Abstract
Tumor cells exhibit altered lipid metabolism compared with normal cells. Cell signaling kinases are important for regulating lipid synthesis and energy storage. How upstream kinases regulate lipid content, versus direct targeting of lipid-metabolizing enzymes, is currently unexplored. We evaluated intracellular lipid concentrations in prostate and breast tumor spheroids, treated with drugs directly inhibiting metabolic enzymes fatty acid synthase (FASN), acetyl-CoA carboxylase (ACC), diacylglyceride acyltransferase (DGAT), and pyruvate dehydrogenase kinase (PDHK), or cell signaling kinase enzymes PI3K, AKT, and mTOR with lipidomic analysis. We assessed whether baseline lipid profiles corresponded to inhibitors' effectiveness in modulating lipid profiles in three-dimensional (3D) growth and their relationship to therapeutic activity. Inhibitors against PI3K, AKT, and mTOR significantly inhibited MDA-MB-468 and PC3 cell growth in two-dimensional (2D) and 3D spheroid growth, while moderately altering lipid content. Conversely, metabolism inhibitors against FASN and DGAT altered lipid content most effectively, while only moderately inhibiting growth compared with kinase inhibitors. The FASN and ACC inhibitors' effectiveness in MDA-MB-468, versus PC3, suggested the former depended more on synthesis, whereas the latter may salvage lipids. Although baseline lipid profiles did not predict growth effects, lipid changes on therapy matched the growth effects of FASN and DGAT inhibitors. Several phospholipids, including phosphatidylcholine, were also upregulated following treatment, possibly via the Kennedy pathway. As this promotes tumor growth, combination studies should include drugs targeting it. Two-dimensional drug screening may miss important metabolism inhibitors or underestimate their potency. Clinical studies should consider serial measurements of tumor lipids to prove target modulation. Pretherapy tumor classification by de novo lipid synthesis versus uptake may help demonstrate efficacy.
Introduction
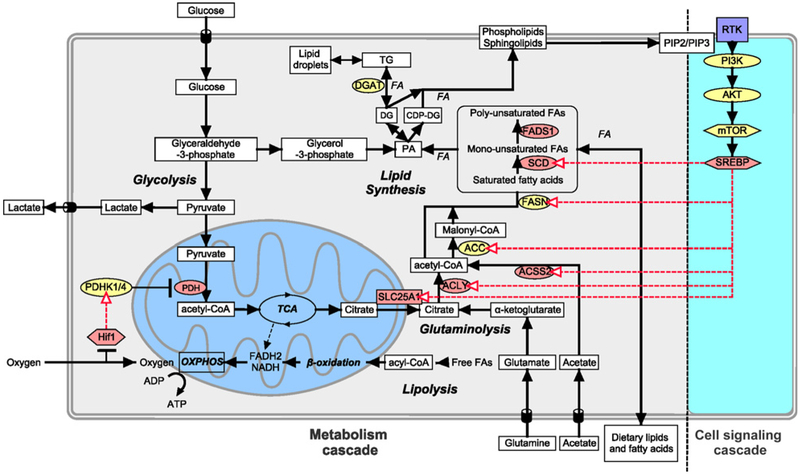
In solid tumors, cancer cells often reside in a hypoxic and metabolically compromised environment where oxygen and nutrient delivery, and acid clearance are reduced. Cancer cells adapt metabolically in many ways to these conditions (1, 2), including changes in fatty acid (FA) metabolism. The substrate for de novo FA biosynthesis is cytoplasmic acetyl-CoA, which can be produced from glucose, glutamine, and acetate (3, 4). Glucose undergoes glycolysis to produce pyruvate, which enters the mitochondria and is converted to acetyl-CoA by pyruvate dehydrogenase (PDH). This mitochondrial acetyl-CoA is converted to citrate by the enzyme citrate synthase (CS) and can be either further oxidized through the tricarboxylic acid cycle (TCA) or transported to the cytosol, where it is cleaved by ATP-citrate lyase (ACLY) to produce oxaloacetate and acetyl-CoA, the precursor for FAs (Fig. 1).
Figure 1.
Lipid biosynthesis in cancer cells. FAs are required by cells to build cell membrane as phospholipids and for energy storage as TG, which is stored in LDs and catabolized in the mitochondria through β-oxidation to provide energy. Lipids can be transported from extracellular source or synthesized by cells using glucose, glutamine, and acetate as carbon source. Cell signaling pathways can also increase lipid biosynthesis. Ligand binding to RTK, which activates PI3K phosphorylation of PIP2 to PIP3, activates AKT and mTOR pathway resulting in activation of the transcription factor SREBP. SREBP can upregulate citrate transporter SLC25A1 and lipid synthesis enzymes ACLY, ACSS2, ACC, FASN, and SCD. Hypoxia also plays a part in lipid biosynthesis, where Hif-1α upregulates glucose uptake, several enzymes in lipid biosynthesis, and reduces flux of glucose to acetyl-CoA through the mitochondria by upregulating PDHK1/4, kinase that inhibits PDH. Enzymes colored in yellow were those targeted with inhibitors. Metabolites are in square white boxes, enzymes are in oval boxes, transcription factors are in hexagonal boxes, and metabolic pathways are in bold and italics. Red arrows represent upregulation of expression by transcription factors.
To produce FAs, such as palmitic acid, acetyl-CoA is carboxylated to produce malonyl-CoA by Acetyl-CoA carboxylase (ACC), which is then condensed with acetyl-CoA to produce FAs by fatty acid synthase (FASN). Three of the major enzymes involved in FA biosynthesis, ACLY, ACC, and FASN, have been implicated in malignant transformation (3, 5-7). Many cancers, including breast and prostate cancer, show an increased expression of FASN, and this is associated with poor prognosis in various types of cancer (8-10). FAs are the substrates utilized to produce complex lipid species. For example, esterification of diacylglycerol (DG) with FAs, by diacylglyceride acyltransferase (DGAT), leads to the formation of triacylglycerides (TG), which is used for energy storage. This process occurs in the endoplasmic reticulum where TG are packaged into lipid droplets (LD), which has been shown to accumulate under hypoxic condition. LD is associated with higher tumor aggressiveness (11) and chemotherapy resistance (12), and protects cells from reoxygenation phase from ROS-induced cell death (13).
Intermediates of the FA pathway can also be converted into different phosphoglycerides, including phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), and phosphatidylinositol (PI), which build the major structural components of biological membranes (14, 15). Other phosphoglycerides, such as phosphatidic acid (PA), phosphatidylglycerol (PG), and cardiolipin (CL), constitute a relatively minor portion of the total cellular phosphoglycerides, and CL is restricted to the mitochondria where it mainly supports mitochondrial functions. Increased levels of phospholipid content have been shown to correlate with cell transformation and tumor progression (14, 16-18), indicating that the phospholipid synthesis rate increases with oncogenesis and tumor progression. Another class of lipids that are found in cell membranes includes sphingolipids such as Sphingomyelin (SM), which consist of a serine backbone as opposed to the glycerol backbone, and sterols such as cholesterol. Cholesterol acts to modulate the membrane fluidity and permeability via interacting with neighboring lipids, such as PC and SM. Cholesterol and sphingolipid metabolism have been shown to be deregulated in cancer (19, 20). Many lipids in plasma membrane and organelles affect cell signaling, the availability of intrinsic membrane proteins, membrane fluidity, and cell polarization (21).
Some of the metabolic enzymes are regulated by the hypoxia transcription factor-1α (Hif-1α) pathway and/or receptor tyrosine kinase (RTK)–mediated activation of PI3k/Akt and mTOR pathway through the activation of sterol regulatory element-binding proteins (SREBP; refs. 13, 22-24). SREBP upregulates the expression of stearoyl-CoA desaturase (SCD), fatty acid desaturase 1 (FADS1), FASN, ACC, ACCS2, ACLY, and solute carrier family 25 member 1 (SLC25A1; Fig. 1). Hypoxia can also inhibit PDH through the induction of its negative regulator pyruvate dehydrogenase kinase 1 (PDHK1) by Hif-1α (25-27). This decreases the oxidation of pyruvate to acetyl-CoA, increases the reduction of pyruvate to lactate, and reduces the synthesis of acetyl-CoA. Reduced acetyl-CoA levels present a dilemma for cancer cells that continue to proliferate under hypoxic conditions, with respect to FA synthesis, which is normally dependent on glucose-derived acetyl-CoA. Instead, hypoxic cancer cells utilize glutamine and acetate to generate Acetyl-CoA (refs. 3, 4; Fig. 1).
