Abstract
Community-acquired pneumonia (CAP) is a worldwide infectious disease caused by bacteria, viruses, or a combination of these infectious agents. Mycoplasma pneumoniae is an atypical pneumonia pathogen that causes high morbidity and mortality in children, and adenovirus can lead to severe pneumonia. However, the etiology of different types of pneumonia is still unclear. In this study, we selected a total of 52 inpatients with M. pneumoniae pneumonia (MPP) (n = 21), adenovirus pneumonia (AVP) (n = 16), or tracheomalacia (n = 15) to serve as a disease control. Bronchoalveolar lavage fluid (BALF) samples that had been obtained for clinical use were analyzed. We compared the differences in microbiota and the expression of 10 inflammatory cytokines in samples between MPP, AVP, and tracheomalacia. We found that the bacterial diversity in MPP was lower than that in AVP and tracheomalacia. Mycoplasma, Streptococcus, and Pseudomonas were predominant in samples of MPP, AVP, and tracheomalacia, respectively. The expression levels of IL-6, IL-8, and IL-10 were significantly higher in inpatients with AVP compared to children hospitalized with tracheomalacia or MPP. The lung microbiota in MPP was remarkably correlated with IL-2, IL-4, IL-5, IL-6, TNF-α, and IL-1α expressions, while this was not found in tracheomalacia and AVP. Microbiota analysis identified a high load of multi-drug resistant Acinetobacter baumannii in the lung microbiota of several inpatients, which might be associated with the long hospitalization length and intra-group differences at the individual level. This study will help to understand the microbial etiology of tracheomalacia, AVP, and MPP and to identify effective therapies for these diseases.
Keywords: adenovirus, bronchoalveolar lavage, cytokine, microbiota, Mycoplasma pneumonia
Introduction
Pneumonia presents a high risk of morbidity and mortality in children under 5 years (1, 2). Bacterial pathogens are usually the main contributors to pneumonia incidence and progression, but atypical pneumonia and viral pneumonia are often more severe. In China, Mycoplasma pneumoniae pneumonia (MPP) generally results in the highest incidence of atypical pathogen pneumonia (3), whereas adenoviral pneumonia (AVP) represents the highest severity pneumonia in children (4).
A variety of microbial commensals that play an indispensable part in immune education and colonization resistance to respiratory pathogens (5, 6) colonize the respiratory tract. Emerging reports have demonstrated patterns of imbalanced upper respiratory tract microbiota in children hospitalized with pneumonia (7–9). The Haemophilus-dominant nasopharyngeal microbiota of patients infected by respiratory syncytial virus are positively correlated with intensive care use and hospital length-of-stay, while Moraxella-dominant or Streptococcus-dominant microbial profile is associated with chronic or allergic diseases (10). The infrequent Leptotrichia rises to the top five genera in patients infected with influenza virus (11, 12). These results imply that imbalanced respiratory microbiota can be pathogen specific. In addition, there is diverse bronchoalveolar lavage fluid (BALF) microbiota between cystic fibrosis, MPP, and Streptococcus pneunomiae pneumonia (SPP) patients (13, 14).
Several blood biomarkers have been clinically tested in children with community-acquired pneumonia (CAP), but the etiological diagnosis and estimation of potential outcomes remain unsolved in most cases. Among traditional biomarkers, two host protein assays based on C-reactive protein (CRP) and procalcitonin (PCT) are widely used in pneumonia diagnosis, with PCT being more sensitive to bacterial/non-bacterial cases and for evaluating severity (15). Pavord reported that the blood eosinophil count is related to the patient risk of pneumonia (16). However, none of the known biomarkers can precisely separate bacterial from viral infection and mild from severe disease.
The mucosal immune system of the respiratory tract can secrete various pro- and anti-inflammatory cytokines, including various interleukins (IL), tumor necrosis factor (TNF) family members, and interferon (IFN) to regulate host immune activity, eliminate pathogens, and recover pulmonary homeostasis (17, 18) during respiratory infection. Recent studies have indicated that host immune responses differ according to disease severity and infectious agents during acute respiratory infection (10, 19). The expression of cytokines in pneumonia patients was pathogen specific. According to previous studies, IL-6, IL-8, and IFN-γ represent the lung immune to response to adenovirus infection (17, 20), and IL-1α, IL-2, IL-4, IL-5, IL-6, IL-8, IL-17A, IFN-γ, and TNF-α are the major inflammatory cytokines induced by M. pneumoniae infection (17, 21–23). Given the severity and lack of preventive measures in MPP and AVP, it is necessary to explore imbalanced lung microbiota, inflammatory cytokines, and traditional biomarkers to improve clinical intervention.
In this study, we addressed the following two questions: (1) if and how the lung microbiota and immune-related cytokines are altered in children with MPP or AVP, and (2) how the imbalanced lung microecology is associated with pneumonia severity. Fifty-two diseased children were recruited from Shenzhen Children's Hospital, including patients with AVP (n = 16), MPP (n = 21), and tracheomalacia (n = 15), and were analyzed by 16S rDNA and customized cytokine assays.
Materials and Methods
Ethics Statement and Informed Consent
This study was carried out in accordance with the recommendations of Declaration of Helsinki, the Ethical Committee of Shenzhen Children's Hospital. The protocol was approved by the Ethical Committee of Shenzhen Children's Hospital under registration number 2016013. Due to the recruited children were <16 years old, their guardians gave written informed consent in accordance with the Declaration of Helsinki.
Study Subjects and Pathogen Detection
All children with pneumonia were recruited from inpatients in Shenzhen Children's Hospital. Given the disease-specific microbiota (24–26), antibiotic-induced microbiota dysbiosis (27), and the alteration in dominant bacteria by ventilator-associated pneumonia or hospital-acquired pneumonia (28), the exclusion criteria for patients were as follows: (1) other underlying pathologies; (2) antibiotics exposure within 1 month before hospitalization; (3) admitted to a pediatric intensive care unit; or (4) mechanical ventilation during hospitalization. According to the reference standard for the diagnosis of pneumonia (29), the inclusion criteria for children with MPP or AVP were as follows: (1) chest radiograph showing consolidation, atelectasis, or infiltration; (2) diagnosed as M. pneumoniae or adenovirus infection via BALF detection (Table 1; Table S1); and (3) no other common respiratory pathogens detected. Considering ethical issues and the invasive sampling method, BALF collection from healthy children is impossible. Instead, children who were diagnosed with tracheomalacia with physical trachea damage and without pathogen infection were selected as control subjects.
Table 1.
