Abstract
Gene editing following designer nuclease cleavage in the presence of a DNA donor template can revert mutations in disease-causing genes. For optimal benefit, reversion of the point mutation in HBB leading to sickle cell disease (SCD) would permit precise homology-directed repair (HDR) while concurrently limiting on-target non-homologous end joining (NHEJ)-based HBB disruption. In this study, we directly compared the relative efficiency of co-delivery of a novel CRISPR/Cas9 ribonucleoprotein targeting HBB in association with recombinant adeno-associated virus 6 (rAAV6) versus single-stranded oligodeoxynucleotides (ssODNs) to introduce the sickle mutation (GTC or GTG; encoding E6V) or a silent change (GAA; encoding E6optE) in human CD34+ mobilized peripheral blood stem cells (mPBSCs) derived from healthy donors. In vitro, rAAV6 outperformed ssODN donor template delivery and mediated greater HDR correction, leading to both higher HDR rates and a higher HDR:NHEJ ratio. In contrast, at 12–14 weeks post-transplant into recipient, immunodeficient, NOD, B6, SCID Il2rγ−/− Kit(W41/W41) (NBSGW) mice, a ∼6-fold higher proportion of ssODN-modified cells persisted in vivo compared to recipients of rAAV6-modified mPBSCs. Together, our findings highlight that methodology for donor template delivery markedly impacts long-term persistence of HBB gene-modified mPBSCs, and they suggest that the ssODN platform is likely to be most amenable to direct clinical translation.
Keywords: sickle cell disease, gene editing, rAAV6, ssODN, homology-directed repair, Crispr/Cas9, hemoglobin disorders, NHEJ versus HDR, in vivo engraftment, NBSGW41 mice, CD34, hematopoietic stem cells, stem cell cures
Introduction
Sickle cell disease (SCD) is caused by a single-nucleotide transversion that increases the hydrophobicity of adult globin (βA) and renders it susceptible to polymerization. Patients with SCD have increased morbidity and a reduced lifespan.1, 2, 3, 4 While curative treatment can be achieved through human leukocyte antigen (HLA)-matched allogeneic transplant from a healthy donor, the availability of HLA-matched donors is limited, and the outcomes are complicated by the possibility of graft-versus-host disease (GvHD) and short-term and long-term impacts following higher intensity myelo-ablative conditioning.5, 6, 7, 8 Gene editing in autologous stem cells could circumvent the limitation of HLA-matched donor availability, directly correcting the disease-causing mutation in self-renewing stem cells and mitigating the historical risk of random integration posed by early viral vectors.9, 10, 11, 12, 13 Recent advances in lentiviral-based gene therapy have begun to provide significant therapeutic benefit in globin disorders, including SCD, leading to increased transfusion independence and improved quality of life.14, 15, 16 As a next step forward in this therapeutic arena, site-specific gene repair using the delivery of a donor template in association with ribonucleoprotein (RNP)-mediated DNA cleavage comprises an attractive next-generation approach, as it has the advantage of reversing the SCD mutation in a targeted manner.
Gene editing requires a site-specific endonuclease that creates a double-stranded break (DSB) that is resolved by cellular DNA repair machinery as seamless repair, error-prone non-homologous end joining (NHEJ), or precise homology-directed repair (HDR) in the presence of a DNA donor template. These repair outcomes are markedly influenced by the stage of the cell cycle. DSBs in quiescent stem cells in the G0/G1 phase are primarily resolved as NHEJ, whereas resolution by HDR requires entry into the S/G2 phase.17, 18, 19 These repair outcomes are mutually exclusive, and, therefore, they compete for overall outcome within individual hematopoietic stem cells (HSCs) and across the HSC population.
Several groups have shown that the SCD mutation in exon 1 of the HBB gene can be corrected by HDR utilizing designer nucleases, including zinc-finger nuclease (ZFN) mRNA, transcription activator-like effector nucleases (TALENs), and CRISPR/Cas9, in combination with alternative methods for co-delivery of a DNA repair template, including integrase-defective lentiviral vectors (IDLVs), recombinant adeno-associated virus 6 (rAAV6), and single-stranded oligodeoxynucleotides (ssODNs).20, 21, 22, 23 Of these approaches, rAAV6 and ssODNs comprise the most efficient donor template delivery platforms. However, total editing outcomes, including the frequency of precise HDR versus NHEJ, have not been simultaneously compared for rAAV6 and ssODN donor template delivery methods. Importantly, to be clinically relevant and therapeutic, high-fidelity HDR outcomes must proportionately exceed the error-prone NHEJ that improperly repairs DSBs and causes genomic instability at the HBB gene.
To better understand the role of donor template delivery in (1) the proportion of HDR and NHEJ outcomes, (2) preserving the integrity and long-term engraftment potential of the HSC compartment after editing, and (3) altering the longitudinal persistence of edited cells, we directly assessed different methods of donor template delivery in vitro and in vivo in adult CD34+ mobilized peripheral blood stem cells (mPBSCs). This study was designed as a proof of concept, where either a sickle mutation (GTC or GTG; encoding Glutamate to Valine change [E6V]) or a silent change (GAA; encoding Glutamate to Glutamate [E6optE]) was introduced into healthy donor mPBSCs. Following RNP-mediated disruption of exon 1 of HBB and alternative donor template delivery, we assessed the outcome of gene editing using molecular analysis via droplet digital PCR (ddPCR) and globin expression via the induction of sickle globin (βS; in the case of GTC or GTG change, E6V) or restoration of adult globin (βA; in the case of GAA change, E6optE) as a functional outcome. Using these approaches, we directly compared the outcome of alternative delivery platforms.
Our in vitro data demonstrated superiority for rAAV6 delivery, leading to proportionately greater HDR than NHEJ, whereas ssODN donor template delivery introduced significantly more NHEJ than HDR. In parallel, we performed a longitudinal assessment of engraftment and persistence of transplanted HDR-edited HSCs containing the GTC change (E6V). In contrast to our in vitro findings, a much greater percentage of cells modified by ssODN donor template persisted at 12–14 weeks in the bone marrow (BM) of NOD, B6, SCID Il2rγ−/− Kit(W41/W41) (NBSGW) recipient mice. Taken together, our findings provide an important functional assessment of alternative methods for HDR-based gene editing, and they help to inform the pathway to future clinical translation.
Results
Optimizing Nuclease Efficiency in CD34 Cells
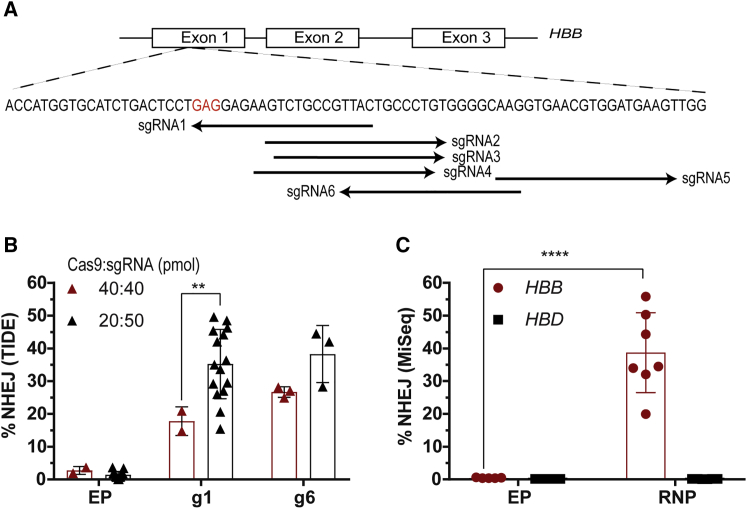
A nuclease screen was conducted to identify methods to efficiently create DSBs within exon 1 of the HBB gene (Figure 1A). The cleavage efficiencies of single-guide RNAs (sgRNAs) delivered as RNP complexes were evaluated in healthy human CD34+ mPBSCs. In the initial nuclease screen, RNP delivery of a series of candidate sgRNAs was tested at a Cas9:sgRNA ratio of 1:1, and it identified guide 4 (g4), g5, g6, and g1 as most efficient at creating DSBs (Figure S1A). Based on these findings, we optimized sgRNA-g1, as it created a DSB adjacent to codon 6, the site of the SCD mutation. sgRNA-g6 (G10), used by other groups,21, 22, 24 was also extensively tested in parallel.
Figure 1.
Screening of Nucleases to Create DSBs in Exon 1 of the HBB Gene
(A) Schematic representation of the genomic HBB gene showing the location of sgRNA-binding sites. A nucleotide substitution from GAG (codon 6 in red) to GTC or GTG changes the amino acid from glutamate to valine and causes SCD. (B) Optimizing the Cas9:sgRNA ratio to maximize editing efficiency in mPBSCs. NHEJ rates were analyzed by TIDE/ICE sequencing (Cas9:sgRNA ratio of 1:1 [40 pmol each], donor n = 2 or ratio of 1:2.5 [20 pmol of Cas9 and 50 pmol of sgRNA], donor n = 15). (C) Evaluating on-target disruption at HBB and possible off-target disruption at HBD by MiSeq analysis in mPBSCs using sgRNA-g1 delivered as RNP (donor n = 7). All bar graphs show mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. p value was calculated by comparing each sample mean with the respective control sample mean by two-way ANOVA with Dunnett’s multiple comparison. See also Figure S1 and Table S2.