These enzymes are therapeutic targets, and a key question is whether direct inhibitors of lipid metabolism would be a more effective strategy than broadly targeting upstream regulators. For this reason, with LC/MS-based lipidomics analysis, we analyzed and compared the effects of inhibition of the PI3K/AKT/mTOR cell signaling pathway to cell metabolism inhibitors that target FASN, ACC, DGAT, and PDHK enzymes in prostate and breast cancer cell lines. This was performed under culture conditions relevant to those encountered within the tumor microenvironment in vivo, i.e., spheroid cultures that reproduce nutritional gradients and oxygen deprivation observed in tumors and could therefore enhance the effectiveness of drugs affecting lipid metabolism. The final aim was to elucidate lipid profile at baseline and after therapy, to assess if this was related to drug effectiveness and could identify other potential targets for synthetic lethality.
Materials and Methods
Cell lines
Cell lines were obtained from the London Research Institute (CRUK) cell services. The cell lines were authenticated using short tandem repeat profiling by CRUK and published in previous publications (3, 28). The PC3 cell line was grown in RPMI supplemented with 10% FCS (PAA Laboratories), 2 mmol/L L-glutamine, and penicillin/streptomycin. The MDA-MB-468 cell lines were grown in DMEM/F12 with 10% FCS (PAA Laboratories), 2 mmol/L L-glutamine, and penicillin/streptomycin. Cells were maintained in a humidified incubator at 5% CO2 and 37°C. For hypoxic exposure, cells were grown at 0.1% O2, 5% CO2, and 37° C. They were regularly checked for mycoplasma and validated.
Drugs
Drugs against metabolic enzymes FASN (AZ22), ACC (AZ52), DGAT (AZ17), and PDHK (AZ30), and cell signaling enzymes PI3K (AZ46), AKT (AZ28), and mTOR (AZ64) were provided by AstraZeneca (S.E. Critchlow, AstraZeneca) and dissolved in DMSO (Table 1 for compound potency on recombinant enzymes, and compound structure in Supplementary Fig. S1). Concentrations used are indicated in the relevant figure legend, and controls were treated with 1% DMSO.
Table 1.
Compounds' potency on recombinant enzymes
| Type | Enzyme target | Compound | Primary enzyme potency (IC50) |
Secondary enzyme potency (IC50) |
|---|---|---|---|---|
| Metabolic targets | FASN | AZ22 | 139 nmol/L FASN | |
| ACC | AZ52 | 202 nmol/L ACC1 | 235 nmol/L ACC2 | |
| DGAT | AZ17 | 7 nmol/L DGAT1 | 450 nmol/L ACAT1 | |
| PDHK | AZ30 | 6 nmol/L PDHK2 | 37 nmol/L PDHK1 | |
| Kinase targets | Pan | AZ46 | 1.2 nmol/L PI3Kα | 3 nmol/L PI3Kβ |
| PI3K | 4 nmol/L PI3Kδ | |||
| 21 nmol/L PI3Kγ | ||||
| 2 nmol/L mTOR | ||||
| AKT | AZ28 | 5 nmol/L AKT1 | 42 nmol/L AKT2 | |
| 21 nmol/L AKT3 | ||||
| mTOR | AZ64 | 4 nmol/L mTOR | 8.8 μmol/L PI3Kα | |
| 5.3 μmol/L PI3Kδ |
Two-dimensional drug viability assay
For two-dimensional (2D) growth, cells were plated in 96-well plates (2,000–5,000 cells per well). After 24 hours, media were replaced with drugs at different concentration or 1% DMSO alone as control. Cells were grown for 3 days in normoxic (20% O2) or hypoxic (0.1% O2) conditions. Cell proliferation was measured using the CyQUANT NF Cell Proliferation Assay Kit (Invitrogen—Life Technologies) following the manufacturer's instructions. Data and growth inhibition by 50% (GI50) calculation was carried out using GraphPad Prism 6 software using Nonlinear regression, log(inhibitor) vs. response – Variable slope calculation (GraphPad Software, Inc.).
Spheroid drug viability assay
For spheroid formation, cells were mixed with 2% Matrigel (Becton Dickinson) in culture medium and plated into 96-well ultralow attachment plates (Costar CLS7007), 10,000 cells per spheroid. Spheroid formation was initiated by centrifugation at 850 × g for 10 minutes, and cultures were incubated for the times indicated. Fresh growth medium, containing drug or 1% DMSO alone, was administered first after 48-hour growth and replaced every 48 hours. Spheroid size was determined at the times indicated using an inverted microscope (Axiovert 100M, Carl Zeiss) where spheroid area was calculated using ImageJ software (29). Spheroid area was used to calculate spheroid radius (r = √(A/π)), from which the diameter (d = 2r) and volume can be calculated (v = (4/3)π r3). Data and growth inhibition by 50% (GI50) calculation was carried out using GraphPad Prism 6 software using Nonlinear regression, log (inhibitor) vs. response – Variable slope calculation (GraphPad Software, Inc.).
Mass spectrometry
Twenty to 25 PC3 and MDA-MB-468 spheroids were treated for 4 days with the inhibitors, collected, and were shipped to Babraham Institute Lipidomics Facility on dry ice for lipid mass spectrometry analysis. Spheroid samples were resuspended in LC-MS grade water. Half of the suspension was extracted, derivatized prior to phosphoinositide UPLC-MS/MS analysis as previously described (30). The other half was used for Folch extraction and analysis of all the other lipids using LC-MS/MS as described previously (3). Lipids in drug-treated spheroids were compared with 1% DMSO-treated control spheroids, and lipid differential abundance was calculated as were their carbon lengths and double-bond numbers. Data were normalized as ng of lipids per 106 total and viable cells. To count cells in spheroids, a sample of 5 spheroids was collected and tryps-inized. Cells were counted using SD100 Cellometer Counting Chamber slides (Nexcelcom) and Cellometer Auto x4 Cell Counter (Nexcelcom). Data uploaded to Metabolights hosted by EBI can be seen athttps://www.ebi.ac.uk/metabolights/.
Statistical analysis and bioinformatics
Lipid abundance data were preprocessed using the R statistical program (v3.2.4). Total and viable cell population was used to normalize the cell lines. Data were transformed to log2 scale for subsequent analysis. Statistical comparison between the treatment and control groups was performed using the paired t test, except for cases where valid abundances were only available for less than three samples, in which case only fold change was reported. Within the treatment groups, the presence of saturated, monounsaturated, and polyunsaturated lipid families was compared using ANOVA followed by the Tukey Honest Significant Differences (HSD) test for pair-wise comparisons. Comparison between saturated, monounsaturated, and polyunsaturated lipid families was performed using the paired t test. Analysis, heatmaps, and barplots were generated by the R statistical program (v3.2.4). For heatmaps and hierarchical clustering, unsupervised exploratory analysis of the metabolomics data (standardized by z-score scale), k-mean, was utilized to determine the optimal number of clusters.
