Highlights
-
•
Composting as bioprospecting source.
-
•
Collection of actinobacteria with potential as biocontrol agents isolated from composted lignocellulosic material.
-
•
Bioactive substances produced by microorganisms against damping-off.
-
•
Family Microbacteriaceae stood out for its biopesticidal and biostimulant capacity.
Keywords: Actinobacteria, Composting, Biological control, Damping-off, Plant growth promoting microorganisms
Abstract
Strains isolated during composting processes of plant waste, and identified as Actinobacteria, proved to be significant producers of compounds that actively participate in the control of phytopathogens, such as those that cause Damping-off disease. Although most of the actinomycetes analyzed showed to be antagonistic strains against common phytopathogens, only some 30% proved to be capable of producing bioactive substances, such as siderophores, salicylic acid, chitinase enzymes or cyanide, so that antibiosis could be considered the most probable antagonistic mechanism for a high proportion of the strains investigated. 6% of the microorganisms identified in this work, were selected as potential strains to be investigated in depth, since they further stimulated plant growth (germination index tests greater than 100%). Microbacteriaceae was one of the most prominent families.
1. Introduction
Damping-off is the term used to describe a significant and dangerous plant disease which is caused by soilborne pathogens [1]. Composting is considered an efficient method to eliminate pathogens, and the actinomycetes that inhabit it contribute remarkably to that [2] using direct approaches, it means producing antibiotics and lytic enzymes, or competing with pathogens for limited nutrients; or indirectly, producing substances which stimulate systemic resistance of plants against pathogens [3]. This work consisted in the bioprospecting, from a composting process, of different bioactive substances released by actinobacteria to suppress damping-off.
2. Materials and methods
During the present study, were used 220 strains belonging to the order Actinomycetales isolated throughout different stages of a composting process of lignocellulosic materials during a period of 189 days. Raw materials, operating conditions and sampling have been previously published [4]. The identities of specific isolates were determined on the basis of partial or nearly full length 16S rRNA gene sequence analysis [5].
Potentially antagonistic strains against damping-off pathogens were identified by dual-culture assays using the following phytopathogenic fungi and oomycetes, Fusarium oxysporum f.sp. melonis CECT 20474 (FOM), Rhizoctonia solani CECT 2824 (RS), Pythium ultimum CECT 2365 (PU) and Phytophthora capsici CECT 20433 (PCAP), supplied by the Spanish Type Culture Collection (CECT). Suppressive effect was demonstrated using the modified techniques of Landa et al. [6]. It consisted in plates prepared with a base of 2% water agar (WA) with a Potato Dextrose Agar (PDA) overlay, containing four 8-mm-diameter steel cylinders placed equidistantly from the edge. Once the cylinders were removed, the wells were filled with 50 μL liquid culture of the antagonist to be assayed and a plug of 5-day-old phytopathogen agent (PA) culture, removed from a PDA plate, was placed at the center of the assay plate. Four replicates for each antagonist-PA combination were prepared. Plates were incubated at 30 °C for 5 days. The inhibition index (I) was expressed as the percentage of PA inhibition in the presence of the antagonistic strain using the formula:
| I = ((100-(Dcr/Dc)) × 100), |
where I is the inhibition index, Dcr is the average diameter of fungal growth in the presence of antagonistic strain (mm), and Dc is the average diameter of fungal growth in the control plate (without antagonistic strain) (mm).
The protocol used to detect microorganisms producing siderophores (SID) was a modified method of Schwyn and Neilands [22]. Actinobacteria were inoculated in 3 mL of King B medium (KBM), consisting of g L−1; proteose-peptone, 20.0; MgSO4, 1.5; K2HPO4, 1.5; and glycerol, 10 mL (pH 7.0). Cultures were incubated at 30 °C during 72 h. Then, cells were removed from the medium by centrifugation (500g, 5 min) and 0.5 ml of supernatant was blended with 0.5 mL of Chrome Azurol S (CAS reagent). The mixture was incubated at room temperature and if colour change since blue to orange-brown in 10 min, it was indicating the presence of siderophores. The absorbance of the mixture was determinated at 630 nm.
