Abstract
Enhancers are non-coding regulatory elements that are distant from their target gene. Their characterization still remains elusive especially due to challenges in achieving a comprehensive pairing of enhancers and target genes. A number of computational biology solutions have been proposed to address this problem leveraging the increasing availability of functional genomics data and the improved mechanistic understanding of enhancer action.
In this review we focus on computational methods for genome-wide definition of enhancer-target gene pairs. We outline the different classes of methods, as well as their main advantages and limitations. The types of information integrated by each method, along with details on their applicability are presented and discussed. We especially highlight the technical challenges that are still unresolved and hamper the effective achievement of a satisfactory and comprehensive solution.
We expect this field will keep evolving in the coming years due to the ever-growing availability of data and increasing insights into enhancers crucial role in regulating genome functionality.
1. Introduction
Enhancers are distal regulatory elements with a crucial role in controlling the expression of genes [1,2]. From many point of views they are analogous to promoters [3], but they are located at a larger distance from the transcription start site (TSS) of the gene they regulate. Enhancers act through the binding of transcription factors just like promoters. However, elucidating the function of enhancers remains more elusive for multiple reasons.
-
1)
The relative location of the enhancer with respect to its target genes can be greatly variable. Enhancers can be present in the vicinity of their target genes but do not necessarily regulate the closest one. They can also be downstream of their target or act across intervening genes to reach their targets [1,4]. Enhancer-target genes (ETG) pairing is further complicated by the fact that one enhancer can in principle act on multiple genes, or one gene can be regulated by multiple enhancers.
-
2)
Enhancers do not have a specific sequence motif or structure for their univocal genome-wide identification. Indeed, a combination of transcription factors can bind to enhancers [[5], [6], [7]], then interacting with other proteins (for example mediator [8]) to initiate transcription of its target gene. This complicates even more the definition of which genomic regions can in fact act as enhancers.
-
3)
The activity of enhancers is extremely cell type-specific. In fact, enhancers are the single genomic feature most variable across tissues and cell types in terms of their activation [9]. While a specific gene may be active in multiple cell types, its activation can be triggered by distinct enhancers in different tissues [10]. As such, this further complicates the definition of a comprehensive set of enhancers, as well as the definition of their target genes.
Among these three challenges, the definition of enhancers-target gene pairs has been gaining growing attention in the field of computational biology and genomics, owing to the increasing availability of genome-wide experimental data that can be exploited to address this problem [11]. Indeed high-throughput genome-wide methodologies to examine transcription factors (TFs) binding, core histone modifications and RNA polymerase II (Pol2) association has drastically altered the perception of how regulatory sequences are distributed in mammalian genomes. With the efforts of large epigenomics consortia like ENCODE [12] and Roadmap Epigenomics [9], genome-wide chromatin mark profiles are available across various cell types.
The regulatory proteins bound at enhancers and those bound at their target gene promoters must get in close physical proximity to regulate their target genes. Physical interaction of enhancers and their distant target genes are facilitated by the formation of loops in chromatin, with the collaboration of various architectural proteins such as mediator or cohesin complexes [5,13]. Thus, one key characteristic feature that can be exploited in refining ETG pairs is their localization within specific chromatin 3D structures, such as topological domains (TADs) [14,15]. The development of new experimental techniques fostered progress in this field by enabling better genome-wide characterization of chromatin architecture. In particular, a number of genomic approaches based on high-throughput sequencing technologies have been derived from chromosome conformation capture (3C) [16]. Namely, the 3C-derived technologies such as Hi-C [17], ChIA-PET [18] and capture Hi-C [19,20] have added more resources to this repertoire of knowledge, while achieving higher and higher resolution over the years [21,22], as reviewed in [23].
Even though enhancers were first described in the ‘80s [24], the recent rise in interest in enhancers has been spurred not only by the growing availability of epigenomics and chromatin architecture data, but also by the increased awareness of their important role as key players in gene regulation and cell identity definition [25]. A growing body of evidence is also corroborating the crucial effect of germline non-coding variants in the general population [26], where disease-associated SNPs are often located in enhancers [27]. Likewise, the role of enhancers in diseases is more and more evident from literature on cancer genomics, where the importance of non-coding mutations has been under-estimated so far [28]. Similarly other genetic diseases are associated with mutations in chromatin modifying enzymes acting on enhancers [29] or ETG pairing being rewired due to structural variations in the genome [30,31]. The recent increase in evidence to the role of enhancer disruption in genetic diseases and cancer [[32], [33], [34]] led to a surge in studies on enhancer-target gene association.
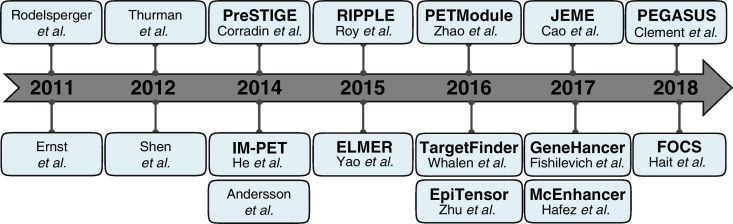
Several computational solutions have been proposed to identify ETG pairs over the past few years (Fig. 1). The increasing number of publications is evidence for the growing interest in this problem and need for bioinformatics solutions. We expect even more methods to be proposed in the coming years, as the number, size and complexity of available functional genomics data used to define enhancers and their targets is rapidly growing.
Fig. 1.
Timeline of the enhancer-target gene pairing algorithms. The main methods described in the review (tool name in bold, if defined) are listed to highlight the timeline of their publication over the years (horizontal axis).
In this review we focus on computational methods for genome-wide definition of ETG pairs. We chiefly discuss how different types of genomics data are leveraged to achieve this goal, as well as the limitations of currently available solutions.
2. Genome-wide Definition of Enhancer Regions
The first practical problem for genome-wide characterization of ETG pairs is actually defining the set of enhancers regions to be considered for the cell type of interest. In spite of efforts by large epigenomic consortia such as ENCODE and FANTOM for enhancer identification, the dynamic and cell type-specific nature of enhancer activity results in the inability to create an exhaustive reference list of enhancers. Although, some studies tried to identify enhancer sequence motifs [26,35,36], there isn't a specific sequence motif that can be generally used for genome-wide identification of enhancers, thus they are usually defined using functional data.
Gene reporter assays in cultured cells is generally employed to identify if a selected sequence can act as an enhancer, but in vivo testing of the reporter or in vivo editing of the enhancer in transgenic animals are considered the definitive proof [37]. In recent years, high throughput versions of gene reporter assays have successfully been adopted for the genome-wide identification of non-coding regulatory sequences, including STARR-sEq. [38] or massively parallel reporter assay (MPRA) [39]. In this regard, it's worth mentioning RAEdb [40], a recent collection of data from high-throughput reporter assays, and the VISTA database [41], a collection of experimentally validated enhancers, including in vivo validated ones.
However, for a genome-wide definition, other types of functional genomics data are usually adopted, including epigenomics or transcriptomics data. As enhancer activity is frequently cell type specific, this approach has the limitation that multiple types of cells must be considered to obtain a comprehensive list. For example, the ENCODE consortium estimated that the human genome comprises hundreds of thousands of enhancers, based on the integrative analysis of 13 chromatin marks across 147 cell types spanning 1640 data sets. It also included 119 transcription factors ChIP-seq and chromatin accessibility data for its prediction [12].
2.1. Epigenomic Data for Defining Enhancers
Chromatin marks-based annotation of enhancer regions relies on functional data correlated with enhancer activation such as binding of transcription factors or other co-factors, specific histone post-translational modifications or chromatin accessibility.
