Abstract
Sequences encoding Olduvai (DUF1220) protein domains show the largest human-specific increase in copy number of any coding region in the genome and have been linked to human brain evolution. Most human-specific copies of Olduvai (119/165) are encoded by three NBPF genes that are adjacent to three human-specific NOTCH2NL genes that have been shown to promote cortical neurogenesis. Here, employing genomic, phylogenetic, and transcriptomic evidence, we show that these NOTCH2NL/NBPF gene pairs evolved jointly, as two-gene units, very recently in human evolution, and are likely co-regulated. Remarkably, while three NOTCH2NL paralogs were added, adjacent Olduvai sequences hyper-amplified, adding 119 human-specific copies. The data suggest that human-specific Olduvai domains and adjacent NOTCH2NL genes may function in a coordinated, complementary fashion to promote neurogenesis and human brain expansion in a dosage-related manner.
Electronic supplementary material
The online version of this article (10.1007/s00439-019-02018-4) contains supplementary material, which is available to authorized users.
Introduction
The increasing availability of primate genomic data is providing a unique opportunity to identify lineage-specific sequence differences among human and other primates. As part of these efforts, genomic factors are beginning to be uncovered that potentially contributed to the evolutionary expansion of the human brain (Sikela 2006; O’Bleness et al. 2012a). While there are a number of genomic mechanisms that may have been critical to brain expansion, several candidate sequences have been reported that involve human-specific gene duplications (Dennis et al. 2012; Florio et al. 2015). A number of these sequences map to 1q21, a region on chromosome 1 that is highly enriched for human lineage-specific gene duplications (Fortna et al. 2004; Dumas et al. 2007). This finding should not be unexpected, as unique evolutionary features of the region have been known to cytogeneticists for decades: it is the site of a human-specific pericentric inversion and is adjacent to a human-specific C-band (Yunis and Prakash 1982).
Among the duplicated genes and coding regions in 1q21 that have been associated with brain evolution, the most dramatically changed are those specifying Olduvai protein domains (formerly DUF1220) (Popesco et al. 2006; Sikela and van Roy 2017). Encoded primarily by the NBPF gene family (Vandepoele et al. 2005), Olduvai sequences have undergone the largest human lineage-specific increase in copy number of any coding region in the genome (~ 300 total copies of which ~ 165 are human-specific) (Popesco et al. 2006; O’Bleness et al. 2012a, b). They have been implicated, in a dose-dependent manner, in brain size [both evolutionarily as well as within the human population (Dumas et al. 2012; Keeney et al. 2014; Zimmer and Montgomery 2015)] and cognitive function (Davis et al. 2014a), and have been shown to promote proliferation in neural stem cells (Keeney et al. 2015). In addition, variation in Olduvai copy number has been associated with cognitive disease: autism, schizophrenia, microcephaly and macrocephaly (Dumas et al. 2012; Davis et al. 2014b, 2015, 2019; Quick et al. 2015). This ability to potentially confer both beneficial and detrimental effects has led to the proposal that the Olduvai family may constitute a cognitive genomic trade-off specific to the human lineage (Dumas and Sikela 2009; Sikela and Quick 2018).
The extreme human lineage-specific increase in Olduvai copy number has been almost entirely due to intragenic tandem expansions of a three-domain unit, termed the Olduvai triplet, that have occurred primarily in four human-specific NBPF genes (NPBF10, NBPF14, NBPF19, and NBPF20) that map to the 1q21.1-2 genomic region (O’Bleness et al. 2012a, b; O’Bleness et al. 2014) (Fig. 1a). Recently, three human-specific NOTCH2NL genes have been implicated in driving brain size in humans and also map to 1q21.1-2 (Fiddes et al. 2018; Suzuki et al. 2018; Florio et al. 2018). These studies provide evidence that the human-specific NOTCH2NL genes expand cortical progenitors and neuronal output and, thus, may have contributed to the expansion of the human cortex. More specifically, they propose that this occurs by increasing the level of self-renewal and/or expansion of radial glial cells, and could thereby facilitate an extended period of neurogenesis and the larger neuronal output that is characteristic of human corticogenesis. Here we provide genomic, phylogenetic, and transcriptional evidence that the new NOTCH2NL genes and adjacent Olduvai sequences may function in a coordinated manner to promote human brain expansion via a dosage-related mechanism.
