Abstract
Not all Salmonella enterica serovars cause the same disease. S. enterica represents an incredibly diverse species comprising >2,600 unique serovars. While some S. enterica serovars are host-restricted, others infect a wide range of hosts. The diseases that nontyphoidal Salmonella (NTS) serovars cause vary considerably, with some serovars being significantly more likely to cause invasive disease in humans than others. Furthermore, while genomic analyses have advanced our understanding of the genetic diversity of these serovars, they have not been able to fully account for the observed clinical differences. One overarching challenge is that much of what is known about Salmonella’s general biology and virulence strategies is concluded from studies examining a select few serovars, especially serovar Typhimurium. As targeted control strategies have been implemented to control select serovars, an increasing number of foodborne outbreaks involving serovars that are less frequently associated with human clinical illness are being detected. Harnessing what is known about the diversity of NTS serovars represents an important factor in achieving the ultimate goal of reducing salmonellosis-associated morbidity and mortality worldwide. In this review we summarize the current understanding of the differences and similarities among NTS serovars, highlighting the virulence mechanisms, genetic differences, and sources that characterize S. enterica diversity and contribute to its success as a foodborne pathogen.
Keywords: nontyphoidal Salmonella, virulence, serovars, foodborne pathogen, food safety
Introduction
Salmonellae are Gram-negative, facultatively anaerobic bacteria. The genus Salmonella, named for Dr. Daniel Salmon, was first described in 1866 by Dr. Theobald Smith (Schultz, 2008). Initially, Salmonella was described as the causative agent of pig cholera (first named Salmonella choleraesuis). However, pig cholera was later discovered to be a viral disease, with Salmonella co-infection being common (Schultz, 2008).
Today, the genus Salmonella includes 2 species: enterica and bongori (Brenner et al., 2000; Figure 1). Within the species enterica, there are 6 subspecies, which are characterized by both a name and a roman numeral: enterica (I), salamae (II), arizonae (IIIa), diarizonae (IIIb), houtenae (IV), and indica (VI) (Brenner et al., 2000). Salmonella bongori was originally classified as Salmonella enterica subspecies V, before being re-classified as a separate species (Brenner et al., 2000). A third species, Salmonella subterranean was proposed (Shelobolina et al., 2004), however, further DNA characterization revealed that this species did not belong in the genus Salmonella (Issenhuth-Jeanjean et al., 2014). Recent analyses suggest there are four major phylogenetic clades within S. enterica subsp. enterica (Worley et al., 2018). The majority of Salmonella isolates infecting warm-blooded hosts belong to subspecies enterica (I), while subspecies II-VI and S. bongori are primarily isolated from environmental sources or cold-blooded hosts, such as reptiles and amphibians.
FIGURE 1.
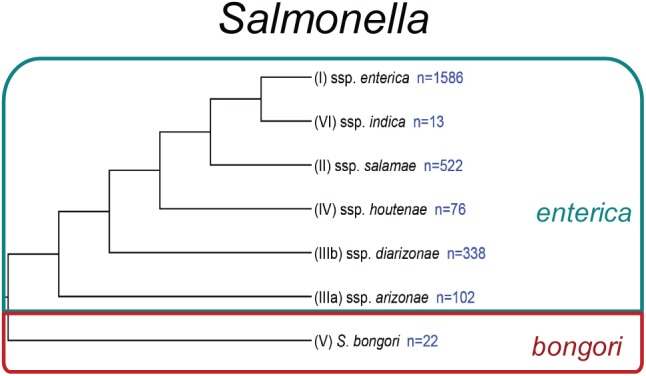
Phylogeny of Salmonella. Phylogenetic analysis reconstructed from Desai et al. (2013). Subspecies identifiers preceding the subspecies (ssp.) name are shown in parentheses. The number of currently recognized serovars (Issenhuth-Jeanjean et al., 2014) within each species or subspecies is listed in blue.
Infection with typhoidal (S. enterica subsp. enterica serovar Typhi) and paratyphoidal serovars (i.e., serovars Paratyphi A, Paratyphi B, Paratyphi C, and Sendai) typically results in an invasive, extra-intestinal disease characterized by a high fever (>39°C), malaise, vomiting, headache, and an elevated pulse rate (Dougan and Baker, 2014; Hiyoshi et al., 2018). Serovar Typhi is host-restricted, and is transmitted human-to-human primarily via fecal contamination of drinking water or improper food handling (Connor and Schwartz, 2005). In contrast, infections with NTS serovars (i.e., all serovars except Typhi, Paratyphi A, Paratyphi B, Paratyphi C, or Sendai) typically result in a self-limiting gastroenteritis that is cleared by the host within 4–7 days (Gal-Mor et al., 2014). Due to these stark discrepancies in disease manifestation, nontyphoidal and typhoid salmonellosis cases are frequently assessed and reported independently.
Serovar Typhimurium is the best-studied serovar for nontyphoidal salmonellosis and has become the “model serovar” for studying NTS (Sabbagh et al., 2010; Tsolis et al., 2011; Fàbrega and Vila, 2013; Rivera-Chavez and Baumler, 2015). The S. Typhimurium strain LT2 has been widely used since the 1940s and has been characterized extensively (Swords et al., 1997). In fact many studies have used LT2 as a model for NTS, despite the fact that this strain encodes a rare start codon (UUG) in its rpoS, rendering this strain avirulent (Lee et al., 1995; Wilmes-Riesenberg et al., 1997). This suggests that conclusions drawn from experiments performed with S. Typhimurium LT2 should be cautiously considered, as this lab strain may not accurately represent NTS virulence. Furthermore, NTS vary significantly with respect to the severity of the illness that they cause, as infection with certain serovars is significantly more likely to result in human invasive disease, hospitalization, and death when compared to the rates of invasive disease observed for S. Typhimurium (Jones et al., 2008). This suggests that while S. Typhimurium was useful as a general model for NTS, there are likely additional virulence factors, adaptations, and other fitness determinants that may not be accurately represented in S. Typhimurium.
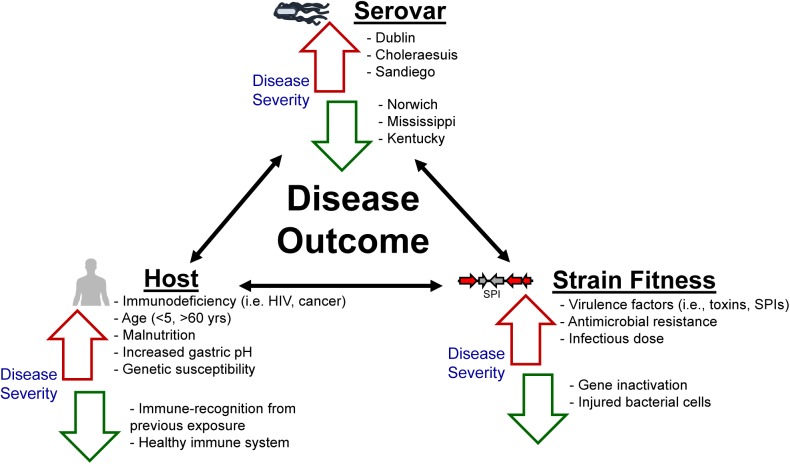
Despite extensive government and industry efforts aimed at reducing the incidence of nontyphoidal salmonellosis, little progress has been made in reducing the number of salmonellosis cases per year (Boore et al., 2015; Ford et al., 2016). The various hosts that NTS serovars encounter and colonize demonstrate their tremendous adaptability. Differences in food consumption, sanitation, cultural traditions, infrastructure, and food safety regulations all influence the global burden of nontyphoidal salmonellosis. Furthermore, differences in host susceptibility, virulence factors/mechanisms specific to select serovars, as well as serovar fitness, contribute to the severity, and outcome of salmonellosis (Figure 2). The role of antimicrobial resistance in the expansion of select NTS subtypes in a novel niche, as well as the potential implications associated with increasing the severity of the outcome of infection due to rapid outgrowth of an antibiotic-resistant Salmonella strain in the GI tract of antibiotic-treated individuals, have been reviewed extensively elsewhere (Rabsch et al., 2001; Furuya and Lowy, 2006; Crump et al., 2015), and therefore will not be covered in this review.
FIGURE 2.
Not all nontyphoidal salmonellosis infections are alike. The morbidity and mortality associated with nontyphoidal salmonellosis exists as an interplay between the host’s immune response as well as the overall fitness of the serovar/strain. Factors listed adjacent to red arrows have been previously established as contributing to increased disease severity, whereas factors listed adjacent to green arrows are associated with a decrease in disease severity.
A number of reviews have detailed the host-response to both nontyphoidal and typhoid salmonellosis (Gal-Mor et al., 2014; Gilchrist et al., 2015; Keestra-Gounder et al., 2015; LaRock et al., 2015; Rivera-Chavez and Baumler, 2015). However, a better understanding of the diversity of NTS serovars and which adaptations make them more suited to colonize different hosts or survive in different environments, is lacking. In this review, we have synthesized studies representing the diversity of NTS serovars to demonstrate how Salmonella uses a variety of virulence factors, and genetic and phenotypic adaptations to become one of the most successful foodborne pathogens worldwide.
Classical and Modern Approaches to Defining and Studying NTS Diversity
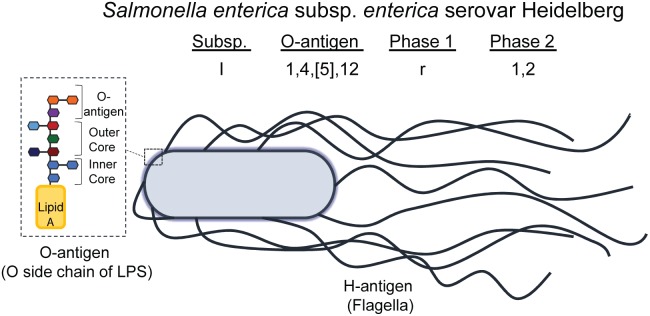
Salmonellae are differentiated based on the reactivity of the O (somatic, or O polysaccharide component of lipopolysaccharide [LPS]) and H (flagellar) antigens to anti-sera, in a classification scheme known as the Kauffman-White-Le Minor scheme (Figure 3; Brenner et al., 2000; Liu et al., 2014). Salmonellae are categorized into 46 unique O groups (Liu et al., 2014), with 114 unique H antigens (McQuiston et al., 2004). Together with the roman numeral abbreviation for the S. enterica subspecies, the combination of O- and H-antigens antigens is what defines the serotype (also known as a serovar) of a given Salmonella strain. Salmonella serovars also have common names that are often the name of the geographical location where they were first isolated (Gossner et al., 2016), to accompany their antigenic formulae (Brenner et al., 2000).
FIGURE 3.
Illustration of NTS O- and H-antigens, comprising the serotyping scheme defined by Kauffman-White-Le Minor. The antigenic formula of a serovar is composed of the subspecies (I -VI), the O-antigen (numbered), and the flagellar antigens called phase 1, phase 2, and phase 3 (if present). Some serovars have lost their phase 1 and/or phase 2 antigen and others have gained additional flagellar antigens (phase 3). The O-antigen illustrated represents the terminal side chain of lipopolysaccharide (LPS) on the cell wall of Gram-negative bacteria, such as Salmonella, which is composed of an inner and outer core of primarily polysaccharides bound to Lipid A; the O side chain is composed of repeating saccharide units, and is the subunit of LPS that differs between serogroups. Salmonella enterica subsp. enterica serovar Heidelberg, the NTS serovar modeled here, is the common name of the serovar having antigenic formula I 1,4,[5],12:r:1,2.
Another traditional classification scheme, called phage typing, involves subjecting Salmonella strains to libraries of phage to determine which phage are able to lyse a given strain. Examples of historically important phage types include S. Typhimurium DT (definitive type) 104, which arose as an important multi-drug resistant (MDR) strain in the United Kingdom in the early 1960s that subsequently spread and became a global epidemic (Threlfall, 2000), and S. Enteritidis PT (phage type) 4, which is associated with contamination of intact chicken eggs (Humphrey, 1994).
Although modern genetic analyses are now typically used to predict the phage type of a given Salmonella isolate, phage typing may still be reported as an additional level of discrimination. Likewise, other in silico typing schemes have increased in popularity, as they provide increased discriminatory power and negate the use for traditional serotyping. Rather, these methods utilize serotype prediction based on sequence typing of the O- and H-antigens (McQuiston et al., 2004), or more recently, using whole genome sequence (WGS) data (Zhang et al., 2015; Yoshida et al., 2016; Robertson et al., 2018). The sequence data are compared to a database of sequences, which compare the O- and H-antigens of known serotypes to predict which anti-sera an isolate would react with, and therefore, the serotype of the isolate. Other identification schemes are based on sequence comparisons of housekeeping genes (Achtman et al., 2012), and more recently on core genome or whole genome multi-locus sequence typing (Alikhan et al., 2018).
Recent phylogenetic analyses have shed light on the fact that many commonly isolated serovars (serovars Newport, Montevideo, Kentucky, Paratyphi B, Derby, Nchanga, Cerro, Bareilly, Stanleyville, Dusseldorf, Livingstone, and others) are polyphyletic (Den Bakker et al., 2011; Cao et al., 2013; Timme et al., 2013; Yoshida et al., 2016; Sévellec et al., 2018; Worley et al., 2018). In fact, a recent study comparing 266 different serovars reported that ∼10% are polyphyletic or paraphyletic (Worley et al., 2018). The polyphyletic nature of some serovars may provide further epidemiological evidence that can assist in outbreak investigations by providing additional discriminatory power, as was recently proposed for S. enterica subsp. enterica serovar Derby among different regions in France (Sévellec et al., 2018). WGS subtyping adds an additional level of discriminatory power that can be used to aid epidemiologic investigations of traceback studies. There are also important clinical implications of polyphyletic Salmonella serovars that arise when different clades of the same serovar differ in the virulence factors that they encode, as is the case for S. Mississippi, where one clade encodes typhoid toxin genes, and the other does not (Miller and Wiedmann, 2016). Genomic analyses are just beginning to identify polyphyletic serovars, and to define key differences associated with the different clinical outcomes observed.
Geographic Diversity – Distribution of NTS Serovars Is Regionally Associated
Infections with NTS account for just over one fifth of all bacterial foodborne illnesses worldwide, causing an estimated 78.7 million cases per year (Havelaar et al., 2015). In the United States NTS is the leading cause of bacterial foodborne illness, resulting in an estimated 1.2 million illnesses, 23,128 hospitalizations, and 452 deaths annually (Scallan et al., 2011). In the US, the 20 NTS serovars most commonly isolated from human clinical cases account for nearly 70% of all NTS clinical cases in humans (Centers for Disease Control and Prevention [CDC], 2016). According to WHO estimates of foodborne disease, the global incidence of nontyphoidal salmonellosis as of 2010 was 1,140 cases per 100,000 people (1.14% of people) (Kirk et al., 2015). However, the burden of nontyphoidal salmonellosis is not equally distributed among different geographic regions. Countries in the Eastern Mediterranean region (e.g., Iran, Kuwait, Saudi Arabia, Egypt, and others) had the highest estimated incidence of nontyphoidal salmonellosis (1,610 cases per 100,000), while the European region (e.g., European Union, Russia, Ukraine, Switzerland, and others) had the lowest (186 cases per 100,000) (Kirk et al., 2015). However, the African region had the highest death rate from nontyphoidal salmonellosis: 1 death per 100,000 persons compared to a global rate of 0.4 deaths per 100,000 persons (Kirk et al., 2015). In contrast to other foodborne bacterial pathogens such as E. coli and Campylobacter spp., the majority of NTS salmonellosis occurs in individuals above 5 years of age (Havelaar et al., 2015).
