Abstract
Gliomas with Isocitrate dehydrogenase 1 (IDH1) mutation have alterations in several enzyme activities, resulting in various metabolic changes. The aim of this study was to determine a mechanism for the better prognosis of gliomas with IDH mutation by performing metabolomic analysis. To understand the metabolic state of human gliomas, we analyzed clinical samples obtained from surgical resection of glioma patients (grades II–IV) with or without the IDH1 mutation, and compared the results with U87 glioblastoma cells overexpressing IDH1 or IDH1R132H. In clinical samples of gliomas with IDH1 mutation, levels of D-2-hydroxyglutarate (D-2HG) were increased significantly compared with gliomas without IDH mutation. Gliomas with IDH mutation also showed decreased intermediates in the tricarboxylic acid cycle and pathways involved in the production of energy, amino acids, and nucleic acids. The marked difference in the metabolic profile in IDH mutant clinical glioma samples compared with that of mutant IDH expressing cells includes a decrease in β-oxidation due to acyl-carnitine and carnitine deficiencies. These metabolic changes may explain the lower cell division rate observed in IDH mutant gliomas and may provide a better prognosis in IDH mutant gliomas.
Subject terms: Cellular neuroscience, Cancer metabolism
Introduction
The number of patients with glioma is increasing annually with the aging of the global population in the US and Japan1,2. Because gliomas develop by infiltrating into normal brain tissue3, complete surgical excision is difficult. Therefore, efforts are underway to improve the degree of surgical excision using various modalities, including navigation and perioperative MRI. Advances have also been made in chemotherapy, immunotherapy4, molecular targeted agents5, intraoperative alkylating agents6,7, and radiotherapy. These treatments have improved the prognosis of glioma. However, even with these therapies, compared to other types of cancer, the median survival time for patients with glioblastoma remains poor at 21.4 months8.
The prognosis for patients with glioma with mutation in Isocitrate dehydrogenase (IDH) was reportedly better than that in patients without the IDH mutation (IDH normal)9,10. In patients with glioblastoma, the median survival in the IDH normal group was approximately 15 months, but surprisingly, the median survival in the IDH mutation group was approximately 31 months10. Clarifying the mechanism of this better prognosis may contribute to development of a new treatment strategy for gliomas.
IDH encodes an enzyme that converts isocitric acid to α-ketoglutaric acid (2-oxoglutaric acid; 2OG) and is categorized into three subtypes. IDH1 is localized in the cytoplasm, whereas IDH2 and IDH3 are localized in mitochondria. Mutation in IDH1 is common in gliomas. Furthermore, mutation of the 132nd arginine to histidine (R132H) is a frequent IDH1 mutation (IDH1R132H)10.
IDH1R132H encodes an enzyme that produces D-2-hydroxyglutarate (D-2HG) from 2OG in the cytoplasm11–15. D-2HG competitively inhibits α-keto acid transaminase activity and alters metabolite levels in the tricarboxylic acid (TCA) cycle16. D-2HG is an oncogenic metabolite that inhibits several 2OG-dependent oxygenases, for example, DNA/RNA-modifying enzymes and JmjC domain-containing enzymes, which modify the epigenetic status of DNA and histones. Several CpG sites in genomic DNA are hypermethylated in glioma patients with IDH mutation17. Because the epigenetic changes occur in gliomas with IDH mutation via TET (Ten-eleven translocation) enzymes that catalyze a key step in the removal of DNA methylation, expression of several enzymes is altered, resulting in various metabolic changes17,18. On the other hand, D-2HG also inhibits ATP synthase and reduces ATP levels in IDH1R132H mutant cells19, suggesting that this enzyme is a target of the growth–suppressive activity of D-2HG. The activity of a variety of enzymes, including those in the TCA cycle and ATP synthase, is thought to change through production of D-2HG due to the IDH1 mutation.
In recent years, the metabolism of cancer cells has attracted much attention. Because the IDH mutation affects metabolism, mainly via the TCA cycle16, we conducted a comprehensive analysis of the metabolites of IDH mutant gliomas with mass spectrometry. In particular, we analyzed clinical glioma samples and compared their metabolism with that of cultured cells. We investigated the TCA cycle, lipid metabolism, amino acid metabolism, and energy metabolism to understand the mechanism of the better prognosis of IDH mutant gliomas.
In this study, we report that levels of D-2HG were significantly increased in the IDH mutant clinical samples similar to cultured glioma cells with IDH mutation, and that levels of intermediate metabolites such as those in the TCA cycle and ATP were altered, most likely through D-2HG activity. Moreover, β-oxidation was decreased only in the IDH mutant glioma samples due to reduced levels of carnitine. These results may explain why gliomas with IDH mutation have a better prognosis. Carnitine may be a better biomarker than D-2HG which is currently a well-known biomarker for IDH mutation. To our knowledge, this is the first report to show comprehensive metabolomic analysis using clinical samples of gliomas with IDH mutation.
Results
Metabolic profile of IDH mutant and IDH normal gliomas
We performed metabolomic analyses of transfected U87 glioblastoma cell lines that expressed mutant IDH or normal IDH and clinical glioma samples with or without IDH mutation using two methods, CE-TOFMS and LC-TOFMS. We detected 187 different metabolites (82 in cation mode and 105 in anion mode) in the U87 cells and 254 different metabolites (cations, 139; anions, 115) in the patient samples from CE-TOFMS analyses. Moreover, we detected 74 substance peaks (41 in positive mode and 33 in negative mode) in the U87 cells and 142 substance peaks (positive, 70; negative, 72) in the patient samples (Supplementary Dataset 1 and 2) from LC-TOFMS analyses. Then, we conducted a heat map analysis using these metabolome data. The clustered heat map shows 396 biochemicals in lysates from five replicates each of IDH mutant glioma tissues and IDH normal tissues (Fig. 1A, left). The clustered heat map for U87 cells showed 261 biochemicals in triplicate lysates from cells expressing IDH1 mutation, IDH1 normal, or vector alone (Fig. 1A, right).