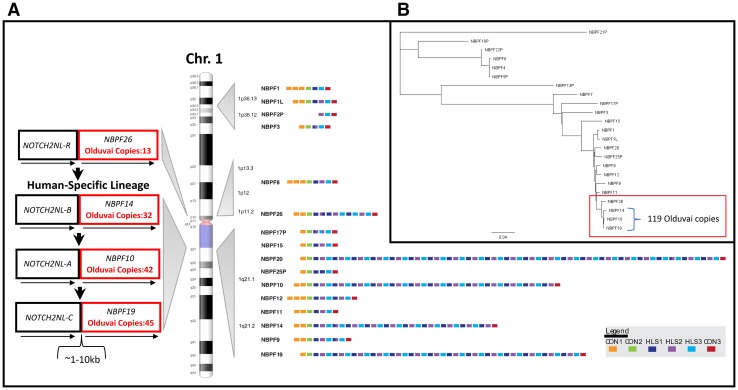
Fig. 1.
Genome organization, phylogeny, and recent evolution of human NBPF genes and Olduvai protein domains. a Shown to the right of chromosome 1 are the genome positions of NBPF genes and the number and arrangement of Olduvai domain subtypes based on the most recent human genome assembly (hg38) (Sikela and Quick 2018). Each block represents an Olduvai protein domain; each of the six primary Olduvai subtypes are depicted by a different color as denoted in the key. Shown to the left of chromosome 1 is the recent evolutionary history of NOTCH2NL/NBPF gene pairs and Olduvai protein domains. Model describes the order and timing of likely duplication events that produced multiple human-specific copies of NOTCH2NL and NBPF genes and human-specific hyper-amplification of Olduvai domains. Arrows below each gene indicate direction of orientation. b Phylogenetic profile of 23 human NBPF genes. Phylogenetic analysis was carried out on human NBPF genes. To avoid confusion resulting from the inclusion of highly similar Olduvai protein domains found in each NBPF gene, sequences were analyzed that only covered the start codon of each gene along with approximately 1 kb of flanking sequence. Phylogenetic profiling was performed using the Genious DNA analysis package and its multiple alignment program. Predicted number of Olduvai domains encoded by three human-specific NBPF genes (NBPF10, NBPF14, and NBPF19) is shown. The red box denotes the part of the NBPF phylogenetic profile which indicates that NBPF26 is older than, and more evolutionarily distant from NBPF10, NBPF14, and NBPF19
Methods
Genomic and phylogenetic analysis
Genome coordinates for the locations of the NBPF and NOTCH genes were obtained from the hg38 human genome assembly at UCSC and also from several publications (O’Bleness et al. 2014; Sikela and Quick 2018; Fiddes et al. 2018; Suzuki et al. 2018). For phylogenetic analyses of NBPF genes, sequences were used that included the predicted start codon and 1 kb of sequence that flanked the start codon. This avoided complications that would arise from inclusion of the highly duplicated human Olduvai sequences, and allowed the analysis to focus specifically on the evolutionary relatedness of NBPF genes. Phylogenetic analysis of NBPF gene sequences was carried out with the Geneious Treebuilder program using the genetic distance model of Tamura-Nei (Tamura and Nei 1993) and the neighbor-joining tree build method (Geneious version 11.1 created by Biomatters. Available from https://www.geneious.com).
Transcriptional analysis of correlations between NBPF and NOTCH2NL genes
Recently generated scRNA-seq data for human developing cortex were obtained from Nowakowski et al. (2017) available through the UCSC cell browser (https://cells.ucsc.edu/?ds=cortex-dev). Because reads mapping to NOTCH2 and NOTCHNL transcripts were not well differentiated, we re-aligned reads to an aggregate NOTCH2NL gene model and to NOTCH2 by mapping to the collapsed hg19 version of NOTCH2NL. We then performed a gene-by-gene Pearson correlation between NOTCH2NL expression and the profiles of all human genes detected across all cells (n = 22,988), and of all genes detected across radial glia only (n = 22,750).
For 3′ pileup studies, expression coverage tracks were obtained from the UCSC human cortex single-cell expression track hub for 572 radial glia cells. For each subtype of radial glia cells, the coverage track was normalized to sum to 1 and then multiplied by the number of cells in that subtype. The resulting cell and sequencing depth normalized coverage tracks were then summed to produce a final radial glial cell expression track.