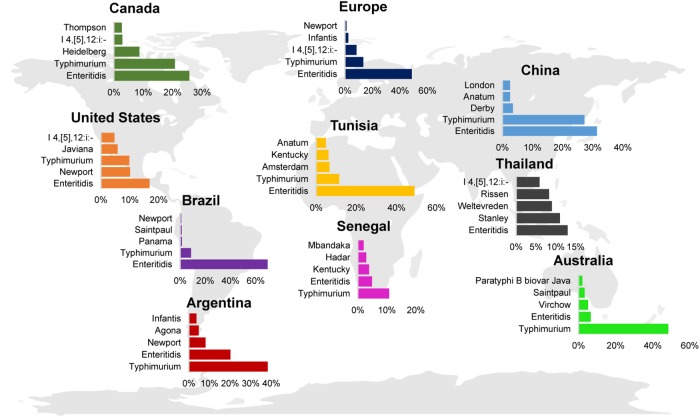
Despite the discrepancy in rates of illness per geographic region, Typhimurium and Enteritidis are consistently reported as the serovars contributing the highest proportion of human clinical salmonellosis worldwide (Figure 4; Hendriksen et al., 2009, 2011; Ran et al., 2011; OzFoodNet Working Group, 2015; Centers for Disease Control and Prevention [CDC], 2016; European Food Safety Authority [EFSA] and European Centre for Disease Prevention, and Control [ECDC], 2017). Interestingly, in some countries the proportion of human clinical cases are dominated by one serovar, such as Typhimurium in Australia or Enteritidis in Brazil, while other countries show a more balanced distribution of cases per serovar (e.g., Enteritidis, Newport, and Typhimurium in the United States). The serovars representing the 3rd, 4th, and 5th most commonly isolated serovars from human clinical infections appear to be more geographically associated. For example, serovars that are found among human cases in Australia (serovars Saintpaul and Virchow) are less common in other countries (Figure 4).
FIGURE 4.
Global distribution of the top 5 NTS serovars associated with human clinical disease. Bar charts represent the top 5 NTS (i.e., serovar Typhi was excluded) serovars reported for Argentina (2007) (Hendriksen et al., 2011), Australia (2011) (OzFoodNet Working Group, 2015), Brazil (2007) (Hendriksen et al., 2011), Canada (2007) (Hendriksen et al., 2011), China (2008) (Ran et al., 2011), the European Union (2016) (European Food Safety Authority [EFSA] and European Centre for Disease Prevention, and Control [ECDC], 2017), Senegal (2007) (Hendriksen et al., 2011), Thailand (2004) (Hendriksen et al., 2009), Tunisia (2007) (Hendriksen et al., 2011), and the United States (2016) (Centers for Disease Control and Prevention [CDC], 2016). Only human clinical cases are reported; data were reported from 2004 to 2016.
The regional differences observed among the incidence of infection by select serovars may be explained by differences in (i) animal hosts that populate the region, (ii) the quality of surveillance and reporting systems (Kirk et al., 2015), which may effectively underestimate the incidence of some serovars, (iii) concurrent immunodeficiencies such as HIV infection or cancer (Okoro et al., 2012), (iv) dietary intake, (v) farming practices or food production practices that might select for specific serovars, or (vi) environmental factors influencing the cultivation, survival, or routes of transmission. In support of differences in environmental exposures, within the United States some serovars are regionally distributed, with some serovars representing the majority of clinical isolates in some geographical regions, but being rarely isolated in other regions (e.g., S. Mississippi) (Boore et al., 2015). This suggests that the majority of human clinical NTS salmonellosis cases in the United States are the result of contamination events that happen at a local level, given that food is often distributed across the country, if not internationally.
Diversity of NTS Virulence Factors and Their Implications in Disease Manifestation
Reflective of an extensively host-adapted lifestyle is the collection of virulence factors possessed by NTS. These include flagella, fimbriae, toxins, pathogenicity islands, and virulence-associated plasmids. These features do not occur in all NTS serovars, and thus their presence or absence influences the virulence and host range of a particular isolate or serovar.
Salmonella Pathogenicity Islands (SPIs)
To date, 24 Salmonella pathogenicity islands have been identified. These horizontally acquired loci encode genes facilitating several virulence mechanisms, including (i) the expression of secretion systems, fimbriae, flagella, and capsules, (ii) serotype conversion, and (iii) host colonization and subsequent survival within the host (Van Asten and Van Dijk, 2005; Fàbrega and Vila, 2013).
Salmonella Pathogenicity Islands 1 and 2 are the best characterized in terms of genetic and phenotypic traits. Of the 24 SPIs (Table 1), SPI-1 is ubiquitous among all Salmonella species and subspecies (Fookes et al., 2011). SPI-1 encodes a type three secretion system (T3SS), which is essential for export of effector proteins required for invasion of host cells. In contrast, SPI-2 is found only in S. enterica, and SPI-22 is only found in S. bongori (Fookes et al., 2011). SPI-2 encodes an additional T3SS, harboring genes that are essential for intracellular survival and for preventing acidification of the Salmonella containing vacuole (SCV). The remaining 21 SPIs are variably present among S. enterica (Kingsley et al., 2003; Shah et al., 2005; Boyen et al., 2006; Saroj et al., 2008; Blondel et al., 2009, 2013; Den Bakker et al., 2011; Shah et al., 2012; Hayward et al., 2013; Lee et al., 2013; Tomljenovic-Berube et al., 2013), with all but three having been found in NTS serovars; SPI-20 and -21 have been found only in S. enterica subsp. arizonae (Blondel et al., 2009), and while SPI-15 occurs in S. enterica subsp. enterica, it has only been identified in S. Typhi to date (Vernikos and Parkhill, 2006; Sabbagh et al., 2010).
Table 1.
Current understanding of the size, function, and distribution of Salmonella pathogenicity islands (SPIs) among Salmonella.
| SPI1 | Size/location2 (kb) | Main genes or gene products3 | Proposed function | Relative distribution4,5 | References |
|---|---|---|---|---|---|
| SPI-1 | 40 | T3SS, effector proteins (e.g., avrA, iacB, invB, sicA, sicP, sipA, sipB, sipC, and sptP) | Host cell invasion and enteropathy | S. enterica S. bongori | Fookes et al., 2011; Cao et al., 2013; Fàbrega and Vila, 2013 |
| SPI-2 | 40 | T3SS, effector proteins (ssaB, ssaE, sscA, sscB, sseA, sseF, sseG, and ttr genes) | Intracellular survival |
S. enterica subsp. enterica S. enterica subsp. salamae S. enterica subsp. diarizonae S. enterica subsp. indica |
Fookes et al., 2011; Fàbrega and Vila, 2013 |
| SPI-3 | Variable (17–36) | Magnesium transport system (mgtCB), misL | Intracellular survival, intestinal colonization |
S. enterica subsp. enterica S. enterica subsp. salamae S. enterica subsp. arizonae (partial) S. enterica subsp. diarizonae (partial) S. enterica subsp. houtenae (partial) S. enterica subsp. indica S. bongori |
Dorsey et al., 2005; Sabbagh et al., 2010; Fookes et al., 2011; Kaur and Jain, 2012 |
| SPI-4 | 25 | T1SS (siiABCDF), non-fimbrial adhesin (siiE) | Adhesion and invasion of epithelial cells, virulence in mice and cows |
S. enterica subsp. enterica S. enterica subsp. houtenae S. enterica subsp. indica S. bongori |
Morgan et al., 2007; Sabbagh et al., 2010; Fookes et al., 2011 |
| SPI-5 | Variable (11–44) | Effectors of SPI-1 and -2 (pipABC, sopB, pipD, and sigDE) | Epithelial invasion, enteric salmonellosis, and chicken colonization | S. enterica subsp. enterica | Fookes et al., 2011; Shah et al., 2012; Fàbrega and Vila, 2013; Cao et al., 2014 |
| SPI-6 | Variable (47–59) | T6SS, atypical fimbriae (safABCD), fimbriae (tcfABCD), and sciS | Invasion, intramacrophage survival, chicken colonization, and virulence in mice |
S. enterica subsp. enterica S. enterica subsp. salamae (partial) S. enterica subsp. arizonae (partial) S. enterica subsp. diarizonae (partial) S. enterica subsp. houtenae (partial) S. enterica subsp. indica (partial) |
Parkhill et al., 2001; Fookes et al., 2011; Cao et al., 2014; Pezoa et al., 2014 |
| SPI-7 | 134 | Vi capsule biosynthesis genes, SopE prophage, and type IVb pilus | Vi exopolysaccharide, host immune modulation, and intramacrophage survival | S. enterica subsp. enterica serovars Typhi, Paratyphi C, Dublin | Pickard et al., 2003; Faucher et al., 2005; Fookes et al., 2011 |
| SPI-8 | 6.8 | Bacteriocin fragment | Unknown | S. enterica subsp. enterica | Faucher et al., 2005; Saroj et al., 2008; Sabbagh et al., 2010; Fookes et al., 2011; Desai et al., 2013 |
| SPI-9 | 16 | T1SS, adhesin | Transport, epithelial adherence |
S. enterica subsp. enterica S. enterica subsp. salamae S. enterica subsp. arizonae S. enterica subsp. diarizonae S. enterica subsp. houtenae S. enterica subsp. indica S. bongori |
Sabbagh et al., 2010; Fookes et al., 2011; Velasquez et al., 2016 |
| SPI-10 | 33 | P4-like prophage, Sef fimbriae | Virulence in mice and chickens, intramacrophage uptake or survival | S. enterica subsp. enterica | Parkhill et al., 2001; Bishop et al., 2005; Sabbagh et al., 2010 |
| SPI-11 | Variable (6–10) | pagCD, envF (some) sRNA RaoN, typhoid toxin gene islet (cdtB, pltA, and pltB) (some) | Intramacrophage survival, serum resistance, and typhoid fever pathology | S. enterica subsp. enterica | Sabbagh et al., 2010; Fookes et al., 2011; Lee et al., 2013 |
| SPI-12 | Variable (6–15) | sspH2 | Actin polymerization, virulence in mice | S. enterica subsp. enterica | Morgan, 2007; Fookes et al., 2011; Tomljenovic-Berube et al., 2013 |
| SPI-13 | 25 | putative lyase, hydrolase, oxidase, and arylsulphatase regulator | Macrophage internalization, virulence in chickens, and mice |
S. enterica subsp. enterica (NTS) S. enterica subsp. arizonae S. enterica subsp. diarizonae S. enterica subsp. houtenae (some) |
Shah et al., 2005; Sabbagh et al., 2010; Fookes et al., 2011; Elder et al., 2016; Espinoza et al., 2017 |
| SPI-14 | 9 | gpiAB, putative acyl-CoA dehydrogenase | Chicken pathogenicity, epithelial invasion |
S. enterica subsp. enterica (NTS) S. enterica subsp. arizonae S. enterica subsp. houtenae (some) |
Shah et al., 2005, 2012; Fookes et al., 2011 |
| SPI-15 | 6.5 | Four putative ORFs | Unknown | S. enterica subsp. enterica serovar Typhi | Vernikos and Parkhill, 2006; Fookes et al., 2011 |
| SPI-16 | 4.5 | Bactoprenol glucosyl transferase and translocase (gtrAB) | LPS modification, seroconversion | S. enterica subsp. enterica | Vernikos and Parkhill, 2006; Fookes et al., 2011 |
| SPI-17 | 5 | Bactoprenol glucosyl transferase and translocase (gtrAB) | LPS modification, seroconversion | S. enterica subsp. enterica | Vernikos and Parkhill, 2006; Fookes et al., 2011 |
| SPI-18 | 2.3 | hlyE hemolysin, taiA invasion-associated protein | Epithelial invasion |
S. enterica subsp. enterica S. enterica subsp. diarizonae |
Den Bakker et al., 2011; Fookes et al., 2011 |
| SPI-19 | 45 | T6SS | Intramacrophage survival, chicken colonization | S. enterica subsp. enterica | Fookes et al., 2011; Blondel et al., 2013; Pezoa et al., 2014 |
| SPI-20 | 34 | T6SS | Unknown | S. enterica subsp. arizonae | Blondel et al., 2009, 2013; Fookes et al., 2011 |
| SPI-21 | 55 | T6SS | Unknown | S. enterica subsp. arizonae | Blondel et al., 2009, 2013; Fookes et al., 2011 |
| SPI-22 | 20 | T6SS | Unknown | S. bongori | Fookes et al., 2011 |
| SPI-23 | 37 | T3SS effectors (sanA, chlR, shaU, and dumE) | Host cell adherence and invasion, invasion of pig epithelial cells | S. enterica subsp. enterica | Hayward et al., 2013, 2014 |
| SPI-24/CS54 | 25 | Outer membrane protein (shdA, sivH, ratAB, sinI, and potR) | Fibronectin binding, murine intestinal colonization, and intramacrophage survival | S. enterica subsp. enterica | Kingsley et al., 2003; Sabbagh et al., 2010 |
1SPI-10 was initially defined in S. Typhi and was thought to be unique to S. Typhi (Sabbagh et al., 2010), although other studies have reported the presence of SPI-10 genes in NTS serovars as well (Den Bakker et al., 2011; Desai et al., 2013). 2In some cases the size varies between serovars depending on the number of genes encoded. 3“some” indicates that not all serovars, or strains within a serovar, carry a given gene(s). 4Distribution based on screening of few representative strains, may not be representative of the entire subspecies as comprehensive studies for all serovars have not been conducted. For SPIs 5 and 11 no data were available for testing in subspecies other than enterica.5“Partial” indicates that strains tested in the labeled subspecies encoded part of the SPI, but not the full SPI.
While SPIs are widespread among a number of S. enterica subsp. enterica serovars, some SPIs have been associated with select serovars, and are proposed to provide fitness advantages for these serovars. For example, SPI-7, encoding the Vi capsule, although classically thought to occur exclusively in S. Typhi (Faucher et al., 2005), has been found in strains of the NTS serovar Dublin as well (Morris et al., 2003; Pickard et al., 2003). It is interesting to note that both of these serovars are associated with invasive disease in humans. While SPI-11 is widespread among S. enterica subsp. enterica (Table 1), some serovars also encode genes for the typhoid toxin at this locus (Den Bakker et al., 2011). The presence of cdtB (a typhoid toxin gene) has been reported to be associated with significantly higher rates of invasive disease (Rodriguez-Rivera et al., 2015). Examples of SPIs associated with the ability to colonize a specific host have also been proposed. Mutation analyses of the T6SS encoded in SPI-19 in S. Gallinarum revealed a role for SPI-19 in the ability to colonize chickens (Blondel et al., 2010). However, introduction of SPI-19 (cloned from S. Gallinarum) in S. Enteritidis negatively impacted this serovar’s ability to colonize chickens (Blondel et al., 2009), therefore suggesting that, depending on the serovar, this SPI may provide a fitness advantage for some serovars, but not for others.
Toxins
Exotoxins constitute toxins that are secreted. Originally identified in serotype Typhi, the typhoid toxin (or Salmonella cytolethal distending toxin) is a genotoxin that has recently been identified in at least 41 NTS serovars as well (Den Bakker et al., 2011), however, it is not found in serovars Typhimurium, Enteritidis, and Newport, which cause many of the clinical infections in the United States, and worldwide. Experiments with S. Typhi have revealed a putative role for this toxin in fine-tuning the host response to infection by targeting specific cell types such as endothelial cells in the brain, and immune cells (Yang et al., 2018). Injection of purified typhoid toxin has been shown to recapitulate some signs of typhoid fever in a mouse model (Song et al., 2013). In NTS, this toxin has been shown to contribute to systemic host colonization by S. Javiana (Miller et al., 2018).
The spv operon, primarily present on the Salmonella virulence plasmid (described below), encodes a toxin that mediates ADP-ribosylation of actin and is implicated in host cell cytoskeletal rearrangements, and finally, apoptosis of the host cell (Libby et al., 1997, 2000; Otto et al., 2000; Lesnick and Guiney, 2001). The spv locus has also been reported to be chromosomally encoded for some isolates in subspecies II, IIIa, IV, and VII (Boyd and Hartl, 1998). SpvC has been shown to contribute to virulence, due to its phosphothreonine lyase activity, which inhibits host MAP kinases (Guiney and Fierer, 2011). SpvB and SpvC are only found in a select number of serovars (see Table 2). Isolates encoding the spv operon are proposed to have enhanced virulence, and are often associated with invasive disease (Libby et al., 1997; Guiney and Fierer, 2011), as spv genes appear to play an important role in suppression of the innate immune response by attenuating the intestinal inflammatory response (Haneda et al., 2012).