Summary of patients' characters and clinical records.
| AVP (n = 16) | MPP (n = 21) | T (n = 15) | p–value | ||||
|---|---|---|---|---|---|---|---|
| Kruskal-Wallis test | AVP-vs.-MPP | AVP-vs.-T | MPP-vs.-T | ||||
| CHARACTERISTICS | |||||||
| Gender | 0.143 | 0.571 | 0.433 | 0.106 | |||
| Female | 6 | 11 | 3 | ||||
| Male | 10 | 10 | 12 | ||||
| Age (years)# | 1.6 (0.4~4.0) | 4.5 (0.8~9.6) | 2.5 (0.3~6.7) | 0.000 | 0.004 | 0.874 | 0.021 |
| Delivery mode | 0.199 | 0.921 | 0.171 | 0.282 | |||
| Cesarean section | 8 | 9 | 3 | ||||
| Vaginally born | 8 | 12 | 12 | ||||
| Feed pattern | 0.163 | 0.680 | 0.045 | 0.385 | |||
| Breast feed | 14 | 16 | 8 | ||||
| Breast feed + Milk feed | 1 | 1 | 1 | ||||
| Milk feed | 1 | 4 | 6 | ||||
| Family history of allergy | 0 | 0 | 1 | 0.289 | 1.000 | 0.483 | 0.417 |
| History of pneumonia | 5 | 1 | 3 | 0.115 | 0.066 | 0.685 | 0.287 |
| Asthma | 0 | 0 | 0 | 1.000 | 1.000 | 1.000 | 1.000 |
| CLINICAL RECORDS | |||||||
| Lung consolidation, atelectasis, infiltration | 16 | 21 | 0 | 0.000 | 1.000 | 0.000 | 0.000 |
| Hospitalization length (days) # | 12.3 (7~21) | 9.1 (4~16) | 7.7 (2~13) | 0.010 | 0.089 | 0.040 | 0.479 |
| Cough | 16 | 19 | 14 | 0.620 | 0.500 | 0.484 | 1.000 |
| Fever | 16 | 16 | 2 | 0.000 | 0.680 | 0.045 | 0.385 |
| Wheezing | 5 | 2 | 6 | 0.024 | 0.066 | 1.000 | 0.029 |
| Fever duration (days) # | 16.6 (4~38) | 11.8 (0~30) | 1 (0~14) | 0.000 | 0.591 | 0.001 | 0.008 |
| Mean of peak temperature (°C) | 40.1 | 39.7 | 39.2 | NA | NA | NA | NA |
| CRP (<0.499 mg/l) | 4 | 5 | 6 | 0.549 | 1.000 | 0.458 | 0.465 |
| PCT (<0.5 ng/ml) | 5/11* | 21 | 3/4 | NA | NA | NA | NA |
| Eosinophil (0.5–5%) | 4 | 12 | 8 | 0.549 | 1.000 | 0.458 | 0.465 |
The normal reference ranges of CRP, PCT and Eosinophil values are <0.499 mg/l, <0.5 ng/ml, and 0.5–5%, according to the laboratory detection. “#,” measure with mean (min ~ max); “a/b
” means normal/detection number; AVP, Adenoviral Pneumonia; MPP, Mycoplasma pneumoniae Pneumonia; T, Tracheomalacia; CRP, C-reactive protein; PCT, Procalcitonin; NA, Not available.
BALF samples (three tubes, 1 ml/tube) were collected through fiber optic bronchoscopy (30). One set of BALF samples was immediately sent to the clinical lab for common pathogen detection. Acinetobacter baumannii, Haemophilus influenzae, Haemophilus parainfluenzae, Moraxella catarrhalis, Staphylococcus aureus, Staphylococcus haemolyticus, and S. pneumoniae were identified by bacterial culture (7). Nucleic acid testing (NAT) was used to diagnose atypical bacterial or virus infection, including that of M. pneumonia, Chlamydia pneumonia, adenovirus, respiratory syncytial virus, Epstein-bar virus, cytomegalovirus, or influenza viruses using specific NAT kits (9). The other two sets of BALF samples were stored at −80°C for cytokine assays and DNA sequencing. Sterile gauze, which was used for wiping the tip of the bronchoscope, served as a negative control (31). Saline solutions and DNA extraction kits were also used for contamination assessment.
Cytokine Immunoassays
We totally select 10 cytokines for detection with the Multiplex Immunoassay Panel (ProcartaPlex, ThermoFisher, Santa Clara, CA, USA), including pro-inflammatory cytokines (IL-1α, IL-2, IL-6, IL-8, IL-17A, IFN-γ, and TNF-α) and anti-inflammatory cytokines (IL-4, IL-5, and IL-10). The supernatant (25 μl) of BALF samples after centrifugation was added to 96-well filter plates and linked to anti-cytokine antibodies following the manufacturer's protocol. The fluorescent signal of each cytokine, indicating its expression level, was identified by a Pixel CCD camera (Luminex Corporation, Austin, TX, USA). The assay results were reported as mean fluorescence intensity (MFI) and were converted to clinically relevant measures (pg/mL) by log transformation according to xPonent® logistic curve fitting (Luminex Corporation, Austin, TX, USA) (32).
DNA Extraction, Library Construction, and Sequencing
Microbial DNA in BALF was extracted using the Power Soil DNA Isolation Kit (Mo Bio Laboratories, Carlsbad, UK) according to the manufacturer's protocol. The V3–V4 regions of the 16S rDNA gene were selected as the target sequence using 338F-806R primers. DNA amplification was carried out by a PCR kit (AP221-02, TransGen Biotech, Beijing, China) and quantified by Qubit (ThermoFisher, Singapore). The target sequence length was qualified through 2% agarose gel electrophoresis. The qualified DNA was concatenated with the sample-specific dual-index sequences, and the unneeded DNA was filtered by magnetic beads. Then, PCR was used to generate enough DNA for the MiSeq sequencing platform using the V3 sequencing reagent kit (Illumina, San Diego, USA) (33).
Sequence Processing, Statistical Analysis, and Visualization
The initial sequencing data were processed by the QIIME suite and in-house programs to filter reads as follows: low-quality, error sequences, abnormal read length, redundancies, and chimeric sequences were removed (9, 34). The qualified paired-reads were connected into tags and then clustered into operational taxonomic units (OTUs). The representative OTUs were aligned to the SILVA reference database (v119), and community diversity calculations were conducted following a previous study (14). Principal coordinate analysis (PCA) was used to inspect the divergence of all samples. The confounding effects of children's characteristics on microbial samples were evaluated by PERMANOVA, with the species annotation referring to previous reports (35, 36). Comparisons of patient characteristics and clinical records were performed with the Wilcoxon rank-sum test (continuous variables) and Chi-square test (categorical variables). Comparisons of microbial composition in two groups were conducted using the Wilcoxon rank-sum test, and all multiple testing results were adjusted by the false discovery rate (FDR). Kruskal–Wallis tests were utilized to compare cytokines and dominant genera in BALF microbiota among the MPP, AVP, and tracheomalacia groups. The correlations among clinical characteristics, cytokines expression, and diversity of the bacterial community were computed by linear regression (37). The correlations between microbiota composition and cytokines/CRP/eosinophils were calculated via PERMANOVA. All graphs were produced by “pheatmap” and “ggplot2” of the R software package (v3.2.3).