Results
Lipidomic analysis of PC3 and MDA-MB-468 spheroids
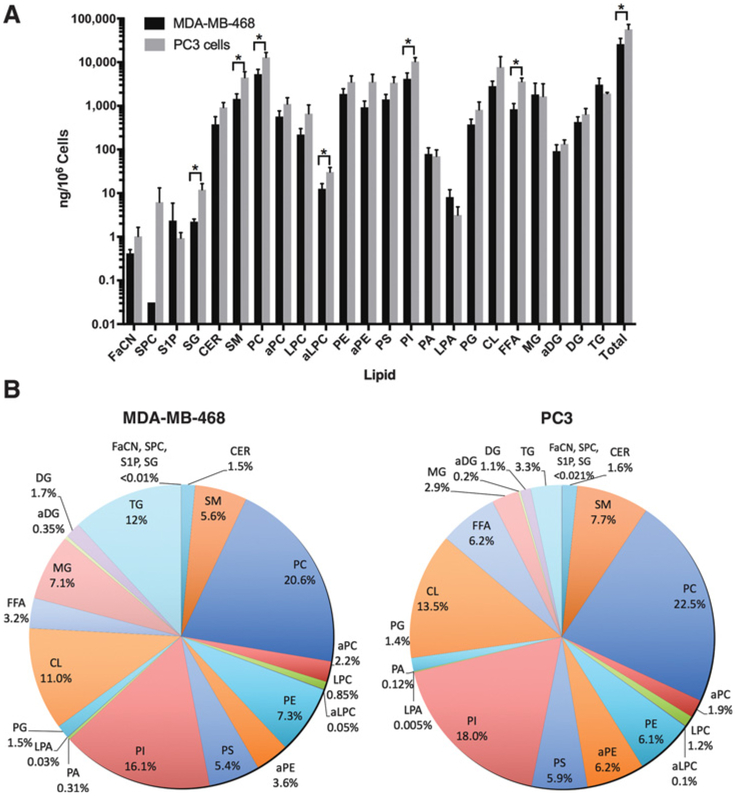
We first generated extracts from spheroid cultures of prostate and breast cancer cells lines and subjected these to lipidomic analysis to analyze total amount and proportion of lipids in the two cell lines (Fig. 2A and B). This revealed that PC3 cells had significantly higher total lipid content than MDA-MB-468 cells (Fig. 2A). A total of 60,000 ng of lipid/106 cells and 26,000 ng of lipid/106 cells were detected in PC3 and MDA-MB-468 spheroids, respectively.
Figure 2.
Lipid levels in MDA-MB-468 and PC3 spheroids. Mass spectrometry analysis of lipids as (A) amount, and (B) percentage in each cell type.
Three cell membrane phospholipids, PC, PI, and CL, were the most abundant lipid species detected in spheroids from both cell types (Fig. 2A and B). Other phospholipids detected included PE, PS, PG, and lyso-PC (LPC). Alkyl-phospholipids, alkyl-PC (aPC), alkyl-PE (aPE), and alkyl-LPC (aLPC), and PA and lyso-PA (LPA) were detected in smaller quantities in MDA-MB-468 and PC3 spheroids. PC3 spheroids had more phospholipids than MDA-MB-468, with PC, aLPC, and PI being significantly higher (Fig. 2A).
SM was the most abundant sphingolipid (Fig. 2B), whereas ceramide (Cer), which is produced de novo from SM, was detected in smaller amounts. PC3 spheroids had more sphingolipids than MDA-MB-468, with SG and SM being significantly higher.
The most abundant neutral lipid detected was TG, followed by monoglyceride (MG) and DG (Fig. 2B). MG and DG are found in lower amounts in cells as they are intermediates in the catabolism or synthesis of TG. The amount of neutral lipids in both PC3 and MDA-MB-468 was similar, but PC3 cells did significantly have more free fatty acid (FFA). Analysis of lipid proportion (Fig. 2A and B) reveals that lipid constituents were in similar proportions in the two cell lines, except for TG, MG, and FFA that were higher in proportion in MDA-MB-468 than PC3.
Inhibition of monolayer and spheroid growth with kinase and metabolic enzyme inhibitors
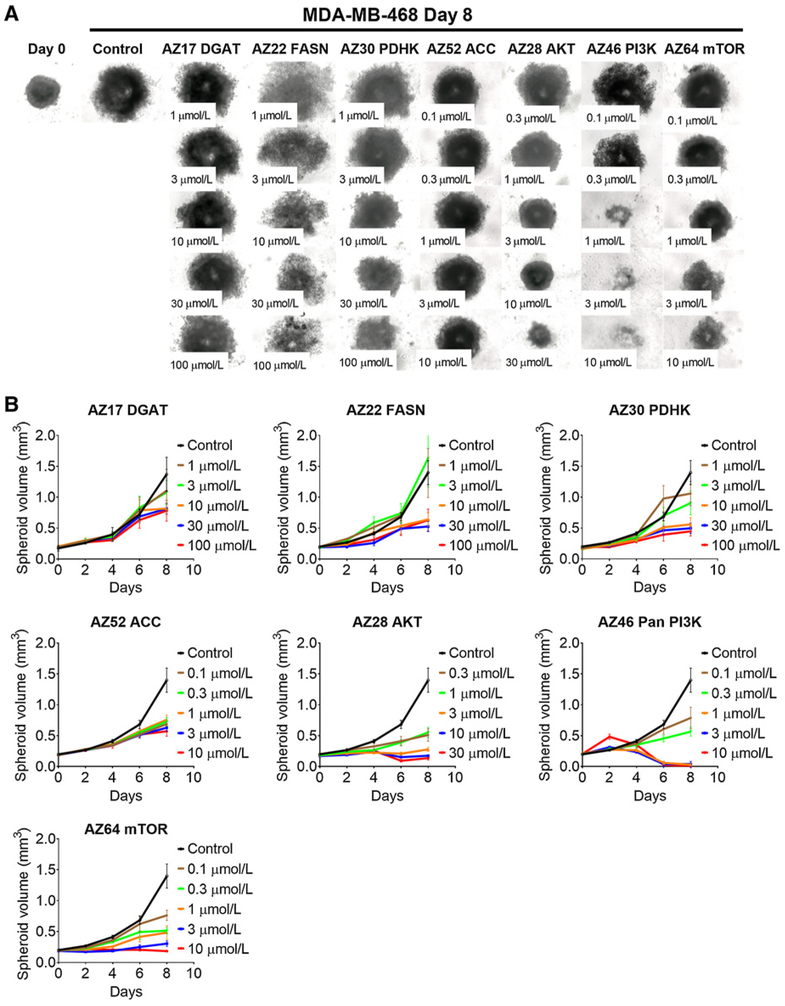
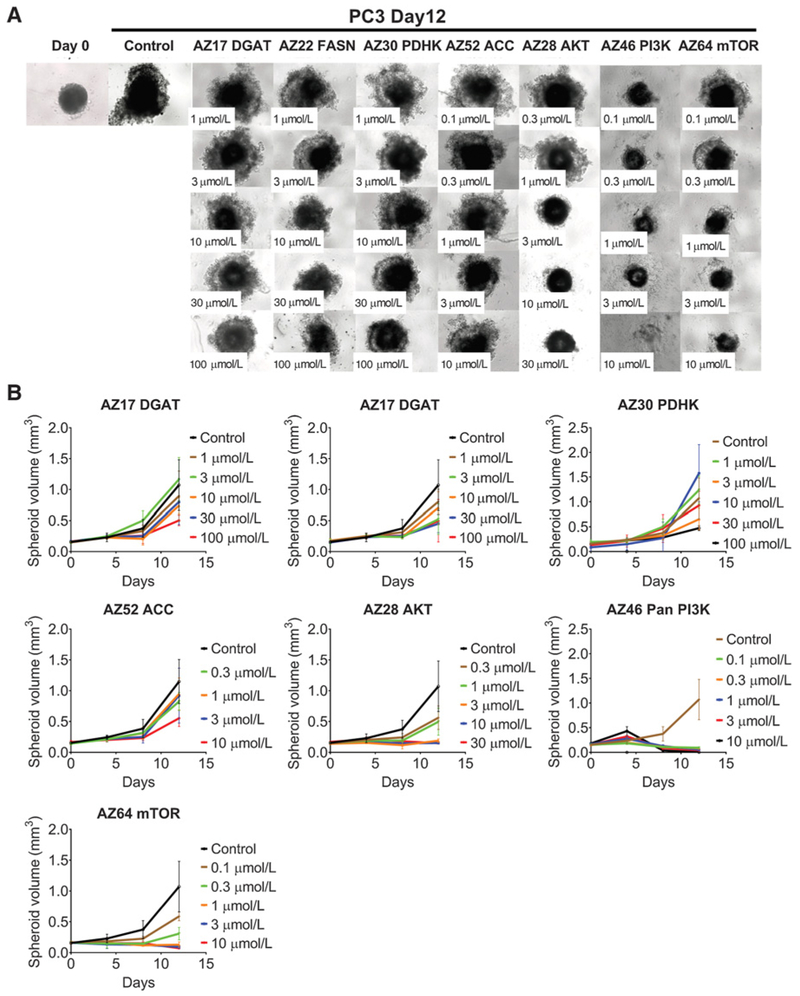
We used inhibitors against the metabolic pathway enzymes PDHK, ACC, FASN, and DGAT, and the kinase enzymes PI3K, AKT, or mTOR on cells cultured as monolayers or as spheroids, and calculated the GI50 (Table 2). In monolayer cultures, after 3 days of exposure in normoxic or hypoxic conditions, all drugs, with the exception of the DGAT inhibitor, attenuated MDA-MB-468 and PC3 cell growth (Supplementary Fig. S2A and S2B; Table 2). The kinase inhibitors acting upon PI3K, AKT, or mTOR had the lowest GI50 for MDA-MB-468 and PC3 cells, while the ACC inhibitor had a limited effect in inhibiting growth of both MDA-MB-468 and PC3 cells under hypoxic conditions (Table 2).