Table 2.
Selected nontyphoidal and paratyphoidal virulence plasmids.
| Serovar | Plasmid name | Size (kb) |
|---|---|---|
| Abortusovis | pSAV | 50–67 |
| Abortusequi | pSTV | 95 |
| Choleraesuis | pSCV | 50–110 |
| Dublin | pSDV | 80 |
| Enteritidis | pSEV | 60 |
| Gallinarum/Pullorum | pSPV | 85 |
| Paratyphi C | pSPCV | 55 |
| Sendai | pSSV | 285 |
| Typhimurium | pSTV/pSLT | 95 |
Virulence plasmids are listed with their corresponding name and size in kilobase pairs (kb).
The ArtAB toxin (for ADP-ribosylating toxin) is encoded by S. Typhimurium DT-104 strains (Saitoh et al., 2005) as well as multiple other NTS serovars (Rodriguez-Rivera et al., 2015). The active subunit, ArtA, ADP-ribosylates host G proteins (Uchida et al., 2009), while ArtB forms the pentameric binding subunit (Tamamura et al., 2017). ArtA and ArtB share homology with the active subunit (S1) and one of the B monomers (S2) of the binding subunit of the pertussis toxin, respectively (Tamamura et al., 2017). In vitro, the ArtAB toxin results in increased production of cAMP in RAW 264.7 cells, and a cell clustering phenotype in CHO cells (Saitoh et al., 2005; Tamamura et al., 2017). In vivo, BALB/c mice injected with 0.5–2 μg of purified ArtAB have significantly higher insulin secretion, and a reduced survival rate compared to mice injected with heat-killed toxin (Saitoh et al., 2005; Tamamura et al., 2017).
Several reports describe heat-labile, trypsin-sensitive cytotoxins produced by several NTS serovars. In some instances, cytotoxic activity is associated with the outer membrane. Cytotoxin production has been reported for extracts of serovars Braenderup, Choleraesuis, Enteritidis, Indiana, Nchanga, Saintpaul, Typhimurium, and Virchow (Reitmeyer et al., 1986; Ashkenazi et al., 1988; Kita et al., 1993; Malik et al., 1996). Considerable work has been conducted to characterize toxic activity in extracts and culture filtrates of multiple serovars (Peterson, 1980; Houston et al., 1981; Gemmell, 1984; Hariharan et al., 1986; Chopra et al., 1987; Khurana et al., 1991; Chary et al., 1993). One gene implicated in the observed cytotoxic activity, stn, has been proposed, although contrasting results about its actual involvement in cytotoxic activity exist, as its protein product has also been implicated in bacterial membrane integrity (Nakano et al., 2012).
Flagella
Most NTS are capable of expressing flagella, which confer motility (Fàbrega and Vila, 2013; Rivera-Chavez and Baumler, 2015). An important exception is serovar Gallinarum biovars Gallinarum and Pullorum, which do not express phase 1 or phase 2 flagella and are therefore non-motile (Foley et al., 2013). Flagella synthesis, assembly, and maintenance requires >50 genes (Bonifield and Hughes, 2003). However, the antigenic subunit, flagellin, is encoded by three genes, fliC (phase 1), fljB (phase 2), and flpA (phase 3; rare and often plasmid-encoded) (McQuiston et al., 2004). For most NTS serovars, 5-10 flagella of peritrichous organization may be observed (Van Asten and Van Dijk, 2005). Salmonella employ phase variation, a reversible genetic rearrangement, to switch between expression of fliC and fljB, a mechanism that is utilized by a number of important bacterial pathogens (Silverman et al., 1979; Bonifield and Hughes, 2003; Garcia-Pastor et al., 2018).
While flagella aid Salmonella in migrating toward host epithelial layers, and thus are important virulence determinants, they are also potent inducers of the host innate immune response (Dos Santos et al., 2018). Flagella have also been shown to allow Salmonella to taxi toward the host-derived nitrate and tetrathionate, which are used as alternate terminal electron acceptors (Rivera-Chavez and Baumler, 2015).
Although there is no obvious evidence linking specific flagellar antigens to differences in virulence or host-adaptations, regulation of flagella upon infection has been established as a mechanism to reduce or prevent activation of a host immune response. For example, in S. Typhimurium, flagellar expression is down-regulated 50–100 fold during infection of RAW 264.7 murine macrophage cells (Srikumar et al., 2015). Furthermore, Spöring et al. (2018) showed that this downregulation is a response to alterations of the cell envelope as a result of cell envelope stress (Spöring et al., 2018). Studies of flagellar regulation in other NTS serovars may reveal key differences in expression, and provide additional roles by which NTS serovars are able to either intentionally trigger an inflammatory immune response, which has been associated with providing a metabolic niche allowing NTS serovars to propagate in the intestine (Rivera-Chavez and Baumler, 2015), or to evade killing by immune cells.
Fimbriae (Pili)
Fimbriae are thin appendages that aid in attachment and adhesion, and are produced by a number of Gram-negative and Gram-positive bacteria. Phenotypic and genomic analyses have identified 39 putative fimbrial operons in Salmonella. Of these, the agf operon is found among isolates of both S. enterica and S. bongori, and encodes the nucleator-dependent curli fimbriae, which are thin, aggregative fimbriae that may aid in bacterial adhesion and invasion (Van Asten and Van Dijk, 2005; Yue et al., 2012). The bfp and pil operons encode type IV fimbriae; the latter operon is found on SPI-7, and its presence is therefore restricted to S. Typhi, Paratyphi C, and NTS serovar Dublin (Morris et al., 2003; Van Asten and Van Dijk, 2005). The remaining 36 fimbrial operons encode chaperone-usher-dependent fimbrial pathways (Clouthier et al., 1994; Van Asten and Van Dijk, 2005; Yue et al., 2012). Of these, 27 have been identified in NTS, with typical serovars containing 5–14 fimbrial gene clusters (Yue et al., 2012). The fim operon is the only chaperone-usher-dependent fimbrial operon found in all S. enterica isolates (Van Asten and Van Dijk, 2005; Yue et al., 2012). While the majority of fimbrial genes are not expressed under standard laboratory culturing conditions, most are expressed in vivo during infection (Laniewski et al., 2017), suggesting a role in adhesion to different tissues, or co-regulation with SPI-2 genes or other genes that are expressed when Salmonella is inside a host cell. The occurrence of other operons in various NTS has been reviewed elsewhere (Van Asten and Van Dijk, 2005). Of interest, the pef (plasmid encoded fimbriae) operon is located on a subset of Salmonella virulence plasmids (Rychlik et al., 2006), and is discussed later in further detail.
Differences in fimbrial gene clusters exist among serovars, and have been proposed as an additional mediator for allowing some serovars to colonize and persist in different hosts/environments (Yue et al., 2012). The number of intact fimbrial gene clusters, as well as the type, varies among serovars. For example, Yue et al. (2012) demonstrated that host adapted/restricted serovars, such as Typhi, Dublin, Paratyphi A, Choleraesuis, and Gallinarum encode multiple non-functional fimbrial genes (Yue et al., 2012). In contrast, serovars demonstrating a broader host range (Typhimurium, Enteritidis, Montevideo, Newport, Tennessee, Kentucky, and others) have relatively few degraded fimbrial genes (Yue et al., 2012). Moreover, allelic variation of fimbrial genes has also been proposed as an adaptation associated with the colonization of certain hosts (Yue et al., 2015). For example, S. Newport isolated from bovine, porcine, equine, or avian hosts was more likely to have allelic variants A/A/A1 (representing allelic combinations of FimH/BcfD/StfH), while S. Newport isolated from humans and the environment had predominately B/B/B1 fimbrial alleles (De Masi et al., 2017). A similar observation was also reported for S. Typhimurium, where different alleles of FimH1 or FimH7 were significantly associated with adherence to different human and bovine intestinal epithelial cell lines (Yue et al., 2015). Furthermore, Yue et al. (2015) also demonstrated inter-serovar differences in the ability of E. coli strains expressing different Salmonella FimH alleles to adhere to host cell lines representing different mammalian species (Yue et al., 2015). For example, expression of porcine-associated Salmonella fimbrial alleles resulted in significantly higher proportions of recombinant E. coli bound to porcine epithelial cell types (Yue et al., 2015). Together, this suggests that maintenance of multiple different fimbrial genes, and allelic variants of these genes, represents a mechanism by which NTS serovars are able to adhere to a variety of surfaces, making them suited to many different environments.
Virulence Plasmids
Nine NTS serovars have been described as harboring a low copy-number virulence plasmid. This plasmid varies among serovars, both in size and in genetic content (Table 2). Furthermore, carriage of the plasmid is not ubiquitous among all isolates of a given serovar (Chu and Chiu, 2006). Isolates that do carry the plasmid, though, generally exhibit increased virulence. The mediator of this increased virulence is the 7.8 kb spvRABCD (Salmonella plasmid virulence) operon, whose effectors alter the host cell cytoskeleton to enhance bacterial survival (Rotger and Casadesús, 1999). While spv is common to all Salmonella virulence plasmids, additional virulence factors or antimicrobial resistance genes may also be encoded (Figure 5).
FIGURE 5.
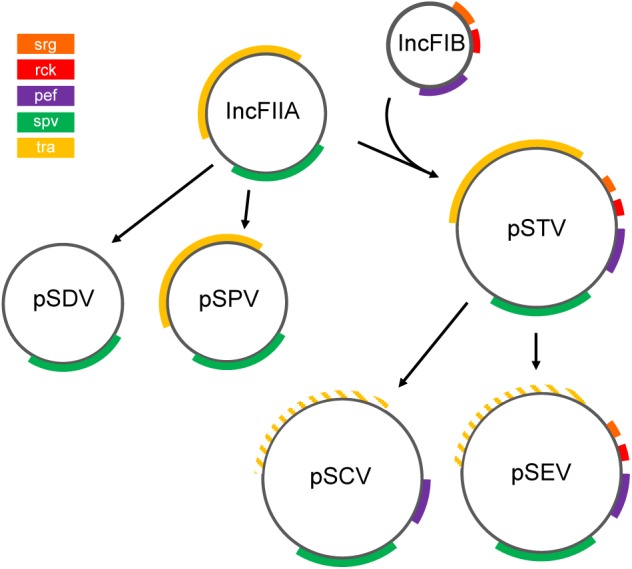
Proposed evolution of NTS virulence plasmids. The ancestral IncFIIA evolved into plasmids pSDV (Dublin) and pSPV (Gallinarum/Pullorum), or fused with replicon IncFIB in the pSTV (Typhimurium) lineage (Rychlik et al., 2006). pSCV (Choleraesuis) and pSEV (Enteritidis) are derived from pSTV (Chu and Chiu, 2006). Colored segments indicate gene clusters. Hatched lines indicate loss of function of the tra locus (Rotger and Casadesús, 1999). Although pSPV encodes tra genes, this plasmid is mobilized via the F-plasmid (Chu and Chiu, 2006). Plasmids and genes are not drawn to scale. This figure is inspired by the theories proposed by Rychlik et al. (2006) and others (Feng et al., 2012).
The variation in content is attributed to the evolution of two distinct plasmid lineages, which have been characterized by sequence analysis, probe hybridization, and compatibility studies (Chu and Chiu, 2006). Serovars Paratyphi C (Liu et al., 2009), Sendai (Silva et al., 2017), Abortusequi (Akiba et al., 1999), and Abortusovis (Uzzau et al., 2000) also encode a virulence plasmid (Table 2), but the evolution of these plasmids is unclear.
The first lineage contains plasmids of S. Gallinarum (Pullorum; pSPV), and S. Dublin (pSDV) (Chu and Chiu, 2006). pSPV and pSDV harbor the replicon repB, which exhibits high similarity to the ancestral replicon IncFIIA (also called RepFIIA) (Rotger and Casadesús, 1999; Rychlik et al., 2006). It has been proposed that the ancestral IncFIIA replicon contained the spv and tra loci, as all Salmonella virulence plasmids encode variants of these genes, although the tra genes in the current pSPV and pSDV virulence plasmids are remnants of a bacterial conjugation system that has undergone variable degradation in several plasmid lineages (Rychlik et al., 2006). By virtue of the tra locus, pSPV may be mobilized in the presence of a F plasmid (Chu and Chiu, 2006; Rychlik et al., 2006), whereas pSDV lacks homology to the tra locus (Rotger and Casadesús, 1999), and cannot be mobilized (Rychlik et al., 2006), suggesting substantial deterioration of the tra locus in pSDV.
The plasmids of serovars Typhimurium (pSTV), Enteritidis (pSEV), and Choleraesuis (pSCV) compose the second lineage (Chu and Chiu, 2006), and contain not only the repB replicon, but also elements of a second replicon, repC (Rotger and Casadesús, 1999). repC exhibits similarity to the IncFIB replicon (Rotger and Casadesús, 1999), which likely encoded the virulence factors pef, srgAB, and rck (Rychlik et al., 2006). The pef operon, or plasmid-encoded fimbriae, is conserved in all three plasmids of this lineage (Rychlik et al., 2006). pef-encoded fimbriae mediate adhesion to cells within the small intestine, and exhibit preferential binding to certain host cells of various species (Baumler et al., 1996). The srgA gene product is important for the biogenesis of the pef-encoded fimbriae (Bouwman et al., 2003), yet this locus has been lost in pSCV (Rychlik et al., 2006), as has rck (Rychlik et al., 2006). rck has been shown to contribute to resistance to complement killing, one of the host’s innate immune responses (Rychlik et al., 2006). As in the first lineage the tra locus exhibits variable degradation among the plasmids pSTV, pSCV, and pSEV. The tra operon of pSTV renders that plasmid capable of conjugative transfer (Rychlik et al., 2006), yet in the S. Enteritidis plasmid (pSEV), and S. Choleraesuis plasmid (pSCV) lineages the tra locus has degraded and is no longer functional (Rotger and Casadesús, 1999; Rychlik et al., 2006). The degradation of tra in pSCV and pSEV may indicate that they descended from pSTV via deletions (Chu and Chiu, 2006).
Disease Diversity-Nontyphoidal Salmonellosis Disease Severity Varies Significantly by Serovar
Although the complete host-pathogen relationship plays an important role in determining the resulting severity of salmonellosis (Figure 2), some NTS serovars excel at causing invasive human clinical infections that are reminiscent of the pathology exhibited by Typhi and paratyphoidal serovars of S. enterica. Invasive infections result from Salmonella’s successful ability to escape the gastrointestinal tract, and subsequently spread to other tissues. These infections are considered to be the most severe, often resulting in hospitalization. Most commonly, invasive salmonellosis is defined as isolation of Salmonella from a “sterile site,” usually blood, joint fluid, or cerebrospinal fluid (Jones et al., 2008). Global estimates of invasive nontyphoidal salmonellosis (regardless of serovar) range from 30 to 227 cases per 100,000 persons (Ao et al., 2015). With the exception of select NTS serovars, most invasive NTS infections are typically associated with individuals from so-called “high risk” populations (i.e.,<5 years old, >65 years old, having an immunodeficiency, or being pregnant). For example, invasive nontyphoidal salmonellosis is more frequently cited in Africa (Okoro et al., 2012; Ao et al., 2015) and Southeast Asia (Lan et al., 2016), and is associated with malnutrition, immunodeficiencies including HIV, and malaria co-infection (Scott et al., 2011; Feasey et al., 2012).