Namely, p300 is a histone acetyltransferase protein acting as transcription co-activator, which is known to be bound at active enhancers [42]. As such, ChIP-seq experiments targeting p300 have often been used for genome-wide annotation of enhancers [43,44]. However, to obtain a more comprehensive and unbiased list of enhancers, ChIP-seq targeting specific histone marks has been adopted as well. The presence of chromatin marks such as high levels of histone H3 lysine 4 mono-methylation (H3K4me1) accompanied by histone H3 lysine 27 acetylation (H3K27ac) is typically found in nucleosomes associated with active enhancers [[45], [46], [47]]. H3K4me1 along with H3K4me3 is usually found also at promoter regions, but the relative enrichment of the two marks is expected to be different at enhancers and promoters [[48], [49], [50]]. The higher ratio of H3K4me1 over H3K4me3 ChIP-seq enrichment signal has been used to discriminate enhancers with respect to other TSS proximal regulatory regions. Nevertheless, a number of reports in literature suggest that H3K4me1 can also be found in enhancers when not in an active state in a specific cell type, sometimes annotated as “weak” [51] or “poised” enhancers [52]. The latter sites may also be associated with histone H3 lysine 27 tri-methylation (H3K27me3) [53]. Thus, ChIP-seq for H3K27ac is usually preferred to focus on enhancers specifically active in the cell type under investigation [52].
Active regulatory regions including enhancers and promoters are generally characterized by chromatin with higher accessibility and are depleted of nucleosomes [54]. Thus, genomics methods probing chromatin accessibility such as DNase-seq and ATAC-seq can be used as alternative approaches for genome-wide identification of regulatory elements. As these techniques do not rely on antibodies, they avoid any potential issues related to specificity or immunoprecipitation efficiency. DNase-seq is based on partial digestion with DNA nucleases such as DNase I, that will cut more frequently in positions of higher accessibility that are not protected by histones or other DNA associated proteins [55]. These regions of increased accessibility are also called DNase hypersensitivity sites (DHS). ATAC-seq instead leverages differential sensitivity to transposase accessibility to identify open chromatin regions [56]. ATAC-seq has a shorter experimental protocol compared to DNase-seq and can be applied on a smaller number of cells, thus making it the technique of choice for rare cell populations. DNase-seq has been used to pinpoint the exact binding site of TFs with a resolution of few base pairs, if the coverage and data quality is high enough to perform DNAse-seq footprinting analysis [57,58]. Footprinting in principle can also be applied on ATAC-seq with some specific adjustments in the analysis [59].
The length and number of putative enhancers are largely affected by the genomic features used to define them. For example, H3K27ac ChIP-seq peaks called with MACS [60] by Roadmap Epigenomics consortium may have sizes ranging between 100 bp to a few kb [61]. DNase-seq peaks may have median size 2.7 kb if called by MACS whereas the same type of data analyzed by the hotspot [62] algorithm yield peaks with median length of 2.5 kb [63]. Both the size and number of enhancers defined in a given cell type will be affected by the methodology adopted. Depending on the experimental setting, in human samples we may expect a number of distal ChIP-seq peaks in the order of few thousands for p300, between 24,566 and 58,023 for H3K4me1 or H3K27ac, and more than 80,000 for chromatin accessibility peaks [53].
While all of these features are generally associated to enhancers, several epigenomics and transcriptomics datasets are combined in a number of computational methods for the identification of enhancers, as reviewed more extensively in [64]. These include, for example, CSI-ANN [65] to define enhancers using cell type-specific data for multiple histone modifications. CSI-ANN algorithm implements artificial neural networks in a two-step process involving data transformation and features extraction to identify chromatin signatures for identification of enhancers. Methods such as Segway [66] or chromHMM [67] classify genomics regions by chromatin states based on segmentation of multiple marks considered simultaneously. A similar approach to define chromatin states, named RFECS [68], is based on a random forest classifier to predict enhancers across different cell types. Instead, DEEP [69] is an enhancer predictor approach incorporating both chromatin marks and sequence features in its model.
2.2. Transcriptomic Data for Defining Enhancers
Recent advances in transcriptomics and genomics highlighted that active enhancers produce enhancer-originating bi-directional non-coding RNAs, also termed eRNAs, which are typically 0.5–2 kb in length [70,71]. Moreover, eRNAs expression level is correlated with the functional activity of the enhancer [72], thus enabling the use of eRNAs as a marker for active enhancers in the genome, as reviewed in [73]. For this purpose total RNA-seq can be used, although the experimental protocol must be tailored to make sure the short non-polyadenylated transcripts are captured and sequenced. Even if such precautions are considered, total RNA-seq will mostly generate reads from mature transcripts, thus leaving few reads to measure eRNAs. As such, a limited coverage and power to detect eRNAs will be available.
For this reason, sequencing protocols for detecting nascent transcripts are often used instead to measure eRNAs transcription. These include, among others, CAGE-sEq. [[74], [75], [76]], GRO-sEq. [77], GRO-cap [77], 5′GRO-sEq. [78], NET-SEq. [79], PRO-cap [80], PRO-SEq. [81], Start-sEq. [82] and TT-SEq. [83]. Hereafter we focus only on CAGE-seq, as it has been used in the ETG pairing methods reviewed here.
CAGE-seq is based on the detection of 5′-capped transcripts and it has been systematically adopted by the FANTOM project, a large-scale collaborative consortium, to map TSS of coding and non-coding genes, as well as the location of enhancers [84,85]. CAGE allows high precision in mapping the position of transcription start sites (TSS), but it has limited sensitivity, thus identifying only a subset of active enhancers in the cell type analyzed. GRO-cap is more sensitive to detect activity at TSS and eRNAs alike [86]. The enhancers identified by the FANTOM project consortium using CAGE-seq were about 40,000 in total across 432 primary cell, 135 tissue and 241 cell line samples from human [87]. This is mostly due to the fact that a large fraction of reads is actually originating from TSS-proximal regions of annotated genes, rather than from distal regulatory elements.
Despite the many technological advancements in genome-wide experimental techniques, the definition of enhancers based on functional genomics data remains challenging because enhancers and promoters have very similar characteristics [88]. In fact, regulatory regions are often first defined based on functional data, then TSS proximal vs distal regulatory elements are distinguished based on their distance from annotated promoters. This wide array of choices has implications in terms of the size of enhancer regions, which can range from a few kb to few base pairs, as well as the number of enhancers.
Likewise, promoter regions can be defined with different parameters. While annotated TSSs are usually derived from reference databases such as RefSEq. [89] or Ensembl [90], then arbitrary choices are made in terms of the window to distinguish between promoter (TSS proximal) and distal regulatory elements. The size of the TSS proximal regions considered to be promoters can be highly variable in the literature as there is no consensus on this parameter either.
3. ETG Pairing Tools Over the Years
Over the past few years a variety of algorithms and bioinformatic tools have been proposed for genome-wide definition of ETG pairs (Fig. 1). To elucidate the complex relationship between distal regulatory regions and their target genes, ETG prediction algorithms have employed a number of different approaches. Due to the inherent complexity of the problem, these methods are generally based on the integrative analysis of multiple genomic features or functional data from genomics techniques (Table 1).
Table 1.
