Abstract
Study Objectives
Sleep spindles are abnormal in several neuropsychiatric conditions and have been implicated in associated cognitive symptoms. Accordingly, there is growing interest in elucidating the pathophysiology behind spindle abnormalities using rodent models of such disorders. However, whether sleep spindles can reliably be detected in mouse electroencephalography (EEG) is controversial necessitating careful validation of spindle detection and analysis techniques.
Methods
Manual spindle detection procedures were developed and optimized to generate an algorithm for automated detection of events from mouse cortical EEG. Accuracy and external validity of this algorithm were then assayed via comparison to sigma band (10–15 Hz) power analysis, a proxy for sleep spindles, and pharmacological manipulations.
Results
We found manual spindle identification in raw mouse EEG unreliable, leading to low agreement between human scorers as determined by F1-score (0.26 ± 0.07). Thus, we concluded it is not possible to reliably score mouse spindles manually using unprocessed EEG data. Manual scoring from processed EEG data (filtered, cubed root-mean-squared), enabled reliable detection between human scorers, and between human scorers and algorithm (F1-score > 0.95). Algorithmically detected spindles correlated with changes in sigma-power and were altered by the following conditions: sleep–wake state changes, transitions between NREM and REM sleep, and application of the hypnotic drug zolpidem (10 mg/kg, intraperitoneal).
Conclusions
Here we describe and validate an automated paradigm for rapid and reliable detection of spindles from mouse EEG recordings. This technique provides a powerful tool to facilitate investigations of the mechanisms of spindle generation, as well as spindle alterations evident in mouse models of neuropsychiatric disorders.
Keywords: automated spindle detection, mouse electroencephalography, polysomnography, sigma, model species, psychiatry
Statement of Significance.
Sleep spindles, an electroencephalogram (EEG) hallmark of NREM sleep, increase following learning and are important for memory consolidation. Sleep spindle abnormalities are emerging as a biomarker and potential therapeutic target of the cognitive deficits of several neuropsychiatric illnesses. However, detecting spindles in a disease model species (the mouse) is difficult and controversial. This is the first article to validate an automated spindle detection method for use with mouse EEG, modified from a clinically validated method used previously with human EEG. Automated spindle detection in mice will facilitate an understanding of the mechanisms of spindle regulation, the functional importance of sleep spindles, and, in turn, potentially inform the development of therapeutic strategies for neuropsychiatric disorders characterized by spindle abnormalities.
Introduction
In humans, sleep spindles are identified in electroencephalographic (EEG) records as transient waxing/waning rhythmic oscillatory events between 8 and 15 Hz. Generated via corticothalamic network activity, these events represent a physiological hallmark of non-rapid eye movement (NREM) sleep [1–5]. Recent studies in humans and rodents show that sleep spindle density or amplitude is increased following learning, and is correlated with the consolidation of memory [6–11]. Spindle abnormalities have been reported across a number of severe neurological and psychiatric disorders, including autism [12], dementia [13], stroke [14], and schizophrenia [15–17], and have been implicated in the associated cognitive deficits characteristic of these disorders. Such findings suggest that spindle abnormalities represent an important endophenotype of neuropsychiatric disorders, and potentially provide a novel target for therapeutic intervention [18]. Accordingly, there is a growing interest in the development of techniques to detect spindles reliably and accurately.
Efforts to elucidate the mechanisms behind the generation of sleep spindles, and disease-related abnormalities, necessitates the use of mouse models, which provide better genetic tractability. While, numerous studies have described the development and optimization of sleep spindle detection techniques of human EEG [19, 20], it remains controversial whether sleep spindles can be reliably detected in mice.
Recent rodent studies have employed a varied array of approaches for detection of spindles [19, 21–28]. However, lacking in these studies is a clear and thorough verification of whether these detection methods reliably assay sleep spindles. This makes reconciliation of the results from different publications difficult and replication of previously published methods challenging. Thus, here we performed a careful comparison of manual and automated spindle detection methods. The overall aim in this study was the development of an automated paradigm for reliable and reproducible detection of spindles from mouse EEG, the first step of which was the systematic investigation of manual and automatic spindle detection techniques. Next, we performed analysis and experiments to confirm that automatically detected mouse spindles responded to experimental conditions as expected based on the literature [29–31]. The frequency and location of spindles were compared to sigma-power across sleep-wakefulness states and at the transitions between wake, NREM and REM sleep. Correlation of detected spindles with sigma-power EEG activity was also evaluated across the vigilance states (wake, NREM, and REM sleep). A final validation verified previously reported sigma-power alterations due to the hypnotic drug zolpidem.
Methods
Animals
Adult male (7) and female (2) C57/BL6J background mice (3–6 months at the time of surgery) from Jackson Laboratory (Bar Harbor, ME, United States) were used in this study. Mice were housed at room temperature with 12-hour light/dark cycle (lights-on at 07:00 am). Food and water were available ad libitum. All procedures were performed in accordance with the National Institutes of Health guidelines and in compliance with the animal protocol approved by the VA Boston Healthcare System Institutional Animal Care and Use Committee (IACUC). Six males were used for unmanipulated EEG recordings; two males and two females were used in the pharmacology work.
Drugs
Zolpidem was obtained from Sigma Aldrich (St. Louis, MO). 1 mg/ml Zolpidem was dissolved in an equimolar tartaric acid solution in 0.9% NaCl (physiological saline) and administered intraperitoneally (IP) at a dosage of 10 mg/kg, as previous reports have shown that this dose reliably promotes NREM sleep [31]. Moreover, zolpidem attenuates sigma-power at this dose and lower [31–33]. A within-subjects approach was used with each animal receiving saline and a single (10 mg/kg, IP) dose of zolpidem at 48-hour intervals to allow recovery from drug administration.
Surgery and EEG recording
EEG screw electrodes were implanted as previously described [34], above the frontal cortex (Anterior-Posterior [AP] Bregma + 1.9 mm, Medial-Lateral [ML] Bregma ± 1.0–1.5 mm) with a reference electrode above the midline cerebellum (AP Lambda −1.0 mm, ML 0.0 mm) and a ground electrode above a distal parietal region (AP Bregma −2.5 mm, ML Bregma −3 mm). Electromyogram (EMG) electrodes were implanted into mice nuchal muscle. Electrodes were connected to EEG/EMG headmounts (Pinnacle Technology Inc., part # 8402-SS, Lawrence, KS), and secured with dental cement. After recovering from surgery for at least one full week, mice were tethered to EEG recording systems (Pinnacle Technology Inc., part # 8200-K1-SL) for >48 hours for habituation in recording chambers. EEG/EMG signals were sampled at 1 KHz, amplified 100× and low-pass filtered at 200 Hz using a two EEG channel, one EMG channel mouse pre-amplifier (Pinnacle Technology Inc., part # 8202-SL).
Sleep scoring
Sleep scoring was performed manually using 4-second epochs as previously described [35] using Sirenia Sleep software (Pinnacle Technology Inc.). Briefly, epochs with desynchronized low amplitude EEG were scored as waking. Epochs with large amplitude slow EEG waves were scored as NREM, and epochs with rhythmic theta waves, paired with low muscle tone were scored as REM. Records with minimal movement artifacts were chosen for the analysis, and data from hours Zeitgeber Time (ZT) 3 hours to ZT 7 hours was analyzed (during the light phase when the animals tend to sleep more). In separate pharmacologic experiments, zolpidem or saline was administered IP at the mid-point of the light phase (ZT 6 hours to ZT 12 hours).