Upon testing both guides at a Cas9:sgRNA ratio of 1:2.5 with the Neon transfection system, total editing rates doubled for sgRNA-g1 (g1, increased from 17.8% ± 4.4% to 35.2% ± 10.6%; g6, 26.7% ± 1.6% to 38.3% ± 8.7%; Figure 1B). The on-target HBB disruption for sgRNA-g1 by MiSeq analysis was 38.7% ± 12.2% (n = 7 donors), and off-target HBD disruption was 0.129% ± 0.01% (Figure 1C). The top 5 off-target genes predicted by consensus constrained topology (CCTOP)25 for sgRNA-g1 showed no insertions or deletions (indels) by T7 endonuclease assay (Table 1; Figure S1B) and tracking of indels by decomposition (TIDE) sequencing (Figure S1C). Overall editing rates increased by ∼2.5-fold with use of a nucleofection system (Lonza; 86% ± 2.6%, n = 3 donors) compared to the electroporation system (Neon; 35.2% ± 10.6%, n = 15 donors; Figure S1D).
Table 1.
Off-Target Genes Predicted by the CCTOP Algorithm
| Gene | Chr | Start | End | Strand | MM | Target Sequence | PAM | Position | Gene | Gene ID |
|---|---|---|---|---|---|---|---|---|---|---|
| OT-1 | 8 | 141103484 | 141103506 | + | 4 | TGAGCGGCAGAGTTCTCCTC | CGG | inter-genic | DENND3 | ENSG00000105339 |
| OT-2 | 19 | 11493685 | 11493707 | + | 4 | CTGACCCCAGACTTCTCCTC | AGG | intronic | MIR7974 | ENSG00000274713 |
| OT-3 | 3 | 182066120 | 182066142 | − | 3 | TTAAAGGAAGACTTCTCCTC | AGG | inter-genic | LINC01206 | ENSG00000242512 |
| OT-4 | 11 | 5234396 | 5234418 | + | 4 | TTGACAGCAGTCTTCTCCTC | AGG | exonic | HBD | ENSG00000223609 |
| OT-5 | 6 | 158475230 | 158475252 | − | 4 | GGAGGGGCAGGCTTCTCCTC | TGG | intronic | TULP4 | ENSG00000130338 |
| HBB | 11 | 5226984 | 5227006 | + | 0 | GTAACGGCAGACTTCTCCTC | AGG | exonic | HBB | ENSG00000244734 |
The prediction of potential off-targets were done using CC-TOP algorithm.25 Chr, chromosome number; MM, mismatch in guide sequence; PAM, protospacer adjacent motif.
Introducing the GTC Change through rAAV6 Donor Template Delivery
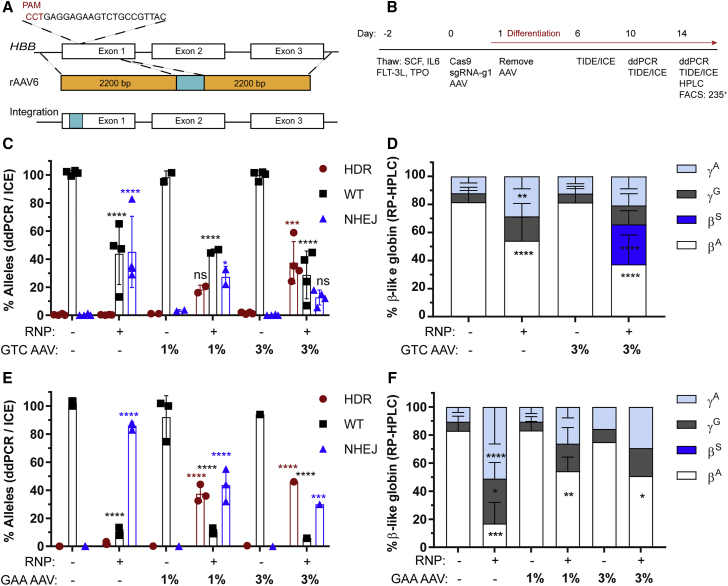
Based on our previous data and work from other groups in achieving efficient HDR,26, 27, 28, 29 we constructed an rAAV6 vector with 2.2-kb homology arms designed to introduce either a GTC (encoding E6V) or a silent change GAA (encoding E6optE) at codon 6 of exon 1 of the HBB gene. The design was focused on preserving intron 1 and native promoter and/or enhancer regions to maximize transcription and translation (Figure 2A). The experimental timeline is shown in Figure 2B.
Figure 2.
Homology-Directed Repair at the HBB Nuclease Target Site Using rAAV6 Donor Template
(A) Schematic representation of rAAV6 cassettes designed to drive either a GTC (E6V) introducing a sickle mutation or a GAA (E6optE) introducing a codon-optimized change at codon 6 by HDR. (B) Experimental timeline for testing gene editing with RNP and rAAV6 delivery followed by erythroid differentiation in mPBSCs. (C) WT (%) and HDR (%) measured by ddPCR and NHEJ (%) measured by ICE sequencing, respectively, following electroporation with RNP alone, transduction with rAAV6 donor template alone, or co-delivery of RNP and GTC (E6V) rAAV6 donor template, at the indicated concentrations (donor n = 4). (D) RP-HPLC analysis of erythroid cells to measure β-globin expression in cells treated with RNP only, rAAV6 only, or RNP plus GTC (E6V) rAAV6 (donor n = 7). (E) WT (%) and HDR (%) measured by ddPCR and NHEJ (%) measured by ICE sequencing, respectively, following electroporation with RNP alone, transduction with rAAV6 donor template alone, or co-delivery of RNP and GAA (E6optE) rAAV6 donor template, at the indicated concentrations (1% rAAV6; donor n = 3). (F) RP-HPLC analysis of erythroid cells to measure β-globin expression in cells treated with RNP only, rAAV6 only, or RNP plus GAA (E6optE) rAAV6 (donor n = 3). βA, adult globin; βS, sickle globin; γG, gamma 2; γA, gamma 1. All bar graphs show mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. p value was calculated by comparing each sample mean of NHEJ (%), HDR (%), WT (%), or globin sub-type (%) with the respective NHEJ (%), HDR (%), WT (%), or globin sub-type (%) of the mock sample by two-way ANOVA with Dunnett’s multiple comparison. Asterisks are color matched to the respective mock sample. See also Figures S2–S4.
Testing the GTC (encoding E6V) rAAV6 donor template (3% culture volume) following RNP-mediated cleavage resulted in HDR rates of 37.5% ± 15% and residual NHEJ rates of 12.7% ± 5.3% (Figure 2C). Testing of GTC (encoding E6V) rAAV6 with both the electroporation and nucleofection systems demonstrated that increases in total editing rates led to increases in rates of both HDR and residual NHEJ (Figure S2A). The HDR and residual NHEJ rates measured when RNP was co-delivered with GTC rAAV6 were additionally validated by colony sequencing (30.8% ± 6.3% HDR, 17.9% ± 7.2% NHEJ, n = 5 donors; Figure S2B). Cell viability after electroporation and transduction with 3% culture volume of GTC rAAV6 was on an average 79.2% (Figure S2C). Globin sub-types were measured in differentiated erythroid precursors by reverse-phase high-performance liquid chromatography (RP-HPLC). Editing alone and editing in the presence of GTC rAAV6 led to a significant decrease in βA (82% to 54% ± 16%, using Neon) and 2-fold increases in γA (HBG1) and γG (HBG2). Co-delivery of RNP and 3% GTC rAAV6 led to 28.4% ± 9.8% βS expression (n = 6 donors; Figure 2D). The globin tetramers in erythroid cells generated following co-delivery of RNP and GTC rAAV6 (encoding E6V) were measured by ion-exchange chromatography (IEC). A decrease in adult hemoglobin (HbA) and a dose-dependent increase in sickle hemoglobin (HbS) tetramers (15.8%) was observed with co-delivery of RNP and 3% GTC rAAV6 (Figure S2D). A sample chromatogram of GTC (encoding E6V) rAAV6-treated cells by RP-HPLC analysis confirmed the presence of a 38.6% βs peak (Figure S3).