Enhancer - Target Gene pairing methods. The table enlists the various ETG algorithms. Their grouping into four main classes is specified: correlation-based (C), supervised learning-based (SL), regression-based (R), score-based (S). Methods with mixed features are specified (e.g. SL + R or C + R). C* is for a method conceptually related to correlation-based solutions. Details on each method and features adopted for ETG pairing are also listed.
| Name | Class | Method details | Features |
|---|---|---|---|
| Correlation-based methods | |||
| Thurman et al. | C | Pearson correlation | DNase-seq |
| Shen et al. | C | Spearman correlation | ChIP-seq for Pol2 and H3K4me1 |
| PreSTIGE | C* | Shannon entropy to select cell type-specific patterns | RNA-seq, ChIP-seq for H3K4me1 |
| ELMER | C | Inverse correlation | RNA-seq, DNA methylation |
| Supervised learning-based methods | |||
| Rodelsperger et al. | SL | Random forest | Distance, conserved synteny, gene ontology, protein-protein interactions |
| Ernst et al. | SL | Logistic regression | Gene expression (microarrays), ChIP-seq for 3 histone marks |
| IM-PET | SL | Random forest | Distance, conserved synteny, correlation between enhancer (CSI-ANN score on 3 histone marks) and target promoter (RNA-seq) activity, TFs binding (sequence motifs) and target promoter correlation |
| PETModule | SL | Random forest | Distance, conserved synteny, DNase-seq |
| TargetFinder | SL | Ensemble of boosted decision trees | DNase-seq, FAIRE-seq, DNA methylation, RNA-seq, ChIP-seq for 32 histone marks, in addition to TFs and architectural proteins |
| McEnhancer | SL | Third-order interpolated Markov chain model in a semi-supervised learning setup via the expectation maximization algorithm | Sequence motifs |
| Regression-based methods | |||
| Andersson et al. | C + R | Pearson correlation, then linear models and lasso shrinkage | DNase-seq |
| RIPPLE | SL + R | Random forest and group lasso | DNase-seq, RNA-seq, ChIP-seq for 8 histone marks and 15 TFs. |
| JEME | R + SL | Multiple linear regression and lasso shrinkage | DNase-seq, RNA-seq, ChIP-seq for 3 histone marks |
| FOCS | R | Ordinary least squares regression | DNase-seq, CAGE-seq |
| Score-based methods | |||
| EpiTensor | S | Higher-order tensors decomposition | DNase-seq, RNA-seq, ChIP-seq for 16 histone marks |
| GeneHancer | S | Additive score with custom weights and data transformations for each quantitative | Distance, TFs co-expression, eRNAs, eQTLs, capture Hi-C |
| PEGASUS | S | Score reflecting the evolutionary sequence and synteny conservation | Conserved synteny and sequence conservation |
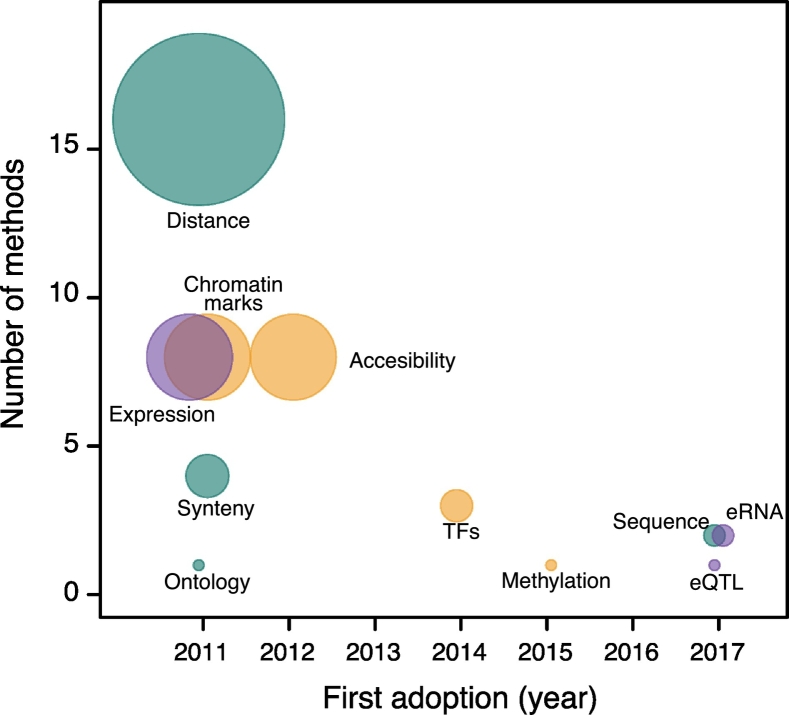
The features considered have evolved with the availability of novel technologies or improved knowledge of enhancer functional mechanism (Fig. 2). The approach adopted by the earlier studies was to assign the nearest gene as putative target of any given enhancer. More recent ones have integrated diverse types of genomic data, information on the chromatin 3D architecture, expression quantitative trait loci (eQTLs) and eRNAs expression.
Fig. 2.
Features used in ETG pairing tools. The figure summarizes the main types of features used to define ETG pairs by the tools discussed in this review. For each feature, its respective frequency (y-axis, number of methods) and first adoption by the tools discussed in this review (x-axis, year) is reported. The size of each dot is also proportional to the frequency (number of methods). The colors represent the category of the data: genomic annotations independent to cell type (dark green); epigenomics data (orange); transcriptomic data (mauve).
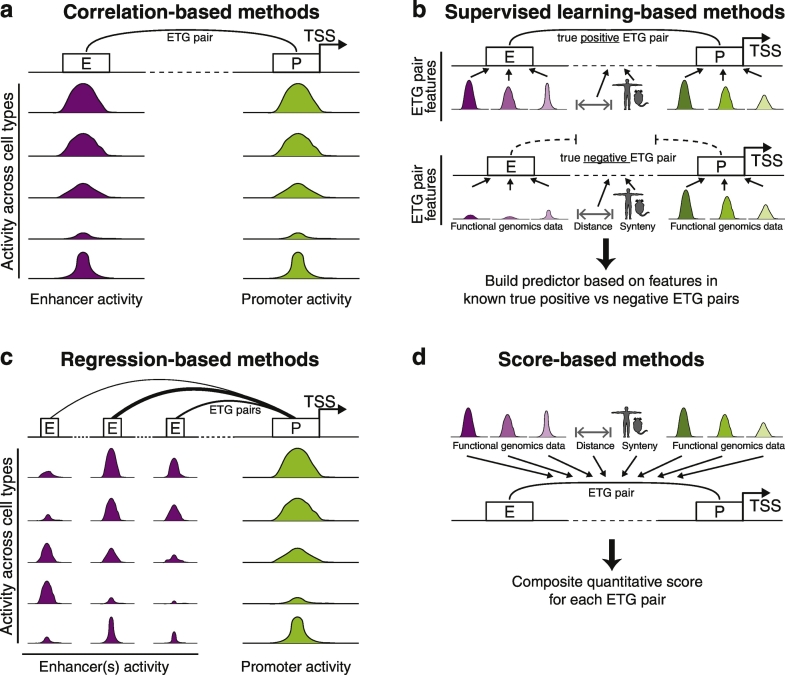
Despite the many methodological differences, the ETG methods proposed so far can be grouped into four main groups: 1) Correlation-based; 2) Supervised learning-based; 3) Regression-based and 4) methods based on other scores (Fig. 3).
Fig. 3.
Main classes of ETG pairing methods. The cartoon highlights the main principles underlying the four main classes of ETG pairing methods as discussed in this review. (a) Correlation-based methods are centered on assessing the correlation between activity of individual enhancer-promoter pairs across multiple cell types. Their activity is measured by one or more types of functional epigenomics or transcriptomics data. (b) Supervised learning-based methods instead build a predictor based on a known set of true positive and negative ETG pairs. For each of these, several features (e.g. functional genomics data) are considered to describe enhancers and promoters activity across multiple cell types. These can also be enriched with other features directly associated to the ETG pair, such as their genomic distance or synteny conservation. (c) Regression-based methods are simultaneously assessing the quantitative contribution to a promoter activity by multiple enhancers within the considered genomic window. These methods leverage a large number of genomic features and functional data. Regression methods can provide a weight for the contribution of individual enhancers (represented by ETG pairing lines of different thickness in the cartoon). (d) Score-based methods integrate into a single quantitative score information from a large set of genomic features and functional data. The score is quantifying the strength of individual ETG pairs. In the cartoons for all methods enhancers and promoters are represented as boxes labelled as “E” or “P”, respectively. TSS is marked with an arrow. Colored (purple or green) curves are used to represent quantitative functional genomics data used to infer the activity level of enhancers or promoters, respectively. They are meant to hint the peaks of various intensity that would be associated to such features in genomics data such as ChIP-seq.