The methodology used for the study of salicylic acid (SAL) producing microorganisms was based on universal testing [7], with the modifications introduced by Gil and Martínez-Merino [8]. From the same culture supernatant than in the case above, extracts were acidified to pH 2 with 2 N HCl. Thereafter, 1 mL of the microbial extract was mixed with 1 mL of distilled water and 2 mL of a 0.1% ferric chloride solution. The formation of a purple complex was observed for producers of salicylic acid, caused by the reaction that occurs between this molecule and iron. To carry out the quantitative assay, after incubation time, the cultures with cells removes were acidified to pH 2 and then extracted whit ethyl acetate by the protocol described by Visca et al. [9]. Absorbance was measured at 527 nm.
Hydrogen cyanide (HCN) released was estimated by a modified picrate/Na2CO3 method [10]. Cultures of all strains in 5 mL of Nutritive Broth (NB) were mixed, after 72 h, with 165 μL saturated picric acid (neutralized with Na2CO3). The reaction mixture was incubated for 5 h to allow colour development. Change in colour from yellow to light brown, moderate brown or reddish brown showed the presence of HCN production. Cyanide quantification was determined at 492 nm in a Shimadzu UV Spectrophotometer UV-1800, as the rest of determinations.
In addition, microbial isolates were screened on agar plates containing 0.5% colloidal chitin (prepared according to [11]), 0.8% Nutrient Broth, 1% malt extract, 1% peptone, 0.1% NaCl and 2% bacteriological agar (w/v) [12] to observe chitinolytic activity (CHIT). Chitinase quantification was assayed by measuring the release of N-acetyl-D-glucosamine (NAGA) from colloidal chitin [13]. The concentration of NAGA in the supernatant was determined according to Reissig et al. [14].
Finally, the Germination Index (GI) was determined according to Zucconi et al., [15], using cress seeds (Lepidium sativum). The GI was calculates as follow:
| GI = ((GxL)/(GcxLc)) × 100, |
where G is the average of germinated seeds in each sample (standardized 104 ufc/mL actinobacterial culture in nutrient broth), Gc is the average of germinated seeds in control plates (sterile distilled water was used), L is the average length of the radicle in the sample (mm), and Lc is the average length of the radicle in the negative control (mm).
Three independent replicates were used in most analyses, except for inhibition indices that were used four. Data obtained were subjected to statistical analysis using Statgraphics Centurion XVI.I (StatPoint, Inc., Virginia). One-way analysis of variance (ANOVA) and multiple comparison tests (Fisher’s Least Significant Difference) were performed to compare mean values for the different levels (P < 0.05) in dual cultures assays to correctly discriminate between controls and samples and in GI assays, to discriminate between blank controls and strain cultures. To select the most effective strains from a suppressive point of view, dispersion diagrams were obtained.