Automated spindle detection
We developed a custom MATLAB (Mathworks, Natick, MA) script which is roughly based on the method used for clinical spindle analysis in Ferrarelli et al. [20]. Briefly, the raw EEG record was first band-pass filtered between 10 and 15 Hz; we focused on this range because it encompasses the peak mouse spindle of ~11 Hz [27] and previous reports implicate 10–15 Hz as the sigma/spindle frequency in mice [28, 29, 36]. In our analysis we observed a separate accumulation of power in the 7–8 Hz range as the NREM period ends; this signal intensified with the transition to REM and continued throughout REM, indicating this separate peak was REM-related theta activity. Additionally, we examined wake–NREM transitions, and observed an increase in power specifically within the 10 Hz to 15 Hz range. Here we employed a Butterworth filter with the following parameters using the MATLAB designfilt function: First stopband frequency = 3 Hz, First passband frequency = 10 Hz [27], Second passband frequency 15 Hz, Second stopband frequency = 22 Hz, with stopband attenuation levels of 24 dB. Next, the root-mean-square (RMS) of the filtered EEG data was computed using a 750 ms window to smooth the trace and generate a signal envelope. After a careful manual inspection of automated detection results using various window sizes, the 750 ms window size was chosen to minimize the erroneous detection of short, spike-like artifactual signals as spindles, as a 500 ms RMS window yielded 10.8 ± 0.6 spindles/minute with average durations of 1.08 ± 0.005 seconds, 250 ms yielded 20.9 ± 1.2 spindles/minute with average durations of 0.53 ± 0.007 seconds and 100 ms yielded 62.04 ± 2.4 spindles/minute with average durations of 0.168 ± 0.003 seconds. RMS values were then cubed, to enhance the separation between noise and signal on the y-axis and facilitate the placement of thresholds. Finally, we used a two-threshold approach to establish inclusion criteria for spindle detection, which employed several user-defined parameters. These threshold values were derived from the mean cubed RMS transform value of the entire trace (all behavioral states) and included both a lower threshold (1.2× mean cubed RMS; default) and upper threshold (3.5× mean cubed RMS; default). Our algorithm also has the option to base threshold settings on SD values. However, we found no significant differences between detections from mean-based or SD-based threshold settings. Our detection algorithm first utilized the upper threshold value to identify putative spindle peaks in the cubed RMS (envelope) record. Next, the lower threshold was used to determine the start and end of each event, to determine if these putative spindles met our temporal criteria (>0.5 seconds and <10 seconds; default) for inclusion in detection [10, 19, 20, 28, 37]. Basis of these thresholds on the mean of the cubed RMS signal, allowed the calculated thresholds to scale appropriately to account for changes in signal variability/amplitude between animals. The above multipliers used to define these thresholds were determined via systematic modification to optimize spindle detection agreement with human scorers in preliminary work. Additionally, we explored various inter-spindle intervals (ISI; range: 0.001–0.5 seconds evaluated here; 0.01–0.2 seconds recommended; see Results). Manual scoring was performed on raw EEG followed by step-wise levels of data processing, as described in the results section, to optimize reliable scoring by humans.
Comparison of manual scoring versus automated scoring
As a quality control step we first compared our automated spindle detection results to manual human scoring of detected spindles using the same processed data. Manual scoring was ultimately performed after processing of raw EEG data using the following three steps: (1) bandpass-filter (10–15 Hz), (2) cubed RMS-transform, (3) plot thresholds. Scorers were instructed to record a time-point representing the center of the spindle that they had detected.
For the purposes of manual scoring, the threshold used need not have any physiological relevance, as its primary purpose was to provide the scorer with a stable landmark allowing determination of the relevance of peaks in the cubed RMS. Without such a landmark, the amplitude of peaks deemed relevant appears to drift for manual scorers depending on the baseline/variance of the RMS trace. Regarding the physiological relevance of these peaks, scorers can refer to the band-filtered, and ultimately the raw trace, to ensure that the cubed RMS peak was resultant from an event that could conceivably be considered to be a spindle, and not noise/artifacts.
This procedure resulted in high agreement of spindle identification between the algorithm and human scorers. The time points of spindles, as determined by algorithmic detection and manual scoring, were plotted side-by-side to visually assess agreement between methods. An agreement matrix was generated whereby every time-point bearing a spindle in the human-scored record was compared to the same pair-wise time-point in the algorithm-scored record “human-vs-algorithm agreement”. Thus, this process answered the question: if the human has scored a spindle, then has the algorithm also done so? Adopting the human scores as the point of reference, this comparison revealed false negatives. The reverse comparison was made whereby we assessed whether there was a spindle detected by the human at the time-point at which the algorithm had made a detection, to determine “algorithm-vs-human agreement” (if the algorithm has scored a spindle, then has the human also done so?). This reverse comparison revealed false positives. False positives and false negatives were used to determine recall and precision, which in turn were used to determine F1-scores which we have used to avoid the misleadingly high measure of “true negatives” (Warby et al [19].). We have identified an additional source of algorithmic detection error which we term unresolved clustered spindles. To accommodate this, we report percentage agreement and percentages of three types of error: False positives, false negatives and unresolved clustered spindles.
Comparison of EEG power analysis and spindle detection
Power spectral density (PSD) analysis was determined for NREM, REM, and wakefulness using the MATLAB pwelch function. We did not remove hybrid epochs, where one epoch overlapped two sleep-wake states, to minimize loss of data from state transitions. A Hanning (2048) window function with 50% overlap was employed to bin frequency transformed data. Power within defined frequency bands, delta (1–4 Hz), sigma (10–15 Hz) or gamma (30–80 Hz), was determined as the sum of all the values across the relevant frequency bins. Behavioral state transition spectrograms were produced by selecting 1 minute of NREM (or wake) followed by 1 minute of REM (or NREM). Each frequency bin was then normalized to the average power from its corresponding bin in wakefulness. Spectra were computed using the multi-taper approach (tapers were 5 and 9, 10-second window, 100 ms steps) with the MATLAB mtspecgramc function. Spectra were first calculated for each transition and averaged for each mouse, then the resulting average spectra were averaged between mice for a grand average spectrogram and plotted. Behavioral state transition power analysis was performed on band-filtered data in four steps as previously described [30]. (1) We extracted the transitions, (2) separated the data into 12-second epoch, (3) computed the PSD for each epoch, (4) normalized the result as a percentage of the total band power. We then counted the number of spindles in each epoch for comparison with the power analysis.
Statistics
Pearson’s correlation was used uniformly and computed in MATLAB. For pharmacological experiments, a two-tailed paired t-test was used to establish significant difference between treatment groups saline versus zolpidem. All averaged data and error bars represented as mean ± standard error of mean. Recall was defined as the fraction of true spindle events that were detected, precision was the fraction of detections that are correct. The F1-score is the harmonic mean of precision and recall scaled from 0 to 1, as previously described [19].
Results
Quality control and refinement of automated spindle detection method
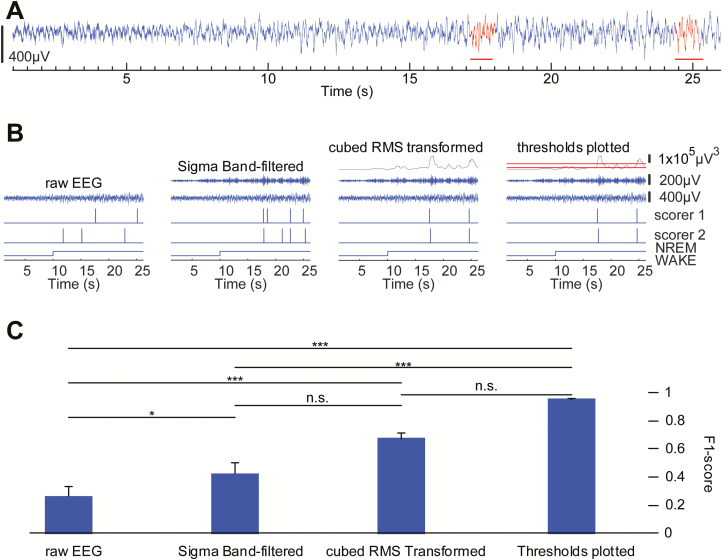
In order to confirm the reliability of our spindle detection method, we performed a comparison of automated spindle detection results with the results of manual scoring which allowed us to rule out the possibility that the algorithm was detecting artifacts as spindles. Before validating the automated spindle detection results with the manual spindle detection results, we first developed a way of reliably scoring spindles by human scorers. Manual spindle detection of mouse EEG recordings was performed by three individuals who were proficient at scoring EEG data for the behavioral vigilance states (wake, NREM, and REM sleep). Thus, each scorer was familiar with the appearance of mouse EEG data. To manually count spindles, a period of 30 minutes was selected from the light phase, when mice were more likely to be in NREM sleep. The initial set of manual scoring was performed on raw EEG data. Three human scorers visually assessed a half hour per mouse of raw EEG (n = 4), identifying waxing and waning transient events that had an appearance discernible from the flanking regions of EEG signal and were ~10–15 Hz (peak power of mouse spindles) and at least 0.5 seconds long (Figure 1A, red). To facilitate identification of spindle frequency events, half-second tick-marks were plotted along raw EEG traces and scorers counted 5–7 peaks between tick-marks. Signals were visualized on a 25-second time-scale which allowed space to be seen between waves at sigma frequencies (Figure 1A). Although this scoring protocol has been used in human EEG [19], we found this method unreliable for accurate and reliable identification of sleep spindles in mouse EEG (Figure 1, B and C, “raw EEG”), leading to low agreement between scorers as determined by F1-scores of 0.26 (±0.07) (Table 1; Figure 1C). Thus, we concluded it is not possible to reliably score mouse spindles manually when using unprocessed EEG data.