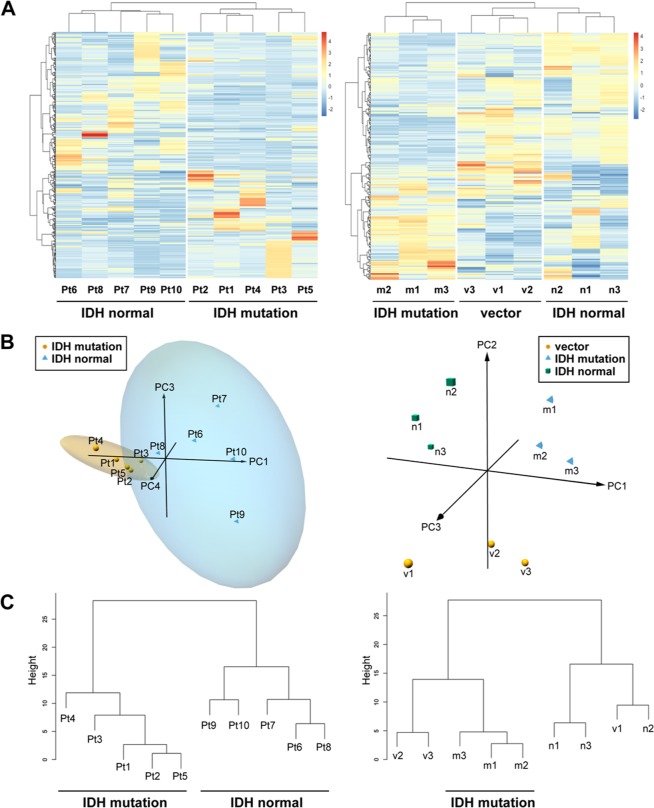
Figure 1.
Heat map, Principal component analysis and Hierarchical clustering dendrogram in IDH mutant/normal gliomas. (A) The clustered heat map shows 396 biochemicals in lysates from five replicates each of IDH mutant glioma tissues (Pt1–Pt5) and normal IDH tissues (Pt6–Pt10) (Left). The heat map for U87 glioblastoma cells shows 261 biochemicals in triplicate lysates from cells expressing IDH1 mutation (m1–m3), normal IDH1 (n1–n3), or vector alone (v1–v3) (Right). (B) 3D plot of principal component analysis (PC Analysis) with fitted ellipses for the biochemicals in the major class category of The Human Metabolome Database (HMDB 3.0) (151 and 115 biochemicals in glioma tissues and U87 cells, respectively). PC1, PC3 and PC4 for glioma tissues (left, PC2 was omitted from the analysis as it did not show any relevant clustering information), and PC1, PC2 and PC3 for U87 cells (right) were plotted. In each group of glioma tissues, 95% confidence interval ellipsoid was illustrated. (C) Hierarchical clustering dendrogram obtained from the three principal components described in B.
One hundred fifty-one biochemicals for glioma tissues (Supplementary Dataset 2) and 115 biochemicals for U87 cells (Supplementary Dataset 1) were selected from The Human Metabolome Database (HMDB 3.0)20 and the principal component (PC) analysis for the biochemicals was conducted. The three-dimensional plot of PC using PC1, PC3, and PC4 for glioma tissues and PC1, PC2, and PC3 for U87 cells were shown in Fig. 1B. The PC analysis of IDH normal samples and samples with IDH mutation formed different PC clusters. This trend was further confirmed by hierarchical clustering dendrogram in Fig. 1C. Importantly, this trend could be observed even in samples from mutant IDH expressing U87 cells. This result clearly indicated that gliomas with IDH mutation had a different metabolic profile compared with that in IDH normal gliomas.
Metabolic differences associated with IDH mutation
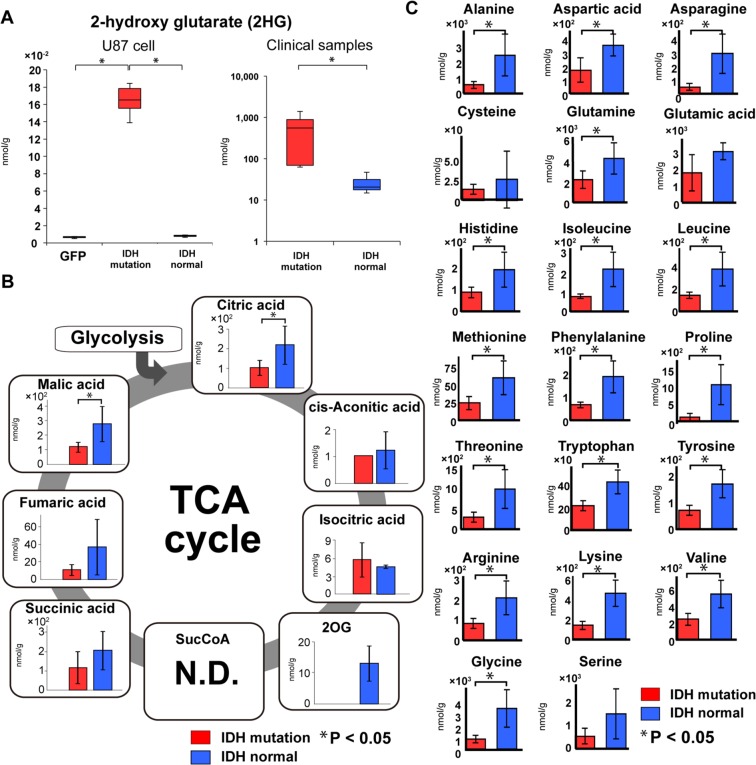
IDH1R132H produces D-2HG from 2OG in the cytoplasm. D-2HG accumulated in gliomas with IDH1 mutation in the previous reports11–15. Our experiments also showed that D-2HG levels were significantly increased in brain tumors bearing IDH1R132H and U87 cells expressing IDH1R132H (t-test, p < 0.05) (Fig. 2A). D-2HG is predicted to inhibit competitively α-keto acid transaminase, which results in inhibition of 2OG synthesis and inhibition of the TCA cycle16. Our experiments with U87 cells showed similar results as a previous study reporting that IDH1R132H expression inhibits the production of 2OG and downstream intermediates in the TCA cycle (Supplementary Fig. S1)21. Experiments with the patient samples also showed similar results for intermediates in the TCA cycle (Fig. 2B). These results indicate that our sample preparation method and metabolomic data from the clinical samples are very reliable. The levels of cis-aconitic acid and isocitric acid, components of TCA cycle, in glioma samples with IDH1 mutation were comparable with those in IDH1 normal glioma samples compared with other components of TCA cycle. These biochemicals could be supplied by the anaplerotic reaction from pyruvate to oxaloacetic acid (OAA) by pyruvate carboxylase (PC) reaction not to stop TCA cycle22.