A study by Jones et al. (2008) found that, compared to S. Typhimurium infections, of which just 5.7% resulted in invasive disease, infections with serovars Choleraesuis (56.4%), Dublin (64.0%), Sandiego (18.9%), and Panama (18.0%) were associated with significantly higher rates of invasive disease among human clinical infections in the US (Jones et al., 2008). While the total number of salmonellosis cases contributed by these serovars is considerably lower than those of other serovars, the number of infections resulting in invasive disease is significantly higher, suggesting that either (i) most of these infections arise from individuals in high risk populations (i.e., only select sub-populations are susceptible to infection with these serovars, and when infection occurs it is often more severe), (ii) the majority of the cases are asymptomatic, and therefore only severe cases are reported, or (iii) these serovars have virulence factors or adaptations that make them inherently more invasive. While further research is needed to conclusively determine which components of the host-pathogen interaction specifically account for the observed invasive phenotype of these serovars, the current understanding is summarized below.
NTS Serovars Associated With High Rates of Invasive Disease in Humans
Salmonella enterica subsp. enterica serovar Choleraesuis (S. Choleraesuis) is a host-adapted serovar, causing swine paratyphoidal disease (Chiu C.-H. et al., 2004; Pedersen et al., 2015). In humans, S. Choleraesuis often causes septicemia with minimal GI-tract inflammation (Chiu C.-H. et al., 2004), resulting in a disease more similar to typhoid fever (Sabbagh et al., 2010). This suggests that, similar to S. Typhi, the severity of the salmonellosis caused by this serovar results from successful evasion of host defenses in the gut, and thus failure of the immune system to detect and control S. Choleraesuis early in the infection. In support of this, most human clinical cases of S. Choleraesuis occur in patients with a pre-existing health condition, such as an immunosuppressive condition (Wang et al., 2006), or other chronic disease (Chiu S. et al., 2004). Like other serovars that are frequently associated with invasive human disease, S. Choleraesuis encodes a T6SS, which plays a role in virulence in vivo (Schroll et al., 2019). S. Choleraesuis strain SC-B67, originally isolated from a human patient with sepsis, has a high proportion of pseudogenes, most of which arose from SNPs resulting in premature stop codons in genes related to metabolism, fimbriae, and the chemotactic response (Chiu et al., 2005). Inactivation of several ancestral genes may limit the adaptability of S. Choleraesuis in several niches including animal hosts and environmental sites, which would account for the lack of clinical cases in animals other than pigs and humans for this serovar. While S. Choleraesuis is rarely isolated from human clinical cases in the US [∼10–20 confirmed cases per year (Centers for Disease Control and Prevention [CDC], 2016)] and the EU (Pedersen et al., 2015), cases of S. Choleraesuis are relatively more common in Southeast Asia such as in Taiwan and Thailand, although control efforts have been very successful at reducing the incidence of S. Choleraesuis infections in these areas (Hendriksen et al., 2009; Hendriksen et al., 2011).
Salmonella enterica subsp. enterica serovar Dublin (S. Dublin) is a bovine host-adapted serovar (Langridge et al., 2015). Genetic analyses have shown that S. Dublin shares a common ancestor with another host-adapted serovar, S. Gallinarum (Langridge et al., 2015), although S. Dublin has substantially fewer pseudogenes (Langridge et al., 2015). S. Dublin encodes two T6SSs located in SPI-6 and SPI-19 (Mohammed et al., 2017). In the United States, confirmed human clinical cases of S. Dublin salmonellosis contribute ∼150 reported cases per year (Centers for Disease Control and Prevention [CDC], 2016), with the majority of S. Dublin infections occurring in adults (median age 55 years, compared to median age of 23 years for infections with all other Salmonella) (Harvey et al., 2017). This suggests that either the incidence of exposure to S. Dublin is higher among older individuals, or S. Dublin is asymptomatic in younger individuals, but causes clinical disease when it encounters a host with a weakened immune system, as is common for other opportunistic pathogens. While S. Dublin infections in humans are significantly more likely to result in hospitalization, invasive disease, prolonged hospital stays, and an increased likelihood of death (Harvey et al., 2017), the true incidence of exposure is unknown, and is likely much higher than the number of clinical infections reported each year. Nevertheless, it is intriguing that this serovar, when successful in colonizing humans, often results in invasive disease.
Infections with S. enterica subsp. enterica serovar Sandiego (S. Sandiego) and S. enterica subsp. enterica serovar Panama (S. Panama) are also associated with high rates of invasive disease (Jones et al., 2008; Crump et al., 2011). In contrast to S. Dublin and S. Choleraesuis, these serovars have not been characterized as being host-adapted. S. Sandiego has been linked to several outbreaks from handling small turtles (Walters et al., 2015), and has also been isolated from livestock including pigs (Oliveira et al., 2002), goats (El Tom et al., 1999), and cows (Boqvist and Vågsholm, 2005). S. Panama has been isolated from pigs, poultry, cows, and goats (Cordano and Virgilio, 1996). Both S. Panama and S. Sandiego encode S. Typhi-associated SPI-18 genes hlyE and taiA, and the typhoid toxin genes (cdtB, pltA, and pltB) (Den Bakker et al., 2011), although the role that these genes, and their corresponding gene products, play in invasive infections involving these serovars has not been assessed. Unlike S. Choleraesuis and S. Dublin, infections with S. Panama and S. Sandiego are more commonly diagnosed among children <5 years old (Boore et al., 2015). Therefore, either environmental exposure to S. Panama and S. Sandiego is more common in young children, perhaps as a result of animal contact, or these serovars possess virulence factors that enable them to successfully cause disease in young children, but not as frequently in adults, as is the case for serovars Choleraesuis and Dublin.
NTS Serovars Associated With Reduced Rates of Human Clinical Disease: Evidence of Loss of Virulence
In contrast to NTS serovars associated with high rates of invasive human clinical disease, there also exist a number of serovars that are commonly isolated from agricultural reservoirs, but which cause a disproportionately low number of human clinical cases. The primary reasons for this include (i) effective kill steps such as heating or other inactivation techniques for contaminated food commodities (i.e., cooking of meat or pasteurization of milk), (ii) mutations in select serovars that effectively reduce their ability to cause disease in select hosts, and (iii) genetic adaptations that allow the serovar to colonize an animal host, but do not impart a fitness advantage allowing for disease manifestation in humans.
S. Cerro is one of the most frequently isolated serovars among dairy clinical isolates (Tewari et al., 2012; Valenzuela et al., 2017), although this serovar is responsible for just ∼30 reported human clinical cases per year in the United States (Centers for Disease Control and Prevention [CDC], 2016). In 2016, S. Cerro was the 4th most common clinical isolate among animal clinical cases reported to the USDA, and represented the 2nd most common serovar reported among clinical cases in cattle (Morningstar-Shaw et al., 2016). Despite S. Cerro being detected in bulk tank raw milk samples, outbreaks from Salmonella-contamination of raw milk in the United States usually involve serovars Typhimurium (Mungai et al., 2015), Montevideo, and Newport (Robinson et al., 2014), and the cow-adapted serovar Dublin (Harvey et al., 2017; Vignaud et al., 2017). S. Cerro isolates have a premature stop-codon in sopA (Rodriguez-Rivera et al., 2014; Kovac et al., 2017), a virulence factor shown to play an important role in Salmonella entry into host cells (Raffatellu et al., 2005). Indeed, S. Cerro isolates have a significantly lower rate of invasion in the Caco-2 cell line, compared to S. Typhimurium and S. Newport strains in a standard gentamicin protection assay (Rodriguez-Rivera et al., 2014). The comprehensive set of reasons behind the discrepant ability of S. Cerro to colonize and amplify within dairy cattle, and its low likelihood of causing clinical disease in both humans and most animals, remains unknown.
In the United States, S. Kentucky represents just 0.1% of reported human clinical cases of salmonellosis (Centers for Disease Control and Prevention [CDC], 2016), yet it is the most commonly isolated serovar from broiler chickens in the United States (United States Department of Agriculture [USDA], 2014). Although experimental evidence suggests that S. Kentucky may persist for longer periods of time in chickens than S. Typhimurium, the reasons why S. Kentucky is infrequently associated with human clinical cases of salmonellosis have yet to be confirmed (Cheng et al., 2015). One possibility for the lack of human clinical cases is that S. Kentucky isolates lack the virulence genes grvA, sseI, sopE, and sodCI (Beutlich et al., 2011; Cheng et al., 2015), which may play a role in human disease but are not necessary for colonizing chickens. Another study by Tasmin et al. (2017) using a phenotypic array found that S. Kentucky ST152 strain SK222_32B differed significantly in its ability to utilize a number of common metabolites, namely 1,2-propanediol (Tasmin et al., 2017), which has previously been cited for enabling S. Typhimurium to successfully outcompete the resident microbiota in the mammalian gut (Faber et al., 2017). S. Kentucky SK222_32B also failed to replicate in macrophages, implying that S. Kentucky has an impaired ability to resist immune-killing, possibly as a result of its lacking super oxide dismutase (sodCI) (Tasmin et al., 2017). Indeed, other studies have shown that, when exposed to media at pH 2.5, S. Kentucky was more sensitive to acid stress than serovars Enteritidis, Hadar, Mbandaka, Senftenberg, Typhimurium, and Worthington (Joerger et al., 2009). In contrast, human clinical cases caused by S. Kentucky are increasing in Europe. Interestingly, S. Kentucky type ST198 is more commonly isolated in human clinical cases (primarily in Europe and Northern Africa), but S. Kentucky ST152 is more commonly isolated from poultry (Le Hello et al., 2013; Haley et al., 2016). While studies suggest that ciprofloxacin resistance encoded by S. Kentucky ST198 explains its sudden expansion in human clinical cases (Le Hello et al., 2011, 2013), future studies examining the genetic and metabolic differences between the chicken-associated (ST152) and human associated (ST198) STs may reveal more conclusive evidence for why certain S. Kentucky STs are associated with different hosts.
Understanding why certain serovars are able to asymptomatically colonize a host represents an important gap in our current understanding of nontyphoidal salmonellosis. In the case of S. Cerro, understanding the importance of mutations enabling this serovar to frequently colonize cattle, but rarely cause clinical symptoms in humans represents a key gap in our understanding of NTS virulence. For S. Kentucky, increases in human clinical infections involving a specific sequence type suggest either a new route of transmission, or possibly a novel adaptation of ST198 that may be absent in other S. Kentucky STs, which might be an important model for how other serovars that are frequently associated with animal reservoirs can adapt to cause human clinical disease.
Adapting to the Situation – Foodborne, Zoonotic, and Environmental Sources of NTS Serovars
While the vast majority of NTS cases are foodborne, infections resulting from animal contact and environmental exposures have been reported. Owing to their ability to successfully survive in a number of different environments, NTS serovars have adapted to achieve a number of different routes of transmission to cause human clinical infections. Traceback investigations aimed at linking a Salmonella isolate to its source represent a persistent challenge. Further complicating this, the majority of infections represent sporadic incidents, which cannot be linked to a common source. Here we review the foodborne, animal-contact, and environmental sources of NTS salmonellosis.
Foodborne Nontyphoidal Salmonellosis
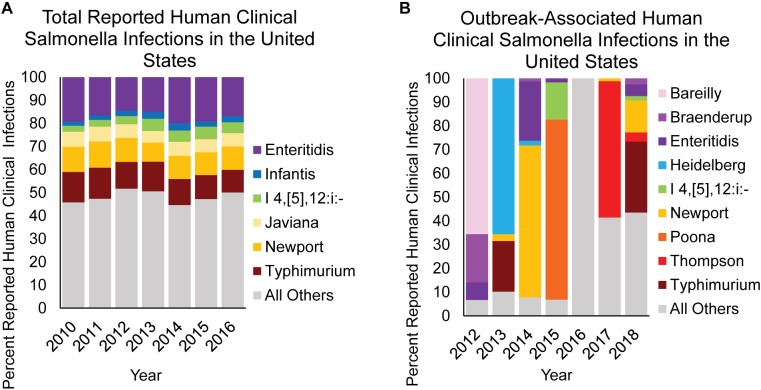
In the United States, approximately 94% of domestically acquired salmonellosis cases are acquired through the consumption of contaminated food (Scallan et al., 2011), with most representing sporadic cases (Ebel et al., 2016). In fact, it has been reported that just 5.9% of NTS infections in the United States are linked to an outbreak (Ebel et al., 2016). This is best represented by comparing the relative contribution of reported NTS serovars isolated from human clinical infections (Figure 6A) with the serovars causing outbreaks traced back to a single food item (Figure 6B). Data compiled between 2012 and 2018 show that in the United States, the primary NTS serovars associated with outbreaks vary significantly from year to year, and are often the result of a few large outbreaks (Figure 6B). The overall trend in human clinical infections remains constant however, as infections reported between 2010 and 2016 in the United States show very little fluctuation by serovar, with the top 6 serovars accounting for ∼50% of all infections each year. Therefore, the accumulation of NTS sporadic infections is likely the result of low-level Salmonella contamination from a wide range of foods, primarily by serovars Typhimurium, Newport, Enteritidis, Javiana, Infantis, and I 4,[5],12:i:-, and not due to large outbreaks involving these serovars.
FIGURE 6.
NTS causing overall vs. confirmed outbreak-associated infections in humans. The contribution of select NTS serovars to (A) overall human clinical infections vs. (B) human clinical infections confirmed from a foodborne outbreak are shown as percent infection for a given category. Data are modified from Centers for Disease Control and Prevention [CDC] (2016, 2018).
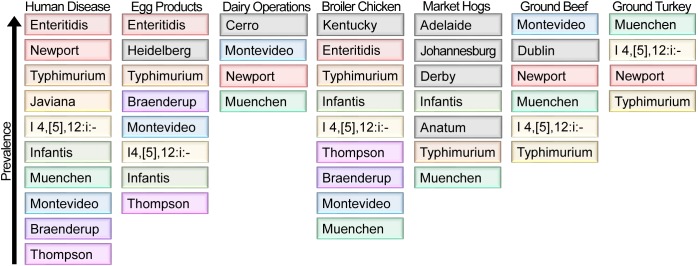
Each food commodity presents its own challenges, including differences in pH, available water, temperature, nutrient composition, osmolarity, and the presence of antimicrobial compounds; combined these represent unique environmental stresses to which Salmonella must adapt. An analysis by the CDC of the food commodities most frequently associated with outbreaks of salmonellosis by serovar reveals that for certain food products, there is a strong association between select serovars and food commodities of animal origin (Jackson et al., 2013). For example, for serovars Enteritidis, Heidelberg, I 4,[5],12:i:-, and Hadar, the majority of outbreaks are associated with poultry (either chicken or turkey meat, or eggs), while serovars Infantis, Montevideo, and Uganda are more commonly associated with outbreaks linked to consumption of pork and beef (Jackson et al., 2013). This suggests that these serovars possess genetic and phenotypic traits that promote their ability to adapt to these environments. The USDA conducts routine surveillance in the United States to monitor retail-bound meat (United States Department of Agriculture [USDA], 2014), egg products (United States Department of Agriculture [USDA], 2017), and raw dairy foods (Sonnier et al., 2018) for contamination by NTS. An interesting observation is that the serovars most frequently isolated from these high-risk food sources do not mirror those predominantly responsible for infection in humans (Figure 7). This is readily apparent from the frequent isolation of S. Kentucky and S. Cerro from poultry meat products and dairy cows, respectively, which does not result in an equivalent predominance among human clinical infections (Figure 7).
FIGURE 7.
NTS serovar prevalence from reported human clinical cases of salmonellosis and surveillance of different animal food sources. The ten serovars most frequently identified in human disease (colored) are listed in order of their prevalence for each of the indicated sources (United States Department of Agriculture [USDA], 2014, 2017; Centers for Disease Control and Prevention [CDC], 2016; Sonnier et al., 2018). When a serovar represents ≥10% of Salmonella identified from a source other than humans, it is shown in gray under that source.