Results
Different Characteristics and Clinical Records in AVP, MPP, and Tracheomalacia Groups
The average age of AVP children (1.6 years) was younger than that of MPP (4.5 years, p-value = 0.004), but there was no significant difference with tracheomalacia patients (2.5 years, p-value = 0.874) (Table 1). Other characteristics showed no statistical significance between the three groups. AVP children had a higher ratio of abnormal PCT (54.5%) and eosinophils (75.0%) and suffered longer hospitalization length and fever duration as well as higher peak temperature compared to MPP (Table 1; Table S1). When comparing AVP/MPP with tracheomalacia patients, fever duration showed a significant difference (Table 1).
Sequencing Data Summary; the Disease Group Was the Most Important Factor Associated With Microbiota
A total of 1,379,984 high-quality tags were produced, averaging 26,504 (24,356–28,227), 26,905 (25,392–29,381), and 26,061 (16,894–28,186) for the AVP, MPP, and tracheomalacia groups, respectively. The average OTUs in AVP, MPP, and tracheomalacia groups numbered 616, 469, and 718, respectively. The concentration of DNA extracted from negative controls was lower than 0.01 ng/μl, whereas it was higher than 80 ng/μl in collected BALF samples. In addition, 16S rDNA gene amplification indicated <0.01 nmol/l bacterial DNA in enveloped DNA extraction materials.
Confounder analysis signified that the disease group was the most significant factor to explain variations in microbial samples (p-value <0.001), followed by feed pattern (p-value = 0.012), gender (p-value = 0.018), age (p-value = 0.022), and cough (p-value = 0.033) (Table S2). The other patient characteristics and clinical records were not strongly related to the microbiota.
The Lung Microbiota Structure Differed Dramatically Among MPP, AVP, and Tracheomalacia Groups
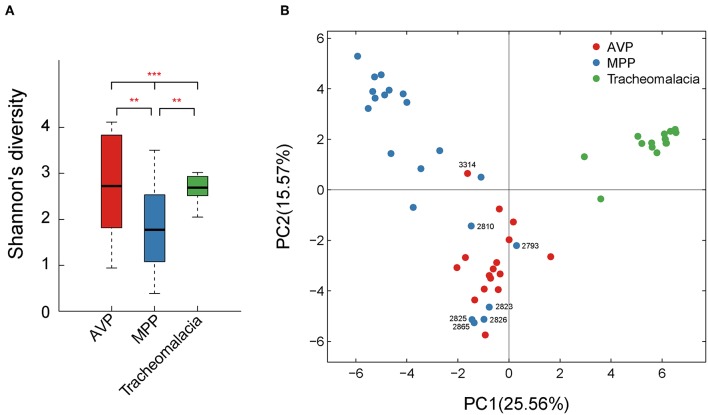
Shannon's index showed that the diversity of BALF microbiota differed significantly (p-value = 0.002) in the three groups, and the diversity of the MPP group was lower than those of the AVP group (p-value = 0.023) or tracheomalacia group (p-value = 0.013), while the lung microbiota diversity in tracheomalacia children was less dynamic than those in the other two groups (Figure 1A). PCA also revealed a divergence among microbial samples in MPP, AVP, and tracheomalacia groups (Figure 1B).
Figure 1.
Diversity and principal coordinate analysis (PCA) of lung microbial community differed significantly in adenovirus pneumonia (AVP), M. pneumoniae pneumonia (MPP) and tracheomalacia. (A) The microbial diversity (Shannon's index) showed significant difference among MPP, AVP, and trancheomalacia. (B) PCA exhibited the divergence of microbial samples in three groups. Red, blue, and green represent the microbial samples in AVP, MPP, and tracheomalacia, respectively. ** and *** represent the p-value ≤ 0.01 and ≤ 0.001, respectively.
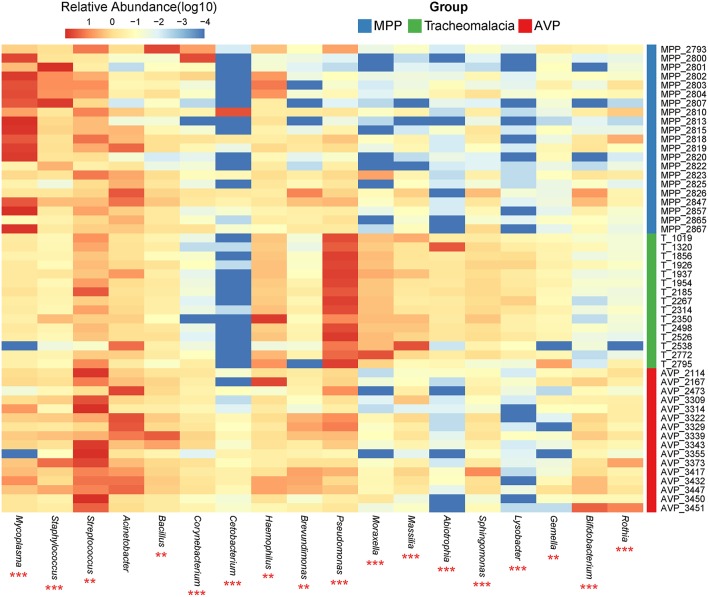
At the phylum level, Proteobacteria and Tenericutes dominated the lung microbiota in tracheomalacia (74.74 ± 12.20%) and MPP (36.98 ± 32.90%) groups, respectively (Table S3). In contrast, Firmicutes represented the highest abundance in the lung microbiota of AVP children (47.14 ± 32.93%). A comparison of lung-dominating genera among the three groups showed statistically significant differences, except for Acinetobacter (q-value = 0.053) (Table 2). Mycoplasma, Streptococcus, and Pseudomonas dominated the BALF microbiota in children with MPP (36.98 ± 32.90%), AVP (36.74 ± 34.59%), and tracheomalacia (43.21 ± 16.84%), respectively (Figure 2; Table 2).
Table 2.