Introducing the GAA Change through rAAV6 Donor Template Delivery
Introduction of the sickle mutation in normal cells does not assess the potential to revert the mutation in patient cells, and it might also alter the fitness of edited erythroid progenitors. In lieu of studies using HSCs from SCD subjects, we next tested the introduction of a silent change (GAA, encoding E6optE). Testing the GAA (encoding E6optE) rAAV6 donor template (1% culture volume) co-delivered with RNP resulted in HDR rates of 37.5% ± 6% and NHEJ rates of 43.7% ± 11.5% (Figure 2E). Of note, the increase in total NHEJ events using co-delivery of RNP and GAA (encoding E6optE) rAAV6 compared with the co-delivery of RNP and GTC (encoding E6V) rAAV6 cassettes likely reflects an increase in overall editing rates using the nucleofection system. While our data for GTC (encoding E6V) rAAV6 editing included experiments using both Neon (n = 3) and Lonza (n = 1), GAA (encoding E6optE) rAAV6 co-delivery was tested exclusively with the Lonza system (n = 3).
Cell viability after electroporation and transduction with 1% GAA (encoding E6optE) rAAV6 was on an average 60% (Figure S2C). RP-HPLC analysis identified a marked decrease in βA levels (from 82% in control cells to 16.9% ± 15% in RNP-edited cells) and 3-fold increases in γA (HBG1) and γG (HBG2) in the RNP-edited samples. In contrast, co-delivery of RNP and 1% GAA (encoding E6optE) rAAV6 led to a less robust reduction in βA levels (54.2% ± 10% βA expression, n = 3; Figure 2F) and less prominent increases in γA (HBG1) and γG (HBG2). The retention of βA expression following co-delivery of RNP and GAA (encoding E6optE) rAAV6 can be ascribed to AAV-mediated HDR. Consistent with this conclusion, RP-HPLC analysis of cells treated with co-delivery of RNP and GAA (encoding E6optE) rAAV6 showed 64.7% βA after HDR (Figure S4). Taken together, our findings demonstrate the capacity of RNP and rAAV6 co-delivery to promote high levels of HDR in exon 1 of HBB, leading to either an introduction of sickle mutation or a silent mutation designed to revert the sickle mutation in patient cells.
Introducing the GTC Change through ssODN Donor Template Delivery
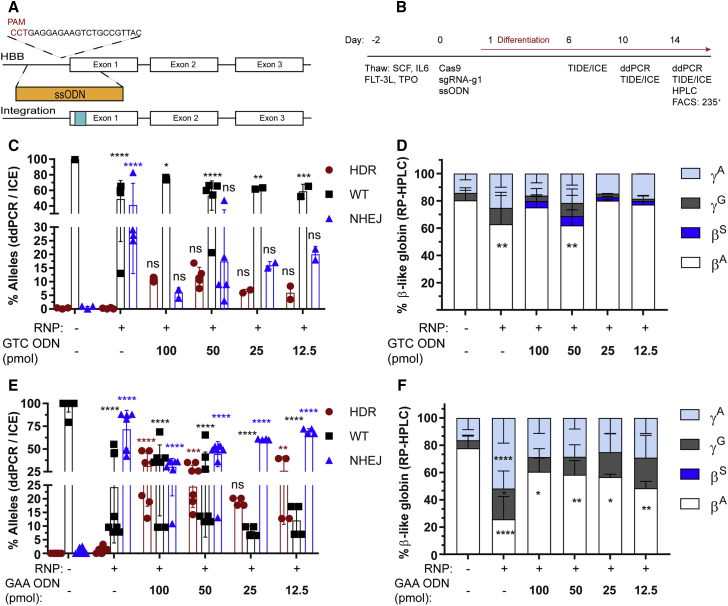
We next performed experiments assessing the efficiency of co-delivery of RNP and ssODNs to introduce the identical nucleotide changes achieved using rAAV6 in mPBSCs. Alternative 168-bp ssODNs were designed to generate a GTG (encoding E6V), GTC (encoding E6V), or GAA (encoding E6optE) nucleotide change (Figure 3A). The experimental timeline is shown in Figure 3B.
Figure 3.
Homology-Directed Repair at the HBB Nuclease Target Site Using ssODN Donor Template
(A) Schematic representation of ssODN cassette designed to drive either a GTC (E6V) introducing a sickle mutation or a GAA (E6optE) introducing a codon-optimized change at codon 6 by HDR. (B) Experimental timeline for testing gene editing with RNP and ssODN delivery followed by erythroid differentiation in mPBSCs. (C) WT (%) and HDR (%) measured by ddPCR and NHEJ (%) measured by ICE sequencing, respectively, following electroporation with RNP alone or co-delivery of RNP and GTC (E6V) ssODN donor template, at the indicated concentrations (50 pmol ssODNs, donor n = 5). (D) RP-HPLC analysis of erythroid cells to measure β-globin expression in cells treated with RNP only or RNP plus GTC (E6V) ssODNs (50 pmol ssODNs, donor n = 5). (E) WT (%) and HDR (%) measured by ddPCR and NHEJ (%) measured by ICE sequencing, respectively, following electroporation with RNP alone or co-delivery of RNP and GAA (E6optE) ssODNs, at the indicated concentrations (50 pmol ssODNs, donor n = 8). (F) RP-HPLC analysis of erythroid cells to measure β-globin expression in cells treated with RNP only or RNP plus GAA (E6optE) ssODNs (50 pmol ssODNs, donor n = 6). α, alpha; βA, adult; βS, sickle; γG, gamma 2; γA, gamma 1. All bar graphs show mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. p value was calculated by comparing each sample mean of NHEJ (%), HDR (%), WT (%), or globin sub-type (%) with the respective NHEJ (%), HDR (%), WT (%), or % globin sub-type (%) of the mock sample by two-way ANOVA with Dunnett’s multiple comparison. Asterisks are color matched to the respective mock sample. See also Figures S5–S8.
There was a dose-dependent increase in cytotoxicity with increasing concentrations of ssODN tested (Figure S5A). The HDR gene conversion rate following co-delivery of RNP and 50 pmol ssODN was 11.9% ± 3.4% for the GTC ODN and 17% ± 4.3% for the GTG ODN, and the residual NHEJ was 17.4% ± 17.5% and 20.0% ± 1.7%, respectively (Figures 3C and S5B). The HDR and NHEJ rates for 50 pmol GTG ssODN (encoding E6V) were further validated by colony sequencing and were 12.6% ± 8.8% and 30.1% ± 12.4%, respectively (Figure S5C).
Globin sub-types were assessed by RP-HPLC. We observed a significant decrease in βA and 1.5-fold increases in γA (HBG1) and γG (HBG2) following RNP-mediated disruption. The co-delivery of RNP and varying concentrations of ssODN led to a dose-dependent increase in sickle globin expression, with optimal βS expression with 50 pmol GTC (encoding E6V) ssODN (Figure 3D) and GTG ssODN (Figure S5D). Editing with GTC (encoding E6V) ssODN resulted in 5.2% βS expression (50 pmol, n = 5; Figure 3D), and editing with GTG (encoding E6V) ssODN resulted in 5.3% βS expression (50 pmol, n = 3), respectively (Figure S5D). Consistent with these averages, a sample chromatogram derived from differentiated erythroid cells demonstrated 8.9% βS expression with GTC (encoding E6V) ssODN (Figure S6) and 9.2% βS with GTG (encoding E6V) ssODN (Figure S7). A direct comparison of editing in mPBSCs from the same donor using GTC ssODN versus rAAV6 demonstrated 8.9% versus 24.5% βS expression, respectively (Figure S6).
Introducing the GAA Change through ssODN Donor Template Delivery
Consistent with our experiments using rAAV6 donor templates, we also tested the introduction of a silent change (GAA, encoding E6optE). Co-delivery of RNP and 50 pmol GAA (encoding E6optE) ssODN resulted in an HDR gene conversion rate of 24.5% ± 7.6% with residual NHEJ rates of 44% ± 13.8% (Figure 3E). With increasing concentrations of ssODN, a dose-dependent increase in HDR and a corresponding decrease in NHEJ were observed with both the Neon and Lonza systems (Figure S5E). Editing outcomes following use of the Neon electroporation system were also validated by colony sequencing, demonstrating HDR rates of 10.6% ± 2.8% and residual NHEJ rates of 35.5% ± 8.6% (Figure S5C).
Globin sub-types in differentiated erythroid pellets were measured, and a significant decrease in βA (25.7%, n = 6 donors) and 1.5- to 3-fold increases in γA (HBG1) and γG (HBG2) were observed with RNP-mediated disruption. In contrast, use of the GAA (encoding E6optE) ssODN donor template retained βA expression at 58.4% (n = 6 donors; Figure 3F). A sample chromatogram, showing globin sub-types in edited differentiated erythroid cells, demonstrates an increase from 0% HbA in RNP-disrupted samples to 75.6% following the co-delivery of RNP and GAA (encoding E6optE) ssODN (Figure S9). A direct comparison of RNP-only edited mPBSCs versus cells edited using the co-delivery of RNP and HDR donor template showed an increase in HbA expression from 0% to 75.6% and 64.7% for GAA ssODN and rAAV6 donor templates, respectively (Figure S9).
