Abstract
Introduction
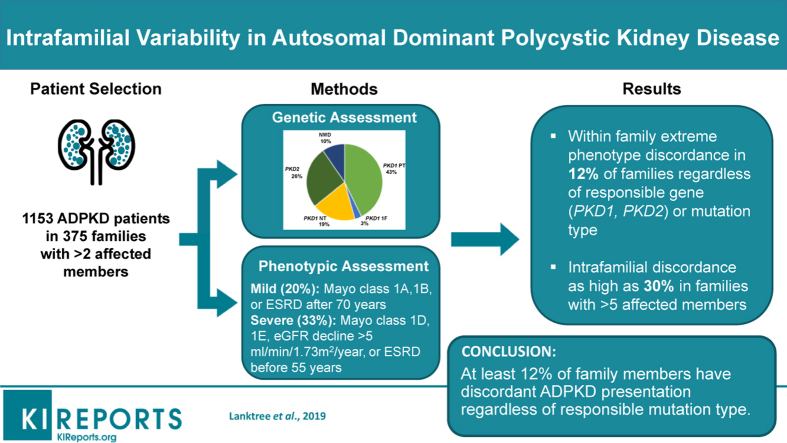
Discordance in kidney disease severity between affected relatives is a recognized feature of autosomal dominant polycystic kidney disease (ADPKD). Here, we report a systematic study of a large cohort of families to define the prevalence and clinical features of intrafamilial discordance in ADPKD.
Methods
The extended Toronto Genetic Epidemiology Study of Polycystic Kidney Disease (eTGESP) cohort includes 1390 patients from 612 unrelated families with ADPKD ascertained in a regional polycystic kidney disease center. All probands underwent comprehensive PKD1 and PKD2 mutation screening. Total kidney volume by magnetic resonance imaging (MRI) was available in 500 study patients.
Results
Based on (i) rate of estimated glomerular filtration rate (eGFR) decline, (ii) age at onset of end-stage renal disease (ESRD), and (iii) Mayo Clinic Imaging Classification (MCIC), 20% of patients were classified as having mild disease, and 33% as having severe disease. Intrafamilial ADPKD discordance with at least 1 mild and 1 severe case was observed in 43 of 371 (12%) families, at a similar frequency regardless of the responsible gene (PKD1/PKD2/no mutation detected) or mutation type (protein-truncating versus nontruncating). Intrafamilial discordance was more common in larger families and was present in 30% of families with more than 5 affected members. The heritability of age at onset of ESRD was similar between different mutation types.
Conclusion
Extreme kidney disease discordance is present in at least 12% of families with ADPKD, regardless of the underlying mutated gene or mutation class. Delineating genetic and environmental modifiers underlying the observed intrafamilial ADPKD variability will provide novel insights into the mechanisms of progression in ADPKD.
Keywords: polycystic kidney disease, ADPKD, genetics
Graphical abstract
ADPKD is the most common hereditary kidney disease worldwide, characterized by bilateral kidney enlargement with numerous cysts and a variable rate of chronic kidney disease (CKD) progression. A large proportion of patients affected with ADPKD also suffer from hypertension, kidney stones, and urinary tract infections, as well as other extrarenal complications, including polycystic liver disease and intracranial artery aneuryms.1 Up to 70% of patients with ADPKD develop ESRD by the age of 70 years.2 However, some patients die at an elderly age with intact renal function whereas others develop ESRD in early or middle adulthood, illustrating the wide spectrum of kidney disease severity in ADPKD.2 Assessing the risk of progression in ADPKD has become of great clinical importance since the approval of tolvaptan, the first mechanism-based drug treatment approved for patients with high risk for progression.3, 4 In this regard, a family history of an older affected relative who developed ESRD at or before age 55 years is thought to be predictive of severe disease in the family.5 By contrast, a positive history of an affected relative who remained renal sufficient at age 70 is thought to be predictive of mild disease in the family.5 However, intrafamilial renal disease variability is a well-documented feature of ADPKD, making risk prediction at the level of the individual patient challenging, even among affected relatives. Comparison of 56 sibships with ADPKD showed a 6.9-year mean difference in the age at onset of ESRD, and 9 sets of monozygotic twins had a 2.1-year mean difference.6 In addition, a significant heritability for both creatinine clearance and age at onset of ESRD was found in 406 affected relatives from 66 PKD1-linked families.7 Taken together, these findings suggest that genetic and environmental modifiers may affect within-family renal disease variability in ADPKD.
In clinically ascertained samples, mutations in PKD1 and PKD2 are responsible for 60%–78% and 15%–26% of ADPKD, respectively.8, 9 About 10%–15% of patients with apparent ADPKD have no identifiable PKD1 or PKD2 mutation,10 and whole-exome sequencing studies have identified additional genes (i.e., GANAB, DNAJB11) mutated in a small proportion of patients (<1%).11, 12 Both genic and allelic heterogeneity contribute to phenotype severity in ADPKD. Mutations in PKD1 lead to more-severe disease with larger kidneys and earlier development of ESRD compared to mutations in PKD2.8, 9 Protein-truncating mutations, caused by nonsense, frameshift, and canonical splice-site mutations, and large insertions/deletions in general, lead to more-severe disease than do nontruncating mutations, including inframe insertion/deletions (Indels), missense mutations, and atypical splicing mutations.8, 9
In addition to family history of renal disease severity and ADPKD mutation class for prognostication, the MCIC employs age-adjusted total kidney volume (TKV) to provide risk assessment of kidney disease progression in ADPKD and identification of high-risk patients for enrollment into clinical trials.13, 14 The MCIC approach is based on the findings of the Consortium for Radiological Imaging Studies of Polycystic Kidney Disease (CRISP), which documented that TKV on average increases at approximately 5% per year during adulthood in patients with ADPKD and predicts future CKD progression.15, 16 A TKV-based prognostic biomarker is particularly useful for identifying patients at high risk for CKD progression before the onset of kidney failure, which typically occurs late in the clinical course.15
Although intrafamilial renal disease discordance among affected relatives is a recognized feature of ADPKD, defining its prevalence is challenging, in part due to a lack of large, family-based cohort studies and robust risk assessment tools to identify patients with severe disease before the onset of CKD. In the current study, we used the following measures: (i) the rate of eGFR decline, (ii) age at onset of ESRD, and/or (iii) MCIC to assess ADPKD kidney disease severity in 1390 patients from 612 unrelated families ascertained through the eTGESP. We identified all families with 2 or more affected relatives with available clinical data on kidney disease severity, and determined the prevalence and clinical features of intrafamilial kidney disease discordance.