Comparison of dominant genera (combined top 10 genera) among three disease groups.
| q-value | Mean | ||||||
|---|---|---|---|---|---|---|---|
| Dominant genera | Kruskal-Wallis test | AVP-vs.-MPP | AVP-vs.-T | MPP-vs.-T | AVP | MPP | T |
| Mycoplasma | <0.001 | 0.126 | 0.003 | 0.001 | 1.922 | 36.979 | 0.378 |
| Staphylococcus | 0.001 | 0.570 | 0.019 | <0.001 | 2.256 | 9.391 | 0.348 |
| Streptococcus | 0.003 | 0.063 | 0.009 | 0.793 | 36.742 | 4.775 | 5.315 |
| Acinetobacter | 0.053 | 0.241 | 0.045 | 1.000 | 12.291 | 4.483 | 2.557 |
| Bacillus | 0.007 | 0.178 | 0.003 | 0.551 | 2.965 | 3.080 | 0.364 |
| Corynebacterium | 0.001 | 0.326 | <0.001 | 0.045 | 0.470 | 2.485 | 0.056 |
| Cetobacterium | <0.001 | 0.182 | <0.001 | 0.113 | 0.113 | 2.011 | 0.001 |
| Haemophilus | 0.009 | 0.493 | 0.023 | 0.019 | 4.243 | 1.873 | 6.315 |
| Brevundimonas | 0.011 | 0.175 | 0.007 | 0.997 | 1.586 | 0.799 | 0.151 |
| Pseudomonas | <0.001 | 0.166 | <0.001 | <0.001 | 2.906 | 0.786 | 43.214 |
| Moraxella | <0.001 | 0.239 | <0.001 | <0.001 | 0.100 | 0.247 | 4.655 |
| Massilia | <0.001 | 0.166 | 0.017 | <0.001 | 0.474 | 0.162 | 3.747 |
| Abiotrophia | <0.001 | 0.970 | <0.001 | <0.001 | 0.022 | 0.025 | 2.583 |
| Sphingomonas | 0.001 | 0.296 | 0.015 | 0.002 | 0.962 | 0.341 | 0.902 |
| Lysobacter | <0.001 | 0.511 | <0.001 | <0.001 | 0.017 | 0.009 | 0.513 |
| Gemella | 0.005 | 0.959 | 0.034 | 0.003 | 0.143 | 0.083 | 0.507 |
| Bifidobacterium | <0.001 | 0.126 | <0.001 | 0.291 | 2.499 | 0.601 | 0.048 |
| Rothia | 0.001 | 0.136 | <0.001 | 0.608 | 1.277 | 0.364 | 0.055 |
AVP, Adenoviral Pneumonia; MPP, Mycoplasma pneumoniae Pneumonia; T, Tracheomalacia; q < 0.05 are shown in bold.
Figure 2.
Dominating genera of lung microbiota were discrepant in AVP, MPP, and tracheomalacia. The abundance of dominated genera combined top 10 genera of each group were significant different, except for Acinetobacter. The relative abundance was transformed by log10 and proportional to the color from blue to red. Samples from MPP, tracheomalacia and AVP groups were colored blue, green, and red, respectively. ** and *** represent q-value of Kruskal-Wallis test ≤0.01 and ≤0.001, respectively.
Expressions of Inflammatory Cytokines Differed in MPP, AVP, and Tracheomalacia Children and Were Correlated With Lung Microbiota
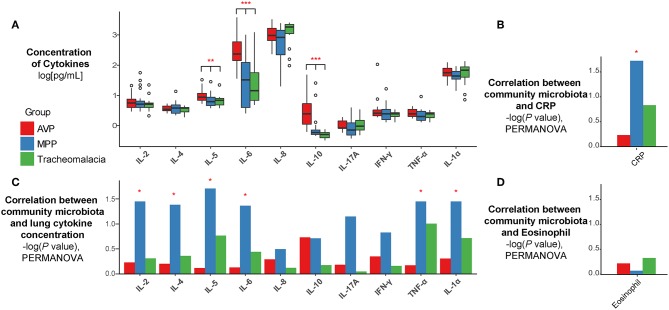
The expressions of IL-5, IL-6, and IL-10 in BALF of AVP were significantly higher than those of MPP and tracheomalacia patients (Figure 3A, p-value: 0.016, <0.001, <0.001, respectively). The average expressions of IL-8, IL-17A, and IL-1α were higher, but showed no significant difference in the lungs of children with tracheomalacia compared to those with AVP and MPP. Nevertheless, the expression of IL-8 alone in the tracheomalacia group was significantly different compared to the MPP group (p-value = 0.032).
Figure 3.
The concentration of cytokines and their correlations with microbiota were disease-specific. (A) The expression of cytokines were disease-specific, concentration of cytokines were recalculated by log10 and drawn as boxplot. (B–D) The correlation between microbiota and cytokines/CRP/Eosinophil were also disease-specific. Statistical differences were marked as p-value on the top of cytokines/CRP. Red, blue, and green represent the patients from AVP, MPP, and tracheomalacia, respectively. *, **, and *** represent q-value of Kruskal-Wallis test ≤0.05, ≤0.01, and ≤0.001, respectively.
Based on all samples, the lung microbiota diversity and concentrations of inflammatory cytokines showed no notable correlation (Figure S1). Hence, we then explored the association between the lung microbial community and cytokine expression in AVP, MPP, and tracheomalacia groups. Our results indicated a strong correlation in the MPP group between the lung microbial community and the concentration of six cytokines, IL-2, IL-4, IL-5, IL-6, TNF-α, and IL-1α (p-values = 0.036, 0.042, 0.020, 0.044, 0.036, and 0.036, respectively) (Figure 3B). Additionally, the lung microbiota in MPP was also correlated with serum CRP levels, which were detected within 24 h after hospitalization (Figure 3C). In contrast, we ascertained no significant correlation between lung microbiota and inflammatory cytokines in AVP and tracheomalacia patients (Figure 3B), or between lung microbiota and eosinophil levels (Figure 3D).