Figure 2.
D-2-hydroxyglutarate, TCA cycle, and Amino acid in IDH mutant gliomas. (A) A box-whisker, the levels of D-2-hydroxyglutarate (D-2HG) were specifically elevated in IDH mutant glioma cells and clinical tumor specimens (red bars). For the U87 glioblastoma cell experiment, the vertical scale shows relative quantification based on the internal standard. (B) 2OG was considerably reduced in the IDH mutant glioma group (red bars) in clinical tumor specimens, and downstream intermediates of 2OG in the TCA cycle were also reduced. Each vertical scale shows the absolute quantification (nmol/g). (C) Amino acid production was reduced in the IDH mutant glioma group (red bars). The vertical scale for each amino acid shows the absolute quantification (nmol/g). Green bar, empty vector-transfected cells; red bars, IDH1R132H-transfected cells or IDH1R132H mutant glioma tissues; blue bars, normal IDH1-transfected cells or normal IDH1 glioma tissues; N.D., not detected; error bar, standard error of mean. *P < 0.05.
Non-essential amino acids are produced from or degraded to intermediate products of glycolysis and the TCA cycle, including 2OG, pyruvate, acetyl-CoA, fumaric acid, succinyl-CoA, and OAA. Therefore, the production of non-essential amino acids is affected considerably by metabolism in the TCA cycle. Interestingly, the amount of all 20 amino acids including essential amino acids was reduced in IDH1 mutant brain tumors (Fig. 2C) and IDH1R132H-expressing U87 cells (Supplementary Fig. S2). These results were more pronounced in clinical samples than in the cell culture model. It is likely that the supply of essential amino acids is limited in tumors with IDH mutation or they are used up to compensate for the limited access of other metabolites such as non-essential amino acids.
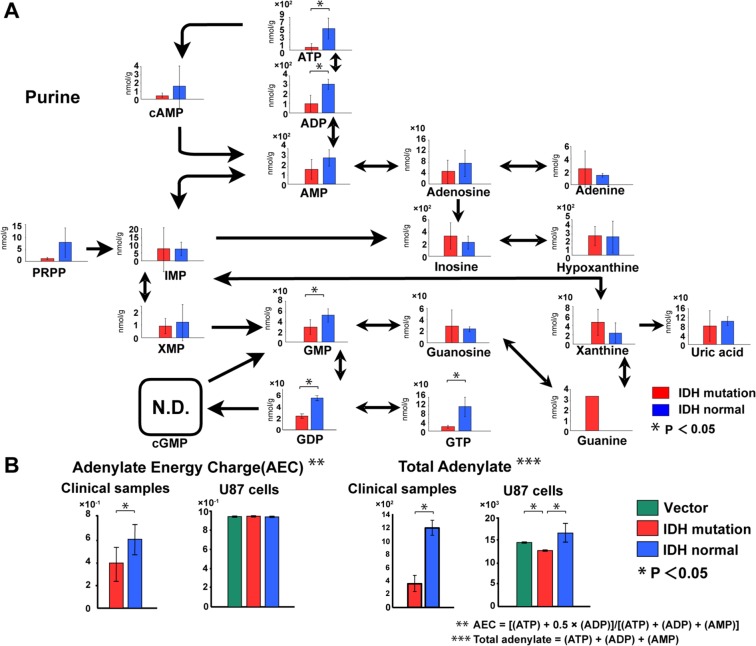
The purine production pathway, which is responsible for nucleic acid production (ATP, ADP, AMP, etc.) and is closely related to energy production, was negatively affected in U87 cells expressing IDH1R132H (Supplementary Fig. S3). Recently, Fu et al. showed that D-2HG directly inhibits ATP synthase and contributes to reduced levels of ATP in IDH1R132H mutant cells19. Our experiments with patient samples also showed reduced levels of ATP in IDH1 mutant gliomas (Fig. 3A). This result confirmed that a reduction in ATP production actually occurred in naturally occurring tumors, as well as the cell culture model that expressed mutant IDH. The adenylate energy charge (AEC) and total adenylate were reduced in IDH mutant gliomas from patients (Fig. 3B). In the cell line experiments, we found no difference in AEC among the groups, but total adenylate was slightly reduced in the IDH mutant cells (Fig. 3B).
Figure 3.
The purine pathway and energy production in IDH mutant gliomas. (A) The purine metabolic pathway was reduced in the IDH mutant glioma group (red bars). Each vertical scale shows the absolute quantification (nmol/g). (B) In the brain tumor tissue experiment, adenylate energy charge (AEC) and total adenylate were reduced. In the U87 glioblastoma cell experiment, we found no clear difference in AEC in cells expressing the IDH mutant (red bars), with each of the three groups having a value of 0.94. Adenylate production was reduced in IDH mutant glioma cells (red bars). Each vertical scale in the U87 cell graphs shows the calculated value relative to the internal standard. The vertical scales in AEC and total adenylate of brain tumors shows the calculated value. Green bars, empty vector-transfected cells; red bars, IDH1R132H-transfected cells or IDH1R132H mutant glioma tissues; blue bars, IDH1 normal-transfected cells or normal IDH1 glioma tissues; N.D., not detected; error bar, standard error of mean. *P < 0.05.