A number of produce (fruits and vegetables) outbreaks have also occurred (Hanning et al., 2009), demonstrating NTS serovars’ ability to survive in acidic and dry environments, as well as at a wide range of temperatures associated with the produce production chain. For example, tomatoes have been associated with several outbreaks involving serovars Montevideo, Newport, Braenderup, and Javiana (Hanning et al., 2009). Shi et al. (2007) observed differences among serovars that were able to grow on ripened tomatoes, likely due to the high acidity associated with this fruit (Shi et al., 2007). Differences in acid tolerance among NTS serovars, may therefore represent one mechanism allowing some serovars to contaminate and grow in select food commodities.
Zoonotic Transmission of Salmonella
Several reports implicate a role for animal contact in human infection with NTS serovars. Not surprisingly, handling of chicks is responsible for a considerable number of salmonellosis cases (Figure 8), in which serovars Typhimurium, Johannesburg, Braenderup, Thompson, and Montevideo are frequently implicated with human outbreaks of salmonellosis (Loharikar et al., 2012; Pabilonia et al., 2014; Habing et al., 2015; Nakao et al., 2015; Anderson et al., 2016; Sharma et al., 2018). Zoonotic infection may also result from exposure to infected companion animals (kittens, guinea pigs, hedgehogs, and turtles) (Figure 8; Centers for Disease Control and Prevention [CDC], 2001; Wright et al., 2005; Bartholomew et al., 2014; Anderson et al., 2017; Gambino-Shirley et al., 2018). Interestingly, NTS infections arising from contact with mammals have been largely linked to serovars Typhimurium and Enteritidis (Hoelzer et al., 2011) (the two serovars causing the majority of foodborne salmonellosis in humans), while zoonoses contracted from turtles included infection with serovars Sandiego, Poona, and Pomona (Gambino-Shirley et al., 2018), which infrequently cause human clinical cases.
FIGURE 8.
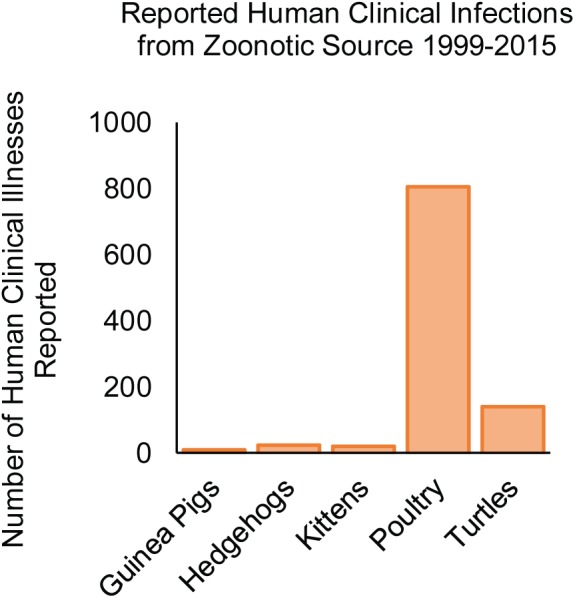
NTS serovars associated with zoonotic infections in the United States from 1999 to 2015. The number of human clinical infections resulting from animal contact is shown according to the source of the infection (Centers for Disease Control and Prevention [CDC], 2001; Wright et al., 2005; Loharikar et al., 2012; Bartholomew et al., 2014; Nakao et al., 2015; Anderson et al., 2016, 2017; Gambino-Shirley et al., 2018).
Environmental Transmission of Salmonella
Although less common, nontyphoidal serovars have been isolated from soil, water, contaminated floors, carts, and other equipment-related sources (Cummings et al., 2014; Strawn et al., 2014; Liu et al., 2018). Environmental sources of NTS serovars often arise from the introduction of NTS-contaminated animal feces, either via contaminated water used to irrigate crops, or from direct contact with feces from the Salmonella-secreting animal. In the case of several teaching hospitals, nosocomial transmission has been confirmed between animals, including different species, implicating the ability of NTS serovars to survive and transit from different environmental sources, to a susceptible host (Cummings et al., 2014). Furthermore, albeit rare, human-human cases of NTS transmission have also been documented (Kariuki et al., 2006).
Outbreaks of foodborne salmonellosis have also been documented in which biofilms harboring NTS serovars were linked to contamination of food products. In 2004 and 2005, two outbreaks involving S. Agona in powdered infant formula were traced back to environmental contamination in the production plant where the formula was produced (Brouard et al., 2007). S. Agona has also been linked to two outbreaks in dry cereal, in 1998 and 2008 (Russo et al., 2013), resulting from S. Agona persistence in the food processing plant. Importantly, these outbreaks highlight the potential for NTS serovars to persist in food processing plants for extended periods of time, and then contaminate food products.
Depending on the environment, transmission of NTS serovars from the environment onto a food surface represents an important, yet indirect, mechanism of transmission for a number of food commodities. While adaptations that allow NTS serovars to persist in different environments, such as Salmonella’s ability to survive in dry foods/environments, are largely dependent on the specific food (i.e., fat content and moisture content, etc.), there are significant differences in the abilities of different serovars to survive in foods.
Reducing NTS Incidence Will Require a Diversified Approach
The broad host-range displayed by a number of NTS serovars represents a key challenge in reducing nontyphoidal salmonellosis, as interventions aimed at targeting a single reservoir are often unsuccessful at eliminating Salmonella from other reservoirs, many of which are unknown. Furthermore, successful control of one serovar has historically been associated with increases in the number of human clinical cases and outbreaks caused by other serovars due to the vacancy of a previously occupied niche. For example, targeted control efforts aimed at eliminating S. Gallinarum (biovars Gallinarum and Pullorum) infections among chickens paralleled the sudden increase in S. Enteritidis, which is able to colonize chickens without causing overt clinical disease (Baumler et al., 2000). Therefore, successful control of one serovar in a given niche may enable the expansion of a new serovar. Complicating this, little is known about the natural reservoirs of the less-studied NTS serovars.
The vast majority of what is known about the virulence factors and mechanisms of NTS serovars is derived from experiments performed with NTS serovar Typhimurium, which have provided beneficial data that have expanded our understanding of how Salmonella cause disease. However, the genetic differences among serovars suggest that a number of mechanisms are serovar-specific, and therefore, improved approaches should consider the importance of using specific serovars (Jones et al., 2008; Suez et al., 2013). Identifying what makes certain serovars better adapted to persist in different environments also represents a key piece in resolving the NTS-contamination puzzle, as many serovars have yet to be associated with their reservoir(s).
The collective effort to reduce-foodborne nontyphoidal salmonellosis will require a collaboration of both laboratory science, generating experimental evidence about how diverse NTS serovars are able to overcome unfavorable environmental conditions to establish infection, and public health authorities utilizing novel methods for enhanced outbreak detection and source tracking. Furthermore, increases in the diversity and number of publicly available WGS data will enable the use of genetic tools (i.e., CRISPR, Lambda-Red mutagenesis, etc.) necessary to carry out genetic and phenotypic analyses among non-Typhimurium NTS serovars, allowing for the development of novel control and treatment strategies that will aid in reducing the overall morbidity and mortality associated with NTS on a global level.
As foodborne transmission represents the most common route of infection with NTS serovars, risk-based research approaches aimed at identifying and targeting specific serovars likely to contaminate a given food product represents an attractive approach to reducing human clinical cases. For example, the synthesis of data regarding contamination of a specific food commodity, such as S. Enteritidis and eggs, can be used to identify key sources of contamination so that research and regulatory initiatives may be focused to control contamination with specific serovars in these niches. Alternatively, assessments based on disease severity may be used to inform new regulatory decisions, such as imposing more stringent limits for some serovars (e.g., those associated with high rates of invasive disease), or relaxed limits for less virulent serovars (e.g., Cerro) or for specific strains showing virulence-attenuating mutations.
Conclusion
While the global burden of nontyphoidal salmonellosis remains as one of the key challenges, novel methods for studying Salmonella have changed the way that the research community detects, investigates, and prevents salmonellosis. Reflecting the diversity of virulence factors, genetic and phenotypic adaptations among the more than 2,600 serovars, reducing contamination with NTS serovars represents a multi-faceted challenge that will require collaborations of industry, government, and academia to achieve the ultimate goal of reducing NTS human clinical infections worldwide.
Author Contributions
RC, CE, and MW conceived and revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Dr. Craig Altier for his comments and suggestions during the preparation of this manuscript, Dr. Renato Orsi for assistance in the preparation of figures, and Dr. Laura Carroll for helpful discussions about S. Kentucky.
Footnotes
Funding. CE was supported by the NIH/USDA-NIFA Dual Purpose with Dual Benefit program; grant number 2014-67015-21697 awarded to Dr. Craig Altier. This material is based upon work that was supported by the National Institute of Food and Agriculture, United States Department of Agriculture, under the award number 2016-67012-25184 (to CE).
References
- Achtman M., Wain J., Weill F.-X., Nair S., Zhou Z., Sangal V., et al. (2012). Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 8:e1002776. 10.1371/journal.ppat.1002776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiba M., Sameshima T., Anzai T., Wada R., Nakazawa M. (1999). Salmonella Abortusequi strains of equine origin harbor a 95kb plasmid responsible for virulence in mice. Vet. Microbiol. 68 265–272. 10.1016/s0378-1135(99)00078-4 [DOI] [PubMed] [Google Scholar]
- Alikhan N.-F., Zhou Z., Sergeant M. J., Achtman M. (2018). A genomic overview of the population structure of Salmonella. PLoS Genet. 14:e1007261. 10.1371/journal.pgen.1007261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson T. C., Marsden-Haug N., Morris J. F., Culpepper W., Bessette N., Adams J. K., et al. (2017). Multistate outbreak of human Salmonella typhimurium infections linked to pet hedgehogs - United States, 2011-2013. Zoonoses Public Health 64 290–298. 10.1111/zph.12310 [DOI] [PubMed] [Google Scholar]
- Anderson T. C., Nguyen T. A., Adams J. K., Garrett N. M., Bopp C. A., Baker J. B., et al. (2016). Multistate outbreak of human Salmonella Typhimurium infections linked to live poultry from agricultural feed stores and mail-order hatcheries, United States 2013. One Health 2 144–149. 10.1016/j.onehlt.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao T. T., Feasey N. A., Gordon M. A., Keddy K. H., Angulo F. J., Crump J. A. (2015). Global burden of invasive nontyphoidal Salmonella disease, 2010. Emerg. Infect. Dis. 21:941. 10.3201/eid2106.140999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazi S., Cleary T. G., Murray B. E., Wanger A., Pickering L. K. (1988). Quantitative analysis and partial characterization of cytotoxin production by Salmonella strains. Infect. Immun. 56 3089–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew M. L., Heffernan R. T., Wright J. G., Klos R. F., Monson T., Khan S., et al. (2014). Multistate outbreak of Salmonella enterica serotype enteritidis infection associated with pet guinea pigs. Vector Borne Zoonotic Dis. 14 414–421. 10.1089/vbz.2013.1506 [DOI] [PubMed] [Google Scholar]
- Baumler A. J., Hargis B. M., Tsolis R. M. (2000). Tracing the origins of Salmonella outbreaks. Science 287 50–52. 10.1126/science.287.5450.50 [DOI] [PubMed] [Google Scholar]
- Baumler A. J., Tsolis R. M., Bowe F. A., Kusters J. G., Hoffmann S., Heffron F. (1996). The pef fimbrial operon of Salmonella typhimurium mediates adhesion to murine small intestine and is necessary for fluid accumulation in the infant mouse. Infect. Immun. 64 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutlich J., Jahn S., Malorny B., Hauser E., Hühn S., Schroeter A., et al. (2011). Antimicrobial resistance and virulence determinants in European “Salmonella genomic Island 1 (SGI1)” positive Salmonella enterica isolates from different origins. Appl. Environ. Microbiol. 77 5655–5664. 10.1128/AEM.00425-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A. L., Baker S., Jenks S., Fookes M., Gaora P. Ó, Pickard D., et al. (2005). Analysis of the hypervariable region of the Salmonella enterica genome associated with tRNAleuX. J. Bacteriol. 187 2469–2482. 10.1128/jb.187.7.2469-2482.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel C. J., Jimenez J. C., Contreras I., Santiviago C. A. (2009). Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC Genom. 10:354. 10.1186/1471-2164-10-354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel C. J., Jimenez J. C., Leiva L. E., Alvarez S. A., Pinto B. I., Contreras F., et al. (2013). The type VI secretion system encoded in Salmonella pathogenicity island 19 is required for Salmonella enterica serotype Gallinarum survival within infected macrophages. Infect. Immun. 81 1207–1220. 10.1128/IAI.01165-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel C. J., Yang H. J., Castro B., Chiang S., Toro C. S., Zaldivar M., et al. (2010). Contribution of the type VI secretion system encoded in SPI-19 to chicken colonization by Salmonella enterica serotypes Gallinarum and Enteritidis. PLoS One 5:e11724. 10.1371/journal.pone.0011724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifield H. R., Hughes K. T. (2003). Flagellar phase variation in Salmonella enterica is mediated by a posttranscriptional control mechanism. J. Bacteriol. 185 3567–3574. 10.1128/jb.185.12.3567-3574.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore A. L., Hoekstra R. M., Iwamoto M., Fields P. I., Bishop R. D., Swerdlow D. L. (2015). Salmonella enterica infections in the United States and assessment of coefficients of variation: a novel approach to identify epidemiologic characteristics of individual serotypes, 1996–2011. PLoS One 10:e0145416. 10.1371/journal.pone.0145416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boqvist S., Vågsholm I. (2005). Risk factors for hazard of release from Salmonella-control restriction on Swedish cattle farms from 1993 to 2002. Prev. Vet. Med. 71 35–44. 10.1016/j.prevetmed.2005.05.003 [DOI] [PubMed] [Google Scholar]
- Bouwman C. W., Kohli M., Killoran A., Touchie G. A., Kadner R. J., Martin N. L. (2003). Characterization of SrgA, a Salmonella enterica serovar Typhimurium virulence plasmid-encoded paralogue of the disulfide oxidoreductase DsbA, essential for biogenesis of plasmid-encoded fimbriae. J. Bacteriol. 185 991–1000. 10.1128/jb.185.3.991-1000.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd E. F., Hartl D. L. (1998). Salmonella virulence plasmid. Modular acquisition of the spv virulence region by an F-plasmid in Salmonella enterica subspecies I and insertion into the chromosome of subspecies II, IIIa, IV and VII isolates. Genetics 149 1183–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyen F., Pasmans F., Donne E., Van Immerseel F., Morgan E., Adriaensen C., et al. (2006). The fibronectin binding protein ShdA is not a prerequisite for long term faecal shedding of Salmonella typhimurium in pigs. Vet. Microbiol. 115 284–290. 10.1016/j.vetmic.2006.02.006 [DOI] [PubMed] [Google Scholar]
