Abstract
Dopamine modulation in the prefrontal cortex (PFC) mediates diverse effects on neuronal physiology and function, but the expression of dopamine receptors at subpopulations of projection neurons and interneurons remains unresolved. Here, we examine D1 receptor expression and modulation at specific cell types and layers in the mouse prelimbic PFC. We first show that D1 receptors are enriched in pyramidal cells in both layers 5 and 6, and that these cells project to intratelencephalic targets including contralateral cortex, striatum, and claustrum rather than to extratelencephalic structures. We then find that D1 receptors are also present in interneurons and enriched in superficial layer VIP-positive (VIP+) interneurons that coexpresses calretinin but absent from parvalbumin-positive (PV+) and somatostatin-positive (SOM+) interneurons. Finally, we determine that D1 receptors strongly and selectively enhance action potential firing in only a subset of these corticocortical neurons and VIP+ interneurons. Our findings define several novel subpopulations of D1+ neurons, highlighting how modulation via D1 receptors can influence both excitatory and disinhibitory microcircuits in the PFC.
Keywords: dopamine, interneuron, neuromodulation, prefrontal cortex, projection neuron
Introduction
Dopamine modulation in the prefrontal cortex (PFC) plays a key role in cognitive, motivational, and emotional behavior (Fuster 2000; Miller and Cohen 2001). Dopamine can signal through multiple receptors, which are subdivided into D1-like (D1 and D5) or D2-like (D2, D3, and D4) (Tritsch and Sabatini 2012). While all these subtypes (D1–5) are present in the PFC (Weiner et al. 1991; Mrzljak et al. 1996; Oda et al. 2010; Clarkson et al. 2017), D1 receptors (D1-Rs) are the most abundant (Lidow et al. 1991; Gaspar et al. 1995; Santana et al. 2009). The importance of D1-R signaling is highlighted by its requirement for PFC-dependent behaviors (Sawaguchi and Goldman-Rakic 1994; Williams and Goldman-Rakic 1995; Seamans et al. 1998), and its disruption in many neuropsychiatric disorders (Grace 2016). Determining which cell types express D1-Rs is therefore essential for understanding how dopamine modulates the PFC in health and disease.
The ability of the PFC to mediate executive control ultimately depends on the diverse long-range projections it sends to other brain regions (Miller and Cohen 2001; Gabbott et al. 2005). For example, intratelencephalic (IT) neurons project within the cortex, including between cerebral hemispheres, and are distinct from pyramidal tract (PT) neurons that project subcortically (Gabbott et al. 2005; Dembrow et al. 2010; Anastasiades et al. 2018). Recent studies indicate that dopamine receptors may differentially segregate between these 2 broad populations of layer 5 (L5) projection neurons in the PFC (Gee et al. 2012; Seong and Carter 2012; Clarkson et al. 2017). Interestingly, D1-Rs are also expressed in layer 6 (L6), where they may modulate corticothalamic (CT) projections (Gaspar et al. 1995). However, there is currently no consensus on which projection neurons primarily express D1-Rs, with reports varying significantly (Vincent et al. 1993; Gaspar et al. 1995). Given that the activity of defined projection neurons can have distinct effects on behavior (Land et al. 2014; Jenni et al. 2017; Murugan et al. 2017; Otis et al. 2017), this represents a significant gap in our understanding of how dopamine modulates PFC outputs.
Like other cortices, the PFC also contains a diverse array of GABAergic interneurons (Markram et al. 2004; Petilla Interneuron Nomenclature et al. 2008; Anastasiades and Butt 2011; Rudy et al. 2011). Interneurons are segregated into distinct subtypes based on their intrinsic electrophysiology, morphology, and immunohistochemical markers (Kubota and Kawaguchi 1994; Kawaguchi and Kubota 1996; Butt et al. 2005; Gonchar et al. 2007; Xu et al. 2010; Anastasiades et al. 2016). Moreover, dopamine receptors are thought to be expressed in several populations of interneurons (Muly et al. 1998; Glausier et al. 2009; Santana et al. 2009), where they can mediate diverse effects (Gonzalez-Islas and Hablitz 2001; Kroner et al. 2007; Towers and Hestrin 2008; Karunakaran et al. 2016). However, as with projection neurons, there are conflicting reports on which interneuron subtypes express D1-Rs in the PFC (Le Moine and Gaspar 1998; Muly et al. 1998; Paspalas and Goldman-Rakic 2005; Santana et al. 2009). While neuromodulation of inhibition plays a key role in cortical circuits (Kruglikov and Rudy 2008; Letzkus et al. 2011; Wester and McBain 2014; Froemke 2015), it is currently unclear whether D1-Rs primarily regulate inhibitory or disinhibitory networks, which is critical to informing models of PFC function (Wang et al. 2004; Glausier et al. 2009).
Here, we examine D1-receptor expressing (D1+) neurons in the mouse prelimbic PFC, combining ex vivo electrophysiology, selective pharmacology, 2-photon microscopy, immunohistochemistry, and retrograde anatomy in multiple transgenic mouse lines to selectively label different populations of projection neurons and interneurons. We find that D1-Rs are strongly expressed in subpopulations of IT neurons found in both L5 and L6. Surprisingly, D1-Rs are absent from parvalbumin (PV+) and somatostatin (SOM+) expressing interneurons but are selectively enriched in a subpopulation of superficial interneurons that express vasoactive intestinal peptide (VIP+). Activation of D1-Rs enhances firing in both D1+ pyramidal cells and VIP+ interneurons, indicating that D1-Rs enhance both excitatory and disinhibitory microcircuits in the PFC.
Materials and Methods
Animals
Experiments used either heterozygous D1-tdTomato mice (Ade et al. 2011) (JAX ID: 016 204) or heterozygous D1-tdTomato mice crossed with either homozygous GAD-Cre (Taniguchi et al. 2011) (JAX ID: 010 802), PV-Cre (Hippenmeyer et al. 2005) (JAX ID: 008 069), SOM-Cre (Taniguchi et al. 2011) (JAX ID: 013 044), VIP-Cre (Taniguchi et al. 2011) (JAX ID: 010 908), or heterozygous 5HT3a-Cre mice (Gerfen et al. 2013) (generously provided by Prof. Gord Fishell). Mice were bred on a C57BL/6 J background with the exception of D1-tdTomato x VIP-Cre mice, which were mixed background. Mice of both sexes were used, and no differences were found. All experimental procedures were approved by the University Animal Welfare Committee of New York University.
Stereotaxic Injections
Mice aged 4–7 weeks were deeply anesthetized with a mixture of ketamine (10 mg/mL) and xylazine (0.1 mg/mL) and head fixed in a stereotax (Kopf Instruments). A small craniotomy was made over the injection site, using coordinates relative to Bregma (dorsoventral, mediolateral, and rostrocaudal, respectively): PFC = −2.1, ± 0.4, +2.2 mm; claustrum (CLA) = −3.6, −3.2, +1.6 mm (injected at 5° to the upright); mediodorsal thalamus (MD) = −3.6, −0.3, −0.5 mm; ventromedial thalamus (VM) = −3.4, −2.7, −0.4 mm (injected at 30° to the upright); ventral tegmental area (VTA) = −4.5, −0.5, −2.95 mm; basolateral amygdala (BLA) = −4.9, −3.2, −1.2 mm, dorsomedial striatum (STR) = −3.2, −1.1, +1.1 mm, pontine nucleus (pons) = −4.7, +0.5, −4.0 mm. For retrograde labeling, pipettes were filled with Cholera Toxin Subunit B (CTB) conjugated to either Alexa-488 or −647 (Life Technologies). Virus varied between experiment: Cre-dependent labeling of interneurons = AAV9-CAG-FLEX-EGFP-WPRE (UPenn), or AAV1-CAG-FLEX-tdTomato-WPRE (UPenn); labeling putative pyramidal cells = AAV1-CaMKII-EGFP-WPRE (UPenn); axon anatomy = AAVDJ-hSyn1-mCherry-IRES-EGFP-Syb2 (SynaptoTag, Stanford); retrograde-Cre and associated axon anatomy = AAVrg-EF1a-mCherry-IRES-Cre (Addgene) and AAV1-EF1a-DIO-EYFP-WPRE (UPenn); MD axon anatomy = AAV1-CB7-mCherry-WPRE (UPenn). Borosilicate pipettes with 5–10 μm tip diameters were backfilled, and between 130–550 nL of solution was pressure injected using a Nanoject III (Drummond), with 30 s of spacing between injections. The pipette was subsequently left in place for an additional 5 min, allowing time to diffuse away from the pipette tip, before being slowly retracted from the brain. Animals were returned to their cages for between 1 and 3 weeks before being used for recording or anatomy, or for 4–6 weeks in the case of the SynaptoTag virus injections.
Slice Preparation
Mice aged 6–8 weeks were anesthetized with a lethal dose of ketamine (25 mg/mL) and xylazine (0.25 mg/mL) and perfused intracardially with ice-cold external solution containing the following (in mM): 65 sucrose, 76 NaCl, 25 NaHCO3, 1.4 NaH2PO4, 25 glucose, 2.5 KCl, 7 MgCl2, 0.4 Na-ascorbate, and 2 Na-pyruvate (295–305 mOsm), and bubbled with 95% O2/5% CO2. Coronal slices (300 μm thick) were cut on a VS1200 vibratome (Leica) in ice-cold external solution, before being transferred to ACSF containing the following (in mM): 120 NaCl, 25 NaHCO3, 1.4 NaH2PO4, 21 glucose, 2.5 KCl, 2 CaCl2, 1 MgCl2, 0.4 Na-ascorbate, and 2 Na-pyruvate (295–305 mOsm), bubbled with 95% O2/5% CO2. Slices were kept for 30 min at 35 °C, before being allowed to recover for 30 min at room temperature. Intrinsic properties were recorded at 30−32°C. To facilitate stable recordings of cells with very high input resistance, modulation of VIP+ interneurons was performed at room temperature. Modulation of pyramidal cells was performed at both 30−32°C and room temperature, with no differences observed across these conditions, so results were pooled for analysis.
Electrophysiology
Whole-cell recordings were obtained from neurons across all layers of the prelimbic subdivision of PFC. Neurons were identified by infrared-differential interference contrast, as previously described (Chalifoux and Carter 2010). Neuronal identity was established by the presence or absence of tdTomato, EGFP and Alexa-conjugated CTB under fluorescent illumination. Borosilicate pipettes (2–6 MΩ) were filled with internal solution comprising (in mM): 135 K-gluconate, 7 KCl, 10 HEPES, 10 Na-phosphocreatine, 4 Mg2-ATP, 0.4 Na-GTP and 0.5 EGTA, 290–295 mOsm, pH 7.3, with KOH. For a subset of experiments, 30 μM Alexa Fluor 594 was included for 2-photon imaging, in which case dye was allowed to diffuse throughout the dendrites and axons for at least 20 min before imaging.
Electrophysiology recordings were made with a Multiclamp 700B amplifier (Axon Instruments), filtered at 4 kHz, and sampled at 10 kHz. Series resistance was typically <20 MΩ for pyramidal cells and <30 MΩ for VIP+ interneurons. Current-clamp recordings were performed in the presence of the synaptic blockers CPP (10 μM), NBQX (10 μM), and Gabazine (10 μM). Dopamine firing modulation was measured in response to a depolarizing square current step of 500 ms duration. The magnitude of the current step was adjusted to reliably evoke 5 action potentials in the recorded neuron, as previously described (Seong and Carter 2012). Recordings were made at resting membrane potential, with no additional holding current applied to adjust for changes in membrane potential throughout the recording. Dopamine receptor pharmacology was performed using wash-in of the selective D1-type dopamine receptor agonist SKF-81 297 (10 μM) and the selective antagonist SCH-23 390 (10 μM). Modulation experiments involved 5 min of baseline firing, either in the presence or absence of SCH-23 390, followed by bath application of SKF-81 297. Firing modulation was calculated by comparing the average number of action potentials evoked per stimulus in this baseline epoch with a 5-min window starting 10 min after initial SKF-81 297 application. All chemicals were purchased from Sigma or Tocris Bioscience.
Two-photon Microscopy
Two-photon imaging was performed on a custom microscope, as previously described (Chalifoux and Carter 2010). Briefly, a Ti:Sapphire laser (Coherent) tuned to 810 nm was used to excite Alexa Fluor 594 to image morphology with a 60× 1.0 NA objective (Olympus). The 3D reconstructions of dendritic morphologies were performed using NeuronStudio (Wearne et al. 2005), while 2D tracing of dendrites and axons for figures was performed using Neurolucida (MBF Bioscience). Dendrite analysis was performed by summing the total, apical, or basal dendrite length.
Histology and Fluorescence Microscopy
Mice were anesthetized with a lethal dose of ketamine (25 mg/mL) and xylazine (0.25 mg/mL) and perfused intracardially with 0.01 M phosphate-buffered saline (PBS) followed by 4% paraformaldehyde (PFA) in 0.01 M PBS. Brains were fixed in 4% PFA in 0.01 M PBS for 4–12 h at 4°C. Slices were prepared at a thickness of 40–60 μm (Leica VT 1000 S vibratome). For enhanced detection of tdTomato signal in D1-tdTomato mice slices were stained with antibodies against RFP. For antibody labeling, slices were washed once in PBS (0.01 M), once in PBS-T (0.2 % Triton-X100), then blocked in PBS-T with 1% w/v bovine serum albumin (BSA) for 1 h at room temperature (except for GAD staining where Triton-X100 was omitted throughout). Primary antibody incubation (rabbit anti-red fluorescent protein, 600-401-379, Rockland, 1:1000; mouse anti-calretinin, MAB1568, Millipore, 1:1000; mouse anti-parvalbumin, MAB1572, Millipore, 1:2000; rat anti-somatostatin, MAB354, Millipore, 1:400; rabbit anti-GAD 65/67 AB1511, Millipore/Sigma 1:1000) was performed at 4°C overnight or for 48 h for GAD. Slices were then washed 4× in PBS at RT before incubating with secondary antibody (goat anti-rabbit Alexa 594, ab150080, AbCam, 1:400; goat anti-rat Alexa 647, 21 247, Fisher-Invitrogen, 1:200; goat anti-mouse Alexa 647, ab150119, Abcam, 1:200; goat anti-rabbit Alexa 647, A-21 244, Fisher-Invitrogen, 1:1000) in PBS-T + BSA for 1 h at room temperature, or overnight at 4°C for goat anti-rabbit Alexa 647 amplification of anti-GAD primary antibody. Slices were washed a further 3× in PBS before being mounted under glass coverslips on gelatin-coated slides using ProLong Gold antifade reagent with DAPI (Invitrogen). Whole-brain images were acquired using a slide-scanning microscope (Olympus VS120) with a 10× 0.25 NA or 20× 0.75 NA objective. Excitation wavelengths were 387, 485, 560, and 650 nm for DAPI, FITC, TRITC, and Cy5, respectively. PFC images were acquired using a confocal microscope (Leica SP8) with 10× 0.4 NA, 20× 0.75 NA, or 40× 1.3 NA oil immersion objective. Excitation wavelengths were 405, 488, 552, and 638 nm for DAPI, FITC, TRITC, and Cy5, respectively. Image processing involved adjusting brightness and contrast using ImageJ (NIH). Cell counting was performed in a 400 × 1000 μm region of interest across the depth of the prelimbic PFC.
