Abstract
The location of GABAergic synapses on dendrites is likely key for neuronal integration. In particular, inhibitory inputs on dendritic spines could serve to selectively veto or modulate individual excitatory inputs, greatly expanding the computational power of individual neurons. To investigate this, we have undertaken a combined functional, molecular, and ultrastructural mapping of the location of GABAergic inputs onto dendrites of pyramidal neurons from upper layers of juvenile mouse somatosensory cortex. Using two-photon uncaging of GABA, intracellular labeling with gerphyrin intrabodies, and focused ion beam milling with scanning electron microscopy, we find that most (96–98%) spines lack GABAergic synapses, although they still display GABAergic responses, potentially due to extrasynaptic GABA receptors. We conclude that GABAergic inputs, in practice, contact dendritic shafts and likely control clusters of excitatory inputs, defining functional zones on dendrites.
Keywords: gerphyrin, intrabodies, two-photon, uncaging, focused ion beam milling and scanning electron microscopy, 3D electron microscopy
Introduction
Synaptic inhibition is a critical element of neural circuits and is mostly mediated by different types of GABAergic interneurons (Fairen et al. 1984; Houser et al. 1984). Interneuron subtypes synapse on different neuronal compartments of pyramidal cells. In general, three general groups of GABAergic neurons can be distinguished: Axo-dendritic cells or cells whose axons form synapses primarily with dendrites (shafts and spines); Axo-somatodendritic cells or cells whose axons form multiple synapses with both dendrites and the somata, but with a variable preference for somata or dendrites; and Axo-axonic cells, or chandelier cells whose axons synapse with the axon initial segment. In addition, there are a variety of types of axo-dendritic cells and axo-somatodendritic cells, with diverse molecular characteristics and patterns of connection (both between the cells themselves and with the pyramidal neurons) (Ascoli et al. 2008). This subcellular specificity, which must be difficult to achieve developmentally, likely has a functional significance (Atallah et al. 2012; Woodruff et al. 2010; Gentet et al. 2012). However it is still unclear what is the effect of selectively inhibiting particular neuronal subcompartments. In particular, dendritic inhibition has been traditionally assumed to target dendritic shafts (Hersch and White 1981), yet there have been also reports in the literature of symmetric synapses (SSs) on dendritic spines (Peters et al. 1991; for a recent review see Kubota et al. 2016). These observations led to the hypothesis that individual spines could implement Boolean logical operations with their inputs (Shepherd and Brayton 1987; Koch et al. 1983). A spine inhibition, as “point inhibition”, could selectively silence or modulate the excitatory input to that same spine, leaving inputs onto other spines intact. On the other hand, dendritic shaft inhibition, as “regional inhibition”, could enable inhibitory input to influence and silence multiple spines simultaneously, and the spatial position of multiple inhibitory synapses may help create various silencing zones (Bloss et al. 2016; Prakash et al. 2012; Gidon and Segev 2012).
Recent studies have reexamined the existence of GABAergic inputs on spines, using fluorescent synaptic markers (Chen et al. 2012; Villa et al. 2016), or their functional role by glutamate and GABA uncaging experiments (Chiu et al. 2013). In the Chen et al. study, GABAergic synapses were identified by labeling of gephyrin, a marker of inhibitory synapses that tethers synaptic GABAA receptors (Tyagarajan and Fritschy 2014; Chen et al. 2012). With this method, the authors measured the distribution of inhibitory synapses with combined fluorescence and electron microscopic imaging, concluding that 30–43% of inhibitory synapses were located on spines and 10–16% of spines had inhibitory synapses. These numbers were significantly larger than estimates from the previous literature and implied that spine inhibition maybe quite significant and have a major functional role. Indeed, the Chiu et al. study reported significant effect of activating GABAergic receptors during single spine glutamate uncaging (Chen et al. 2012).
To understand inhibitory innervation of spines in more detail, we have studied neocortical dendrites of pyramidal neurons from postnatal (P14) juvenile mice. To map GABAergic synapses, we used two-photon uncaging of RuBi-GABA, intrabody labeling of gephryin, and combined focused ion beam milling and scanning electron microscopy (FIB/SEM). Our functional, molecular, and ultrastructural datasets agree well with each other and reveal that only a very small number (2–4%) of spines have GABAergic synapses, even though most spines respond to GABA, presumably through extrasynaptic receptors. Our results validate the regional model of dendritic inhibition, demonstrating the strong preponderance of GABAergic synapse on dendritic shafts and indicating that spines with GABAergic synapses are very rare, at least in our samples of upper layer neurons from juvenile somatosensory cortex.
Materials and Methods
Animals
All procedures involving animals were in accordance with the US National Institutes of Health Guide for the care and use of laboratory animals and approved by the Institutional Animal Care and Use Committees of Columbia University, Animal Care and Use Review Office from Army Research Office, the European Community Directive 2010/63/EU, and the local ethics committee of the Spanish National Research Council.
Slice Preparation and Electrophysiology
About 300-μm thick coronal slices from visual cortex of 14-day-old CD-1 mice were prepared using a Leica VT1000-S vibratome with cutting solution containing (in mM): 27 NaHCO3, 1.5 NaH2PO4, 222 Sucrose, 2.6 KCl, 3 MgSO4, and 0.5 CaCl2. The slices were incubated at 32 °C for 30 min and then kept at room temperature for at least 30 min before transferring them to the recording chamber. The recording chamber was bathed in ACSF (pH 7.4), kept at room temperature and saturated with 95% O2 and 5% CO2, containing (in mM): 126 NaCl, 3 KCl, 2 MgSO4, 2 CaCl2, 1.1 NaH2PO4, 26 NaHCO3, and 10 dextrose. Unless indicated, neurons were held at 0 mV membrane potential. Whole-cell electrodes (3–5 MΩ) were used. All voltage clamp recordings were done with intracellular solution (pH 7.4), containing (in mM): 132 Cs-methanesulfonate, 3 CsCl, 10 HEPES, 4 Mg-ATP, 0.4 Na-GTP, 10 Na2-phosphocreatine, and 0.1 Alexa Fluor 594.
We performed recordings from layer 2/3 pyramidal cells using MultiClamp 700B (Molecular Devices) amplifiers, and acquired the signals through a National Instruments PCI 6259 board with digitized at 10 kHz and recorded using custom software written in LabView (www.packio.org, National Instruments) (Packer et al. 2012). All the analyses were done using custom Matlab-based software.
mIPSCs were recorded at 0 mV of holding potential in cells voltage clamped with cesium-based intracellular solution. mIPSCs were recorded at room temperature (22–25 °C).
Two-Photon RuBi-GABA Uncaging and Mapping
Images were acquired using a custom-made two-photon laser scanning microscope based on the Olympus FV-300 system (FV-300 side-mounted to a BX50WI microscope with a 60x, 1.1NA, water immersion objective) and a tunable Ti:Sapphire laser (Chameleon Ultra, Coherent). Fluorescence was detected with a top-mounted Hamamatsu H7422-P40 PMT connected to a Hamamatsu C7319 preamplifier whose output was connected to the ScanImage system (Pologruto et al. 2003).
For two-photon RuBi-GABA uncaging, the Ti:Sapphire laser was used. Images of the soma or dendrite were first acquired, using 950 nm excitation light to prevent RuBi-GABA uncaging, at 9x or 10x digital zoom with the ScanImage software in XY scan mode. An uncaging point was selected and its uncaging laser power/duration were determined in the point stimulation mode of the ScanImage software. For RuBi-GABA mapping, a group of uncaging points with 1.5 μm gap in X and Y axes were selected and the points were stimulated every 2 s. In performing uncaging, laser wavelength was tuned into 800 nm.