3.1. Correlation-based Algorithms
The correlation-based ETG pairing algorithms originate on the rationale that the activity status of an enhancer and its target gene would be correlated across multiple cell types. As such, these algorithms rely on a large panel of epigenomics or transcriptomic data covering multiple conditions to estimate a quantitative score describing the enhancers or genes activity status. The activity of enhancers for example could be measured by ChIP-seq H3K4me1 and the activity of target genes by Pol2 ChIP-seq as in Shen et al [91], where a panel of 19 mouse cell types were analyzed. Thurman et al instead used DNase-seq read coverage at 1,454,901 distal DHS regions and correlated them to DNase-seq signal at promoters across 79 cell types to identify target genes, which were then validated using the 5C technique [92]. A slight variation over this approach was adopted by the same authors in [93], by correlating DNase-seq at distal regulatory regions with gene expression. Instead, ELMER [94,95] identifies enhancers as distal regulatory elements with differential methylation pattern across a large compendium of cancer samples.
The correlation-based approach assumes that enhancers activity changes across cell-types, which is in fact expected based on literature in this field, as a certain degree of variance in their activity scores is required to identify putative ETG pairs based on correlation. A key advantage of the correlation-based approaches is that they can identify multiple targets of an enhancer and can directly derive a quantitative measure of the strength of association. Another advantage of these methods is that correlation can be measured also between regulatory elements and genes within a short distance from each other, thus potentially achieving high spatial resolution in ETG pairing. However, there may be confounding correlation patterns in case enhancer regions are defined at a resolution higher than that of the functional chromatin mark data used to measure their activity.
The major limitation of correlation-based methods is the availability of genomics data over a large panel of cells, with comparable quality and resolution across all conditions. Moreover, using correlation of functional genomics data across cell types for potential ETG pairs may overlook the cell type and time specificity of such interactions, thus missing relevant connections when extending to a new cell type or time point. It's worth remarking that, even though eRNA expression provides a reliable estimation of enhancers activity, there are still open problems related to sensitivity in their detection [86]. Thus, there is not a complete consensus about which epigenomic or transcriptomic data type is the best solution to assess enhancers activity. Extending this uncertainty to the next level of calling ETG pairs could then be problematic. Correlation-based algorithms may also be confounded by enhancers that are active only in one or few cell types, thus resulting in a high correlation score actually based on a few data points with high signal. While this may cause some concerns about false positives, it is also a relevant feature allowing the detection of cell specific elements with single data points of high activity. Finally, the correlation between individual enhancers and target genes does not directly consider the fact that multiple enhancers can act on a gene in a cooperative fashion as the correlation is measured independently for each ETG pair.
A different but conceptually related approach is adopted by the PreSTIGE method [96] which uses H3K4me1 ChIP-seq and RNA-seq to estimate the activity of enhancers and target genes, respectively. However, it does not measure correlation directly. Instead PreSTIGE focuses on selecting cell type-specific patterns both at enhancers and genes based on Shannon entropy. Then ETG pairs are called if there's also a match in the cell type where both enhancer and gene are active. Thus, PreSTIGE is different from regular correlation-based methods as it explicitly prioritizes cell type-specific patterns. PreSTIGE delineates enhancer gene interactions focusing on enhancer with variants associated to specific diseases, as such this may be considered a limitation as it does not aim to provide a comprehensive characterization of ETG pairs.
Differently from all other correlation-based methods, ELMER looks for patterns of inverse correlation between the methylation level at enhancers and the expression level at the closest 20 genes (10 upstream and 10 downstream of each candidate differentially methylated enhancer). The significantly inverse correlation is identified based on a non-parametric Mann-Whitney U test, for each enhancer-gene pair, comparing the expression level in the cancer patients grouped by methylation level in the enhancer (highest vs lowest 20% of patients by enhancer methylation level).
3.2. Supervised Learning-based Algorithms
Supervised learning (SL)-based algorithms leverage a selected set of known true-positive ETG pairs to identify their associated patterns in genomic annotations or functional data to build a machine learning classifier (Table 1). The predictor will typically incorporate features derived from epigenomics and transcriptomics data in the training set of different cell types. In principle, the predictor would then be applicable to call ETG pairs in other independent cell types.
The number and type of features used by individual prediction tools are greatly variable (Table 1 and Fig. 2), but most of them take into account a combination of gene expression, chromatin accessibility (DHS) and histone marks. Moreover, all of them are considering to some degree the distance between enhancers and genes. Among the other SL-based algorithms, IM-PET [97] uses a mixed approach as the features considered include correlation of enhancer and promoter activity, i.e. the idea at the core of correlation-based ETG pairing methods. This actually implies that a panel of cell types would also be required to apply the model on a completely independent set of cells. Indeed, in the original publication the model is tested by cross validation.
In this group, McEnhancer uses a different solution as the predictor of ETG pairs is based on sequence composition in sets of co-regulated genes and enhancers [98]. The rationale behind this approach is that co-regulated genes will share at least part of their regulators in terms of transcription factors. As such, similar transcription factors binding sites may be included in their enhancers. This was also the rationale underlying earlier works on sequence motifs co-occurrence in cis-regulatory regions, but centered on TSS surrounding windows and not properly focusing on ETG pairing [99]. The rationale of focusing on sequence motifs for enhancers is also in line with results from methods scoring the impact of SNPs on non-coding regulatory sequences in independent studies [26].
A key advantage of SL-based methods is that, once the classifier is trained, in principle it could predict ETG pairs in other cell types. However, given the very cell type-specific nature of distal regulatory elements, the reliability of the classifiers can vary greatly when applied to different cell types, as shown by Cao et al. [100].
The main limitation of these methods, is that the training of the classifier requires a set of known positive as well as negative interactions. The SL-based tools proposed so far have used a variety of approaches to define positive and negative sets of ETG pairs. An earlier approach adopted by Ernst et al. was actually considering all enhancers up to 125 kb from the TSS [101]. Most of the methods listed in (Table 1) rely instead on some type of chromatin conformation capture data to define the true set of ETG pairs. Namely, IM-PET and JEME [100] use ChIA-PET data, whereas PETModule [102] uses both Hi-C and ChIA-PET data to define the true positive pairs. Instead TargetFinder [103] uses only Hi-C to define the training set of true positives.
Even if it's commonly accepted that enhancers and target genes need to come in close physical proximity to regulate transcription, the persistence and frequency of such interactions are still a matter of investigation. A number of reports suggests that enhancer promoter loops may be detectable also in cell types where the target gene is not active [21], or proposed that interactions may precede the activation of target genes [[104], [105], [106]]. As such, the detection of contacts with 3C derived methods does not unambiguously prove the presence of an active regulatory interaction in a given cell type. These studies suggests that the presence of a physical interaction between two regulatory regions alone doesn't necessarily imply active transcription of the target gene [107].
The definition of true negative sets of ETG pairs is complicated by the need to rely on even more assumptions. Indeed, even if Hi-C or other 3C-derived methods do not have the statistical power to detect significantly high interaction signal, this does not ensure that there's no contact between two loci. The lack of strong Hi-C signal may be due to a number of technical reasons [108]. Nevertheless, all the tools mentioned above build the negative set by selecting pairs of loci with distance distributions similar to true ETG pairs and ensuring they are not detected as interaction in chromatin conformation capture data.
In conclusion, the SL-based algorithms are hampered by the lack of comprehensive genome-wide definition of known true positive and negative ETG pairs. As such, the selection of the training set is expected to affect the performance of the algorithms.
3.3. Regression-based Algorithms
Regression-based methods work on the rationale that multiple enhancers can regulate a single gene, thus they use a combinatorial rather than pair-wise approach for ETG pairing. Regression-based methods identify significant relationships between enhancers and target genes, while at the same time assessing the strength of impact of multiple enhancers on their target.
As the number of variables in the regression model grows quickly with the addition of more candidate enhancers paired with each gene, limiting the starting set of ETG candidates is crucial for the regression-based methods.