Figure 1.
Manual sleep spindle detection becomes more reliable with progressive levels of data processing. Agreement between human scorers on the identification of sleep spindles is greater than F-score of 0.95 only when the EEG data is processed using the algorithm’s procedures. (A) Representative example of mouse cortical EEG data illustrates the difficulty in identifying sleep spindle activity (red horizontal line) in raw data. (B) Representative example of comparison of manual detection of sleep spindles between two experienced scorers. Agreement between detected events between scorers was very low using only the raw EEG trace but was greatly improved with availability of additional data processing, including (1) band-pass filter, (2) cubed RMS transform, and (3) thresholds plotted. (C) Interscorer agreement improved across each level of additional data processing. Agreement between human scorers on the identification of sleep spindles is greater than 0.95 only after providing scorers with all three levels of data processing to confirm detected events. *p < 0.05, ***p < 0.001.
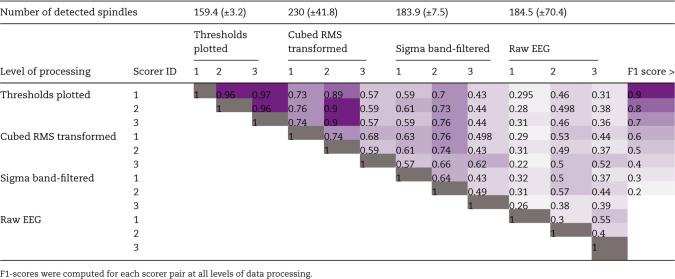
Table 1.
F1-scores show reliability of spindle detections between three human scorers increase as mouse EEG data is processed using the algorithm’s procedures
Guided by our work with the automated spindle detection paradigms, we next aimed to determine a minimal set of data processing steps that would provide consistent manual spindle detection results; allowing human scorers to agree on spindle locations with F1-scores above 0.9. Manual scoring was repeated on the same periods of mouse EEG as above, with incremental levels of data processing, to optimize between-scorer agreement. First, EEG data was band-pass filtered across a frequency range corresponding to the peak frequency of rodent spindles (sigma; 10–15 Hz). Plotting the filtered data aligned with raw EEG for scoring improved human agreement, but only to 0.42(±0.08) (Figure 1, B and C, “sigma band-filtered”). We next included the cubed RMS of the band-pass filtered data. This further aided visual detection of transient events, reflecting the amplitude of sigma filtered data, and leading to interscorer agreement of 0.67(±0.04) (Figure 1, B and C, “cubed RMS transformed”).
We speculated that remaining disagreement was largely due to inconsistency in determining which cubed RMS peaks were sufficient to be counted as spindles, across the duration of the signal, and between scorers. Thus, we additionally plotted a threshold line to provide scorers with stable landmark for consistent determination of the relevance of peaks in the transformed data. At this level of data processing, three scorers could agree above 0.95 in all cases (n = 4; 0.95 ± 0.003; Figure 1, B and C, “threshold plotted”). Finally, we determined F1 scores between human scorers across the levels of processing to determine levels of inter-rater agreement (Table 1).
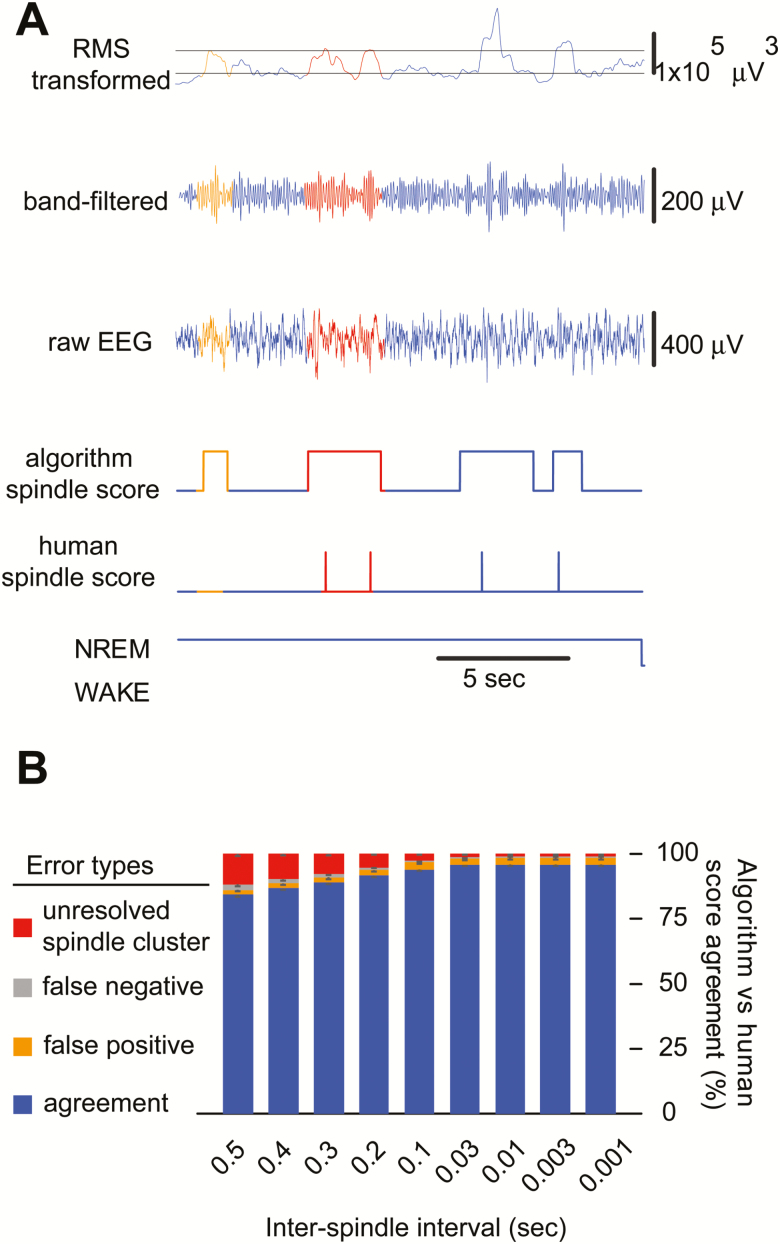
Next, we refined our algorithmic detection criteria by comparing the automated spindle detection results with a large set of manual scoring by six human scorers evaluating EEG records from six mice, 4 hours per mouse. Specifically, informed by one of the detection algorithm (a5) described in Warby et al. [19], our method takes into account the time between successive events which we have termed the ISI [38, 39]. We found in our first comparison between human scores and automatic scores using an ISI of 0.5 seconds, agreement was 84.4 ± 0.8% (F1-score = 0.92 ± 0.005) (Figure 2B). We determined that false negatives and false positives were uncommon (<2.1% each, Figure 2B). However, we found our largest source of error originated from unresolved clustered spindles (11.8%; Figure 2A, red), where a human scorer could discern multiple spindles close together, but the algorithm counted only one. We attributed this to the length of the ISI and found that setting the ISI to 0.5 seconds, based on previous work in human EEG, resulted in poor resolution of closely positioned spindles (Figure 2, A and B). We thus tested various ISI values starting from 0.4 second and continuously reduced the value until we reached the shortest possible time point, determined by our sampling rate. We found agreement continued to improve up to an ISI of 0.01 second at which point agreement was 95.9 ± 0.5% (F1-score = 0.98 ± 0.003) (Figure 2B). An ISI of 0.2 second yielded agreement above 90% (F1-score = 0.96 ± 0.003). Additionally, a one-way ANOVA with Bonferroni’s post-hoc comparison was used to determine significant differences between the ISI lengths. 0.1 second was the longest ISI not significantly different from the shortest ISI of 0.001. Therefore, we used an ISI of 0.1 second for all subsequent analysis.
Figure 2.
Spindle detection errors can be reduced by shortening the ISI. (A) A representative stretch of EEG data comparing manual versus automated spindle scoring demonstrates that unresolved spindle clusters are a major source of spindle detection disagreements between the algorithm and human scorers, including: (1) false positives (orange), which were events not scored by human scorer but scored by automated detection, such discrepancies are typical when the algorithm determines the trace to reach the upper threshold but the human cannot; (2) unresolved spindle clusters (red), which were events scored as multiple events by human scorer but as a single event by automated detection. These discrepancies arise from long ISI settings. Minimal error was observed due to false negatives (gray). (B) Bar graph shows percent agreement between manual and algorithmic spindle detections and denotes error type contributing to inconsistency. Reducing the minimum time interval used for the ISI criteria in automated detection reduced human-algorithm error from unresolved spindle clusters (4-hour data, six mice).