Reduced carnitine levels in IDH mutant glioma clinical samples
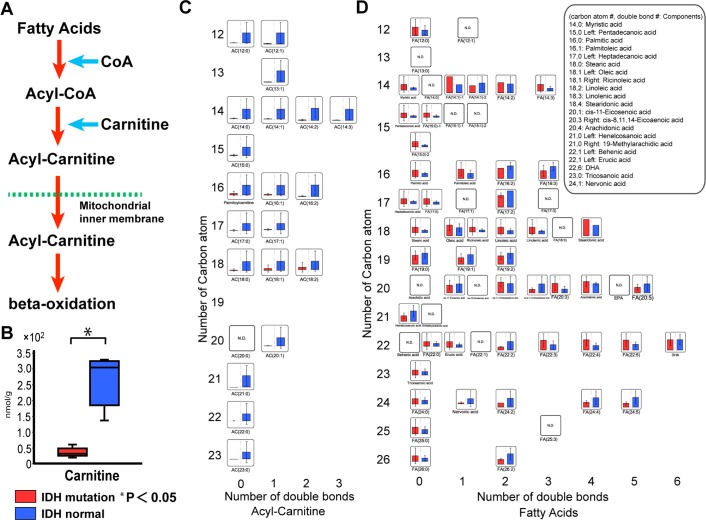
Beta-oxidation takes place in a metabolic pathway that produces energy via oxidation of fatty acids, which bind carnitine in the mitochondrial intermembrane space and are converted to acyl-carnitine. They then cross the mitochondrial inner membrane and enter the mitochondrial matrix where they are converted to acetyl-CoA through β-oxidation. Acetyl-CoA is then used as a substrate in the TCA cycle and in other metabolic reactions (Fig. 4A). Interestingly, carnitine levels were significantly reduced in the IDH mutant group, especially in clinical samples (t-test, p < 0.05) (Fig. 4B and Supplementary Fig. S4). The degree of the difference in the level of carnitine between both groups was much clearer than that of D-2HG (Fig. 4B vs Fig. 2A). As a result of reduced levels of carnitine, levels of all acyl-carnitines were also reduced in the clinical brain tumor samples with IDH mutation (Fig. 4C), although fatty acid levels in IDH mutant samples were similar to those in normal IDH tumor samples (Fig. 4D). In the clinical brain tumor tissues, carnitine and acyl-carnitine were reduced strikingly in the IDH1 mutant group, indicating suppression of β-oxidation. Because the levels of malonyl-CoA were not increased in glioma samples with IDH1 mutation (Supplementary Dataset 2), fatty acid synthesis may not be activated. On the other hand, the levels of acyl-carnitine did not change in U87 cells expressing mutant IDH1 and β-oxidation may not be suppressed in these cells (Supplementary Fig. S4). Although β-oxidation has been thought to be less efficient in the brain compared with other tissues, β-oxidation may be more efficient in IDH1 normal than IDH1 mutant gliomas. Markedly reduced β-oxidation activity in IDH1 mutant gliomas may be involved in the reduction of energy production.
Figure 4.
Analysis of carnitine and lipid metabolism in IDH mutation gliomas. (A) Schematic of the β-oxidation pathway: the lipid metabolism pathway. (B) Carnitine was reduced in the IDH mutant glioma group (red bars). The vertical scale shows the absolute quantification (nmol/g). (C) Acyl-carnitine was reduced in the IDH mutant glioma group (red bars). (D) Fatty acids were not obviously elevated in the IDH mutant glioma group (red bars). In graphs C and D, the vertical scales showed the number of carbon atoms. The horizontal axis scale shows the number of double bonds. The vertical scale shows the relative quantification based on the internal standard. Red bars, IDH1R132H glioma; blue bars, normal IDH1 glioma; N.D., not detected; error bar, standard error of mean. *P < 0.05.
Discussion
In this study, we performed a comprehensive metabolomic analysis of clinical glioma samples with and without IDH mutation and compared the results with an IDH mutant-expressing U87 glioblastoma cell line. We showed a significant increase in D-2HG and altered levels of intermediate metabolites such as those in the TCA cycle, energy production, amino acid production, and nucleic acid production in both the IDH mutant clinical samples and mutant IDH-expressing cells (Figs 2–4 and Supplementary Figs S1–S4). Most importantly, we could obtain statistically significant results from this metabolomic analysis although sample size in our current study using clinical samples is relatively small. Our detailed data analysis of biochemicals could clearly separate glioma samples with IDH1 mutation from normal IDH1 glioma samples (Fig. 1). This indicates that IDH1 mutation causes metabolic reprogramming in cells and these metabolic changes may contribute glioma pathogenesis. Our metabolomics analysis of mutant IDH tumor samples indicated the significant reduction of carnitine and suppression of β-oxidation, which were the marked differences between the clinical samples and the cell lines (t-test, p < 0.05).
Mutant IDH1R132H encodes an enzyme that aberrantly produces D-2HG from 2OG in the cytoplasm11–15. D-2HG is a competitive inhibitor of 2OG-dependent dioxygenases and affects downstream events such as DNA methylation and histone methylation. D-2HG is also predicted to competitively inhibit 2OG transaminase, which results in inhibition of 2OG synthesis, thereby inhibiting the TCA cycle16. Therefore, D-2HG is thought to affect many cellular processes through the activity of 2OG-dependent enzymes. In this study, we showed that the D-2HG level was significantly increased in tumor samples with IDH mutation and mutant IDH-expressing cells compared with control samples (Fig. 2A). 2OG and downstream metabolic intermediates in the TCA cycle were reduced in the mutatnt IDH-expressing glioblastoma cell line (Supplementary Fig. S1). This result matched well with a past study that used a different cell line16. Importantly, our metabolomic analyses using clinical tumor samples showed similar or more pronounced results (Fig. 2B), suggesting that our metabolomic analyses using clinical tumor samples with IDH mutation are highly reliable. Although a report was published that examined levels of D-2HG in clinical brain tumor samples with IDH mutation, this study is the first comprehensive metabolomic analysis of clinical glioma samples with IDH mutation16.