In this category, JEME uses all enhancers within 1 Mb of each TSS as the starting set of candidates. Then it uses multiple linear regression coupled with lasso shrinkage to assess the errors terms in predicting TSS activity based on the activity of all candidate enhancers considered at once. JEME is actually a hybrid method as after the regression step a random forest classifier is trained based on each cell type-specific data to predict ETG pairs. The predictor is based on the cell type-specific error terms estimated by regression models, the chromatin marks data at enhancers, TSS and intervening window, as well as the distance between enhancer and candidate target. The true positive set of ETG pairs is defined based on ChIA-PET, Hi-C or eQTL data. As such JEME incorporates both features of regression and predictor-based methods.
Similarly, RIPPLE [109] is a SL-based approach that incorporates regression methods for features selection. It trains cell type-specific random forests on 5C data, i.e. chromatin loops, from Sanyal et al. [4] as positive set and in addition uses an approach based on multi-task learning and group lasso to perform joint feature selection across four cell lines.
FOCS [110] instead can be considered only regression-based. It starts by predicting each promoter activity based on k closest enhancers, using ordinary least squares (OLS) regression models. The activity of enhancers and promoters is initially estimated based on DNase-seq data by ENCODE. Then FOCS is applied on alternative datasets including: DNase-seq data by Roadmap Epigenomic, CAGE-seq data by FANTOM5 and a custom compendium of publicly available GRO-seq datasets. The regression models are trained on multiple cell types with leave-one-out cross validation. The reliability of each prediction is tested against the observed promoter activity in the left-out sample. Then, for refined predictions, the full model is trained and elastic-net shrinkage is performed to select the enhancers more relevant for regulating the target gene.
An earlier regression-based approach was adopted by the FANTOM project consortium as described in Andersson et al. [87]. In this case a mixed solution was proposed as Pearson correlation was first used for an initial selection of ETG pairs, then linear models and lasso shrinkage were adopted to further select the most informative pairs. The correlation is measured on CAGE-based estimations of enhancer and promoter activity. The authors claim that using CAGE yields a higher fraction of ETG pairs validated by ChIA-PET, as compared to correlation based on DNase-seq. These solutions combine the advantages of correlation and regression approaches, but still have the main limitation of regression methods that is the need to limit ETG pairing to a pre-defined window (500Kb from TSS in this case).
Regression-based methods have in principle the ability to determine the relative influence of one or more predictor variables. Thus, multiple enhancers that are candidate regulators of a given gene can be ranked to select the most informative ones. The main limitations with these methods are that they rely on some arbitrarily chosen parameters, most notably the definition of the window or maximum number of enhancers considered around each TSS. They also generally need a large compendium of cell types with functional data used to build the models. Thus, they suffer from a combination of the limitations already discussed for correlation and SL-based methods.
3.4. Other Score-based Methods
A few algorithms have implemented other custom quantitative scores to assign target genes to enhancers. The common idea underlying these approaches is to use a single quantitative score to define the strength of association between enhancers and target genes, taking into account multiple types of information.
For example GeneHancer [111] uses a score accounting for eQTLS, TF-target gene co-expression, eRNAs, capture Hi-C and genomic distance between enhancer and target gene. As these metrics have all different distributions and ranges of values they are combined together with various data transformations and weights.
EpiTensor [112] instead combines together 16 chromatin marks, RNA-seq and DNase-seq across 5 cell types by leveraging higher order tensors decomposition, from which eigenlocus vectors are derived and used to compute the “spatial association score”. This score basically accounts for similarities in patterns of functional genomics data over distant genomic loci and is used to call associated peaks. While this score is shown to have good concordance with Hi-C derived interactions for enhancers-promoter pairs, the same score has lower area under the curve (AUC) in a receiver operating characteristic (ROC) curve built on other interactions called by Hi-C (e.g. promoter-exon). While these results confirm the method can call ETG pairs, the selective discrepancy with other Hi-C interactions is not explained.
Conversely, PEGASUS [113,114] does not rely on any functional genomics data, as it completely relies instead on evolutionary conservation. Namely PEGASUS first defines regulatory elements based on sequence conservation, then links them to target genes using a synteny conservation score.
The major advantage of having a single quantitative score for enhancer-gene pairing is that it enables a more flexible prioritization of ETG pairs by adjusting a single threshold on the score. Moreover, all the possible interactions of any enhancer or gene can be obtained. For example, if an enhancer has the potential to regulate multiple genes, or a gene is regulated by several enhancers, these multi-way relationships can be explored and scored.
The main limitation of the score-based approaches is that they rely on a number of assumptions and arbitrarily defined parameters or weights to be able to combine a heterogenous set of information into a single quantitative value.
4. Other Key Differences Between the Algorithms
ETG pairing methods are different not only in the algorithmic details, but also in the way multiple parameters and information are used to define enhancers and promoters. Some methods are focused on single histone modifications, e.g. PreSTIGE defines putative enhancers as H3K4me1 enriched sites, whereas IM-PET uses a more sophisticated algorithm (CSI-ANN) described above [65]. FOCS and Thurman et al. rely instead on non-promoter DNase-seq peaks to define enhancers. Other methods, such as TargetFinder or JEME, use chromatin states definitions as obtained by Segway or chromHMM applied on data from ENCODE or Roadmap Epigenomics consortia. RFECS [68] instead is adopted by EpiTensor to define enhancers. Finally, GeneHancer uses an even more mixed enhancer set based on ENCODE, Ensembl, FANTOM and VISTA data [41].
As such, in most cases the results coming from different methods are not directly comparable due to the differences in enhancer regions definition itself.
Another key difference between the algorithms is the way true positive ETG pairs are defined. While this is a crucial feature especially for SL-based methods, to some degree all of the articles in this field assess the concordance with an expected true positive set. 3C and its high-throughput derivatives including 5C, Hi-C, capture Hi-C and ChIA-PET are used by many tools to define true positives, as detailed above, due to the role of chromatin 3D organization in ETG interaction. eQTLs are also a popular alternative to demonstrate a connection between a regulatory sequence and a gene expression, in particular using data from the GTEx project is a popular choice [115]. However, all of these options have different resolutions in defining functional or physical connections between distant loci. Namely, due to the limited coverage, Hi-C data are usually summarized at the level of genomic bins with sizes in the order of few or several kb, thus larger than the enhancers definitions used by many tools. Moreover, interactions which are just 1 or 2 bins apart can hardly be resolved by Hi-C interactions calling algorithms, thus loosing many possible true positive ETG pairs [116]. Conversely, eQTLs narrow down the core of the regulatory region by restricting its range to the SNPs contained in a linkage disequilibrium region. The linkage disequilibrium region might actually extend up to a few thousand base pairs, even if the reported eQTL SNPs cover one or few base pairs. This resolution is also not achievable for the ETG pairing tools which rely on functional genomics techniques. For example, in ChIP-seq the ultimate resolution limit is the chromatin fragmentation size, which is usually in the order of a few hundred base pairs [117].
5. Applicability
While most of the ETG tools presented aim to be a generalizable approach, they have been developed using specific study models, which may affect their applicability to other conditions. As summarized more in details in Table 2, these may range from 4 cell lines (TargetFinder) to as many as 935 cell and tissue types (JEME) or 2630 samples (FOCS). Starting from a large collection of cell or tissue types for the characterization of ETG is certainly a strength as it allows to capture the cell type-specific nature of enhancers. However, the optimal number of cell types to be considered in the ETG pairs annotation is yet to be determined. Moreover, the tools were applied to different organisms, including human, mouse, fruitfly and zebrafish, as detailed in (Table 2). These different organisms also imply different genome sizes and complexity of the ETG regulatory network, which may affect the method's performance.
Table 2.