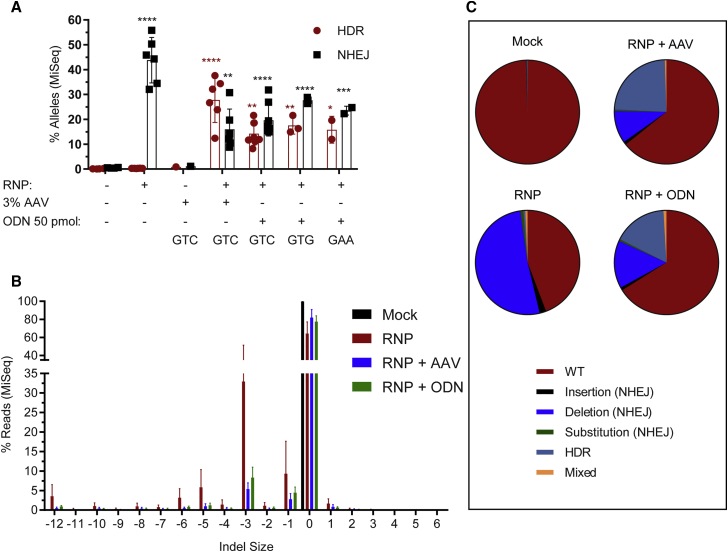
Comparison of ssODN and rAAV6 Donor Template Delivery Methods by MiSeq Analysis
To further assess gene-editing efficiencies achieved using our alternative platforms, we used MiSeq analysis to validate the editing outcomes. The HDR and NHEJ rates achieved using the Neon electroporation system for RNP co-delivery in association with all ssODN donor templates versus GTC (encoding E6V) rAAV6 donor templates were assessed. An average of 107,799 pairwise-aligned reads was obtained from each in vitro sample (Figure S9A). The data clearly demonstrate that the rAAV6 donor template drives higher levels of HDR than NHEJ (GTC rAAV6: 27.8% HDR and 16% NHEJ) and that ssODN delivery drives higher levels of NHEJ than HDR (GTC ssODN: 14.3% HDR and 19.6% NHEJ) in vitro (Figures 4A–4C).
Figure 4.
Comparison of Outcomes of ssODN and rAAV6 Donor Template Delivery Methods by MiSeq Analysis
(A) Quantification of HDR versus NHEJ edits by MiSeq analysis in cells treated with GTC (E6V) rAAV6 (n = 6) versus ssODNs (using GTC [E6V, n = 8], GTG [E6V, n = 3], or GAA [E6optE, n = 2] ssODNs). (B) Indel spectrum analysis by MiSeq comparing RNP-mediated editing alone to residual indels present in cells after the promotion of HDR with either rAAV6 or ssODN delivery (donor n = 6). (C) The various gene-editing outcomes, WT, NHEJ (insertion, substitution, deletion), and HDR, measured in the following samples: mock, RNP alone, and co-delivery of RNP with rAAV6 and RNP with ssODNs. The samples analyzed were the pre-transplant input and in vitro-edited samples analyzed on day 14 post-editing (n = 6). All bar graphs show mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. p value was calculated by comparing each sample mean of NHEJ (%) and HDR (%) with the respective NHEJ (%) and HDR (%) of the mock sample by two-way ANOVA with Dunnett’s multiple comparison. Asterisks are color matched to the respective mock sample. See also Figure S9.
Analyzing the indel spectrum produced, RNP alone resulted in 60.4% deletions (primarily −3-, −1-, −5-, −6-, and −12-bp deletions) along with 2% insertions. Co-delivery of a donor template with RNP decreased the indel spectrum to primarily −3- and −1-bp deletions (Figures 4B and S9B). Wild-type (WT), NHEJ with deletions, and HDR alleles were observed in both rAAV6-edited and ssODN-edited samples (Figure 4C). Crispresso30 analysis identified that rAAV6 donor template delivery resulted in fewer frameshift mutations (8.6% in vitro) compared to ssODN donor template delivery (12.2% in vitro; Figure S9C).
Impact of ssODN versus rAAV6 Delivery on Sustained Engraftment of HDR-Edited Cells In Vivo
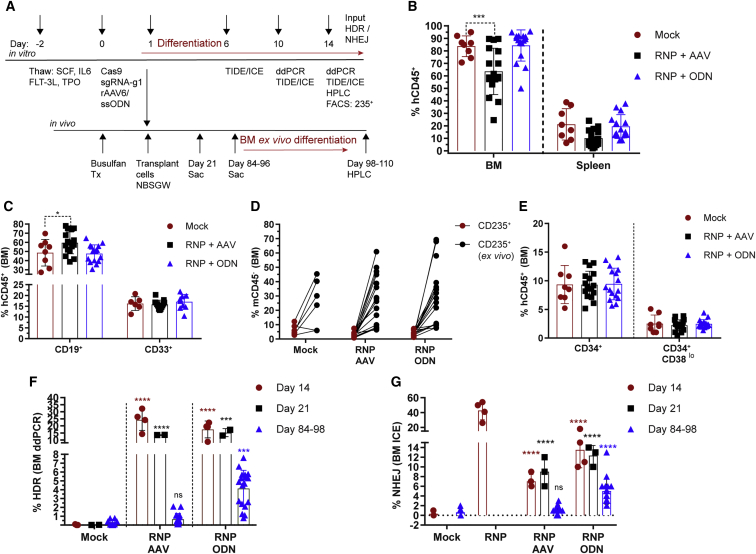
To understand the role of alternative donor template platforms in altering the long-term engraftment potential of HDR-edited CD34+ cells, healthy control mPBSCs edited to introduce the GTC (encoding E6V) change were transplanted into busulfan-conditioned (12.5–25 mg/kg) NBSGW recipient mice, an immunodeficient strain that permits the development of a human erythroid compartment. Cells, derived from identical donors, edited with each platform (2 × 106 cells) were transplanted at day 1 following electroporation. Transplanted animals were assessed over time and evaluated at 3 or 12–14 weeks for human cell engraftment in the BM and spleen (Figure 5A).
Figure 5.
Engraftment Potential of rAAV6- versus ssODN-Edited HSCs
(A) Experimental timeline for testing gene editing with GTC (E6V) rAAV6- or ssODN-treated cells in vitro in mPBSCs and in vivo in the NBSGW mouse model. Red lines indicate placement of cells in erythroid differentiation conditions. (B) Human cell (hCD45+) chimerism in the BM and spleen at days 84–96, with gating based on forward scatter (FSC), side scatter (SSC), and single cells. (C) Human CD19+ and CD33+ subsets within the BM hCD45+ population. (D) Human CD235+ cells in the BM gated on the mCD45− population. The BM cells were cultured ex vivo for 14 days in erythroid differentiation media, and CD235+ (ex vivo) was measured by flow cytometry. (E) Proportion of human CD34+ and CD34+CD38lo cells within the BM hCD45+ population. (F) HDR rates determined by ddPCR within the GTC (E6V) rAAV6- or ssODN-treated input cells (day 14, n = 4 transplants, single donor), at 3 weeks post-transplant (day 21, n = 2), and at 12–14 weeks post-transplantation (days 84–96; mock n = 8, RNP + rAAV6 n = 17, RNP + ssODN n = 18). (G) NHEJ rates determined by ICE sequencing for GTC (E6V) rAAV6- or ssODN-treated input cells (day 14), at 3 weeks (day 21) post-transplant, and at 12–14 weeks (days 84–96) post-transplant. All bar graphs show mean ± SD. ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. p value was calculated by comparing each sample mean of NHEJ (%), HDR (%), and WT (%) with the respective NHEJ (%), HDR (%), and WT (%) of the mock sample by two-way ANOVA with Dunnett’s multiple comparison. Asterisks are color matched to the respective mock sample. See also Figures S10–S15.
Human chimerism was comparable for NBSGW recipients of mock- or ssODN-edited cells. In contrast, a significant decrease in human (h)CD45+ cell engraftment was observed in recipients of rAAV6-edited cells (Figure 5B). No decline in hCD45+ cell engraftment was observed at 14 weeks in animals that received cells transduced with rAAV6 alone (without RNP co-delivery; data not shown). Additionally, the proportion of CD19+ B cells was modestly increased in the rAAV6-edited group, suggesting skewing toward more differentiated progeny (Figure 5C). Other lineages, including myeloid (CD33+), T (CD3+), and erythroid (CD235+) cells, were represented equivalently across cohorts (Figures 5C, 5D, and S10A–S10C).
Cells isolated from the BM were cultured in erythroid differentiation media for 2 weeks after harvest to permit expansion of CD235+ cells (with an increase from 4.01% at harvest to 27.6% CD235+ in ex vivo cultures; Figure 5D). Representative flow plots of edited donor cells pre- and post-transplant revealed equivalent proportions of primitive HSC sub-populations, including CD34+, CD34+CD38lo, and CD34+CD38loCD133+CD90+ cells (Figures 5E, S11A, and S11B).
