol deviations) to build models and conduct simulations, and compare their modeling and simulation outputs against those of the authors; resulting in an additional test of reproducibility. To comprehend a modeler's art, one needs to understand the justifications behind modeling and simulation decisions. Extraction of these justifications from the documents provided by each individual team can facilitate qualitative comparisons at any given step of the workflow. It is important to understand the burden of modeling and simulation as accessibility to modeling software, expertise level, engineering labor, and computational cost may influence modelers' decisions. This information can be inferred from the documents of individual teams to quantify the economics of modeling and simulation.
Credibility and criticality of using knee models for scientifically and clinically relevant investigations need to be assessed in a holistic manner. This assessment relies on the final results of the simulation studies (noted in model reuse phase) and depends on the extent of documentation and the relevance of calibration and benchmarking to reuse cases. It requires a comprehensive and diligent examination of specifications, protocol deviations, and intermediate and final outputs of modeling and simulation. The computational modeling community has been actively developing guidance on management of workflows [39], standards [56], verification and validation [57], and reporting [58]. A third-party reviewer can assess the responsiveness and compliance of each team to follow such guidance. The reviewer can also provide a critique of the trustworthiness and utility of each team's models, processes, and simulation predictions. To this end, the project relies on a collaboration with the U.S. Food and Drug Administration (FDA), where regulatory and scientific personnel can act as an external and independent authority to evaluate and provide an unbiased comparison of individual modeling and simulation workflows, their outcomes, and trends across the groups.
3. Up-to-Date Progress
The project website provides an entry point for all activities related to the comprehensive study [42]. The project workflow is organized there and communications among the five contributing teams are also summarized. Curation and dissemination of project resources, specifically source data, each individual team's documentation and models, and outreach materials, are conducted through various sections of the web-based platform, which is supported by SimTK3.
Contributing teams decided to use data from specimen oks003 of Open Knee(s) and specimen DU02 of Natural Knee Data. Specimen oks003 represents a left knee of a 25 yr old female donor (height: 1.73 m; mass: 68 kg; body mass index: 22.8). Specimen DU02 represents a right knee of a 44 yr old male donor (height: 1.83 m; mass: 70.31 kg; body mass index: 21.02). Data from each specimen have been compartmentalized and designated for different stages of modeling. Staged dissemination of earmarked data have been conducted through the project website [42].
The model development phase has started upon agreement of target outcomes by contributing teams [59]. Specific goals are (i) to develop two initial working knee models (inclusive of tibiofemoral and patellofemoral joints) (one using Open Knee(s) data, another using Natural Knee Data), (ii) to conduct simulations of passive flexion as a sample case, (iii) to document and disseminate the model and model components, modeling and simulation processes, and simulation results. The term “initial working model” refers to a model that is completely defined to carry out preliminary simulations. The teams are allowed to interpret the meaning of “passive flexion” to decide upon the loading and boundary conditions of their models for the simulation case. The earmarked data are limited to only imaging data for oks003 of Open Knee(s) (general purpose, cartilage, and axial, coronal, and sagittal soft tissue magnetic resonance image volumes) and both imaging and anatomical landmark data for DU02 of Natural Knee Data (a T2-weighted magnetic resonance image volume and a computed tomography volume, digitized landmarks on the femur, tibia, and patella).
Each team has already submitted separate specifications for processing each data set, before starting the execution of model development activities. These a-priori submissions were aimed to demonstrate their chosen path for modeling and simulation. By providing their responses in relation to different data sets, modeling nuances based on variable data content and quality can be captured. The teams were asked to provide this documentation in a detail such that other modeling teams can follow the steps to reproduce the modeling and simulation outputs. They were also asked to provide justifications to support their decisions and to note software, hardware, and labor requirements. All these documents have been disseminated at the project site [60]. Gross evaluation of these documents already indicates the diversity of software packages selected for generation of models and for simulation (Table 1). Overlaps and differences in anatomical and mechanical representations of tissue structures can be observed (Table 2). Subtleties of specific processes to obtain the same information, i.e., when segmenting the boundaries of tissues (Fig. 5), are documented.
Table 1.
Contributing teams propose to use a variety of software (proprietary or free and open source) for modeling and simulation (M & S) activities during the model development phase
| M & S Team 1 CC | M & S Team 2 DU | M & S Team 3 ABI | M & S Team 4 CSU | M & S Team 5 HSS | |
|---|---|---|---|---|---|
| Anatomical reconstruction | 3d slicer [61] meshlab [62] | simpleware scanip [63] | Custom in matlab [64] | 3d slicer [61] meshlab [62] | mimics [65] geomagic [66] |
| Meshing | salome [67] | hypermesh [68] | MAP Client [69] | ia-femesh [70] | — |
| Scripting | pythona | matlab [64] | matlab [64] | pythona | matlab [64] |
| Simulation | febio [71] | abaqus [72] | febio [71] | abaqus [72] | adams [73] |
Note: Anatomical reconstruction includes activities for image segmentation and for generation of tissue surfaces. Meshing primarily refers to the generation of three-dimensional volumetric finite element meshes. Scripting includes programing to assist model development and customization steps. CC, Cleveland Clinic; DU, University of Denver; ABI, Auckland Bioengineering Institute; CSU, Cleveland State University; HSS, Hospital for Special Surgery.
Table 2.