In Situ Hybridization
Mice were anesthetized with a lethal dose of ketamine (25 mg/mL) and xylazine (0.25 mg/mL) and perfused intracardially with chilled 0.01 M PBS. The brain was extracted and immediately submerged in isopentane cooled on dry ice. Tissue was coated in O.C.T. media (Tissue Tek) and stored in an airtight container at −80 °C until sectioning. 10-μm sections were taken on a cryostat at −20°C and mounted on Superfrost Plus microscope slides (Fisher) and stored at −80°C. In situ hybridization of Mm-Drd1a-C2 and tdTomato-C3 probes was performed using a standard RNAscope protocol for flash frozen tissue from ACD bio. Slides were mounted under glass coverslips using ProLong Gold antifade reagent with DAPI (Invitrogen). Images were acquired using a confocal microscope (Leica SP8) with 20× 0.75 NA or 40× 1.3 NA oil immersion objective.
Data Analysis
Electrophysiology and imaging data were acquired using National Instruments boards and custom software written in MATLAB (MathWorks). Off-line analysis was performed using custom software written in Igor Pro (WaveMetrics). Input resistance was measured using the steady-state response to a −50 pA current injection for pyramidal cells and −10 or −20 pA for interneurons. The membrane time constant (tau) was measured using exponential fits to these same hyperpolarizations. Voltage sag due to h-current was calculated by taking the minimum voltage in the first 200 ms, subtracting the average voltage over the final 100 ms, and dividing by the steady-state value. Spike frequency adaptation was calculated as the ratio of the initial interspike interval (ISI) and final ISI in response to a 500 ms depolarizing current pulse which evoked >5 action potentials. For cell counting as a function of layer, individual cells were assigned a distance from the midline (top of layer 1), binned in 25 μm increments across the depth of PFC, and then assigned into individual layers. Layers were defined based on peaks in neuron density (Table 1), which gave defined ranges for each layer (Table 2). Data were collected from at least 3 slices per animal, with a minimum of 3 mice per projection class/interneuron subtype. Cell-by-cell mRNA puncta analysis was performed using a circular region of interest (17.5 μm in diameter) placed over the soma of individual neurons and manual counting of puncta for Drd1a and tdTomato.
Table 1.
Projection neuron boundaries
| L1/L2 border | Distance from midline (μm) |
| Start of BLA cells | 125 ± 8 |
| L2/L3 border | Distance from midline (μm) |
| End of the first peak of BLA cells | 219 ± 3 |
| End of the first peak D1 cells | 231 ± 5 |
| L3/L5a border | Distance from midline (μm) |
| Start of the second peak of BLA cells | 321 ± 9 |
| End of the second peak of BLA cells | 376 ± 7 |
| L5a/L5b upper border | Distance from midline (μm) |
| Start of Pons cells | 345 ± 3 |
| Start of VTA cells | 375 ± 10 |
| L5b upper/L5b lower border | Distance from midline (μm) |
| End of peak cSTR cells | 530 ± 8 |
| End of peak CT/PT dual labeleda | 492 ± 27 |
| L5b lower/L6 border | Distance from midline (μm) |
| End of VTA cells | 664 ± 16 |
| End of Pons cells | 627 ± 8 |
Note: Designating individual layer boundaries based on the start and end of peak projection neuron density across multiple projection neuron populations.
aData reanalyzed from Collins et al. (2018).
Table 2.
Layer boundaries
| Lower bound (μm from midline) | Analysis range (μm from midline) | |
|---|---|---|
| L1 | 125 ± 8 | 0–125 |
| L2 | 227 ± 3 | 126–225 |
| L3 | 321 ± 9 | 226–325 |
| L5a | 376 ± 7 | 326–375 |
| L5b upper | 511 ± 5 | 376–500 |
| L5b lower | 648 ± 7 | 501–650 |
| L6 | WM | 651–1000 |
Note: Designating lower layer boundaries after averaging across projection neuron populations shown in Table 1. Analysis range indicates distances from midline used to divide data into individual layers using 25 μm bins. WM = white matter.
Experimental Design and Statistical Analysis
Summary data are reported in the text and shown in figures as arithmetic mean ± SEM, unless otherwise stated. Statistical comparisons were performed in GraphPad Prism (version 7.0c) using a 2-tailed nonparametric Mann–Whitney U test. Significance was defined as P < 0.05.
Results
D1 Receptors are Expressed in Excitatory and Inhibitory Cells in the PFC
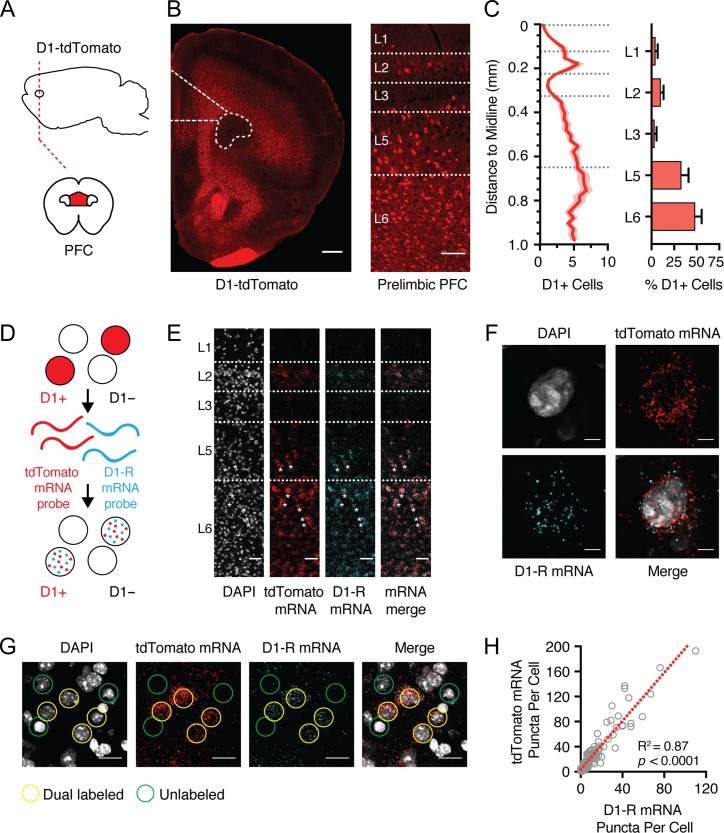
We studied D1-receptor expressing (D1+) neurons using D1-tdTomato mice, in which expression of the red fluorescent protein tdTomato is driven by the D1 receptor promoter (Fig. 1A) (Ade et al. 2011). D1+ neurons were prominent in the prelimbic (PL) PFC, with most cells residing in L5 and L6 (Fig. 1B,C; L5 = 33 ± 3% of all D1+; L6 = 48 ± 3%, n = 8 mice). We also observed a smaller population of neurons located in superficial layers 1 (L1), 2 (L2), and 3 (L3) (Fig. 1B,C; L1 = 5 ± 1% of all D1+; L2 = 10 ± 1%; L3 = 4 ± 1%). Within each layer, D1+ neurons represented a subpopulation of cells, consistent with restricted D1-R expression in the PFC (Seong and Carter 2012). D1+ neurons were also present in regions adjacent to PL PFC, including the infralimbic (IL) PFC and primary motor cortex (M1) (Supplementary Fig. S1). However, the distribution of D1+ neurons in PL and IL PFC was distinct to M1, which lacked superficial D1+ cells and had a lower density of D1+ cells (Supplementary Fig. S1). For all future analyses, we focused on the PL PFC, where D1 receptors are important for sustained activity (Seamans and Yang 2004).
Figure 1.
D1-tdTomato mice replicate endogenous D1-R expression. (A) Schematic of coronal slice through the medial PFC (red). (B) Left, coronal slice showing D1-tdTomato expression in the PFC, highlighting the prelimbic subdivision in dotted outline. Right, expanded view showing distribution of D1-tdTomato-positive (D1+) neurons across layers of prelimbic PFC. Scale bars = 500 and 100 μm, respectively. (C) Summary of D1+ neuron distribution as a function of distance from midline (left) and as a function of different layers (right). Bin size = 25 μm. Dashed lines represent laminar boundaries. (D) Schematic of in situ hybridization process to distinguish tdTomato mRNA (red) and D1-R mRNA (blue) in single PFC neurons. (E) Left to right, distribution of DAPI-labeled cells (gray), tdTomato mRNA (red), D1-R mRNA (blue), and merged image, showing colocalization of mRNAs across layers of prelimbic PFC. Scale bars = 50 μm. Asterisks indicate a subset of colabeled neurons. (F) Zoomed image showing colocalization of tdTomato and D1-R mRNA within an individual L5 PFC neuron. Scale bars = 5 μm. (G) Colocalization of tdTomato and D1-R mRNA within a subset of deep layer PFC neurons. Examples of cells enriched for both mRNAs are highlighted in yellow, while poorly enriched cells are shown in green. Scale bars = 20 μm. (H) Correlation of number of D1-R and tdTomato mRNA puncta within individual neurons. Dashed red line shows linear fit to the data. Values shown as mean ± SEM.
To validate that tdTomato faithfully replicates endogenous D1-R expression, we probed for D1-R (Drd1a) and tdTomato mRNAs using multiplex fluorescent in situ hybridization (Fig. 1D) (Wang et al. 2012). The distributions of mRNA puncta for Drd1a and tdTomato were similar and mirrored that observed for D1-tdTomato cells (Fig. 1B,E). On a cell-by-cell basis, there was strong overlap in expression levels of Drd1a and tdTomato mRNA (Fig. 1F,G), with strong correlation between the number of Drd1a and tdTomato-positive puncta in individual neurons (Fig. 1H; n = 105 cells, from 3 mice,R2 = 0.87, P < 0.0001). These findings support the use of D1-tdTomato mice to explore D1+ neurons in the PFC.
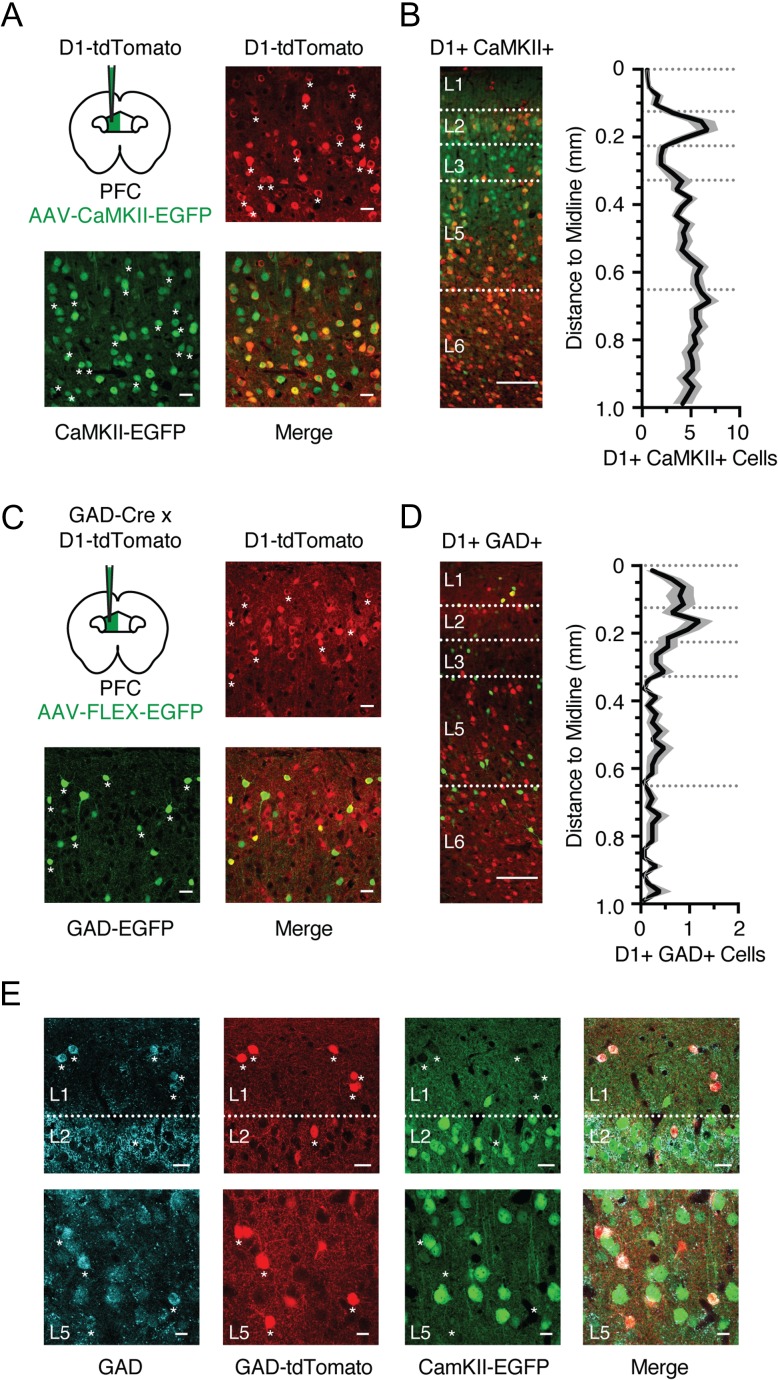
Cortical neurons are broadly divided into either glutamatergic pyramidal cells or GABAergic interneurons. To label glutamatergic neurons, we injected AAV-CaMKII-EGFP virus into the PFC of D1-tdTomato mice (Fig. 2A). We found that most D1+ neurons were also CaMKII+ (87 ± 2% of D1+ cells, n = 5 mice), indicating the majority are glutamatergic. Accordingly, most colabeled neurons were found in deep layers, similar to the overall D1+ population (Fig. 2B). Despite this strong overlap, we also observed some D1+ CaMKII- neurons, which could be cells that avoided viral transfection with the CaMKII virus, or alternatively a population of D1+ GABAergic interneurons that coexpress GAD (glutamate decarboxylase). To test for the latter possibility, we crossed D1-tdTomato x GAD-Cre transgenic mice and injected Cre-dependent AAV-FLEX-EGFP virus into the PFC (Fig. 2C). Although most GAD+ neurons were D1-R negative (D1−), a population of colabeled interneurons was present in superficial layers (Fig. 2D; 7.8 ± 0.1% of D1+ cells, n = 3 mice). To further validate this labeling strategy, we injected AAV-CaMKII-EGFP and AAV-FLEX-tdTomato into the PFC of GAD-Cre mice, while staining for GAD using antibody labeling. We found the 2 populations were nonoverlapping, with GAD colocalized to tdTomato+ but not EGFP+ neurons (Fig. 2E). Moreover, EGFP+ neurons were absent from layer 1, which only contained GAD+ tdTomato+ neurons, as expected (Fig. 2E). Together, these findings indicate that the majority of D1+ CaMKII+ neurons are located in L5 and L6, whereas D1+ GAD+ interneurons are found in L1 and L2.