Details on methods applicability. The table lists details concerning each method usability and applicability. Namely, the organisms, cell and tissue types used to develop the tools are specified, as well as details on the code availability.
| Name | Organism | Samples | Code availability |
|---|---|---|---|
| Rodelsperger et al. | Mouse | Embryonic mouse forebrain and limb | – |
| Ernst et al. | Human | 9 cell lines | – |
| Thurman et al. | Human | 125 cell lines | – |
| Shen et al. | Mouse | 19 cell and tissue types | – |
| PreSTIGE | Human | 12 cell lines | Galaxy module |
| IM-PET | Drosophila and human | Drosophila and 12 human cell lines | Galaxy module and archive with a collection of scripts for PERL, Python, Java, R and others tools |
| Andersson et al. | Human | 432 primary cell, 135 tissue and 241 cell lines | – |
| RIPPLE | Human | 4 cell lines | Bitbucket repository with a collection of C++ programs and MATLAB scripts (current version: 1.0) |
| ELMER | Human | 2841 TCGA samples | Bioconductor version: Release (3.9) R package |
| PETModule | Mouse and human | 2 mouse cell lines and 8 human cell lines | Archive with a collection of scripts for Python, Java and other tools |
| TargetFinder | Human | 4 cell lines | GitHub repository with a collection of Python scripts |
| EpiTensor | Human | 5 cell lines | Archive with a collection of scripts for Bash, R, MATLAB and other tools (current version: v0.9) |
| JEME | Human | 935 human primary cell and tissue types | GitHub repository with a collection of scripts for Bash, R and other tools |
| GeneHancer | Human | Cell lines from multiple compendiums | GeneCards web portal |
| McEnhancer | Drosophila | Drosophila embryo development stages | GitHub repository with a collection of scripts for Bash, PERL, R, Python, Java and others tools |
| PEGASUS | Zebrafish and human | Human embryonic stem cells and zebrafish developmental stages | – |
| FOCS | Human | 2630 samples | GitHub repository with a collection of scripts for R |
Another crucial practical consideration about the applicability of each ETG method is related to the availability of a user-friendly implementation. To this end, some tools are implemented as a Galaxy module, but in most cases an heterogenous set of scripts is shared directly through a git-based source code repository or simply a compressed archive file (see Table 2 and Supplementary Table 1 for details). GeneHancer instead has implemented its prediction in the GeneCards web portal [118], where candidate enhancers and their annotations are displayed on relevant GeneCard entries.
Finally, in term of applicability it's worth mentioning that just a few tools discussed in this review performed an ad hoc experimental validation of the ETG pairs, whereas most of them relied on comparisons to previously published data or tools to assess performances. Namely, EpiTensor performed 3C-qPCR in IMR90 cell line to validate 14 randomly selected pairs, of which they achieved a 93% validation rate. IM-PET has also performed 3C-qPCR on 9 randomly selected predictions in two cell types and achieved 81% validation rate. JEME selected three genes (TERT, PSRC1 and RBM24) for its experimental validation, confirmed their corresponding enhancer activity using luciferase assay and showed the CRISPR-Cas9 deletion of the enhancers diminished the transcriptional levels of the respective genes. McEnhancer went as far as testing enhancers and target genes coordinated tissues specificity in vivo in Drosophila embryos.
6. Summary and Outlook
In conclusion, a large number of computational biology solutions have been proposed in the past few years to achieve a comprehensive matching of enhancers and putative target genes. This surge in publications in this field has been motivated on one hand by the ever increasing availability of functional genomics data that can be used for this purpose, and on the other hand by an increased understanding of the central role of distal regulatory elements in physiological and pathological processes. For the same reasons we expect a further increase in available solutions for ETG pairing in the coming years.
Despite the variety in methodological solutions proposed so far by literature in this field, a few crucial limitations hamper an effective and complete genome-wide ETG pairing, as summarized in (Table 3) for the four main classes of methods. There are two primary issues affecting all methods: 1) the lack of a genome-wide exhaustive reference list of all non-coding regions in a given organism genome that can act as enhancers, and 2) the lack of a large set of experimentally validated true positive and true negative ETG pairs to be used as reference gold standard for methods development and benchmarking. Reaching a consensus on these two crucial aspects will be instrumental to advance the field in the coming years.
Table 3.
Pros and cons of ETG pairing approaches. Table summarizes the advantages and limitations of the methodology.
| Method | Correlation | Machine learning | Regression | Score based |
|---|---|---|---|---|
| Pros |
|
|
|
|
| Cons |
|
|
|
|
The following are the supplementary data related to this article
URLs for tools. Table lists the URL of the tools of ETG pairing methods.
Declaration of Competing Interest
The authors claim no conflict of interest.
Acknowledgements
We thank Mattia Forcato and Pierre-Luc Germain for critical feedback on an earlier version of the manuscript. We acknowledge support by AIRC Start-up grant 2015 n.16841 to F·F and AIRC fellowship n. 22416 to J.M.H.
References
- 1.De Laat W., Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506. doi: 10.1038/nature12753. [DOI] [PubMed] [Google Scholar]
- 2.Pennacchio L.A., Bickmore W., Dean A., Nobrega M.A., Bejerano G. Enhancers: five essential questions. Nat Rev Genet. 2013;14:288–295. doi: 10.1038/nrg3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim T.-K., Shiekhattar R. Architectural and functional commonalities between enhancers and promoters. Cell. 2015;162:948–959. doi: 10.1016/j.cell.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanyal A., Lajoie B.R., Jain G., Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ong C.T., Corces V.G. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yip K.Y., Cheng C., Bhardwaj N., Brown J.B., Leng J., Kundaje A. Classification of human genomic regions based on experimentally determined binding sites of more than 100 transcription-related factors. Genome Biol. 2012;13:R48. doi: 10.1186/gb-2012-13-9-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joshi A. Mammalian transcriptional hotspots are enriched for tissue specific enhancers near cell type specific highly expressed genes and are predicted to act as transcriptional activator hubs. BMC Bioinform. 2014;15:412. doi: 10.1186/s12859-014-0412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen B.L., Taatjes D.J. The mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol. 2015;16:155–166. doi: 10.1038/nrm3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yen A., Kheradpour P., Zhang Z., Heravi-moussavi A., Liu Y., Amin V. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Visel A., Akiyama J.A., Shoukry M., Afzal V., Rubin E.M., Pennacchio L.A. Functional autonomy of distant-acting human enhancers. Genomics. 2009;93:509–513. doi: 10.1016/j.ygeno.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mora A., Sandve G.K., Gabrielsen O.S., Eskeland R. In the loop: promoter–enhancer interactions and bioinformatics. Brief Bioinform. 2016;17 doi: 10.1093/bib/bbv097. bbv097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kagey M.H., Newman J.J., Bilodeau S., Zhan Y., Orlando D.A., van Berkum N.L. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin F., Li Y., Dixon J.R., Selvaraj S., Ye Z., Lee A.Y. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 17.Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fullwood M.J., Liu M.H., Pan Y.F., Liu J., Xu H., Mohamed Y Bin, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoenfelder S., Furlan-magaril M., Mifsud B., Tavares-cadete F., Sugar R., Javierre B. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 2015;25:582–597. doi: 10.1101/gr.185272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mifsud B., Tavares-Cadete F., Young A.N., Sugar R., Schoenfelder S., Ferreira L. Mapping long-range promoter contacts in human cells with high-resolution capture hi-C. Nat Genet. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- 21.Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonev B., Mendelson Cohen N., Szabo Q., Fritsch L., Papadopoulos G.L., Lubling Y. Multiscale 3D genome rewiring during mouse neural development. Cell. 2017;171 doi: 10.1016/j.cell.2017.09.043. [557–572.e24] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pal K., Forcato M., Ferrari F. Hi-C analysis: from data generation to integration. Biophys Rev. 2019;11:67–78. doi: 10.1007/s12551-018-0489-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- 25.Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee D., Gorkin D.U., Baker M., Strober B.J., Asoni A.L., McCallion A.S. A method to predict the impact of regulatory variants from DNA sequence. Nat Genet. 2015;47:955–961. doi: 10.1038/ng.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rheinbay E., Parasuraman P., Grimsby J., Tiao G., Engreitz J.M., Kim J. Recurrent and functional regulatory mutations in breast cancer. Nature. 2017;547:55–60. doi: 10.1038/nature22992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirabella A.C., Foster B.M., Bartke T. Chromatin deregulation in disease. Chromosoma. 2016;125:75–93. doi: 10.1007/s00412-015-0530-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lupiáñez D.G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spielmann M., Lupiáñez D.G., Mundlos S. Structural variation in the 3D genome. Nat Rev Genet. 2018;19:453–467. doi: 10.1038/s41576-018-0007-0. [DOI] [PubMed] [Google Scholar]