Model development specifications indicate variations in gross anatomical and mechanical representations of tissue structures in models proposed by the modeling and simulation (M&S) teams
| Anatomy | Mechanics | |
|---|---|---|
| Bone | ||
| M & S Team 1—CC | Surface | Rigid |
| M & S Team 2—DU | Surface | Rigid |
| M & S Team 3—ABI | Surface | Rigid |
| M & S Team 4—CSU | Surface | Rigid |
| M & S Team 5—HSS | Surface | Rigid |
| Cartilage | ||
| M & S Team 1—CC | Solid volume | Isotropic, nearly incompressible, hyperelastic |
| M & S Team 2—DU | Solid volume | Isotropic, nearly incompressible, linear elastica |
| M & S Team 3—ABI | Solid volume | Isotropic, nearly incompressible, linear elastic |
| M & S Team 4—CSU | Surface | Elastic foundation, linear |
| M & S Team 5—HSS | Surface | Rigid |
| Ligament | ||
| M & S Team 1—CC | Solid volume | Transversely isotropic, nearly incompressible, hyperelastic, with tension only fibers |
| M & S Team 2—DU | Distributed lines surface | Nonlinear tension only springs fiber reinforced membrane, tension only fibers |
| M & S Team 3—ABI | Distributed lines | Nonlinear tension only springs |
| M & S Team 4—CSU | Distributed lines | Nonlinear tension only springs, crosslinking with transverse springs |
| M & S Team 5—HSS | Distributed lines | Nonlinear tension only springs |
| Meniscus | ||
| M & S Team 1—CC | Solid volume | Transversely isotropic, nearly incompressible, hyperelastic, with tension only fibers |
| M & S Team 2—DU | Not modeled. | Not modeled. |
| M & S Team 3—ABI | Distributed lines | Linear tension only springs, equivalent resistance in anterior-posterior and medial-lateral directions |
| M & S Team 4—CSU | Solid volume | Transversely isotropic, linearly elastic |
| M & S Team 5—HSS | Distributed surfaces | Radially discretized set of rigid objects connected with three-dimensional linear springs |
Note: CC, Cleveland Clinic; DU, University of Denver; ABI, Auckland Bioengineering Institute; CSU, Cleveland State University; HSS, Hospital for Special Surgery.
Italic text is used to emphasize that the tissue structure is “not modeled” by the team.
Reported as addendum to specifications.
Fig. 5.
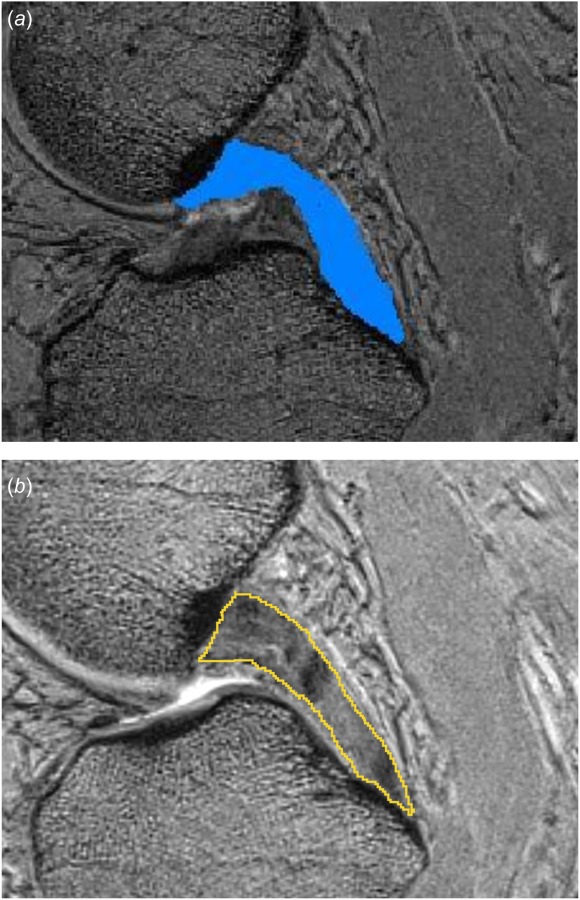
Sample images from proposed model development specifications in regard to segmentation of ligaments: (a) the team from the University of Denver proposes to segment the posterior cruciate ligament using the paint tool in Simpleware ScanIP [63]. The segmentation will be used to determine insertion locations of springs, which will be refined using probed point data and (b) the team from the Cleveland Clinic proposes to use 3D Slicer [61] to manually segment the same ligament in order to generate a full continuum representation of its volume. It is interesting to note that both groups independently and unknowingly chose the same ligament approximately at the same image location to provide an example of ligament segmentation. Image from University of Denver documentation was cropped to match the bounds of the image from Cleveland Clinic documentation.
A more detailed analysis of proposed modeling strategies, e.g., for the medial collateral ligament, demonstrates the diversity of modeling choices. Two schools of thought in modeling and simulation of ligaments [34] also emerge in the individual teams' workflows. Four of the teams utilize a discrete, spring-based representation of the ligament whereas one team relies on a continuum-based representation of the whole ligament volume (Table 2). Variations in anatomical and mechanical interpretation of the medial collateral ligament exist even when the teams belong to the same school of thought. The number of anatomical bundles and the number of fibers to represent individual bundles vary. Three teams model both the superficial and deep bundles; one team only focuses on the superficial medial collateral ligament. The number of fibers for each bundle ranges from three to six. Some teams primarily rely on anatomical imaging data to identify origin and insertion of the ligament while others leverage literature on anatomical descriptions relative to bony landmarks. Mechanical representation of the ligament for spring-based models is usually the same but stiffness properties vary depending on the values cited from the literature. All teams acknowledge the necessity to prescribe and eventually calibrate ligament slack lengths (or in situ strain, its material counterpart). Initial choices of these vary largely depending on the differences in the studies that they rely on. A few of the teams acknowledge modeling decisions that are specific to the medial collateral ligament, e.g., wrapping around the bones (by three teams) and its attachment to the medial meniscus (by two teams). Some of these modeling choices are driven by computational cost, while others are based on the desire to faithfully capture specimen-specific anatomy and to predict full field stress–strain response within the ligament. In some cases, they are based on prior modeling success to mitigate risks associated with convergence of simulations. It is possible that each model yields an equivalent force-deformation response of the medial collateral ligament independent of the modeling strategy. A recent study however, reported that the fidelity of ligament models can lead to large variability in predictions of joint level kinematics and contact even when modeling decisions are made within one research group [74]. Thus, predictions of ligament forces, and their contributions to knee kinematics and kinetics, might vary even more given the diversity of the modeling approaches of the separate teams. The level of agreement across groups is not yet determined and will be a focus of our work.