Figure 2.
D1 receptors are expressed in pyramidal cells and interneurons. (A) Schematic for injection of AAV-CaMKII-EGFP into the prelimbic PFC of D1-tdTomato mouse, and confocal images from L5 showing D1-tdTomato expression (red), CaMKII-EGFP expression (green), and merged image. Asterisks indicate a subset of colabeled neurons. Scale bar = 25 μm. (B) Left, distribution and overlap of D1+ (red) and CaMKII+ (green) neurons across layers of the prelimbic PFC. Right, summary distribution of colabeled D1+ CaMKII+ neurons as a function of distance from midline. Bin size = 25 μm. (C) Similar to (A) for injection of AAV-FLEX-EGFP into GAD-Cre x D1-tdTomato mouse, showing D1-tdTomato expression (red), GAD-EGFP expression (green), and merged image from superficial layers. Asterisks indicate colabeled neurons. Scale bar = 25 μm. (D) Left, distribution and overlap of D1+ (red) and GAD+ (green) neurons across layers of the prelimbic PFC. Right, summary distribution of colabeled D1+ GAD+ neurons of cell density as a function of distance from midline. Bin size = 25 μm. (E) Confocal images from the L1/2 border (top row), or L5 (bottom row) of GAD-Cre mice following injection of AAV-FLEX-tdTomato and AAV-CaMKII-EGFP into PFC and subsequent antibody staining for GAD. Left to right, GAD antibody staining, AAV-FLEX-tdTomato labeling, AAV-CaMKII-EGFP labeling, merged image. Asterisks indicate a subset of colabeled tdTomato+ GAD+ neurons. Scale bar = 25 μm. Values shown as mean ± SEM.
D1+ Pyramidal Cells have Distinct Properties and Respond to D1-R Activation
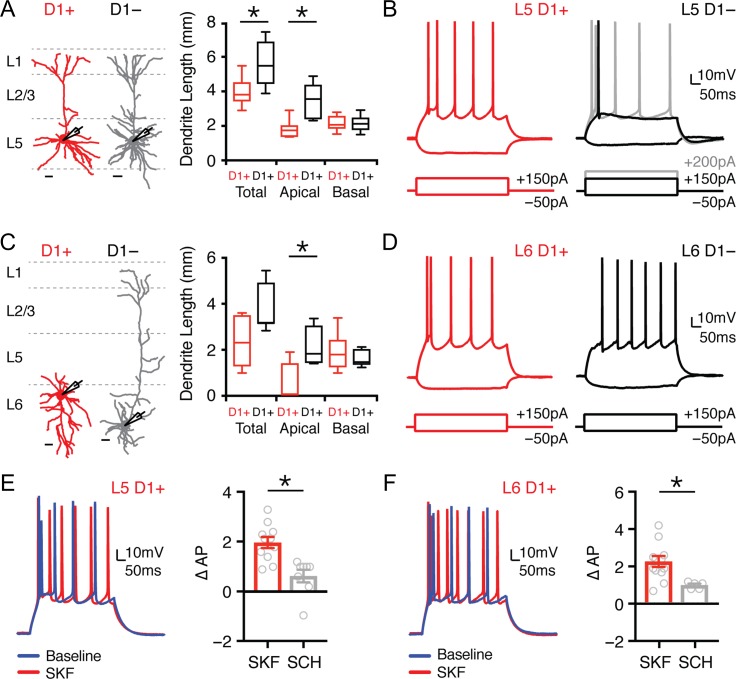
Pyramidal cells in deep layers of cortex segregate into multiple subtypes based on their dendritic morphology and intrinsic physiology (Hattox and Nelson 2007; Dembrow et al. 2010; Thomson 2010; Anastasiades et al. 2018). We previously showed that D1+ and D1− pyramidal cells in L5 of the juvenile PFC differ in their morphology and physiology (Seong and Carter 2012). To extend these findings, we compared D1+ and D1− pyramidal cells in both L5 and L6 of the adult PFC (Fig. 3A,C; D1+: L5 n = 7, L6 n = 8; D1−: L5 n = 8, L6 n = 7). We found sparse apical dendrites in L5 D1+ neurons (D1+ = 1827 ± 219 μm; D1− = 3448 ± 377 μm; P = 0.002) and L6 D1+ neurons (D1+ = 462 ± 323 μm; D1− = 2218 ± 336 μm; P = 0.01), with the latter often having multipolar or inverted dendrites (Fig. 3A,C). We also found different intrinsic properties in L5 D1+ neurons, which are more hyperpolarized, have higher input resistance, and minimal voltage sag (Fig. 3B and Table 3). Differences persisted for L6 D1+ neurons, which showed less-voltage sag but similar resting potential and input resistance (Fig. 3D and Table 3). Interestingly, L6 D1− neurons also displayed minimal spike frequency adaptation, in contrast to the other cell types we recorded (Fig. 3D and Table 3).
Figure 3.
L5 and L6 D1+ pyramidal cells are modulated by D1 receptors. (A) Left, dendrite reconstructions from 2-photon images of L5 D1+ (red) and L5 D1− (gray) pyramidal cells. Scale bars = 50 μm. Right, quantification of total, apical, and basal dendrite length for D1+ and D1− L5 pyramidal cells. (B) Intrinsic properties and AP firing of L5 D1+ (red) and L5 D1− (black/gray) pyramidal cells in response to depolarizing and hyperpolarizing current steps. (C,D) Similar to (A,B) for L6 D1+ (red) and L6 D1− (black) pyramidal cells. (E) Left, AP firing of L5 D1+ pyramidal cells in response to depolarizing current step during baseline (blue) and after wash-in of the D1-R agonist SKF-81 297 (10 μM) (red). Right, summary of change in number of evoked APs (∆AP) recorded from L5 D1+ pyramidal cells after application of either SKF or SCH + SKF. (F) Similar to (E) for L6 D1+ pyramidal cells. Values shown as median ± quartiles (A andC) or mean ± SEM (E and F). *P < 0.05. See also Table 3.
Table 3.
Intrinsic properties of PFC pyramidal cells
| Cell type | RMP (mV) | Rin (MΩ) | Sag (%) | Adaptation |
|---|---|---|---|---|
| L5 D1+ (n = 10) | −73.4 ± 1.4* | 202.0 ± 15.6* | 2.5 ± 0.6* | 0.17 ± 0.02 |
| L5 D1− (n = 11) | −68.3 ± 1.0* | 105.8 ± 8.7* | 11.4 ± 3.1* | 0.21 ± 0.03 |
| L6 D1+ (n = 10) | −70.8 ± 1.0 | 270.6 ± 37.0 | 3.8 ± 0.8* | 0.26 ± 0.07* |
| L6 D1− (n = 9) | −71.3 ± 0.8 | 205.8 ± 15.0 | 9.2 ± 1.4* | 0.87 ± 0.04* |
| L5 D1+ CC+ (n = 11) | −76.5 ± 1.2 | 220 ± 15.3 | 2.3 ± 0.6 | 0.20 ± 0.03 |
| L5 D1− CC+ (n = 9) | −74.7 ± 1.1 | 209.0 ± 22.8 | 1.5 ± 0.5 | 0.16 ± 0.02 |
Note: Resting membrane potential (RMP), input resistance (Rin), voltage sag due to h-current (Sag %) and adaptation ratio for L5 and L6 D1+ and D1− pyramidal cells. *P < 0.05.
Having explored the intrinsic properties of D1+ projection neurons, we assessed if they are modulated by D1-Rs. Dopamine receptors are known to regulate action potential (AP) firing in many brain regions, including the PFC (Gulledge and Jaffe 1998; Henze et al. 2000; Gorelova et al. 2002; Seamans and Yang 2004). To rule out network effects, we studied modulation of AP firing in the presence of synaptic blockers. In L5 D1+ neurons, we found the D1-R agonist SKF-81 297 (10 μM) increased firing, which was blocked by preincubation with the D1-R antagonist SCH-23 390 (10 μM) (Fig. 3E; ∆AP: SKF = 2.0 ± 0.2, n = 11; SKF + SCH = 0.6 ± 0.3, n = 8; P = 0.0015). Similar regulation was seen for L6 D1+ neurons, with enhanced firing following SKF-81 297, but not with SCH-23 390 (Fig. 3F; ∆AP: SKF = 2.2 ± 0.3, n = 11; SKF + SCH = 0.9 ± 0.1, n = 5; P = 0.01). Together, these results indicate that L5 and L6 D1+ neurons are morphologically and physiologically distinct from adjacent D1− neurons, and that functional D1-Rs in these cells robustly enhance AP firing. Together, these results suggest that D1+ and D1− cells may be able to differentially sample and respond to their synaptic inputs (Baker et al. 2018). Interestingly, similar intrinsic physiology and morphology are also observed for retrogradely labeled IT neurons (Hattox and Nelson 2007; Dembrow et al. 2010; Thomson 2010; Anastasiades et al. 2018), suggesting that projection neuron identity may help classify D1+ and D1− neurons in mouse PFC.
Multiple Classes of Projection Neurons Contact Long-range Targets
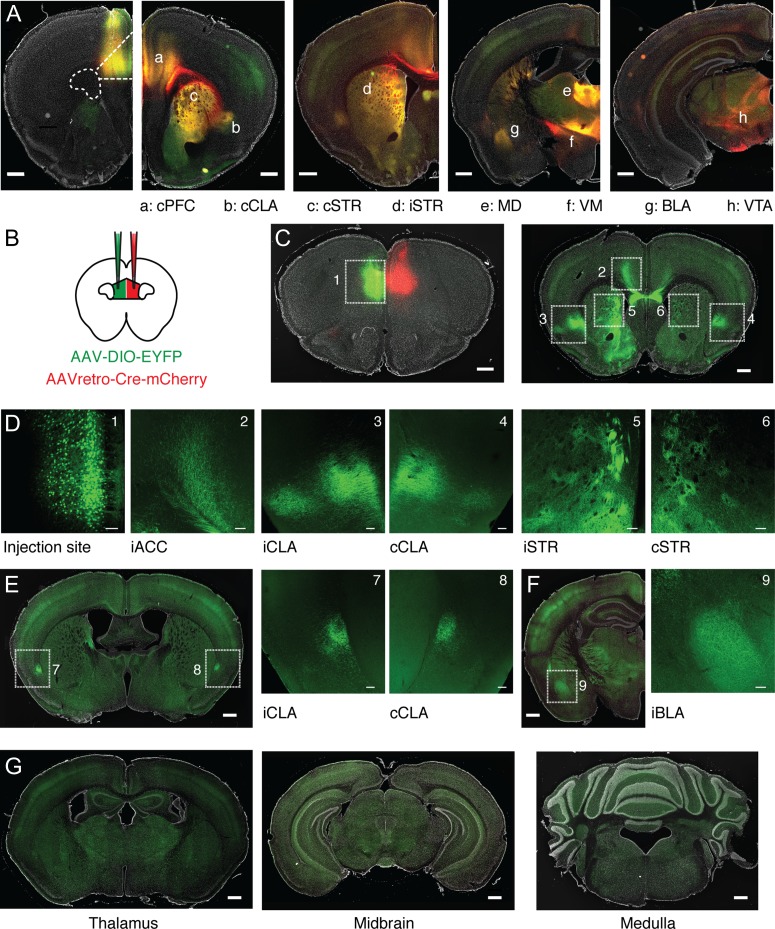
To begin to explore projection neurons, we first determined the long-range targets of the PFC by injecting AAV-SynaptoTag, which labels axons in red and synapses in green at different target regions (Xu and Sudhof 2013) (n = 3 mice). Viral expression occurred across all layers of PL PFC, with some spread along the injection tract into adjacent cingulate cortex (Fig. 4A). Monosynaptic target regions were distinguished from passing axons (red) by the colocalization of axonal and synaptic labeling (red and green). These regions included the contralateral PFC (cPFC), contralateral and ipsilateral claustrum (cCLA and iCLA), contralateral and ipsilateral striatum (cSTR and iSTR), mediodorsal (MD) and ventromedial (VM) thalamus, BLA, and VTA (Fig. 4A). These various output pathways have different functional roles and represent potential targets for D1+ projection neurons in the PFC.
Figure 4.