Comparing algorithmically detected spindles to putative proxies of spindle activity: percentage sigma-power at transitions from NREM to REM
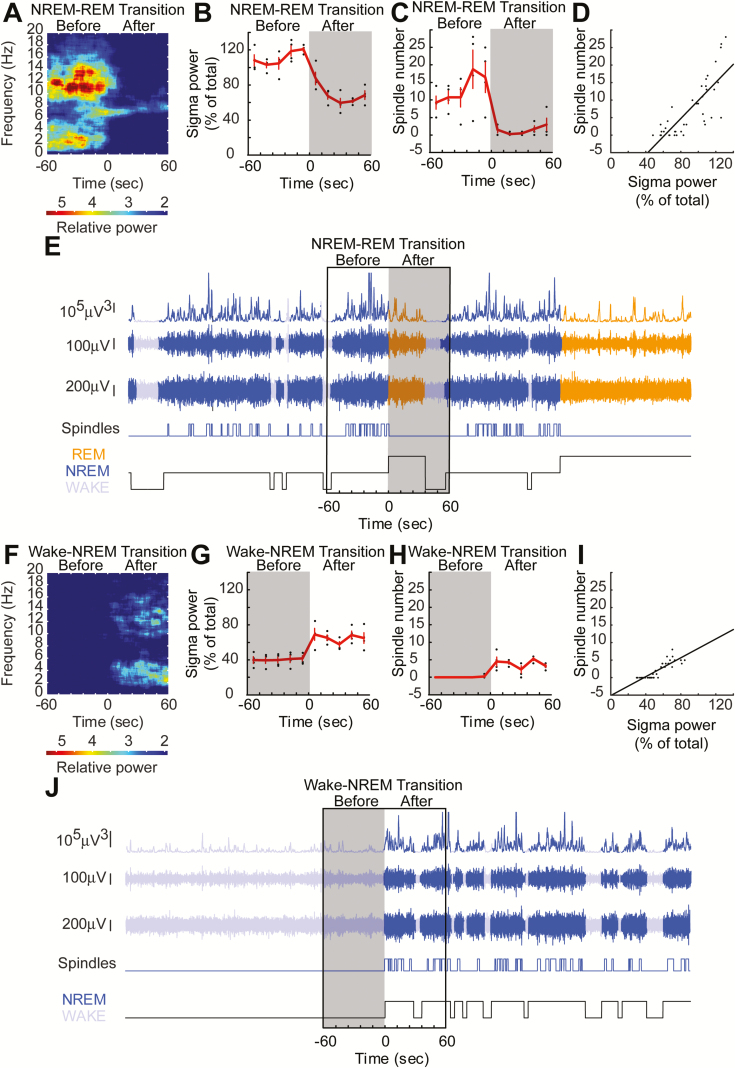
Previous studies in mice, have employed measures of sigma-power as a proxy of spindle activity [29, 30, 40]. In particular, the percentage of baseline sigma-power occurring at transitions from NREM into REM or from wake into NREM, when spindle activity would be expected to be high. We replicated this measure in our mice and compared findings to automated spindle detection results. A spectrogram of NREM–REM transitions revealed that power in the sigma range accumulated when NREM was close to transitioning into REM; this sigma surge decayed rapidly with the onset of REM sleep (Figure 3A). This surge in power was between 10 and 15 Hz, consistent with previous reports [28, 29, 36]. Detected spindles were observed to surge prior to NREM–REM transition, similar to sigma-power (Figure 3, B and C). Further, the number of spindles that we detected were highly correlated with the sigma-power around NREM–REM transitions (Figure 3D; Pearson’s correlation, r = 0.93, p = 1.1 × 10−4). This result is perhaps unsurprising as it is likely to be state dependent. That is, when the animal is in NREM, both spindles and sigma-power are high, but when the animal is in REM, both spindles and sigma-power are low. However, the correlation between percent sigma-power and spindle number in the NREM period alone was also significantly correlated (Pearson’s correlation, r = 0.89, p = 0.04), removing behavioral state dependence from this effect. A similar relationship was seen with the wake–NREM sigma-power surge (Figure 3, F and G) and spindles (Figure 3H). Both spindles and sigma-power were highly correlated (Figure 3I; Pearson’s correlation, r = 0.98, p = 3.7 × 10−7). These results increased our confidence that the events algorithmically detected as spindles were indeed a product of the same underlying physiology previously reported as a measure of mouse spindle activity. However, with our spindle detection method, we now have the advantage of being able to examine spindles across the whole record rather than just at behavioral state transitions (Figure 3, E and J).
Figure 3.
Automated spindle detection correlates with sigma-power (10–15 Hz) at sleep-wake transitions. (A) Sigma-power (10–15 Hz) surges can be observed prior to NREM–REM state transitions. Theta band (5–9 Hz) power increases prior to NREM–REM transitions and continues on into REM sleep. Spectrogram at NREM–REM transition. (B) Sigma-power surges can be observed at NREM–REM state transitions, calculated as previously described [29]. (C) Spindle numbers aligned at the same periods as shown in (A) followed a similar trajectory. (D) Spindle number correlated with the sigma-power surge. (E) Example stretch of EEG data showing that many detected spindles occur outside of the state-transition window. (F–J) The same comparisons are depicted for wake–NREM transitions showing that spindle number correlates highly with sigma-power.
Comparing algorithmically detected spindles to a broad measure of spindle rhythmogenesis: spontaneously fluctuating sigma-power
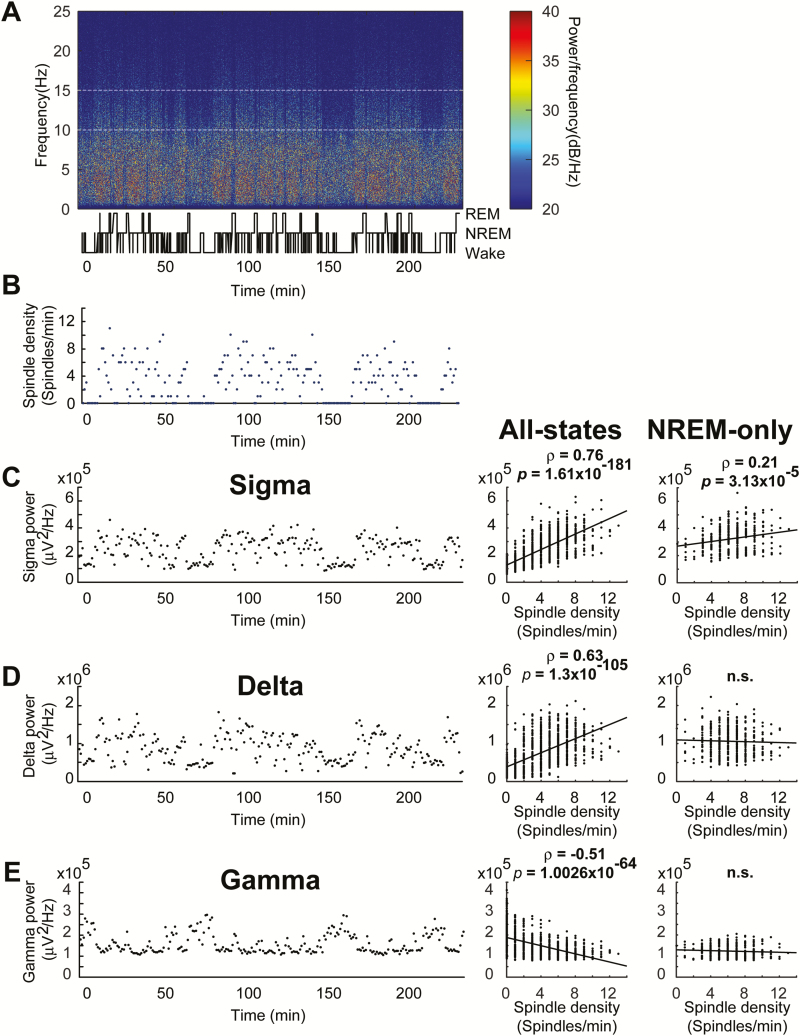
Total sigma-power over long periods of time (e.g. periods 30 minutes or longer) did not correlate with spindles (Table 2). To overcome the problem of between-animal sigma-power variability, we used a within-animal comparison of spindles and sigma-power. Figure 4A depicts a representative sleep-wake profile, comparing the fluctuation of the number of NREM spindles (Figure 4B) to the fluctuation of sigma-power (Figure 4C, left). These parameters were closely correlated (Figure 4C, middle; Pearson’s correlation, r = 0.76, p = 1.61 × 10−181). However, we considered the possibility that this relationship might be behavioral vigilance state dependent as opposed to spindles and sigma-power correlating per se. To test this, we analyzed the relationship between NREM spindles and other sleep-wakefulness state associated oscillations. When spindles occur during NREM sleep, with accompanying high delta-power, the relationship between spindles and delta-power (Figure 4D, left) was closely correlated (Figure 4D, middle; r = 0.63, p = 1.3 × 10−105). Gamma-power is associated with waking and is diminished during NREM sleep. Thus, we predicted a sleep-wakefulness state dependent negative association between spindles and gamma-power (Figure 4E, left), which was evident (Figure 4E, middle; r = −0.51, p = 1.0026 × 10−64). These results raised the question of whether there was a true association between sigma-power and spindles, or if it was secondary to a sleep-wakefulness state relationship. That is, sigma-power is high in NREM as are spindles, and vice versa. Thus, we extracted all the NREM data and performed a time course correlation between spindles and sigma-power. In the absence of states other than NREM, spindles and sigma-power correlated significantly (Figure 4C, right; r = 0.21, p = 3.13 × 10−5). The r value is lower than those observed in the correlations in which sleep-wake state had not been controlled, possibly due to removal of a major source of variance in sigma-power by removing wake and REM sleep. NREM spindle density (spindles/minute), on the other hand, was less sensitive to this, for only NREM spindles were used for both all-states and NREM-only analysis, and thus, the impact of removing wake and REM here was negligible. The positive association between delta-power and spindles, and the negative association between gamma-power and spindles were both lost when only NREM data were analyzed (Figure 4D, right and Figure 4E, right; r = 0.037, p = 0.48 & r = 0.08, p = 0.13, respectively).