Methods
Study Population
The eTGESP cohort comprised consecutive patients seen at a regional PKD center at the Toronto General Hospital from December 1, 2006 to December 30, 2017. Patients were referred from more than 100 academic and community nephrologists at the Greater Toronto Area and from St. Joseph’s Healthcare in Hamilton, Ontario, Canada. Patients were referred for early risk stratification and genetic assessment, not at the point of ESRD for transplant assessment. All except 2% of patients seen participated in the current study and provided consent according to a prespecified protocol approved by the institutional review board at the University Health Network in Toronto, Ontario, Canada.
Measurement of Exposures and Outcomes
Date of birth, age at recruitment, and last follow-up was available for all patients. Age of death and development of ESRD was obtained from patient history or medical chart when available. Using age, sex, race, and serum creatinine measurements, eGFR was estimated using the Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) equation.17 Rate of decline of kidney function was determined using linear regression of available CKD-EPI eGFR values in all patients with more than 3 measurements over more than 1 year. Ultrasound- or MRI-based criteria confirmed the ADPKD diagnosis in all probands.18 All MR images were reviewed and classified as typical (class 1) or atypical (class 2).15 MRI-derived TKV was estimated by an experienced radiologist using the ellipsoid method and coupled with age at MRI to generate the MCIC risk class.15 All cases with atypical PKD on renal imaging (i.e., MCIC class 2) were excluded from the current study. Genetic testing was performed in a single research laboratory in Toronto using a validated long-range polymerase chain reaction protocol and bidirectional sequencing of coding region and splice junctions of PKD1 and PKD2.8, 9 As previously reported, all nonsense, frameshift, and canonical splice-site mutations were grouped as protein-truncating mutations, and nonsynonymous missense or atypical splice-site mutations were grouped as non-truncating mutations. All nontruncating mutations and inframe insertions/deletions were evaluated for their potential pathogenicity using prediction algorithms (Align GVGD, PolyPhen-2, SIFT, PROVEAN, and Human Splicing Finder), review of the PKD mutation database (http://pkdb.mayo.edu), and segregation analysis with additional affected family members when possible.8, 9 All mutation-negative patients were re-screened by multiplex ligation–dependent probe amplification for detection of large gene rearrangements.19
Assessment of ADPKD Severity
In order to identify families with discordant disease severity, we first evaluated all patients individually to score their disease severity using stringent criteria. Mild disease was defined as (i) ESRD or renal sufficiency at the age of 70 or more years, or (ii) an MCIC risk class of 1A or 1B. Severe disease was defined as presence of at least one of the following criteria: (i) development of ESRD before the age of 55 years; (ii) an MCIC risk class of 1D or 1E; and (iii) >5 ml/min per 1.73 m2 per-year loss in eGFR with at least 3 measurements over at least 1 year. Patients who did not satisfy any of the above criteria had intermediate or indeterminate disease severity.
Statistical Analysis
We compared the prevalence of intrafamilial discordance between mutation types using Pearson’s χ2, and trends by family size were determined using the Cochrane-Armitage test for trend. The 95% confidence intervals (95% CIs) for binomial proportions were calculated using the normal approximation (Wald) interval. We compared age of ESRD onset or age at censoring between mutation classes (PKD1 protein-truncating, PKD1 nonprotein-truncating, PKD1 in frame insertion/deletion, PKD2, and no mutation detected) by Kaplan-Meyer survival analysis. To account for the familial relationship and biological sex in the analysis, we used a mixed-effects Cox proportional hazard model, as implemented in the R package “coxme” (https://cran.r-project.org/web/packages/coxme/index.html). Each sample was assumed to have an individual random effect/risk of reaching ESRD, but those risks were correlated between family members according to the distance of the familial relationship.
Heritability was defined as the proportion of trait variability attributable to additive genetic effects. We estimated heritability using SOLAR-Eclipse (v.8.1.1, http://www.solar-eclipse-genetics.org/), which implements the variance components method. All familial ADPKD patients were connected by creation of common ancestors with phenotypes coded as unknown. We first fitted an intercept-only Cox proportional hazard model, with age at ESRD or censoring as the survival outcome variable, and biological sex as a covariate. The deviance residuals were used as the quantitative trait variable in SOLAR-Eclipse. Mutation-specific heritability was compared using 2-sided 2-sample t-tests.