Characterization of PFC projection targets across the brain. (A) From left to right, representative injection of AAV-SynaptoTag, showing labeling of PFC axons (red), synaptic terminals (green), and their overlap (yellow) in (a) contralateral PFC (cPFC), (b) contralateral claustrum (cCLA), (c) contralateral dorsomedial striatum (cSTR), (d) ipsilateral dorsomedial striatum (iSTR), (e) mediodorsal thalamus (MD), (f) ventromedial thalamus (VM), (g) basolateral amygdala (BLA), and (h) ventral tegmental area (VTA). Scale bar = 500 μm. (B) Schematic for injection of AAVretro-Cre-mCherry into the cPFC and AAV-DIO-EYFP into the ipsilateral (i)PFC. (C) Left, representative injection sites, showing EYFP-labeled CC neurons in iPFC, and location of AAVretro-Cre-mCherry in cPFC. Right, distribution of CC neuron axons throughout multiple IT projection targets, including cortex, striatum, and claustrum. Numbered boxes indicate zoomed imaging regions shown in (D). Scale bars = 500 μm. (D) Confocal images of boxed regions in (C), highlighting labeled cell bodies and axons. Left to right, the AAV-DIO-EYFP injection site, iACC, iCLA, cCLA, iSTR, cSTR. Scale bars = 100 μm. (E) Left, distribution of CC neuron axons in caudal claustrum. Scale bar = 500 μm. Right, confocal images of boxed regions highlighting labeled axons in both iCLA and cCLA. Scale bars = 100 μm. (F) Left, distribution of CC neuron axons in iBLA. Scale bar = 500 μm. Right, confocal image of boxed region highlighting labeled axons in iBLA. Scale bar = 100 μm. (G) Left to right, absence of CC neuron axons from thalamus, midbrain, and medulla. Scale bars = 500 μm.
Our physiology experiments indicate that D1+ neurons display IT properties, similar to corticocortical (CC) neurons projecting via the corpus callosum (Hattox and Nelson 2007; Dembrow et al. 2010; Thomson 2010; Anastasiades et al. 2018). However, pyramidal cells within this broad subclass can also send branching projections to multiple long-range targets (Wilson 1987; Cowan and Wilson 1994). To determine any additional targets of CC neurons, we next injected AAVretro-Cre-mCherry into the cPFC (Tervo et al. 2016), along with AAV-DIO-EYFP into the ipsilateral (i)PFC (Fig. 4B,C). EYFP+ CC neurons were distributed across layers (Fig. 4D), with labeled axons found in several distant brain regions, including the ipsilateral anterior cingulate cortex (iACC), iCLA, cCLA, iSTR, and cSTR (Fig. 4C,D). CC neurons also projected to more caudal claustral regions (Fig. 4E) and sent axons to the iBLA (Fig. 4F), but not extratelencephalic (ET) targets such as thalamus, midbrain and medulla (Fig. 4G) (Baker et al. 2018). Interestingly, these findings are similar to recent reports of a genetically defined population of deep layer IT neurons (Nakayama et al. 2018). Taken together, these data suggest that D1+ neurons may be IT cells that target the cortex, claustrum, and striatum, motivating us to examine these projection neurons in more detail, including how these cell types segregate across different layers of the PFC.
Projection Neuron Subtypes Define Distinct Layers in Prelimbic PFC
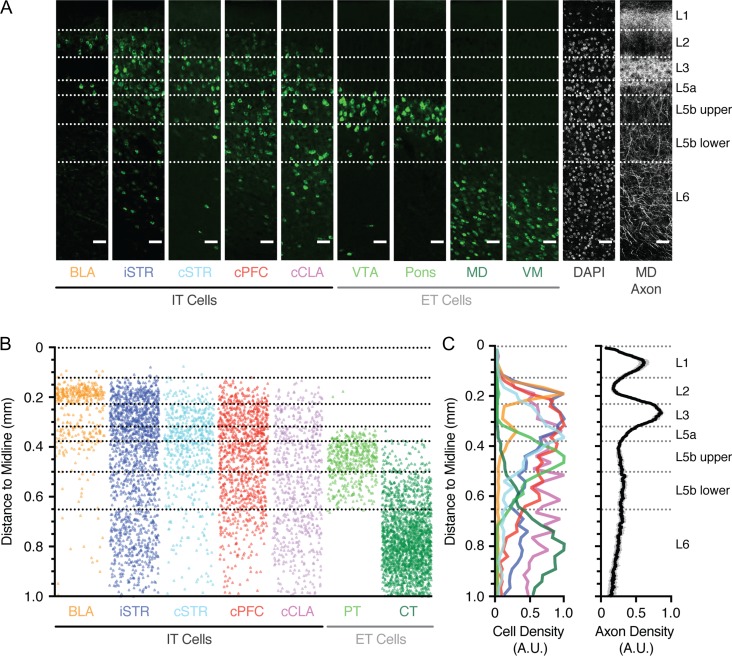
Throughout cortex, subclasses of projection neurons display specific distributions across different layers (Gabbott et al. 2005; Oberlaender et al. 2012; Oswald et al. 2013; Harris and Shepherd 2015). To thoroughly determine these distributions in PFC, we independently injected Alexa-conjugated cholera toxin subunit B (CTB) into identified target regions (BLA n = 4, iSTR n = 3, cSTR n = 3, cPFC n = 3, cCLA n = 3, VTA n = 4, Pons n = 3, MD n = 4, VM n = 3). We observed retrogradely labeled projection neurons across layers 2–6 of PFC, with different classes distributed in a laminar-specific manner (Fig. 5A). To further assist with the classification of layers, we injected AAV-mCherry into the MD nucleus of thalamus and imaged the distribution of thalamic axon in PFC (Fig. 5A) (Ueta et al. 2013; Collins et al. 2018). Based on these bands of axon and cell density, we were able to designate individual layers across the depth of PFC (Fig. 5B,C). Overall, we found that: L1 contained a peak in MD axon, but lacked projection neurons; L2 possessed a high density of cortico-amygdala (CA) neurons, but also a diverse array of other IT cells, including corticostriatal (CS) neurons projecting to iSTR; L3 is the main thalamorecipient layer and comprises IT cells, with a greater density of CS neurons projecting to cSTR and CC neurons. L5 could be divided into 3 distinct sublayers (Lorente de No 1992): L5a contained a second band of CA neurons, the peak of CS neurons projecting to cSTR, and increased density of corticoclaustral (CCL) neurons; L5b possessed ET populations, including PT and CT neurons, with PT neurons biased to upper L5b and less dense in lower L5b. Finally, L6 lacked PT neurons and contained both IT cells and a higher density of CT neurons. Together, these findings highlight the distribution and laminar structure of different projection neurons in the prelimbic PFC (Fig. 5B,C), extending on previous work in rat (Gabbott et al., 2005). Moreover, they allowed us to define specific boundaries for individual layers and sublayers in this agranular region of cortex (Table 1) and suggest that D1+ neurons located in L5 and L6 are likely to be CC or CCL cells.
Figure 5.
Laminar distributions of different projection neurons in the PFC. (A) Confocal images showing the distribution of retrogradely labeled neurons, DAPI-labeled somas, and thalamic axon across the depth of prelimbic PFC, following injection of fluorescently tagged Cholera Toxin subunit B (CTB) into BLA, iSTR, cSTR, cPFC, cCLA, VTA, pons, MD, VM, or AAV-mCherry into MD. Projection neurons are divided into intratelencephalic (IT) and extratelencephalic (ET) populations. Scale bars = 50 μm. (B) Overlay of the position of all counted retrogradely labeled projection neurons as a function of distance to the midline. VTA and Pons are combined as pyramidal tract (PT) neurons, MD and VM are combined as corticothalamic (CT) neurons. (C) Left, summary of peak-normalized cell density for projection neurons as a function of distance to the midline. Right, summary of peak-normalized MD axon density as a function of distance to the midline. See also Table 1.
D1 Receptors are Found in a Subset of IT Neurons
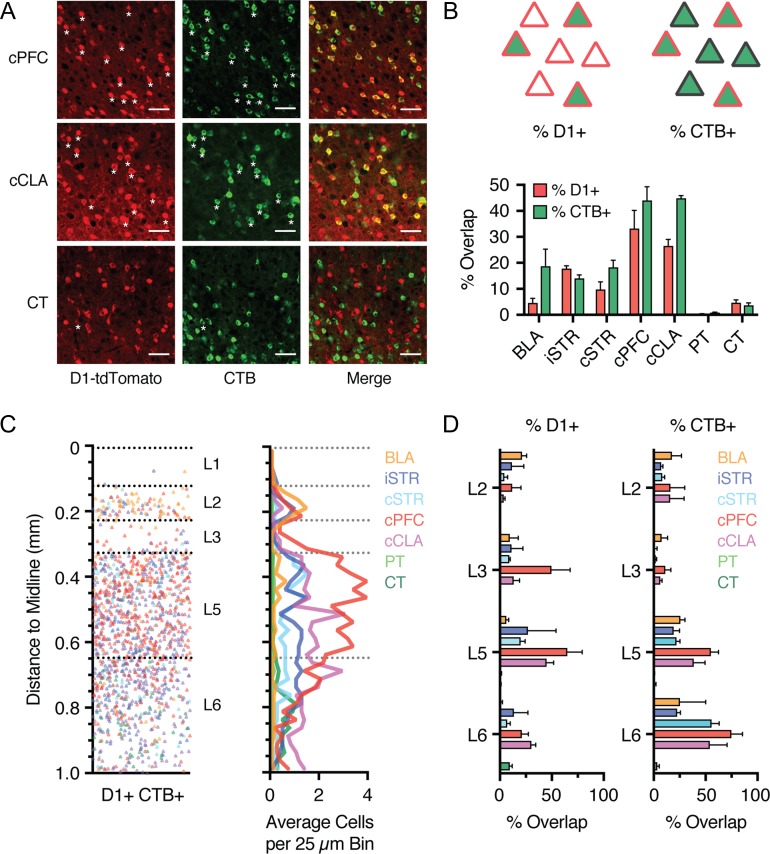
To determine which projection neuron populations express D1-R, we next examined the colocalization of D1+ neurons and different populations of CTB-labeled (CTB+) neurons (Fig. 6A). For each of the retrogradely labeled populations (Fig. 4), we first quantified the percentage of D1+ neurons that are also CTB+ across all layers (% D1, calculated as % (D1+ CTB+)/D1+). We found strong colabeling for cells projecting to cPFC and cCLA (Fig. 6A,B; cPFC = 33 ± 7%, n = 3 mice; cCLA = 27 ± 4%, n = 3 mice), less colabeling for cells projecting to BLA and striatum (Fig. 6A,B; BLA = 4.6 ± 1.8%, n = 4 mice; iSTR = 17.8 ± 1.1%, n = 3 mice, cSTR = 9.8 ± 2.9%, n = 3 mice), essentially no colabeling for PT neurons projecting to pons or VTA, and a small yet consistently colabeled population of L6 CT neurons projecting to MD and VM (Fig. 6A,B; PT = 0.4 ± 0.1%, VTA n = 4 mice, pons n = 3 mice; CT = 4.7 ± 1.1%, MD n = 4 mice, VM n = 3 mice) (Hoerder-Suabedissen et al. 2018). Plotting % D1 for each population across cortical depth revealed projection-specific distributions (Fig. 6C), with CC and CCL neurons showing the strongest overlap across several layers, and highest colabeling in deep layers (Fig. 6D).
Figure 6.
D1+ neurons in L5 and L6 are predominantly IT neurons. (A) Confocal images showing overlap of D1+ neurons (red) and retrogradely labeled CTB+ neurons (green) that project to cPFC (top), cCLA (middle), or thalamus (CT, bottom). Scale bar = 50 μm. Asterisks indicate a subset of colabeled neurons. Images taken from layer 5 (cPFC and cCLA) and layer 6 (CT). (B) Top left, schematic indicating the percentage of D1+ neurons across all layers that are retrogradely labeled (% D1, calculated as % (D1+ CTB+)/D1+). Top right, schematic indicating the percentage of CTB+ neurons that are also D1+ neurons (% CTB, calculated as % (D1+ CTB+)/CTB+). Bottom, summary of both the percentage of D1+ neurons (% D1) and the percentage of CTB+ neurons (% CTB), respectively, for each of the projection classes shown in Figure 5. (C) Left, distribution of all retrogradely labeled D1+ CTB+ colabeled projection neurons as a function of distance to the midline. Right, Summary of the binned and averaged distribution of D1+ CTB+ colabeled projection neurons as a function of distance to the midline. Bin size = 25 μm. (D) Left, summary of percentage of D1+ neurons that are retrogradely labeled (% D1+) as a function of layer. Right, summary of percentage of CTB+ neurons that are also D1+ neurons (% CTB+) as a function of layer. Values shown as mean ± SEM.
Although this analysis identifies the main projection targets of D1+ neurons, it could miss cells that represent only a small proportion of the overall population, but themselves possess a high degree of D1-R enrichment. To account for this possibility, we also quantified the percentage of CTB+ neurons that are D1+ (% CTB, calculated as % (D1+ CTB+)/CTB+). Approximately half of all neurons projecting to cPFC or cCLA were D1+ (Fig. 6B; cPFC = 44 ± 5%, cCLA = 45 ± 1%). However, this quantification could in principle underestimate overlap in deep layers, given that many cPFC and cCLA neurons are located in L3, where D1+ neurons are largely absent. Accordingly, plotting % CTB for individual layers revealed that a high percentage of CC and CCL neurons in L5 and L6 were D1+ (Fig. 6D; cPFC: L5 = 55 ± 7%, L6 = 75 ± 11%; cCLA: L5 = 39 ± 11%, L6 = 54 ± 17 %). Furthermore, around 20% of neurons that project to either BLA or striatum are also D1+ (BLA = 19 ± 7%, iSTR = 14 ± 1%, cSTR = 18 ± 3%), but very few neurons projecting via the PT or to thalamus (PT = 0.9 ± 0.1%, CT = 3.7 ± 1%). These findings indicate that we did not overlook a key D1+ subtype (Fig. 6C,D). Together, these results confirm that D1+ pyramidal cells are primarily IT neurons located in deep layers of the PFC, with prominent projections to cPFC and cCLA.