Results
Genomic and phylogenetic evidence for paired evolution
Current human genomic data indicate that there are four human-specific NOTCH2NL genes: NOTCH2NL-A, NOTCH2NL-B and NOTCH2NL-C, located on 1q21.1, and NOTCH2NL-R located on 1p11.2 (Fiddes et al. 2018; Suzuki et al. 2018). While chimpanzee and gorilla have copies of NOTCH2NL, none are functional (Fiddes et al. 2018). Immediately adjacent to, and downstream of, each of these four NOTCH paralogs is an NBPF gene in the same orientation as its NOTCH2NL partner (O’Bleness et al. 2014; Fiddes et al. 2018; Suzuki et al. 2018) (Fig. 1a). This striking genomic arrangement suggests that each of the additional copies of NOTCH2NL that appeared in the human genome did not duplicate as a single gene, but rather did so as a two-gene module, composed of one NOTCH2NL gene and one NBPF gene.
While there remains some ambiguity about the precise steps by which the NOTCH2NL paralogs and adjacent NBPF genes appeared, several solutions have been proposed (Fig. 1a, Supplemental Fig. 1). Due to the rapid evolution of this region in both humans and other great apes, as well as the observed gene conversion between paralogs, it is difficult to conclusively determine which scenario happened. However, there are some lines of evidence in the human lineage that favor scenarios where NOTCH2NL-A/-B/-C derived from NOTCH2L-R. Because all NOTCH2NL genes in the human lineage are associated with PDE4DIP, and both chimpanzee and gorilla have one PDE4DIP-associated copy, the four human copies must derive from that copy. The previous analysis of the Simons Diversity Project (Fiddes et al. 2018) showed gene conversion between NOTCH2NL-A and NOTCH2NL-B is prevalent in the population, and gene conversion between NOTCH2NL-C and NOTCH2NL-A/-B happens infrequently. No gene conversion was observed between NOTCH2 and NOTCH2NL-R and the three paralogs on 1q21.1. This is supported by the sequence identity observed between pairwise comparisons of NOTCH2 and NOTCH2NL paralogs. In GRCh38, NOTCH2 has 98.19% sequence similarity to NOTCH2NL-R, while NOTCH2NL-A/-B/-C have 98.89% to 98.91% identity to each other.
These lines of evidence suggest that a likely model for NOTCH2NL evolution in hominids involves first a gene conversion event between NOTCH2NL-R and NOTCH2 restoring a functional 5′ end, and then, a subsequent duplication event sometime either shortly before or after the pericentric inversion of chromosome 1 (Yunis and Prakash 1982). Following this event, NOTCH2NL-R ceased gene converting with NOTCH2, while the new NOTCH2NL in 1q21.1 quickly gave rise to the other two paralogs. These three paralogs continue to have segregating gene conversion events happening to this day, which makes determining the exact order of their appearance difficult. Further supporting this model is that in the human assembly GRCh38 NOTCH2NL-R is 1.8% diverged from NOTCH2, while NOTCH2NL-A/-B/-C are 1.1% diverged, representing 61.1% fewer accumulated mutations, which is in line with expectation if the three loci are undergoing continual gene conversion and as a result evolving together.
If the four human-specific NOTCH2NL genes and four adjacent human-specific NBPF genes duplicated together as two-gene units, the phylogenetic history of these two sets of genes should be identical. To examine the relationships among the four NBPF genes that are paired with NOTCH2NL genes, we focused on sequence near the start codon that did not contain repeated Olduvai domains and was, therefore, less likely to be subject to gene conversion. These comparisons indicate that the three NBPF genes on 1q21.1-2 (NBPF10, NBPF14, and NBPF19) are more similar to one another than they are to NBPF26 on 1p12 (Fig. 1b). In addition, NBPF26 is the most related to ancestral NBPF genes, suggesting that the most plausible history involves NOTCH2NL-R/NBPF26 being the original PDE4DIP-associated copy that survived from the most recent common ancestor with chimpanzee. Taken together, the data indicate that the NOTCH2NL-R/NBPF26 gene pair on 1p12 duplicated as a unit, with the resulting new pair (NOTCH2NL-B/NBPF14) moving to the 1q21.1 region (Fig. 1a). Subsequently, NOTCH2NL-B/NBPF14 duplicated two more times in the 1q21.1-2 region, generating NOTCH2NL-A/NBPF10 and NOTCH2NL-C/NBPF19.