Association of Lung Microbiota and Inflammatory Cytokine Expression With Disease Severity at the Individual Level
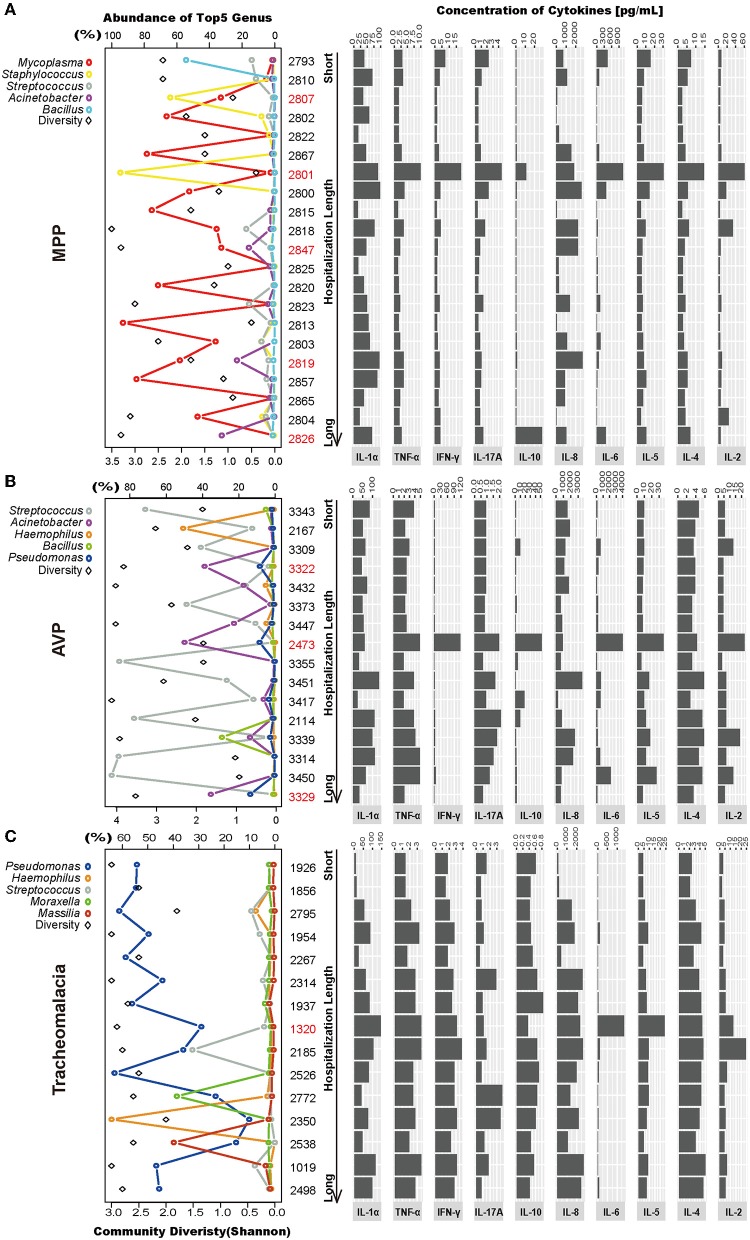
Lung microbiota diversity exhibited no significant correlation with hospitalization length or fever duration (Figure S2). In addition, there was no marked association of CRP, PCT, and eosinophils detected at 24 h after hospitalization with hospitalization length or fever duration (Figure S3). Moreover, at the individual level, we observed wide variability in the lung microbial community and cytokine concentrations in each group (Figure 4).
Figure 4.
Intragroup microbial structure and cytokines concentration showed individual difference. Top 5 genera and community diversity were used to show community structure in each group (on the left). The concentrations of 10 cytokines were drawn on the right. The patients were sorted by hospitalization length. (A–C) Represent the MPP, AVP, and tracheomalacia group, respectively. The top five genera were painted with specific colors and circle, the diversity was shaped rectangle and in black color. Special samples in each group were colored red.
In the MPP group, the lung microbiota of patients 2,807 and 2,801 was dominated by Staphylococcus (64.04 and 94.64%) (Figure 4A; Table S3), which could be assigned as Staphylococcus warneri (58.42 and 85.80%, respectively) (Table S4). The bacterial diversity in the lungs was low in both patients 2,807 and 2,801 (Figure 4A). Except for IL-8, IL-10, and IL-1α, the expression of the other seven inflammatory cytokines was the highest in patient 2,801 compared to the other patients (Figure 4A). In the lung microbiota of patients 2,847, 2,819, and 2,826, we identified 15.84, 23.19, and 32.48% abundance of Acinetobacter (Figure 4A; Table S3), which was assigned as A. baumannii (13.18, 20.76, and 25.20%, respectively) (Table S4). With the increasing load of A. baumannii in the lung microbiota, patients 2,847, 2,819, and 2,826 tended to suffer longer hospitalization length (9, 12, and 16 days, respectively) and fever duration (0, 12, and 30 days, respectively) (Table S1). The lung concentration of IL-10 in patient 2,826 (27.89 pg/ml) was dramatically higher than that in the other MPP patients (average of 2.51 pg/ml).
A. baumannii was also expected to dominate the lung microbiota in AVP patients 3,322, 2,473, and 3,329 (31.62, 44.55, and 29.76%, respectively) (Figure 4B; Table S4). The expression of five inflammatory cytokines was overwhelmingly higher in patient 2,473 compared to the other AVP patients, whereas patient 3,329 suffered the longest hospitalization length (21 days) (Figure 4B; Table S1). In the tracheomalacia group, patients with Pseudomonas dominance (mainly Pseudomonas veronii) were inclined to suffer shorter hospitalization lengths (average of 7.1 days) compared to those with other dominant microbial genera in the lung microbiota (9–10 days) (Figure 4C). For patient 1,320, who had the highest lung concentration of IL-6 (1,363.00 pg/ml), the lung microbiota was dominated by Abiotrophia defectiva (32.78%) and P. veronii (27.26%) (Figure 4C).
Discussion
A number of recent studies have demonstrated that various microbial commensals colonize the respiratory tract, serving as gatekeepers to respiratory health (38–40). In cases of lower respiratory tract infection, various immune responses can be activated through different pathogens (41), such as pathogen-associated molecular patterns (PAMP) and damage-associated molecular patterns (DAMP) (42, 43). M. pneumoniae is a bacterial pathogen that can deplete other bacteria through direct competition or activation of bacterial clearance (17, 18). In contrast, virus infections impair the lung epithelial layer and suppress the immune response, which promotes bacterial outgrowth and frequently leads to secondary bacterial infection (43). These results in lower bacterial diversity in lung microbiota in MPP compared to AVP. Several studies have also reported specific lung microbiota associations, such as Streptococcus being enriched in SPP patients and S. aureus being predominant in patients with methicillin-resistant S. aureus (MRSA) (14, 44). Furthermore, the lung microbiota of children with tracheomalacia is dominated by Pseudomonas, which is consistent with pediatric cystic fibrosis (13). The diversity in lung bacterial compositions can be partly explained by specific mechanisms of pathogen clearance between M. pneumoniae and adenovirus.
Though current clinical biomarkers are beneficial for CAP diagnosis, CRP, PCT, and eosinophil counts still have difficulty separating bacterial from viral infections or mild from severe cases. The etiological diagnosis of pediatric CAP and estimations of the outcome remain unsolved problems in most cases. Several cytokines can be induced by M. pneumoniae, giving it the potential to predict severity (17, 45). During viral infection, the host immune system is activated through various intercellular signaling cascades (46, 47). IL-6 and IFN-γ are the two major cytokines that inhibit the viral replication and virus-associated inflammation (17, 42, 48). Excessive expression of IL-10 suppresses the clearance of pathogens, which leads to secondary bacterial lung infection (49). In general, clinical biomarkers and cytokines expression can explain the severity of disease in AVP compared to MPP and tracheomalacia. For children with tracheomalacia, combined clinical tests with microbiota and cytokines suggest that lung ecology is shaped by chronic inflammation rather than acute respiratory infection (50, 51). Comparing MPP/AVP with tracheomalacia, the expression of cytokines has the potential to indicate lung microbiota disequilibrium.