D1 Receptors Modulate a Subpopulation of CC Neurons
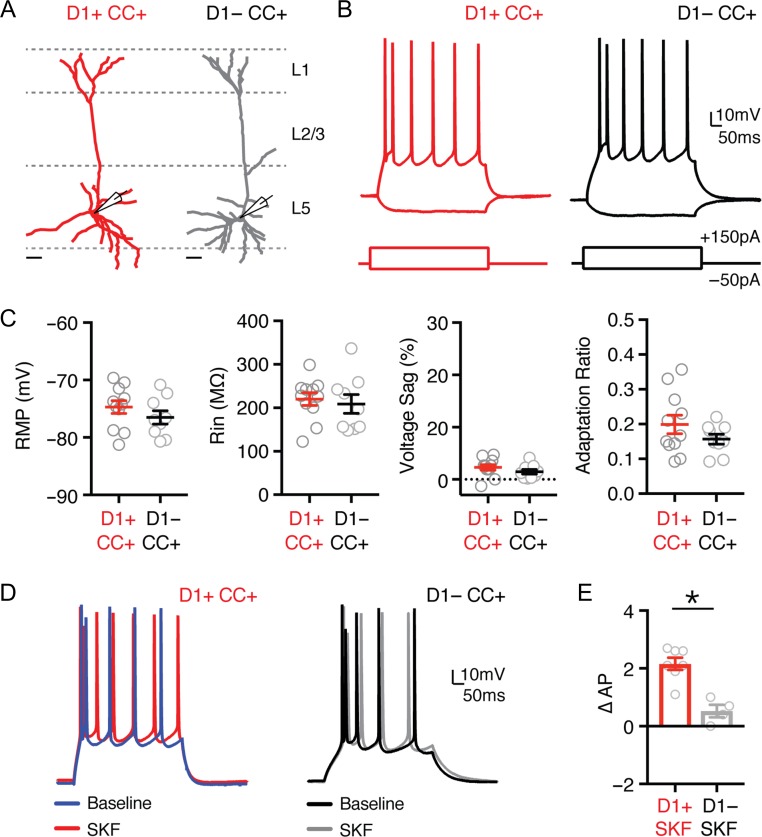
Our data indicate that in L5 the majority of D1+ neurons are IT cells, while most D1− neurons are PT cells. However, despite significant overlap between D1-R expression and cPFC projecting cells, some CC neurons were observed to be D1-negative. This suggested modulation can be uncoupled from projection target, motivating us to compare D1+ and D1− CC neurons. Whole-cell physiology and dendritic reconstructions revealed similar morphology and physiology, with indistinguishable RMP, Rin, voltage sag, and adaptation (Fig. 7A–C and Table 3;CC+ D1+ n = 11, CC+ D1− n = 9). However, while bath application of SKF enhanced the firing of CC+ D1+ neurons, it had no effect on CC+ D1− neurons (Fig. 7D,E; ∆AP: CC+ D1+ = 2.2 ± 0.4, n = 7; CC+ D1− = 0.5 ± 0.3, n = 4; P = 0.003). These results confirm there are at least 2 populations of CC neurons in L5, with only a subset modulated by D1-Rs. They also indicate that D1-R expression and projection target are not synonymous, such that not all CC neurons are D1+ neurons, and vice versa.
Figure 7.
Heterogeneous modulation of CC neurons by D1 receptors. (A) Dendrite reconstructions from 2-photon images of representative retrogradely labeled D1+ and D1− corticocortical (CC) neurons (D1+ CC+ in red, D1− CC+ in gray) whose cell bodies are located in layer 5 (L5). Scale bars = 50 μm. (B) Intrinsic properties and AP firing of D1+ CC+ (red) and D1− CC+ (black) neurons, in response to depolarizing and hyperpolarizing current steps. (C) Summary of resting membrane potential (RMP), input resistance (Rin), voltage sag (Sag %), and adaptation ratio in the 2 cell types. (D) AP firing of D1+ CC (left) and D1− CC (right) neurons at baseline (blue/black) and in response to wash-in of the D1-R agonist SKF-81 297 (10 μM) (red/gray). (E) Summary of change in number of evoked APs (∆AP) recorded from D1+ CC+ and D1− CC+ neurons after application of SKF-81 297 (10 μM). Values shown as mean ± SEM. *P < 0.05. See also Table 3.
D1 Receptors are Expressed in VIP+ Interneurons
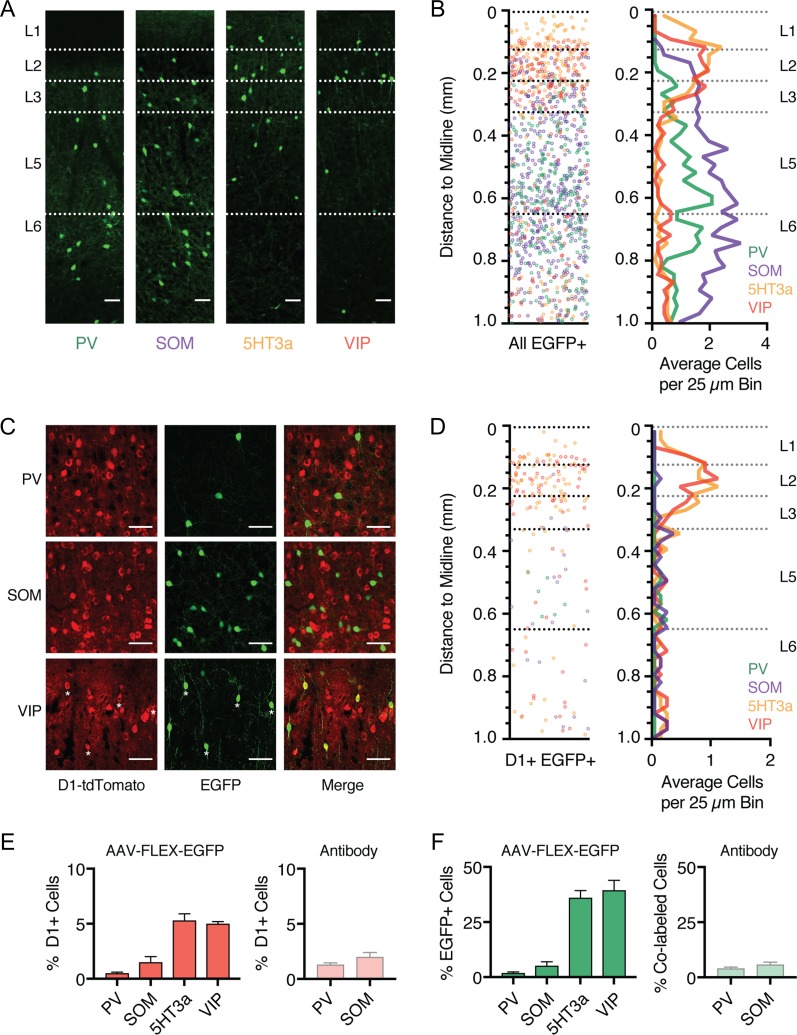
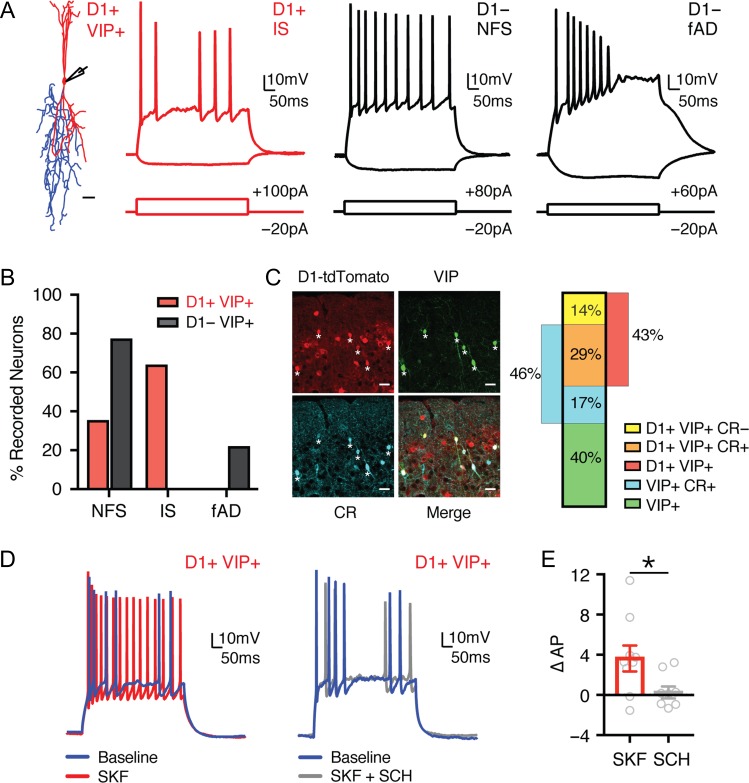
Our initial results indicated that, in addition to projection neurons, D1-Rs are expressed in a subset of GABAergic interneurons. Cortical interneurons are often segregated into distinct subtypes based on their physiology, morphology, and expression of histochemical markers (Kubota and Kawaguchi 1994; Kawaguchi and Kubota 1996; Cauli et al. 1997; Anastasiades et al. 2016). Three of these markers, parvalbumin (PV+), somatostatin (SOM+), and the serotonin receptor 3a (5HT3a+), label almost 100% of cortical GABAergic interneurons (Rudy et al. 2011). The 5HT3a+ population is particularly diverse and contains a further major subset of interneurons that express vasoactive intestinal peptide (VIP+) (Lee et al. 2010; Rudy et al. 2011). PV+ and SOM+ interneurons primarily inhibit pyramidal cells, while VIP+ neurons inhibit other interneurons and are engaged in disinhibitory networks. To determine D1-R overlap within these populations, we first crossed PV−, SOM−, 5HT3a−, and VIP-Cre lines with the D1-tdTomato line to produce interneuron specific double transgenic mice. To selectively label cortical interneuron subtypes, we then injected AAV-FLEX-EGFP into the PFC of each mouse line (Fig. 8A), a strategy that we and others have previously shown selectively labels distinct interneuron subtypes in the PFC (Kuhlman and Huang 2008; Marlin and Carter 2014; McGarry and Carter 2016; Anastasiades et al. 2018). Consistent with studies from other regions of cortex (Gonchar et al. 2007; Lee et al. 2010), we found that PV+ and SOM+ interneurons were distributed across layers 2–6, whereas 5HT3a+ and VIP+ interneurons were largely biased to superficial layers (Fig. 8A,B). This anatomical characterization provides insight into the laminar distributions of the main GABAergic interneuron populations in the mouse PFC. Given that overlap of D1+ and GAD+ is most prominent in superficial layers, these results suggest that D1-Rs are selectively expressed in VIP+ and/or 5HT3a+ interneurons.
Figure 8.
D1 receptors are expressed in a subpopulation of interneurons. (A) Confocal images of EGFP expression in the prelimbic PFC of PV-, SOM-, 5HT3a-, and VIP-Cre x D1-tdTomato double transgenic mouse lines injected with AAV-FLEX-EGFP virus. Scale bar = 100 μm. (B) Left, overlay of the position of all EGFP+ interneurons as a function of distance to the midline. Right, summary of the binned and averaged distribution of labeled PV+, SOM+, 5HT3a+, and VIP+ interneurons as a function of distance from the midline. Bin size = 25 μm. (C) Confocal images showing D1+ (red), EGFP+ (green), and merged image, showing overlap in VIP+ interneurons (bottom) but not PV+ interneurons (top), or SOM+ interneurons (middle). Scale bar = 50 μm. Asterisks indicate colabeled neurons. Images taken from layer 5 (PV+ and SOM+) and layer 2 (VIP+). (D) Left, distribution of the position of all D1+ EGFP+ colabeled interneurons as a function of distance to the midline. Right, summary of the binned and averaged distribution of D1+ EGFP+ colabeled interneurons as a function of distance from the midline. Bin size = 25 μm. (E) Left, summary of the percentage of D1+ neurons which are colabeled with EGFP (% D1, calculated as (D1+ EGFP+)/D1+) as a function of interneuron subtype. Right, similar but for antibody staining against PV+ and SOM+ interneurons. (F) Similar to (E), but for percentage of EGFP+ interneurons that are also D1+ (% EGFP, calculated as (D1+ EGFP+/EGFP+)) (left), or % of antibody-stained PV+ and SOM+ interneurons that were colabeled as D1+ (right). Values shown as mean ± SEM.
We next examined the colabeling of D1+ neurons within each interneuron population (Fig. 8C,D), using a similar analysis to that described for projection neurons above. Surprisingly, we observed minimal colabeling of D1+ neurons and either PV+ or SOM+ interneurons (Fig. 8C–E; % D1, calculated as % (D1+ EGFP+)/D1+): PV+ = 0.5 ± 0.1%, n = 3 mice; SOM+ = 1.5 ± 0.5%, n = 3 mice). In contrast, we observed substantial colabeling of D1+ neurons and both 5HT3a+ and VIP+ interneurons (Fig. 8C–E; % D1: 5HT3a+ = 5.3 ± 0.6%, n = 3 mice; VIP+ = 5.0 ± 0.2%, n = 3 mice). Plotting the distribution of dual-labeled cells as a function of distance to midline revealed pronounced 5HT3a+ and VIP+ colabeling in superficial layers (Fig. 8D). Furthermore, the percentage of D1+ VIP+ interneurons was similar to that of D1+ GAD+ cells in our initial pan-interneuron experiments (Fig. 2). Given that the VIP+ population is a major subset of 5HT3a+ interneurons (Rudy et al., 2011), these findings suggest VIP+ interneurons comprise the majority of D1+ interneurons in the PFC. Interestingly, quantifying the number of EGFP+ interneurons that are also D1+ (% EGFP, calculated as % (D1+ EGFP+)/EGFP+) showed that less than half of 5HT3a+ and VIP+ interneurons are D1+ (Fig. 8F; % EGFP: 5HT3a+ = 36.1 ± 3.2%, VIP+ = 39.5 ± 4.4%), while also confirming minimal colabeling of PV+ and SOM+ interneurons (Fig. 8F; % EGFP: PV+ = 1.9 ± 0.5%, SOM+ = 5.2 ± 1.8%). This was not an artifact of using Cre lines, as similar results were obtained with antibodies against PV and SOM (Fig. 8E,F; % (D1+ PV+)/PV+ = 4.1 ± 0.6%, n = 3 mice; % (D1+ SOM+)/SOM+ = 5.9 ± 1.0%, n = 3 mice). These findings indicate that D1-Rs are expressed in a subset of VIP+ interneurons, which are primarily located in superficial layers of PFC.