A striking difference becomes evident, however, when one compares the human-specific NOTCH2NL/NBPF gene increases with the Olduvai increases encoded by these NBPF genes. While the NOTCH2NL paralogs (and their NBPF partners) went from one gene to four in humans, Olduvai copies encoded by these NBPF genes underwent human-specific hyper-amplification, increasing from 13 copies (encoded by NBPF26) to 132 (i.e., adding 119 copies encoded by NBPF10, NBPF14, and NBPF19) (Fig. 1a, b).
The two most plausible scenarios for how these expansions occurred are the following: The long tandem Olduvai expansions found on NBPF10, NBPF14, and NBPF19 either had to occur independently on each of the three new human-specific NBPF genes after the genes appeared in 1q21.1-2, or a duplicated copy of NBPF26 appeared in 1q21.1-2 and, after adding many Olduvai copies (i.e., becoming NBPF14), duplicated twice more (i.e., producing NBPF10 and NBPF19). The latter scenario is the most parsimonious and if, true, implies that each time one of these two new expanded duplicate genes appeared it would have instantaneously added large numbers of Olduvai copies to the human genome. In either case, the human-specific Olduvai hyper-amplification would have had to take place very recently (within the past 3 million years as has been proposed for the three NOTCH2NL genes on 1q21.1) and, thus, would correlate with the extreme enlargement of the human brain that occurred during this time (e.g., a threefold increase in size over the past 1.8 million years) (Florio et al. 2017).
Transcriptional evidence for coordinated regulation
The fact that the three expanded human-specific NBPF genes are adjacent to (within 1–10 kb), and in the same orientation as, the three human-specific NOTCH2 NL genes, raises the possibility that the expression of the NBPF genes and NOTCH2NL genes may be under shared transcriptional control. This possibility is further supported by transcriptional profiling using scRNA-seq (Pollen et al. 2014; Nowakowski et al. 2017), which indicates that NBPF10 and NOTCH2NL show very similar expression patterns in developing human cortex, with especially high expression in radial glial cells (Fig. 2a–c). Radial glia are thought to be central contributors to neurogenesis and brain expansion (Hansen et al. 2010; Pollen et al. 2015). To more precisely examine whether NOTCH2NL and NBPF genes could be regulated by common mechanisms, we first quantified NOTCH2NL expression separately from NOTCH2 by realigning single-cell reads to an aggregate gene model of NOTCH2NL paralogs (methods). We then examined the Pearson correlation of NOTCH2NL to all genes (n = 22,988) across diverse cortical cell types. The top correlates of NOTCH2NL included canonical radial glia markers such as VIM, HES1, and SLC1A3 (Pollen et al. 2015), as well as NBPF14, NOTCH2, and NBPF10 (Fig. 2d; Table S1). These strong correlations could reflect shared regulatory control, but they might simply reflect specificity for radial glia and the shared absence of expression in other cell populations. Therefore, we further examined the correlation of NOTCH2NL to all genes among radial glia cells only. Remarkably, the top gene correlated with NOTCH2NL out of all genes was NBPF14 (Fig. 2e, Table S1). Therefore, NOTCH2NL and NBPF genes not only share radial glia specificity, but are also very tightly co-expressed at the level of single cells. This extremely strong correlation could be driven by a single fusion transcript recently observed with a NOTCH2NL paralog at the 5′ end and an NBPF paralog at the 3′ end (Dougherty et al. 2018). However, we see limited transcriptional evidence for this combined transcript as 3′ biased scRNA-seq reads are concentrated at the 3′ end of NOTCH2NL, suggesting that the majority of NOTCH2NL transcripts terminate as predicted (Supplementary Fig. 2A–C). Consistent with shared transcriptional regulation, the correlation between NOTCH2NL and NBPF genes is further supported across adult human tissues (Supplementary Fig. 2D). Notably, expression of these genes is relatively low in the adult brain compared with other tissue types and with radial glia in the developing cortex. These findings are consistent with other expression studies which show that NBPF (i.e., Olduvai) sequences are expressed at the right time and place to be important to cortical neurogenesis (Keeney et al. 2015), and are among a small set of human-specific genes that show high expression in neural progenitor cells (Suzuki et al. 2018; Florio et al. 2018).
Fig. 2.