Results
Assessing Kidney Disease Severity in the eTGESP Cohort
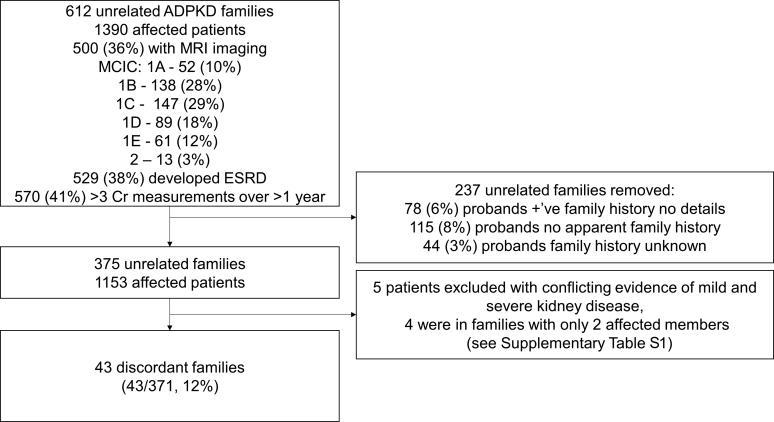
In total, in this study we examined 1390 patients from 612 unrelated families from the eTGESP cohort (Figure 1). The probands of all study families were screened for PKD1 and PKD2 mutations. Five-hundred patients had MRI-derived TKV and MCIC risk assessment; 529 patients had reached ESRD; and 89 patients died without development of ESRD. A total of 237 (17%) patients were excluded due to: (i) positive family history of ADPKD but no available clinical data in any of the affected relatives (78 cases [6%]); (ii) no apparent family history of ADPKD (115 cases [8%]); or (iii) unknown and unavailable family history (44 cases [3%]). Of the remaining 375 unrelated families containing at least 2 affected relatives, 241 (64%) had a PKD1 mutation (protein-truncating: 160 of 375 [43%]; inframe Indel: 11 of 375 [3%]; nontruncating: 70 of 375 [19%]), 98 (26%) had a PKD2 mutation, and 36 (10%) had no detected mutation (Supplementary Figure S1). Two families had evidence of biallelic inheritance: one with both a PKD1 missense mutation (c.6395T>G; p.F2132C) and frameshift insertion (c.11359_11360delAG; p.P3788fs), and a second with a PKD2 truncating mutation (c.2159_2160InsA; p.N720fs) and a PKD1 missense variant (c.8299C>T; p.R2767C). Families with more affected members were not enriched for severe mutations (P = 0.84; Supplementary Figure S2).
Figure 1.
Study flow diagram of the extended Toronto Genetic Epidemiology Study of Polycystic Kidney Disease cohort. ADPKD, autosomal dominant polycystic kidney disease; Cr, creatinine; ESRD, end-stage renal disease; MCIC, Mayo Clinic Imaging Classification; MRI, magnetic resonance imaging.
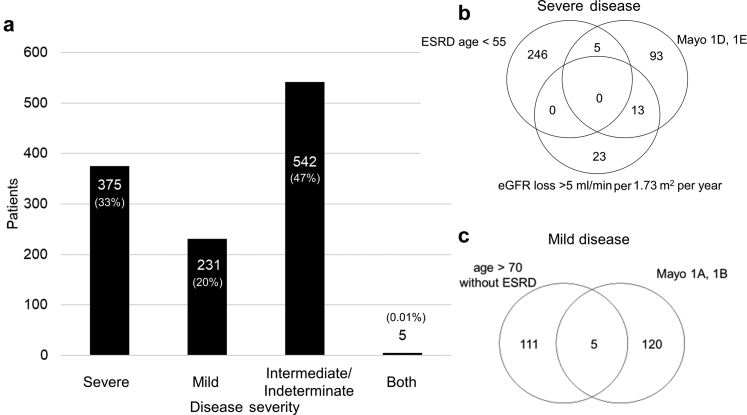
Overall, 231 of 1153 (20%) patients were classified as having mild disease; 375 of 1153 (33%) patients had severe disease; and 542 of 1153 (47%) did not have evidence of either mild or severe disease and were classified as intermediate or indeterminate (Figure 2). A total of 302 of 1153 (26%) patients had intermediate disease, including 220 who reached ESRD between the ages of 55 and 70 years, and 84 who had an MCIC of 1C; 2 patients had both an MCIC 1C and reached ESRD between age 55 and 70 years. A total of 240 of 1153 (21%) patients had indeterminate disease, including: 63 who died before age 70 years without renal risk stratification but without ESRD, 30 of whom were above age 55 years; and 177 patients between the ages of 18 and 70 years with renal sufficiency and no imaging available, 53 of whom were above age 55 years. The mean age at last follow-up, and gender, for those with severe, intermediate or indeterminate, and mild disease were, respectively, 45.7 years (SD = 9.9), 54% male; 53.6 years (SD = 14.0), 45% male; and 61.4 years (SD = 19.8), 39% male (Supplementary Figure S3).
Figure 2.
Kidney disease severity in the extended Toronto Genetic Epidemiology Study of Polycystic Kidney Disease cohort. Proportion of patients with mild, intermediate or indeterminate, or severe kidney disease (a). Five patients had conflicting assessment of kidney disease severity (i.e., evidence of both mild and severe disease; see Supplementary Table S1). Venn diagrams showing criteria used for defining the severe (b) and mild (c) cases. eGFR, estimated glomerular filtration rate; ESRD, end-stage renal disease; Mayo, Mayo Clinic Imaging Classification.
Five patients (0.4%) showed conflicting evidence of both mild and severe disease (Supplementary Table S1). All 5 were young women (mean age: 29.6 years) with small kidneys (1A or 1B) and eGFR >90 ml/min per 1.23 m2, but with an average eGFR decline >5 ml/min per 1.73 m2 per year over 1.4–3.5 years of follow-up. Three had PKD1 protein-truncating mutations, one had a PKD1 nontruncating mutation (p.G3651S), and one had a PKD2 mutation. All had a family history of intermediate-to-severe ADPKD. Due to the conflict in rating their renal disease severity, they were excluded from further analysis. Four of the excluded patients were in families with 2 affected members, thus reducing the total number of families with at least 2 affected members from 375 to 371. Fifteen patients had both MCIC and age at ESRD available, and none had discrepant risk classifications.