The decrease of ATP level was pronounced in glioma samples of IDH mutant group rather than cell culture system. Non-essential amino acids and acyl-carnitine as an energy source were also decreased in the clinical samples with IDH mutation. ATP production via lipid metabolism relies on β-oxidation. Carnitine plays an important role in the initiation of β-oxidation. Reitman’s study16 and our previous study21 using IDH1R132H-transfected cells did not show that carnitine levels were reduced. In this study, we showed that carnitine was significantly reduced in the clinical glioma samples with IDH mutation (Fig. 4B). Acyl-carnitine was also strikingly reduced in IDH mutant tissues, most likely because of carnitine deficiency.
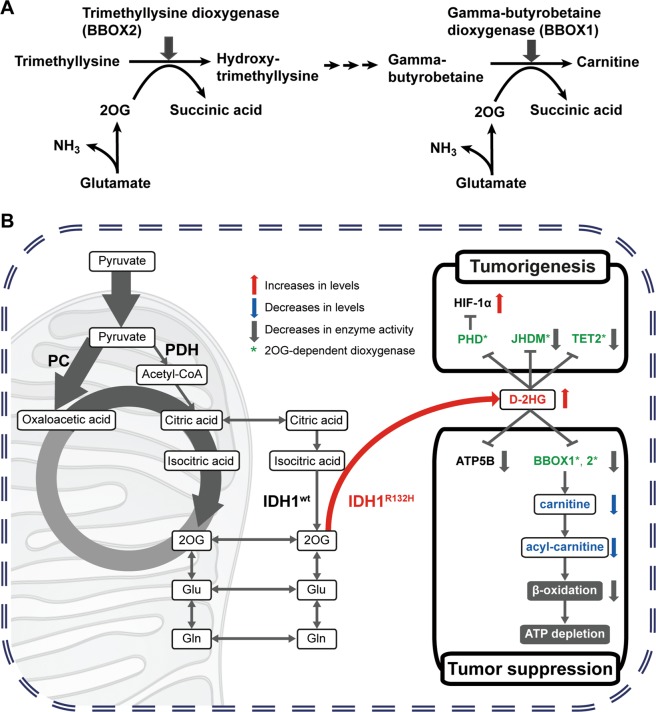
There are two 2OG-dependent dioxygenases in the carnitine synthetic pathway. Gamma-butyrobetaine dioxygenase (BBOX 1), which is the last enzyme in the carnitine synthetic pathway, produces carnitine using gamma-butyrobetaine and 2OG (Fig. 5A). Trimethyllysine dioxygenase (BBOX 2), which is the first enzyme in the carnitine synthetic pathway, converts trimethyllysine into hydroxytrimethyllysine. D-2HG may inhibit the activity of these enzymes and reduce carnitine levels, because D-2HG is a competitive inhibitor of 2OG-dependent dioxygenases23,24. In fact, some authors have speculated that BBOX 1 and BBOX 2 are inhibited by D-2HG24. Indeed, there is a report that BBOX 1 is inhibited by D-2HG at a relatively high IC50 in vitro25. Moreover, 2OG is a cofactor for both BBOX 1 and BBOX 2. The levels of 2OG in clinical glioma samples with IDH mutation are low, which may contribute to reduced levels of carnitine. Although carnitine synthesis mainly occurs in the liver, its synthesis may occur locally in the brain at lower levels26,27. The decrease in carnitine in IDH mutant tumors may result in suppression of β-oxidation, contribute to decreased ATP production, and relate to a better prognosis.
Figure 5.
The hypothesis of the mechanism of IDH mutant gliomas. (A) 2OG-dependent dioxygenases in carnitine biosynthesis. There are two 2OG-dependent dioxygenases in the carnitine synthetic pathway. Carnitine is produced from gamma-butyrobetaine by gamma-butyrobetaine dioxygenase (BBOX 1) that is the last enzyme in the carnitine synthetic pathway. Trimethyllysine is converted into hydroxytrimethyllysine by trimethyllysine dioxygenase (BBOX 2) that is the first enzyme in the carnitine synthetic pathway. In these chemical reactions, 2OG acts as a cofactor. Reduced levels of 2OG in IDH mutant clinical tissues may lead to decreased carnitine synthesis through inhibition of these enzyme activities. (B) The proposed hypothesis for the mechanism of metabolism in IDH mutant tissues. The IDH mutation has two effects: a tumorigenic effect and a tumor suppressive effect. IDH1 mutant gliomas produce D-2-hydroxyglutarate (D-2HG), resulting in inhibition of 2OG synthesis and downstream metabolic intermediates in the TCA cycle. As a tumorigenic effect, D-2HG produced by mutant IDH causes activation of HIF-1α through the inhibition of PHD (prolyl hydroxylases) activity and the direct inhibition of JHDM (Jumonji C-domain-containing histone demethylases) and TET2 activities. As a tumor suppressive effect, D-2HG inhibits ATP production through at least two ways. D-2HG directly inhibits ATP synthase activity by interaction with ATP5B (ATP synthase β subunit) and reduces ATP production. In addition, β-oxidation in clinical gliomas with IDH mutation is suppressed due to the reduction of carnitine levels. BBOX 1 and BBOX 2 in the carnitine synthetic pathway may be inhibited by high levels of D-2HG and lower levels of 2OG. Oxaloacetic acid could be supplied from pyruvate by pyruvate carboxylase (PC) as an anaplerotic reaction.