Given different genetic backgrounds and living environments (26, 37, 52), homeostasis between microbiota and the immune system in the lungs is individual-specific (37). Current pathogen tests are limited to typically known respiratory pathogens (53) and take a long time to culture, which might mislead or delay pneumonia diagnosis. Microbiota analysis could identify various kinds of bacteria in the meantime, including uncommon respiratory pathogens (54). In this study, several patients were identified with high proportions of A. baumannii, S. warneri, or A. defectiva, which are not detected in conventional respiratory pathogen tests. These highly drug-resistant and pathogenic bacteria (55–57) lead to the overexpression of cytokines and are related to high disease severity (58, 59).
Many studies have sought more robust biomarkers for pneumonia diagnosis. Sonal reports that no single clinical symptom is strongly associated with pneumonia (29). Principi reported that three host protein assays combining CRP, TNF-related apoptosis-inducing ligand (TRAIL), and plasma IFN-γ protein-10 (IP-10) have advantages for pathogen diagnosis and severity assessment over former biomarkers (15). Due to the small sample size and various individual differences, we did not find a strong correlation between clinical information, microbiota, and cytokines in pneumonia.
Several other limitations should be taken into account. Given that no effective medicines exist for MPP/AVP, the patients were provided with empirical treatments, which could disturb the airway ecology slightly (60). Because fiberoptic bronchoscopy is an invasive operation, it is restricted to severe disease cases. Hence, the time points for sample collection were not simultaneous and within a short period after hospitalization. In addition to immune responses, host gene expression should be considered in understanding the pathogen-microbiota-host response axis.
In conclusion, this study provides a significant reference for lung microbiota and immunity in MPP, AVP, and tracheomalacia children. This will fill some knowledge gaps in exploring effective therapies for AVP, MPP, as well as tracheomalacia.
Data Availability
Sequencing data have been deposited in GenBank under accession number: SRP130820 (Table S5) and SRP138709.
Ethics Statement
This study was carried out in accordance with the recommendations of Declaration of Helsinki, the Ethical Committee of Shenzhen Children's Hospital. The protocol was approved by the Ethical Committee of Shenzhen Children's Hospital under registration number 2016013. Due to the recruited children were <16 years old, their guardians gave written informed consent in accordance with the Declaration of Helsinki.
Author Contributions
YZ and DL: conception and design. HW, ZL, GX, and YS: specimens collection and common pathogens detection. XF, DL, and XX: data acquisition and analysis. QZ and WD: result interpretation and manuscript written. YaL, ZY, and YiL: result visualization and revision. YY, KS, and SL: final approval. All authors reviewed this manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mr. Xiaofeng Lin from EasyPub for polishing language.
Footnotes
Funding. This study was supported by the Key Medical Disciplines Building Project of Shenzhen (SZXJ2017005), Sanming Project of Medicine in Shenzhen (SZSM201512030), Shenzhen public service platform for clinical drug trials (20151964) and Shenzhen Science and Technology Project (JCYJ20170303155012371).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fped.2019.00265/full#supplementary-material
Lung concentrations of cytokines shown no significant association with the diversity of lung microbial community.
No significant correlations between the diversity of lung microbial community with four important clinical records.
No significant correlations between hospitalization length/fever duration and three infection representative blood indictors.
The clinical information and cytokines expression of patients with MPP (n = 21), AVP (n = 16) and Tracheomalacia (n = 15).
The confounding factors between clinical characters and microbiota by PERAMONVA method.
The relative abundance of combined top 10 phylum and genera in MPP, AVP and Tracheomalacia.
The relative abundance (%) of species belong to the top 5 genera in MPP, AVP and Tracheomalacia.
Released data from our previous study (n = 17).
References
- 1.Liu L, Oza S, Hogan D, Perin J, Rudan I, Lawn JE, et al. Global, regional, and national causes of child mortality in 2000-13, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet. (2015) 385:430–40. 10.1016/S0140-6736(14)61698-6 [DOI] [PubMed] [Google Scholar]
- 2.Dagan R, Bhutta ZA, de Quadros CA, Garau J, Klugman KP, Khuri-Bulos N, et al. The remaining challenge of pneumonia: the leading killer of children. Pediatr Infect Dis J. (2011) 30:1–2. 10.1097/INF.0b013e3182005389 [DOI] [PubMed] [Google Scholar]
- 3.Ning G, Wang X, Wu D, Yin Z, Li Y, Wang H, et al. The etiology of community-acquired pneumonia among children under 5 years of age in mainland China, 2001-2015: a systematic review. Hum Vaccin Immunother. (2017) 13:2742–50. 10.1080/21645515.2017.1371381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu MP, Ma LY, Zheng Q, Dong LL, Chen ZM. Clinical characteristics of adenovirus associated lower respiratory tract infection in children. World J Pediatr. (2013) 9:346–9. 10.1007/s12519-013-0431-3 [DOI] [PubMed] [Google Scholar]
- 5.Zhou Y, Gao H, Mihindukulasuriya KA, La Rosa PS, Wylie KM, Vishnivetskaya T, et al. Biogeography of the ecosystems of the healthy human body. Genome Biol. (2013) 14:R1. 10.1186/gb-2013-14-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Human Microbiome Project C Structure, function and diversity of the healthy human microbiome. Nature. (2012) 486:207–14. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pettigrew MM, Gent JF, Kong Y, Wade M, Gansebom S, Bramley AM, et al. Association of sputum microbiota profiles with severity of community-acquired pneumonia in children. BMC Infect Dis. (2016) 16:317. 10.1186/s12879-016-1670-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakwinska O, Bastic Schmid V, Berger B, Bruttin A, Keitel K, Lepage M, et al. Nasopharyngeal microbiota in healthy children and pneumonia patients. J Clin Microbiol. (2014) 52:1590–4. 10.1128/JCM.03280-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu Z, Dai W, Liu Y, Zhou Q, Wang H, Li D, et al. The alteration of nasopharyngeal and oropharyngeal microbiota in children with MPP and non-MPP. Genes. (2017) 8:e380. 