RuBi-GABA was added to the bath at 300 μM concentration. For perisomatic uncaging experiments in Figure 1a, RuBi-GABA was applied through a glass pipette filled with a solution of 1.6 mM RuBi-GABA in ACSF. As RuBi-GABA is light-sensitive, all the experiments were conducted with minimal light. The computer screens and video monitor were covered with two layers of Rosco #27 medium red filters. To establish whole-cell access the cells were subject to oblique illumination with and IR-pass filter on top of the microscope field diaphragm, and visualized through a CMOS camera (Thorlabs DCC1545M). Although GABA uncaging may also activate slower GABAB receptors, we did not see any long-term effects in our voltage clamp experiments after GABA uncaging.
Figure 2.
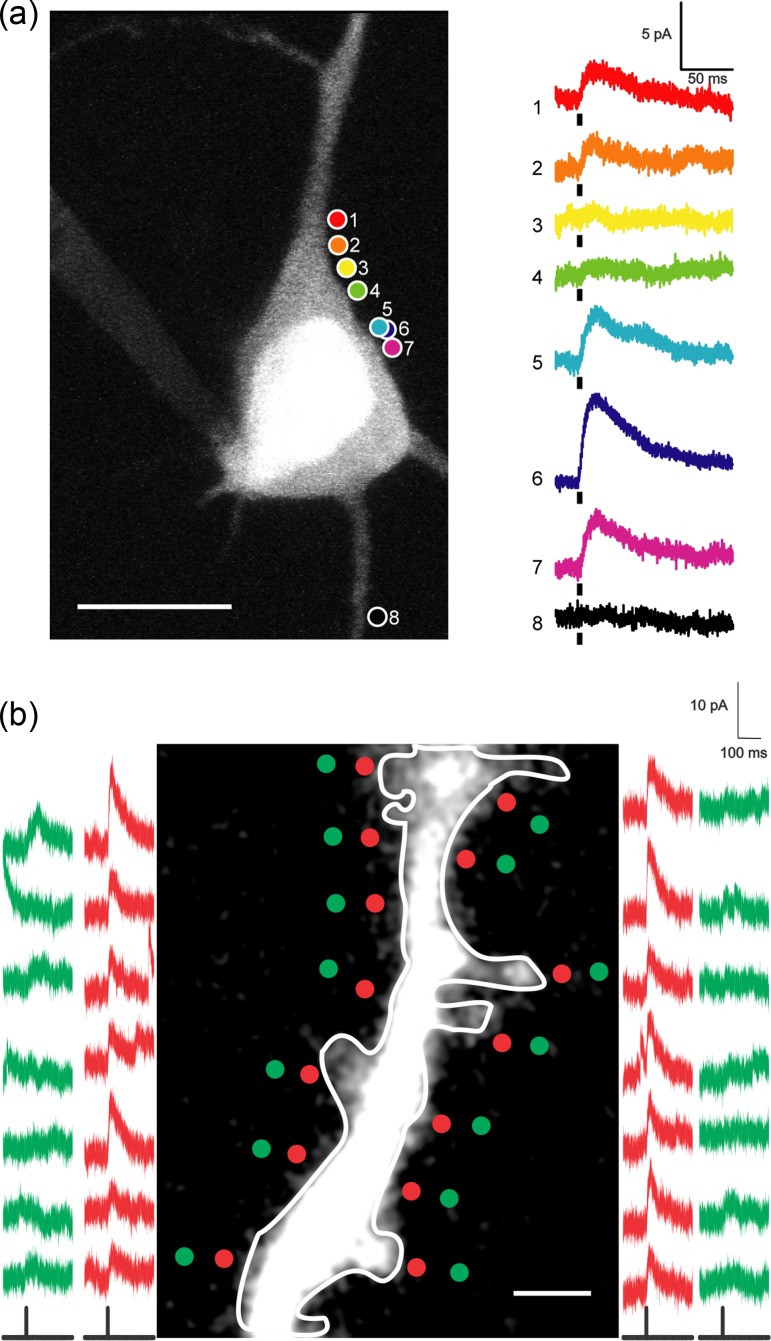
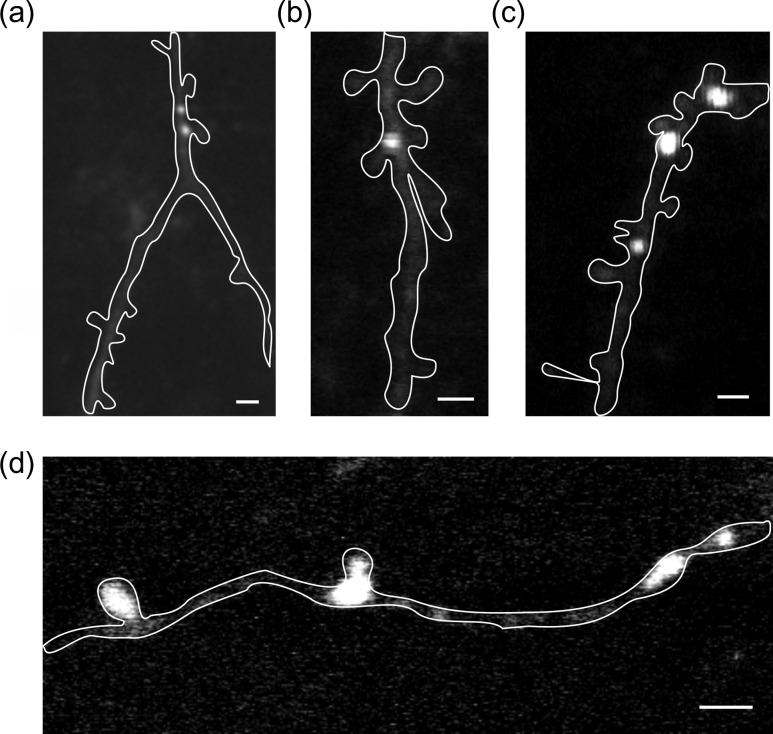
Functional GABA receptors mapping in soma and dendrites with two-photon RuBi-GABA uncaging. (a) Somatic mapping. This example shows eight different uncaging sites on the same cell. Points 5 and 6 are at the same XY coordinates, but at different Z planes. Scale bars: 5 pA, 50 ms, 10 μm. Black bars under traces indicate uncaging laser pulse. RuBi-GABA was applied through a glass pipette. (b) Dendritic mapping. Red traces are uncaging responses from stimulation points at red dots, which are on same level and side from dendrite. Green traces are uncaging responses from control stimulations at green dots, ~1 μm away from respective red dots. These reduced control responses reveal a spatial resolution of two-photon RuBi-GABA uncaging is ~1 μm. We observed RuBi-GABA uncaging response everywhere in dendrite with variability in response amplitude. Scale bar: 2 μm.
In Utero Electroporation
In utero electroporation experiments were conducted as previously described with some modifications (dal Maschio et al. 2012; Bando et al. 2016). Mouse embryos at embryonic day 15.5 (E15.5) were in utero electroporated with pCAG-GPHN.FingR-eGFP-CCR5TC (Gross et al. 2013). Pregnant mice (CD-1; ICR, Charles River) were anesthetized with isoflurane 11 (induction, 3% v/v; surgery, 2% v/v) and the uterine horns were exposed by laparotomy. The plasmid (GPHN.FingR-eGFP, 1.0 μg/μL in final concentration) mixed with the dye Fast Green (0.05% w/v; Sigma-Aldrich) for visually tracking injection location was injected through the uterine wall into the left lateral ventricle of each embryo through the uterine wall using a pulled glass capillary. After soaking the uterine horn with warm phosphate-buffered saline (PBS, 37 °C), each embryo’s head was carefully held between tweezers with platinum disk electrodes (CUY650P5, NepaGene), while positive side of the electrodes was accurately positioned at the somatosensory cortex. Subsequently, electric pulses (three 10-ms poring pulses at 50 V, followed by three 50-ms transfer pulses at 15 V with 50-ms intervals) were delivered through these triple-electrodes using an electroporator (NEPA21, NepaGene). After the electroporation, the uterine horns were returned into the abdominal cavity and the skin was closed with sutures. Electroporated mice were used for brain slices in 2 weeks after birth.