The input HDR rates (day 14 in culture) across 4 transplants were 24.28% ± 7.5% and 17.5% ± 6% for rAAV6 and ssODN delivery methods, respectively. HDR-edited cells in the BM at 3 weeks post-transplant declined to 13.58% ± 0.16% and 15.19% ± 2.8% (n = 2) for rAAV6 and ssODN delivery, respectively. At 12–14 weeks, the HDR rates declined precipitously to 0.66% ± 0.66% (n = 17) in recipients of rAAV6-edited cells. Strikingly, HDR rates also declined, but to a much lesser extent, to 4.136% ± 2.1% (n = 18) in recipients of ssODN-edited cells (Figure 5F). The input NHEJ was 7% ± 1.4% and 13.5% ± 3.7% for rAAV6 and ssODN donor template delivery methods, respectively, and that remained unchanged at 3 weeks post-transplant (rAAV6, 9% ± 3%; ssODN, 12.3% ± 2.1%) and declined at 12–14 weeks (NHEJ rAAV6, 1.3% ± 0.85%; ssODN, 5% ± 2.7%; Figure 5G). HDR and NHEJ rates in the BM of NBSGW mice in vivo were verified by MiSeq analysis (HDR rAAV6, 0.65% ± 0.65%; ssODN, 3.84% ± 2.1%; NHEJ rAAV6, 2.5% ± 2.5%; ssODN, 9.9% ± 5.3%; Figures S12A and S12B). An average of 120,534 pairwise-aligned MiSeq reads was obtained from each in vivo BM sample (Figure S12C). Crispresso30 analysis identified that the BM samples of rAAV6-edited animals had fewer frameshift mutations (1.4% in vivo) compared to BM samples from ssODN-edited animals (5.3% in vivo; Figure S12D).
A subset of each initial edited input CD34+ cell population was maintained in vitro in erythroid culture conditions and analyzed for globin sub-types. rAAV6-edited cells exhibited 16.4% ± 6.8% and ssODN-edited cells exhibited 12.42% ± 4.4% βS expression (Figure S12A). The ex vivo BM cultures analyzed by HPLC expressed 3.8% βS (n = 3 animals) in the ssODN-edited group. In contrast, βS was not detected in the rAAV6- (n = 4 animals) or mock-edited samples (n = 2 animals; Figure S13B). HPLC of the 69 burst-forming unit-erythroid (BFU-E) colonies revealed 3/35 colonies derived from the ssODN-edited group expressing βS, resulting in an average of 5.13% βS expression. In contrast, βS expression was not detected in rAAV6-edited (n = 26 colonies) or mock-edited colonies (n = 8 colonies; Figures S13C and S13D). The HPLC profile of single colonies for mock samples contained 97% HbA, whereas the edited groups had a decrease in HbA and an increase in HbF (HbF rAAV6, 17.4%; ssODN, 17.9%) and/or HbS (Figures S13C and S13D). Chromatograms of single-erythroid colonies derived from the ssODN-edited group demonstrated βS expression levels of 38.7%, 84.5%, and 56.3% (Figures S14 and S15). Taken together, these data demonstrate that ssODN-modified cells outperformed rAAV6-modifed cells in NBSGW mice in vivo, leading to both higher sustained engraftment of HDR-edited cells and sickle globin expression.
Discussion
Delivery of a DNA donor template comprises a crucial step in achieving precise gene correction following targeted gene cleavage in human HSCs. Importantly, the overall ratio of HDR to NHEJ significantly impacts the potential clinical benefit of gene correction in SCD. While significant previous work has separately assessed alternative nuclease and donor template delivery methods, no direct in vitro and in vivo comparison of the most efficient methods has been performed to date. In the current study, we assessed the role of alternative donor template delivery methods to achieve initial gene conversion events in vitro as well as the impact on the survival, stem-like potential, and sustained engraftment of edited cells in vivo. Our combined data demonstrate the complexity and address some of the challenges in achieving long-term clinical gene correction in SCD. We observed no major differences in HSC viability, phenotype, or expansion in vitro using rAAV6 compared with ssODN delivery. However, we show that rAAV6 donor templates mediate consistently higher HDR:NHEJ ratios. In contrast, in transplant experiments, we show that higher levels of sustained HDR are achieved using HSCs edited with ssODN donor templates.
We performed an initial screening of multiple candidate guide RNAs spanning a 53-bp region around the sickle mutation site. As shown here, sgRNA-g1 efficiently creates DSBs immediately adjacent to the sickle mutation site (between 21 and 22 bp), and it was, therefore, chosen as a potentially more useful guide than sgRNA-g6 (G10)21, 22, 24 that generates a DSB between 37 and 38 nt in exon 1 of HBB, which is 16 bp away from the mutation site. Use of Cas9:sgRNA at a ratio of 1:2.5 promoted the highest levels of editing in human mPBSCs, with no demonstrable off-target effects (Figures 1B, S1B, and S1C). Of note, total editing rates increased 2.5-fold when using the nucleofection system (Figure S1D), with increases in both HDR as well as residual indels in vitro (Figures S2A and S5E). The nucleofection system is now widely used to edit the HBB locus.22, 31, 32 Although the increase in HDR using the nucleofection system is a favorable outcome, it is associated with a parallel, clinically undesirable increase in NHEJ that could be impactful when trying to preferentially promote exonic repair. Following delivery of RNPs containing sgRNA-g1, we tested the capacity of alternative rAAV6 cassettes versus a series of ssODNs to drive nucleotide changes in the sixth codon of exon 1 of HBB. Our in vitro studies demonstrated that rAAV6 promotes greater rates of HDR than NHEJ (GTC rAAV6, 37.5% ± 15% HDR and 12.7% ± 5.3% NHEJ; Figure 2C); in contrast, ssODN delivery drives more NHEJ than HDR (GTC ssODN, 11.9% ± 3.4% HDR and 17.4% ± 17.5% NHEJ; Figures 3C and 4A).
Importantly, in parallel with our in vitro analysis of gene editing, we directly assessed the potential impact of alternative donor template delivery methods on the sustained engraftment of HDR-edited cells. We utilized the NBSGW humanized model that permits high levels of human HSC engraftment and facilitates studies of erythroid cells.33, 34, 35 Tracking of edited cells in the BM and spleen at 12–14 weeks post-transplant demonstrated that multi-lineage engraftment was achieved in all recipients. However, overall human chimerism was lower in recipients of rAAV6-edited cells compared to recipients transplanted with mock- or ssODN-edited populations (Figure 5B). Relative to input levels, HDR rates were lower in cells within the BM for both donor template delivery platforms. This overall decline in engraftment of HDR-edited cells is consistent with multiple previous reports using each of these donor template delivery approaches.20, 22, 29, 31, 32, 36, 37 Notably, however, despite the loss of HDR-edited cells in both settings, the proportion of HDR-edited cells remained ∼6-fold higher (4.14%, n = 18 versus 0.66%, n = 17) in recipients of ssODN-edited mPBSCs compared to cells modified with rAAV6. Similarly, 5% NHEJ was detected in animals that were engrafted with ssODN-edited cells, while only 1.3% NHEJ was detected in recipients of cells modified with rAAV6.
Our in vivo data suggest that, in comparison with ssODN-edited cells, rAAV6-edited cells fail to engraft efficiently and/or are rapidly outcompeted by unedited cells. The negative impact of rAAV6 likely reflects a synergistic response to combined assaults to the HSC compartment through DSBs and virally mediated template delivery. These combined events and the presence of episomal AAV could alter HSC metabolism, thereby limiting proliferation, survival, and/or self-renewal.38 Alternatively, a smaller proportion of long-term HSCs (LT-HSCs) could have been modified using rAAV6 delivery, leading to a decline in long-term gene-modified cells. Importantly, consistent with our molecular data showing very low HDR rates in engrafted cells with rAAV6 delivery, βS expression was identified only when engrafted BM cells were enriched for pooled 235+ erythroid cells (data not shown). In contrast, we observed βS expression via RP-HPLC analysis at levels (∼5%) equivalent to our molecular data in both pooled- and single-erythroid colonies in recipients of ssODN-edited mPBSCs.
As a proof-of-concept study to compare donor template delivery methods in driving gene correction, we introduced the sickle mutation, thereby contributing to an overall lower engraftment of HDR-edited cells than would be achieved following gene correction in SCD patient cells. We introduced the sickle allele to provide a clear functional phenotype for HDR, to compare different donor delivery methods, to evaluate sustained HDR rates in vivo, and to assess globin expression (sickle globin) in the BM. The E6V GTC change introduces a disease mutation that is likely to confer a survival disadvantage that affects the persistence of edited human cells in the BM, a feature not present in most other studies. Of note, compared with our findings, other groups utilizing co-delivery of RNP and rAAV6 donors targeting the HBB and CD40L loci, respectively, in mPBSCs have reported higher sustained HDR rates in vivo (3.5%–4.4%).22, 39 In both models, rAAV6-edited cells have equal survival opportunity compared with wild-type cells to re-constitute the BM.