Table 2.
Growth inhibition of PC3 and MDA-MB-468 cells in 2D and 3Dgrowth following treatment with inhibitors against metabolic enzymes PDHK, ACC, FASN, and DGAT, and kinases PI3K, AKT, and mTOR
| Prostate cancer (PC3) |
Breast cancer (MDA-MB-468) |
||||
|---|---|---|---|---|---|
| Type | Enzyme target | 2D | 3D | 2D | 3D |
| Metabolic targets | FASN | N = 26 μmol/L | 7.2 μmol/L | N = 19 μmol/L | 17 μmol/L |
| H = 84 μmol/L | H = 22 μmol/L | ||||
| ACC | N = NC | 11 μmol/L | N = NC | NC | |
| H = 11 μmol/L | H = 60 μmol/L | ||||
| DGAT | N = NC | 46 μmol/L | N = NC | NC | |
| H = NC | H = NC | ||||
| PDHK | N = 54 μmol/L | NC | N = 17 μmol/L | 5.4 μmol/L | |
| H = NC | H = 46 μmol/L | ||||
| Kinase targets | Pan PI3K | N = 0.1 μmol/L | 0.02 μmol/L | N = 0.2 μmol/L | 0.14 μmol/L |
| H = 0.2 μmol/L | H = 0.3 μmol/L | ||||
| AKT | N = 20 μmol/L | 0.36 μmol/L | N = 8.3 μmol/L | 0.17 μmol/L | |
| H = 29 μmol/L | H = 8.6 μmol/L | ||||
| mTOR | N = 0.8 μmol/L | 0.1 μmol/L | N = 1 μmol/L | 0.12 μmol/L | |
| H = 0.7 μmol/L | H = 1.4 μmol/L | ||||
NOTE: In 2D growth, cells were incubated under normoxia (N) and hypoxia (H). Results are shown as concentration of the drug that inhibited 50% growth (growth inhibition 50%–GI50), calculated using GraphPrism nonlinear regression (curve fit).
Abbreviation: NC, not calculated due to poor curve fit or no loss in cell viability with highest concentration.
All drugs inhibited spheroid growth following 8 days of exposure for MDA-MB-468 spheroids (Fig. 3A and B; Table 2) and 12 days for PC3 spheroids (Fig. 4A and B). As with monolayer cultures, a greater effect on growth was observed with kinase inhibitors acting on AKT, PI3K, and mTOR (Figs. 3 and 4; Supplementary Fig. S3; Table 2). As with monolayers, the metabolic inhibitors were less effective in inhibiting spheroid growth (Figs. 3 and 4; Supplementary Fig. S3; Table 2). MDA-MB-468 spheroids were more sensitive to FASN and PDHK inhibitors (Fig. 3; Supplementary Fig. S3A), whereas PC3 spheroids were sensitive to FASN and ACC inhibitors (Fig. 4; Supplementary Fig. S3B). Unlike 2D growth, DGAT did inhibit PC3 spheroid growth, whereas MDA-MB-468 spheroids were more resistant to this inhibitor (Supplementary Fig. S3; Table 2).
Figure 3.
MDA-MB-468 spheroid growth inhibition with inhibitors. Microscope images (A) and (B) growth curves of spheroids treated with inhibitors against metabolic enzymes PDHK, ACC, FASN, and DGAT, and kinases PI3K, AKT, and mTOR following 8 days of growth. Between 5 and 15 spheroids were measured per condition, and errors bars represent SD.
Figure 4.
PC3 spheroid growth inhibition with inhibitors. Microscope images (A) and (B) growth curves of spheroids treated with inhibitors against metabolic enzymes PDHK, ACC, FASN, and DGAT, and kinases PI3K, AKT, and mTOR following 12 days of growth. Between 5 and 15 spheroids were measured per condition, and errors bars represent SD.
Effects of metabolic and kinase inhibitors on cellular lipids
To establish the effect of inhibitors on cellular lipids, we used two different concentrations of inhibitors, and lipid changes were analyzed by trend (Supplementary Figs. S4-S6) and significance (Fig. 5; Supplementary Fig. S7). Drug concentrations were selected based on data from previous experiments. Drugs that inhibited PI3K, AKT, and mTOR kinases altered levels of many lipid species in spheroids from both cell types (Supplementary Fig. S4), but only a small proportion of lipid species were significantly altered (Fig. 5; Supplementary Fig. S7; Table 3). In contrast, inhibitors against metabolic enzymes DGAT and FASN were more effective in changing lipid levels in both cell types (Supplementary Fig. S5) with many being significant (Fig. 5; Supplementary Fig. S7; Table 3).
Figure 5.
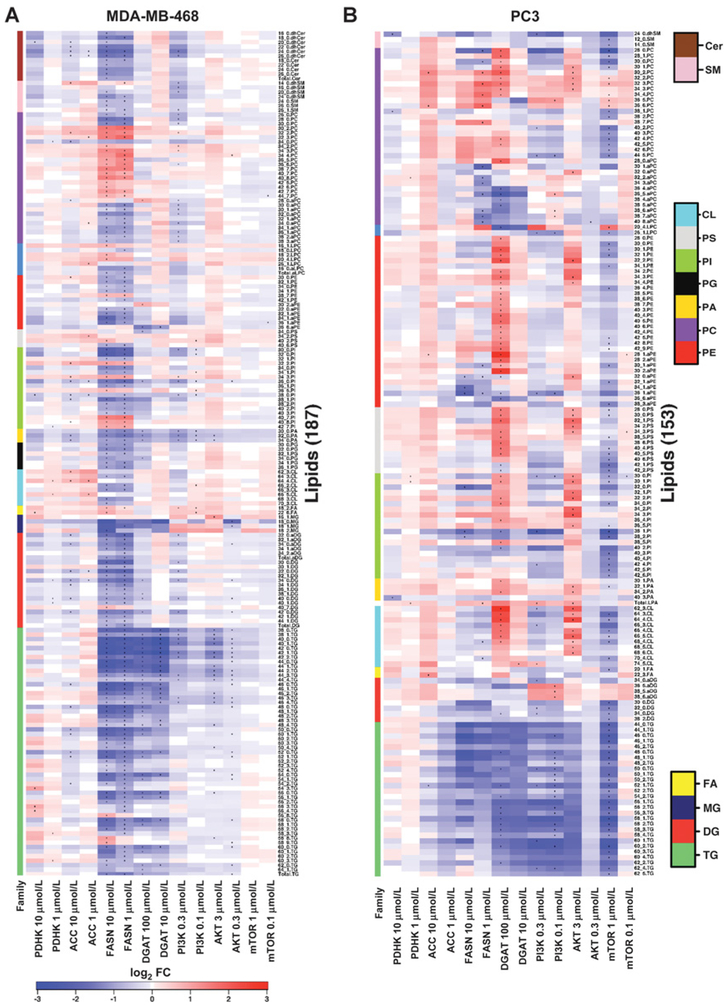
Significant differential abundance analysis of lipids as a heat map. (A) MDA-MB-468 and (B) PC3 spheroids were treated for 4 days with inhibitors against metabolic enzymes PDHK, ACC, FASN, and DGAT, and kinases PI3K, AKT, and mTOR at two different concentrations. All lipids were compared against DMSO control, normalized to total cells. Only lipids that were differentially abundant (P < 0.05) following treatment by at least one drug (see dots in heat map) are represented. Side covariates indicate lipid families, and lipids within each family are sorted by name. The paired t test was used where three replicates were available. Only fold change is reported where two samples were available (P < 0.05 and absolute log2FC > 1).