3. Results and discussion
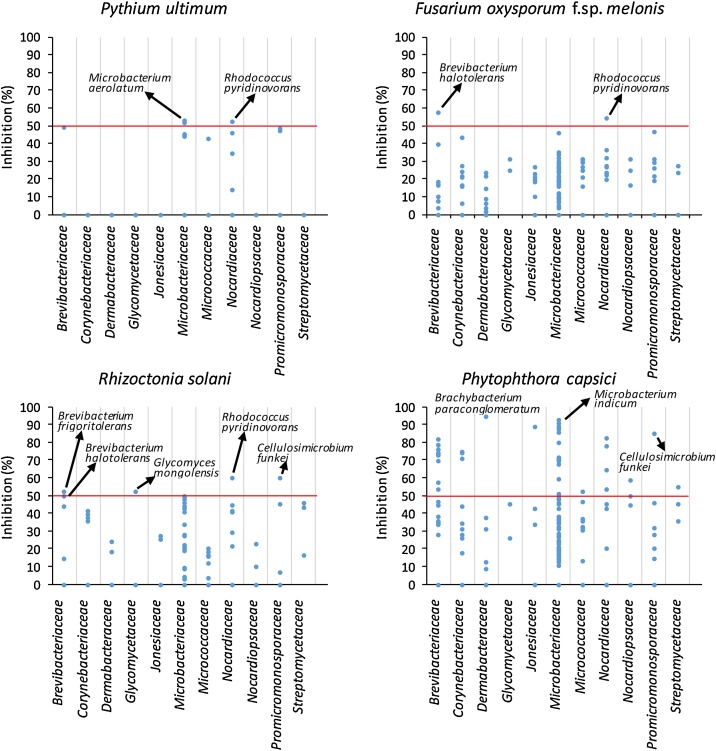
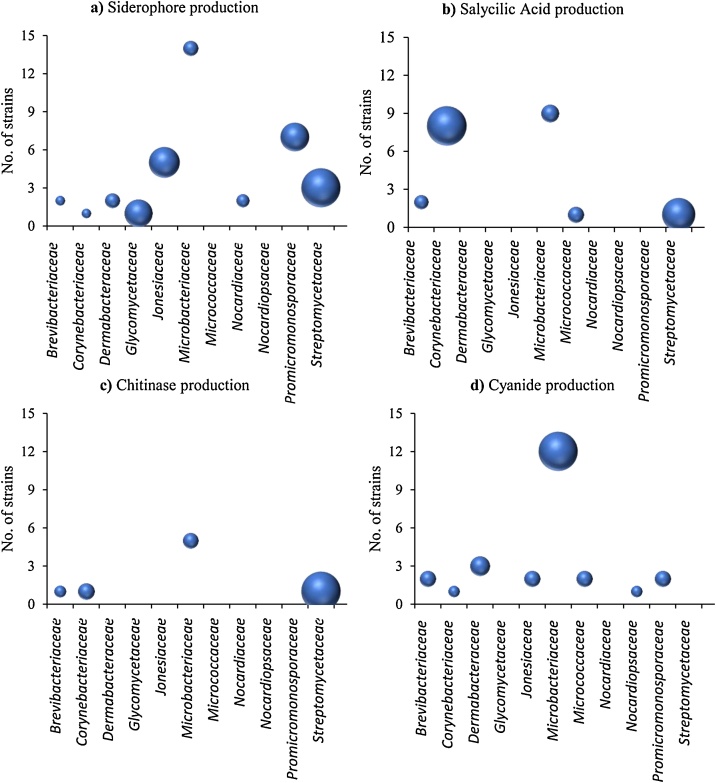
From 220 actinobacterial strains tested against PU, FOM, RS, PCAP, 171 were suppressive at least against one of the four phytopathogens assayed (Fig.1). However, it was significant the case of some species capable of suppressing the presence of more than one pathogen (for example, Rhodococcus pyridinovorans was effective against PU, FOM and RS causing above 50% inhibition). On the other hand, there were cases of reduced but very potent spectrum such as Brachybacterium paraconglomeratum and Microbacterium indicum with inhibition indices almost 100% versus PCAP. In general, most of the strains tested were able to inhibit the in vitro growth of RS and PCAP. On the contrary, the least susceptible phytopathogenic agent to the biocontrol strains tested was PU, which coincides with similar works [16]. Antagonism can usually be explained as a form of directed antibiosis from antagonistic strains towards phytopathogens. However, in addition to (or instead of) antibiotic production, some other bioactive substances can be released by antagonists. In this sense, the production of different substances of interest (SID, SAL, CHIT and HCN) was also investigated in the 171 potentially antagonistic strains (Fig. 2). In all, 31 were recognized as producers of SID (Fig. 2a), substances representing an indirect mechanism against pathogens by iron competition. Analogously, 14 strains produced SAL (Fig. 2b), a key metabolite involved on the induction of systemic resistance in plants [17]; 6 strains were found to be CHIT producers (Fig. 2c) and 20 generated HCN (Fig. 2d), both substances directly related to pathogen inhibition. These results would explain how these 71 strains, with positive results, could avoid damping-off disease by the production of bioactive substances (SID, CHIT, SAL or CNH), in addition or instead of other antibiosis mechanisms, while the remaining 100 strains are likely to induce phytopathogen inhibition through the production of other diffusible enzymes or antibiotic-like substances. The possibility that the same strain uses 2 or more combined control mechanisms cannot be ruled out. Nevertheless, El-Tarabily [18] showed how the application of a consortium of actinobacteria increased the effect of suppression of seedling damping off and root- and crown-rot diseases by Pythium aphanidermatum. Then, the combination of interesting microbes could be a useful option in the formulation of biocontrol agents.