In keeping with previous observations, patients with severe disease were more likely to have a PKD1 mutation (275 of 375 [73%]) than those with intermediate/indeterminate disease severity (218 of 542 [40%]) or mild disease (27 of 231 [12%]; P < 1 x 10–5; Supplementary Figure S4).8 Those with mild disease were more likely to have a PKD2 mutation (114 of 231 [49%]) than those with intermediate/indeterminate disease severity (156 of 542 [29%]) or those with severe disease (29 of 375 [8%]). Patients with severe disease were also more likely to have a protein-truncating mutation, whereas those with mild disease were more likely to have a nontruncating mutation (P < 1 x 10–5; Supplementary Figure S4). Similar to our previous study, survival analysis showed that mutation type was a significant predictor of the age at onset of ESRD (likelihood ratio test P < 2.2 x 10–16; Supplementary Figure S5).8 Moderate heritability (35%–70%) was observed for age at ESRD onset across major mutation classes but did not significantly differ between the classes (P > 0.05; Table 1).
Table 1.
Heritability of age at onset of end-stage renal disease is similar regardless of gene or mutation type
| Mutation type | h2 (SE) | P comparison versus not heritable | P comparison versus PKD1-PT | P comparison versus PKD1-NT |
|---|---|---|---|---|
| PKD1-PT | 0.45 (0.12) | 0.00001 | — | 0.28 |
| PKD1-NT | 0.70 (0.19) | 0.00009 | 0.28 | — |
| PKD2 | 0.35 (0.20) | 0.03 | 0.65 | 0.23 |
h2, heritability; NT, nontruncating; PT, protein-truncating.
Intrafamilial Kidney Disease Variability
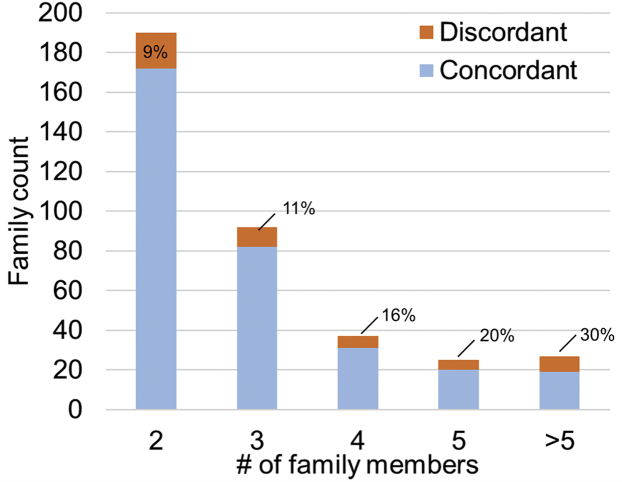
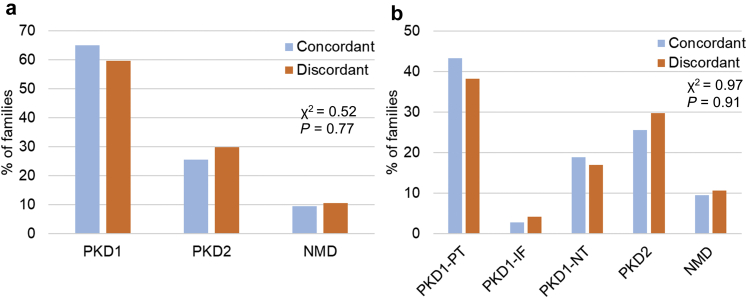
Next, we determined the minimal prevalence and associated clinical features of intrafamilial renal disease discordance in our study cohort. Of 371 families with data on at least 2 affected family members, 43 (12%, 95% CI: 8%–15%) had evidence of intrafamilial discordance of their kidney disease severity, defined as having at least 1 affected relative with mild disease and 1 affected relative with severe disease. If indeterminate patients were excluded from the analysis, we observed intrafamilial discordance in 43 of 292 (15%, 95% CI: 11%–19%) families. Only 45 families had at least 2 affected relatives with MCIC risk assessment, and 7 of these 45 (16%, 95% CI: 5%–26%) displayed discordant renal disease severity (1 family member with MCIC A or B, and another family member with MCIC D or E). As expected, the prevalence of intrafamilial renal disease discordance increased with family size: 14 of 190 (7%, 95% CI: 4%–11%) families with 2 affected relatives, increasing to 8 of 27 (30%, 95% CI: 12%–47%) of families with more than 5 affected relatives displayed disease discordance (Figure 3). There was no significant difference in the prevalence of discordant families by the gene mutated (i.e., PKD1 vs. PKD2) or by mutation type (i.e., PKD1 PT mutations, PKD1 inframe indels, PKD1 NT mutations, PKD2 mutations, and no mutation detected, P > 0.5; Figure 4).
Figure 3.
Large families are more likely to display discordance in kidney disease severity. Families were defined as discordant if they had at least 1 affected member with mild kidney disease and 1 member with severe kidney disease. Cochrane-Armitage test for trend: Z = 2.82; P = 0.005.
Figure 4.
Presence of PKD1, PKD2, or no mutation detected (NMD) is not associated with intrafamilial discordance of kidney disease severity in autosomal dominant polycystic kidney disease (ADPKD). Comparison of gene (a) and mutation type (b) responsible for ADPKD in families with concordant and discordant intrafamilial kidney disease severity. IF, inframe insertion/deletion; NT, nontruncating; PT, protein-truncating.