4. Potential Implications and Moving Ahead
This work reflects broader efforts by federal research agencies including the National Institute of Health to ensure reproducibility in the biomedical sciences [75]. The outcomes of a study to understand the art of modeling and simulation in knee biomechanics (and its influence on reproducibility and credibility) have implications that are broadly applicable in simulation-based research and medicine. In one scenario, simulations conducted by different models, which are based on the same data, will result in the same conclusions. This outcome will confirm that the modelers art does not have a substantial impact on scientific and clinical interpretation. Such a finding implies that a modeling and simulation strategy that is convenient and responsive to available expertise, resources, and protocols can be adapted. More divergent outcomes are also possible, i.e., knee models built by different teams may result in different conclusions in simulation studies that are scientifically and clinically relevant. This finding would reflect concerns of the scientific community regarding reproducibility in biomedical research. This outcome would provide the knee modeling community a valuable learning opportunity to increase the reproducibility of modeling and simulation workflows. When investigators model the same knee and rely on the same anatomical and mechanical data, they cannot use inherent variability in modeled samples as a justification for heterogeneity in model predictions [76]. Instead, a comprehensive examination of modeling and simulation protocols and their intermediate products need to be conducted. The origins of deviations can be pinpointed by evaluating geometric and mechanical representations of individual joint components, e.g., ligaments, cartilage, menisci, and loading and boundary conditions.
An in-depth examination of modeling and simulation processes, their data needs, and their outputs will have important implications on the economics of modeling and simulation, standards and good practice guidelines, and data and model exchange requirements. If protocols, e.g., anatomy segmentation, are interchangeable, one that requires lower amount of resources can be recommended as a cost-effective strategy. Overlaps between approaches may point toward a consensus for unification of knee modeling. Differences may be objectively assessed to identify minimum required fidelity of tissue representations, i.e., for approximation of ligament function. A modeler may want to substitute a tissue representation, e.g., meniscus, with that of another's to enhance his/her virtual interpretation of the same knee. This will motivate development of tools to facilitate switching between different data structures [77] and software formats [78], therefore enabling model sharing in a grass roots manner.
A complete record of modeling and simulation activities along with input data, resultant models, and simulation predictions has important implications for reproducibility and credibility in computational biomechanics. Journal articles may be limited to an abridged description of simulation studies and models that they depend on, even when performed in a systematic and comprehensive manner [6]. When models are shared, simulations can be repeated with ease [79]. Yet, reproducibility of model development, calibration, and benchmarking processes can still be questionable. Completion of the proposed study will provide the opportunity for anyone to test reproducibility by using the same shared data, following the disseminated documentation (specification and protocol deviations) to ideally reach the same simulation results and/or conclusions. The multitude of competing implementations of the same virtual knee will likely enhance confidence in the modeling practice, as also noted by broadly applicable guidance initiatives in the biomedical modeling and simulation community [39].
The deliverables of the proposed study can support regulatory use of computational modeling in knee biomechanics and serve as data and metadata to support regulatory science. The FDA is leading regulatory science initiatives, those that help to establish tools, methods and approaches to assess the safety, efficacy, quality, and performance of FDA-regulated products [80]. The FDA calls out for specific uses of modeling and simulation to better predict product safety and efficacy, including computational representation of an anatomical or physiological structure, such as the human knee, where medical devices can be virtually implanted and evaluated for performance. Within the last few years, the Center for Devices and Radiological Health (CDRH) has recognized computational modeling as a regulatory science priority [81], where simulation evidence would be used to support the marketing application for the medical device (relying on the FDA-recommended specific format for the report) [58]. A modeling and simulation platform might be considered a “nonclinical assessment model” as part of the medical device development tools program [82], which supports the qualification of tools that aid medical device evaluation. Synergistically, the Office of Science and Engineering Laboratories, the research arm of CDRH, has dozens of ongoing research projects to advance scientific aspects of computing, and verification and validation (V & V) methodologies to assess the credibility of modeling and simulation [83]. The FDA has provided leadership to support the advancements of credibility methods, such as V & V through the ASME V & V 40 subcommittee [84], which focuses V & V for modeling and simulation applied to medical devices. The new ASME V & V 40 standard has been published recently [57], providing a framework for determining the amount of rigor (in terms of V & V) needed to support using a computational model for a specific context of use. Outcomes of knee modeling and simulation processes and the way they are described herein can be used as exemplars of existing regulatory guidance and may support mock reviews aimed for training.
The immediate impact of deciphering the art of simulation-based knee biomechanics is founded on the need for increased dependability and quality of knee models. Through public dissemination, different types of knee models and related simulation workflows will become available to scientists, clinicians, and trainees for any particular use. Detailed documentation of implementation details and motivation and justification behind the preferences of modelers will eventually lead to standardization and best practices of knee modeling, facilitating exchange, and repurposing.
Description of a variety of knee modeling workflows and delivery of their outcomes are ongoing at the public project site [42]. The collaborating teams look forward to engaging with the broader knee biomechanics community to establish a collective understanding of the art and reproducibility of knee modeling. Any interested party is invited to submit their own modeling and simulation workflows, i.e., their art, to build models and conduct simulations, from start to end, using the disseminated data and simulation goals. Knee modelers are encouraged to follow protocols already submitted by the teams to test their reproducibility. This exercise may be particularly useful for trainees. While learning the art of modeling, they can contribute to the science of modeling and simulation in knee biomechanics. Of course, this approach and the insights gained in the practice of modeling and simulation can be applied in other musculoskeletal joints and organs, and to different modeling modalities.
Acknowledgment
AE outlined the document, wrote the first full draft and acquired input from co-authors. All co-authors added content in relevant sections, reviewed, and edited the draft and confirmed their agreement with the contents of the document.