Table 3.
List of lipids in 3D spheroids significantly altered by the drugs
| Prostate cancer (PC3) |
Breast cancer (MDA-MB-468) |
||||
|---|---|---|---|---|---|
| Type | Enzyme target | Increased | Decreased | Increased | Decreased |
| Metabolic targets | FASN | PC | CL, PI, aPE, aPC, PC | TG, DG, FFA, PI, PS,PE, LPC, PC, dhSM | TG, DG, aDG, MG, CL, PG, PA,PI, aPE, PE, aLPC, LPC, aPC, PC, SM, Cer, dhCer |
| ACC | FFA, aPE, PC | TG | CL, PI, PS, PE, LPC,aPC, PC, dhSM | TG, DG, aDG, PA, PI, aPC, PC,dhCer | |
| DGAT | CL, PA, PI, PS, aPE, PE,aPC, PC | TG, DG, PI, PS, aPE, aPC | aPE, aPC | TG, DG, PA, PI, PS | |
| PDHK | LPA, PI, aPC | PA, PC, dhSM | TG, FFA, CL | PI, DG, PC | |
| Kinase targets | Pan PI3K | aDG, CL, LPA, aPC, PC | TG, DG, CL, PI, aPE, LPC,PC, dhSM | FFA, CL, PI, PS, PC | TG, DG, aDG, PA, PI, aPC, PC,SM, dhSM, dhCer |
| AKT | CL, PA, PI, PS, aPE, PE,aPC, PC | aPC | MG, PC | TG, DG, aDG, MG, PA, PI | |
| mTOR | PS, aPE, LPC, PC | TG, DG, FFA, CL, PA, PI, PS,aPE, PE, LPC, PC, SM | aPE, PC | ||
Neutral lipids.
The PI3K inhibitor at 0.3 μmol/L and the AKT inhibitor at 3 μmol/L and 0.3 μmol/L significantly reduced several TG species in MDA-MB-468 spheroids, whereas no significant changes were observed with the mTOR inhibitor (Fig. 5A). DG and MG concentrations were not significantly altered, but a trend in increasing levels was observed following AKT inhibition (Supplementary Fig. S4A), which suggests that targeting the PI3K/AKT pathway inhibits DGAT. In PC3 spheroids, the PI3K inhibitor at 0.3 and 0.1 μmol/L, and the mTOR inhibitor at 1 μmol/L reduced several TG and DG species, whereas no significant changes in TG levels were seen with the AKT inhibitor (Fig. 5B), showing contrasting and reciprocal effects of the mTOR and AKT inhibitors between the cell lines.
The most significant changes in neutral lipids were observed following treatment with inhibitors against FASN and DGAT (Fig. 5; Supplementary Fig. S7). A trend and a significant reduction in total TG and several TG species were observed in MDA-MB-468 spheroids following treatment with FASN and DGAT inhibitors (Supplementary Fig. S5A and S5B; Fig. 5; Supplementary Fig. S7A). FASN inhibition also significantly reduced levels of total DG and MG in MDA-MB-468 spheroids, confirming the role of FASN in FA synthesis, which is required to make MG, DG, and TG (Fig. 5A). In contrast, FASN inhibition did not significantly reduce TGs in PC3 spheroids, but a trend was observed (Supplementary Fig. S5A), whereas 100 μmol/L DGAT inhibitor in PC3 spheroids significantly reduced several TGs (Fig. 5B). It is possible that PC3 rely less on lipid synthesis than MDA-MB-468 and acquire FA via uptake.
No clear trend in lipid changes was observed following treatment with ACC and PDHK inhibition (Supplementary Fig. S6). However, inhibition of ACC (10 μmol/L) had a small and significant effect on lowering several TGs and DGs in MDA-MB-468 cells, but only affected 52:1 TG in PC3 spheroids (Fig. 5). The PDHK inhibitor treatment at 10 and 1 μmol/L resulted in a significant increase in several TGs in MDA-MB-468 spheroids, but not in PC3 spheroids (Fig. 5).
FFAs.
Very few significant changes in FFA levels were observed. The levels of poly-unsaturated 18:2 and 22:6 FFA were significantly increased in MDA-MB-468 spheroids following treatment with PI3K and FASN inhibitors (Fig. 5A). The FFA 22:6 was also significantly increased in MDA-MB-468 spheroids following treatment with 10 μmol/L PDHK inhibitor. In PC3 spheroids, FFA 20:1 significantly decreased following treatment with 1 μmol/L mTOR inhibitor, whereas the ACC inhibitor at 10 μmol/L significantly increased FFA 22:3 (Fig. 5B), and an increasing trend was observed following treatment with 100 μmol/L DGAT inhibitor (Supplementary Fig. S5B). Again, this may suggest that PC3 rely less on lipid synthesis than MDA-MB-468 and acquire FA via uptake.
Phospholipids.
With the cell signaling inhibitors, phospholipids in PC3 spheroids were more sensitive to AKT and mTOR inhibition than MDA-MB-468 spheroids (Fig. 5A and B; Supplementary Fig. S4A and S4C). Levels of CL, PI, PS, aPE, PE, and PC were significantly increased following treatment with 3 μmol/L AKT inhibitor in PC3 spheroids, and an increasing trend was observed in many phospholipids in both PC3 and MDA-MB-468 spheroids (Supplementary Fig. S4A). Contrasting effects were observed in MDA-MB-468 and PC3 spheroids following mTOR inhibition, with many phospholipids showing a decreasing trend in PC3 spheroids, whereas levels increased in MDA-MB-468 spheroids (Supplementary Fig. S4C). No clear trend on phospholipid levels was observed following treatment with PI3K inhibitor (Supplementary Fig. S4B), but PI3K inhibitor did significantly decrease some PA, PI, PC, and aPC species, and significantly increase several PC, PI, PS, and CL in MDA-MB-468 spheroids (Fig. 5A). In PC3 spheroids, the PI3K inhibitor was less effective but significantly reduced the levels of some CL, PI, aPE, LPC, and PC species, whereas total LPA and some aPC and PC species were significantly increased (Fig. 5B).
Analysis of phospholipids following treatment with lipid metabolism inhibitors against DGAT and FASN revealed no clear trend (Supplementary Fig. S5), but did show that DGAT and FASN inhibitors were the most effective in significantly altering levels, with PC3 being more sensitive to DGAT inhibition, whereas MDA-MB-468 were more sensitive to FASN inhibition (Fig. 5A and B). This again may suggest that PC3 rely less on lipid synthesis than MDA-MB-468 and acquire FA via uptake.
In PC3 spheroids treated with 100 μmol/L DGAT inhibitor, many CL, PA, PI, PS, aPE, PE, and PC species and 28:0 aPC were significantly upregulated, suggesting increase in both the CDP-DG pathway and the Kennedy pathway, whereas LPC, aPC, and some PI, PS, and aPE species were significantly decreased (Fig. 5B). In contrast, fewer significant changes in phospholipid levels were observed in MDA-MB-468 spheroids treated with DGAT inhibitor, where 36.6 aPE, 34:0 PS, 36:0 PI, 30:0, 32:0 and 34:0 PA, and 34:0 PG were significantly reduced following treatment with 100 μmol/L DGAT (Fig. 5A).
MDA-MB-468 spheroids were more sensitive than PC3 spheroids to FASN inhibition. PA, PI, PS, PE, PG, and CL were significantly reduced following FASN inhibition, and this coincided with a reduced MDA-MB-468 spheroid growth (Fig. 5A). Some polyunsaturated PI and PC species were significantly upregulated in MDA-MB-468 spheroids, whereas shorter saturated PC (e.g., 26:0, 28:0, and 30:0) decreased following treatment (Fig. 5A). Increases in polyunsaturated PCs levels were also observed in PC3 spheroids treated with 1 μmol/L FASN inhibitor (Fig. 5B). FASN inhibition in MDA-MB-468 spheroids significantly reduced total saturated PC, PI, and PE, whereas no changes were observed in total saturated phospholipids in PC3 spheroids (Supplementary Fig. S8). Unlike PC, some aPC species significantly decreased in MDA-MB-468 and PC3 spheroids following FASN inhibition.