Electron Microscopy
For ultrastructural identification of inhibitory synapses, combined FIB/SEM was used (Merchan-Perez et al. 2009). Animals were administered a lethal intraperitoneal injection of sodium pentobarbital (40 mg/kg) and were intracardially perfused with 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M phosphate buffer (PB). The brain was then extracted from the skull and processed for electron microscopy according to a previously described protocol (Santuy et al. 2018). Two stacks of 241 and 344 serial sections were obtained from layer 2/3 of somatosensory cortex of a P14 mouse. The size of individual images was 2048 × 1536 pixels, with a spatial resolution of 3.722 nm/pixel in the Xand Y axes, so that the field of view was 7.62 × 5.72 μm, or 43.58 μm2. Resolution in the Z axis, equivalent to section thickness, was 20 nm. The volume of neuropil sampled was 210.05 μm3 and 299.82 μm3, respectively, totaling 509.87 μm3. We used Espina software for the identification and segmentation of synapses (Morales et al. 2011). When synaptic junctions were sectioned obliquely or en face, they were digitally resliced with Espina so they could be examined from different angles (Merchán-Pérez et al. 2009). Synapses showing a prominent postsynaptic density were classified as asymmetric, while those having a thin postsynaptic density, similar to the presynaptic density, were classified as symmetric (Colonnier 1968; Gray 1959).
Results
To map the distribution of GABAergic synapses in dendrites from pyramidal neurons we used functional, molecular and ultrastructural methods, combining two-photon RuBi-GABA uncaging, fluorescent labeling of gephyrin by intrabodies and FIB/SEM.
Activating GABAergic Synapses by Two-Photon Uncaging of RuBi-GABA
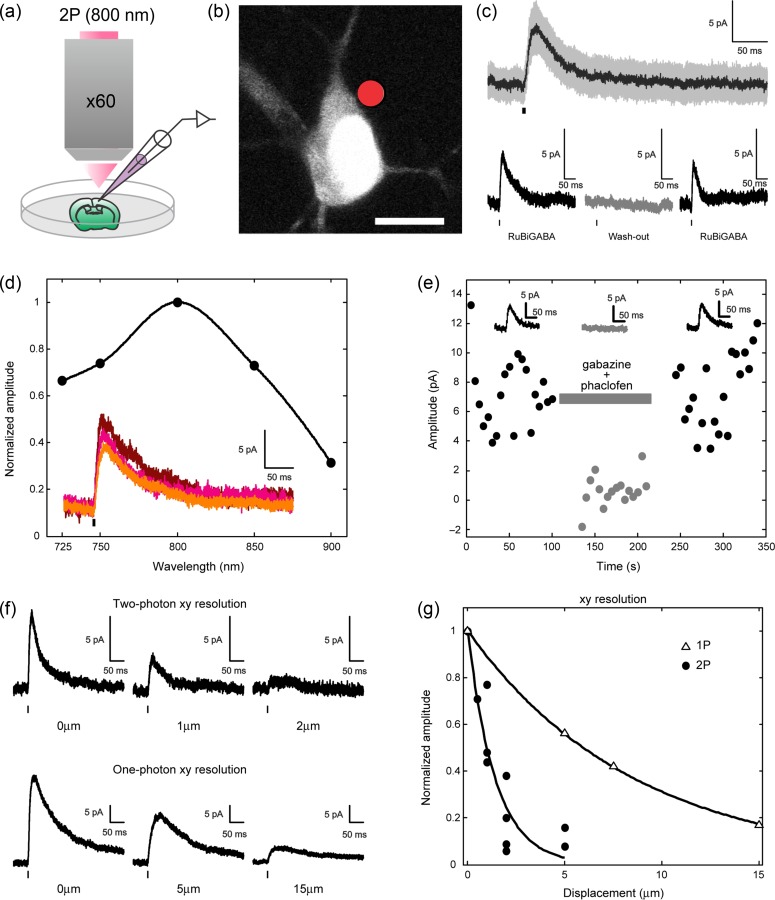
We first carried out two-photon uncaging of the caged GABA compound RuBi-GABA (Rial-Verde et al. 2008) by applying it to brain slices from juvenile (P14) mouse primary visual cortex while performing whole-cell patch recordings from layer 2/3 pyramidal cells (Fig. 1a–b). We have previously demonstrated the use of RuBi-GABA to activate GABAergic synapses using one-photon uncaging (Rial-Verde et al. 2008) and now we repeated these control experiments, but with two-photon excitation. A 5-ms laser pulse (800 nm; 35–55 mW at the sample), focused next to the cell somatic membrane, elicited outward currents in the recorded cell (Fig. 1c). We ruled out the possibility that the laser pulse, rather than GABA uncaging, elicited the recorded responses by removing the positive pressure from the pipette focally applying RuBi-GABA. In the absence of RuBi-GABA, the laser alone was unable to induce responses (Fig. 1c). After this, we tuned the laser to different wavelengths, while matching the average power at the sample. In this manner, we obtained response amplitude versus wavelength curves for RuBi-GABA (Fig. 1d). These data indicated that 800 nm was an optimal wavelength for two-photon GABA uncaging so all the following experiments were performed at this wavelength.
Figure 1.
Two-photon RuBi-GABA uncaging. (a) Experimental setup for two-photon RuBi-GABA uncaging (b) Typical two-photon uncaging experiment with RuBi-GABA, showing a cortical layer 2/3 pyramid loaded with Alexa Fluor 594. The red dot indicates site of two-photon uncaging. Scale bar: 10 μm. (c) (top panel) Average (black) and standard deviation (gray) of 20 successive two-photon uncaging responses. (bottom panel) Average response induced by two-photon uncaging with application of RuBi-GABA (left), after washing RuBi-GABA (middle), and recovery of the response in reapplication of RuBi-GABA (right). (d) Normalized amplitude of responses induced by two-photon uncaging at different excitation wavelengths. Inset shows an example of responses from the same cell at 800 nm (top, brown), 750 nm (middle, magenta), and 725 nm (bottom, orange). (e) Peak two-photon uncaging response amplitude before, during (gray bar), and after the application of GABA antagonists (gabazine and phaclofen). Insets show the average trace for each condition. Black bars under traces indicate the uncaging laser pulse. (f) (top panel) XY-axis resolution of two-photon uncaging: initial response with perisomatic uncaging point (left), response after 1 μm lateral displacement of the uncaging point away from the cell (middle), and response after 2 μm lateral displacement (right). (bottom panel) XY-axis resolution of one-photon uncaging: initial response with perisomatic uncaging point (left), response after 5 μm lateral displacement of the uncaging point away from the cell (middle), and response after 15 μm lateral displacement (right). One-photon RuBi-GABA uncaging was performed as described in Rial Verde et al. (2008). (g) Normalized amplitude of responses induced by two-photon (filled circles) and one-photon (empty triangles) uncaging at different distances from the cell soma. Scale bars: 5 pA, 50 ms. Black bars under traces indicate the uncaging laser pulse.