Even higher sustained HDR rates (10%–12%, and more recently 25.5%) using an rAAV6 donor template have been reported using patient cord blood CD34+ cells in the correction of X-linked severe combined immunodeficiency (SCID-X1).40, 41 A higher engraftment rate of CD34+ cells derived from cord blood (or fetal liver) compared with mPBSCs and the lack of survival disadvantage likely account for some of these differences.29 Additional improvement in engraftment of AAV6-edited cells could potentially be achieved using the Lonza nucleofection system in association with alternative culture conditions, including lower cell seeding density following editing.36, 41 These approaches may improve the survival of HDR-edited cells and/or enrich for editing within long-term HSCs. However, within the scope of this study, comparing introduction of the sickle mutation into mobilized CD34+ cells from the same donor, ssODN delivery substantially outperformed rAAV6 donor delivery based on better sustained engraftment of HDR-edited cells within the BM at 12–14 weeks.
Consistent with our findings introducing the sickle mutation in healthy donor mPBSCs, recent work using SCD patient-derived mPBSCs and ssODN-based gene correction to revert the sickle mutation resulted in sustained HDR rates of ∼20% in the BM at 16–20 weeks post-transplant.32 In contrast to HSCs edited to express the sickle mutation (as in our study), homozygous sickle cells that undergo mono-allelic or bi-allelic correction gain a fitness and/or survival advantage and outcompete cells homozygous for the sickle allele as well as mutant cells with indels. As the next step toward clinical application of HDR-based editing in SCD, we generated codon-optimized sickle correction donor templates encoding an E6optE amino acid change via HDR targeting. We achieved efficient HDR rates in vitro using this approach in mPBSCs (rAAV6, 37.5%; ssODN, 29.6%). Testing these correction templates in patients with SCD will provide important additional information regarding the potential for clinical translation. Ideally, precise correction through HDR should exceed the error-prone NHEJ outcomes when editing at the HBB gene. However, based on selective advantage, the delivery of HSCs with heterogeneous editing outcomes resulting in βA/β0, βA/βS, and βA/βA would be predicted to functionally outperform βS/βS, βS/β0, and β0/β0 alleles in vivo, providing a spectrum of outcomes more desirable than homozygous sickle alleles.
Taken together, our findings provide the first demonstration that ssODN delivery outperforms rAAV6 donor template delivery in permitting sustained gene conversion outcomes in vivo in long-term HSCs. As shown here, rAAV6 mediates more efficient HDR rates in vitro, and it has previously been demonstrated by our group and others to minimally impact in vivo function of gene-edited primary human T or B cells.26, 42 Additional modifications, however, will likely be required to increase the efficiency of AAV6 donor delivery to drive sustained HDR in long-term HSCs to permit effective clinical translation.
Materials and Methods
rAAV6 Production
rAAV6 stocks were produced as previously described.43 The rAAV6 vector, serotype helper, and HgT1-adeno helper plasmids44 were transfected into HEK293T cells. Cells were harvested at 48 h, lysed, and treated with benzonase. An iodixanol density gradient was used to purify the virions with recombinant rAAV6 genomes. The qPCR-based titers of rAAV6 genomes were determined by using inverted terminal repeat (ITR)-specific primers and probe.45 1%, 2%, and 3% of the culture volume were used for transducing rAAV6 into mPBSCs.
CD34+ HSCs
Frozen mPBSCs were purchased from Cooperative Center for Excellence in Hematology (CCEH) at Fred Hutchinson Cancer Research Institute, Seattle, WA.
sgRNA and TALEN Designs
A search of the literature identified guide RNA sequences that are widely used across groups to edit the HBB gene.22, 24, 31, 32 We further designed guides that were predicted to cut close to the sickle mutation using CRISPR design tools (https://zlab.bio/guide-design-resources; http://crispor.tefor.net/). All guides were synthesized as chemically modified 2′-O-methyl analogs with 3′ phosphorothioate inter-nucleotide linkages in the first three 5′ and 3′ terminal residues (Synthego, CA).
Electroporation, Transduction of Cells, and Erythroid Differentiation Culture
Alt-R S.p Cas9 Nuclease 3NLS protein was used for all studies (Integrated DNA Technologies, Coralville, IA). The CD34+ cells were cultured in stem cell growth medium (SCGM; CellGenix, NH) with 100 ng/mL each of fms-like tyrosine kinase 3 ligand (FLT-3L), thrombopoietin (TPO), human stem cell factor (hSCF), and interleukin-6 (IL-6) (PeproTech, Rocky Hill, NJ). Cells were electroporated 48 h after thaw, using the Neon electroporation system (Thermo Fisher Scientific, Waltham, MA) at 1,300 V, 20 ms, and 1 pulse or the Lonza 4-D nucleofector (Lonza, Basel, Switzerland; CM149 protocol). The Cas9 RNP was made right before electroporation or nucleofection by mixing 20 pmol Cas9 and 50 pmol sgRNA (per 2 × 105 cells, ratio of 1:2.5 of Cas9:sgRNA). The RNP mixture was made fresh and incubated at room temperature for 15 min. ssODN donor templates were used at 100, 50, 25, and 12.5 pmol for every 2 × 105 cells, and they were added into the mixture of RNP right before electroporation or nucleofection. Cells after electroporation or nucleofection were added to either rAAV6 containing SCGM media with cytokines (at a 1%, 2%, or 3% culture volume; 3% GTC rAAV6 ∼MOI of 4,500–5,100, 1% GAA rAAV6 ∼MOI of 2,190) or to plain SCGM media with cytokines for ssODN-treated and control cells. The cells were incubated in media overnight at 37°C for 18 h. After 18 h, the cells were transferred to tissue culture non-treated plates containing Iscove's modified Dulbecco's medium (IMDM) with 1 ng/mL hIL-3, 2 IU/mL erythropoietin (EPO), 20 ng/mL hSCF, 20% heat-inactivated fetal bovine serum (FBS), and 1% penicillin and streptomycin. (Fisher Scientific, Hampton, NH and PeproTech, Rocky Hill, NJ). The cell density was kept between 5 × 105 and 1 × 106 cells/mL to minimize fetal hemoglobin induction due to proliferative stress or overcrowding.46, 47 CD235 expression was monitored at day 14 by flow cytometry (Table S3).
Measuring HDR Events with rAAV6 and ssODN Using ddPCR
gDNA was extracted (from cells cultured in vitro in differentiation media on day 14) with DNeasy blood and tissue kit (QIAGEN, Germantown, MD), and it was RNase treated. 100 ng gDNA was treated with 6 units of EcoRV high-fidelity restriction enzyme (EcoRV-HF; New England Biolabs, Ipswich, MA) at 37°C for 15 min to digest the gDNA outside of the amplicon region. ddPCR forward and reverse primers (ddPCR F/R) were used to amplify a 210-bp amplicon. The assay was designed as a dual-probe assay, with WT-hexachlorofluorescein (HEX) and HDR-6-carboxyfluorescein (FAM) probe31 run together, and the reference-HEX probe was run in parallel in a separate well with the same ddPCR F/R primers (Table S1), using ddPCR supermix for probes (no deoxyuridine-5'-triphosphate [dUTP], Bio-Rad). The droplets were generated and amplified on a Bio-Rad thermocycler (95°C, 5 min; 94°C, 30 s; 56°C, 1 min; 72°C, 1 min; step 2, 49 cycles of 98°C, 10 min and 12°C, ∞). The FAM and HEX fluorescence intensities were measured on the Bio-Rad QX200 machine (Bio-Rad, Hercules, CA). The HDR (%) events (HDR-FAM+) and WT (WT-HEX+) events were calculated after correction for the reference gene (REF-HEX+; Table S1).
Measuring Indel Frequencies
gDNA from day 10 post-electroporation was used to amplify the 1,250-bp amplicon around the cut site with forward and reverse primers (HBB-F/R-1250; Table S1). The PCR products were cleaned using NucleoSpin gel and PCR clean-up kit (Machery Nagel, Bethlehem, PA), and they were subject to Sanger sequencing with the sequencing primer (SCL-F/R-386; Table S1). The sequences were analyzed using TIDE or inference of CRISPR edits (ICE) algorithm to measure INDELs following editing.48, 49
MiSeq Analysis
The HBB (386-bp) and HBD (301-bp) gene-specific amplicons were amplified from 200 ng gDNA using PrimeSTAR GXL DNA polymerase (TaKaRa, Kusatsu, Japan) with MiSeq primers (Table S1). The primers added an overhang adaptor sequence onto the amplicons. Nextera 96-index kit (FC-121-1012, Illumina, San Diego, CA) was used to add a 5′ and 3′ unique index to each sample. The samples were purified with Agencourt AMPure XP (Beckman Coulter, Brea, CA), and the band was verified on a PAGE gel. The samples were measured and pooled to make libraries, and quality control was done on Qubit (Thermo Fisher Scientific, Waltham, MA) and analyzed on MiSeq 500 CycleV2 kit (Illumina, San Diego, CA) at the Genomics core, Fred Hutchinson Cancer Research Institute, Seattle, WA. The data were mined using the Crispresso2 algorithm.30 Analysis of HBB gene was used for quantitation of on-target gene modification and HBD gene was used for quantitation of off-target analysis.