- 32.Chen Han, Li Chunyan, Peng Xinxin, Zhou Zhicheng, Weinstein John N. The Cancer genome atlas research network HL. A Pan-Cancer analysis of enhancer expression in nearly 9000 patient samples. Cell. 2018:386–399. doi: 10.1016/j.cell.2018.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith E., Shilatifard A. Enhancer biology and enhanceropathies. Nat Struct Mol Biol. 2014;21:210–219. doi: 10.1038/nsmb.2784. [DOI] [PubMed] [Google Scholar]
- 34.Murakawa Y., Yoshihara M., Kawaji H., Nishikawa M., Zayed H., Suzuki H. Enhanced identification of transcriptional enhancers provides mechanistic insights into diseases. Trends Genet. 2016;32:76–88. doi: 10.1016/j.tig.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 35.Kleftogiannis D., Ashoor H., Bajic V.B. TELS: a novel computational framework for identifying motif signatures of transcribed enhancers. Genomics Proteomics Bioinformatics. 2018;16:332–341. doi: 10.1016/j.gpb.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colbran L.L., Chen L., Capra J.A. Short DNA sequence patterns accurately identify broadly active human enhancers. BMC Genomics. 2017;18:536. doi: 10.1186/s12864-017-3934-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Catarino R.R., Stark A. Assessing sufficiency and necessity of enhancer activities for gene expression and the mechanisms of transcription activation. Genes Dev. 2018;32:202–223. doi: 10.1101/gad.310367.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arnold C.D., Gerlach D., Stelzer C., Boryn L.M., Rath M., Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- 39.Melnikov A., Murugan A., Zhang X., Tesileanu T., Wang L., Rogov P. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol. 2012;30:271–277. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai Z., Cui Y., Tan Z., Zhang G., Tan Z., Zhang X. RAEdb: a database of enhancers identified by high-throughput reporter assays. Database. 2019 doi: 10.1093/database/bay140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Visel A., Minovitsky S., Dubchak I., Pennacchio L.A. VISTA enhancer browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raisner R., Kharbanda S., Jin L., Jeng E., Chan E., Merchant M. Enhancer activity requires CBP/P300 Bromodomain-dependent histone H3K27 acetylation. Cell Rep. 2018;24:1722–1729. doi: 10.1016/j.celrep.2018.07.041. [DOI] [PubMed] [Google Scholar]
- 43.Cotney J., Leng J., Oh S., DeMare L.E., Reilly S.K., Gerstein M.B. Chromatin state signatures associated with tissue-specific gene expression and enhancer activity in the embryonic limb. Genome Res. 2012;22:1069–1080. doi: 10.1101/gr.129817.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Visel A., Blow M.J., Li Z., Zhang T., Akiyama J a, Plajzer-frick I, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2010;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonn S., Zinzen R.P., Girardot C., Gustafson E.H., Perez-Gonzalez A., Delhomme N. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- 46.Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S.A., Flynn R.A., Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2010;470:279. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heintzman N.D., Hon G.C., Hawkins R.D., Kheradpour P., Stark A., Harp L.F. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koch F, Andrau J-C. Initiating RNA Polymerase II and TIPs as hallmarks of enhancer activity and tissue-specificity. Transcription 2011;2:263–8. doi: 10.4161/trns.2.6.18747. [DOI] [PMC free article] [PubMed]
- 49.Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 50.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ernst J., Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zentner G.E., Scacheri P.C. The chromatin fingerprint of gene enhancer elements*. J Biol Chem. 2012;287:30888–30896. doi: 10.1074/jbc.R111.296491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsompana M., Buck M.J. Chromatin accessibility: a window into the genome. Epigenetics Chromatin. 2014;7:33. doi: 10.1186/1756-8935-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boyle A.P., Davis S., Shulha H.P., Meltzer P., Margulies E.H., Weng Z. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buenrostro J.D., Wu B., Chang H.Y., Greenleaf W.J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vierstra J., Stamatoyannopoulos J.A. Genomic footprinting. Nat Methods. 2016;13:213–221. doi: 10.1038/nmeth.3768. [DOI] [PubMed] [Google Scholar]
- 58.Brenowitz M., Senear D.F., Kingston R.E. DNase I footprint analysis of protein-DNA binding. Curr Protoc Mol Biol. 2001 doi: 10.1002/0471142727.mb1204s07. [Chapter 12:Unit 12.4] [DOI] [PubMed] [Google Scholar]
- 59.Li Z., Schulz M.H., Look T., Begemann M., Zenke M., Costa I.G. Identification of transcription factor binding sites using ATAC-seq. Genome Biol. 2019;20:45. doi: 10.1186/s13059-019-1642-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bernstein B.E., Stamatoyannopoulos J.A., Costello J.F., Ren B., Milosavljevic A., Meissner A. The NIH roadmap Epigenomics mapping consortium. Nat Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.John S., Sabo P.J., Thurman R.E., Sung M.-H., Biddie S.C., Johnson T.A. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koohy H., Down T.A., Spivakov M., Hubbard T. A comparison of peak callers used for DNase-Seq data. PLoS One. 2014;9 doi: 10.1371/journal.pone.0096303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kleftogiannis D., Kalnis P., Bajic V.B. Progress and challenges in bioinformatics approaches for enhancer identification. Brief Bioinform. 2016;17:967–979. doi: 10.1093/bib/bbv101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Firpi H.A., Ucar D., Tan K. Vol. 26. 2010. Discover regulatory DNA elements using chromatin signatures and artificial neural network; pp. 1579–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hoffman M.M., Buske O.J., Wang J., Weng Z., Bilmes J.A., Noble W.S. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat Methods. 2012;9:473–476. doi: 10.1038/nmeth.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ernst J., Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rajagopal N., Xie W., Li Y., Wagner U., Wang W., Stamatoyannopoulos J. RFECS: a random-Forest based algorithm for enhancer identification from chromatin state. PLoS Comput Biol. 2013;9 doi: 10.1371/journal.pcbi.1002968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kleftogiannis D., Kalnis P., Bajic V.B. DEEP: a general computational framework for predicting enhancers. Nucleic Acids Res. 2014;43:e6. doi: 10.1093/nar/gku1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim T.-K., Hemberg M., Gray J.M., Costa A.M., Bear D.M., Wu J. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.De Santa F., Barozzi I., Mietton F., Ghisletti S., Polletti S., Tusi B.K. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arner E., Weinhold N., Jacobsen A., Schultz N., Sander C., Lee W. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science. 2015;347:1010–1015. doi: 10.1126/science.1259418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li W., Notani D., Rosenfeld M.G. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 2016;17:207–223. doi: 10.1038/nrg.2016.4. [DOI] [PubMed] [Google Scholar]
- 74.Kodzius R., Kojima M., Nishiyori H., Nakamura M., Fukuda S., Tagami M. CAGE: cap analysis of gene expression. Nat Methods. 2006;3:211–222. doi: 10.1038/nmeth0306-211. [DOI] [PubMed] [Google Scholar]