Footnotes
Contributor Information
Ahmet Erdemir, Department of Biomedical Engineering and Computational Biomodeling (CoBi) Core, Lerner Research Institute, Cleveland Clinic, 9500 Euclid Avenue (ND20), Cleveland, OH 44195 e-mail: erdemira@ccf.org.
Thor F. Besier, Department of Engineering Science, Auckland Bioengineering Institute, University of Auckland, Auckland 1010, New Zealand
Jason P. Halloran, Department of Mechanical Engineering, Center for Human Machine Systems, Cleveland State University, Cleveland, OH 44115
Carl W. Imhauser, Department of Biomechanics, Hospital for Special Surgery, New York, NY 10021
Peter J. Laz, Department of Mechanical and Materials Engineering, Center for Orthopaedic Biomechanics, University of Denver, Denver, CO 80210
Tina M. Morrison, Division of Applied Mechanics, Office of Science and Engineering Laboratories, Center for Devices and Radiological Health, Food and Drug Administration, Silver Spring, MD 20993
Kevin B. Shelburne, Department of Mechanical and Materials Engineering, Center for Orthopaedic Biomechanics, University of Denver, Denver, CO 80210
Funding Data
National Institute of Biomedical Imaging and Bioengineering (Funder ID: 10.13039/100000070).
National Institutes of Health (Grant Nos. R01EB024573, R01GM104139, and R01EB015497; Funder ID: 10.13039/100000002).
References
- [1]. Maas, S. A. , Ateshian, G. A. , and Weiss, J. A. , 2017, “ FEBio: History and Advances,” Annu. Rev. Biomed. Eng., 19, pp. 279–299. 10.1146/annurev-bioeng-071516-044738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2]. Seth, A. , Hicks, J. L. , Uchida, T. K. , Habib, A. , Dembia, C. L. , Dunne, J. J. , Ong, C. F. , DeMers, M. S. , Rajagopal, A. , Millard, M. , Hamner, S. R. , Arnold, E. M. , Yong, J. R. , Lakshmikanth, S. K. , Sherman, M. A. , Ku, J. P. , and Delp, S. L. , 2018, “ OpenSim: Simulating Musculoskeletal Dynamics and Neuromuscular Control to Study Human and Animal Movement,” PLoS Comput. Biol., 14(7), p. e1006223. 10.1371/journal.pcbi.1006223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3]. Kazemi, M. , Dabiri, Y. , and Li, L. P. , 2013, “ Recent Advances in Computational Mechanics of the Human Knee Joint,” Comput. Math. Methods Med., 2013, p. 718423. 10.1155/2013/718423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4]. Peters, A. E. , Akhtar, R. , Comerford, E. J. , and Bates, K. T. , 2018, “ Tissue Material Properties and Computational Modelling of the Human Tibiofemoral Joint: A Critical Review,” PeerJ., 6, p. e4298. 10.7717/peerj.4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5]. Taylor, M. , and Prendergast, P. J. , 2015, “ Four Decades of Finite Element Analysis of Orthopaedic Devices: Where are We Now and What Are the Opportunities?,” J. Biomech., 48(5), pp. 767–778. 10.1016/j.jbiomech.2014.12.019 [DOI] [PubMed] [Google Scholar]
- [6]. Erdemir, A. , Guess, T. M. , Halloran, J. , Tadepalli, S. C. , and Morrison, T. M. , 2012, “ Considerations for Reporting Finite Element Analysis Studies in Biomechanics,” J. Biomech., 45(4), pp. 625–633. 10.1016/j.jbiomech.2011.11.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]. Erdemir, A. , 2016, “ Open Knee: Open Source Modeling and Simulation in Knee Biomechanics,” J. Knee Surg., 29(2), pp. 107–116. 10.1055/s-0035-1564600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. Harris, M. D. , Cyr, A. J. , Ali, A. A. , Fitzpatrick, C. K. , Rullkoetter, P. J. , Maletsky, L. P. , and Shelburne, K. B. , 2016, “ A Combined Experimental and Computational Approach to Subject-Specific Analysis of Knee Joint Laxity,” ASME J. Biomech. Eng., 138(8), p. 081004. 10.1115/1.4033882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9]. Kia, M. , Schafer, K. , Lipman, J. , Cross, M. , Mayman, D. , Pearle, A. , Wickiewicz, T. , and Imhauser, C. , 2016, “ A Multibody Knee Model Corroborates Subject-Specific Experimental Measurements of Low Ligament Forces and Kinematic Coupling During Passive Flexion,” ASME J. Biomech. Eng., 138(5), p. 051010. 10.1115/1.4032850 [DOI] [PubMed] [Google Scholar]
- [10]. Balci, O. , 2012, “ A Life Cycle for Modeling and Simulation,” Simulation, 88(7), pp. 870–883. 10.1177/0037549712438469 [DOI] [Google Scholar]
- [11]. Henak, C. R. , Anderson, A. E. , and Weiss, J. A. , 2013, “ Subject-Specific Analysis of Joint Contact Mechanics: Application to the Study of Osteoarthritis and Surgical Planning,” ASME J. Biomech. Eng., 135(2), p. 021003. 10.1115/1.4023386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]. Maletsky, L. , Shalhoub, S. , Fitzwater, F. , Eboch, W. , Dickinson, M. , Akhbari, B. , and Louie, E. , 2016, “ In Vitro Experimental Testing of the Human Knee: A Concise Review,” J. Knee Surg., 29(2), pp. 138–148. 10.1055/s-0035-1566739 [DOI] [PubMed] [Google Scholar]