- Brenner F., Villar R., Angulo F., Tauxe R., Swaminathan B. (2000). Salmonella nomenclature. J. Clin. Microbiol. 38 2465–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouard C., Espié E., Weill F.-X., Kérouanton A., Brisabois A., Forgue A.-M., et al. (2007). Two consecutive large outbreaks of Salmonella enterica serotype Agona infections in infants linked to the consumption of powdered infant formula. Pediatr. Infect. Dis. 26 148–152. 10.1097/01.inf.0000253219.06258.23 [DOI] [PubMed] [Google Scholar]
- Cao G., Allard M., Strain E., Stones R., Zhao S., Brown E., et al. (2014). Genetic diversity of Salmonella pathogenicity islands SPI-5 and SPI-6 in Salmonella Newport. Foodborne Pathog. Dis. 11 798–807. 10.1089/fpd.2014.1784 [DOI] [PubMed] [Google Scholar]
- Cao G., Meng J., Strain E., Stones R., Pettengill J., Zhao S., et al. (2013). Phylogenetics and differentiation of Salmonella Newport lineages by whole genome sequencing. PLoS One 8:e55687. 10.1371/journal.pone.0055687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention [CDC] (2001). Outbreaks of multidrug-resistant Salmonella typhimurium associated with veterinary facilities—idaho, minnesota, and Washington,. (1999). MMWR Morb. Mortal Wkly Rep. 50 701–704. [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention [CDC] (2016). National Enteric Disease Surveillance: Salmonella Annual Report, 2016. Atlanta, GA: Centers for Disease Control and Prevention. [Google Scholar]
- Centers for Disease Control and Prevention [CDC] (2018). Reports of Selected Salmonella Outbreak Investigations. Available at: https://www.cdc.gov/salmonella/outbreaks.html (accessed April 26, 2018). [Google Scholar]
- Chary P., Prasad R., Chopra A. K., Peterson J. W. (1993). Location of the enterotoxin gene from Salmonella typhimurium and characterization of the gene products. FEMS Microbiol. Lett. 111 87–92. 10.1016/0378-1097(93)90187-7 [DOI] [PubMed] [Google Scholar]
- Cheng Y., Pedroso A. A., Porwollik S., Mcclelland M., Lee M. D., Kwan T., et al. (2015). rpoS-Regulated core genes involved in the competitive fitness of Salmonella enterica serovar Kentucky in the intestines of chickens. Appl. Environ. Microbiol. 81 502–514. 10.1128/AEM.03219-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C.-H., Su L.-H., Chu C. (2004). Salmonella enterica serotype Choleraesuis: epidemiology, pathogenesis, clinical disease, and treatment. Clin. Microbiol. Rev. 17 311–322. 10.1128/cmr.17.2.311-322.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu S., Chiu C. H., Lin T. Y. (2004). Salmonella enterica serotype Choleraesuis infection in a medical center in northern Taiwan. J. Microbiol. Immunol. Infect. 37 99–102. [PubMed] [Google Scholar]
- Chiu C.-H., Tang P., Chu C., Hu S., Bao Q., Yu J., et al. (2005). The genome sequence of Salmonella enterica serovar Choleraesuis, a highly invasive and resistant zoonotic pathogen. Nucleic Acids Res. 33 1690–1698. 10.1093/nar/gki297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra A. K., Houston C. W., Peterson J. W., Prasad R., Mekalanos J. J. (1987). Cloning and expression of the Salmonella enterotoxin gene. J Bacteriol. 169 5095–5100. 10.1128/jb.169.11.5095-5100.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C., Chiu C. H. (2006). Evolution of the virulence plasmids of non-typhoid Salmonella and its association with antimicrobial resistance. Microbes Infect. 8 1931–1936. 10.1016/j.micinf.2005.12.026 [DOI] [PubMed] [Google Scholar]
- Clouthier S. C., Collinson S. K., Kay W. W. (1994). Unique fimbriae-like structures encoded by sefD of the SEF14 fimbrial gene cluster of Salmonella enteritidis. Mol. Microbiol. 12 893–901. [DOI] [PubMed] [Google Scholar]
- Connor B. A., Schwartz E. (2005). Typhoid and paratyphoid fever in travellers. Lancet Infect. Dis. 5 623–628. 10.1016/s1473-3099(05)70239-5 [DOI] [PubMed] [Google Scholar]
- Cordano A. M., Virgilio R. (1996). Evolution of drug resistance in Salmonella panama isolates in Chile. Antimicrob. Agents Chemother. 40 336–341. 10.1128/aac.40.2.336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump J. A., Medalla F. M., Joyce K. W., Krueger A. L., Hoekstra R. M., Whichard J. M., et al. (2011). Antimicrobial resistance among invasive nontyphoidal Salmonella enterica in the United States, National Antimicrobial Resistance Monitoring System, 1996-2007. Antimicrob. Agents Chemother. 55 1148–1154. 10.1128/AAC.01333-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump J. A., Sjölund-Karlsson M., Gordon M. A., Parry C. M. (2015). Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin. Microbiol. Rev. 28 901–937. 10.1128/CMR.00002-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings K. J., Rodriguez-Rivera L. D., Mitchell K. J., Hoelzer K., Wiedmann M., Mcdonough P. L., et al. (2014). Salmonella enterica serovar Oranienburg outbreak in a veterinary medical teaching hospital with evidence of nosocomial and on-farm transmission. Vector Borne Zoonotic Dis. 14 496–502. 10.1089/vbz.2013.1467 [DOI] [PubMed] [Google Scholar]
- De Masi L., Yue M., Hu C., Rakov A. V., Rankin S. C., Schifferli D. M. (2017). Cooperation of adhesin alleles in Salmonella-host tropism. mSphere 2:e00066-17. 10.1128/mSphere.00066-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Bakker H. C., Switt A. I. M., Govoni G., Cummings C. A., Ranieri M. L., Degoricija L., et al. (2011). Genome sequencing reveals diversification of virulence factor content and possible host adaptation in distinct subpopulations of Salmonella enterica. BMC Genom. 12:425. 10.1186/1471-2164-12-425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai P. T., Porwollik S., Long F., Cheng P., Wollam A., Clifton S. W., et al. (2013). Evolutionary genomics of Salmonella enterica subspecies. MBio 4:e00198-13. 10.1128/mBio.00579-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsey C. W., Laarakker M. C., Humphries A. D., Weening E. H., Baumler A. J. (2005). Salmonella enterica serotype Typhimurium MisL is an intestinal colonization factor that binds fibronectin. Mol. Microbiol. 57 196–211. 10.1111/j.1365-2958.2005.04666.x [DOI] [PubMed] [Google Scholar]
- Dos Santos A. M. P., Ferrari R. G., Conte-Junior C. A. (2018). Virulence factors in Salmonella Typhimurium: the sagacity of a bacterium. Curr Microbiol. 76 762–773. 10.1007/s00284-018-1510-4 [DOI] [PubMed] [Google Scholar]
- Dougan G., Baker S. (2014). Salmonella enterica serovar Typhi and the pathogenesis of typhoid fever. Annu. Rev. Microbiol. 68 317–336. 10.1146/annurev-micro-091313-103739 [DOI] [PubMed] [Google Scholar]
- Ebel E. D., Williams M. S., Cole D., Travis C. C., Klontz K. C., Golden N. J., et al. (2016). Comparing characteristics of sporadic and outbreak-associated foodborne illnesses. United States, 2004-2011. Emerg. Infect. Dis. 22 1193–1200. 10.3201/eid2207.150833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Tom G., Abdel Rahman S., Elamin E., Yassin T. (1999). Isolation of the Salmonella serotype San-Diego from lymph nodes of slaughtered goats. Sudan J. Vet. Res. 16 61–65. [Google Scholar]
- Elder J. R., Chiok K. L., Paul N. C., Haldorson G., Guard J., Shah D. H. (2016). The Salmonella pathogenicity island 13 contributes to pathogenesis in streptomycin pre-treated mice but not in day-old chickens. Gut Pathog. 8:16. 10.1186/s13099-016-0098-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza R. A., Silva-Valenzuela C. A., Amaya F. A., Urrutia I. M., Contreras I., Santiviago C. A. (2017). Differential roles for pathogenicity islands SPI-13 and SPI-8 in the interaction of Salmonella enteritidis and Salmonella Typhi with murine and human macrophages. Biol. Res. 50:5. 10.1186/s40659-017-0109-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Food Safety Authority [EFSA] and European Centre for Disease Prevention, and Control [ECDC] (2017). The European Union summary report on trends, and sources of zoonoses, zoonotic agents, and food. (-)borne outbreaks in 2016. Euro. Surveill. 15:e05077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber F., Thiennimitr P., Spiga L., Byndloss M. X., Litvak Y., Lawhon S., et al. (2017). Respiration of microbiota-derived 1,2-propanediol drives Salmonella expansion during colitis. PLoS Pathog. 13:e1006129. 10.1371/journal.ppat.1006129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fàbrega A., Vila J. (2013). Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin. Microbiol. Rev. 26 308–341. 10.1128/CMR.00066-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucher S. P., Curtiss R., III, Daigle F. (2005). Selective capture of Salmonella enterica serovar Typhi genes expressed in macrophages that are absent from the Salmonella enterica serovar Typhimurium genome. Infect. Immun. 73 5217–5221. 10.1128/iai.73.8.5217-5221.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feasey N. A., Dougan G., Kingsley R. A., Heyderman R. S., Gordon M. A. (2012). Invasive nontyphoidal salmonella disease: an emerging and neglected tropical disease in Africa. Lancet 379 2489–2499. 10.1016/S0140-6736(11)61752-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Liu J., Li Y. G., Cao F. L., Johnston R. N., Zhou J., et al. (2012). Inheritance of the Salmonella virulence plasmids: mostly vertical and rarely horizontal. Infect. Genet. Evol. 12 1058–1063. 10.1016/j.meegid.2012.03.004 [DOI] [PubMed] [Google Scholar]
- Foley S. L., Johnson T. J., Ricke S. C., Nayak R., Danzeisen J. (2013). Salmonella pathogenicity and host adaptation in chicken-associated serovars. Microbiol. Mol. Biol. Rev. 77 582–607. 10.1128/MMBR.00015-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fookes M., Schroeder G. N., Langridge G. C., Blondel C. J., Mammina C., Connor T. R., et al. (2011). Salmonella bongori provides insights into the evolution of the Salmonellae. PLoS Pathog. 7:e1002191. 10.1371/journal.ppat.1002191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford L., Glass K., Veitch M., Wardell R., Polkinghorne B., Dobbins T., et al. (2016). Increasing incidence of Salmonella in Australia, 2000-2013. PloS One 11:e0163989. 10.1371/journal.pone.0163989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya E. Y., Lowy F. D. (2006). Antimicrobial-resistant bacteria in the community setting. Nat. Rev. Microbiol. 4 36–45. 10.1038/nrmicro1325 [DOI] [PubMed] [Google Scholar]
- Gal-Mor O., Boyle E. C., Grassl G. A. (2014). Same species, different diseases: how and why typhoidal and nontyphoidal Salmonella enterica serovars differ. Front. Microbiol. 5:391 10.3389/fmicb.2014.00391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambino-Shirley K., Stevenson L., Concepcion-Acevedo J., Trees E., Wagner D., Whitlock L., et al. (2018). Flea market finds and global exports: four multistate outbreaks of human Salmonella infections linked to small turtles, United States-2015. Zoonoses Public Health 65 560–568. 10.1111/zph.12466 [DOI] [PubMed] [Google Scholar]
- Garcia-Pastor L., Puerta-Fernandez E., Casadesus J. (2018). Bistability and phase variation in Salmonella enterica. Biochim. Biophys. Acta Gene Regul. Mech. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Gemmell C. G. (1984). Comparative study of the nature and biological activities of bacterial enterotoxins. J. Med. Microbiol. 17 217–235. 10.1099/00222615-17-3-217 [DOI] [PubMed] [Google Scholar]
- Gilchrist J. J., Maclennan C. A., Hill A. V. (2015). Genetic susceptibility to invasive Salmonella disease. Nat. Rev. Immunol. 15 452–463. 10.1038/nri3858 [DOI] [PubMed] [Google Scholar]
- Gossner C. M., Le Hello S., De Jong B., Rolfhamre P., Faensen D., Weill F.-X., et al. (2016). Around the world in 1,475 Salmonella geo-serotypes. Emerg. Infect. Dis. 22 1298–1302. 10.3201/eid2207.141678 [DOI] [Google Scholar]
- Guiney D. G., Fierer J. (2011). The role of the spv genes in Salmonella pathogenesis. Front. Microbiol. 2:129. 10.3389/fmicb.2011.00129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habing G. G., Kessler S. E., Mollenkopf D. F., Wittum T. E., Anderson T. C., Barton Behravesh C., et al. (2015). Distribution and diversity of Salmonella strains in shipments of hatchling poultry, United States, 2013. Zoonoses Public Health 62 375–380. 10.1111/zph.12157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B. J., Kim S. W., Pettengill J., Luo Y., Karns J. S., Van Kessel J. A. S. (2016). Genomic and evolutionary analysis of two Salmonella enterica serovar Kentucky sequence types isolated from bovine and poultry sources in North America. PLoS One 11:e0161225. 10.1371/journal.pone.0161225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haneda T., Ishii Y., Shimizu H., Ohshima K., Iida N., Danbara H., et al. (2012). Salmonella type III effector SpvC, a phosphothreonine lyase, contributes to reduction in inflammatory response during intestinal phase of infection. Cell. Microbiol. 14 485–499. 10.1111/j.1462-5822.2011.01733.x [DOI] [PubMed] [Google Scholar]
- Hanning I. B., Nutt J., Ricke S. C. (2009). Salmonellosis outbreaks in the United States due to fresh produce: sources and potential intervention measures. Foodborne Pathog. Dis. 6 635–648. 10.1089/fpd.2008.0232 [DOI] [PubMed] [Google Scholar]
- Hariharan H., Booth B. A., Brickman T. J., Katt W. C., Boesman-Finkelstein M., Finkelstein R. A. (1986). Competitive enzyme-linked immunosorbent assay for cholera-related enterotoxins in Salmonella typhimurium. J. Clin. Microbiol. 24 298–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey R. R., Friedman C. R., Crim S. M., Judd M., Barrett K. A., Tolar B., et al. (2017). Epidemiology of Salmonella enterica serotype Dublin infections among humans, United States, 1968–2013. Emerg. Infect. Dis. 23 1493–1501. 10.3201/eid2309.170136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havelaar A. H., Kirk M. D., Torgerson P. R., Gibb H. J., Hald T., Lake R. J., et al. (2015). World health organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med. 12:e1001923. 10.1371/journal.pmed.1001923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward M. R., Abuoun M., La Ragione R. M., Tchórzewska M. A., Cooley W. A., Everest D. J., et al. (2014). SPI-23 of S. Derby: role in adherence and invasion of porcine tissues. PLoS One 9:e107857. 10.1371/journal.pone.0107857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward M. R., Jansen V. A., Woodward M. J. (2013). Comparative genomics of Salmonella enterica serovars Derby and Mbandaka, two prevalent serovars associated with different livestock species in the UK. BMC Genom. 14:365. 10.1186/1471-2164-14-365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriksen R. S., Bangtrakulnonth A., Pulsrikarn C., Pornruangwong S., Noppornphan G., Emborg H.-D., et al. (2009). Risk factors and epidemiology of the ten most common Salmonella serovars from patients in Thailand: 2002–2007. Foodborne Pathog. Dis. 6 1009–1019. 10.1089/fpd.2008.0245 [DOI] [PubMed] [Google Scholar]
- Hendriksen R. S., Vieira A. R., Karlsmose S., Lo Fo Wong D. M., Jensen A. B., et al. (2011). Global monitoring of Salmonella serovar distribution from the World Health Organization Global Foodborne Infections Network Country Data Bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathog. Dis. 8 887–900. 10.1089/fpd.2010.0787 [DOI] [PubMed] [Google Scholar]
- Hiyoshi H., Tiffany C. R., Bronner D. N., Baumler A. J. (2018). Typhoidal Salmonella serovars: ecological opportunity and the evolution of a new pathovar. FEMS Microbiol. Rev. 42 527–541. 10.1093/femsre/fuy024 [DOI] [PubMed] [Google Scholar]