Table 2.
Spindle density only correlated with sigma-power when data were analyzed in time bins of 15 minutes duration or shorter
| Bin width | 1 minute | 15 minutes | 30 minutes | 4 hours |
|---|---|---|---|---|
| Pearson’s correlation coefficient (r) | 0.21 | 0.60 | 0.57 | −0.57 |
| p-value | 3.13 × 10−5 | 0.01 | 0.14 | 0.43 |
Pearson’s correlation coefficient and p-values were given for comparisons of spindle density and sigma-power in 15-minute bins and 30-minute bins. Both cases reflect the full 4-hour period used throughout the rest of the study.
Figure 4.
During NREM sleep, spindle density correlates with sigma-power, but not with delta or gamma-power (right column of C–E). (A) Representative spectrogram with temporally aligned hypnogram shows the gross mouse sleep-wake patterns across the 4-hour recording period. (B) The number of NREM-spindles (blue) in each 1-minute block from the same representative case across the 4-hour recording is depicted; spindles occurring in REM or wakefulness are omitted. (C, D, E Left) Absolute sigma-power (C), delta-power (D), and gamma-power (E) in 1-minute blocks from a representative case are depicted. (C Middle) Spindle density (3.4 ± 0.1 spindles/minute) and sigma-power correlated in four animals. (C Right) Spindle density (6.3 ± 0.1 spindles/minute) and sigma-power correlated when only NREM was considered in four animals. (D Middle) Spindle numbers and delta-power were correlated in four animals. (D Right) Spindle numbers and delta-power were no longer correlated when only NREM was considered in four animals. (E Middle) Spindle numbers and gamma-power were negatively correlated in four animals. (E Right) Spindle numbers and gamma-power were no longer negatively correlated when only NREM was considered in four animals.
Upon realizing that controlling for sleep–wake state revealed sigma, but not delta or gamma, correlated with spindles, we revisited the behavioral state transitional power surge analysis for evaluation of delta- and gamma-power. Analysis revealed a state-dependent positive correlation between spindles and delta-power at NREM to REM (Pearson’s correlation, r = 0.88, p = 7.3 × 10−4) and wake to NREM transitions (Pearson’s correlation, r = 0.92, p = 1.8 × 10−4). Furthermore, a state-dependent negative correlation between spindles and gamma-power was observed at the NREM to REM (Pearson’s correlation, r = −0.79, p = 0.007) and wake to NREM transitions (Pearson’s correlation, r = −0.92, p = 1.9 × 10−4). However, these relationships were not apparent within the NREM-only period of the transition. Only sigma remained significantly correlated with spindles in the NREM period of the transition (Pearson’s correlation, r = 0.91, p = 0.03), further confirming that sigma alone bears a state-independent relationship with NREM spindles.
Finally, we found that the vast majority (Table 3) of detected events fell clearly within NREM, suggesting that spindles are indeed a physiological aspect of NREM. 75.9 ± 9.8% of the spindle events detected during wakefulness occurred during periods when muscle tone, as measured by EMG, was in the NREM range (lack of overt activity, decreased muscle tone compared to wake), suggesting either short transient moments of NREM emerge within epochs scored as wake or that the spindle occurred during quiet wakefulness (Table 3). Further, more than 40% of “wake spindles” occurred at transitions between NREM and wake (Table 3), as NREM from the neighboring epoch contaminated the wake-scored epoch. Except for one event in one mouse, all wake spindles found at sleep–wake state transitions were paired with NREM-like EMG amplitude. Thus, detected spindles are less likely to occur in wake epochs. An additional possibility to explain spindles that occur in wake epochs could be that the spindle has come from a region of the brain exhibiting local sleep patterns. Thus, it has been reported that EEG derived measures of sleep in the cortex of rats and humans can be regionalized, a phenomenon referred to as “local sleep”. Indeed, localized spindles have been reported in humans [41, 42] and this phenomenon could underlie sleep spindles that occur in epochs containing drowsiness that were identified as wake.
Table 3.
Spindle numbers and percentage for each behavioral state across 4-hour records in four mice
| Total spindles | NREM spindles | “Wake” spindles | “Wake” spindles with NREM-like EMG | “Wake” to NREM spindles | NREM to “Wake” spindles | REM spindles | |
|---|---|---|---|---|---|---|---|
| Mean ± SEM | 902.5 ± 14.0 | 805.8 ± 20.8 | 44.0 ± 19.4 | 29.0 ± 9.1 | 4.3 ± 2.4 | 13.8 ± 4.6 | 52.8 ± 9.3 |
| % Total spindles ± SEM | 89.3 ± 1.4 | 4.9 ± 2.2 | 3.2 ± 1.1 | 0.5 ± 0.3 | 1.5 ± 0.5 | 0.1 ± 0.01 |
Spindles detected during wakefulness were compared to their temporally aligned EMG signal. NREM-like EMG is defined as values less than three standard deviations above the mean of RMS transformed EMG activity during NREM.