To further confirm that two-photon uncaging produces responses mediated by GABA receptors, we applied GABA receptor antagonists. A mixture of the GABAA antagonist gabazine (40 μM) and the GABAB antagonist phaclofen (1 mM) reversibly and completely blocked the currents obtained in response to two-photon uncaging (Fig. 1e). We concluded that RuBi-GABA uncaging can be used to optically activate GABA receptors with two-photon excitation.
To characterize the spatial resolution of two-photon RuBi-GABA uncaging, we repeated experiments measuring uncaging responses but now systematically varying the X–Y distance from the somatic membrane. A 2-μm lateral shift practically abolished the responses to two-photon uncaging (Fig. 1f, top panel). In comparison, responses to one-photon uncaging with a visible CW laser source persisted, albeit reduced, after a 15-μm lateral shift of the focal point (Fig. 1f, bottom panel). Overall, the spatial resolution of two-photon RuBi-GABA uncaging was several fold better than that of one-photon uncaging (Fig. 1g). Under these conditions, these results indicate that two-photon uncaging of RuBi-GABA can be used as a tool to locate and stimulate individual GABAergic synapses in living tissue, with a spatial resolution of ~2 μm.
Functional GABAA Receptor Mapping by RuBi-GABA Uncaging Reveals Hotspots
After these calibration experiments, we used two-photon RuBi-GABA to functionally map GABAergic inhibitory synapses onto pyramidal neurons. For these experiments, we uncaged RuBi-GABA with 2-ms laser pulses, and, first explored responses in the perisomatic membrane of layer 2/3 pyramidal neurons (Fig. 2a). In several neurons, we observed clear heterogeneities in the uncaging responses of different spatial positions, with local “hotspots” where RuBi-GABA uncaging generated strong outward currents (Fig. 2a, positions 1, 6), surrounded by membrane territories where the responses were weaker or absent (Fig. 2a, positions 4, 8). Although we did not independently confirm the existence of GABAergic synapses in those experiments, the fact that the perisomatic membrane has abundant GABAergic innervation (DeFelipe et al. 1997) suggests that the heterogeneity maybe due to differences in axo-somatic synapse density or receptor availability in them.
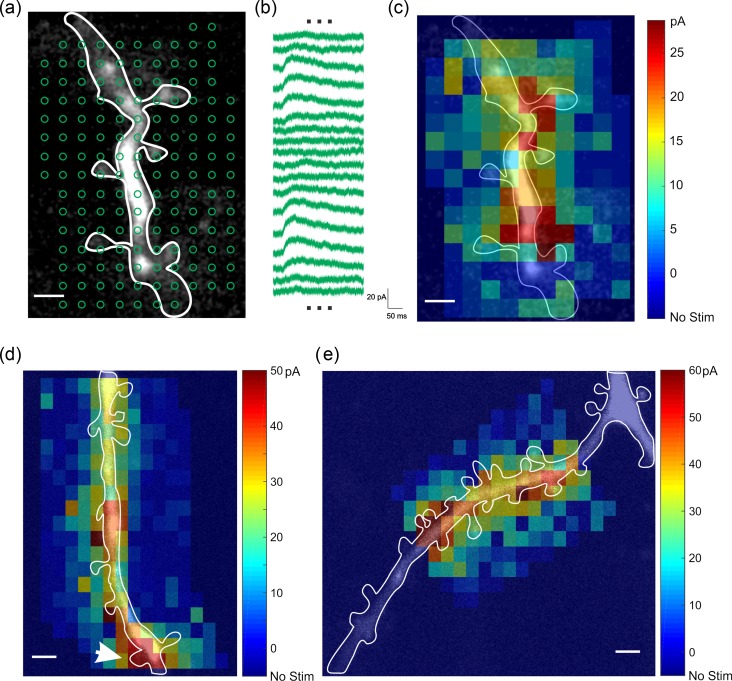
We then proceeded to use two-photon RuBi-GABA uncaging to map functional GABAergic synapses on dendrites, which was the objective of our study. Our dendritic sample was biased towards proximal dendrites, as they were experimentally more accessible, and we stimulated multiple locations on dendrites to map GABAergic responses with spatial resolution that approached that of individual spines (~1 μm; Fig. 2b). To achieve this resolution, we reduced the laser power until we did not detect any response when stimulating points 1 μm away from the uncaging location on a dendritic spine (Fig. 2b). In these experiments, we observed that dendrites also had hotspots, with clear uncaging responses along their surface, but that those responses varied in amplitude in different positions (Fig. 2b). To identify these hotspots, we performed systematic RuBi-GABA mapping by grid uncaging with a 1.5-μm spatial pitch (Fig. 3a). As expected, we found that uncaging responses increased by approaching to dendritic membrane but decreased by moving away from dendritic membrane (Fig. 3b). Using a pseudocolor scale based on the peak amplitude of uncaging responses, we mapped hotspots along the dendrite (n = 16 hotspots, nine dendrites, eight cells, eight slices, six animals; Fig. 3c–e). These functional hotspots were mostly on dendritic shafts (Fig. 3c–e) although we also occasionally found some on spines (Fig. 3d; white arrow). The spatial resolution of functional RuBi-GABA mapping was often not good enough to identify the core of a hotspot. However, when we measured the center coordinates of hotspots, the ratio of hotspots on dendritic spines over total hotspots was 16.6% (Table 1), indicating that most putative GABAergic synapses were on dendritic shafts.
Figure 3.
RuBi-GABA uncaging reveals GABAergic hotspots. (a) Two-photon RuBi-GABA uncaging grid map of dendritic GABAergic responses. 153 uncaging pulses were done with 2 s intervals. (b) Representative uncaging responses with stimulations indicated in (a). (c) Pseudocolor map of peak uncaging response peak amplitudes. (d–e) Examples of functional GABA receptor mapping in dendrite. Note functional hotspots of GABA receptors on dendritic shafts. Spine with a white arrow in (d) had clear uncaging response. Scale bar: 2 μm.
Table 1.