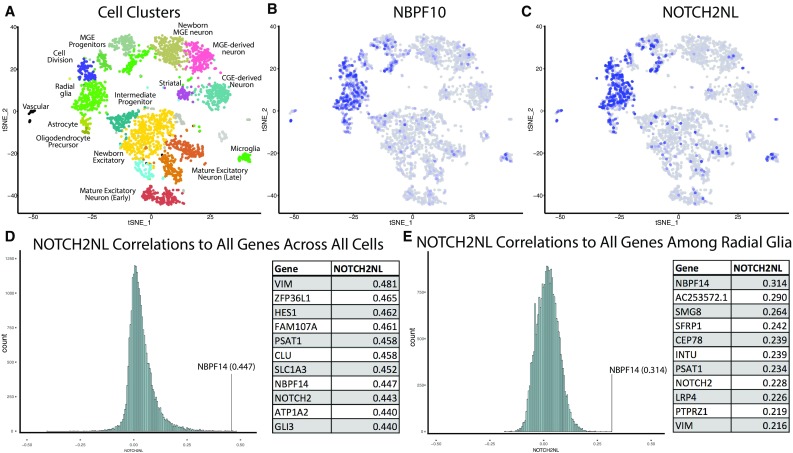
Radial glia-specific expression of NBPF10 and NOTCH2NL in developing human telencephalon samples. a–c Scatterplot of 3412 cells from developing telencephalon after single-cell RNA-seq principal component analysis and t-stochastic neighbor embedding (tSNE) as described in Nowakowski et al. (2017). Cells are colored by annotated cell type clusters (a), NBPF10 expression (b), and NOTCH2NL expression (c). Histograms of Pearson correlation coefficients are shown representing results of a comparison of scRNA-seq expression profiles of NOTCH2NL vs. all human genes from all cells (d) and from radial glia (e) with tables showing the top correlated genes
Discussion
The most probable model of how the four human-specific NBPF/NOTCH2NL gene pairs appeared in the human genome (Fig S1C) not only fits well with available phylogenetic data, but also may help explain the existence of two well-established cytogenetic landmarks associated with human chromosome 1: the human-specific pericentric inversion and the human-specific C-band at 1q12. In this model, the NPBF26/NOTCHNL-R gene pair appeared on 1p12 and then duplicated forming NBPF14/NOTCH2NL-B, which was then moved to the 1q21.1-2 region as a result of the human-specific pericentric inversion event. This placed NBPF14/NOTCH2NL-B next to the large human-specific band of constitutive heterochromatin (C-band) at 1q12. The C-band is rich with highly repeated sequences and a strong promoter of non-allelic homologous recombination (NAHR) events. Thus, proximity to the C-band could have driven both the production of the two additional human-specific 1q21.1 copies (NBPF14/NOTCH2NL-A and NBPF19/NOTCH2NL-C), as well as the extreme expansions of the human-specific tandemly repeated copies of Olduvai that are found in NBPF10, NBPF14, and NBPF19).
In addition to these three expanded NBPF genes, there is a fourth gene in 1q21.1-2, NBPF20, that also contains long tandemly arranged human-specific copies of Olduvai. However, NBPF20 does not have an adjacent NOTCH2NL gene and, while the other expanded NBPF genes (NBPF10, NBPF14, and NBPF19) show high sequence relatedness to one another, NBPF20 is phylogenetically more distant (Fig. 1b). Given these findings, it is likely that the intragenic Olduvai expansions found on NBPF20 arose independently, separate from the expansions found on NBPF10, NBPF14, and NBPF19. However, the fact that all four of the most highly expanded human-specific NBPF genes are found in the 1q21.1 region (and no highly expanded NBPF genes are found outside of this region) is also consistent with the possibility that proximity to the human-specific C-band at 1q12 may have promoted these extreme tandem expansions of Olduvai copy number in humans.
The tandemly expanded Olduvai sequences are found typically as Olduvai triplets in NBPF10, NBPF14, NBPF19, and NBPF20 genes and, while triplet sequences are different between these genes, they are highly similar within each gene. This is likely the result of gene conversion and concerted evolution, which tends to homogenize tandemly repeated sequences within a gene (Ganley and Kobayashi 2007). In addition, the fact that, within an NBPF gene, the triplet sequences, as a unit, are more highly conserved than the individual domains supports the idea that homogenization is occurring at the level of the triplets within a gene.