- [13]. Guan, S. , Gray, H. A. , Keynejad, F. , and Pandy, M. G. , 2016, “ Mobile Biplane X-Ray Imaging System for Measuring 3D Dynamic Joint Motion During Overground Gait,” IEEE Trans. Med. Imaging, 35(1), pp. 326–336. 10.1109/TMI.2015.2473168 [DOI] [PubMed] [Google Scholar]
- [14]. Liukkonen, M. K. , Mononen, M. E. , Tanska, P. , Saarakkala, S. , Nieminen, M. T. , and Korhonen, R. K. , 2017, “ Application of a Semi-Automatic Cartilage Segmentation Method for Biomechanical Modeling of the Knee Joint,” Comput. Methods Biomech. Biomed. Eng., 20(13), pp. 1453–1463. 10.1080/10255842.2017.1375477 [DOI] [PubMed] [Google Scholar]
- [15]. Erdemir, A. , McLean, S. , Herzog, W. , and van den Bogert, A. J. , 2007, “ Model-Based Estimation of Muscle Forces Exerted During Movements,” Clin. Biomech., 22(2), pp. 131–154. 10.1016/j.clinbiomech.2006.09.005 [DOI] [PubMed] [Google Scholar]
- [16]. Giambini, H. , Dragomir-Daescu, D. , Nassr, A. , Yaszemski, M. J. , and Zhao, C. , 2016, “ Quantitative Computed Tomography Protocols Affect Material Mapping and Quantitative Computed Tomography-Based Finite-Element Analysis Predicted Stiffness,” ASME J. Biomech. Eng., 138(9), p. 091003. 10.1115/1.4034172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17]. Westover, L. M. , Sinaei, N. , Küpper, J. C. , and Ronsky, J. L. , 2016, “ Quantifying In Vivo Laxity in the Anterior Cruciate Ligament and Individual Knee Joint Structures,” Comput. Methods Biomech. Biomed. Eng., 19(14), pp. 1567–1577. 10.1080/10255842.2016.1170122 [DOI] [PubMed] [Google Scholar]
- [18]. Razu, S. S. , and Guess, T. M. , 2018, “ Electromyography-Driven Forward Dynamics Simulation to Estimate In Vivo Joint Contact Forces During Normal, Smooth, and Bouncy Gaits,” ASME J. Biomech. Eng., 140(7), p. 071012. 10.1115/1.4038507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19]. Schafer, K. A. , Tucker, S. , Griffith, T. , Sheikh, S. , Wickiewicz, T. L. , Nawabi, D. H. , Imhauser, C. W. , and Pearle, A. D. , 2016, “ Distribution of Force in the Medial Collateral Ligament Complex During Simulated Clinical Tests of Knee Stability,” Am. J. Sports Med., 44(5), pp. 1203–1208. 10.1177/0363546515623510 [DOI] [PubMed] [Google Scholar]
- [20]. Bates, N. A. , Schilaty, N. D. , Nagelli, C. V. , Krych, A. J. , and Hewett, T. E. , 2018, “ Validation of Noncontact Anterior Cruciate Ligament Tears Produced by a Mechanical Impact Simulator Against the Clinical Presentation of Injury,” Am. J. Sports Med., 46(9), pp. 2113–2121. 10.1177/0363546518776621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. Ewing, J. A. , Kaufman, M. K. , Hutter, E. E. , Granger, J. F. , Beal, M. D. , Piazza, S. J. , and Siston, R. A. , 2016, “ Estimating Patient-Specific Soft-Tissue Properties in a TKA Knee,” J. Orthop. Res., 34(3), pp. 435–443. 10.1002/jor.23032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22]. Kiapour, A. , Kiapour, A. M. , Kaul, V. , Quatman, C. E. , Wordeman, S. C. , Hewett, T. E. , Demetropoulos, C. K. , and Goel, V. K. , 2013, “ Finite Element Model of the Knee for Investigation of Injury Mechanisms: Development and Validation,” ASME J. Biomech. Eng., 136(1), p. 011002. 10.1115/1.4025692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23]. Dhaher, Y. Y. , Kwon, T.-H. , and Barry, M. , 2010, “ The Effect of Connective Tissue Material Uncertainties on Knee Joint Mechanics Under Isolated Loading Conditions,” J. Biomech., 43(16), pp. 3118–3125. 10.1016/j.jbiomech.2010.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. Sharifi, M. , Shirazi-Adl, A. , and Marouane, H. , 2017, “ Computational Stability of Human Knee Joint at Early Stance in Gait: Effects of Muscle Coactivity and Anterior Cruciate Ligament Deficiency,” J. Biomech., 63, pp. 110–116. 10.1016/j.jbiomech.2017.08.004 [DOI] [PubMed] [Google Scholar]
- [25]. Ali, A. A. , Harris, M. D. , Shalhoub, S. , Maletsky, L. P. , Rullkoetter, P. J. , and Shelburne, K. B. , 2017, “ Combined Measurement and Modeling of Specimen-Specific Knee Mechanics for Healthy and ACL-Deficient Conditions,” J. Biomech., 57, pp. 117–124. 10.1016/j.jbiomech.2017.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26]. Erdemir, A. , Hunter, P. J. , Holzapfel, G. A. , Loew, L. M. , Middleton, J. , Jacobs, C. R. , Nithiarasu, P. , Löhner, R. , Wei, G. , Winkelstein, B. A. , Barocas, V. H. , Guilak, F. , Ku, J. P. , Hicks, J. L. , Delp, S. L. , Sacks, M. , Weiss, J. A. , Ateshian, G. A. , Maas, S. A. , McCulloch, A. D. , and Peng, G. C. Y. , 2018, “ Perspectives on Sharing Models and Related Resources in Computational Biomechanics Research,” ASME J. Biomech. Eng., 140(2), p. 024701. 10.1115/1.4038768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27]. Sibole, S. C. , and Erdemir, A. , 2012, “ Chondrocyte Deformations as a Function of Tibiofemoral Joint Loading Predicted by a Generalized High-Throughput Pipeline of Multi-Scale Simulations,” PLoS One, 7(5), p. e37538. 10.1371/journal.pone.0037538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28]. Mesfar, W. , and Shirazi-Adl, A. , 2006, “ Biomechanics of Changes in ACL and PCL Material Properties or Prestrains in Flexion Under Muscle Force-Implications in Ligament Reconstruction,” Comput. Methods Biomech. Biomed. Eng., 9(4), pp. 201–209. 10.1080/10255840600795959 [DOI] [PubMed] [Google Scholar]