Functional mapping of GABAergic synapses on dendrites
| On spines | On shafts | Total |
|---|---|---|
| 3 | 15 | 18 |
| 16.7% | 83.3% | 100% |
Molecular Hotspots Revealed by Gephyrin Intrabodies Correlate with Functional Hotspots
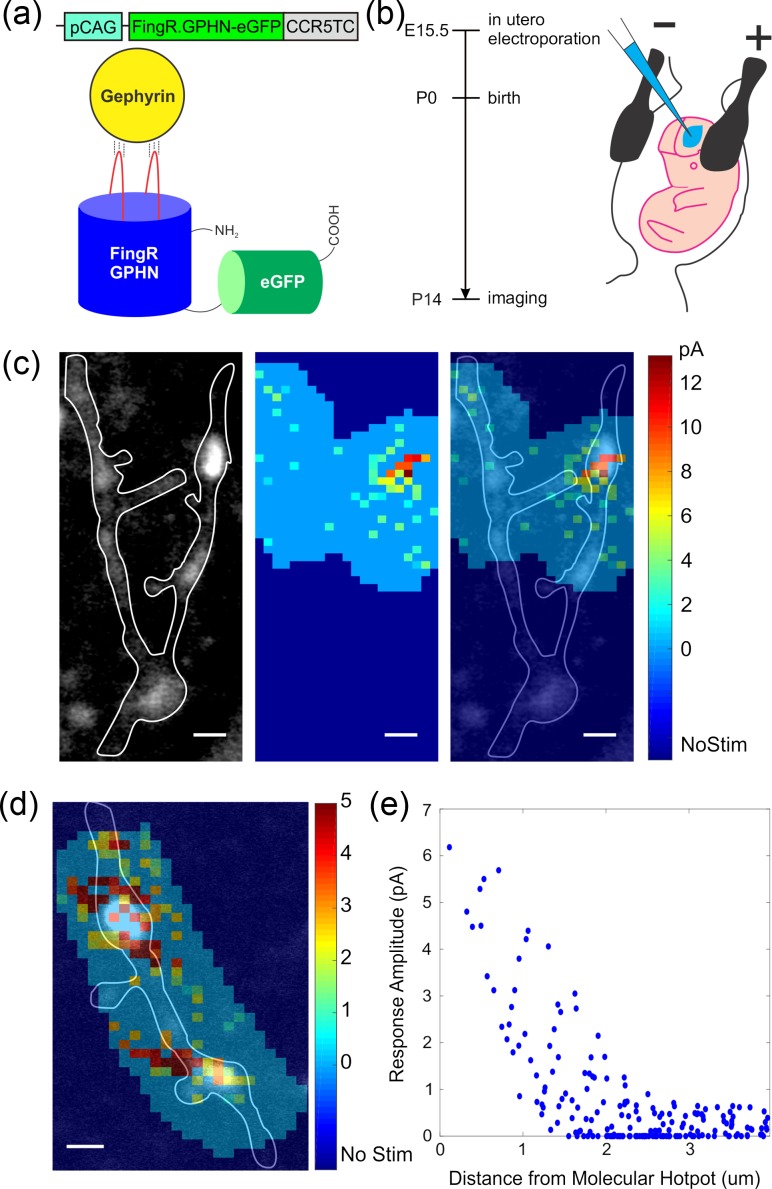
To test whether functional hotspots were GABAerigc synapses, we used intrabody labeling of gephyrin. The gephyrin intrabody is a fusion protein of fibronectin and eGFP (Fig. 4a) (Gross et al. 2013). When the intrabody plasmid is transfected into neurons and expressed, the intrabody protein binds to gephyrin and fluoresces as green puncta (Gross et al. 2013). This intrabody method minimally interferes with endogenous gephyrin expression, an advantage versus transfecting fluorescently labeled gephyrin, which labels exogenous gephyrin and may have a different spatial distribution. In addition, intrabody labeling enables real-time detection of inhibitory synapses in living tissue, as opposed to gephyrin antibody staining, performed post hoc in fixed samples.
Figure 4.
GABAergic hotspots are correlated with gerphyrin hotspots. (a) Schematic drawing of DNA construct and protein structure of gephyrin intrabody. (b) (left) Experimental time line for molecular mapping of GABA receptors by performing in utero electroporation of gephyrin intrabody. (right) Schematic image how in utero electroporation was performed. (c) (left) Molecular labeling of gephyrin intrabody. A gephyrin punctum (or a molecular hotspot for a putative inhibitory synapse) is shown in the right dendritic branch of image. (middle) Functional GABA receptor mapping was done and a functional hotspot is shown on the right side of image. (right) Overlaid images of (left) and (middle) panels show molecular hotspot and functional hotspot are at the same location. Scale bar: 2 μm. (d) another example overlaying molecular hotspot and function hotspot (e) spatial correlation between molecular and functional hotspots in (c) shown as uncaging response as a function of distance from center of mass spatial coordinates of molecular hotspot.
To express gephyrin intrabodies, we performed in utero electroporation of gephyrin intrabody plasmids at embryonic age 15.5 (Fig. 4b) and carried out RuBi-GABA mapping of pyramidal neurons in brain slices at P14 as before, but now while also imaging gephyrin intrabody puncta. In these experiments, we found that gephyrin intrabody puncta (Fig. 4c, left) and functional RuBi-GABA hotspots (Fig. 4c, middle) had overlapping spatial patterns (n = 5/5, four dendrites, four cells, four slices, four animals; Fig. 4c, right). Also, the functional response plot as a function of distance from center of molecular hotspot showed a strong spatial correlation between both maps (Fig. 4d; ~1 μm length constant). These experiments confirmed that RuBi-GABA hotspots corresponded to the position of GABAergic synapses. This correspondence also demonstrated that gephyrin intrabody puncta were functionally active GABAergic synapses, confirming the validity of the intrabody staining.
Molecular Gephyrin Hotspot Distribution in Dendrites
After validating that gephyrin intrabody puncta were functional, we investigated GABAergic synapse distribution using this molecular mapping method. As before, we electroporated gephyrin intrabody plasmids in utero and performed fluorescence imaging of the green intrabody puncta in brain slices from layer 2/3 of mouse somatosensory cortex (n = 737 hotspots, 306 dendrites, 10 slices, 5 animals). We found gephyrin puncta locations in dendritic shafts (Fig. 5a–c, d/right), spines (Fig. 5d/left), and in both spine/shafts (Fig. 5d/middle). To analyze this, we calculated two ratios: (1) the proportion of gephyrin puncta in spines/total gephyrin puncta and (2) the proportion of spines with gephyrin puncta/total spines (Table 2). While 18.0% of gephyrin puncta were in spines, a very small proportion of spines had gephyrin puncta (3.62%, 133/3764 spines). Furthermore, we compared gephyrin puncta in layer 1 and layer 2/3 to explore if gephyrin puncta ratio changes between proximal and distal dendrites (Table 3). The spines/total ratios showed only small difference between layers (18.3% in layer 1 and 23.0% in layer 2/3).
Figure 5.
Gerphyrin intrabodies mapping of inhibitory synapses. (a–d) Molecular hotspots are mainly in dendritic shafts. Occasionally, molecular hotspots are found in dendritic spines or in both dendritic spines and shaft (see middle spot in d). Scale bars: 2 μm.
Table 2.
Molecular mapping of GABAergic synapses on dendrites
| On spines | On shafts | Total |
|---|---|---|
| 133 | 604 | 737 |
| 18.0% | 82.0% | 100% |
Table 3.
Comparison of molecular mapping of GABAergic synapses on different layers
| On spines | On shafts | Total | |
|---|---|---|---|
| Dendrites in Layer 1 | 56 | 250 | 306 |
| 18.3% | 81.7% | 100% | |
| Dendrites in Layer 2/3 | 111 | 371 | 482 |
| 23.0% | 77.0% | 100% |
Ultrastructural Mapping of GABAergic Synapses in Dendrites
As a complementary investigation of GABAergic synapse distribution, we performed an ultrastructural analysis of layer 2/3 neuropil from P14 mouse somatosensory cortex, the same age as the functional and molecular mapping study. We used stacks of serial sections obtained by FIB/SEM (Merchan-Perez et al. 2009) to identify asymmetric and symmetric synaptic profiles, as evidence of excitatory or inhibitory synapses, respectively (Colonnier 1968; Gray 1959; Houser et al. 1984; Peters et al. 1991). The identification of synapses was not based on single images but on the examination of the full sequence of sections where the synapse was visible. We used “Espina” software (Morales et al. 2011) for the 3D visualization, segmentation, and classification of synaptic junctions (Fig. 6; Supplemental videos 1 and 2). When synaptic junctions were sectioned obliquely or “en face”, the stack of images was digitally resliced to examine the synapse from different angles. This greatly facilitated the classification of synapses since three orthogonal sections can be viewed simultaneously, thereby allowing us to classify synapses as asymmetric (AS; when they had a prominent postsynaptic density), or symmetric (SS; when they had a thin postsynaptic density). Although it has been described that some terminals that establish AS and SS can synthesize neurotransmitters other than glutamate and GABA, respectively, such as acetylcholine, serotonin, noradrenaline or dopamine, a large proportion of these axonal systems are non-synaptic (Beaulieu and Somogyi 1990; DeFelipe and Jones 1988; Descarries and Mechawar 2000). Therefore, AS and SS that use acetylcholine, serotonin, noradrenaline, or dopamine must represent a very small proportion of the total number of AS and SS.