10.3390/genes8120380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasegawa K, Mansbach JM, Ajami NJ, Espinola JA, Henke DM, Petrosino JF, et al. Association of nasopharyngeal microbiota profiles with bronchiolitis severity in infants hospitalised for bronchiolitis. Eur Respir J. (2016) 48:1329–39. 10.1183/13993003.00152-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu HF, Li A, Zhang T, Ren ZG, He KX, Zhang H, et al. Disordered oropharyngeal microbial communities in H7N9 patients with or without secondary bacterial lung infection. Emerg Microbes Infect. (2017) 6:e112 10.1038/emi.2017.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wen Z, Xie G, Zhou Q, Qiu C, Li J, Hu Q, et al. Distinct nasopharyngeal and oropharyngeal microbiota of children with influenza A virus compared with healthy children. Biomed Res Int. (2018) 2018:6362716. 10.1155/2018/6362716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zemanick ET, Wagner BD, Robertson CE, Ahrens RC, Chmiel JF, Clancy JP, et al. Airway microbiota across age and disease spectrum in cystic fibrosis. Eur Respir J. (2017) 50:1700832. 10.1183/13993003.00832-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Dai W, Qiu C, Li S, Wang W, Xu J, et al. Mycoplasma pneumoniae and Streptococcus pneumoniae caused different microbial structure and correlation network in lung microbiota. J Thorac Dis. (2016) 8:1316–22. 10.21037/jtd.2016.04.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Principi N, Esposito S. Biomarkers in pediatric community-acquired pneumonia. Int J Mol Sci. (2017) 18:447. 10.3390/ijms18020447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pavord ID, Lettis S, Anzueto A, Barnes N. Blood eosinophil count and pneumonia risk in patients with chronic obstructive pulmonary disease: a patient-level meta-analysis. Lancet Respir Med. (2016) 4:731–41. 10.1016/S2213-2600(16)30148-5 [DOI] [PubMed] [Google Scholar]
- 17.Yang J, Hooper WC, Phillips DJ, Talkington DF. Cytokines in Mycoplasma pneumoniae infections. Cytokine Growth Factor Rev. (2004) 15:157–68. 10.1016/j.cytogfr.2004.01.001 [DOI] [PubMed] [Google Scholar]
- 18.Peteranderl C, Sznajder JI, Herold S, Lecuona E. Inflammatory responses regulating alveolar ion transport during pulmonary infections. Front Immunol. (2017) 8:446. 10.3389/fimmu.2017.00446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasegawa K, Perez-Losada M, Hoptay CE, Epstein S, Mansbach JM, Teach SJ, et al. RSV vs. rhinovirus bronchiolitis: difference in nasal airway microRNA profiles and NFkappaB signaling. Pediatr Res. (2018) 83:606–14. 10.1038/pr.2017.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu W, Booth JL, Duggan ES, Patel KB, Coggeshall KM, Metcalf JP. Human lung innate immune cytokine response to adenovirus type 7. J Gen Virol. (2010) 91:1155–63. 10.1099/vir.0.017905-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandez-Botran R, Uriarte SM, Arnold FW, Rodriguez-Hernandez L, Rane MJ, Peyrani P, et al. Contrasting inflammatory responses in severe and non-severe community-acquired pneumonia. Inflammation. (2014) 37:1158–66. 10.1007/s10753-014-9840-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Chen Q, Shi C, Lv H, Xu X, Yu L. Changes of serum TNF-alpha, IL-5 and IgE levels in the patients of mycoplasma pneumonia infection with or without bronchial asthma. Int J Clin Exp Med. (2015) 8:3901–6. [PMC free article] [PubMed] [Google Scholar]
- 23.Saghafian-Hedengren S, Mathew JL, Hagel E, Singhi S, Ray P, Ygberg S, et al. Assessment of cytokine and chemokine signatures as potential biomarkers of childhood community-acquired pneumonia severity: a nested cohort study in India. Pediatr Infect Dis J. (2017) 36:102–8. 10.1097/INF.0000000000001364 [DOI] [PubMed] [Google Scholar]
- 24.de Steenhuijsen Piters WA, Heinonen S, Hasrat R, Bunsow E, Smith B, Suarez-Arrabal MC, et al. Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med. (2016) 194:1104–15. 10.1164/rccm.201602-0220OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gollwitzer ES, Marsland BJ. Microbiota abnormalities in inflammatory airway diseases–potential for therapy. Pharmacol Ther. (2014) 141:32–9. 10.1016/j.pharmthera.2013.08.002 [DOI] [PubMed] [Google Scholar]
- 26.Frayman KB, Armstrong DS, Carzino R, Ferkol TW, Grimwood K, Storch GA, et al. The lower airway microbiota in early cystic fibrosis lung disease: a longitudinal analysis. Thorax. (2017) 72:1104–12. 10.1136/thoraxjnl-2016-209279 [DOI] [PubMed] [Google Scholar]
- 27.Ferrer M, Mendez-Garcia C, Rojo D, Barbas C, Moya A. Antibiotic use and microbiome function. Biochem Pharmacol. (2017) 134:114–26. 10.1016/j.bcp.2016.09.007 [DOI] [PubMed] [Google Scholar]
- 28.Hotterbeekx A, Xavier BB, Bielen K, Lammens C, Moons P, Schepens T, et al. The endotracheal tube microbiome associated with Pseudomonas aeruginosa or Staphylococcus epidermidis. Sci Rep. (2016) 6:36507 10.1038/srep36507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shah SN, Bachur RG, Simel DL, Neuman MI. Does this child have pneumonia?: the rational clinical examination systematic review. JAMA. (2017) 318:462–71. 10.1001/jama.2017.9039 [DOI] [PubMed] [Google Scholar]
- 30.Bao Y, Li Y, Qiu C, Wang W, Yang Z, Huang L, et al. Bronchoalveolar lavage fluid microbiota dysbiosis in infants with protracted bacterial bronchitis. J Thorac Dis. (2018) 10:168–74. 10.21037/jtd.2017.12.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prevaes SM, de Steenhuijsen Piters WA, de Winter-de Groot KM, Janssens HM, Tramper-Stranders GA, Chu ML, et al. Concordance between upper and lower airway microbiota in infants with cystic fibrosis. Eur Respir J. (2017) 49:1602235. 10.1183/13993003.02235-2016 [DOI] [PubMed] [Google Scholar]
- 32.Caffrey AK, Lehmann MM, Zickovich JM, Espinosa V, Shepardson KM, Watschke CP, et al. IL-1alpha signaling is critical for leukocyte recruitment after pulmonary Aspergillus fumigatus challenge. PLoS Pathog. (2015) 11:e1004625. 10.1371/journal.ppat.1004625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. (2013) 79:5112–20. 10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. (2010) 7:335–6. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelly BJ, Imai I, Bittinger K, Laughlin A, Fuchs BD, Bushman FD, et al. Composition and dynamics of the respiratory tract microbiome in intubated patients. Microbiome. (2016) 4:7. 10.1186/s40168-016-0151-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, et al. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. (2014) 42:D643–8. 10.1093/nar/gkt1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dickson RP, Erb-Downward JR, Falkowski NR, Hunter EM, Ashley SL, Huffnagle GB. The lung microbiota of healthy mice are highly variable, cluster by environment, and reflect variation in baseline lung innate immunity. Am J Respir Crit Care Med. (2018) 198:497–508. 10.1164/rccm.201711-2180OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lanaspa M, Bassat Q, Medeiros MM, Munoz-Almagro C. Respiratory microbiota and lower respiratory tract disease. Expert Rev Anti Infect Ther. (2017) 15:703–11. 10.1080/14787210.2017.1349609 [DOI] [PubMed] [Google Scholar]