- Hoelzer K., Moreno Switt A. I., Wiedmann M. (2011). Animal contact as a source of human nontyphoidal salmonellosis. Vet. Res. 42:34. 10.1186/1297-9716-42-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston C. W., Koo F. C., Peterson J. W. (1981). Characterization of Salmonella toxin released by mitomycin C-treated cells. Infect. Immun. 32 916–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey T. (1994). Contamination of egg shell and contents with Salmonella enteritidis: a review. Int. J. Food Microbiol. 21 31–40. 10.1016/0168-1605(94)90197-x [DOI] [PubMed] [Google Scholar]
- Issenhuth-Jeanjean S., Roggentin P., Mikoleit M., Guibourdenche M., De Pinna E., Nair S., et al. (2014). Supplement 2008–2010 (no. 48) to the White–Kauffmann–Le Minor scheme. Res. Microbiol. 165 526–530. 10.1016/j.resmic.2014.07.004 [DOI] [PubMed] [Google Scholar]
- Jackson B. R., Griffin P. M., Cole D., Walsh K. A., Chai S. J. (2013). Outbreak-associated Salmonella enterica serotypes and food commodities, United States, 1998-2008. Emerg. Infect. Dis. 19 1239–1244. 10.3201/eid1908.121511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger R. D., Sartori C. A., Kniel K. E. (2009). Comparison of genetic and physiological properties of Salmonella enterica isolates from chickens reveals one major difference between serovar Kentucky and other serovars: response to acid. Foodborne Pathog. Dis. 6 503–512. 10.1089/fpd.2008.0144 [DOI] [PubMed] [Google Scholar]
- Jones T. F., Ingram L. A., Cieslak P. R., Vugia D. J., Tobin-D’angelo M., Hurd S., et al. (2008). Salmonellosis outcomes differ substantially by serotype. J. Infect. Dis. 198 109–114. 10.1086/588823 [DOI] [PubMed] [Google Scholar]
- Kariuki S., Revathi G., Kariuki N., Kiiru J., Mwituria J., Muyodi J., et al. (2006). Invasive multidrug-resistant nontyphoidal Salmonella infections in Africa: zoonotic or anthroponotic transmission? J. Med. Microbiol. 55 585–591. 10.1099/jmm.0.46375-0 [DOI] [PubMed] [Google Scholar]
- Kaur J., Jain S. K. (2012). Role of antigens and virulence factors of Salmonella enterica serovar Typhi in its pathogenesis. Microbiol. Res. 167 199–210. 10.1016/j.micres.2011.08.001 [DOI] [PubMed] [Google Scholar]
- Keestra-Gounder A. M., Tsolis R. M., Bäumler A. J. (2015). Now you see me, now you don’t: the interaction of Salmonella with innate immune receptors. Nat. Rev. Microbiol. 13 206–216. 10.1038/nrmicro3428 [DOI] [PubMed] [Google Scholar]
- Khurana S., Ganguly N. K., Khullar M., Panigrahi D., Walia B. N. (1991). Studies on the mechanism of Salmonella typhimurium enterotoxin-induced diarrhoea. Biochim. Biophys. Acta 1097 171–176. 10.1016/0925-4439(91)90031-4 [DOI] [PubMed] [Google Scholar]
- Kingsley R. A., Humphries A. D., Weening E. H., De Zoete M. R., Winter S., Papaconstantinopoulou A., et al. (2003). Molecular and phenotypic analysis of the CS54 island of Salmonella enterica serotype Typhimurium: identification of intestinal colonization and persistence determinants. Infect. Immun. 71 629–640. 10.1128/iai.71.2.629-640.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk M. D., Pires S. M., Black R. E., Caipo M., Crump J. A., Devleesschauwer B., et al. (2015). World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med. 12:e1001921. 10.1371/journal.pmed.1001921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita E., Kamikaidou N., Nakano A., Kashiba S. (1993). Isolation of a cytotoxin from L-form Salmonella typhimurium. FEMS Microbiol. Lett. 109 179–184. 10.1016/0378-1097(93)90016-u [DOI] [PubMed] [Google Scholar]
- Kovac J., Cummings K. J., Rodriguez-Rivera L. D., Carroll L. M., Thachil A., Wiedmann M. (2017). Temporal genomic phylogeny reconstruction indicates a geospatial transmission path of Salmonella Cerro in the United States and a clade-specific loss of hydrogen sulfide production. Front. Microbiol. 8:737. 10.3389/fmicb.2017.00737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan N. P. H., Phuong T. L. T., Huu H. N., Thuy L., Mather A. E., Park S. E., et al. (2016). Invasive nontyphoidal Salmonella infections in Asia: clinical observations, disease outcome and dominant serovars from an infectious disease hospital in Vietnam. PLoS Negl. Trop. Dis. 10:e0004857. 10.1371/journal.pntd.0004857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge G. C., Fookes M., Connor T. R., Feltwell T., Feasey N., Parsons B. N., et al. (2015). Patterns of genome evolution that have accompanied host adaptation in Salmonella. Proc. Natl. Acad. Sci. U.S.A. 112 863–868. 10.1073/pnas.1416707112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laniewski P., Baek C. H., Roland K. L., Curtiss R., III (2017). Analysis of spleen-induced fimbria production in recombinant attenuated Salmonella enterica serovar Typhimurium vaccine strains. MBio 8:e01189-17. 10.1128/mBio.01189-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRock D. L., Chaudhary A., Miller S. I. (2015). Salmonellae interactions with host processes. Nat. Rev. Microbiol. 13 191–205. 10.1038/nrmicro3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hello S., Bekhit A., Granier S. A., Barua H., Beutlich J., Zajac M., et al. (2013). The global establishment of a highly-fluoroquinolone resistant Salmonella enterica serotype Kentucky ST198 strain. Front. Microbiol. 4:395. 10.3389/fmicb.2013.00395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hello S., Hendriksen R. S., Doublet B., Fisher I., Nielsen E. M., Whichard J. M., et al. (2011). International spread of an epidemic population of Salmonella enterica serotype Kentucky ST198 resistant to ciprofloxacin. J. Infect. Dis. 204 675–684. 10.1093/infdis/jir409 [DOI] [PubMed] [Google Scholar]
- Lee I. S., Lin J., Hall H. K., Bearson B., Foster J. W. (1995). The stationary-phase sigma factor σS (RpoS) is required for a sustained acid tolerance response in virulent Salmonella typhimurium. Mol. Micro. 17 155–167. 10.1111/j.1365-2958.1995.mmi_17010155.x [DOI] [PubMed] [Google Scholar]
- Lee Y. H., Kim S., Helmann J. D., Kim B. H., Park Y. K. (2013). RaoN, a small RNA encoded within Salmonella pathogenicity island-11, confers resistance to macrophage-induced stress. Microbiology 159 1366–1378. 10.1099/mic.0.066688-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesnick M. L., Guiney D. G. (2001). The best defense is a good offense - Salmonella deploys an ADP-ribosylating toxin. Trends Microbiol. 9 2–4. 10.1016/s0966-842x(00)01902-8 [DOI] [PubMed] [Google Scholar]
- Libby S. J., Adams L. G., Ficht T. A., Allen C., Whitford H. A., Buchmeier N. A., et al. (1997). The spv genes on the Salmonella dublin virulence plasmid are required for severe enteritis and systemic infection in the natural host. Infect. Immun. 65 1786–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby S. J., Lesnick M., Hasegawa P., Weidenhammer E., Guiney D. G. (2000). The Salmonella virulence plasmid spv genes are required for cytopathology in human monocyte-derived macrophages. Cell. Microbiol. 2 49–58. 10.1046/j.1462-5822.2000.00030.x [DOI] [PubMed] [Google Scholar]
- Liu B., Knirel Y. A., Feng L., Perepelov A. V., Senchenkova S. Y. N., Reeves P. R., et al. (2014). Structural diversity in Salmonella O antigens and its genetic basis. FEMS Microbiol. Rev. 38 56–89. 10.1111/1574-6976.12034 [DOI] [PubMed] [Google Scholar]
- Liu H., Whitehouse C. A., Li B. (2018). Presence and persistence of Salmonella in water: the impact on microbial quality of water and food safety. Front. Public Health 6:159–159. 10.3389/fpubh.2018.00159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. Q., Feng Y., Wang Y., Zou Q. H., Chen F., Guo J. T., et al. (2009). Salmonella paratyphi C: genetic divergence from Salmonella choleraesuis and pathogenic convergence with Salmonella typhi. PLoS One 4:e4510. 10.1371/journal.pone.0004510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loharikar A., Briere E., Schwensohn C., Weninger S., Wagendorf J., Scheftel J., et al. (2012). Four multistate outbreaks of human Salmonella infections associated with live poultry contact, United States, 2009. Zoonoses Public Health 59 347–354. 10.1111/j.1863-2378.2012.01461.x [DOI] [PubMed] [Google Scholar]
- Malik P., Sharma V. D., Thapliyal D. C. (1996). Partial purification and characterization of Salmonella cytotoxin. Vet. Microbiol. 49 11–19. 10.1016/0378-1135(95)00151-4 [DOI] [PubMed] [Google Scholar]
- McQuiston J. R., Parrenas R., Ortiz-Rivera M., Gheesling L., Brenner F., Fields P. I. (2004). Sequencing and comparative analysis of flagellin genes fliC, fljB, and flpA from Salmonella. J. Clin. Microbiol. 42 1923–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. A., Betteken M. I., Guo X., Altier C., Duhamel G. E., Wiedmann M. (2018). The typhoid toxin produced by the nontyphoidal Salmonella enterica serotype Javiana is required for induction of a DNA damage response in vitro and systemic spread in vivo. mBio 9 e00467-18. 10.1128/mBio.00467-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. A., Wiedmann M. (2016). The cytolethal distending toxin produced by nontyphoidal Salmonella serotypes Javiana, Montevideo, Oranienburg, and Mississippi induces DNA Damage in a manner similar to that of serotype Typhi. mBio 7 e02109–e02116. 10.1128/mBio.02109-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed M., Hello S., Leekitcharoenphon P., Hendriksen R. (2017). The invasome of Salmonella Dublin as revealed by whole genome sequencing. BMC Infect. Dis. 17:544. 10.1186/s12879-017-2628-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan E. (2007). “Salmonella pathogenicity islands,” in Salmonella Molecular Biology and Pathogenesis, eds Rhen M., Maskell D., Mastroeni P., Threlfall J. (Norfolk: Horizon Bioscience; ), 67–87. 10.1007/978-3-642-56031-6_5 [DOI] [Google Scholar]
- Morgan E., Bowen A. J., Carnell S. C., Wallis T. S., Stevens M. P. (2007). SiiE is secreted by the Salmonella enterica serovar Typhimurium pathogenicity island 4-encoded secretion system and contributes to intestinal colonization in cattle. Infect. Immun. 75 1524–1533. 10.1128/iai.01438-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morningstar-Shaw B. R., Mackie T. A., Barker D. K., Palmer E. A. (2016). Salmonella Serotypes Isolated from Animals and Related Sources. Ames, IA: Diagnostic Bacteriology Laboratory. [Google Scholar]
- Morris C., Tam C. K., Wallis T. S., Jones P. W., Hackett J. (2003). Salmonella enterica serovar Dublin strains which are Vi antigen-positive use type IVB pili for bacterial self-association and human intestinal cell entry. Microb. Pathog. 35 279–284. 10.1016/j.micpath.2003.08.001 [DOI] [PubMed] [Google Scholar]
- Mungai E. A., Behravesh C. B., Gould L. H. (2015). Increased outbreaks associated with nonpasteurized milk, United States, 2007–2012. Emerg. Infect. Dis. 21 119–122. 10.3201/eid2101.140447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M., Yamasaki E., Ichinose A., Shimohata T., Takahashi A., Akada J. K., et al. (2012). Salmonella enterotoxin (Stn) regulates membrane composition and integrity. Dis. Model Mech. 5 515–521. 10.1242/dmm.009324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao J. H., Pringle J., Jones R. W., Nix B. E., Borders J., Heseltine G., et al. (2015). ‘One Health’ investigation: outbreak of human Salmonella Braenderup infections traced to a mail-order hatchery - United States, 2012-2013. Epidemiol. Infect. 143 2178–2186. 10.1017/S0950268815000151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoro C. K., Kingsley R. A., Connor T. R., Harris S. R., Parry C. M., Al-Mashhadani M. N., et al. (2012). Intracontinental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat. Gen. 44 1215–1221. 10.1038/ng.2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira C. J. B., Carvalho L. F. O. S., Fernandes S. A., Tavechio A. T., Menezes C. C. P., Domingues F. J., Jr. (2002). Antimicrobial resistance of Salmonella serotypes isolated from slaughter-age pigs and environmental samples. Microb. Drug Resist. 8 407–411. 10.1089/10766290260469697 [DOI] [PubMed] [Google Scholar]
- Otto H., Tezcan-Merdol D., Girisch R., Haag F., Rhen M., Koch-Nolte F. (2000). The spvB gene-product of the Salmonella enterica virulence plasmid is a mono(ADP-ribosyl)transferase. Mol. Microbiol. 37 1106–1115. 10.1046/j.1365-2958.2000.02064.x [DOI] [PubMed] [Google Scholar]
- OzFoodNet Working Group (2015). Monitoring the incidence and causes of diseases potentially transmitted by food in Australia: annual report of the OzFoodNet network, 2011. Commun. Dis. Intell. Q. Rep. 39E236–E264. [PubMed] [Google Scholar]
- Pabilonia K. L., Cadmus K. J., Lingus T. M., Bolte D. S., Russell M. M., Van Metre D. C., et al. (2014). Environmental Salmonella in agricultural fair poultry exhibits in Colorado. Zoonoses Public Health 61 138–144. 10.1111/zph.12056 [DOI] [PubMed] [Google Scholar]
- Parkhill J., Dougan G., James K., Thomson N., Pickard D., Wain J., et al. (2001). Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413 848–852. [DOI] [PubMed] [Google Scholar]
- Pedersen K., Sørensen G., Löfström C., Leekitcharoenphon P., Nielsen B., Wingstrand A., et al. (2015). Reappearance of Salmonella serovar Choleraesuis var. Kunzendorf in Danish pig herds. Vet. Microbiol. 176 282–291. 10.1016/j.vetmic.2015.01.004 [DOI] [PubMed] [Google Scholar]
- Peterson J. W. (1980). Salmonella toxin. Pharmacol. Ther. 11 719–724. 10.1016/0163-7258(80)90046-7 [DOI] [PubMed] [Google Scholar]
- Pezoa D., Blondel C. J., Silva C. A., Yang H. J., Andrews-Polymenis H., Santiviago C. A., et al. (2014). Only one of the two type VI secretion systems encoded in the Salmonella enterica serotype Dublin genome is involved in colonization of the avian and murine hosts. Vet. Res. 45:2. 10.1186/1297-9716-45-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard D., Wain J., Baker S., Line A., Chohan S., Fookes M., et al. (2003). Composition, acquisition, and distribution of the Vi exopolysaccharide-encoding Salmonella enterica pathogenicity island SPI-7. J. Bacteriol. 185 5055–5065. 10.1128/jb.185.17.5055-5065.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabsch W., Tschäpe H., Bäumler A. J. (2001). Nontyphoidal salmonellosis: emerging problems. Microbes Infect. 3 237–247. 10.1016/s1286-4579(01)01375-2 [DOI] [PubMed] [Google Scholar]
- Raffatellu M., Wilson R. P., Chessa D., Andrews-Polymenis H., Tran Q. T., Lawhon S., et al. (2005). SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype Typhimurium invasion of epithelial cells. Infect. Immun. 73 146–154. 10.1128/iai.73.1.146-154.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran L., Wu S., Gao Y., Zhang X., Feng Z., Wang Z., et al. (2011). Laboratory-based surveillance of nontyphoidal Salmonella infections in China. Foodborne Pathog. Dis. 8 921–927. 10.1089/fpd.2010.0827 [DOI] [PubMed] [Google Scholar]
- Reitmeyer J. C., Peterson J. W., Wilson K. J. (1986). Salmonella cytotoxin: a component of the bacterial outer membrane. Microb. Pathog. 1 503–510. 10.1016/0882-4010(86)90012-4 [DOI] [PubMed] [Google Scholar]
- Rivera-Chavez F., Baumler A. J. (2015). The pyromaniac inside you: Salmonella metabolism in the host gut. Annu. Rev. Microbiol. 69 31–48. 10.1146/annurev-micro-091014-104108 [DOI] [PubMed] [Google Scholar]