While the 1q21 region is enriched for human-specific Olduvai sequences, it has also been linked with a number of disease-associated copy number variations. Among these, 1q21-associated duplications have been implicated in autism and macrocephaly, while reciprocal deletions have been linked with schizophrenia and microcephaly (Dumas et al. 2012; Brunetti-Pierri et al. 2008; Girirajan et al. 2013; Mefford et al. 2008; Stefansson et al. 2008). It has been suggested that the NOTCH2NL-A and NOTCH2NL-B genes provide the breakpoints for this syndrome, and, thus, have been instrumental in the recombination events that led to these gains/losses (Fiddes et al. 2018). However, because of the highly duplicated nature of the Olduvai sequences encoded by the adjacent NBPF genes (NBPF10 and NBPF14), the involvement of Olduvai sequences in providing the breakpoints for this 1q21-associated duplication/deletion syndrome has not yet been directly examined. Given that highly duplicated, tandemly arranged sequences, such as the Olduvai repeats on NBPF10 and NBPF14, are especially prone to recombination, it is likely that, in some cases, the breakpoints are within these NBPF genes. Higher resolution analyses may be able to more definitively address this issue in the future.
Because human-specific Olduvai sequences are found primarily as long tandemly arranged repeats, one might suspect that they serve only to promote recombination and, thus, are functionally unimportant. While such a recombinogenic architecture may have been instrumental in the generation of the newly duplicated NOTCH2NL paralogs and, as mentioned above, may be a significant contributor to the 1q21-duplication/deletion syndrome, there are several reasons why it is unlikely that Olduvai sequences are functionally silent partners of the NOTCH2NL genes. The Olduvai sequences found in the three NBPF genes adjacent to the NOTCH2NL genes encode long open-reading frames (O’Bleness et al. 2012a, b; O’Bleness et al. 2014) express a protein product that in brain is restricted to neurons (Popesco et al. 2006), and show strong signals of positive selection at the protein sequence level (Popesco et al. 2006). Functional data also support a more active role for Olduvai sequences in corticogenesis, as introduction of Olduvai sequences has been shown to promote proliferation of neural stem cells (Keeney et al. 2015). These observations, when combined with the findings reported here, suggest that human-specific Olduvai domains and adjacent NOTCH2NL genes may function in a coordinated, complementary fashion to promote neurogenesis and brain expansion in a dosage-related manner.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. S1. Possible NBPF/NOTCH2NL phylogenies. Three possible phylogenies are presented in increasing order of likelihood. A NBPF/NOTCH2NL pair present in 1p12 is moved to 1q21.1 via pericentric inversion. This copy is then duplicated back to 1p12 in addition to being duplicated further in 1q21.1. B NBPF/NOTCH2NL in 1p12 is duplicated to 1q21.1 prior to the pericentric inversion, then the inversion swaps the two copies. C NBPF/NOTCH2NL is duplicated a second time in 1p12 prior to the pericentric inversion, which moves only one of the two copies to 1q21.1 where it duplicates further. Scenario C is most likely because it involves a linear phylogeny, which is supported by the data (PDF 811 kb)
Supplementary Fig. S2.Normalized read coverage for NOTCH2NL-A/-B/-C and NBPF10/14/19 was analyzed in 572 radial glia cells from a scRNA-seq method with a parital 3′ bias (A–C). A large spike in coverage is observed past the 3′ UTR of NOTCH2NL suggesting that expression of NOTCH2NL and NBPF mainly occurs through two separate transcripts in this cell type. D. Heatmap of gene expression values expressed as transcripts per million reads (TPM) across adult human tissue samples from the GTEx resource (https://gtexportal.org). Genes are clustered by expression across tissue types, and tissues are clustered based on expression of labeled genes. Because the dataset does not include NBPF19 and NBPF26, only NBPF10 and NBPF14 are shown (PDF 456 kb)
Table data uploaded as Auxiliary Supplementary Materials. Table S1 contains Pearson correlation coefficients of the brain expression profiles of NOTCH2 and NOTCH2NL vs. all genes (n = 22,988) among all cells (first two columns) and among radial glia only (third and fourth columns, n = 22,750 genes) (XLSX 1442 kb)
Acknowledgements
We thank Nate Anderson for help with figures and manuscript preparation and Ilea Heft for providing NBPF gene sequences. We also thank Bruce Appel, Santhosh Girirajan, Kirk Hansen, David Haussler, Aaron Issaian, Mark Johnston, Kent Riemondy, and Debby Silver for helpful discussions. This research was supported by NIH award R01MH108684 to JMS.