We found a discrepancy in carnitine and acyl-carnitine levels between the clinical glioma samples with IDH mutation and the IDH1R132H-transfected cells (Fig. 4 and Supplementary Fig. S4). Two explanations for this are possible. One is that sufficient amounts of carnitine may be supplied by the serum in the culture medium in the cell culture system, but not in IDH mutant tissues. We examined the effect of carnitine in the culture medium by growth rate of U87 cells. Supplementation of carnitine into the culture medium containing 10% dialyzed-fetal bovine serum (dialyzed-FBS) had a positive effect on cell growth of U87 cells. In addition, meldonium, a carnitine transporter and BBOX 1 inhibitor, supplemented to the 10% FBS-containing culture medium reduced proliferation of U87 cells (Supplementary Fig. S5). These data strongly suggested that the medium supplemented with FBS contains carnitine, which is important for proliferation and β-oxidation at least in part on cell culture system. The other is that 2OG was strongly decreased in the IDH mutant tissues, but only slightly decreased in the cell culture system (Fig. 2B and Supplementary Fig. S1). 2OG is replenished through the conversion from glutamine to glutamate, as well as the TCA cycle. A sufficient amount of 2OG may be synthesized from glutamine that is present in the culture medium in the cell culture system. However, a sufficient amount of glutamine may not be present in IDH mutant tissues. In fact, our metabolomic analyses showed that glutamine levels were reduced in IDH mutant tissues, but not in cell culture. As a result, we conclude that β-oxidation is decreased strikingly in the clinical glioma samples with IDH mutation. The supply of metabolites is likely altered locally in solid tumors, indicating the importance of investigations using clinical samples as in our current study.
The proposed hypothesis for the mechanism of metabolism in IDH mutant tissues is shown in Fig. 5B. The IDH mutation has two effects: a tumorigenic effect and a tumor suppressive effect. D-2HG produced by mutant IDH causes activation of HIF-1α through the inhibition of PHD (prolyl hydroxylases) activity and the direct inhibition of JHDM (Jumonji C-domain-containing histone demethylases) and TET2 activity18,28–30. These modulations of the activities are related to the tumorigenic effect. On the other hand, D-2HG inhibits ATP production through at least two mechanisms. D-2HG directly inhibits ATP synthase activity and reduces ATP production19. In addition to the reduction of ATP, we elucidated that β-oxidation in clinical gliomas with IDH mutation is suppressed due to the reduction of carnitine levels. BBOX 1 and BBOX 2, which are involved in carnitine synthesis, are 2OG-dependent dioxygenase and their activities may also be inhibited by D-2HG24. In addition to D-2HG production, because 2OG is maintained at very low levels in clinical gliomas with IDH mutation, BBOX 1 and BBOX 2 activities might be strongly inhibited. These are related to the tumor suppressive effect of IDH mutation.
For cell division, not only energy, but also amino acids and nucleic acids must be constantly supplied and are required at higher levels in cancer cells compared to normal cells. Our results suggest that cell division in IDH mutant gliomas is negatively affected and may explain why IDH mutant gliomas have a better prognosis. Pyruvate kinase isoform M2 (PKM2) is glycolytic enzyme catalyzing the conversion of phosphoenolpyruvate (PEP) to pyruvate. Serine binds to and activates PKM2, and that PKM2 activity in cells is reduced in response to serine deprivation31. Therefore, serine is related to glycolysis and the activity of glycolytic reaction affects the production of biochemicals. Methionine is related to DNA and RNA methylation32. Our study showed that serine, glycine, and methionine were decreased in gliomas with IDH mutation (Fig. 2C). The decrease of serine resulted in the suppression of folate metabolism and glutathione production. Folate is related to cell division and glutathione is related to oxidative stress. These results may also link to the better prognosis of gliomas with the IDH mutation (Supplementary Fig. S6).
The main purpose of this study was to understand the change of metabolism in clinical glioma samples. Thus, we just used U87 cell culture model as reference for some aspects such as D-2HG production because it does not sometimes reflect aspects of clinical samples. Furthermore, we verified the appropriate size of the clinical samples for the number of detected metabolites using MetSizeR33 and PASS16 (NCSS LLC, Kaysville, USA). It was presumed that this clinical study was needed to more samples (Supplementary Fig. S7). Collection of glioma clinical samples with IDH mutation is not easy due to low incidence of patients with this mutation in only one hospital. We will collect more samples, conduct metabolomic analysis, and further confirm our current results in the future. Regarding to the carnitine in the group sample sizes of 5 and 5, 70.4% power was calculated even when considering the Bonferroni’s correction for multiple testing, alpha = 0.00033 (Supplementary Table S1). Thus, regardless of the small sample size, it is thought that carnitine is sufficiently reliable.
In summary, we obtained a large amount of information by conducting metabolomics on IDH mutant glioma cells and clinical tissues. Our results demonstrate that, in IDH mutant gliomas, (1) the TCA cycle is inhibited, (2) β-oxidation is suppressed, especially in clinical samples due to carnitine deficiency, (3) energy production is reduced, and (4) molecules required for cell division (amino acids and nucleic acids) are reduced. These results may explain the mechanism for the better prognosis of gliomas with IDH mutation. We also propose that carnitine may be a better biomarker for IDH mutant gliomas than D-2HG. The carnitine synthesis pathway might become a therapeutic target for IDH normal glioma with poor prognosis.
Methods
Cell line
The human glioblastoma cell line U87 was obtained from ATCC and cultured as described previously21. U87 cells do not harbor the IDH mutation.
Specimens from patient’s brain tumors
We obtained specimens from 10 patients who underwent surgery for glioma between 2006 and 2012 at Jichi Medical University. The clinical samples were used after obtaining informed consent from the patients and approval from the Jichi Medical University Ethical Board (approval number 11–31). This study was performed in accordance with the ethical standards of the 1964 Declaration of Helsinki, as well as the GCP guidelines. The tumor samples were collected during surgery, immediately frozen in liquid nitrogen in the operating room, and stored at −80 °C until metabolomic analysis. No anticancer drugs or medications (mercaptopurine, fluorouracil, etc.) that could modify the metabolism were administered prior to surgery. Patient clinical information is listed in Supplementary Table S2 and Fig. S8). Histopathology was performed in the Department of Pathology of the University. All tumors were classified according to the current WHO classification of tumors of the CNS34.