Figure 6.
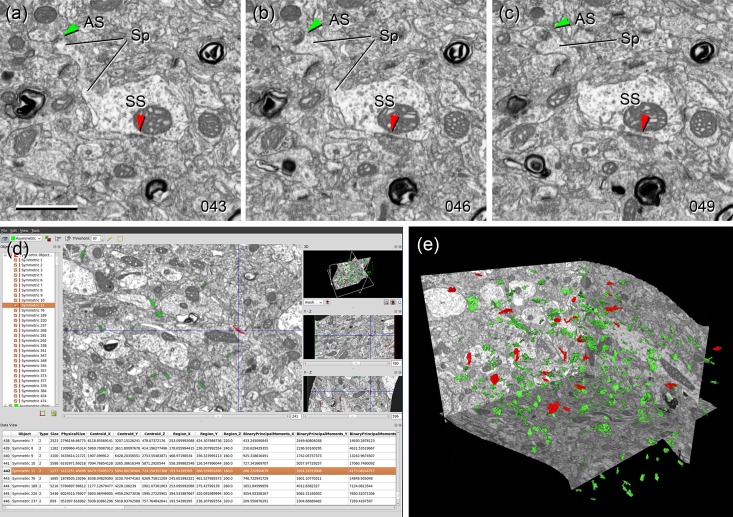
Ultrastructural mapping of synapses by FIB/SEM. (a–c) Three nonconsecutive serial sections of the neuropil of the mouse somatosensory cortex (layer 2/3) showing examples of asymmetric (excitatory) and symmetric (inhibitory) synapses. The numbers at the right bottom corners indicate the order of each photomicrograph in the series. Individual section thickness was 20 nm, so the examples shown here are separated by 60 nm. An asymmetric synapse (AS, green arrowheads) is established on a dendritic spine (Sp). A symmetric synapse (SS, red arrowheads) is established on the shaft of the parent dendrite. (d) Snapshot of Espina, the software tool that has been used for the automatic segmentation of synaptic junctions based on the gray level of synaptic densities. Each individual synaptic junction is given a unique number (left panel) and is color-coded as asymmetric (green) or symmetric (red)(central panel). The stack of images can be navigated in 3D to identify the postsynaptic target of each individual synapse as either a dendritic spine or shaft. To help with this task, the program also performs interactive digital reslicing through orthogonal planes (right middle and bottom panels) and three-dimensional reconstructions of synaptic junctions (upper right panel). (e) Three-dimensional reconstruction of the synaptic junctions present inside a stack of neuropil. Asymetric synaptic junctions are represented in red and symmetric synaptic junctions in green. See also Supplemental videos 1 and 2. Scale bar in (a), 1000 nm for (a–c).
A total number of 804 synapses were identified within the EM stacks. The synaptic target (i.e., a spine or a dendritic shaft) was identified in 692 cases. Of these, 633 (91.47%) were AS and 59 (8.53%) were SS. Synapses whose target could not be identified (mainly because postsynaptic structures were truncated by the stack boundaries) were discarded for further analysis. The location of AS and SS on dendritic spines and shafts is summarized in Table 4. We found that most synapses (83.96%) were AS targeting dendritic spines, followed by AS on dendritic shafts (7.51%) and SS on dendritic shafts (7.08%). The lowest percentage was SS on spines (1.45%). Synapses on spines were mostly AS (98.31%), with only rare SS profiles (1.69%). Most synapses on spines were made on the spine head, although 1.55% of AS and 30% of SS were found on the spine neck. Finally, the majority of AS were on dendritic spines (AS in spines/total AS = 92%). However, the majority of SS were on dendritic shafts (SS in spines/total SS = 17%; Table 4). These results corresponded well with the gephyrin intrabody mapping, further validating both methods (Table 5). The percentage of spines that had two synapses was very low (12 spines with two synapses/680 total spines = 1.76%). Four of these spines established one AS and one SS, while the remaining established two AS. We did not find any spine with two SS, or with more than two synapses.
Table 4.
Ultrastructural mapping of excitatory and inhibitory synapses on dendrites
| On spines | On shafts | Total | |
|---|---|---|---|
| Asymmetric synapses (Excitatory) | 581 | 52 | 633 |
| 91.8% | 8.2% | 100% | |
| Symmetric synapses (Inhibitory) | 10 | 49 | 59 |
| 16.9% | 83.1% | 100% |
Table 5.
Comparison of the results in present study and Chen et al. (2012)
| Kwon et al. (gephyrin intrabody) | Kwon et al. (FIB/SEM of WT) | Chen et al. (Teal-GHN) | Chen et al. (EM of Teal- GPHN) | |
|---|---|---|---|---|
| Spine inhibitory synapses/total inhibitory synapses (%) | 18.0 | 16.9 | 29.7 | 40.0 |
| Spine inhibitory synapses/total spines (%) | 3.62 | 1.69 | 13.6 | 12.5 |
Discussion
In this study, we mapped the distribution of GABAergic synapses on dendrites of pyramidal neurons using functional, molecular and ultrastructural methods. Using two-photon RuBi-GABA uncaging, we find functional GABAergic hotspots and show that they correspond to intrabody gephyrin puncta, confirming that they are functional GABAergic synapses. We then use intrabody gerphyrin labelling to map the position of GABAergic synapses on dendrites and spines. The systematic mapping of those gephyrin pucnta and tabulation of their position agree very well with ultrastructural reconstructions and identification of SS onto spines and dendritic shafts. Our combined results indicate that in the sample studied, most GABAergic contacts (80%) are made on dendritic shafts, with around 20% on spines (Tables 1–4). It is well established that the axons of GABAergic neurons form synapses with all regions of the pyramidal cell (i.e., the dendritic shaft, soma, axon initial segment, and dendritic spines) and that some GABAergic interneurons such as double bouquet cells, Martinotti cells, and basket cells establish synapses on spines (Jones and Powell 1969; Somogyi and Cowey 1981; Kisvarday et al. 1985; DeFelipe et al. 1989; Kawaguchi and Kubota 1998; Wang et al. 2004; DeFelipe et al. 2006; for a review see Kubota et al. 2016). However, in our samples, only very few spines (~2%) had GABAergic synapses, perhaps explained by the fact that there are relatively few GABAergic interneurons and an extraordinary abundance of spines. At the same time, while GABAergic synapses on spines are very scarce, in our functional data, most spines have GABAergic responses, probably due to extrasynaptic receptors, as previously suggested (Afroz et al. 2016).