- 39.Dickson RP, Erb-Downward JR, Huffnagle GB. Towards an ecology of the lung: new conceptual models of pulmonary microbiology and pneumonia pathogenesis. Lancet Respir Med. (2014) 2:238–46. 10.1016/S2213-2600(14)70028-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Man WH, de Steenhuijsen Piters WA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol. (2017) 15:259–70. 10.1038/nrmicro.2017.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin C, Burgel PR, Lepage P, Andrejak C, de Blic J, Bourdin A, et al. Host-microbe interactions in distal airways: relevance to chronic airway diseases. Eur Respir Rev. (2015) 24:78–91. 10.1183/09059180.00011614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Troy NM, Bosco A. Respiratory viral infections and host responses; insights from genomics. Respir Res. (2016) 17:156. 10.1186/s12931-016-0474-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brealey JC, Sly PD, Young PR, Chappell KJ. Viral bacterial co-infection of the respiratory tract during early childhood. FEMS Microbiol Lett. (2015) 362:fnv062. 10.1093/femsle/fnv062 [DOI] [PubMed] [Google Scholar]
- 44.Kawanami T, Yatera K, Yamasaki K, Noguchi S, Fukuda K, Akata K, et al. Clinical impact of methicillin-resistant staphylococcus aureus on bacterial pneumonia: cultivation and 16S ribosomal RNA gene analysis of bronchoalveolar lavage fluid. BMC Infect Dis. (2016) 16:155. 10.1186/s12879-016-1493-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chkhaidze I, Kapanadze N. Cytokines as the predictors of severe Mycoplasma pneumoniae pneumonia in children (review). Georgian Med News. (2017) 267:89–95. 10.1038/srep37037 [DOI] [PubMed] [Google Scholar]
- 46.Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. (2009) 326:257–63. 10.1126/science.1179050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chevrier N, Mertins P, Artyomov MN, Shalek AK, Iannacone M, Ciaccio MF, et al. Systematic discovery of TLR signaling components delineates viral-sensing circuits. Cell. (2011) 147:853–67. 10.1016/j.cell.2011.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lauder SN, Jones E, Smart K, Bloom A, Williams AS, Hindley JP, et al. Interleukin-6 limits influenza-induced inflammation and protects against fatal lung pathology. Eur J Immunol. (2013) 43:2613–25. 10.1002/eji.201243018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goulding J, Godlee A, Vekaria S, Hilty M, Snelgrove R, Hussell T. Lowering the threshold of lung innate immune cell activation alters susceptibility to secondary bacterial superinfection. J Infect Dis. (2011) 204:1086–94. 10.1093/infdis/jir467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gu W, Jiang W, Zhang X, Chen Z, Yan Y, Huang L, et al. Refractory wheezing in Chinese children under 3 years of age: bronchial inflammation and airway malformation. BMC Pediatr. (2016) 16:145. 10.1186/s12887-016-0680-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hysinger EB, Panitch HB. Paediatric Tracheomalacia. Paediatr Respir Rev. (2016) 17:9–15. 10.1016/j.prrv.2015.03.002 [DOI] [PubMed] [Google Scholar]
- 52.Lloyd CM, Marsland BJ. Lung homeostasis: influence of age, microbes, and the immune system. Immunity. (2017) 46:549–61. 10.1016/j.immuni.2017.04.005 [DOI] [PubMed] [Google Scholar]
- 53.Murdoch DR, O'Brien KL, Driscoll AJ, Karron RA, Bhat N, Pneumonia Methods Working G, et al. Laboratory methods for determining pneumonia etiology in children. Clin Infect Dis. (2012) 54(Suppl 2):S146–52. 10.1093/cid/cir1073 [DOI] [PubMed] [Google Scholar]
- 54.Noguchi S, Yatera K, Kawanami T, Yamasaki K, Fukuda K, Naito K, et al. Pneumonia and empyema caused by Streptococcus intermedius that shows the diagnostic importance of evaluating the microbiota in the lower respiratory tract. Intern Med J. (2014) 53:47–50. 10.2169/internalmedicine.53.0971 [DOI] [PubMed] [Google Scholar]
- 55.Wilhelm N, Sire S, Le Coustumier A, Loubinoux J, Beljerd M, Bouvet A. First case of multiple discitis and sacroiliitis due to Abiotrophia defectiva. Eur J Clin Microbiol Infect Dis. (2005) 24:76–8. 10.1007/s10096-004-1265-7 [DOI] [PubMed] [Google Scholar]
- 56.Purcell K, Fergie J. Concurrent serious bacterial infections in 2396 infants and children hospitalized with respiratory syncytial virus lower respiratory tract infections. JAMA Pediatr. (2002) 156:322–4. 10.1001/archpedi.156.4.322 [DOI] [PubMed] [Google Scholar]
- 57.Tommasi R, Brown DG, Walkup GK, Manchester JI, Miller AA. ESKAPEing the labyrinth of antibacterial discovery. Nat Rev Drug Discov. (2015) 14:529–42. 10.1038/nrd4572 [DOI] [PubMed] [Google Scholar]
- 58.Garcia-Nunez M, Millares L, Pomares X, Ferrari R, Perez-Brocal V, Gallego M, et al. Severity-related changes of bronchial microbiome in chronic obstructive pulmonary disease. J Clin Microbiol. (2014) 52:4217–23. 10.1128/JCM.01967-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dickson RP, Singer BH, Newstead MW, Falkowski NR, Erb-Downward JR, Standiford TJ, et al. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol. (2016) 1:16113. 10.1038/nmicrobiol.2016.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. (2018) 555:623–8. 10.1038/nature25979 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lung concentrations of cytokines shown no significant association with the diversity of lung microbial community.
No significant correlations between the diversity of lung microbial community with four important clinical records.
No significant correlations between hospitalization length/fever duration and three infection representative blood indictors.
The clinical information and cytokines expression of patients with MPP (n = 21), AVP (n = 16) and Tracheomalacia (n = 15).
The confounding factors between clinical characters and microbiota by PERAMONVA method.
The relative abundance of combined top 10 phylum and genera in MPP, AVP and Tracheomalacia.
The relative abundance (%) of species belong to the top 5 genera in MPP, AVP and Tracheomalacia.
Released data from our previous study (n = 17).
Data Availability Statement
Sequencing data have been deposited in GenBank under accession number: SRP130820 (Table S5) and SRP138709.