Experimental manipulation of spindles and sigma-power: analysis of z-drug induced changes in EEG
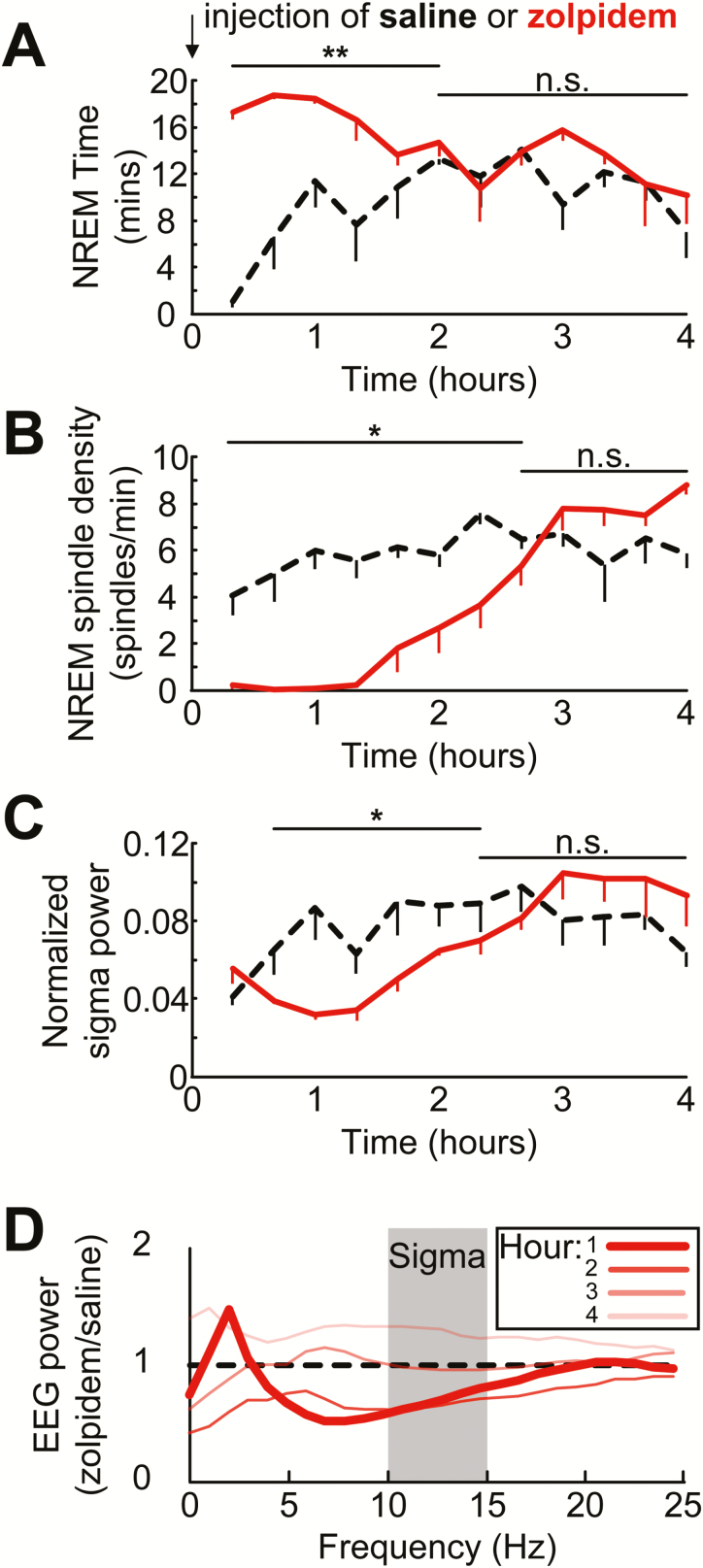
Having described the relationship between sigma-power and sleep spindles in spontaneous EEG records, we considered this sigma correlation as a secondary measure and wanted to add rigor to the association by establishing a causal relationship between these two measures. For this, we used the hypnotic drug zolpidem, which has been shown to decrease sigma-power in rodents [31–33]. The drug was administered to animals (10 mg/kg; IP), and EEG measures of spindle and sigma-power were compared to saline control injections. Figure 5 depicts the time spent in NREM, NREM spindle density and sigma-power, following drug injection in 20-minute intervals. As predicted, zolpidem induced a short latency onset of NREM sleep (1.5 ± 0.5 minutes) with significantly more NREM in the first 2 hours (paired t-test; t (3) = 10.3, p = 0.002). Compared to vehicle injection, zolpidem (Figure 5, B and C) treated mice presented with fewer sleep spindles (paired t-test; t (3) = 5.6, p = 0.01) and lower sigma-power (paired t-test; t (3) = 3.4, p = 0.04). Comparing drug versus saline power profiles over 4 hours (Figure 5D) revealed that the strongest spindle and sigma-power suppressing effects occurred within the first hour of drug treatment (Figure 5D, bold lines) consistent with previous findings, showing that receptor occupancy in the brain is maximal (75%) at 5–10 minutes following an IP injection of zolpidem in mice [43], and falls below 10% after 1 hour [43]. The effect of zolpidem on sigma-power was also analyzed only in NREM periods. This analysis revealed a suppression of sigma-power lasted at least 4 hours (data not shown), which is consistent with the literature [33].
Figure 5.
Spindle density and sigma-power decrease in parallel after hypnotic drug treatment despite an elevation of NREM sleep. Average time spent in NREM (A) aligned with NREM spindle density (spindles/minute) (B) and normalized sigma-power (C) are depicted for 4 hours following an intraperitoneal injection of saline (black dotted line) or zolpidem (10 mg/kg; red solid line). Data depicted as the mean of four animals, error bars represent SEM; 20-minute bins. (D) EEG power after an injection of zolpidem. Dark represents the first hour producing the largest effect, while subsequent hours are shown by lighter shades. *p < 0.05,**p < 0.01, n.s. not significant.
Discussion
As far as we are aware, this is the first article to systematically validate an automated spindle detection method for use with mouse EEG. Our results suggest that this automated method provides a valid representation of sleep spindle activity for the following reasons: (1) Spindle density correlated well with sigma-power, a measure which itself is sensitive to alterations in spindle activity [37, 44]; (2) detected spindles occurred primarily in NREM sleep; (3) spindles increased during transitions into or out of NREM sleep; (4) Within the state of NREM sleep, spindle density correlated with sigma-power but not delta or gamma-power (right column Figure 4); (5) the hypnotic drug zolpidem caused parallel changes in spindle density and sigma-power. Overall, our automated detection method provides more information about the properties of the spindles than the use of sigma-power alone, such as spindle frequency and duration, allowing assessment of coupling with other EEG signals. Thus, we believe our automated method will be extremely useful for researchers in the field.
One of the major findings of this work is that manual detection of sleep spindles in unprocessed mouse EEG is not possible. This is consistent with the anecdotal observations of many other researchers in the field who have questioned the ability to measure sleep spindles in mice. While it is possible to score sleep spindles in raw EEG from human subjects, several steps of EEG data processing were required in mouse EEG before high agreement between human scorers was observed (agreement above 0.95). We utilized these optimized human scores to determine the reliability of our custom MATLAB-based algorithm. This manual validation of algorithmic results provided a level of validation akin to that which had been performed in automated spindle detection methods of human EEG [19].
Our automated mouse spindle detection method was modified from the algorithm employed in Ferrarelli et al. [20] for use with human EEG data. We chose to base our detection method on this algorithm because Warby et al. [19] found that this method provided the best precision of the six human spindle detection methods they evaluated. Optimal precision is more favorable than recall for mouse EEG, because intracranial electrodes have better signal-to-noise than scalp electrodes used in human studies. Additionally, the MATLAB based code was intuitive and easy to modify for our needs with mouse EEG. Furthermore, this method had been previously modified for use in a mouse EEG study [27], although systematic validation was not conducted. Thus, our validation of a modified Ferrarelli spindle detection method compliments the existing mouse literature and allows comparison with human studies which use the same methodology.
For this study amplitude threshold settings were calculated from cubed RMS data including epochs from all behavioral states. It is important to note that this data represented normal light period sleep–wake behavior and was free of movement induced artifacts. While generally not an issue, an unusually high proportion of wakefulness, or excessive locomotor activity-induced EEG artifacts, have the potential to negatively impact the amplitude threshold calculation. Since pristine recordings may not always be available, we suggest several possible ways to deal with these issues: (1) Score epochs containing movement artifacts separately from wakefulness. Our MATLAB script can exclude such epochs from the threshold calculation. (2) Thresholds can be based on NREM epochs alone. For this, alternative multipliers must be determined empirically using exemplar recordings. When using NREM EEG data only for baseline calculations, we have found that an upper threshold multiplier of 2.5 and a lower threshold multiplier of 1 provides similar threshold values as the default settings with artifact-free records. Using NREM only threshold settings may also be used if the EEG record does not include sleep-wakefulness state proportions that are typical of natural undisturbed behavior in mice. (3) Finally, if possible amplitude thresholds can be calculated from longer duration EEG records, when available, to mitigate spindle detection issues arising from atypical sleep-wake in a particular section of the EEG record.
Measuring individual spindle events has several advantages over the use of surrogates of sleep spindle activity such as sigma-power including the ability to describe the properties of spindles such as their frequency, duration, and amplitude. Sigma-power has been commonly used in publications that explore sleep spindle-generating neurocircuitry [29, 30]. Here we show that total NREM sigma-power is a poor marker of spindle activity for several reasons. As shown in Table 2 spindle density only correlated with sigma-power if short sample time periods were used (<15-minute bins). Any data binning above 15 minutes did not reveal a correlation between spindle density and sigma-power. Further, sigma-power used alone to assess spindles can be difficult to interpret, providing conflicting outcomes depending on differing analysis techniques. For example, in three mouse studies, overall NREM sigma-power was unchanged across EEG recording sessions, comparing measures in experimental and control mice [29, 30, 40]. However, measures of sigma-power at state transitions between NREM to REM sleep differed between the three studies. These reports highlight the need to maintain fine temporal resolution within the EEG recording, and thus demonstrate the importance of detecting individual sleep spindles rather than just sigma-power.