Comparison with Previous Studies
Our results are in agreement with the traditional view from ultrastructural sampling that most inhibitory synapses occur in dendritic shafts and that spine inhibitory synapses are rare (Peters et al. 1991; for a recent review see Kubota et al. 2016). Recently, Chen et al. used exogenous fluorescent gephyrin expression combined with electron microscopy to map GABAergic synapses in adult mouse visual cortex (Chen et al. 2012). This study reported a relatively large proportion of GABAergic synapses in spines (30–40% of all GABAergic synapses in dendrites), which is significantly larger that our measurements (17–18%). One reason for this difference might be due to the different brain regions (visual vs. somatosensory cortex), or layers (2/3 vs. 4) or maturity of the animal (juvenile vs. adult) studied, as the inhibition onto spines might be different in different parts of the cortex, different neurons, or at different developmental stages. To evaluate these possibilities and to find out the general rules of the distribution of inhibitory synapses in dendrites, future comparative studies are required on other brain areas, layers, ages, and species. However, an additional reason for this difference could be methodological: exogenous gephyrin overexpression may significantly increase the number of puncta or synapses in spines and lead to an artifactual overestimation of the numbers of GABAergic synapses. In fact, gephyrin stabilizes GABAergic synapses (Tyagarajan and Fritschy 2014) and it is possible that additional exogenous gephyrin molecules could stabilize GABAergic synapses in spines and increase the proportion of spine GABAergic synapses. Furthermore, the EM data provided by Chen et al. were obtained from a single dendritic segment where 10 inhibitory synapses were found. In the present study, we sampled a much larger tissue volume (510 μm3), and the sample size of inhibitory synapses is much larger (n = 59). Consistent with this, data obtained from juvenile rat somatosensory cortex also suggest that the percentage of inhibitory synapses on dendritic spines is very low, with numbers similar to the ones we obtained (Santuy et al. 2018). Specifically, that study showed that 75.55% of all synapses were AS located on spines, 14.73% were AS located on dendritic shafts, 7.08% were SS located on shafts, and 2.65% were SS located on spines. Other quantitative studies have reported different percentages of SS on spines but, in all cases, the proportion of SS on spines is much lower than SS on dendritic shafts, and very small when compared with the percentage of AS on spines. For example, a study on the distribution of synapses in rat barrel cortex showed that, at P15, AS were mainly located on spines (82%), followed by dendritic shafts (17%), with <1% of the AS on somas (Micheva and Beaulieu 1996). They also reported that, at this age, 15% of the synapses were SS that were mainly located on dendritic shafts (54%), followed by spines (39%), and cell somata (8%). In the visual cortex of the adult cat (Beaulieu and Colonnier 1985) 66.4% of all synapses were AS located on spines, 17.6% were AS located on dendritic shafts, 10.6% were SS located on shafts, and 5.3% were SS located on spines. In addition, our data shed new light on the interpretation of the uncaging results from Chiu et al. (Chiu et al., 2013; Villa et al. 2016), who used one-photon RuBi-GABA uncaging to explore the functional effects of GABAergic synapses on excitatory inputs and concluded that spines had GABAergic synapses. While we did not perform the same experiments, our two-photon uncaging maps show however a paucity of hotspots on spines. But because one-photon RuBi-GABA uncaging has poorer spatial resolution, as compared with two-photon uncaging (Fig. 2c) and since we find that spines without GABAergic synapses also respond to GABA uncaging, we would interpret their results as being caused by the activation of nearby GABAergic synapses in dendritic shafts or of extrasynaptic GABAergic receptors in spines, rather than by GABAergic synapses on spines.
Functional Implications
It has been suggested that GABAergic inputs onto spines could suppress the excitatory inputs selectively (Chiu et al. 2013) and lead to selective point inhibition (Villa et al. 2016). Also, inhibitory synapses could veto the thalamic excitatory input in the cases where spines are innervated by one excitatory and one inhibitory synapse (Kubota et al. 2007, 2015). However, our results indicate that only a small minority of GABAergic synapses are on spines (Table 5) and only a fraction of them are established on dually innervated spines. Therefore, although we cannot rule out the possibility that a small population of inhibitory synapses on spines may exert a local veto, or that different cortical neurons may possess significant GABAergic synapses on spines, we would propose instead that dendritic inhibition in practice occurs on the shaft, consistent with the regional inhibition model. Dendritic inhibition, in our view, would therefore lack sufficient spatial specificity to modulate individual excitatory inputs in spines, whereas it may affect clusters of inputs in a larger segment of dendrite, instead. This regional effect could be used, for example, to build inhibitory zones that are specific to individual dendritic branches, defining individual branches, rather than individual spines, as functional units of the dendrite. In addition, shaft inhibition could also effectively block the transmission of excitatory potentials propagating from distal dendrites to soma, or shunt backpropagating action potentials from soma to dendrites. Future experiments, perhaps using two-photon holographic uncaging, which can generate spatial uncaging patterns (Nikolenko et al. 2008) could be used to discern the functional effects of inhibition on dendritic integration.
Supplementary Material
Notes
We are grateful to members of the laboratory for help, Darcy Peterka for optical engineering, Masayuki Sakamoto, and Yuki Bando for in utero electroporation; and Reka Lesto, Azadeh Hamzehei Sichani, and Samuel Kim for molecular biology. Conflict of Interest: None declared.
Funding
Supported by the NIMH (R01MH101218, R01MH100561). This material is based upon work supported by, or in part by, the US Army Research Laboratory and the US Army Research Office under contract number W911NF-12-1-0594 (MURI) and the European Union’s Horizon 2020 Research and Innovation Programme under grant agreements no. 720270 and 785907 (Human Brain Project).
References
- Ascoli GA, Alonso-Nanclares L, Anderson SA, Barrionuevo G, Benavides-Piccione R, Burkhalter A, Buzsaki G, Cauli B, DeFelipe J, Fairén A, et al. 2008. Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat Neurosci. 9:557–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afroz S, Parato J, Shen H, Smith SS. 2016. Synaptic pruning in the female hippocampus is triggered at puberty by extrasynaptic GABAA receptors on dendritic spines. Elife. 5:e15106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atallah BV, Bruns W, Carandini M, Scanziani M. 2012. Parvalbumin-expressing interneurons linearly transform cortical responses to visual stimuli. Neuron. 73:159–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando Y, Irie K, Shimomura T, Umeshima H, Kushida Y, Kengaku M, Fujiyoshi Y, Hirano T, Tagawa Y. 2016. Control of spontaneous Ca2+ transients is critical for neuronal maturation in the developing neocortex. Cereb Cortex. 26:106–117. [DOI] [PubMed] [Google Scholar]
- Beaulieu C, Colonnier M. 1985. A laminar analysis of the number of round-asymmetrical and flat-symmetrical synapses on spines, dendritic trunks, and cell bodies in area 17 of the cat. J Comp Neurol. 231:180–189. [DOI] [PubMed] [Google Scholar]
- Beaulieu C, Somogyi P. 1990. Targets and quantitative distribution of GABAergic synapses in the visual cortex of the cat. Eur J Neurosci. 2:296–303. [DOI] [PubMed] [Google Scholar]
- Bloss EB, Cembrowski MS, Karsh B, Colonell J, Fetter RD, Spruston N. 2016. Structured dendritic inhibition supports branch-selective integration in CA1 pyramidal cells. Neuron. 89:1016–1030. [DOI] [PubMed] [Google Scholar]