- [29]. Shirazi, R. , and Shirazi-Adl, A. , 2009, “ Analysis of Partial Meniscectomy and ACL Reconstruction in Knee Joint Biomechanics Under a Combined Loading,” Clin. Biomech., 24(9), pp. 755–761. 10.1016/j.clinbiomech.2009.07.005 [DOI] [PubMed] [Google Scholar]
- [30]. Bates, N. A. , McPherson, A. L. , Nesbitt, R. J. , Shearn, J. T. , Myer, G. D. , and Hewett, T. E. , 2017, “ Robotic Simulation of Identical Athletic-Task Kinematics on Cadaveric Limbs Exhibits a Lack of Differences in Knee Mechanics Between Contralateral Pairs,” J. Biomech., 53, pp. 36–44. 10.1016/j.jbiomech.2016.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31]. Rao, C. , Fitzpatrick, C. K. , Rullkoetter, P. J. , Maletsky, L. P. , Kim, R. H. , and Laz, P. J. , 2013, “ A Statistical Finite Element Model of the Knee Accounting for Shape and Alignment Variability,” Med. Eng. Phys., 35(10), pp. 1450–1456. 10.1016/j.medengphy.2013.03.021 [DOI] [PubMed] [Google Scholar]
- [32]. Nuño, N. , and Ahmed, A. M. , 2001, “ Sagittal Profile of the Femoral Condyles and Its Application to Femorotibial Contact Analysis,” ASME J. Biomech. Eng., 123(1), pp. 18–26. 10.1115/1.1339819 [DOI] [PubMed] [Google Scholar]
- [33]. Guo, H. , Santner, T. J. , Lerner, A. L. , and Maher, S. A. , 2017, “ Reducing Uncertainty When Using Knee-Specific Finite Element Models by Assessing the Effect of Input Parameters,” J. Orthop. Res., 35(10), pp. 2233–2242. 10.1002/jor.23513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34]. Galbusera, F. , Freutel, M. , Dürselen, L. , D'Aiuto, M. , Croce, D. , Villa, T. , Sansone, V. , and Innocenti, B. , 2014, “ Material Models and Properties in the Finite Element Analysis of Knee Ligaments: A Literature Review,” Front Bioeng. Biotechnol., 2, p. 54. 10.3389/fbioe.2014.00054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35]. Rachmat, H. H. , Janssen, D. , Zevenbergen, W. J. , Verkerke, G. J. , Diercks, R. L. , and Verdonschot, N. , 2014, “ Generating Finite Element Models of the Knee: How Accurately Can We Determine Ligament Attachment Sites From MRI Scans?,” Med. Eng. Phys., 36(6), pp. 701–707. 10.1016/j.medengphy.2014.02.016 [DOI] [PubMed] [Google Scholar]
- [36]. Donahue, T. L. H. , Hull, M. L. , Rashid, M. M. , and Jacobs, C. R. , 2002, “ A Finite Element Model of the Human Knee Joint for the Study of Tibio-Femoral Contact,” ASME J. Biomech. Eng., 124(3), pp. 273–280. 10.1115/1.1470171 [DOI] [PubMed] [Google Scholar]
- [37]. Halloran, J. P. , Petrella, A. J. , and Rullkoetter, P. J. , 2005, “ Explicit Finite Element Modeling of Total Knee Replacement Mechanics,” J. Biomech., 38(2), pp. 323–331. 10.1016/j.jbiomech.2004.02.046 [DOI] [PubMed] [Google Scholar]
- [38]. Li, G. , Gil, J. , Kanamori, A. , and Woo, S. L. , 1999, “ A Validated Three-Dimensional Computational Model of a Human Knee Joint,” ASME J. Biomech. Eng., 121(6), pp. 657–662. 10.1115/1.2800871 [DOI] [PubMed] [Google Scholar]
- [39]. Erdemir, A. , Mulugeta, L. , and Lytton, W. W. , 2015, “ Ten ‘Not So’ Simple Rules for Credible Practice of Modeling and Simulation in Healthcare: A Multidisciplinary Committee Perspective,” Biomedical Engineering Society/Food and Drug Administration Frontiers in Medical Devices Conference: Innovations in Modeling and Simulation, Washington, DC, May 18–20, 2015. [Google Scholar]
- [40]. Anderson, A. E. , Ellis, B. J. , and Weiss, J. A. , 2007, “ Verification, Validation and Sensitivity Studies in Computational Biomechanics,” Comput. Methods Biomech. Biomed. Eng., 10(3), pp. 171–184. 10.1080/10255840601160484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41]. Fregly, B. J. , Besier, T. F. , Lloyd, D. G. , Delp, S. L. , Banks, S. A. , Pandy, M. G. , and D'Lima, D. D. , 2012, “ Grand Challenge Competition to Predict In Vivo Knee Loads,” J. Orthop. Res., 30(4), pp. 503–513. 10.1002/jor.22023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].SimTK, 2018, “ SimTK: Reproducibility in Simulation-Based Prediction of Natural Knee Mechanics: Project Home,” SimTK, accessed Dec. 1, 2018, https://simtk.org/projects/kneehub
- [43].NIH, 2018, “ Project Information—NIH RePORTER—NIH Research Portfolio Online Reporting Tools Expenditures and Results,” National Institutes of Health, Bethesda, MD, accessed Dec. 1, 2018, https://projectreporter.nih.gov/project_info_description.cfm?aid=9366122&icde=41076822