Fig. 1.
Dispersion graphs of the biocontrol ability of 220 actinobacterial strains 2 grouped by family, against phytopathogens tested (PU, FOM, RS, PCAP). Some 3 representative strains with Inhibition Indices above 50% are labeled in the graphs. All 4 results are means (n = 4 repetitions).
Fig. 2.
Number of positive strains for the production of bioactive substances, 6 grouped by family. SID (a); SAL (b); CHIT (c), and HCN (d). The size of the bubbles 7 represents the number of positive strains related to the total number of strains within 8 each family. All results are means (n = 3 repetitions).
In the selection process, SID, SAL, CHIT and HCN producers were further investigated in order to quantify, so that the best final actinobacterial strains could be selected. The concentrations achieved for each bioactive substance were as follows (range of concentrations and the identified strain responsible for maximum production are both indicated): SID (0.16–1.98 μg/mL, Rhodococcus rhodochrous); SAL (4.40–20.11 μg/mL, Microbacterium esteraromaticum); CHIT (0.12–1.18 μg/mL, Microbacterium ginsengiterrae); HCN (0.11–1.05 μg/mL Brevibacterium epidermidis).
Khabbaz et al. [19] indicated that bacteria of the genera Pseudomonas and Bacillus can produce secondary metabolites, antibiotics, siderophores, lytic enzymes and phytohormones that enable them to suppress diseases and promote plant growth. Actinobacterial strains included in the present study exhibited the same abilities. These authors also attributed the production of different types of antibiotics to the capacity of Pseudomonas and Bacillus to inhibit the growth of P. capsici and R. solani (two of the phytopathogens included in this study) and considered the production of hydrogen cyanide as a clear mechanism of pathogen inhibition, since HCN is highly toxic against pathogens at very low concentrations and therefore it can also contribute to antibiosis against phytopathogens.
Fig. 2 show the number of strains in each actinobacterial family with the ability of producing a given bioactive substance, while the sphere diameters indicate the abundance of producers related to the total number of isolates within a family. Most of the isolates were grouped in the Microbacteriaceae family (95 strains). This family also accounted for the highest number of members with observable production of any of the bioactive substances analyzed. Nevertheless, as bubble sizes show, Microbacteriaceae only showed the greatest relative abundance of HCN producers; other families proved to be more representative in SID (Streptomycetaceae, Fig. 2a), SAL (Corynebacteriaceae, Fig. 2b) and CHIT (Streptomycetaceae, Fig. 2c) production. As expected, Streptomycetaceae was a prominent family, inasmuch as there was not a large quantity of their members in the actinobacterial collection, but they were importantly involved in the production of SID and CHIT. Some of the most outstanding strains were identified as Streptomyces albus, S. aureofaciens, and S. lividans. Finally, the last set of experiments was related to test the toxicity of microorganisms over radicular plant system, so that strains selected as good candidates to become biological control agents were those which gave rise to GI equal to 100% or above. Altogether, just 13 out of 71 selected in prior assay, met the established requirement. These strains were identified as (see the number of strains of a given species before the species name and the GI% in parenthesis): 1 Brevibacterium epidermidis (104.12%); 2 Cellulosimicrobium funkei (115.47% and 107.81%); 1 Microbacterium aerolatum (101.56%); 2 M. barkeri (120.31% and 132.00%); 2 M. esteraromaticum (111.87% and 107.81%); 1 M. oxydans (121.88%); 1 M. sediminis (104.00%); 2 M. xinjiangensis (104.69% and 160.94%); 1 Rhodococcus rhodochrous (100.00%).