Engraftment Studies in NBSGW Mice
The NBSGW mice were purchased from Jackson Laboratories and maintained in a designated pathogen-free facility at the Seattle Children’s Research Institute (SCRI). All animal studies were performed according to the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) standards and were approved by the SCRI Institutional Animal Care and Use Committee (IACUC). In our experiments, 6- to 7-week-old NBSGW33, 34, 35 mice were busulfan (Selleckchem) treated 24 h before transplant of edited cells. 2 × 106 edited cells were infused by tail vein 24 h after editing. The animals in the mock treatment group received 2 × 106 cells that were cultured for 48 h under identical conditions but were not electroporated. The animals were monitored regularly. The BM and spleen from these animals were harvested at 3 weeks and 12–14 weeks after transfer, and the cells were analyzed for human chimerism of hCD45+ and mCD45+ and multi-lineage engraftment of CD19+, CD33+, CD235+, CD3+, CD34+, and CD38+ cells (Table S3). The gDNA from BM cells was harvested and analyzed by ddPCR to determine HDR (%) and WT (%). The indels were analyzed by ICE sequencing. The BM cells were cultured in erythroid differentiation media for 2 weeks after harvest. The cells from ex vivo differentiation cultures were measured for CD235+ expression by flow cytometry. The cells were also pelleted, washed, and analyzed by RP-HPLC at 2 weeks post-harvest to look for globin expression. BM cells (30,000 cells/plate/3 mL methylcellulose) were added to MethoCult complete media (STEMCELL Technologies, Vancouver, Canada) and plated for colony-forming unit (CFU) analysis. Single BFU-E colonies were picked at 14 days post-harvest, lysed in water, and analyzed by IEC for globin expression.
Statistical Analysis
The data collected from experiments were analyzed on Graph Pad Prism 7 using two-way ANOVA with Dunnett’s multiple comparisons test. All samples across groups were compared to control or mock-treated cells to evaluate significance (ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001).
Author Contributions
Conceptualization, S.P., D.J.R., A.M.S., and K.J.; Methodology, S.P., D.J.R., A.M.S., and K.J.; Investigation, S.P., S.N.L., M.P.B., O.N., C.L., and S.S.; Writing – Original Draft, S.P.; Writing – Review & Editing, S.P. and D.J.R.; Funding Acquisition, D.J.R. and A.M.S.; Resources, D.J.R.; Supervision, S.P., C.T.L., D.J.R., and A.M.S.
Conflicts of Interest
A.M.S. is an employee of Casebia Therapeutics, Cambridge, MA, and receives equity. O.N. and C.L. are employees and shareholders of bluebird bio, Cambridge, MA. The other authors declare no competing interests.
Acknowledgments
We would like to thank Iram Khan, King Hung and Ezra Lopez for making rAAV6 viruses. All the sequencing support was provided by the Seattle Children’s Research Institute sequencing core. We would like to thank Garrett Heffner for scientific discussions and expertise. Synthego Inc., CA, provided chemically modified sgRNA for screening. We are grateful for their assistance. MiSeq data were processed by Seattle Children’s Research Institute, Bioinformatics Core. We would like to thank Andrew Timms and Paul Hodor for their assistance and support with processing the MiSeq data. This work was supported by the Seattle Children’s Research Institute, Program for Cell and Gene Therapy; the NIH (under award 1R01HL136135-02); bluebird bio, Inc.; and the Children’s Guild Association Endowed Chair in Pediatric Immunology (D.J.R.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.05.025.
Supplemental Information
References
- 1.Kato G.J., Piel F.B., Reid C.D., Gaston M.H., Ohene-Frempong K., Krishnamurti L., Smith W.R., Panepinto J.A., Weatherall D.J., Costa F.F., Vichinsky E.P. Sickle cell disease. Nat. Rev. Dis. Primers. 2018;4:18010. doi: 10.1038/nrdp.2018.10. [DOI] [PubMed] [Google Scholar]
- 2.Piel F.B., Steinberg M.H., Rees D.C. Sickle Cell Disease. N. Engl. J. Med. 2017;376:1561–1573. doi: 10.1056/NEJMra1510865. [DOI] [PubMed] [Google Scholar]
- 3.Sankaran V.G., Weiss M.J. Anemia: progress in molecular mechanisms and therapies. Nat. Med. 2015;21:221–230. doi: 10.1038/nm.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serjeant G.R. The natural history of sickle cell disease. Cold Spring Harb. Perspect. Med. 2013;3:a011783. doi: 10.1101/cshperspect.a011783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoban M.D., Orkin S.H., Bauer D.E. Genetic treatment of a molecular disorder: gene therapy approaches to sickle cell disease. Blood. 2016;127:839–848. doi: 10.1182/blood-2015-09-618587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson F.L., Look A.T., Gockerman J., Ruggiero M.R., Dalla-Pozza L., Billings F.T., 3rd Bone-marrow transplantation in a patient with sickle-cell anemia. N. Engl. J. Med. 1984;311:780–783. doi: 10.1056/NEJM198409203111207. [DOI] [PubMed] [Google Scholar]
- 7.Gluckman E. Allogeneic transplantation strategies including haploidentical transplantation in sickle cell disease. Hematology (Am. Soc. Hematol. Educ. Program) 2013;2013:370–376. doi: 10.1182/asheducation-2013.1.370. [DOI] [PubMed] [Google Scholar]
- 8.Bauer D.E., Brendel C., Fitzhugh C.D. Curative approaches for sickle cell disease: A review of allogeneic and autologous strategies. Blood Cells Mol. Dis. 2017;67:155–168. doi: 10.1016/j.bcmd.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 9.Braun C.J., Boztug K., Paruzynski A., Witzel M., Schwarzer A., Rothe M., Modlich U., Beier R., Göhring G., Steinemann D. Gene therapy for Wiskott-Aldrich syndrome--long-term efficacy and genotoxicity. Sci. Transl. Med. 2014;6:227ra33. doi: 10.1126/scitranslmed.3007280. [DOI] [PubMed] [Google Scholar]
- 10.Hacein-Bey-Abina S., Garrigue A., Wang G.P., Soulier J., Lim A., Morillon E., Clappier E., Caccavelli L., Delabesse E., Beldjord K. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hacein-Bey-Abina S., von Kalle C., Schmidt M., Le Deist F., Wulffraat N., McIntyre E., Radford I., Villeval J.L., Fraser C.C., Cavazzana-Calvo M., Fischer A. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- 12.Modlich U., Navarro S., Zychlinski D., Maetzig T., Knoess S., Brugman M.H., Schambach A., Charrier S., Galy A., Thrasher A.J. Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors. Mol. Ther. 2009;17:1919–1928. doi: 10.1038/mt.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein S., Ott M.G., Schultze-Strasser S., Jauch A., Burwinkel B., Kinner A., Schmidt M., Krämer A., Schwäble J., Glimm H. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat. Med. 2010;16:198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- 14.Thompson A.A., Walters M.C., Kwiatkowski J., Rasko J.E.J., Ribeil J.A., Hongeng S., Magrin E., Schiller G.J., Payen E., Semeraro M. Gene Therapy in Patients with Transfusion-Dependent β-Thalassemia. N. Engl. J. Med. 2018;378:1479–1493. doi: 10.1056/NEJMoa1705342. [DOI] [PubMed] [Google Scholar]
- 15.Marktel S., Scaramuzza S., Cicalese M.P., Giglio F., Galimberti S., Lidonnici M.R., Calbi V., Assanelli A., Bernardo M.E., Rossi C. Intrabone hematopoietic stem cell gene therapy for adult and pediatric patients affected by transfusion-dependent ß-thalassemia. Nat. Med. 2019;25:234–241. doi: 10.1038/s41591-018-0301-6. [DOI] [PubMed] [Google Scholar]
- 16.Ribeil J.A., Hacein-Bey-Abina S., Payen E., Magnani A., Semeraro M., Magrin E., Caccavelli L., Neven B., Bourget P., El Nemer W. Gene Therapy in a Patient with Sickle Cell Disease. N. Engl. J. Med. 2017;376:848–855. doi: 10.1056/NEJMoa1609677. [DOI] [PubMed] [Google Scholar]