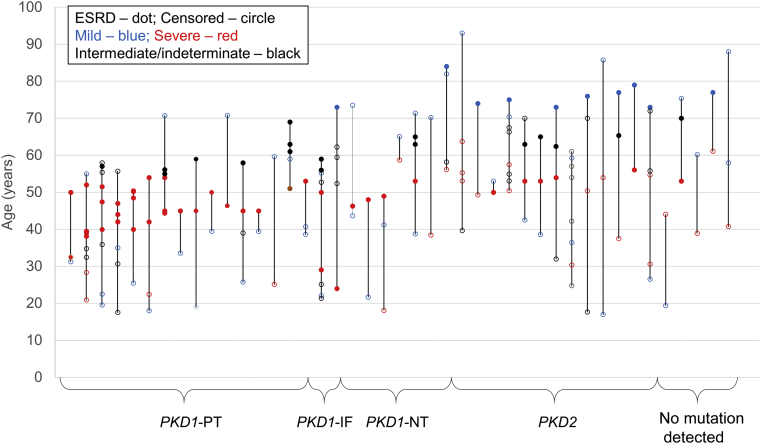
Families with PKD1 PT mutations on average have the most-severe renal disease compared to all the other mutation classes (Supplementary Figure S5). In these families with severe mutations, the use of MCIC (1A and 1B) identified 88% (14 of 16) of the mildly affected relatives, and a history of “renal sufficiency at the age of 70 years or older” identified the remaining 2 (Figure 5). Conversely, families with PKD2 mutations on average have the mildest renal disease compared to all the other mutation classes (Supplementary Figure S5). In these families, the use of MCIC (1D and 1E) identified 62% (8 of 13) of the severely affected relatives, and a history of “ESRD before the age of 55 years” identified the remaining 5 (Figure 5).
Figure 5.
Families display intrafamilial disease discordance regardless of responsible underlying mutation type. Each vertical line represents 1 family with intrafamilial discordance, and each dot represents a single patient. Filled dots represent age at end-stage renal disease (ESRD), and empty circles are age of censor (death or last follow-up with renal sufficiency). IF, in-frame insertion/deletion; NT, nontruncating; PT, protein-truncating.
Discussion
In this large prospective study of clinically ascertained families with ADPKD recruited from a single geographic region, we found a minimal prevalence estimate of intrafamilial kidney disease discordance of 12%, defined by the presence of at least 1 mildly affected relative and 1 severely affected relative, using stringent criteria. Among the study patients, 20% had mild disease, 33% had severe disease, and 47% had intermediate or indeterminate disease severity. In keeping with previous observations, those with severe disease were more likely to have PKD1 truncating mutations, whereas those with mild disease were more likely to have PKD2 mutations and nontruncating PKD1 mutations. Heritability estimates of the age at ESRD onset were similar regardless of ADPKD mutation type, indicating that additional genetic and environmental factors contribute to and modify the kidney disease severity, regardless of the main ADPKD mutation type. Families with kidney disease discordance between affected relatives were equally likely to have PKD1 or PKD2 mutations, and equally likely to have protein-truncating or nontruncating mutations.
The use of multiple criteria (i.e., age at ESRD, MCIC risk class, and slope of eGFR decline) to define disease severity allowed us to increase the number of informative families for evaluation and include patients from multiple generations. For younger patients, the TKV-based MCIC risk class provides a robust measure of disease severity, as eGFR often remains within the normal range for the first 3 to 4 decades of life. In contrast, late-onset ESRD or continued renal sufficiency despite old age (above 70 years) provides a simple means to identify patients with mild disease, whereas early-onset ESRD (before age 55 years) is indicative of severe disease. Including all 3 measures produces an effective method to examine intergenerational discordance in kidney disease severity over a wide range of CKD stages. As would be expected, larger families, by providing additional affected relatives for comparison, yielded a higher prevalence of intrafamilial kidney disease discordance than did small families. Complete evaluation of all affected relatives and deep phenotyping, such as TKV in all participants, may uncover additional families with intrafamilial kidney disease discordance. We put great effort into obtaining clinical records for family members of probands, but complete records were unavailable at times. Thus, our estimate of intrafamilial kidney disease discordance should be viewed as a lower-bound estimate.
Study of intrafamilial kidney disease discordance provides an opportunity to delineate genetic and environmental modifiers and improve our ability to predict the rate of kidney disease progression in ADPKD. One potential explanation for intrafamilial disease discordance is compound heterozygosity or digenic inheritance of an additional mutation in a cystogenic gene, including PKD1 and PKD2. Our previous work described a family with bilineal PKD1 inframe indel and PKD1 missense mutations, resulting in intrafamilial discordance similar to that described here.8 Overall, bilineal mutations in PKD1 and PKD2 appear uncommon, accounting for only 2 families identified here. Mosaicism, in which 2 populations of cells with different genotypes exist in the same person, can lead to intrafamilial kidney disease discordance. We previously described a family in eTGESP with somatic mosaicism, including an affected mother with germline transmission of a PKD1 frameshift deletion to her daughter.10 All the daughter’s cells carried the heterozygous PKD1 frameshift mutation, leading to a 1:1 ratio of wild type to mutant alleles and a severe phenotype, whereas her mother had a 10:1 ratio of wild type to mutant alleles in blood and a mild phenotype. Mosaicism is difficult to detect with Sanger (capillary) sequencing, owing to a low signal-to-noise ratio, and the prevalence of mosaicism in ADPKD remains poorly defined. However, the recognition of somatic mosaicism will improve with the advance of next-generation sequencing and likely accounts for a proportion of cases displaying intrafamilial kidney disease discordance in ADPKD.20
Mutations in additional disease modifiers, including COL4A1 and HNF1B,21, 22 have been described and could create intrafamilial variation if 1 member carries the additional mutation and the other does not. Mutations in genes responsible for other cystic disease, such as autosomal dominant polycystic liver disease and tuberous sclerosis complex, are also possible candidates. In our recent analyses of whole genome and exome sequencing of the general population, 1% of the population was found to carry a truncation mutation, and 23% of the population was found to carry rare missense mutations in genes with potential to modify the kidney disease phenotype in ADPKD.23 Common genetic variants may also modify the kidney disease severity of ADPKD, as exemplified by the variant association observed with DKK3,24 and a polygenic component with numerous genetic variants with small effects cumulatively contributing to disease progression is possible.25 On the other hand, concomitant clinical conditions, including obesity, diabetes, vascular disease, and acute kidney injury, and environmental factors such as cigarette smoking, diet, and water intake could potentially alter the rate of progression and contribute to intrafamilial kidney disease discordance in ADPKD. Changes in intervention rates (i.e., prescription of angiotensin-converting enzyme inhibitors) or secular changes, such as increasing prevalence of obesity, could also lead to differences in disease severity between generations of affected relatives from the same family.