D1 Receptors Modulate a Subpopulation of VIP+ Interneurons
VIP+ interneurons are highly diverse, comprising multiple distinct morphological and electrophysiological subtypes (Kawaguchi and Kubota 1996; Miyoshi et al. 2010; Pronneke et al. 2015; He et al. 2016). To further characterize D1+ VIP+ interneurons, we next injected AAV-FLEX-EGFP into D1-tdTomato x VIP-Cre double transgenic mice, and performed targeted current-clamp recordings from cells in superficial layers (Fig. 9A). When considering all VIP+ interneurons, we observed many of the previously described firing patterns, including irregular-spiking (IS), non-fast-spiking (NFS), and fast-adapting (fAD) subtypes (Fig. 9A and Table 4) (Miyoshi et al. 2010). Interestingly, IS neurons were exclusively contained within the D1+ VIP+ population, forming a substantial proportion of D1+ VIP+ but not D1− VIP+ cells (Fig. 9B; D1+ VIP+: n = 9/14, D1− VIP+: n = 0/9). VIP+ interneurons that have IS firing properties typically have bipolar morphologies, coexpress calretinin (CR), and target other interneurons to mediate disinhibition (Acsady et al. 1996; Lee et al. 2013; He et al. 2016). Consistent with these findings, D1+ VIP+ interneurons had bipolar morphologies (Fig. 9A) and were frequently colabeled with calretinin (Fig. 9C; n = 3). The proportion of D1+ VIP+ interneurons that were CR+ (67 ± 6 %) was very similar to the proportion of D1+ VIP+ interneurons with IS firing properties (64% of total). These findings indicate that D1-Rs are particularly enriched in a specific subpopulation of VIP+ interneurons, which mediate disinhibition across cortex.
Figure 9.
A specific subclass of D1+ VIP+ interneurons is modulated by D1-Rs. (A) Left, representative morphology of a D1+ VIP+ interneuron, with soma and dendrites in red and axon in blue. Scale bar = 25 μm. Right, summary of responses to depolarizing and hyperpolarizing current steps for D1+ VIP+ and D1− VIP+ interneurons in superficial layers of prelimbic PFC. Intrinsic properties showing D1+ irregular-spiking (IS), D1− non-fast-spiking (NFS) and D1− fast-adapting (fAD) firing patterns. (B) Summary of percentage of D1+ VIP+ (red) and D1− VIP+ (black) interneurons that exhibit NFS, IS, or fAD firing patterns. (C) Left, confocal images from superficial layers of D1+ (red), VIP+ (green), calretinin antibody staining (CR, cyan), and merged image. Scale bar = 25 μm. Asterisks indicate triple-labeled neurons. Right, summary of percentages of labeled cells. (D) Firing properties of D1+ VIP+ interneurons in baseline (blue) and following wash-in of either SKF alone (red, left) or SKF with SCH (gray, right). (E) Summary of change in AP firing in D1+ VIP+ interneurons following wash-in of either SKF alone (red) or SKF with SCH (gray). Values shown as mean ± SEM. *P < 0.05. See also Table 4.
Table 4.
Intrinsic properties of PFC VIP+ interneurons
| Cell type | RMP (mV) | Rin (MΩ) | Sag (%) | Tau (ms) | AP height (mV) | Max Freq. (Hz) | AHP (mV) | AHP (ms) |
|---|---|---|---|---|---|---|---|---|
| IS (n = 9) | −65.5 ± 2.3 | 650.9 ± 72.3 | 1.5 ± 1.3 | 26.9 ± 3.1 | 64.3 ± 2.4 | 28.6 ± 4.3 | 8.8 ± 1.6 | 3.5 ± 0.1 |
| NFS (n = 12) | −68.2 ± 1.3 | 620.8 ± 48.3 | 0.8 ± 1.0 | 27.2 ± 3.1 | 63.0 ± 2.7 | 32.2 ± 2.3 | 8.0 ± 1.5 | 4.3 ± 0.2 |
| fAD (n = 2) | −69.1 | 702.6 | 2.7 | 29.3 | 58.6 | 13 | 3.0 | 4.5 |
Note: Resting membrane potential (RMP), input resistance (Rin), voltage sag due to h-current (Sag), membrane time constant (Tau), action potential (AP) height, maximum firing frequency (Max Freq.), afterhyperpolarization (AHP) amplitude, and AHP time to peak, for individual VIP+ interneuron populations in superficial layers of PFC.
Finally, we examined D1-R modulation of D1+ VIP+ interneurons, in the presence of synaptic blockers to prevent network activity. We found that wash-in of SKF-81 297 (10 μM) alone strongly enhanced the firing of these cells, whereas wash-in of SKF-81 297 in the presence of SCH-23 390 (10 μM) had no effect on firing (Fig. 9D,E; ∆AP: SKF = 3.8 ± 1.4, n = 9, SKF + SCH = 0.4 ± 0.6, n = 8; P = 0.04). Together, these findings indicate that D1-Rs also strongly enhance the firing properties of D1+ VIP+ interneurons.
Discussion
We have determined the cell- and layer-specific expression of D1 dopamine receptors in the mouse PFC. We found that D1-Rs robustly modulate subsets of pyramidal cells and GABAergic interneurons. We showed significant overlap between deep layer D1+ neurons and cells that project throughout the telencephalon. Additionally, we observed a subpopulation of superficial D1+ VIP+ GABAergic interneurons, which are known to inhibit other interneurons. Together, our results highlight the specificity with which D1-Rs exert influence on both excitatory and disinhibitory microcircuits in the mouse PFC.
Our data indicate that most D1+ neurons reside in L5 and L6, which parallel the increased density of both dopaminergic axon terminals and dopamine receptors in deeper layers of PFC (Berger et al. 1976; Santana et al. 2009; Van De Werd et al. 2010). Most D1+ neurons are also glutamatergic, consistent with ultrastructural observations, which indicate the presence of D1-Rs at glutamatergic presynaptic terminals and postsynaptic spines (Smiley et al. 1994; Paspalas and Goldman-Rakic 2005), where they may also function to regulate synaptic responses (Gao et al. 2001; Urban et al. 2002). Importantly, our work complements a recent study that observed presynaptic D1-R modulation in PFC and hippocampal, but not subcortical, inputs to PFC (Burke et al. 2018), suggesting preferential modulation of specific subnetworks by dopamine. Moreover, L5 and L6 D1+ neurons are modulated by D1-Rs, leading to enhanced AP firing, consistent with our previous work in juvenile mice (Seong and Carter 2012). However, while D1-Rs have been proposed to enhance persistent firing of L3 pyramidal cells in primates (Paspalas et al. 2013), they are conspicuously absent from L3 in mouse. This may represent differences in cortical architecture across species, with L3 functioning as the primary thalamorecipient layer in mouse (Collins et al., 2018). Alternatively, the enhancement in firing in superficial layers may be mediated by alternative mechanisms, for example, VIP-mediated disinhibition.
Defining the laminar boundaries in the PFC and other frontal cortices has been challenging, because landmarks like L4 are absent (Uylings et al. 2003). Our anatomical results revealed the distribution of numerous cell types in the PFC, extending on work in rats (Gabbott et al. 2005). Superficial layers (L1–3) contain most of the 5HT3a+ and VIP+ interneurons, which also populate superficial layers in sensory cortices (Gonchar et al. 2007; Lee et al. 2010). They are also enriched in CA neurons (Gabbott et al. 2005; Little and Carter 2013) and contain a high density of IT neurons (Wilson 1987; Otsuka and Kawaguchi 2011; Oswald et al. 2013; Anastasiades et al. 2018). L2 has a higher density of CA neurons, while L3 is defined by a dense band of thalamic axon (Ueta et al. 2013; Collins et al. 2018). Deep layers of PL can be delineated by the increased density of subcortical projection neurons (Molnar and Cheung 2006; Oswald et al. 2013; Harris and Shepherd 2015). Interestingly, L5 can be subdivided into 3 distinct sublayers based on the relative density and identity of PT neurons. PT neurons are less prevalent in L5a, which forms a narrow layer containing a second peak of CA neurons and a high density of cSTR cells; upper L5b has a high density of PT cells, including those projecting to thalamus; in contrast PT cells in lower L5b typically lack thalamic projections (Lorente de No 1992; Gabbott et al. 2005; Ueta et al. 2013; Collins et al. 2018). This analysis elaborates on recent studies (DeNardo et al. 2015; Clarkson et al. 2017) and provides clarity on the extent of L3 and sublayers of L5. Together, these findings highlight the complexity of the mouse PFC, detailing its laminar structure (Tables 1 and 2).
Using retrograde tracing, we identified L5 and L6 D1+ neurons as IT cells, which project to both ipsilateral and contralateral cortices (Mercer et al. 2005; Brown and Hestrin 2009; Morishima et al. 2011). Interestingly, we also observed a high percentage of deep layer D1+ neurons projecting laterally, toward the claustrum. Consistent with D1-R expression in CCL neurons, L6 D1+ neurons have inverted pyramidal morphologies (Bueno-Lopez et al. 1991; Mendizabal-Zubiaga et al. 2007). In frontal cortices, projections to claustrum are more prevalent (Atlan et al. 2017; Brown et al. 2017) and distributed more broadly across layers than those from sensory cortices (Smith and Alloway 2010), which are typically restricted to L6. Projections from frontal cortex to claustrum may play an important role in cognitive functions (Smith and Alloway 2014; White et al. 2018). Given that both dopamine and the claustrum are implicated in hallucinations and attention deficits (Goll et al. 2015; Brown et al. 2017), the expression of D1-Rs in CCL neurons may be significant for schizophrenia and ADHD (Goll et al. 2015; Grace 2016). In addition to D1+ CC and CCL neurons, we observed some D1+ CA neurons, which regulate feeding (Land et al. 2014). Finally, we observed a subset of D1+ CS neurons, consistent with IT neurons sending collaterals to iSTR and cSTR (Wilson 1987; Cowan and Wilson 1994). The high percentage of colabeled CC, CCL, and CS populations in deep layers can be explained by our AAVretro-Cre axon tracing experiments (Fig. 4), with projections to multiple targets yielding an effectual overlap > 100%. However, it should also be noted that in some instances we cannot account for all D1+ neurons in each layer. This is expected, as many PFC target structures extend along the rostrocaudal axis, making it challenging to ensure that each injection site overlaps with the entire PFC axonal field in a given target. Together, these findings suggest that D1-Rs impact a distributed IT network projecting to multiple targets, potentially leading to diverse effects on cognition and behavior.
Previous studies using D1-Cre mice indicate diffuse projections mediated by D1+ neurons, including cortical and subcortical targets (Han et al. 2017). Although the broad conclusion that D1-Rs are expressed in IT cells is supported by both studies, there are notable differences. In particular, we observed very few D1-expressing L6 CT neurons, and projections via the PT were rarely observed. One explanation comes from our in situ hybridization analysis, which shows varying levels of D1-R expression across a wide range of cells in PFC. Because Cre-dependent expression acts like a switch, projections from subpopulations of cells that express only low levels of D1-R could also contribute to axon profiles when using the D1-Cre mice. Therefore, if L6 CT cells express some D1-Rs at much lower levels than IT cells (Gaspar et al. 1995), their projection would still be labeled. However, it should be noted that D1-Rs do not modulate D1− neurons, suggesting those cells are not regulated (Seong and Carter 2012).
Our results indicate multiple populations of IT neurons exist within PFC, with distinct dopamine receptor expression profiles. The highest density of IT neurons is in L3, the only layer where D1-Rs were largely absent. Within L5, not all IT cells express D1-Rs, with L5 D1− CC neurons insensitive to D1-R agonists. One possibility is that these cells may be D3+ neurons, whose firing is suppressed by D3 receptors (Clarkson et al. 2017). Alternatively, D1-R expression may fluctuate within the IT cell class, either over the course of development, in response to activity, or salient stimuli (Brenhouse et al. 2008; Zhao et al. 2017). In the future, it will be interesting to assess the functional significance of individual IT neuron subclasses within PFC and determine the importance of dopamine modulation on these diverse populations of projection neurons (Otsuka and Kawaguchi 2011; Hirai et al. 2012; Ueta et al. 2013; Yamashita et al. 2013). Moreover, PT neurons are another class of D1− projection neurons whose physiology is influenced by D2 receptors (Gee et al. 2012). These findings highlight the importance of both cell type and layer in interpreting the impact of neuromodulators and may account for some of the heterogeneity that was initially observed for dopamine modulation in unlabeled neurons (Penit-Soria et al. 1987; Geijo-Barrientos and Pastore 1995; Gulledge and Jaffe 1998, 2001; Zhou and Hablitz 1999; Seamans and Yang 2004). They may also explain some of the varied effects of dopamine receptor agonists and antagonists on PFC-dependent behaviors (Floresco and Magyar 2006; St Onge et al. 2011; Jenni et al. 2017).
In addition to pyramidal cells, we observed that superficial layers also contain a population of D1+ GABAergic interneurons. Previous reports from primate PFC suggest that PV+ interneurons may express D1-Rs (Muly et al. 1998; Glausier et al. 2009), but the degree of colabeling reported varies widely (Le Moine and Gaspar 1998; Paspalas and Goldman-Rakic 2005; Tritsch and Sabatini 2012). Surprisingly, we observed minimal colabeling between D1 receptors and either PV+ or SOM+ interneurons, suggesting these inhibitory circuits are not directly modulated by D1-Rs. One possibility is that there are pronounced differences in D1-R expression between species. Another interesting possibility is that PV+ interneurons may instead express the D5 receptor, as observed in the striatum (Centonze et al. 2003; Oda et al. 2010; Tritsch and Sabatini 2012).
Instead, D1-Rs are primarily found in superficial VIP+ interneurons, which show irregular-spiking firing properties, coexpress calretinin, and are strongly modulated by D1-Rs, leading to enhanced AP firing. Throughout cortex, VIP+ interneurons mediate disinhibition by inhibiting SOM+ interneurons in the local circuit (Pfeffer et al. 2013; Pi et al. 2013; Karnani et al. 2016). In the PFC, recent studies highlight an important role for VIP+ activity in short-term memory tasks (Kamigaki and Dan 2017), consistent with previous modeling studies (Wang et al. 2004). Because dopamine levels increase during cognitive tasks (Watanabe et al. 1997; Phillips et al. 2004), our findings provide a mechanism linking elevated PFC dopamine with VIP+ interneuron activity through D1-Rs. Moreover, engaging disinhibitory circuits could explain why superficial networks are enhanced by D1-Rs, even though receptors are not prominent in pyramidal cells. Previous studies indicate that VIP+ interneurons are also under the control of other neuromodulators, including serotonin, acetylcholine, and noradrenaline (Beaulieu and Somogyi 1991; Smiley and Goldman-Rakic 1996; Paspalas and Papadopoulos 1999; Lee et al. 2010; Letzkus et al. 2011; Rudy et al. 2011). Our findings provide further support for the idea that VIP+-mediated disinhibition may be a common circuit mechanism utilized by many neuromodulatory systems (Wester and McBain 2014).