- 17.Branzei D., Foiani M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 18.Chiruvella K.K., Liang Z., Wilson T.E. Repair of double-strand breaks by end joining. Cold Spring Harb. Perspect. Biol. 2013;5:a012757. doi: 10.1101/cshperspect.a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgan R.A., Gray D., Lomova A., Kohn D.B. Hematopoietic Stem Cell Gene Therapy: Progress and Lessons Learned. Cell Stem Cell. 2017;21:574–590. doi: 10.1016/j.stem.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoban M.D., Cost G.J., Mendel M.C., Romero Z., Kaufman M.L., Joglekar A.V., Ho M., Lumaquin D., Gray D., Lill G.R. Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood. 2015;125:2597–2604. doi: 10.1182/blood-2014-12-615948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeWitt M.A., Corn J.E., Carroll D. Genome editing via delivery of Cas9 ribonucleoprotein. Methods. 2017;121-122:9–15. doi: 10.1016/j.ymeth.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dever D.P., Bak R.O., Reinisch A., Camarena J., Washington G., Nicolas C.E., Pavel-Dinu M., Saxena N., Wilkens A.B., Mantri S. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539:384–389. doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoban M.D., Lumaquin D., Kuo C.Y., Romero Z., Long J., Ho M., Young C.S., Mojadidi M., Fitz-Gibbon S., Cooper A.R. CRISPR/Cas9-Mediated Correction of the Sickle Mutation in Human CD34+ cells. Mol. Ther. 2016;24:1561–1569. doi: 10.1038/mt.2016.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hendel A., Bak R.O., Clark J.T., Kennedy A.B., Ryan D.E., Roy S., Steinfeld I., Lunstad B.D., Kaiser R.J., Wilkens A.B. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015;33:985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dobson L., Reményi I., Tusnády G.E. CCTOP: a Consensus Constrained TOPology prediction web server. Nucleic Acids Res. 2015;43(W1) doi: 10.1093/nar/gkv451. W408-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sather B.D., Romano Ibarra G.S., Sommer K., Curinga G., Hale M., Khan I.F., Singh S., Song Y., Gwiazda K., Sahni J. Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Sci. Transl. Med. 2015;7:307ra156. doi: 10.1126/scitranslmed.aac5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bak R.O., Dever D.P., Reinisch A., Cruz Hernandez D., Majeti R., Porteus M.H. Multiplexed genetic engineering of human hematopoietic stem and progenitor cells using CRISPR/Cas9 and AAV6. eLife. 2017;6:e27873. doi: 10.7554/eLife.27873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J., DeClercq J.J., Hayward S.B., Li P.W., Shivak D.A., Gregory P.D., Lee G., Holmes M.C. Highly efficient homology-driven genome editing in human T cells by combining zinc-finger nuclease mRNA and AAV6 donor delivery. Nucleic Acids Res. 2016;44:e30. doi: 10.1093/nar/gkv1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J., Exline C.M., DeClercq J.J., Llewellyn G.N., Hayward S.B., Li P.W., Shivak D.A., Surosky R.T., Gregory P.D., Holmes M.C., Cannon P.M. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat. Biotechnol. 2015;33:1256–1263. doi: 10.1038/nbt.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinello L., Canver M.C., Hoban M.D., Orkin S.H., Kohn D.B., Bauer D.E., Yuan G.C. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol. 2016;34:695–697. doi: 10.1038/nbt.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeWitt M.A., Magis W., Bray N.L., Wang T., Berman J.R., Urbinati F., Heo S.J., Mitros T., Muñoz D.P., Boffelli D. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci. Transl. Med. 2016;8:360ra134. doi: 10.1126/scitranslmed.aaf9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magis W., DeWitt M.A., Wyman S.K., Vu J.T., Heo S.-J., Shao S.J., Hennig F., Romero Z.G., Campo-Fernandez B., McNeill M. In vivo selection for corrected β-globin alleles after CRISPR/Cas9 editing in human sickle hematopoietic stem cells enhances therapeutic potential. bioRxiv. 2018 [Google Scholar]
- 33.Fiorini C., Abdulhay N.J., McFarland S.K., Munschauer M., Ulirsch J.C., Chiarle R., Sankaran V.G. Developmentally-faithful and effective human erythropoiesis in immunodeficient and Kit mutant mice. Am. J. Hematol. 2017;92:E513–E519. doi: 10.1002/ajh.24805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McIntosh B.E., Brown M.E., Duffin B.M., Maufort J.P., Vereide D.T., Slukvin I.I., Thomson J.A. Nonirradiated NOD,B6.SCID Il2rγ-/- Kit(W41/W41) (NBSGW) mice support multilineage engraftment of human hematopoietic cells. Stem Cell Reports. 2015;4:171–180. doi: 10.1016/j.stemcr.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahmig S., Kronstein-Wiedemann R., Fohgrub J., Kronstein N., Nevmerzhitskaya A., Bornhäuser M., Gassmann M., Platz A., Ordemann R., Tonn T., Waskow C. Improved Human Erythropoiesis and Platelet Formation in Humanized NSGW41 Mice. Stem Cell Reports. 2016;7:591–601. doi: 10.1016/j.stemcr.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Charlesworth C.T., Camarena J., Cromer M.K., Vaidyanathan S., Bak R.O., Carte J.M., Potter J., Dever D.P., Porteus M.H. Priming Human Repopulating Hematopoietic Stem and Progenitor Cells for Cas9/sgRNA Gene Targeting. Mol. Ther. Nucleic Acids. 2018;12:89–104. doi: 10.1016/j.omtn.2018.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Ravin S.S., Li L., Wu X., Choi U., Allen C., Koontz S., Lee J., Theobald-Whiting N., Chu J., Garofalo M. CRISPR-Cas9 gene repair of hematopoietic stem cells from patients with X-linked chronic granulomatous disease. Sci. Transl. Med. 2017;9:eaah3480. doi: 10.1126/scitranslmed.aah3480. [DOI] [PubMed] [Google Scholar]
- 38.Cromer M.K., Vaidyanathan S., Ryan D.E., Curry B., Lucas A.B., Camarena J., Kaushik M., Hay S.R., Martin R.M., Steinfeld I. Global Transcriptional Response to CRISPR/Cas9-AAV6-Based Genome Editing in CD34+ Hematopoietic Stem and Progenitor Cells. Mol. Ther. 2018;26:2431–2442. doi: 10.1016/j.ymthe.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuo C.Y., Long J.D., Campo-Fernandez B., de Oliveira S., Cooper A.R., Romero Z., Hoban M.D., Joglekar A.V., Lill G.R., Kaufman M.L. Site-Specific Gene Editing of Human Hematopoietic Stem Cells for X-Linked Hyper-IgM Syndrome. Cell Rep. 2018;23:2606–2616. doi: 10.1016/j.celrep.2018.04.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schiroli G., Ferrari S., Conway A., Jacob A., Capo V., Albano L., Plati T., Castiello M.C., Sanvito F., Gennery A.R. Preclinical modeling highlights the therapeutic potential of hematopoietic stem cell gene editing for correction of SCID-X1. Sci. Transl. Med. 2017;9:eaan0820. doi: 10.1126/scitranslmed.aan0820. [DOI] [PubMed] [Google Scholar]
- 41.Pavel-Dinu M., Wiebking V., Dejene B.T., Srifa W., Mantri S., Nicolas C.E., Lee C., Bao G., Kildebeck E.J., Punjya N. Gene correction for SCID-X1 in long-term hematopoietic stem cells. Nat. Commun. 2019;10:1634. doi: 10.1038/s41467-019-09614-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hung K.L., Meitlis I., Hale M., Chen C.Y., Singh S., Jackson S.W., Miao C.H., Khan I.F., Rawlings D.J., James R.G. Engineering Protein-Secreting Plasma Cells by Homology-Directed Repair in Primary Human B Cells. Mol. Ther. 2018;26:456–467. doi: 10.1016/j.ymthe.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khan I.F., Hirata R.K., Russell D.W. AAV-mediated gene targeting methods for human cells. Nat. Protoc. 2011;6:482–501. doi: 10.1038/nprot.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rutledge E.A., Halbert C.L., Russell D.W. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J. Virol. 1998;72:309–319. doi: 10.1128/jvi.72.1.309-319.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aurnhammer C., Haase M., Muether N., Hausl M., Rauschhuber C., Huber I., Nitschko H., Busch U., Sing A., Ehrhardt A., Baiker A. Universal real-time PCR for the detection and quantification of adeno-associated virus serotype 2-derived inverted terminal repeat sequences. Hum. Gene Ther. Methods. 2012;23:18–28. doi: 10.1089/hgtb.2011.034. [DOI] [PubMed] [Google Scholar]
- 46.Wojda U., Noel P., Miller J.L. Fetal and adult hemoglobin production during adult erythropoiesis: coordinate expression correlates with cell proliferation. Blood. 2002;99:3005–3013. [PubMed] [Google Scholar]
- 47.Xiang J., Wu D.C., Chen Y., Paulson R.F. In vitro culture of stress erythroid progenitors identifies distinct progenitor populations and analogous human progenitors. Blood. 2015;125:1803–1812. doi: 10.1182/blood-2014-07-591453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brinkman E.K., Chen T., Amendola M., van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsiau T., Maures T., Waite K., Yang J., Kelso R., Holden K., Stoner R. Inference of CRISPR Edits from Sanger Trace Data. bioRxiv. 2018 doi: 10.1089/crispr.2021.0113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.