Having validated our spindle detection method, we now have more confidence that the sigma-power surge at NREM to REM transitions is a valid proxy of spindle activity in mice; this had previously not been established. In our hands, increased sigma-power was found at sleep-wakefulness state transitions (NREM–REM and wake–NREM) similar to previous reports in mice [29, 30, 40]. Importantly, detected spindles and sigma-power measures were well correlated. Thus, detecting individual spindles provides greater sensitivity and temporal detail than is possible by studying sigma-power alone.
Seminal work by Steriade and colleagues [45] investigated the interplay between spindles and slow-waves, alluding to functionally relevant interactions. However, this work was performed in cats, in which spindles are more clearly visible in raw EEG. With recent advances in mouse genetic approaches and signal processing techniques, future experiments may reveal whether new therapeutic strategies are more efficacious by targeting the coupling of spindles to their physiologically relevant position with other oscillations, such as delta oscillations and/or hippocampal sharp-wave-ripples, rather than simplified analysis of the spindle number or sigma-power. With our method, we resolved spindles as individual transient events, which enabled estimates of start-points and end-points of spindles, allowing alignment or superimposition of spindles with other transient events, such as slow waves [26]. This provided the level of fine-grain detail required for studying spindles in temporal relation to other oscillations.
The frequency band that constitutes the spindle range has been defined inconsistently between different studies [26–29, 46] Lüthi et al. [47] discuss 10–15 Hz as the sigma/spindle frequency. Vyazovskiy et al. [28] focused on 10–13 Hz, Kim et al. [27] used 10–16 Hz, Astori et al. used 10–12 Hz [29], Franken et al. [46] cited 11–15 Hz, and Latchoumane et al. [26] designated 7–10 Hz as the sigma range, each of these studies were performed using mice. Broader ranges have been used in classic work in cats which focused on 7–14 Hz [48] or 7–16 Hz [5]. Consistent with other reports [28, 29, 36], we evaluated NREM to REM transitions and observed a distinct area of high energy between the frequencies of ~10 Hz and ~15 Hz which decayed rapidly with the onset of REM sleep, and a separate theta peak of ~7 Hz which continued into REM.
Here we also replicate the finding that zolpidem attenuates sigma-power [31–33] in mice. We further demonstrate for the first time that a decrease in spindle density parallels the decrease in sigma-power. Furthermore, as shown in Figure 5, the decrease in spindle density appears more robust than the decrease in sigma-power. One consideration, however, is that these drugs in rodents produce the opposite effect on sleep spindles in humans, that is, increasing both spindle number and sigma-power [18, 49, 50]. The reason for this between-species discrepancy is not known but is likely because doses given to mice are usually ~100-fold higher (mg/kg) than in humans. Mice require at least 5–10 mg/kg to induce NREM sleep reliably [31–33], whereas human studies typically administer a 3 mg or 10 mg total dose per participant regardless of body weight [18, 49, 50].
A number of approaches have been explored for automated detection of sleep spindles (Warby et al. [19]). While these methods are in general quite similar, subtle variations in data processing methods make it difficult to compare spindle findings across different studies. Such methods principally vary based on the method employed to provide a quantitative measure of power in the spindle frequency range; including RMS or Hilbert transform, and wavelet-based approaches. Moreover, threshold settings are often based on mean values as a baseline [20, 27, 51], whereas other methods employ alternatives such as standard deviation values [26, 52] or percentiles [37, 53]. While each of these approaches are utilized to the same end, different techniques may have certain advantages, depending upon the measure of interest (temporal localization, accuracy of spindle frequency, etc.). For example, wavelet-based approaches may be more sensitive to peak frequencies (detecting narrow frequency ranges) whereas band-filter based approaches are more sensitive to broader frequency bands. We favored a band-filtered approach here, because spindles are classically described as oscillatory events existing within a defined frequency bandwidth. Other significant sources of variability include the frequency band used to define spindles as well as event duration criteria. Our detection approach was designed to be flexible and is easily adjustable to account for differing detection criteria; it has a graphical user interface for adjustment of the main detection parameters. For example, users can vary the duration settings to detect only spindles that last longer than a second, or they can vary the threshold settings in order to selectively detect large spindles. Spindle amplitude is a parameter of particular interest because large spindles may reflect events generated closer to the recording electrode and/or those most likely to induce synaptic plasticity in cortical circuits [54].
There has also been some variation in reported spindle density in mice, our automated method detected sleep spindle density in the same range as previous reports. Most previous studies report mouse NREM spindle density in the range of 3–11 spindles per minute [28, 37, 55]. Our average NREM spindle density during the light phase was 6.3 spindles per minute (Figure 4C, right). One study reported a lower value of 1.66 spindles per minute [27]; using more stringent detection criteria. Hence, this study possibly excluded many lower amplitude events and thus detected fewer spindles overall compared to other studies. Moving forward, it would be extremely helpful for the field to come to a better consensus on the frequency and duration of sleep spindles. To facilitate this effort, we will provide the MATLAB script for our detection algorithm to investigators upon request.
In conclusion, despite mice becoming a preferred model species for the study of sleep, no validated method for detecting sleep spindles has been previously reported. This study validated an automated spindle detection method in mouse EEG recordings, which will hopefully facilitate future work directed at understanding the mechanisms of spindle generation and functional impact of sleep spindles.
Acknowledgments
We thank Felipe L. Schiffino, Elena R. Tilli, and Leana K. Radzik for their contributions to the data analysis.
This work was conducted at VA Boston Healthcare System, 1400 VFW Parkway, West Roxbury, MA 02132.
Funding
U.S. Department of Veterans Affairs BX002130 (J.M.M.), U.S. Department of Veterans Affairs BX001356 (R.E.B.), U.S. Department of Veterans Affairs BX002774 (R.E.S.), U.S. Department of Veterans Affairs BX001404 (R.B.), and National Institute of Mental Health R01 MH039683 (R.E.B.), National Heart, Lung, and Blood Institute T32 HL007901 (D.S.U.), National Institute of Mental Health T32 MH016259 (D.D.A.), and National Heart, Lung, and Blood Institute P01 HL095491 (R.E.S.). J.T.M. received partial salary compensation and funding from Merck but has no conflict of interest with this work. J.T.M., J.M.M., R.E.S., R.W.M., R.E.B., and R.B. are Research Health Scientists at VA Boston Healthcare System, West Roxbury, MA. The contents of this work do not represent the views of the U.S. Department of Veterans Affairs or the United States Government.
Conflict of interest statement. None declared.