Together, our data provide a detailed overview of D1-R expression in both excitatory and inhibitory neurons of the mouse PFC. By increasing the firing of L5 D1+ IT cells projecting across the corpus callosum, D1-R activation may increase communication between hemispheres, which plays an important role in delay period activity (Li et al. 2016). Activating these neurons may also regulate activity within the local network, where CC neurons make contacts onto other CC neurons, as well as CT and PT cells (Mercer et al. 2005; Brown and Hestrin 2009). Moreover, by increasing the activity of VIP+ interneurons, D1-Rs can disinhibit the local network, which is known to play an important role in cortical function, including within the PFC (Wang et al. 2004; Garcia Del Molino et al. 2017; Kamigaki and Dan 2017). Thus, D1-Rs can activate both excitatory circuits in deep layers and disinhibitory circuits in superficial layers, both of which should bias the network toward increased excitation and may play an important role in gating processes such as selective attention (Kamigaki 2018). These findings help explain the role of D1-Rs in the PFC, with important implications for understanding dopamine modulation in cognitive processing and related neuropsychiatric disorders.
Supplementary Material
Funding
This work was supported by National Institute of Health NIH R01 MH085974 (to A.G.C).
Notes
The authors thank members of the Carter lab for helpful discussions and comments on the manuscript. We thank Susan Sheng, Mian Hou, Virginia Garcia-Marin, and Claudia Farb for help with immunocytochemistry and histology. Conflict of Interest: None declared.
References
- Acsady L, Gorcs TJ, Freund TF. 1996. Different populations of vasoactive intestinal polypeptide-immunoreactive interneurons are specialized to control pyramidal cells or interneurons in the hippocampus. Neuroscience. 73:317–334. [DOI] [PubMed] [Google Scholar]
- Ade KK, Wan Y, Chen M, Gloss B, Calakos N. 2011. An improved BAC transgenic fluorescent reporter line for sensitive and specific identification of striatonigral medium spiny neurons. Front Syst Neurosci. 5:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiades PG, Butt SJ. 2011. Decoding the transcriptional basis for GABAergic interneuron diversity in the mouse neocortex. Eur J Neurosci. 34:1542–1552. [DOI] [PubMed] [Google Scholar]
- Anastasiades PG, Marlin JJ, Carter AG. 2018. Cell-type specificity of callosally evoked excitation and feedforward inhibition in the prefrontal cortex. Cell Rep. 22:679–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiades PG, Marques-Smith A, Lyngholm D, Lickiss T, Raffiq S, Katzel D, Miesenbock G, Butt SJ. 2016. GABAergic interneurons form transient layer-specific circuits in early postnatal neocortex. Nat Commun. 7:10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlan G, Terem A, Peretz-Rivlin N, Groysman M, Citri A. 2017. Mapping synaptic cortico-claustral connectivity in the mouse. J Comp Neurol. 525:1381–1402. [DOI] [PubMed] [Google Scholar]
- Baker A, Kalmbach B, Morishima M, Kim J, Juavinett A, Li N, Dembrow N. 2018. Specialized subpopulations of deep-layer pyramidal neurons in the neocortex: bridging cellular properties to functional consequences. J Neurosci. 38:5441–5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu C, Somogyi P. 1991. Enrichment of cholinergic synaptic terminals on GABAergic neurons and coexistence of immunoreactive GABA and choline acetyltransferase in the same synaptic terminals in the striate cortex of the cat. J Comp Neurol. 304:666–680. [DOI] [PubMed] [Google Scholar]
- Berger B, Thierry AM, Tassin JP, Moyne MA. 1976. Dopaminergic innervation of the rat prefrontal cortex: a fluorescence histochemical study. Brain Res. 106:133–145. [DOI] [PubMed] [Google Scholar]
- Brenhouse HC, Sonntag KC, Andersen SL. 2008. Transient D1 dopamine receptor expression on prefrontal cortex projection neurons: relationship to enhanced motivational salience of drug cues in adolescence. J Neurosci. 28:2375–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SP, Hestrin S. 2009. Intracortical circuits of pyramidal neurons reflect their long-range axonal targets. Nature. 457:1133–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SP, Mathur BN, Olsen SR, Luppi PH, Bickford ME, Citri A. 2017. New breakthroughs in understanding the role of functional interactions between the neocortex and the claustrum. J Neurosci. 37:10877–10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno-Lopez JL, Reblet C, Lopez-Medina A, Gomez-Urquijo SM, Grandes P, Gondra J, Hennequet L. 1991. Targets and laminar distribution of projection neurons with ‘inverted’ morphology in rabbit cortex. Eur J Neurosci. 3:415–430. [DOI] [PubMed] [Google Scholar]
- Burke KJ Jr., Keeshen CM, Bender KJ. 2018. Two forms of synaptic depression produced by differential neuromodulation of presynaptic calcium channels. Neuron. 99:969–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt SJ, Fuccillo M, Nery S, Noctor S, Kriegstein A, Corbin JG, Fishell G. 2005. The temporal and spatial origins of cortical interneurons predict their physiological subtype. Neuron. 48:591–604. [DOI] [PubMed] [Google Scholar]
- Cauli B, Audinat E, Lambolez B, Angulo MC, Ropert N, Tsuzuki K, Hestrin S, Rossier J. 1997. Molecular and physiological diversity of cortical nonpyramidal cells. J Neurosci. 17:3894–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centonze D, Grande C, Usiello A, Gubellini P, Erbs E, Martin AB, Pisani A, Tognazzi N, Bernardi G, Moratalla R, et al. 2003. Receptor subtypes involved in the presynaptic and postsynaptic actions of dopamine on striatal interneurons. J Neurosci. 23:6245–6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalifoux JR, Carter AG. 2010. GABAB receptors modulate NMDA receptor calcium signals in dendritic spines. Neuron. 66:101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson RL, Liptak AT, Gee SM, Sohal VS, Bender KJ. 2017. D3 receptors regulate excitability in a unique class of prefrontal pyramidal cells. J Neurosci. 37:5846–5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins DP, Anastasiades PG, Marlin JJ, Carter AG. 2018. Reciprocal circuits linking the prefrontal cortex with dorsal and ventral thalamic nuclei. Neuron. 98:366–379 e364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan RL, Wilson CJ. 1994. Spontaneous firing patterns and axonal projections of single corticostriatal neurons in the rat medial agranular cortex. J Neurophysiol. 71:17–32. [DOI] [PubMed] [Google Scholar]
- Dembrow NC, Chitwood RA, Johnston D. 2010. Projection-specific neuromodulation of medial prefrontal cortex neurons. J Neurosci. 30:16922–16937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNardo LA, Berns DS, DeLoach K, Luo L. 2015. Connectivity of mouse somatosensory and prefrontal cortex examined with trans-synaptic tracing. Nat Neurosci. 18:1687–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floresco SB, Magyar O. 2006. Mesocortical dopamine modulation of executive functions: beyond working memory. Psychopharmacology (Berl). 188:567–585. [DOI] [PubMed] [Google Scholar]
- Froemke RC. 2015. Plasticity of cortical excitatory-inhibitory balance. Annu Rev Neurosci. 38:195–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuster JM. 2000. Prefrontal neurons in networks of executive memory. Brain Res Bull. 52:331–336. [DOI] [PubMed] [Google Scholar]
- Gabbott PL, Warner TA, Jays PR, Salway P, Busby SJ. 2005. Prefrontal cortex in the rat: projections to subcortical autonomic, motor, and limbic centers. J Comp Neurol. 492:145–177. [DOI] [PubMed] [Google Scholar]
- Gao WJ, Krimer LS, Goldman-Rakic PS. 2001. Presynaptic regulation of recurrent excitation by D1 receptors in prefrontal circuits. Proc Natl Acad Sci U S A. 98:295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Del Molino LC, Yang GR, Mejias JF, Wang XJ. 2017. Paradoxical response reversal of top-down modulation in cortical circuits with three interneuron types. Elife. 6:e29742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar P, Bloch B, Le Moine C. 1995. D1 and D2 receptor gene expression in the rat frontal cortex: cellular localization in different classes of efferent neurons. Eur J Neurosci. 7:1050–1063. [DOI] [PubMed] [Google Scholar]
- Gee S, Ellwood I, Patel T, Luongo F, Deisseroth K, Sohal VS. 2012. Synaptic activity unmasks dopamine D2 receptor modulation of a specific class of layer V pyramidal neurons in prefrontal cortex. J Neurosci. 32:4959–4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geijo-Barrientos E, Pastore C. 1995. The effects of dopamine on the subthreshold electrophysiological responses of rat prefrontal cortex neurons in vitro. Eur J Neurosci. 7:358–366. [DOI] [PubMed] [Google Scholar]
- Gerfen CR, Paletzki R, Heintz N. 2013. GENSAT BAC cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits. Neuron. 80:1368–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glausier JR, Khan ZU, Muly EC. 2009. Dopamine D1 and D5 receptors are localized to discrete populations of interneurons in primate prefrontal cortex. Cereb Cortex. 19:1820–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll Y, Atlan G, Citri A. 2015. Attention: the claustrum. Trends Neurosci. 38:486–495. [DOI] [PubMed] [Google Scholar]
- Gonchar Y, Wang Q, Burkhalter A. 2007. Multiple distinct subtypes of GABAergic neurons in mouse visual cortex identified by triple immunostaining. Front Neuroanat. 1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Islas C, Hablitz JJ. 2001. Dopamine inhibition of evoked IPSCs in rat prefrontal cortex. J Neurophysiol. 86:2911–2918. [DOI] [PubMed] [Google Scholar]
- Gorelova N, Seamans JK, Yang CR. 2002. Mechanisms of dopamine activation of fast-spiking interneurons that exert inhibition in rat prefrontal cortex. J Neurophysiol. 88:3150–3166. [DOI] [PubMed] [Google Scholar]
- Grace AA. 2016. Dysregulation of the dopamine system in the pathophysiology of schizophrenia and depression. Nat Rev Neurosci. 17:524–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulledge AT, Jaffe DB. 1998. Dopamine decreases the excitability of layer V pyramidal cells in the rat prefrontal cortex. J Neurosci. 18:9139–9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulledge AT, Jaffe DB. 2001. Multiple effects of dopamine on layer V pyramidal cell excitability in rat prefrontal cortex. J Neurophysiol. 86:586–595. [DOI] [PubMed] [Google Scholar]
- Han SW, Kim YC, Narayanan NS. 2017. Projection targets of medial frontal D1DR-expressing neurons. Neurosci Lett. 655:166–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KD, Shepherd GM. 2015. The neocortical circuit: themes and variations. Nat Neurosci. 18:170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattox AM, Nelson SB. 2007. Layer V neurons in mouse cortex projecting to different targets have distinct physiological properties. J Neurophysiol. 98:3330–3340. [DOI] [PubMed] [Google Scholar]
- He M, Tucciarone J, Lee S, Nigro MJ, Kim Y, Levine JM, Kelly SM, Krugikov I, Wu P, Chen Y, et al. 2016. Strategies and tools for combinatorial targeting of gabaergic neurons in mouse cerebral cortex. Neuron. 91:1228–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henze DA, Gonzalez-Burgos GR, Urban NN, Lewis DA, Barrionuevo G. 2000. Dopamine increases excitability of pyramidal neurons in primate prefrontal cortex. J Neurophysiol. 84:2799–2809. [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S. 2005. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 3:e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai Y, Morishima M, Karube F, Kawaguchi Y. 2012. Specialized cortical subnetworks differentially connect frontal cortex to parahippocampal areas. J Neurosci. 32:1898–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerder-Suabedissen A, Hayashi S, Upton L, Nolan Z, Casas-Torremocha D, Grant E, Viswanathan S, Kanold PO, Clasca F, Kim Y, et al. 2018. Subset of cortical layer 6b neurons selectively innervates higher order thalamic nuclei in mice. Cereb Cortex. 28:1882–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenni NL, Larkin JD, Floresco SB. 2017. Prefrontal dopamine D1 and D2 receptors regulate dissociable aspects of decision making via distinct ventral striatal and amygdalar circuits. J Neurosci. 37:6200–6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamigaki T. 2018. Prefrontal circuit organization for executive control. Neurosci Res. 10.1016/j.neures.2018.08.017 [DOI] [PubMed] [Google Scholar]
- Kamigaki T, Dan Y. 2017. Delay activity of specific prefrontal interneuron subtypes modulates memory-guided behavior. Nat Neurosci. 20:854–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani MM, Jackson J, Ayzenshtat I, Tucciarone J, Manoocheri K, Snider WG, Yuste R. 2016. Cooperative subnetworks of molecularly similar interneurons in mouse neocortex. Neuron. 90:86–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunakaran S, Chowdhury A, Donato F, Quairiaux C, Michel CM, Caroni P. 2016. PV plasticity sustained through D1/5 dopamine signaling required for long-term memory consolidation. Nat Neurosci. 19:454–464. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kubota Y. 1996. Physiological and morphological identification of somatostatin- or vasoactive intestinal polypeptide-containing cells among GABAergic cell subtypes in rat frontal cortex. J Neurosci. 16:2701–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroner S, Krimer LS, Lewis DA, Barrionuevo G. 2007. Dopamine increases inhibition in the monkey dorsolateral prefrontal cortex through cell type-specific modulation of interneurons. Cereb Cortex. 17:1020–1032. [DOI] [PubMed] [Google Scholar]
- Kruglikov I, Rudy B. 2008. Perisomatic GABA release and thalamocortical integration onto neocortical excitatory cells are regulated by neuromodulators. Neuron. 58:911–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Kawaguchi Y. 1994. Three classes of GABAergic interneurons in neocortex and neostriatum. Jpn J Physiol. 44(Suppl 2):S145–148. [PubMed] [Google Scholar]
- Kuhlman SJ, Huang ZJ. 2008. High-resolution labeling and functional manipulation of specific neuron types in mouse brain by Cre-activated viral gene expression. PLoS One. 3:e2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land BB, Narayanan NS, Liu RJ, Gianessi CA, Brayton CE, Grimaldi DM, Sarhan M, Guarnieri DJ, Deisseroth K, Aghajanian GK, et al. 2014. Medial prefrontal D1 dopamine neurons control food intake. Nat Neurosci. 17:248–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Moine C, Gaspar P. 1998. Subpopulations of cortical GABAergic interneurons differ by their expression of D1 and D2 dopamine receptor subtypes. Brain Res Mol Brain Res. 58:231–236. [DOI] [PubMed] [Google Scholar]