Inhibition of ACC and PDHK resulted only in a small number of significant changes in phospholipids in MDA-MB-468 and PC3 spheroids (Fig. 5). However, an increasing trend in phospholipids was observed in MDA-MB-468 following treatment with 10 μmol/L ACC inhibitor (Supplementary Fig. S6A).
Sphingolipids.
The levels of different sphingolipids were also significantly altered by PI3K, PDHK, ACC, and FASN inhibition in MDA-MB-468 spheroids and by PDHK, PI3K, and mTOR inhibition in PC3 spheroids, with the majority being downregulated. The PI3K inhibitor at 0.3 μmol/L significantly downregulated 26:1 SM, 14:0, 16:0 and 20:0 dhSM, and 24:0 and 26:0 dhCer in MDA-MB-468 spheroids (Fig. 5A). Only 24:0 dhSM species were significantly downregulated in PC3 spheroids (Fig. 5B). The levels of 12:0 and 14:0 SM were significantly reduced in PC3 spheroids by the mTOR inhibitor (Fig. 5B).
Addition of the PDHK inhibitor at 10 μmol/L caused a reduction in 20:0 dhCer in MDA-MB-468 spheroids and 24:0 dhSM in PC3 spheroids (Fig. 5A and B). The ACC inhibitor downregulated some dhCer species and upregulated 14:0 dhSM species in MDA-MB-468 spheroids, whereas no change was observed in PC3 spheroids (Fig. 5A and B).
Finally, the FASN inhibitor at 10 μmol/L significantly reduced total Cer and several SM, shSM, and dhCER species, whereas in contrast, 14:0 dhSM was significantly upregulated in MDA-MB-468 spheroids (Fig. 5A). However, no changes in sphingolipids were seen in PC3 spheroids after treatment with the FASN inhibitor.
Discussion
A key aim of the study was to asses if baseline lipid profiles were related to growth delay produced by agents inhibiting major pathways of lipid metabolism. The initial lipid profiles in the PC3 versus MDA-MB-468 spheroids differed significantly. PC3 cells had higher total lipid content and had a higher proportion of membrane lipids compared with TGs, whereas MDA-MB-468 had higher proportion of TGs compared with membrane lipids. Thus, the effects of specific inhibitors of key enzymes that can regulate these pathways might be predicted by the predominance of the substrates and products. PC3 were more sensitive to DGAT. TGs comprised 12% of lipids in MDA-MB-468 but only 3% in PC3. MG similarly were elevated in MDA-MB-468 (7%) versus PC3 (2.9%). Although no flux analysis was performed, the data suggest that there was much greater storage in MDA-MB-468, and great utilization or turnover in PC3, reflected in the increased sensitivity to DGAT in the latter cell line. Cancer cells, especially breast cancer, have larger number of LD containing TG in their cytoplasm than normal cells (31-33). LD role in cancer is only beginning to be explored, and recent evidence suggests that higher levels of LDs are associated with higher tumor aggressiveness (11) and chemotherapy resistance (12), and protect cells from reoxygenation phase from ROS-induced cell death (13). Reducing LD in cancer cells may help in cancer treatment by removing protective effects of LD.
In PC3 cells, there was a higher phospholipid proportion. Inhibition of ACC, which regulates the first and rate-limiting step in the biosynthesis of FAs, has been shown to change phospholipid composition, reducing PG and altering cell membrane properties (34), and thus might be expected to have a greater effect on cells with a higher proportion of phospholipids.
The in vitro results for metabolic inhibitors on purified enzyme assays were much more effective than on intact cells, in the nanomolar range in the former and micromolar in the latter (Tables 1 and 2). This is most likely due to cellular uptake and excretion, as the tool compounds were selected for specificity and inhibitory potency. However, the 2D and threedimensional (3D) results are both in the micromolar range, and in many cases, the drugs were 5 or more fold more effective against 3D growth. Thus, it is unlikely that uptake is the main cause for differences between cell lines and drug activity. The data support the importance of analyzing 3D growth, particularly for drugs directly affecting metabolic pathways, which would allow metabolic gradients and exchanges to be targeted.
The inhibitor effects on lipids were studied only in 3D for the above reasons. A further key aim was to assess if the changes induced in lipid profiles were related to baseline levels or the antiproliferative effects. DGAT inhibitor did significantly reduce a greater range of neutral lipids in PC3 than MDA-MB-468, potentially correlating with growth-inhibitory effects and baseline profile.
For ACC inhibition, the overall effects on lipids were small and showed no selective effects on phospholipids in PC3 cells. In this case, there was no prediction based on effects on lipids, but it is possible that the cells depend on different isoforms of ACC, cytoplasmic ACC1, and mitochondrial ACC2 both inhibited by AZ52 (34). Inhibiting the latter would affect beta-oxidation as opposed to lipid synthesis and be unlikely to show major changes in lipids. Inhibition of ACC did show more lipid changes in MDA-MB-468 cells and spheroid growth. The difference in effects on lipids and growth may reflect greater reliance on synthesis in MDA-MB-468, whereas PC3 may adapt by upregulation of lipid transport and uptake, e.g., by FABP regulation (13). Studies have demonstrated that prostate cancer cells have low rate of glycolysis and glucose uptake, and tend to uptake FA (35). The preferential uptake of lipids over glucose in prostate cancer has previously been assessed for potential therapeutic targeting (35-37).
Most effective metabolic targeting drugs that altered lipid profiles in MDA-MB-468 and PC3 cells were inhibitors that target FASN and DGAT (Fig. 6). FASN inhibition was more effective on the growth of MDA-MB-468 cells and produced greater reduction in lipid profiles, particularly TG, DG, and MGs, with much less effects on PC3 lipids. The MDA-MB-468 has lower total lipids, so baseline was not predictive. Again, the difference in effects on lipids and growth may reflect greater reliance on synthesis in MDA-MB-468. DGAT inhibition, PC3 growth more than MDA-MB-468 and produced greater and more significant TG reduction in the former cell line.
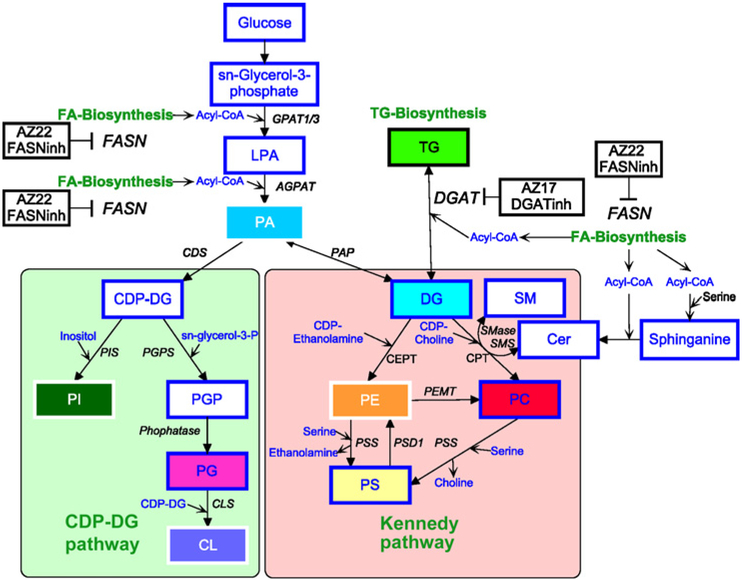
Figure 6.
Phospholipid and TG synthesis pathway. Phospholipids in cells can be synthesized via the CDP-DG and Kennedy pathway using PA and TG. Inhibitors against FASN and DGAT have an impact on both pathways. FASN inhibitor inhibits FA biosynthesis, whereas DGAT catalyzes the synthesis of TG from DG and Acyl-CoA. AGPAT, 1-acylglycerol-3-phosphate O-acyltransferase 1; CDS, cytidylyltransferase; CLS, Cardiolipin synthase; CPET, choline/ ethanolamine phophotransferase; CPT, choline phosphotransferase; GPAT1/3, glycerol-3-phosphate acyltransferase 1/3; PAP, phosphatidic acid phosphatase; PEMT, phosphatidylethanolamine N-methyltransferase; PGPS, phosphatidylglycerolphosphate synthase; PIS, phosphatidylinositol synthase; PSD1, phosphatidylserine decarboxylase; PSS, phosphatidylserine synthase.