- [44].SimTK, 2018, “ SimTK: Open Knee(s): Virtual Biomechanical Representations of the Knee Joint: Project Home,” SimTK, accessed Dec. 1, 2018, https://simtk.org/projects/openknee
- [45].University of Denver, 2018, “Natural Knee Data,” Center for Orthopaedic Biomechanics, University of Denver, Denver, CO, accessed Dec. 1, 2018, https://digitalcommons.du.edu/natural_knee_data/
- [46].SimTK, 2018, “ Specifications/ExperimentationAnatomicalImaging—Openknee,” SimTK, accessed Dec. 1, 2018, https://simtk.org/plugins/moinmoin/openknee/Specifications/ExperimentationAnatomicalImaging
- [47].SimTK, 2018, “ Specifications/ExperimentationJointMechanics—Openknee,” SimTK, accessed Dec. 1, 2018, https://simtk.org/plugins/moinmoin/openknee/Specifications/ExperimentationJointMechanics
- [48]. Ali, A. A. , Shalhoub, S. S. , Cyr, A. J. , Fitzpatrick, C. K. , Maletsky, L. P. , Rullkoetter, P. J. , and Shelburne, K. B. , 2016, “ Validation of Predicted Patellofemoral Mechanics in a Finite Element Model of the Healthy and Cruciate-Deficient Knee,” J. Biomech., 49(2), pp. 302–309. 10.1016/j.jbiomech.2015.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49]. Henninger, H. B. , Reese, S. P. , Anderson, A. E. , and Weiss, J. A. , 2010, “ Validation of Computational Models in Biomechanics,” Proc. Inst. Mech. Eng. H, 224(7), pp. 801–812. 10.1243/09544119JEIM649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50]. Wilson, D. R. , Feikes, J. D. , Zavatsky, A. B. , and O'Connor, J. J. , 2000, “ The Components of Passive Knee Movement are Coupled to Flexion Angle,” J. Biomech., 33(4), pp. 465–473. 10.1016/S0021-9290(99)00206-7 [DOI] [PubMed] [Google Scholar]
- [51]. Arilla, F. V. , Yeung, M. , Bell, K. , Rahnemai-Azar, A. A. , Rothrauff, B. B. , Fu, F. H. , Debski, R. E. , Ayeni, O. R. , and Musahl, V. , 2015, “ Experimental Execution of the Simulated Pivot-Shift Test: A Systematic Review of Techniques,” Arthroscopy, 31(12), pp. 2445–2454. 10.1016/j.arthro.2015.06.027 [DOI] [PubMed] [Google Scholar]
- [52]. Felson, D. T. , Nevitt, M. C. , Yang, M. , Clancy, M. , Niu, J. , Torner, J. C. , Lewis, C. E. , Aliabadi, P. , Sack, B. , McCulloch, C. , and Zhang, Y. , 2008, “ A New Approach Yields High Rates of Radiographic Progression in Knee Osteoarthritis,” J. Rheumatol., 35(10), pp. 2047–2054.http://www.jrheum.org/content/35/10/2047.long [PMC free article] [PubMed] [Google Scholar]
- [53]. Caruthers, E. J. , Thompson, J. A. , Chaudhari, A. M. W. , Schmitt, L. C. , Best, T. M. , Saul, K. R. , and Siston, R. A. , 2016, “ Muscle Forces and Their Contributions to Vertical and Horizontal Acceleration of the Center of Mass During Sit-to-Stand Transfer in Young, Healthy Adults,” J. Appl. Biomech., 32(5), pp. 487–503. 10.1123/jab.2015-0291 [DOI] [PubMed] [Google Scholar]
- [54]. Bland, J. M. , and Altman, D. G. , 2007, “ Agreement Between Methods of Measurement With Multiple Observations per Individual,” J. Biopharm. Stat., 17(4), pp. 571–582. 10.1080/10543400701329422 [DOI] [PubMed] [Google Scholar]
- [55]. Taha, A. A. , and Hanbury, A. , 2015, “ An Efficient Algorithm for Calculating the Exact Hausdorff Distance,” IEEE Trans. Pattern Anal. Mach. Intell., 37(11), pp. 2153–2163. 10.1109/TPAMI.2015.2408351 [DOI] [PubMed] [Google Scholar]
- [56].NASA, 2016, “ NASA Technical Standards System (NTSS),” National Aeronautics and Space Administration, Washington, DC, Document No. NASA-STD-7009.https://standards.nasa.gov/standard/nasa/nasa-std-7009
- [57].ASME, 2018, “ Assessing Credibility of Computational Modeling Through Verification and Validation: Application to Medical Devices,” American Society of Mechanical Engineers, New York, Standard No. V V 40 - 2018.https://www.asme.org/products/codes-standards/vv-40-2018-assessing-credibility-computational
- [58].U.S. FDA, 2018, “ Reporting of Computational Modeling Studies in Medical Device Submissions—Guidance for Industry and Food and Drug Administration Staff,” U.S. Food & Drug Administration, Silver Spring, MD, accessed Dec. 1, 2018, https://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/UCM381813.pdf
- [59].SimTK, 2018, “ ModelDevelopment—Kneehub,” SimTK, accessed Dec. 1, 2018, https://simtk.org/plugins/moinmoin/kneehub/ModelDevelopment
- [60].SimTK, 2018, “ SimTK: Reproducibility in Simulation-Based Prediction of Natural Knee Mechanics: Documents,” SimTK, accessed Dec. 1, 2018, https://simtk.org/docman/?group_id=1061
- [61].3D Slicer, 2018, “ 3D Slicer,” accessed Dec. 1, 2018, https://www.slicer.org/
- [62].MeshLab, 2018, “ MeshLab,” accessed Dec. 1, 2018, http://www.meshlab.net/
- [63].Synopsis, 2018, “ Simpleware ScanIP,” Synopsis, Mountain View, CA, accessed Dec. 1, 2018, https://www.synopsys.com/simpleware/products/software/scanip.html
- [64].MathWorks, 2018, “ MATLAB—MathWorks—MATLAB & Simulink,” The Mathworks, Natick, MA, accessed Dec. 1, 2018, https://www.mathworks.com/products/matlab.html