- Lee S, Hjerling-Leffler J, Zagha E, Fishell G, Rudy B. 2010. The largest group of superficial neocortical GABAergic interneurons expresses ionotropic serotonin receptors. J Neurosci. 30:16796–16808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kruglikov I, Huang ZJ, Fishell G, Rudy B. 2013. A disinhibitory circuit mediates motor integration in the somatosensory cortex. Nat Neurosci. 16:1662–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzkus JJ, Wolff SB, Meyer EM, Tovote P, Courtin J, Herry C, Luthi A. 2011. A disinhibitory microcircuit for associative fear learning in the auditory cortex. Nature. 480:331–335. [DOI] [PubMed] [Google Scholar]
- Li N, Daie K, Svoboda K, Druckmann S. 2016. Robust neuronal dynamics in premotor cortex during motor planning. Nature. 532:459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidow MS, Goldman-Rakic PS, Gallager DW, Rakic P. 1991. Distribution of dopaminergic receptors in the primate cerebral cortex: quantitative autoradiographic analysis using [3H]raclopride, [3H]spiperone and [3H]SCH23390. Neuroscience. 40:657–671. [DOI] [PubMed] [Google Scholar]
- Little JP, Carter AG. 2013. Synaptic mechanisms underlying strong reciprocal connectivity between the medial prefrontal cortex and basolateral amygdala. J Neurosci. 33:15333–15342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorente de No R. 1992. The cerebral cortex of the mouse (a first contribution—the “acoustic” cortex). Somatosens Mot Res. 9:3–36. [DOI] [PubMed] [Google Scholar]
- Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. 2004. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 5:793–807. [DOI] [PubMed] [Google Scholar]
- Marlin JJ, Carter AG. 2014. GABA-A receptor inhibition of local calcium signaling in spines and dendrites. J Neurosci. 34:15898–15911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry LM, Carter AG. 2016. Inhibitory gating of basolateral amygdala inputs to the prefrontal cortex. J Neurosci. 36:9391–9406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendizabal-Zubiaga JL, Reblet C, Bueno-Lopez JL. 2007. The underside of the cerebral cortex: layer V/VI spiny inverted neurons. J Anat. 211:223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer A, West DC, Morris OT, Kirchhecker S, Kerkhoff JE, Thomson AM. 2005. Excitatory connections made by presynaptic cortico-cortical pyramidal cells in layer 6 of the neocortex. Cereb Cortex. 15:1485–1496. [DOI] [PubMed] [Google Scholar]
- Miller EK, Cohen JD. 2001. An integrative theory of prefrontal cortex function. Annu Rev Neurosci. 24:167–202. [DOI] [PubMed] [Google Scholar]
- Miyoshi G, Hjerling-Leffler J, Karayannis T, Sousa VH, Butt SJ, Battiste J, Johnson JE, Machold RP, Fishell G. 2010. Genetic fate mapping reveals that the caudal ganglionic eminence produces a large and diverse population of superficial cortical interneurons. J Neurosci. 30:1582–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar Z, Cheung AF. 2006. Towards the classification of subpopulations of layer V pyramidal projection neurons. Neurosci Res. 55:105–115. [DOI] [PubMed] [Google Scholar]
- Morishima M, Morita K, Kubota Y, Kawaguchi Y. 2011. Highly differentiated projection-specific cortical subnetworks. J Neurosci. 31:10380–10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrzljak L, Bergson C, Pappy M, Huff R, Levenson R, Goldman-Rakic PS. 1996. Localization of dopamine D4 receptors in GABAergic neurons of the primate brain. Nature. 381:245–248. [DOI] [PubMed] [Google Scholar]
- Muly EC 3rd, Szigeti K, Goldman-Rakic PS. 1998. D1 receptor in interneurons of macaque prefrontal cortex: distribution and subcellular localization. J Neurosci. 18:10553–10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan M, Jang HJ, Park M, Miller EM, Cox J, Taliaferro JP, Parker NF, Bhave V, Hur H, Liang Y, et al. 2017. Combined social and spatial coding in a descending projection from the prefrontal cortex. Cell. 171:1663–1677 e1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama H, Ibanez-Tallon I, Heintz N. 2018. Cell-type-specific contributions of medial prefrontal neurons to flexible behaviors. J Neurosci. 38:4490–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlaender M, de Kock CP, Bruno RM, Ramirez A, Meyer HS, Dercksen VJ, Helmstaedter M, Sakmann B. 2012. Cell type-specific three-dimensional structure of thalamocortical circuits in a column of rat vibrissal cortex. Cereb Cortex. 22:2375–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda S, Funato H, Adachi-Akahane S, Ito M, Okada A, Igarashi H, Yokofujita J, Kuroda M. 2010. Dopamine D5 receptor immunoreactivity is differentially distributed in GABAergic interneurons and pyramidal cells in the rat medial prefrontal cortex. Brain Res. 1329:89–102. [DOI] [PubMed] [Google Scholar]
- St Onge JR, Abhari H, Floresco SB. 2011. Dissociable contributions by prefrontal D1 and D2 receptors to risk-based decision making. J Neurosci. 31:8625–8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald MJ, Tantirigama ML, Sonntag I, Hughes SM, Empson RM. 2013. Diversity of layer 5 projection neurons in the mouse motor cortex. Front Cell Neurosci. 7:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otis JM, Namboodiri VM, Matan AM, Voets ES, Mohorn EP, Kosyk O, McHenry JA, Robinson JE, Resendez SL, Rossi MA, et al. 2017. Prefrontal cortex output circuits guide reward seeking through divergent cue encoding. Nature. 543:103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka T, Kawaguchi Y. 2011. Cell diversity and connection specificity between callosal projection neurons in the frontal cortex. J Neurosci. 31:3862–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paspalas CD, Goldman-Rakic PS. 2005. Presynaptic D1 dopamine receptors in primate prefrontal cortex: target-specific expression in the glutamatergic synapse. J Neurosci. 25:1260–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paspalas CD, Papadopoulos GC. 1999. Noradrenergic innervation of peptidergic interneurons in the rat visual cortex. Cereb Cortex. 9:844–853. [DOI] [PubMed] [Google Scholar]
- Paspalas CD, Wang M, Arnsten AF. 2013. Constellation of HCN channels and cAMP regulating proteins in dendritic spines of the primate prefrontal cortex: potential substrate for working memory deficits in schizophrenia. Cereb Cortex. 23:1643–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penit-Soria J, Audinat E, Crepel F. 1987. Excitation of rat prefrontal cortical neurons by dopamine: an in vitro electrophysiological study. Brain Res. 425:263–274. [DOI] [PubMed] [Google Scholar]
- Petilla Interneuron Nomenclature, Ascoli GA, Alonso-Nanclares L, Anderson SA, Barrionuevo G, Benavides-Piccione R, Burkhalter A, Buzsaki G, Cauli B, Defelipe J, Fairen A, et al. 2008. Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat Rev Neurosci. 9:557–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer CK, Xue M, He M, Huang ZJ, Scanziani M. 2013. Inhibition of inhibition in visual cortex: the logic of connections between molecularly distinct interneurons. Nat Neurosci. 16:1068–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AG, Ahn S, Floresco SB. 2004. Magnitude of dopamine release in medial prefrontal cortex predicts accuracy of memory on a delayed response task. J Neurosci. 24:547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi HJ, Hangya B, Kvitsiani D, Sanders JI, Huang ZJ, Kepecs A. 2013. Cortical interneurons that specialize in disinhibitory control. Nature. 503:521–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronneke A, Scheuer B, Wagener RJ, Mock M, Witte M, Staiger JF. 2015. Characterizing VIP neurons in the barrel cortex of VIPcre/tdTomato mice reveals layer-specific differences. Cereb Cortex. 25:4854–4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudy B, Fishell G, Lee S, Hjerling-Leffler J. 2011. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev Neurobiol. 71:45–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santana N, Mengod G, Artigas F. 2009. Quantitative analysis of the expression of dopamine D1 and D2 receptors in pyramidal and GABAergic neurons of the rat prefrontal cortex. Cereb Cortex. 19:849–860. [DOI] [PubMed] [Google Scholar]
- Sawaguchi T, Goldman-Rakic PS. 1994. The role of D1-dopamine receptor in working memory: local injections of dopamine antagonists into the prefrontal cortex of rhesus monkeys performing an oculomotor delayed-response task. J Neurophysiol. 71:515–528. [DOI] [PubMed] [Google Scholar]
- Seamans JK, Floresco SB, Phillips AG. 1998. D1 receptor modulation of hippocampal-prefrontal cortical circuits integrating spatial memory with executive functions in the rat. J Neurosci. 18:1613–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seamans JK, Yang CR. 2004. The principal features and mechanisms of dopamine modulation in the prefrontal cortex. Prog Neurobiol. 74:1–58. [DOI] [PubMed] [Google Scholar]
- Seong HJ, Carter AG. 2012. D1 receptor modulation of action potential firing in a subpopulation of layer 5 pyramidal neurons in the prefrontal cortex. J Neurosci. 32:10516–10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiley JF, Goldman-Rakic PS. 1996. Serotonergic axons in monkey prefrontal cerebral cortex synapse predominantly on interneurons as demonstrated by serial section electron microscopy. J Comp Neurol. 367:431–443. [DOI] [PubMed] [Google Scholar]
- Smiley JF, Levey AI, Ciliax BJ, Goldman-Rakic PS. 1994. D1 dopamine receptor immunoreactivity in human and monkey cerebral cortex: predominant and extrasynaptic localization in dendritic spines. Proc Natl Acad Sci U S A. 91:5720–5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JB, Alloway KD. 2010. Functional specificity of claustrum connections in the rat: interhemispheric communication between specific parts of motor cortex. J Neurosci. 30:16832–16844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JB, Alloway KD. 2014. Interhemispheric claustral circuits coordinate sensory and motor cortical areas that regulate exploratory behaviors. Front Syst Neurosci. 8:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Fu Y, Lu J, Lin Y, et al. 2011. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 71:995–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tervo DG, Hwang BY, Viswanathan S, Gaj T, Lavzin M, Ritola KD, Lindo S, Michael S, Kuleshova E, Ojala D, et al. 2016. A designer AAV variant permits efficient retrograde access to projection neurons. Neuron. 92:372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson AM. 2010. Neocortical layer 6, a review. Front Neuroanat. 4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers SK, Hestrin S. 2008. D1-like dopamine receptor activation modulates GABAergic inhibition but not electrical coupling between neocortical fast-spiking interneurons. J Neurosci. 28:2633–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritsch NX, Sabatini BL. 2012. Dopaminergic modulation of synaptic transmission in cortex and striatum. Neuron. 76:33–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueta Y, Hirai Y, Otsuka T, Kawaguchi Y. 2013. Direction- and distance-dependent interareal connectivity of pyramidal cell subpopulations in the rat frontal cortex. Front Neural Circuits. 7:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban NN, Gonzalez-Burgos G, Henze DA, Lewis DA, Barrionuevo G. 2002. Selective reduction by dopamine of excitatory synaptic inputs to pyramidal neurons in primate prefrontal cortex. J Physiol. 539:707–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uylings HB, Groenewegen HJ, Kolb B. 2003. Do rats have a prefrontal cortex? Behav Brain Res. 146:3–17. [DOI] [PubMed] [Google Scholar]
- Van De Werd HJ, Rajkowska G, Evers P, Uylings HB. 2010. Cytoarchitectonic and chemoarchitectonic characterization of the prefrontal cortical areas in the mouse. Brain Struct Funct. 214:339–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent SL, Khan Y, Benes FM. 1993. Cellular distribution of dopamine D1 and D2 receptors in rat medial prefrontal cortex. J Neurosci. 13:2551–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, Luo Y. 2012. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 14:22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Tegner J, Constantinidis C, Goldman-Rakic PS. 2004. Division of labor among distinct subtypes of inhibitory neurons in a cortical microcircuit of working memory. Proc Natl Acad Sci U S A. 101:1368–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Kodama T, Hikosaka K. 1997. Increase of extracellular dopamine in primate prefrontal cortex during a working memory task. J Neurophysiol. 78:2795–2798. [DOI] [PubMed] [Google Scholar]
- Wearne S.L., Rodriguez A., Ehlenberger D.B., Rocher A.B., Henderson S.C., and Hof P.R. (2005). New techniques for imaging, digitization and analysis of three-dimensional neural morphology on multiple scales. Neuroscience 136, 661–680. [DOI] [PubMed] [Google Scholar]
- Weiner DM, Levey AI, Sunahara RK, Niznik HB, O’Dowd BF, Seeman P, Brann MR. 1991. D1 and D2 dopamine receptor mRNA in rat brain. Proc Natl Acad Sci U S A. 88:1859–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wester JC, McBain CJ. 2014. Behavioral state-dependent modulation of distinct interneuron subtypes and consequences for circuit function. Curr Opin Neurobiol. 29:118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MG, Panicker M, Mu C, Carter AM, Roberts BM, Dharmasri PA, Mathur BN. 2018. Anterior cingulate cortex input to the claustrum is required for top-down action control. Cell Rep. 22:84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GV, Goldman-Rakic PS. 1995. Modulation of memory fields by dopamine D1 receptors in prefrontal cortex. Nature. 376:572–575. [DOI] [PubMed] [Google Scholar]
- Wilson CJ. 1987. Morphology and synaptic connections of crossed corticostriatal neurons in the rat. J Comp Neurol. 263:567–580. [DOI] [PubMed] [Google Scholar]
- Xu X, Roby KD, Callaway EM. 2010. Immunochemical characterization of inhibitory mouse cortical neurons: three chemically distinct classes of inhibitory cells. J Comp Neurol. 518:389–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Sudhof TC. 2013. A neural circuit for memory specificity and generalization. Science. 339:1290–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, Pala A, Pedrido L, Kremer Y, Welker E, Petersen CC. 2013. Membrane potential dynamics of neocortical projection neurons driving target-specific signals. Neuron. 80:1477–1490. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhang J, Yang H, Cui D, Song J, Ma Q, Luan W, Lai B, Ma L, Chen M, et al. 2017. Memory retrieval in addiction: a role for miR-105-mediated regulation of D1 receptors in mPFC neurons projecting to the basolateral amygdala. BMC Biol. 15:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FM, Hablitz JJ. 1999. Dopamine modulation of membrane and synaptic properties of interneurons in rat cerebral cortex. J Neurophysiol. 81:967–976. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.