Sánchez-San Fulgencio et al. [16] reported that thermotolerant bacteria isolated during plant waste composting processes exhibited a variety of capabilities, such as phosphate solubilization and production of siderophores, cyanide, salicylic acid, chitinase-like enzymes and antimicrobial lipopeptides. Although the high temperatures generated during composting are considered an important factor of sanitation, it seems that the above mentioned biotic factors can significantly influence the elimination or deactivation of harmful agents, not only during the composting process, but also after the application of compost as a substrate to soils as well. With the specific case of actinobacteria, the ability to produce compounds with useful features in pharmacology, agriculture and biotechnology is widely known [20]. These products are currently expensive, so consider the ability of actinomycetes producing interesting substances as siderophores or chitinases to promote plant growth and enhance antagonism to phytopathogens, opens new and sustainable possibilities. The potential of this group of bacteria has been previously reported [21], and is currently gaining importance, since is related to a remarkable inhibitory activity against phytopathogenic fungi.
In conclusion, the results illustrate that 13 strains belonging to the order Actinomycetales, with special mention to Microbacteriaceae family, could be stated as an important group of microorganisms with biological control potential against damping-off and surely other plant diseases, due to the production of bioactive substances. Since these microorganisms were isolated from compost samples, this makes it an adequate environment for the bioprospecting of biopesticides and biostimulants substances.
We confirm that the manuscript has been read and approved by all named authors and that there are no other persons who satisfied the criteria for authorship but are not listed. We further confirm that the order of authors listed in the manuscript has been approved by all of us.
We confirm that we have given due consideration to the protection of intellectual property associated with this work and that there are no impediments to publication, including the timing of publication, with respect to intellectual property. In so doing we confirm that we have followed the regulations of our institution concerning intellectual property.
Acknowledgment
This work was supported by the Spanish “Ministerio de Economía y Competitividad” Project AGL2012-36434.
References
- 1.Gravel V., Martinez C., Antoun H., Tweddell R.J. Antagonist microorganisms with the ability to control Pythium damping-off of tomato seeds in rockwool. Biocontrol. 2005;50:771–786. [Google Scholar]
- 2.Mehta C.M., Palni U., Franke-Whittle I.H., Sharma A.K. Compost: its role, mechanism and impact on reducing soil-borne plant diseases. Waste. Manage. 2014;34:607–622. doi: 10.1016/j.wasman.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Hwangbo H., Kim K., Choi H. Effects of biocontrol agents on suppression of damping-off in Cucumis sativus L. caused by Rhizoctonia solani. Hortic. Environ. Biote. 2016;57(2):191–196. [Google Scholar]
- 4.Jurado M.M., Suárez-Estrella F., Vargas-García M.C., López M.J., López-González J.A., Moreno J. Evolution of enzymatic activities and carbon fractions throughout composting of plant waste. J. Environ. Manage. 2014;133:355–364. doi: 10.1016/j.jenvman.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 5.López-González J.A., Suárez-Estrella F., Vargas-García M.C., López M.J., Jurado M.M., Moreno J. Dynamics of bacterial microbiota during lignocellulosic waste composting: studies upon its structure, functionality and biodiversity. Bioresour. Technol. 2015;175:406–416. doi: 10.1016/j.biortech.2014.10.123. [DOI] [PubMed] [Google Scholar]