Plasmids and transfection
Plasmids expressing normal IDH1 or IDH1R132H under the control of the CMV promoter were described previously21. The plasmid was transfected into the U87 glioblastoma cell line using Lipofectamine 2000 (Invitrogen). We confirmed the expression of IDH1 by western blotting as described previously21.
Detection of mutations in IDH1 and IDH2
Total DNA was extracted from U87 cells and tumor samples using the DNeasy Blood & Tissue Kit (Qiagen). A DNA fragment of IDH1 or IDH2 was amplified by PCR with a pair of primers as described10, and mutations were confirmed by direct sequencing.
Metabolomic analysis
Twenty-four hours after transfection, to inactivate enzymes, triplicate wells of U87 cells were quenched with methanol containing an internal standard (Solution ID: H3304-1002, Human Metabolome Technologies, Tsuruoka, Japan). Next, the cells were scraped off and stored at −80 °C until analysis. The number of transfected U87 cells for analysis is described in Supplementary Table S3A. The amounts of tumor samples that were used for analyses are described in Supplementary Table S3B. Tumor samples were quenched with 1500 μl acetonitrile and 20 μl D-camphor-10-sulfonic acids as an internal reference standard, and were homogenized on ice.
We normalized the data using sample weight for clinical samples and the number of cells for cultured cells. As the statistical test to compare the two groups, we use two-tailed Welch’s t-test. The Welch’s t-test is used when two samples have unequal variances (the violation of the homoscedasticity assumption) but fit to the normal distribution. In case of the violation of the normality assumption, the Mann-Whitney test was performed. The fitness to a normal distribution was examined before Welch’s t-test. Bonferroni adjustment was performed to correct P value for the multiple comparison.
Furthermore, to extract the candidates of biochemical compounds that attribute to either IDH1 normal or IDH1 mutant, the principal component analysis was performed after all data values were normalized with mean 0 and variance 1 by using a peak value of each compound to fit a normal distribution.
Mass spectrometry
Capillary electrophoresis time-of-flight mass spectrometry (CE-TOFMS)
Metabolomics were measured and data were processed with an Agilent CE-TOFMS system (Agilent Technologies) equipped with a fused silica capillary with an internal diameter of 50 µm × 80 cm. Measurement of extracted metabolites in both the positive and negative modes was performed by using a commercially available electrophoresis buffer. Alignment of detected peaks was performed according to the mass-to-charge ratio (m/z) value and normalized migration time (Supplementary Table S4A). The glycolysis system, pentose phosphate cycle, TCA cycle, purine metabolic pathway, pyrimidine metabolic pathway, nicotinic acid, nicotinamide metabolic pathway, and various amino acid metabolic pathways were analyzed. We then performed the relative quantification of these data with the internal reference standard. We also performed measurements by adding the absolute quantitative value for 116 metabolites (Supplementary Dataset 1 and 2).
Liquid chromatography time-of-flight mass spectrometry (LC-TOFMS)
The positive and negative modes were performed using an Agilent 1200 series Rapid Resolution LC system SL (Agilent Technologies) equipped with an Octa Decyl Silyl analytical column of 2 mm × 50 mm and 2 µm particle size (Supplementary Table S4B).
Metabolome measurements were carried out by a facility service at Human Metabolome Technology Inc. (Tsuruoka, Japan).
Data processing and analysis
The detected peaks were automatically extracted using the automatic integration software MasterHands ver.2.9.0.9 (Keio University, Tokyo, Japan), and the m/z, peak area as the peak information, migration time (MT) with CE-TOFMS, and retention time (RT) with LC-TOFMS were obtained. The obtained peak area was converted to the relative peak area with the following formula: Relative peak area = target peak area/ (internal reference standard peak area × sample volume).
The detected peaks were compared with all substances registered on the Human Metabolome Technologies Metabolic Substance Database based on the m/z and MT or RT values. The error tolerance for the search was ± 0.5 min for MT or RT and ± 10 ppm for m/z (Supplementary Dataset 1 and 2). The metabolome pathway mapping is shown as a bar graph with the obtained peaks. Abbreviations used in figures and tables are shown in Supplementary Table S5.
Heat map, PCA, and hierarchical dendrogram
All analysis was performed using R35 (Version 3.4.3) with R Studio36 (Version 1.1.383). Heat map was generated using the R package “pheatmap” under Euclidean distance with ward. D2 linkage method for each row and column. Blue color represents lower concentration; red color represents higher concentration. PCA was performed using the R “prcomp” function, and the 3D plot was created using the R package “pca3d”. The cluster for hierarchical dendrogram was calculated using the R “hclust” function under Euclidean distance with ward. D2 method.
The adenylate energy charge
The adenylate energy charge (AEC) and total adenylate, which serve as indicators of the energy status inside the cell, were obtained as follows: AEC = [(ATP) + 0.5 × (ADP)]/ [(ATP) + (ADP) + (AMP)]; total adenylate = [(ATP) + (ADP) + (AMP)].
Supplementary information
Acknowledgements
This research was supported by the FY2012 Jichi Medical School Research Encouragement Award, the FY2011 Grand Prize for Promotion of Human Metabolome Technologies Metabolomics Cutting Edge Research, the MEXT-Supported Program for the Strategic Research Foundation at Private Universities 2013–2017, and the Japan Society for the Promotion of Sciences Grant-in-Aid for Scientific Research (KAKENHI) Grant 17K16659 (to S.M.), 17K08497 (to K.T.), and 17K11248 (to H.E.).