In conclusion, by using multiple stringent criteria to define ADPKD severity, we found a minimal prevalence of kidney disease discordance in 11% of our study families regardless of their underlying mutated gene or mutation class. Potential causes for the observed variability include bilineal ADPKD and somatic mosaicism for some families, but much of the intrafamilial kidney disease discordance remains unexplained. Future studies of families with intrafamilial kidney disease discordance by next-generation sequencing targeting a panel of potential cystic or CKD modifier genes have the potential to uncover genetic contributors to kidney disease progression in ADPKD.
Disclosure
MBL and YP received compensation for participating in advisory and consultancy boards with Otsuka pharmaceuticals. All the other authors declared no competing interests.
Acknowledgments
MBL received a Ben Lipps Post-Doctoral Fellowship from the American Society of Nephrology and is a fellow of the Krescent Program, a national kidney research training partnership of the Kidney Foundation of Canada, the Canadian Society of Nephrology, and the Canadian Institutes of Health Research. The Canadian Institutes of Health Research Strategy for Patient Oriented Research in Chronic Kidney Disease program grant to YP supported this work.
MBL, EG, and YP designed the study; MBL and EG analyzed the data, produced the figures, and drafted the paper; WL and AP provided statistical support; the remaining authors contributed data collection and validation, and edited and approved the final version of the manuscript. The authors thank the patients and their families, as well as the administrative and research staff of the Center for Innovative Management of PKD at Toronto General Hospital and St. Joseph’s Healthcare, Hamilton, Toronto, Canada.
Footnotes
Figure S1. Prevalence of different mutation types in extended Toronto Genetic Epidemiology Study of Polycystic Kidney Disease (eTGESP) families with clinical information available for 2 or more affected relatives.
Figure S2. Mild mutations were equally prevalent in large families.
Figure S3. Age of patients with severe, intermediate/indeterminate, and mild ADPKD.
Figure S4. Protein-truncating PKD1 mutations are more common in patients with severe kidney disease.
Figure S5. Survival analysis by responsible ADPKD mutation type.
Table S1. Cases with conflicting evidence for both mild and severe kidney disease.
Supplementary Material
Figure S1. Prevalence of different mutation types in extended Toronto Genetic Epidemiology Study of Polycystic Kidney Disease (eTGESP) families with clinical information available for 2 or more affected relatives.
Figure S2. Mild mutations were equally prevalent in large families.
Figure S3. Age of patients with severe, intermediate/indeterminate, and mild ADPKD.
Figure S4. Protein-truncating PKD1 mutations are more common in patients with severe kidney disease.
Figure S5. Survival analysis by responsible ADPKD mutation type.
Table S1. Cases with conflicting evidence for both mild and severe kidney disease.
References
- 1.Lanktree M.B., Chapman A.B. New treatment paradigms for ADPKD: moving towards precision medicine. Nat Rev Nephrol. 2017;13:750–768. doi: 10.1038/nrneph.2017.127. [DOI] [PubMed] [Google Scholar]; Lanktree MB, Chapman AB. New treatment paradigms for ADPKD: moving towards precision medicine. Nat Rev Nephrol. 2017;13:750-768. [DOI] [PubMed]
- 2.Spithoven E.M., Kramer A., Meijer E. Renal replacement therapy for autosomal dominant polycystic kidney disease (ADPKD) in Europe: prevalence and survival─an analysis of data from the ERA-EDTA Registry. Nephrol Dial Transplant. 2014;29(suppl 4):iv15–iv25. doi: 10.1093/ndt/gfu017. [DOI] [PMC free article] [PubMed] [Google Scholar]; Spithoven EM, Kramer A, Meijer E, et al. Renal replacement therapy for autosomal dominant polycystic kidney disease (ADPKD) in Europe: prevalence and survival─an analysis of data from the ERA-EDTA Registry. Nephrol Dial Transplant. 2014;29(suppl 4):iv15-iv25. [DOI] [PMC free article] [PubMed]
- 3.Torres V.E., Chapman A.B., Devuyst O. Tolvaptan in later-stage autosomal dominant polycystic kidney disease. N Engl J Med. 2017;377:1930–1942. doi: 10.1056/NEJMoa1710030. [DOI] [PubMed] [Google Scholar]; Torres VE, Chapman AB, Devuyst O, et al. Tolvaptan in later-stage autosomal dominant polycystic kidney disease. N Engl J Med. 2017;377:1930-1942. [DOI] [PubMed]
- 4.Torres V.E., Chapman A.B., Devuyst O. Tolvaptan in patients with autosomal dominant polycystic kidney disease. N Engl J Med. 2012;367:2407–2418. doi: 10.1056/NEJMoa1205511. [DOI] [PMC free article] [PubMed] [Google Scholar]; Torres VE, Chapman AB, Devuyst O, et al. Tolvaptan in patients with autosomal dominant polycystic kidney disease. N Engl J Med. 2012;367:2407-2418. [DOI] [PMC free article] [PubMed]
- 5.Barua M., Cil O., Paterson A.D. Family history of renal disease severity predicts the mutated gene in ADPKD. J Am Soc Nephrol. 2009;20:1833–1838. doi: 10.1681/ASN.2009020162. [DOI] [PMC free article] [PubMed] [Google Scholar]; Barua M, Cil O, Paterson AD, et al. Family history of renal disease severity predicts the mutated gene in ADPKD. J Am Soc Nephrol. 2009;20:1833-1838. [DOI] [PMC free article] [PubMed]