For PDHK inhibition, we observed little effect on lipid profile, and spheroid growth was moderately affected. This was unexpected, as hypoxia should render cancer cells more sensitive to this inhibitor, by reactivating Krebs cycle activity under adverse conditions.
Thus, for two of the drugs, against ACC and FASN, it appeared that if a cell line was more dependent on lipid synthesis, there would be change in lipids and growth on treatment, but if lipid uptake was more important, there would be no lipid change and growth. For three targets, the reduction in lipid profiles correlated with a reduction in growth (ACC, FASN, and DGAT).
Whereas all the above discussion relates to 3D growth in normoxia, we conducted 2D growth experiments in normoxia and hypoxia. Overall, there was little change in drug sensitivity in hypoxia, showing that using hypoxia to try to replicate in vivo conditions was not sufficient to generate the sensitivity found in 3D growth. This supports the use of 3D models for metabolism inhibitors, where more severe hypoxia and gradients for many metabolites occur.
Although not novel, our results show that inhibitors of the kinases PI3K, AKT, and mTOR are about 10-fold more potent than lipid metabolism inhibitors in blocking the growth of cancer cells in spheroid cultures, likely due to multiple cellular processes they inhibit (38-40). Each of the signaling kinase inhibitors modified lipid profiles in both PC3 and MDA-MB-468 cancer cell lines. However, for these inhibitors, the effect on lipid composition did not correlate with their effect on cell growth. For example, the pan PI3K inhibitor was more active in changing lipid profiles in MDA-MB-468 than in PC3 cells, but was 7-fold less potent in reducing MDA-MB-468 spheroid growth when compared with PC3 cells.
Both cells have a functional PI3K and a mutant PTEN and p53 (41, 42), and are sensitive to inhibitors against PI3K and AKT, but PC3 cells were more sensitive to pan PI3K inhibition. Similarly, the AKT inhibitor produced a stronger effect on lipid metabolism in PC3 cells compared with MDA-MB-468 cells. However, the drug effects on spheroid growth were equivalent in both cell types.
For mTOR inhibition, there were major effects on lipid profile in PC3 cells but only a minimal response in MDA-MB-468, showing that mTOR has different functions in these cells, yet the GI50 for this drug was similar.
Thus, for these signaling kinase inhibitors, the direction and extent of change in lipid profiles were not related to their growth effects. Our data show that, in spite of the importance of these kinases for lipid metabolism, their therapeutic efficacy is likely to be due to their effects on other targets (38-40).
Although inhibitors of lipid metabolism can prevent the formation of some lipid species, they can also increase the formation of others with potential protumorigenic properties. In our study, we observed that DGAT inhibition leads to increased levels of several phospholipids (e.g., PC, PE, PS, CL, and the majority of PI species), most likely due to the inhibition of the metabolic step converting DG into TG (Fig. 6). As DG is the main metabolic substrate for the synthesis of phospholipids via the Kennedy/CDP-DG pathway, inhibition of DGAT can enhance phospholipid synthesis. The most abundant phospholipid that increased following DGAT inhibition was PC, which is known to contribute to increased growth in cancer cells (14, 16-18). The second most abundant phospholipid that was increased following DGAT inhibition was PI, which is involved in lipid signaling, cell signaling, and membrane trafficking (30, 43). The third accumulating lipid was CL, which is located mainly on the inner membrane of mitochondria and activates enzymes involved in oxidative phosphorylation (44). PE and PS were detected at lower amounts but still increased following DGAT inhibition. These lipids have key roles in metabolism and apoptosis (21, 45).
FASN inhibition was also associated with a relative increase of polyunsaturated FA, which could be attributed to enhanced FA uptake or the increased hydrolysis of TG, DG, and MG, thus increasing the pool of unsaturated and polyunsaturated FA, which can serve as substrate for the synthesis of polyunsaturated PC.
The clinical relevance of these data relates to screening and developing effective drugs targeting lipid metabolism, and the development of pharmacodynamic markers to select appropriate patients for clinical trial and prove the target has been inhibited in the tumor tissue. Thus, screening in 3D rather than 2D will be important, due to its more complex environment that is similar to a tumor environment, which will give a more relevant drug dose response. Baseline lipid profiles are unlikely to predict the effectiveness of the drugs, but a way to classify tumors which have a large component of lipid uptake versus de novo synthesis may be helpful. This could be done by PET imaging with agents that include acetate, choline and palmitate (46, 47). Most relevant would be serial analysis of biopsies to detect the changes in profiles on therapy and prove that dosage regimen at least modified the target.
Many of these drugs tested here reduced the levels of TG, found in LD, which may reduce tumor aggressiveness, chemotherapy resistance, and protection against ROS-induced cell death (11-13). With regards to upregulation of lipids on therapy, the Kennedy pathway has been extensively investigated as a drug target (14, 16-18, 48). Inhibition of choline kinase alpha (CKα), the first committed step to PC synthesis, by the selective small-molecule ICL-CCIC-0019, suppressed growth of a panel of cancer cell lines and inhibited tumor xenograft growth in mice (48), and one CKα inhibitor has been used in phase I clinical trials against advanced solid tumors (http://clinicaltrials.gov/show/NCT01215864). Our data suggest that combination strategies including such drugs should be tested, as single modifiers alone are unlikely to be effective.
Supplementary Material
Acknowledgments
This work was funded by Cancer Research UK C602/A18974 (to A.L. Harris) and AstraZeneca, as part of the Lipid Metabolism Consortium. The authors would like to thank Neil Jones and Angus Lauder from Cancer Research UK for the management of the consortium and scientific advice. A.L. Harris and this work were also funded by the Breast Cancer Research Foundation (BCRF) 2018.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Disclosure of Potential Conflicts of Interest
S.E. Critchlow is Director at, and has an ownership interest (including stock, patents, etc.) in, AstraZeneca. E. Gottlieb has an ownership interest (including stock, patents, etc.) in, and is a consultant/advisory board member for, MetaboMed Ltd. No potential conflicts of interest were disclosed by the other authors.