References
- 1. Fuentealba P, et al. . The reticular nucleus revisited: intrinsic and network properties of a thalamic pacemaker. Prog Neurobiol. 2005;75(2):125–141. [DOI] [PubMed] [Google Scholar]
- 2. Fuentealba P, et al. . Prolonged hyperpolarizing potentials precede spindle oscillations in the thalamic reticular nucleus. Proc Natl Acad Sci U S A. 2004;101(26):9816–9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nuñez A, et al. . Intracellular evidence for incompatibility between spindle and delta oscillations in thalamocortical neurons of cat. Neuroscience. 1992;48(1):75–85. [DOI] [PubMed] [Google Scholar]
- 4. Steriade M, et al. . Abolition of spindle oscillations in thalamic neurons disconnected from nucleus reticularis thalami. J Neurophysiol. 1985;54(6):1473–1497. [DOI] [PubMed] [Google Scholar]
- 5. Steriade M, et al. . The deafferented reticular thalamic nucleus generates spindle rhythmicity. J Neurophysiol. 1987;57(1):260–273. [DOI] [PubMed] [Google Scholar]
- 6. Diekelmann S, et al. . The memory function of sleep. Nat Rev Neurosci. 2010;11(2):114–126. [DOI] [PubMed] [Google Scholar]
- 7. Fogel SM, et al. . The function of the sleep spindle: a physiological index of intelligence and a mechanism for sleep-dependent memory consolidation. Neurosci Biobehav Rev. 2011;35(5):1154–1165. [DOI] [PubMed] [Google Scholar]
- 8. Miyamoto H, et al. . Reciprocal interaction of sleep and synaptic plasticity. Mol Interv. 2003;3(7):404–417. [DOI] [PubMed] [Google Scholar]
- 9. Schiffelholz T, et al. . Novel object presentation affects sleep-wake behavior in rats. Neurosci Lett. 2002;328(1):41–44. [DOI] [PubMed] [Google Scholar]
- 10. Eschenko O, et al. . Elevated sleep spindle density after learning or after retrieval in rats. J Neurosci. 2006;26(50):12914–12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mölle M, et al. . The influence of learning on sleep slow oscillations and associated spindles and ripples in humans and rats. Eur J Neurosci. 2009;29(5):1071–1081. [DOI] [PubMed] [Google Scholar]
- 12. Limoges E, et al. . Atypical sleep architecture and the autism phenotype. Brain. 2005;128(Pt 5):1049–1061. [DOI] [PubMed] [Google Scholar]
- 13. Latreille V, et al. . Sleep spindles in Parkinson’s disease may predict the development of dementia. Neurobiol Aging. 2015;36(2):1083–1090. [DOI] [PubMed] [Google Scholar]
- 14. Hermann DM, et al. . Evolution of neurological, neuropsychological and sleep-wake disturbances after paramedian thalamic stroke. Stroke. 2008;39(1):62–68. [DOI] [PubMed] [Google Scholar]
- 15. Keshavan MS, et al. . Sleep correlates of cognition in early course psychotic disorders. Schizophr Res. 2011;131(1-3):231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Astori S, et al. . Synaptic plasticity at intrathalamic connections via CaV3.3 T-type Ca2+ channels and GluN2B-containing NMDA receptors. J Neurosci. 2013;33(2): 624–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Davies G, et al. . A systematic review of the nature and correlates of sleep disturbance in early psychosis. Sleep Med Rev. 2017;31:25–38. [DOI] [PubMed] [Google Scholar]
- 18. Wamsley EJ, et al. . The effects of eszopiclone on sleep spindles and memory consolidation in schizophrenia: a randomized placebo-controlled trial. Sleep. 2013;36(9):1369–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Warby SC, et al. . Sleep-spindle detection: crowdsourcing and evaluating performance of experts, non-experts and automated methods. Nat Methods. 2014;11(4):385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ferrarelli F, et al. . Reduced sleep spindle activity in schizophrenia patients. Am J Psychiatry. 2007;164(3):483–492. [DOI] [PubMed] [Google Scholar]
- 21. Halassa MM, et al. . State-dependent architecture of thalamic reticular subnetworks. Cell. 2014;158(4):808–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wells MF, et al. . Thalamic reticular impairment underlies attention deficit in Ptchd1(Y/-) mice. Nature. 2016;532(7597):58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Phillips KG, et al. . Decoupling of sleep-dependent cortical and hippocampal interactions in a neurodevelopmental model of schizophrenia. Neuron. 2012;76(3):526–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gardner RJ, et al. . Differential spike timing and phase dynamics of reticular thalamic and prefrontal cortical neuronal populations during sleep spindles. J Neurosci. 2013;33(47):18469–18480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rothschild G, et al. . A cortical-hippocampal-cortical loop of information processing during memory consolidation. Nat Neurosci. 2017;20(2):251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Latchoumane C-FV, et al. . Thalamic spindles promote memory formation during sleep through triple phase-locking of cortical, thalamic, and hippocampal rhythms. Neuron. 2017;95(2):424–435.e6. [DOI] [PubMed] [Google Scholar]
- 27. Kim D, et al. . Characterization of topographically specific sleep spindles in mice. Sleep. 2015;38(1):85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vyazovskiy VV, et al. . The dynamics of spindles and EEG slow-wave activity in NREM sleep in mice. Arch Ital Biol. 2004;142(4):511–523. [PubMed] [Google Scholar]
- 29. Astori S, et al. . The Ca(V)3.3 calcium channel is the major sleep spindle pacemaker in thalamus. Proc Natl Acad Sci U S A. 2011;108(33):13823–13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Winsky-Sommerer R, et al. . Normal sleep homeostasis and lack of epilepsy phenotype in GABA A receptor alpha3 subunit-knockout mice. Neuroscience. 2008;154(2):595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alexandre C, et al. . Sleep-stabilizing effects of E-6199, compared to zopiclone, zolpidem and THIP in mice. Sleep. 2008;31(2):259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Uygun DS, et al. . Bottom-up versus top-down induction of sleep by Zolpidem acting on histaminergic and neocortex neurons. J Neurosci. 2016;36(44):11171–11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kopp C, et al. . Sleep EEG changes after zolpidem in mice. Neuroreport. 2004;15(14):2299–2302. [DOI] [PubMed] [Google Scholar]
- 34. Kim T, et al. . Cortically projecting basal forebrain parvalbumin neurons regulate cortical gamma band oscillations. Proc Natl Acad Sci U S A. 2015;112(11):3535–3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tobler I, et al. . Sleep and sleep regulation in normal and prion protein-deficient mice. J Neurosci. 1997;17(5):1869–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ang G, et al. . Absent sleep EEG spindle activity in GluA1 (Gria1) knockout mice: relevance to neuropsychiatric disorders. Transl Psychiatry. 2018;8(1):154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim A, et al. . Optogenetically induced sleep spindle rhythms alter sleep architectures in mice. Proc Natl Acad Sci U S A. 2012;109(50):20673–20678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ray LB, et al. . Expert and crowd-sourced validation of an individualized sleep spindle detection method employing complex demodulation and individualized normalization. Front Hum Neurosci. 2015;9:507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tsanas A, et al. . Stage-independent, single lead EEG sleep spindle detection using the continuous wavelet transform and local weighted smoothing. Front Hum Neurosci. 2015;9:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wimmer RD, et al. . Sustaining sleep spindles through enhanced SK2-channel activity consolidates sleep and elevates arousal threshold. J Neurosci. 2012;32(40):13917–13928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vyazovskiy VV, et al. . Local sleep in awake rats. Nature. 2011;472(7344):443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Andrillon T, et al. . Sleep spindles in humans: insights from intracranial EEG and unit recordings. J Neurosci. 2011;31(49):17821–17834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Benavides J, et al. . In vivo interaction of zolpidem with central benzodiazepine (BZD) binding sites (as labeled by [3H]Ro 15-1788) in the mouse brain. Preferential affinity of zolpidem for the omega 1 (BZD1) subtype. J Pharmacol Exp Ther. 1988;245(3):1033–1041. [PubMed] [Google Scholar]
- 44. Ni K-M, et al. . Selectively driving cholinergic fibers optically in the thalamic reticular nucleus promotes sleep. Elife. 2016;5:e10382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Steriade M, et al. . Intracellular analysis of relations between the slow (< 1 Hz) neocortical oscillation and other sleep rhythms of the electroencephalogram. J Neurosci. 1993;13(8):3266–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Franken P, et al. . Genetic variation in EEG activity during sleep in inbred mice. Am J Physiol. 1998;275(4 Pt 2):R1127–R1137. [DOI] [PubMed] [Google Scholar]
- 47. Lüthi A. Sleep spindles: where they come from, what they do. Neuroscientist. 2014;20(3):243–256. [DOI] [PubMed] [Google Scholar]
- 48. Steriade M, et al. . Reticularis thalami neurons revisited: activity changes during shifts in states of vigilance. J Neurosci. 1986;6(1):68–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lundahl J, et al. . EEG spectral power density profiles during NREM sleep for gaboxadol and zolpidem in patients with primary insomnia. J Psychopharmacol. 2012;26(8):1081–1087. [DOI] [PubMed] [Google Scholar]
- 50. Kaestner EJ, et al. . Pharmacologically increasing sleep spindles enhances recognition for negative and high-arousal memories. J Cogn Neurosci. 2013;25(10):1597–1610. [DOI] [PubMed] [Google Scholar]
- 51. Wamsley EJ, et al. . Reduced sleep spindles and spindle coherence in schizophrenia: mechanisms of impaired memory consolidation? Biol Psychiatry. 2012;71(2):154–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mölle M, et al. . Grouping of spindle activity during slow oscillations in human non-rapid eye movement sleep. J Neurosci. 2002;22(24):10941–10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Martin N, et al. . Topography of age-related changes in sleep spindles. Neurobiol Aging. 2013;34(2):468–476. [DOI] [PubMed] [Google Scholar]
- 54. Rosanova M, et al. . Pattern-specific associative long-term potentiation induced by a sleep spindle-related spike train. J Neurosci. 2005;25(41):9398–9405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Espinosa F, et al. . Ablation of Kv3.1 and Kv3.3 potassium channels disrupts thalamocortical oscillations in vitro and in vivo. J Neurosci. 2008;28(21):5570–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]