Author contributions
JMS conceived and supervised the study. ITF, JMD, and JMS analyzed genomic and phylogenetic data. JMD and AAP carried out statistical analysis of scRNA-seq data. AAP, ITF, and JMS obtained and analyzed scRNA-seq data. All authors discussed the data and contributed to data interpretation. JMS wrote the paper with contributions from ITF, AAP, and JMD. JMS is founder of GATC Science, a biotech company focused on genomics. The other authors declare no competing financial interests. Requests for materials should be addressed to the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ian T. Fiddes and Alex A. Pollen have contributed equally to this study.
Contributor Information
Ian T. Fiddes, Email: Ian.T.Fiddes@gmail.com
Alex A. Pollen, Email: Alex.Pollen@ucsf.edu
Jonathan M. Davis, Email: jackdavis.epi@gmail.com
James M. Sikela, Email: james.sikela@ucdenver.edu
References
- Brunetti-Pierri N, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JM, et al. DUF1220 copy number is linearly associated with increased cognitive function as measured by total IQ and mathematical aptitude scores. Hum Genet. 2014;134:67–75. doi: 10.1007/s00439-014-1489-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JM, et al. DUF1220 dosage is linearly associated with increasing severity of the three primary symptoms of autism. PLoS Genet. 2014;10:e1004241. doi: 10.1371/journal.pgen.1004241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JM, Quick VBS, Sikela JM. Replicated linear association between DUF1220 copy number and severity of social impairment in autism. Hum Genet. 2015;134:569–575. doi: 10.1007/s00439-015-1537-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JM, Heft I, Scherer SW, Sikela JM. A third linear association between Olduvai (DUF1220) copy number and severity of the classic symptoms of inherited autism. Am J Psychiatry. 2019;5:566. doi: 10.1176/appi.ajp.2018.18080993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis MY, et al. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell. 2012;149:912–922. doi: 10.1016/j.cell.2012.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty ML, et al. Transcriptional fates of human-specific segmental duplications in brain. Genome Res. 2018;28:1566–1576. doi: 10.1101/gr.237610.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas L, Sikela JM. DUF1220 domains, cognitive disease, and human brain evolution. Cold Spring Harb Symp Quant Biol. 2009;74:375–382. doi: 10.1101/sqb.2009.74.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas L, et al. Gene copy number variation spanning 60 million years of human and primate evolution. Genome Res. 2007;17:1266–1277. doi: 10.1101/gr.6557307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas LJ, et al. DUF1220-domain copy number implicated in human brain-size pathology and evolution. Am J Hum Genet. 2012;91:444–454. doi: 10.1016/j.ajhg.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiddes IT, et al. Human-specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell. 2018;173:1356–1369. doi: 10.1016/j.cell.2018.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florio M, et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science. 2015;347:1465–1470. doi: 10.1126/science.aaa1975. [DOI] [PubMed] [Google Scholar]
- Florio M, Borrell V, Huttner WB. Human-specific genomic signatures of neocortical expansion. Curr Opin Neurobiol. 2017;42:33–44. doi: 10.1016/j.conb.2016.11.004. [DOI] [PubMed] [Google Scholar]
- Florio M, Heide M, Pinson A, Brandl H, Albert M, Winkler S, Wimberger P, Huttner WB, Hiller M. Evolution and cell-type specificity of human-specific genes preferentially expressed in progenitors of fetal neocortex. eLife. 2018;7:e32332. doi: 10.7554/eLife.32332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortna A, et al. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004;2:E207. doi: 10.1371/journal.pbio.0020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley ARD, Kobayashi T. Highly efficient concerted evolution in the ribosomal DNA repeats: total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Res. 2007;17:184–191. doi: 10.1101/gr.5457707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, et al. Refinement and discovery of new hotspots of copy-number variation associated with autism spectrum disorder. Am J Hum Genet. 2013;92:221–237. doi: 10.1016/j.ajhg.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PRL, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- Keeney JG, Dumas L, Sikela JM. The case for DUF1220 domain dosage as a primary contributor to anthropoid brain expansion. Front Hum Neurosci. 2014;8:1–11. doi: 10.3389/fnhum.2014.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney JG, et al. DUF1220 protein domains drive proliferation in human neural stem cells and are associated with increased cortical volume in anthropoid primates. Brain Struct Funct. 