- Robertson J., Yoshida C., Kruczkiewicz P., Nadon C., Nichani A., Taboada E. N., et al. (2018). Comprehensive assessment of the quality of Salmonella whole genome sequence data available in public sequence databases using the Salmonella in silico Typing Resource (SISTR). Microb. Genom. 4:e000151. 10.1099/mgen.0.000151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson T. J., Scheftel J. M., Smith K. E. (2014). Raw milk consumption among patients with non–outbreak-related enteric infections, Minnesota, USA, 2001–2010. Emerg. Infect. Dis. 20 38–44. 10.3201/eid2001.120920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Rivera L. D., Bowen B. M., Den Bakker H. C., Duhamel G. E., Wiedmann M. (2015). Characterization of the cytolethal distending toxin (typhoid toxin) in nontyphoidal Salmonella serovars. Gut Pathog. 7:19. 10.1186/s13099-015-0065-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Rivera L. D., Switt A. I. M., Degoricija L., Fang R., Cummings C. A., Furtado M. R., et al. (2014). Genomic characterization of Salmonella Cerro ST367, an emerging Salmonella subtype in cattle in the United States. BMC Genom. 15:427. 10.1186/1471-2164-15-427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotger R., Casadesús J. (1999). The virulence plasmids of Salmonella. Int. Microbiol. 2 177–184. [PubMed] [Google Scholar]
- Russo E. T., Biggerstaff G., Hoekstra R. M., Meyer S., Patel N., Miller B., et al. (2013). A recurrent, multistate outbreak of Salmonella serotype Agona infections associated with dry, unsweetened cereal consumption, United States, 2008. J. Food Protect. 76 227–230. 10.4315/0362-028X.JFP-12-209 [DOI] [PubMed] [Google Scholar]
- Rychlik I., Gregorova D., Hradecka H. (2006). Distribution and function of plasmids in Salmonella enterica. Vet. Microbiol. 112 1–10. 10.1016/j.vetmic.2005.10.030 [DOI] [PubMed] [Google Scholar]
- Sabbagh S. C., Forest C. G., Lepage C., Leclerc J.-M., Daigle F. (2010). So similar, yet so different: uncovering distinctive features in the genomes of Salmonella enterica serovars Typhimurium and Typhi. FEMS Microbiol. Lett. 305 1–13. 10.1111/j.1574-6968.2010.01904.x [DOI] [PubMed] [Google Scholar]
- Saitoh M., Tanaka K., Nishimori K., Makino S.-I., Kanno T., Ishihara R., et al. (2005). The artAB genes encode a putative ADP-ribosyltransferase toxin homologue associated with Salmonella enterica serovar Typhimurium DT104. Microbiology 151 3089–3096. 10.1099/mic.0.27933-0 [DOI] [PubMed] [Google Scholar]
- Saroj S. D., Shashidhar R., Karani M., Bandekar J. R. (2008). Distribution of Salmonella pathogenicity island (SPI)-8 and SPI-10 among different serotypes of Salmonella. J. Med. Microbiol. 57 424–427. 10.1099/jmm.0.47630-0 [DOI] [PubMed] [Google Scholar]
- Scallan E., Hoekstra R. M., Angulo F. J., Tauxe R. V., Widdowson M.-A., Roy S. L., et al. (2011). Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17 7–15. 10.3201/eid1701.P11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroll C., Huang K., Ahmed S., Kristensen B. M., Pors S. E., Jelsbak L., et al. (2019). The SPI-19 encoded type-six secretion-systems (T6SS) of Salmonella enterica serovars Gallinarum and Dublin play different roles during infection. Vet. Microbiol. 230 23–31. 10.1016/j.vetmic.2019.01.006 [DOI] [PubMed] [Google Scholar]
- Schultz M. (2008). Theobald Smith. Emerg. Infect. Dis. 14 1940–1942. 10.3201/eid1412.081188 [DOI] [Google Scholar]
- Scott J. A. G., Berkley J. A., Mwangi I., Ochola L., Uyoga S., Macharia A., et al. (2011). Relation between falciparum malaria and bacteraemia in Kenyan children: a population-based, case-control study and a longitudinal study. Lancet 378 1316–1323. 10.1016/S0140-6736(11)60888-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sévellec Y., Vignaud M.-L., Granier S. A., Lailler R., Feurer C., Le Hello S., et al. (2018). Polyphyletic nature of Salmonella enterica serotype Derby and lineage-specific host-association revealed by genome-wide analysis. Front. Microbiol. 9:891. 10.3389/fmicb.2018.00891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah D. H., Lee M. J., Park J. H., Lee J. H., Eo S. K., Kwon J. T., et al. (2005). Identification of Salmonella gallinarum virulence genes in a chicken infection model using PCR-based signature-tagged mutagenesis. Microbiology 151 3957–3968. 10.1099/mic.0.28126-0 [DOI] [PubMed] [Google Scholar]
- Shah D. H., Zhou X., Kim H. Y., Call D. R., Guard J. (2012). Transposon mutagenesis of Salmonella enterica serovar Enteritidis identifies genes that contribute to invasiveness in human and chicken cells and survival in egg albumen. Infect. Immun. 80 4203–4215. 10.1128/IAI.00790-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Erdman M. M., Munoz-Vargas L., Mollenkopf D. F., Habing G. G. (2018). Changes in the prevalence, genotypes and antimicrobial resistance phenotypes of nontyphoidal Salmonella recovered from mail-order hatchling poultry sold at US feed stores, 2013-2015. Zoonoses Public Health. 65 e102–e112. 10.1111/zph.12416 [DOI] [PubMed] [Google Scholar]
- Shelobolina E. S., Sullivan S. A., O’neill K. R., Nevin K. P., Lovley D. R. (2004). Isolation, characterization, and U(VI)-reducing potential of a facultatively anaerobic, acid-resistant Bacterium from Low-pH, nitrate- and U(VI)-contaminated subsurface sediment and description of Salmonella subterranea sp. nov. Appl. Environ. Microbiol. 70 2959–2965. 10.1128/aem.70.5.2959-2965.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Namvar A., Kostrzynska M., Hora R., Warriner K. (2007). Persistence and growth of different Salmonella serovars on pre- and postharvest tomatoes. J. Food Prot. 70 2725–2731. 10.4315/0362-028x-70.12.2725 [DOI] [PubMed] [Google Scholar]
- Silva C., Puente J. L., Calva E. (2017). Salmonella virulence plasmid: pathogenesis and ecology. Pathog. Dis. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Silverman M., Zieg J., Simon M. (1979). Flagellar-phase variation: isolation of the rh1 gene. J. Bacteriol. 137 517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Gao X., Galán J. E. (2013). Structure and function of the Salmonella Typhi chimaeric A2B5 typhoid toxin. Nature 499 350–354. 10.1038/nature12377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnier J. L., Karns J. S., Lombard J. E., Kopral C. A., Haley B. J., Kim S. W., et al. (2018). Prevalence of Salmonella enterica, Listeria monocytogenes, and pathogenic Escherichia coli in bulk tank milk and milk filters from US dairy operations in the National Animal Health Monitoring System Dairy 2014 study. J. Dairy Sci. 101 1943–1956. 10.3168/jds.2017-13546 [DOI] [PubMed] [Google Scholar]
- Spöring I., Felgner S., Preuße M., Eckweiler D., Rohde M., Häussler S., et al. (2018). Regulation of flagellum biosynthesis in response to cell envelope stress in Salmonella enterica serovar Typhimurium. mBio 9:e00736-17. 10.1128/mBio.00736-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikumar S., Kröger C., Hébrard M., Colgan A., Owen S. V., Sivasankaran S. K., et al. (2015). RNA-seq brings new insights to the intra-macrophage transcriptome of Salmonella Typhimurium. PLoS Pathog. 11:e1005262. 10.1371/journal.ppat.1005262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn L. K., Danyluk M. D., Worobo R. W., Wiedmann M. (2014). Distributions of Salmonella subtypes differ between two U.S. produce-growing regions. Appl. Environ. Microbiol. 80 3982–3991. 10.1128/AEM.00348-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suez J., Porwollik S., Dagan A., Marzel A., Schorr Y. I., Desai P. T., et al. (2013). Virulence gene profiling and pathogenicity characterization of nontyphoidal Salmonella accounted for invasive disease in humans. PLoS One 8:e58449. 10.1371/journal.pone.0058449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swords W. E., Cannon B. M., Benjamin W. H., Jr. (1997). Avirulence of LT2 strains of Salmonella typhimurium results from a defective rpoS gene. Infect. Immun. 65 2451–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamura Y., Tanaka K., Uchida I. (2017). Characterization of pertussis-like toxin from Salmonella spp. that catalyzes ADP-ribosylation of G proteins. Sci. Rep. 7:2653. 10.1038/s41598-017-02517-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasmin R., Hasan N. A., Grim C. J., Grant A., Choi S. Y., Alam M. S., et al. (2017). Genotypic and phenotypic characterization of multidrug resistant Salmonella Typhimurium and Salmonella Kentucky strains recovered from chicken carcasses. PLoS One 12:e0176938. 10.1371/journal.pone.0176938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari D., Sandt C. H., Miller D. M., Jayarao B. M., M’ikanatha N. M. (2012). Prevalence of Salmonella Cerro in laboratory-based submissions of cattle and comparison with human infections in Pennsylvania, 2005–2010. Foodborne Pathog. Dis. 9 928–933. 10.1089/fpd.2012.1142 [DOI] [PubMed] [Google Scholar]
- Threlfall E. J. (2000). Epidemic Salmonella typhimurium DT 104–a truly international multiresistant clone. J. Antimicrob. Chemother. 46 7–10. 10.1093/jac/46.1.7 [DOI] [PubMed] [Google Scholar]
- Timme R. E., Pettengill J. B., Allard M. W., Strain E., Barrangou R., Wehnes C., et al. (2013). Phylogenetic diversity of the enteric pathogen Salmonella enterica subsp. enterica inferred from genome-wide reference-free SNP characters. Genome Biol. Evol. 5 2109–2123. 10.1093/gbe/evt159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomljenovic-Berube A. M., Henriksbo B., Porwollik S., Cooper C. A., Tuinema B. R., Mcclelland M., et al. (2013). Mapping and regulation of genes within Salmonella pathogenicity island 12 that contribute to in vivo fitness of Salmonella enterica Serovar Typhimurium. Infect. Immun. 81 2394–2404. 10.1128/IAI.00067-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsolis R. M., Xavier M. N., Santos R. L., Baumler A. J. (2011). How to become a top model: impact of animal experimentation on human Salmonella disease research. Infect. Immun. 79 1806–1814. 10.1128/IAI.01369-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida I., Ishihara R., Tanaka K., Hata E., Makino S., Kanno T., et al. (2009). Salmonella enterica serotype Typhimurium DT104 ArtA-dependent modification of pertussis toxin-sensitive G proteins in the presence of [32P]NAD. Microbiology 155 3710–3718. 10.1099/mic.0.028399-0 [DOI] [PubMed] [Google Scholar]
- United States Department of Agriculture [USDA] (2014). Serotypes Profile of Salmonella Isolates from Meat and Poultry Products January 1998 through December 2014. Available at: https://www.fsis.usda.gov/wps/wcm/connect/3866026a-582d-4f0e-a8ce-851b39c7390f/Salmonella-Serotype-Annual-2014.pdf?MOD=AJPERES (accessed April 26, 2018). [Google Scholar]
- United States Department of Agriculture [USDA] (2017). “FSIS Microbiological Testing Program for Pasteurized Egg Products, Calendar Years 1995–2017”. Food Safety and Inspection Service. Available at: https://www.fsis.usda.gov/wps/portal/fsis/topics/data-collection-and-reports/microbiology/testing-program-for-rte-meat-and-poultry-products/microbiological-testing-program-for-pasteurized-egg-products/pasteurized-egg-products (accessed April 28, 2018). [Google Scholar]
- Uzzau S., Gulig P. A., Paglietti B., Leori G., Stocker B. A., Rubino S. (2000). Role of the Salmonella abortusovis virulence plasmid in the infection of BALB/c mice. FEMS Microbiol. Lett. 188 15–18. 10.1016/s0378-1097(00)00204-4 [DOI] [PubMed] [Google Scholar]
- Valenzuela J., Sethi A., Aulik N., Poulsen K. (2017). Antimicrobial resistance patterns of bovine Salmonella enterica isolates submitted to the Wisconsin veterinary diagnostic laboratory: 2006–2015. J. Dairy Sci. 100 1319–1330. 10.3168/jds.2016-11419 [DOI] [PubMed] [Google Scholar]
- Van Asten A. J., Van Dijk J. E. (2005). Distribution of “classic” virulence factors among Salmonella spp. FEMS Immunol. Med. Microbiol. 44 251–259. [DOI] [PubMed] [Google Scholar]
- Velasquez J. C., Hidalgo A. A., Villagra N., Santiviago C. A., Mora G. C., Fuentes J. A. (2016). SPI-9 of Salmonella enterica serovar Typhi is constituted by an operon positively regulated by RpoS and contributes to adherence to epithelial cells in culture. Microbiology 162 1367–1378. 10.1099/mic.0.000319 [DOI] [PubMed] [Google Scholar]
- Vernikos G. S., Parkhill J. (2006). Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics 22 2196–2203. 10.1093/bioinformatics/btl369 [DOI] [PubMed] [Google Scholar]
- Vignaud M.-L., Cherchame E., Marault M., Chaing E., Le Hello S., Michel V., et al. (2017). MLVA for Salmonella enterica subsp. enterica serovar Dublin: development of a method suitable for inter-laboratory surveillance and application in the context of a raw milk cheese outbreak in France in 2012. Front. Microbiol. 8:295. 10.3389/fmicb.2017.00295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters M. S., Simmons L., Anderson T. C., Dement J., Van Zile K., Matthias L. P., et al. (2015). Outbreaks of salmonellosis from small turtles. Pediatrics 137 1–9. 10.1542/peds.2015-1735 [DOI] [PubMed] [Google Scholar]
- Wang J. Y., Hwang J. J., Hsu C. N., Lin L. C., Hsueh P. R. (2006). Bacteraemia due to ciprofloxacin-resistant Salmonella enterica serotype Choleraesuis in adult patients at a university hospital in Taiwan, 1996-2004. Epidemiol. Infect. 134 977–984. 10.1017/s095026880600608x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes-Riesenberg M. R., Foster J. W., Curtiss R., III (1997). An altered rpoS allele contributes to the avirulence of Salmonella typhimurium LT2. Infect. Immun. 65 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley J., Meng J., Allard M. W., Brown E. W., Timme R. E. (2018). Salmonella enterica phylogeny based on whole-genome sequencing reveals two new clades and novel patterns of horizontally acquired genetic elements. mBio 9 e2303–e2318. 10.1128/mBio.02303-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J. G., Tengelsen L. A., Smith K. E., Bender J. B., Frank R. K., Grendon J. H., et al. (2005). Multidrug-resistant Salmonella Typhimurium in four animal facilities. Emerg. Infect. Dis. 11 1235–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. A., Lee S., Zhao J., Thompson A. J., Mcbride R., Tsogtbaatar B., et al. (2018). In vivo tropism of Salmonella Typhi toxin to cells expressing a multiantennal glycan receptor. Nat. Microbiol. 3 155–163. 10.1038/s41564-017-0076-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida C. E., Kruczkiewicz P., Laing C. R., Lingohr E. J., Gannon V. P., Nash J. H., et al. (2016). The Salmonella in silico typing resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One 11:e0147101. 10.1371/journal.pone.0147101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue M., Han X., De Masi L., Zhu C., Ma X., Zhang J., et al. (2015). Allelic variation contributes to bacterial host specificity. Nat. Commun. 6:8754. 10.1038/ncomms9754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue M., Rankin S. C., Blanchet R. T., Nulton J. D., Edwards R. A., Schifferli D. M. (2012). Diversification of the Salmonella fimbriae: a model of macro- and microevolution. PLoS One 7:e38596. 10.1371/journal.pone.0038596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Yin Y., Jones M. B., Zhang Z., Deatherage Kaiser B. L., Dinsmore B. A., et al. (2015). Salmonella serotype determination utilizing high-throughput genome sequencing data. J. Clin. Microbiol. 53 1685–1692. 10.1128/JCM.00323-15 [DOI] [PMC free article] [PubMed] [Google Scholar]