- Chen JL, Villa KL, Cha JW, So PT, Kubota Y, Nedivi E. 2012. Clustered dynamics of inhibitory synapses and dendritic spines in the adult neocortex. Neuron. 74:361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CQ, Lur G, Morse TM, Carnevale NT, Ellis-Davies GC, Higley MJ. 2013. Compartmentalization of GABAergic inhibition by dendritic spines. Science. 340:759–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonnier M. 1968. Synaptic patterns on different cell types in the different laminae of the cat visual cortex. An electron microscope study. Brain Res. 9:268–287. [DOI] [PubMed] [Google Scholar]
- dal Maschio M, Ghezzi D, Bony G, Alabastri A, Deidda G, Brondi M, Sato SS, Zaccaria RP, Di Fabrizio E, Ratto GM, et al. 2012. High-performance and site-directed in utero electroporation by a triple-electrode probe. Nat Commun. 3:960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFelipe J, Hendry SH, Jones EG. 1989. Synapses of double bouquet cells in monkey cerebral cortex visualized by calbindin immunoreactivity. Brain Res. 503:49–54. [DOI] [PubMed] [Google Scholar]
- DeFelipe J, Jones EG. 1988. A light and electron microscopic study of serotonin-immunoreactive fibers and terminals in the monkey sensory-motor cortex. Exp Brain Res. 71:171–182. [DOI] [PubMed] [Google Scholar]
- DeFelipe J, Marco P, Fairen A, Jones EG. 1997. Inhibitory synaptogenesis in mouse somatosensory cortex. Cereb Cortex. 7:619–634. [DOI] [PubMed] [Google Scholar]
- DeFelipe J, Ballesteros-Yanez I, Inda MC, Munoz A. 2006. Double-bouquet cells in the monkey and human cerebral cortex with special reference to areas 17 and 18. Prog Brain Res. 154:15–32. [DOI] [PubMed] [Google Scholar]
- Descarries L, Mechawar N. 2000. Ultrastructural evidence for diffuse transmission by monoamine and acetylcholine neurons of the central nervous system. Prog Brain Res. 125:27–47. [DOI] [PubMed] [Google Scholar]
- Fairen A, De Felipe J, Regidor J. 1984. In: Peters A, Jones EG, editors. ). Nonpyramidal neurons. In Cerebral cortex. New York: Plenum; p. 201–253. [Google Scholar]
- Gentet LJ, Kremer Y, Taniguchi H, Huang ZJ, Staiger JF, Petersen CCH. 2012. Unique functional properties of somatostatin-expressing GABAergic neurons in mouse barrel cortex. Nat Neurosci. 15:607–612. [DOI] [PubMed] [Google Scholar]
- Gidon A, Segev I. 2012. Principles governing the operation of synaptic inhibition in dendrites. Neuron. 75:330–341. [DOI] [PubMed] [Google Scholar]
- Gray EG. 1959. Axo-somatic and axo-dendritic synapses of the cerebral cortex: an electron microscopic study. J Anat. 83:420–433. [PMC free article] [PubMed] [Google Scholar]
- Gross GG, Junge JA, Mora RJ, Kwon HB, Olson CA, Takahashi TT, Liman ER, Ellis-Davies GC, McGee AW, Sabatini BL, et al. 2013. Recombinant probes for visualizing endogenous synaptic proteins in living neurons. Neuron. 78:971–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch SM, White EL. 1981. Quantification of synapses formed with apical dendrites of Golgi-impregnated pyramidal cells: variability in thalamocortical inputs, but consistency in the ratios of asymmetrical to symmetrical synapses. Neuroscience. 6:1043–1051. [DOI] [PubMed] [Google Scholar]
- Houser CR, Vaughn JE, Hendry SHC, Jones EG, Peters A. 1984. GABA neurons in cerebral cortex. Functional properties of cortical cells In: Jones EG, Peters A, editors. Cerebral Cortex. NewYork: PlenumPress; p. 63–89. [Google Scholar]
- Jones EG, Powell TPS. 1969. Morphological variations in dendritic spines of the neocortex. J Cell Sci. 5:509–529. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kubota Y. 1998. Neurochemical features and synaptic connections of large physiologically-identified GABAergic cells in the rat frontal cortex. Neuroscience. 85:677–701. [DOI] [PubMed] [Google Scholar]
- Kisvarday ZF, Martin KA, Whitteridge D, Somogyi P. 1985. Synaptic connections of intracellularly filled clutch cells: a type of small basket cell in the visual cortex of the cat. J Comp Neurol. 241:111–137. [DOI] [PubMed] [Google Scholar]
- Koch C, Poggio T, Torre V. 1983. Nonlinear interactions in a dendritic tree: Localization, timing and role in information processing. Proc Natl Acad Sci USA. 80:2799–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Hatada S, Kondo S, Karube F, Kawaguchi Y. 2007. Neocortical inhibitory terminals innervate dendritic spines targeted by thalamocortical afferents. J Neurosci. 27:1139–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Karube F, Nomura M, Kawaguchi Y. 2016. The diversity of cortical inhibitory synapses. Front Neural Circuits. 10:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Kondo S, Nomura M, Hatada S, Yamaguchi N, Mohamed AA, Karube F, Lübke J, Kawaguchi Y. 2015. Functional effects of distinct innervation styles of pyramidal cells by fast spiking cortical interneurons. Elife. 4:e07919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchan-Perez A, Rodriguez JR, Alonso-Nanclares L, Schertel A, Defelipe J. 2009. Counting synapses using FIB/SEM microscopy: a true revolution for ultrastructural volume reconstruction. Front Neuroanat. 3:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheva KD, Beaulieu C. 1996. Quantitative aspects of synaptogenesis in the rat barrel field cortex with special reference to GABA circuitry. J Comp Neurol. 373:340–354. [DOI] [PubMed] [Google Scholar]
- Morales J, Alonso-Nanclares L, Rodriguez JR, Defelipe J, Rodriguez A, Merchan-Perez A. 2011. Espina: a tool for the automated segmentation and counting of synapses in large stacks of electron microscopy images. Front Neuroanat. 5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolenko V, Watson BO, Araya R, Woodruff A, Peterka DS, Yuste R. 2008. SLM microscopy: scanless two-photon imaging and photostimulation with spatial light modulators. Front Neural Circuits. 2:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AM, Peterka DS, Hirtz JJ, Prakash R, Deisseroth K, Yuste R. 2012. Two-photon optogenetics of dendritic spines and neural circuits. Nat Methods. 12:1202–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Palay SL, de Webster HF. 1991. The fine structure of the nervous system. Neurons and their Supporting Cells. New York: Oxford University Press. [Google Scholar]
- Pologruto TA, Sabatini BL, Svoboda K. 2003. ScanImage: flexible software for operating laser scanning microscopes. Biomed Eng Online. 2:13–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R, Yizhar O, Grewe B, Ramakrishnan C, Wang N, Goshen I, Packer AM, Peterka DS, Yuste R, Schnitzer MJ, et al. 2012. Two-photon optogenetic toolbox for fast inhibition, excitation and bistable modulation. Nat Methods. 12:1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rial Verde EM, Zayat L, Etchenique R, Yuste R. 2008. Photorelease of GABA with visible light using an inorganic caging group. Front Neural Circuits. 2:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santuy A, Rodriguez JR, DeFelipe J, Merchan-Perez A. 2018. Volume electron microscopy of the distribution of synapses in the neuropil of the juvenile rat somatosensory cortex. Brain Struct Funct. 223:77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd GM, Brayton RK. 1987. Logic operations are properties of computer-simulated interactions between excitable dendritic spines. Neuroscience. 21:151–165. [DOI] [PubMed] [Google Scholar]
- Somogyi P, Cowey A. 1981. Combined Golgi and electron microscopic study on the synapses formed by double bouquet cells in the visual cortex of the cat and monkey. J Comp Neurol. 195:547–566. [DOI] [PubMed] [Google Scholar]
- Somogyi P, Tamas G, Lujan R, Buhl E. 1998. Salient features of synaptic organisation in the cerebral cortex. Brain Res Brain Res Rev. 26:113–135. [DOI] [PubMed] [Google Scholar]
- Tyagarajan SK, Fritschy JM. 2014. Gephyrin: a master regulator of neuronal function? Nat Rev Neurosci. 15:141–156. [DOI] [PubMed] [Google Scholar]
- Villa KL, Berry KP, Subramanian J, Cha JW, Oh WC, Kwon HB, Kubota Y, So PT, Nedivi E. 2016. Inhibitory synapses are repeatedly assembled and removed at persistent sites in vivo. Neuron. 89:756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Toledo-Rodriguez M, Gupta A, Wu C, Silberberg G, Luo J, Markram H. 2004. Anatomical, physiological and molecular properties of Martinotti cells in the somatosensory cortex of the juvenile rat. J Physiol. 561:65–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff AR, Anderson SA, Yuste R. 2010. The enigmatic function of chandelier cells. Front Neurosci. 4:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.