- 6.Persu A., Duyme M., Pirson Y. Comparison between siblings and twins supports a role for modifier genes in ADPKD. Kidney Int. 2004;66:2132–2136. doi: 10.1111/j.1523-1755.2004.66003.x. [DOI] [PubMed] [Google Scholar]; Persu A, Duyme M, Pirson Y, et al. Comparison between siblings and twins supports a role for modifier genes in ADPKD. Kidney Int. 2004;66:2132-2136. [DOI] [PubMed]
- 7.Paterson A.D., Magistroni R., He N. Progressive loss of renal function is an age-dependent heritable trait in type 1 autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2005;16:755–762. doi: 10.1681/ASN.2004090758. [DOI] [PubMed] [Google Scholar]; Paterson AD, Magistroni R, He N, et al.Progressive loss of renal function is an age-dependent heritable trait in type 1 autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2005;16:755-762. [DOI] [PubMed]
- 8.Hwang Y.-H., Conklin J., Chan W. Refining genotype-phenotype correlation in autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2016;27:1861–1868. doi: 10.1681/ASN.2015060648. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hwang Y-H, Conklin J, Chan W, et al. Refining genotype-phenotype correlation in autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2016;27:1861-1868. [DOI] [PMC free article] [PubMed]
- 9.Heyer C.M., Sundsbak J.L., Abebe K.Z. Predicted mutation strength of nontruncating PKD1 mutations aids genotype-phenotype correlations in autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2016;27:2872–2884. doi: 10.1681/ASN.2015050583. [DOI] [PMC free article] [PubMed] [Google Scholar]; Heyer CM, Sundsbak JL, Abebe KZ, et al. Predicted mutation strength of nontruncating PKD1 mutations aids genotype-phenotype correlations in autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2016;27:2872-2884. [DOI] [PMC free article] [PubMed]
- 10.Iliuta I.-A., Kalatharan V., Wang K. Polycystic Kidney Disease without an apparent family history. J Am Soc Nephrol. 2017;28:2768–2776. doi: 10.1681/ASN.2016090938. [DOI] [PMC free article] [PubMed] [Google Scholar]; Iliuta I-A, Kalatharan V, Wang K, et al. Polycystic Kidney Disease without an apparent family history. J Am Soc Nephrol. 2017;28:2768-2776. [DOI] [PMC free article] [PubMed]
- 11.Porath B., Gainullin V.G., Cornec-Le Gall E. Mutations in GANAB, encoding the glucosidase IIα subunit, cause autosomal-dominant polycystic kidney and liver disease. Am J Hum Genet. 2016;98:1193–1207. doi: 10.1016/j.ajhg.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Porath B, Gainullin VG, Cornec-Le Gall E, et al. Mutations in GANAB, encoding the glucosidase IIα subunit, cause autosomal-dominant polycystic kidney and liver disease. Am J Hum Genet. 2016;98:1193-1207. [DOI] [PMC free article] [PubMed]
- 12.Cornec-Le Gall E., Olson R.J., Besse W. Monoallelic mutations to DNAJB11 cause atypical autosomal-dominant polycystic kidney disease. Am J Hum Genet. 2018;102:832–844. doi: 10.1016/j.ajhg.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cornec-Le Gall E, Olson RJ, Besse W, et al. Monoallelic mutations to DNAJB11 cause atypical autosomal-dominant polycystic kidney disease. Am J Hum Genet. 2018;102:832-844. [DOI] [PMC free article] [PubMed]
- 13.Perrone R.D., Mouksassi M.-S., Romero K. A drug development tool for trial enrichment in patients with autosomal dominant polycystic kidney disease. Kidney Int Rep. 2017;2:451–460. doi: 10.1016/j.ekir.2017.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; Perrone RD, Mouksassi M-S, Romero K, et al.A drug development tool for trial enrichment in patients with autosomal dominant polycystic kidney disease. Kidney Int Rep. 2017;2:451-460. [DOI] [PMC free article] [PubMed]
- 14.Irazabal M.V., Abebe K.Z., Bae K.T. Prognostic enrichment design in clinical trials for autosomal dominant polycystic kidney disease: the HALT-PKD clinical trial. Nephro Dial Transplant. 2017;32:1857–1865. doi: 10.1093/ndt/gfw294. [DOI] [PMC free article] [PubMed] [Google Scholar]; Irazabal MV, Abebe KZ, Bae KT, et al. Prognostic enrichment design in clinical trials for autosomal dominant polycystic kidney disease: the HALT-PKD clinical trial. Nephro Dial Transplant. 2017;32:1857-1865. [DOI] [PMC free article] [PubMed]
- 15.Irazabal M.V., Rangel L.J., Bergstralh E.J. Imaging classification of autosomal dominant polycystic kidney disease: a simple model for selecting patients for clinical trials. J Am Soc Nephrol. 2015;26:160–172. doi: 10.1681/ASN.2013101138. [DOI] [PMC free article] [PubMed] [Google Scholar]; Irazabal MV, Rangel LJ, Bergstralh EJ, et al. Imaging classification of autosomal dominant polycystic kidney disease: a simple model for selecting patients for clinical trials. J Am Soc Nephrol. 2015;26:160-172. [DOI] [PMC free article] [PubMed]