2015;220:3053–3060. doi: 10.1007/s00429-014-0814-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science. 2017;358:1318–1323. doi: 10.1126/science.aap8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Bleness M, Searles VB, Varki A, Gagneux P, Sikela JM. Evolution of genetic and genomic features unique to the human lineage. Nat Rev Genet. 2012;13:853–866. doi: 10.1038/nrg3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Bleness, et al. Evolutionary history and genome organization of DUF1220 protein domains. G3 (Bethesda) 2012;2:977–986. doi: 10.1534/g3.112.003061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Bleness M, et al. Finished sequence and assembly of the DUF1220-rich 1q21 region using a haploid human genome. BMC Genom. 2014;15:387. doi: 10.1186/1471-2164-15-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol. 2014;32:1053–1058. doi: 10.1038/nbt.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, et al. Molecular identity of human outer radial glia during cortical development. Cell. 2015;163:55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popesco MC, et al. Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science. 2006;313:1304–1307. doi: 10.1126/science.1127980. [DOI] [PubMed] [Google Scholar]
- Quick VS, Davis JM, Olincy A, Sikela J. DUF1220 copy number is associated with schizophrenia risk and severity: implications for understanding autism and schizophrenia as related diseases. Transl Psychiatry. 2015;5:e697. doi: 10.1038/tp.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikela JM. The jewels of our genome: the search for the genomic changes underlying the evolutionarily unique capacities of the human brain. PLoS Genet. 2006;2:646–655. doi: 10.1371/journal.pgen.0020080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikela JM, Quick VBS. Genomic trade-offs: are autism and schizophrenia the steep price of the human brain? Hum Genet. 2018;137:1–13. doi: 10.1007/s00439-017-1865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikela JM, van Roy F. Changing the name of the NBPF/DUF1220 domain to the Olduvai domain. F1000Research. 2017 doi: 10.12688/f1000research.13586.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki IK, et al. Human-specific NOTCH2NL genes expand cortical neurogenesis through delta/notch regulation. Cell. 2018;173:1370–1384. doi: 10.1016/j.cell.2018.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Vandepoele K, et al. A novel gene family NBPF: intricate structure generated by gene duplications during primate evolution. Mol Biol Evol. 2005;22:2265–2274. doi: 10.1093/molbev/msi222. [DOI] [PubMed] [Google Scholar]
- Yunis JJ, Prakash O. The origin of man: a chromosomal pictorial legacy. Science. 1982;215:1525–1530. doi: 10.1126/science.7063861. [DOI] [PubMed] [Google Scholar]
- Zimmer F, Montgomery SH. Phylogenetic analysis supports a link between DUF1220 domain number and primate brain expansion. Genome Biol Evol. 2015;7:2083–2088. doi: 10.1093/gbe/evv122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1. Possible NBPF/NOTCH2NL phylogenies. Three possible phylogenies are presented in increasing order of likelihood. A NBPF/NOTCH2NL pair present in 1p12 is moved to 1q21.1 via pericentric inversion. This copy is then duplicated back to 1p12 in addition to being duplicated further in 1q21.1. B NBPF/NOTCH2NL in 1p12 is duplicated to 1q21.1 prior to the pericentric inversion, then the inversion swaps the two copies. C NBPF/NOTCH2NL is duplicated a second time in 1p12 prior to the pericentric inversion, which moves only one of the two copies to 1q21.1 where it duplicates further. Scenario C is most likely because it involves a linear phylogeny, which is supported by the data (PDF 811 kb)
Supplementary Fig. S2.Normalized read coverage for NOTCH2NL-A/-B/-C and NBPF10/14/19 was analyzed in 572 radial glia cells from a scRNA-seq method with a parital 3′ bias (A–C). A large spike in coverage is observed past the 3′ UTR of NOTCH2NL suggesting that expression of NOTCH2NL and NBPF mainly occurs through two separate transcripts in this cell type. D. Heatmap of gene expression values expressed as transcripts per million reads (TPM) across adult human tissue samples from the GTEx resource (https://gtexportal.org). Genes are clustered by expression across tissue types, and tissues are clustered based on expression of labeled genes. Because the dataset does not include NBPF19 and NBPF26, only NBPF10 and NBPF14 are shown (PDF 456 kb)
Table data uploaded as Auxiliary Supplementary Materials. Table S1 contains Pearson correlation coefficients of the brain expression profiles of NOTCH2 and NOTCH2NL vs. all genes (n = 22,988) among all cells (first two columns) and among radial glia only (third and fourth columns, n = 22,750 genes) (XLSX 1442 kb)