- [65].Materialise, 2018, “3 D Medical Image Processing Software | Materialise Mimics,” Materialise NV, Leuven, Belgium, accessed Dec. 1, 2018, https://www.materialise.com/en/medical/software/mimics
- [66].3D Systems, 2018, “ Software,” 3D Systems, Rock Hill, SC, accessed Dec. 1, 2018, https://www.3dsystems.com/software
- [67].SALOME, 2018, “ Welcome to the www.Salome-Platform.Org—SALOME Platform,” SALOME, accessed Dec. 1, 2018, http://www.salome-platform.org/
- [68].Altair, 2018, “ Large Model Finite Element Preprocessing—Altair HyperMesh,” Altair, Troy, MI, accessed Dec. 1, 2018, https://altairhyperworks.com/product/hypermesh
- [69].MAP, 2018, “ Musculoskeletal Atlas Project (MAP) Client Documentation—Latest—MAP Client Latest Documentation,” accessed Dec. 1, 2018, https://map-client.readthedocs.io/en/latest/
- [70].CCAD, 2018, “IA-FEMesh,” Center for Computer Aided Design, The University of Iowa, Iowa City, IA, accessed Dec. 1, 2018, https://www.ccad.uiowa.edu/MIMX/projects/IA-FEMesh
- [71].FEBio, 2018, “ FEBio Software Suite,” accessed Dec. 1, 2018, https://febio.org/
- [72].SIMULIA, 2018, “ Abaqus Unified FEA—SIMULIATM by Dassault Systèmes®,” SIMULIA, Johnston, RI, accessed Dec. 1, 2018, https://www.3ds.com/products- services/simulia/products/abaqus/
- [73].MSC, 2018, “ Adams—The Multibody Dynamics Simulation Solution,” MSC Software, Newport Beach, CA, accessed Dec. 1, 2018, http://www.mscsoftware.com/product/adams
- [74]. Naghibi Beidokhti, H. , Janssen, D. , van de Groes, S. , Hazrati, J. , Van den Boogaard, T. , and Verdonschot, N. , 2017, “ The Influence of Ligament Modelling Strategies on the Predictive Capability of Finite Element Models of the Human Knee Joint,” J. Biomech., 65, pp. 1–11. 10.1016/j.jbiomech.2017.08.030 [DOI] [PubMed] [Google Scholar]
- [75]. Collins, F. S. , and Tabak, L. A. , 2014, “ Policy: NIH Plans to Enhance Reproducibility,” Nature, 505(7485), pp. 612–613. 10.1038/505612a [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76]. Dreischarf, M. , Zander, T. , Shirazi-Adl, A. , Puttlitz, C. M. , Adam, C. J. , Chen, C. S. , Goel, V. K. , Kiapour, A. , Kim, Y. H. , Labus, K. M. , Little, J. P. , Park, W. M. , Wang, Y. H. , Wilke, H. J. , Rohlmann, A. , and Schmidt, H. , 2014, “ Comparison of Eight Published Static Finite Element Models of the Intact Lumbar Spine: Predictive Power of Models Improves When Combined Together,” J. Biomech., 47(8), pp. 1757–1766. 10.1016/j.jbiomech.2014.04.002 [DOI] [PubMed] [Google Scholar]
- [77]. Britten, R. D. , Christie, G. R. , Little, C. , Miller, A. K. , Bradley, C. , Wu, A. , Yu, T. , Hunter, P. , and Nielsen, P. , 2013, “ FieldML, A Proposed Open Standard for the Physiome Project for Mathematical Model Representation,” Med. Biol. Eng. Comput., 51(11), pp. 1191–1207. 10.1007/s11517-013-1097-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78]. Meng, Q. , Jin, Z. , Fisher, J. , and Wilcox, R. , 2013, “ Comparison Between FEBio and Abaqus for Biphasic Contact Problems,” Proc. Inst. Mech. Eng. H, 227(9), pp. 1009–1019. 10.1177/0954411913483537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79]. Erdemir, A. , Guess, T. M. , Halloran, J. P. , Modenese, L. , Reinbolt, J. A. , Thelen, D. G. , and Umberger, B. R. , 2016, “ Commentary on the Integration of Model Sharing and Reproducibility Analysis to Scholarly Publishing Workflow in Computational Biomechanics,” IEEE Trans. Biomed. Eng., 63(10), pp. 2080–2085. 10.1109/TBME.2016.2602760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].U.S. FDA, 2011, “ Advancing Regulatory Science at FDA: A Strategic Plan, August 2011,” U.S. Food & Drug Administration, Silver Spring, MD, accessed Dec. 1, 2018, https://www.fda.gov/downloads/ScienceResearch/SpecialTopics/RegulatoryScience/UCM268225.pdf
- [81].U.S. FDA, 2017, “ CDRH Regulatory Science Priorities (FY 2017),” U.S. Food & Drug Administration, Silver Spring, MD, accessed Dec. 1, 2018, https://www.fda.gov/downloads/medicaldevices/scienceandresearch/ucm521503. pdf
- [82].U.S. FDA, 2018, “ Medical Device Development Tools (MDDT),” U.S. Food & Drug Administration, Silver Spring, MD, accessed Dec. 1, 2018, https://www.fda.gov/medicaldevices/scienceandresearch/medicaldevicedevelopment toolsmddt/
- [83]. Morrison, T. M. , Pathmanathan, P. , Adwan, M. , and Margerrison, E. , 2018, “ Advancing Regulatory Science With Computational Modeling for Medical Devices at the FDA's Office of Science and Engineering Laboratories,” Front. Med., 5, p. 241. 10.3389/fmed.2018.00241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].ASME, 2018, “ Committee Pages—V & V 40 Verification and Validation in Computational Modeling of Medical Devices,” American Society of Mechanical Engineers, New York, accessed Dec. 1, 2018, https://cstools.asme.org/csconnect/CommitteePages.cfm?Committee= 100108782