Author Contributions
S.M., K.T., K.O. E.W., K.K. and H.E. conceived the experiments, S.M., K.T., M.U., A.G., T.Y., H.M. and A.K. conducted the experiments, S.M., K.T, E.S., Y.O., M.M. and H.E. analyzed the results. All authors reviewed the manuscript.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Satsuki Miyata, Email: smiyata@jichi.ac.jp.
Kaoru Tominaga, Email: tominaga@jichi.ac.jp.
Hitoshi Endo, Email: hendo@jichi.ac.jp.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-46217-5.
References
- 1.Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2005–2009. Neuro Oncol. 2012;14:v1–v49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Comittee of Brain Tumor Registry of Japan Report of Brain Tumor Registry of Japan (2001–2004). 13th ed. Neurol. Med. Chir. (Tokyo) 2014;54(Suppl):1–102. [Google Scholar]
- 3.Wilson CB. Glioblastoma: the past, the present, and the future. Clin. Neurosurg. 1992;38:32–48. [PubMed] [Google Scholar]
- 4.Pellegatta S, Cuppini L, Finocchiaro G. Brain cancer immunoediting: novel examples provided by immunotherapy of malignant gliomas. Expert Rev. Anticancer Ther. 2011;11:1759–1774. doi: 10.1586/era.11.102. [DOI] [PubMed] [Google Scholar]
- 5.Khasraw M, Simeonovic M, Grommes C. Bevacizumab for the treatment of high-grade glioma. Expert Opin. Biol. Ther. 2012;12:1101–1111. doi: 10.1517/14712598.2012.694422. [DOI] [PubMed] [Google Scholar]
- 6.Brem H, et al. Placebo-controlled trial of safety and efficacy of intraoperative controlled delivery by biodegradable polymers of chemotherapy for recurrent gliomas. Lancet. 1995;345:1008–1012. doi: 10.1016/S0140-6736(95)90755-6. [DOI] [PubMed] [Google Scholar]
- 7.Bexell D, Svensson A, Bengzon J. Stem cell-based therapy for malignant glioma. Cancer Treat. Rev. 2013;39:358–365. doi: 10.1016/j.ctrv.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Muragaki Y, et al. Phase I/IIa trial of autologous formalin-fixed tumor vaccine concomitant with fractionated radiotherapy for newly diagnosed glioblastoma. Clinical article. J. Neurosurg. 2011;115:248–255. doi: 10.3171/2011.4.JNS10377. [DOI] [PubMed] [Google Scholar]
- 9.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu Y, Huang R, Zheng Y, Zhang Z, Liang A. Glioma-derived mutations in isocitrate dehydrogenase 2 beneficial to traditional chemotherapy. Biochem. Biophys. Res. Commun. 2011;410:218–223. doi: 10.1016/j.bbrc.2011.05.108. [DOI] [PubMed] [Google Scholar]
- 13.Gross S, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J. Exp. Med. 2010;207:339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ward PS, et al. Identification of additional IDH mutations associated with oncometabolite R(-)-2-hydroxyglutarate production. Oncogene. 2012;31:2491–2498. doi: 10.1038/onc.2011.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ward PS, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reitman ZJ, et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc. Natl. Acad. Sci. USA. 2011;108:3270–3275. doi: 10.1073/pnas.1019393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christensen BC, et al. DNA methylation, isocitrate dehydrogenase mutation, and survival in glioma. J. Natl. Cancer Inst. 2011;103:143–153. doi: 10.1093/jnci/djq497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guilhamon P, et al. Meta-analysis of IDH-mutant cancers identifies EBF1 as an interaction partner for TET2. Nat. Commun. 2013;4:2166. doi: 10.1038/ncomms3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu X, et al. 2-hydroxyglutarate inhibits ATP synthase and mTOR Signaling. Cell Metab. 2015;22:508–515. doi: 10.1016/j.cmet.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wishart DS, et al. HMDB 3.0-The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:801–807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyata S, et al. An R132H mutation in isocitrate dehydrogenase 1 enhances p21 expression and inhibits phosphorylation of retinoblastoma protein in glioma cells. Neurol. Med. Chir. (Tokyo) 2013;53:645–654. doi: 10.2176/nmc.oa2012-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahn CS, Metallo CM. Mitochondria as biosynthetic factories for cancer proliferation. Cancer Metab. 2015;3:1. doi: 10.1186/s40170-015-0128-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Losman JA, Kaelin WG. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 2013;27:836–852. doi: 10.1101/gad.217406.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chowdhury R, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaz FM, Wanders RJA. Carnitine biosynthesis in mammals. Biochem. J. 2002;361:417–429. doi: 10.1042/bj3610417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Celestino-Soper PBS, et al. A common X-linked inborn error of carnitine biosynthesis may be a risk factor for nondysmorphic autism. Proc. Natl. Acad. Sci. USA. 2012;109:7974–7981. doi: 10.1073/pnas.1120210109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu C, Ward P, Kapoor G, Rohle D. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee HY, et al. HIF-1-dependent induction of Jumonji domain-containing protein (JMJD) 3 under hypoxic conditions. Mol. Cells. 2014;37:43–50. doi: 10.14348/molcells.2014.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burr SP, et al. Mitochondrial Protein Lipoylation and the 2-Oxoglutarate Dehydrogenase Complex Controls HIF1α Stability in Aerobic Conditions. Cell Metab. 2016;8:740–752. doi: 10.1016/j.cmet.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaneton B, et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491:458–462. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maddocks OD, et al. Serine Metabolism Supports the Methionine Cycle and DNA/RNA Methylation through De Novo ATP Synthesis in Cancer Cells. Mol. Cell. 2016;21:210–221. doi: 10.1016/j.molcel.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nyamundanda G, et al. MetSizeR: selecting the optimal sample size for metabolomic studies using an analysis based approach. BMC Bioinformatics. 2013;14:338. doi: 10.1186/1471-2105-14-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Louis DN, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 35.Team RDC. R: A language and environment for statistical computing, http://www.r-project.org/ (2015).
- 36.Team Rs. RStudio: Integrated Development Environment for R, http://www.rstudio.com (2015).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.