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
References
- 1.Semenza GL. Regulation of cancer cell metabolism by hypoxia-inducible factor 1. Semin Cancer Biol 2009;19:12–6. [DOI] [PubMed] [Google Scholar]
- 2.Warburg O On respiratory impairment in cancer cells. Science 1956;124:269–70. [PubMed] [Google Scholar]
- 3.Schug ZT, Peck B, Jones DT, Zhang Q, Grosskurth S, Alam IS, et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell 2015;27:57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 2012;481:380–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene 2005;24:6314–22. [DOI] [PubMed] [Google Scholar]
- 6.Swinnen JV, Vanderhoydonc F, Elgamal AA, Eelen M, Vercaeren I, Joniau S, et al. Selective activation of the fatty acid synthesis pathway in human prostate cancer. Int J Cancer 2000;88:176–9. [DOI] [PubMed] [Google Scholar]
- 7.Yoon S, Lee MY, Park SW, Moon JS, Koh YK, Ahn YH, et al. Up-regulation of acetyl-CoA carboxylase alpha and fatty acid synthase by human epidermal growth factor receptor 2 at the translational level in breast cancer cells. J Biol Chem 2007;282:26122–31. [DOI] [PubMed] [Google Scholar]
- 8.Yoshii Y, Furukawa T, Oyama N, Hasegawa Y, Kiyono Y, Nishii R, et al. Fatty acid synthase is a key target in multiple essential tumor functions of prostate cancer: uptake of radiolabeled acetate as a predictor of the targeted therapy outcome. PLoS One 2013;8:e64570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rysman E, Brusselmans K, Scheys K, Timmermans L, Derua R, Munck S, et al. De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Res 2010;70:8117–26. [DOI] [PubMed] [Google Scholar]
- 10.Liu H, Wu X, Dong Z, Luo Z, Zhao Z, Xu Y, et al. Fatty acid synthase causes drug resistance by inhibiting TNF-alpha and ceramide production. J Lipid Res 2013;54:776–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nieva C, Marro M, Santana-Codina N, Rao S, Petrov D, Sierra A. The lipid phenotype of breast cancer cells characterized by Raman microspectroscopy: towards a stratification of malignancy. PLoS One 2012;7:e46456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rak S, De Zan T, Stefulj J, Kosovic M, Gamulin O, Osmak M. FTIR spectroscopy reveals lipid droplets in drug resistant laryngeal carcinoma cells through detection of increased ester vibrational bands intensity. Analyst 2014;139:3407–15. [DOI] [PubMed] [Google Scholar]
- 13.Bensaad K, Favaro E, Lewis CA, Peck B, Lord S, Collins JM, et al. Fatty acid uptake and lipid storage induced by HIF-1alpha contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep 2014;9:349–65. [DOI] [PubMed] [Google Scholar]
- 14.Gibellini F, Smith TK. The Kennedy pathway–De novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life 2010;62:414–28. [DOI] [PubMed] [Google Scholar]
- 15.Vance JE. Phospholipid synthesis and transport in mammalian cells. Traffic 2015;16:1–18. [DOI] [PubMed] [Google Scholar]
- 16.Aboagye EO, Bhujwalla ZM. Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Res 1999;59:80–4. [PubMed] [Google Scholar]
- 17.Zech SG, Kohlmann A, Zhou T, Li F, Squillace RM, Parillon LE, et al. Novel small molecule inhibitors of choline kinase identified by fragment-based drug discovery. J Med Chem 2016;59:671–86. [DOI] [PubMed] [Google Scholar]
- 18.Glunde K, Bhujwalla ZM, Ronen SM. Choline metabolism in malignant transformation. Nat Rev Cancer 2011;11:835–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorin A, Gabitova L, Astsaturov I. Regulation of cholesterol biosynthesis and cancer signaling. Curr Opin Pharmacol 2012;12:710–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogretmen B Sphingolipid metabolism in cancer signalling and therapy. Nat Rev Cancer 2018;18:33–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 2008;9:112–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valli A, Rodriguez M, Moutsianas L, Fischer R, Fedele V, Huang HL, et al. Hypoxia induces a lipogenic cancer cell phenotype via HIF1alpha-dependent and -independent pathways. Oncotarget 2015;6:1920–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Accioly MT, Pacheco P, Maya-Monteiro CM, Carrossini N, Robbs BK, Oliveira SS, et al. Lipid bodies are reservoirs of cyclooxygenase-2 and sites of prostaglandin-E2 synthesis in colon cancer cells. Cancer Res 2008;68:1732–40. [DOI] [PubMed] [Google Scholar]
- 24.Lewis CA, Brault C, Peck B, Bensaad K, Griffiths B, Mitter R, et al. SREBP maintains lipid biosynthesis and viability of cancer cells under lipid- and oxygen-deprived conditions and defines a gene signature associated with poor survival in glioblastoma multiforme. Oncogene 2015;34:5128–40. [DOI] [PubMed] [Google Scholar]
- 25.SchulzeA HarrisAL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature 2012;491:364–73. [DOI] [PubMed] [Google Scholar]
- 26.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab 2006;3:187–97. [DOI] [PubMed] [Google Scholar]
- 27.Kamphorst JJ, Chung MK, Fan J, Rabinowitz JD. Quantitative analysis of acetyl-CoA production in hypoxic cancer cells reveals substantial contribution from acetate. Cancer Metab 2014;2:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peck B, Schug ZT, Zhang Q, Dankworth B, Jones DT, Smethurst E, et al. Inhibition of fatty acid desaturation is detrimental to cancer cell survival in metabolically compromised environments. Cancer Metab 2016;4:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leek R, Grimes DR, Harris AL, McIntyre A. Methods: using three-dimensional culture (spheroids) as an in vitro model of tumour hypoxia. Adv Exp Med Biol 2016;899:167–96. [DOI] [PubMed] [Google Scholar]
- 30.Clark J, Anderson KE, Juvin V, Smith TS, Karpe F, Wakelam MJ, et al. Quantification of PtdInsP3 molecular species in cells and tissues by mass spectrometry. Nat Methods 2011;8:267–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menendez JA, Lupu R. Fatty acid synthase regulates estrogen receptor-alpha signaling in breast cancer cells. Oncogenesis 2017;6:e299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chalbos D, Joyeux C, Galtier F, Rochefort H. Progestin-induced fatty acid synthetase in human mammary tumors: from molecularto clinical studies. J Steroid Biochem Mol Biol 1992;43:223–8. [DOI] [PubMed] [Google Scholar]
- 33.JarcE KumpA, MalavasicP Eichmann TO,ZimmermannR Petan T. Lipid droplets induced by secreted phospholipase A2 and unsaturated fatty acids protect breast cancer cells from nutrient and lipotoxic stress. Biochim Biophys Acta 2018;1863:247–65. [DOI] [PubMed] [Google Scholar]
- 34.Glatzel DK, Koeberle A, Pein H, Loser K, Stark A, Keksel N, et al. Acetyl-CoA carboxylase 1 regulates endothelial cell migration by shifting the phospholipid composition. J Lipid Res 2018;59:298–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Zuckier LS, Ghesani NV. Dominant uptake of fatty acid over glucose by prostate cells: a potential new diagnostic and therapeutic approach. Anticancer Res 2010;30:369–74. [PubMed] [Google Scholar]
- 36.Pizer ES, Thupari J, Han WF, Pinn ML, Chrest FJ, Frehywot GL, et al. Malonyl-coenzyme-A is a potential mediator of cytotoxicity induced by fatty-acid synthase inhibition in human breast cancer cells and xenografts. Cancer Res 2000;60:213–8. [PubMed] [Google Scholar]
- 37.Ventura R, Mordec K, Waszczuk J, Wang Z, Lai J, Fridlib M, et al. Inhibition of de novo palmitate synthesis by fatty acid synthase induces apoptosis in tumor cells by remodeling cell membranes, inhibiting signaling pathways, and reprogramming gene expression. EBioMedicine 2015;2:808–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dienstmann R, Rodon J, Serra V, Tabernero J. Picking the point of inhibition: a comparative review of PI3K/AKT/mTOR pathway inhibitors. Mol Cancer Ther 2014;13:1021–31. [DOI] [PubMed] [Google Scholar]
- 39.Ward PS, Thompson CB. Signaling in control of cell growth and metabolism. Cold Spring Harb Perspect Biol 2012;4:a006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janku F, Yap TA, Meric-Bernstam F. Targeting the PI3K pathway in cancer: are we making headway? Nat Rev Clin Oncol 2018;15:273–91. [DOI] [PubMed] [Google Scholar]
- 41.Seim I, Jeffery PL, Thomas PB, Nelson CC, Chopin LK. Whole-genome sequence of the metastatic PC3 and LNCaP human prostate cancer cell lines. G3 (Bethesda) 2017;7:1731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith SE, Mellor P, Ward AK, Kendall S, McDonald M, Vizeacoumar FS, et al. Molecular characterization of breast cancer cell lines through multiple omic approaches. Breast Cancer Res 2017;19:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cantley LC. The phosphoinositide 3-kinase pathway. Science 2002; 296:1655–7. [DOI] [PubMed] [Google Scholar]
- 44.Osman C, Voelker DR, Langer T. Making heads or tails of phospholipids in mitochondria. J Cell Biol 2011;192:7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bogdanov M, Dowhan W, Vitrac H. Lipids and topological rules governing membrane protein assembly. Biochim Biophys Acta 2014;1843:1475–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cai Z, Mason NS, Anderson CJ, Edwards WB. Synthesis and preliminary evaluation of an (1)(8)F-labeled oleic acid analog for PET imaging of fatty acid uptake and metabolism. Nucl Med Biol 2016;43:108–15. [DOI] [PubMed] [Google Scholar]
- 47.Challapalli A, Aboagye EO. Positron emission tomography imaging of tumor cell metabolism and application to therapy response monitoring. Front Oncol 2016;6:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trousil S, Kaliszczak M, Schug Z, Nguyen QD, Tomasi G, Favicchio R, et al. The novel choline kinase inhibitor ICL-CCIC-0019 reprograms cellular metabolism and inhibits cancer cell growth. Oncotarget 2016;7:37103–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.