- 75.Takahashi H., Lassmann T., Murata M., Carninci P. 5′ end-centered expression profiling using cap-analysis gene expression and next-generation sequencing. Nat Protoc. 2012;7:542–561. doi: 10.1038/nprot.2012.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Valen E., Pascarella G., Chalk A., Maeda N., Kojima M., Kawazu C. Genome-wide detection and analysis of hippocampus core promoters using DeepCAGE. Genome Res. 2009;19:255–265. doi: 10.1101/gr.084541.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Core L.J., Waterfall J.J., Lis J.T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lam M.T.Y., Cho H., Lesch H.P., Gosselin D., Heinz S., Tanaka-Oishi Y. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Churchman L.S., Weissman J.S. Native elongating transcript sequencing (NET-seq) Curr Protoc Mol Biol. 2012 doi: 10.1002/0471142727.mb0414s98. [Chapter 4:Unit 4.14.1–17] [DOI] [PubMed] [Google Scholar]
- 80.Kwak H., Fuda N.J., Core L.J., Lis J.T. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mahat D.B., Kwak H., Booth G.T., Jonkers I.H., Danko C.G., Patel R.K. Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq) Nat Protoc. 2016;11:1455. doi: 10.1038/nprot.2016.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nechaev S., Fargo D.C., dos Santos G., Liu L., Gao Y., Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schwalb B., Michel M., Zacher B., Fruhauf K., Demel C., Tresch A. TT-seq maps the human transient transcriptome. Science. 2016;352:1225–1228. doi: 10.1126/science.aad9841. [DOI] [PubMed] [Google Scholar]
- 84.FANTOM Consortium and the RIKEN PMI and CLST (DGT) A promoter-level mammalian expression atlas. Nature. 2014;507:462. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hon C.-C., Ramilowski J.A., Harshbarger J., Bertin N., Rackham O.J.L., Gough J. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Adiconis X., Haber A.L., Simmons S.K., Levy Moonshine A., Ji Z., Busby M.A. Comprehensive comparative analysis of 5′-end RNA-sequencing methods. Nat Methods. 2018;15:505–511. doi: 10.1038/s41592-018-0014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Core L.J., Martins A.L., Danko C.G., Waters C.T., Siepel A., Lis J.T. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet. 2014;46:1311–1320. doi: 10.1038/ng.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.O'Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Frankish A., Abdul Salam A.I., Vullo A., Zadissa A., Winterbottom A., Parton A. Ensembl 2019. Nucleic Acids Res. 2018;47:D745–D751. doi: 10.1093/nar/gky1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shen Y., Yue F., Mc Cleary D.F., Ye Z., Edsall L., Kuan S. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sheffield N.C., Thurman R.E., Song L., Safi A., Stamatoyannopoulos J.A., Lenhard B. Patterns of regulatory activity across diverse human cell types predict tissue identity, transcription factor binding, and long-range interactions. Genome Res. 2013;23:777–788. doi: 10.1101/gr.152140.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yao L., Shen H., Laird P.W., Farnham P.J., Berman B.P. Inferring regulatory element landscapes and transcription factor networks from cancer methylomes. Genome Biol. 2015;16:1–21. doi: 10.1186/s13059-015-0668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Silva T.C., Coetzee S.G., Gull N., Yao L., Hazelett D.J., Noushmehr H. ELMER v.2: an R/Bioconductor package to reconstruct gene regulatory networks from DNA methylation and transcriptome profiles. Bioinformatics. 2018 doi: 10.1093/bioinformatics/bty902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Corradin O., Saiakhova A., Akhtar-Zaidi B., Myeroff L., Willis J., Cowper-Sallari R. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 2014;24:1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.He B., Chen C., Teng L., Tan K. Global view of enhancer-promoter interactome in human cells. Proc Natl Acad Sci. 2014;111:E2191–E2199. doi: 10.1073/pnas.1320308111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hafez D., Karabacak A., Krueger S., Hwang Y.C., Wang L.S., Zinzen R.P. McEnhancer: predicting gene expression via semi-supervised assignment of enhancers to target genes. Genome Biol. 2017;18:1–21. doi: 10.1186/s13059-017-1316-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Warner J.B., Philippakis A.A., Jaeger S.A., He F.S., Lin J., Bulyk M.L. Systematic identification of mammalian regulatory motifs' target genes and functions. Nat Methods. 2008;5:347–353. doi: 10.1038/nmeth.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cao Q., Anyansi C., Hu X., Xu L., Xiong L., Tang W. Reconstruction of enhancer-target networks in 935 samples of human primary cells, tissues and cell lines. Nat Genet. 2017;49:1428–1436. doi: 10.1038/ng.3950. [DOI] [PubMed] [Google Scholar]
- 101.Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhao C., Li X., Hu H. PETModule: a motif module based approach for enhancer target gene prediction. Sci Rep. 2016;6:1–10. doi: 10.1038/srep30043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Whalen S., Truty R.M., Pollard K.S., Francisco S., Francisco S., Francisco S. Enhancer-promoter interactions are encoded by complex genomic signatures on looping chromatin. Nat Genet. 2016;48:488–496. doi: 10.1038/ng.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Apostolou E., Ferrari F., Walsh R.M., Bar-Nur O., Stadtfeld M., Cheloufi S. Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming. Cell Stem Cell. 2013;12:699–712. doi: 10.1016/j.stem.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stadhouders R., Vidal E., Serra F., Di Stefano B., Le Dily F., Quilez J. Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat Genet. 2018;50:238–249. doi: 10.1038/s41588-017-0030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wei Z., Gao F., Kim S., Yang H., Lyu J., An W. Klf4 organizes long-range chromosomal interactions with the oct4 locus in reprogramming and pluripotency. Cell Stem Cell. 2013;13:36–47. doi: 10.1016/j.stem.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 107.Kragesteen B.K., Spielmann M., Paliou C., Heinrich V., Schopflin R., Esposito A. Dynamic 3D chromatin architecture contributes to enhancer specificity and limb morphogenesis. Nat Genet. 2018;50:1463–1473. doi: 10.1038/s41588-018-0221-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yaffe E., Tanay A. Probabilistic modeling of hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat Genet. 2011;43:1059–1065. doi: 10.1038/ng.947. [DOI] [PubMed] [Google Scholar]
- 109.Roy S., Siahpirani A.F., Chasman D., Knaack S., Ay F., Stewart R. A predictive modeling approach for cell line-specific long-range regulatory interactions. Nucleic Acids Res. 2015;43:8694–8712. doi: 10.1093/nar/gkv865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hait T., Amar D., Shamir R., Elkon R. FOCS: a novel method for analyzing enhancer and gene activity map, an extensive enhancer–promoter. Genome Biol. 2018;19:1–14. doi: 10.1186/s13059-018-1432-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fishilevich S., Nudel R., Rappaport N., Hadar R., Plaschkes I., Iny Stein T. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database. 2017;2017:1–17. doi: 10.1093/database/bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhu Y., Chen Z., Zhang K., Wang M., Medovoy D., Whitaker J.W. Constructing 3D interaction maps from 1D epigenomes. Nat Commun. 2016;7:1–11. doi: 10.1038/ncomms10812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Clément Y., Torbey P., Gilardi-Hebenstreit P., Roest Crollius H. Genome-wide enhancer - gene regulatory maps in two vertebrate genomes. BioRxiv. 2018;244475 [Google Scholar]
- 114.Naville M., Ishibashi M., Ferg M., Bengani H., Rinkwitz S., Krecsmarik M. Long-range evolutionary constraints reveal cis-regulatory interactions on the human X chromosome. Nat Commun. 2015;6:6904. doi: 10.1038/ncomms7904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S. The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Forcato M., Nicoletti C., Pal K., Livi C.M., Ferrari F., Bicciato S. Comparison of computational methods for hi-C data analysis. Nat Methods. 2017;14:679–685. doi: 10.1038/nmeth.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Park P.J. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Safran M., Dalah I., Alexander J., Rosen N., Iny Stein T., Shmoish M. 2010. GeneCards Version 3: the human gene integrator. Database (Oxford) 2010. [baq020] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
URLs for tools. Table lists the URL of the tools of ETG pairing methods.