- 16.Yu A.S.L., Shen C., Landsittel D.P. Baseline total kidney volume and the rate of kidney growth are associated with chronic kidney disease progression in autosomal dominant polycystic kidney disease. Kidney Int. 2018;93:691–699. doi: 10.1016/j.kint.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yu ASL, Shen C, Landsittel DP, et al. Baseline total kidney volume and the rate of kidney growth are associated with chronic kidney disease progression in autosomal dominant polycystic kidney disease. Kidney Int. 2018;93:691-699. [DOI] [PMC free article] [PubMed]
- 17.Levey A.S., Stevens L.A. Estimating GFR using the CKD Epidemiology Collaboration (CKD-EPI) creatinine equation: more accurate GFR estimates, lower CKD prevalence estimates, and better risk predictions. Am J Kidney Dis. 2010;55:622–627. doi: 10.1053/j.ajkd.2010.02.337. [DOI] [PMC free article] [PubMed] [Google Scholar]; Levey AS, Stevens LA. Estimating GFR using the CKD Epidemiology Collaboration (CKD-EPI) creatinine equation: more accurate GFR estimates, lower CKD prevalence estimates, and better risk predictions. Am J Kidney Dis. 2010;55:622-627. [DOI] [PMC free article] [PubMed]
- 18.Pei Y., Hwang Y.-H., Conklin J. Imaging-based diagnosis of autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2015;26:746–753. doi: 10.1681/ASN.2014030297. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pei Y, Hwang Y-H, Conklin J, et al. Imaging-based diagnosis of autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2015;26:746-753. [DOI] [PMC free article] [PubMed]
- 19.Consugar M.B., Wong W.C., Lundquist P.A. Characterization of large rearrangements in autosomal dominant polycystic kidney disease and the PKD1/TSC2 contiguous gene syndrome. Kidney Int. 2008;74:1468–1479. doi: 10.1038/ki.2008.485. [DOI] [PMC free article] [PubMed] [Google Scholar]; Consugar MB, Wong WC, Lundquist PA, et al. Characterization of large rearrangements in autosomal dominant polycystic kidney disease and the PKD1/TSC2 contiguous gene syndrome. Kidney Int. 2008;74:1468-1479. [DOI] [PMC free article] [PubMed]
- 20.Lanktree MB, Iliuta I-A, Haghighi A, et al. Evolving role of genetic testing for the clinical management of autosomal dominant polycystic kidney disease [e-pub ahead of print]. Nephro Dial Transplant. doi: 10.1093/ndt/gfy261. Accessed May 27, 2019. [DOI] [PubMed]
- 21.Cornec-Le Gall E., Chebib F.T., Madsen C.D. The value of genetic testing in polycystic kidney diseases illustrated by a family with PKD2 and COL4A1 mutations. Am J Kidney Dis. 2018;72:302–308. doi: 10.1053/j.ajkd.2017.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cornec-Le Gall E, Chebib FT, Madsen CD, et al. The value of genetic testing in polycystic kidney diseases illustrated by a family with PKD2 and COL4A1 mutations. Am J Kidney Dis. 2018;72:302-308. [DOI] [PMC free article] [PubMed]
- 22.Bergmann C., von Bothmer J., Ortiz Brüchle N. Mutations in multiple PKD genes may explain early and severe polycystic kidney disease. J Am Soc Nephrol. 2011;22:2047–2056. doi: 10.1681/ASN.2010101080. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bergmann C, von Bothmer J, Ortiz Bruchle N, et al. Mutations in multiple PKD genes may explain early and severe polycystic kidney disease. J Am Soc Nephrol. 2011;22:2047-2056. [DOI] [PMC free article] [PubMed]
- 23.Lanktree M.B., Haghighi A., Guiard E. Prevalence estimates of polycystic kidney and liver disease by population sequencing. J Am Soc Nephrol. 2018;29:2593–2600. doi: 10.1681/ASN.2018050493. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lanktree MB, Haghighi A, Guiard E, et al. Prevalence estimates of polycystic kidney and liver disease by population sequencing. J Am Soc Nephrol. 2018;29:2593-2600. [DOI] [PMC free article] [PubMed]
- 24.Liu M., Shi S., Senthilnathan S. Genetic variation of DKK3 may modify renal disease severity in ADPKD. J Am Soc Nephrol. 2010;21:1510–1520. doi: 10.1681/ASN.2010030237. [DOI] [PMC free article] [PubMed] [Google Scholar]; Liu M, Shi S, Senthilnathan S, et al. Genetic variation of DKK3 may modify renal disease severity in ADPKD. J Am Soc Nephrol. 2010;21:1510-1520. [DOI] [PMC free article] [PubMed]
- 25.Timpson N.J., Greenwood C.M.T., Soranzo N. Genetic architecture: the shape of the genetic contribution to human traits and disease. Nat Rev Genet. 2018;19:110–124. doi: 10.1038/nrg.2017.101. [DOI] [PubMed] [Google Scholar]; Timpson NJ, Greenwood CMT, Soranzo N, et al. Genetic architecture: the shape of the genetic contribution to human traits and disease. Nat Rev Genet. 2018;19:110-124. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Prevalence of different mutation types in extended Toronto Genetic Epidemiology Study of Polycystic Kidney Disease (eTGESP) families with clinical information available for 2 or more affected relatives.
Figure S2. Mild mutations were equally prevalent in large families.
Figure S3. Age of patients with severe, intermediate/indeterminate, and mild ADPKD.
Figure S4. Protein-truncating PKD1 mutations are more common in patients with severe kidney disease.
Figure S5. Survival analysis by responsible ADPKD mutation type.
Table S1. Cases with conflicting evidence for both mild and severe kidney disease.