- 6.Landa B., Hervás A., Bettiol W., Jiménez-Díaz R.M. Antagonistic activity of bacteria from the chickpea rhizosphere against Fusarium oxysporum f.sp. ciceris. Phytoparasitica. 1997;25(4):305–318. [Google Scholar]
- 7.Trinder P. Rapid determination of salicylate in biological fluids. Biochem. J. 1954;57(2):301–303. doi: 10.1042/bj0570301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gil M.J., Martínez-Merino V.J. Practical Studies for Medicinal Chemistry. U.N.R.C.; Río Cuarto: 2011. Determination of the free salicylic acid concentration into aspirin by forming Fe+3 complexes; pp. 101–107. [Google Scholar]
- 9.Visca P., Cievro A., Sanfilippo V., Orsi N. Iron-regulated salicylate synthesis by Pseudomonas spp. J. Gen. Microbiol. 1993;139:1995–2001. doi: 10.1099/00221287-139-9-1995. [DOI] [PubMed] [Google Scholar]
- 10.Lorck H. Production of hydrocyanic acid by bacteria. Physiol. Planta. 1948;1 [Google Scholar]
- 11.Roberts W., Selitrennikoff C.P. Plant and bacterial chitinases differ in antifungal activity. J. Gen. Plant Pathol. 1988;134:169–176. [Google Scholar]
- 12.Zarei M., Aminzadeh S., Zolgharnein H., Safahieh A., Ghoroghi A., Motallebi A., Daliri M., Lotfi A.S. Serratia Marcescens B4A Chitinase product optimization using Taghchi Approach. Iran. J. Biotechnol. 2010;8(4):252–256. [Google Scholar]
- 13.Tweddell R.J., Jabaji-Hare S.H., Charest P.M. Production of chitinases and b-1,3-glucanases by Stachybotrys elegans, a mycoparasite of Rhizoctonia solani. Appl. Environ. Microbiol. 1994;60:489–495. doi: 10.1128/aem.60.2.489-495.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reissig J.L., Strominger J.L., Leloir L.F. A modified colorimetric method for the estimation of N-acetylamino sugars. J. Biol. Chem. 1955;217:959–966. [PubMed] [Google Scholar]
- 15.Zucconi F., Forte M., Monaco A.D.E., De Bertoldi M. Biological evaluation of compost maturity. BioCycle. 1981;22(4):27–29. [Google Scholar]
- 16.Sánchez-San Fulgencio N., Suárez-Estrella F., López M.J., Jurado M., López-González J.A., Moreno J. Biotic aspects involved in the control of damping-off producing agents: the role of the thermotolerant microbiota isolated from composting of plant waste. Biol. Control. 2018;124:82–91. [Google Scholar]
- 17.Palmer I.A., Shang Z., Fu Z.Q. Salicylic acid-mediated plant defense: recent developments, missing links, and future outlook. Front. Biol. 2017;12:258–270. [Google Scholar]
- 18.El-Tarabily K.A. An endophytic chitinase-producing isolate of Actinoplanes missouriensis, with potential for biological control of root rot of lupine caused by Plectosporium tabacinum. Aust. J. Bot. 2003;51:257–266. [Google Scholar]
- 19.Khabbaz S.E., Zhang L., Cáceres L.A., Sumarah M., Wang A., Abbasi P.A. Characterisation of antagonistic Bacillus and Pseudomonas strains for biocontrol potential and suppression of damping-off and root rot diseases. Ann. Appl. Biol. 2015:0003–4746. [Google Scholar]
- 20.Jacob S., Sajjalaguddam R., Kumar K.V., Varshney R., Sudini H.K. Assessing the prospects of Streptomycessp. RP1A-12 in managing groundnut stem rot disease caused by Sclerotium rolfsii Sacc. J. Gen. Plant Pathol. 2016;82:96–104. [Google Scholar]
- 21.El-Shatoury S., El-Kraly O., El-Kazzaz W., Dewedar A. Antimicrobial activities of Actinomycetes inhabiting Achillea fragrantissima (Family: Compositae) Egypt. J. Nat. Toxins. 2009;6:1–15. [Google Scholar]
- 22.Schwyn B., Neilands J.B. Universal chemichal assay for the detection and determination of siderophores. Anal. Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]