Abstract
Hypercholesterolemia is a condition that is characterized by very high levels of cholesterol in the blood and is a major correlating factor with heart disease. Indeed, high levels of the low-density lipoprotein (LDL) have been causally linked to the development of atherosclerotic cardiovascular disease (ASCVD). A method to specifically reduce cholesterol in the blood in a long-term, stable manner could prove therapeutically relevant. Cholesterol is removed from the blood by the LDL receptor (LDLR) in the liver. Others and we have discovered that a long non-coding RNA (lncRNA; BM450697) functions as an endogenous epigenetic regulator of LDLR and that the repression of this lncRNA by the action of small interfering RNAs (siRNAs) results in the activation of LDLR. We found here, through the interrogation of two siRNAs that can target this lncRNA, both in a transcriptional and post-transcriptional manner, that BM450697 functions as a local scaffold for modulating LDLR transcription. Moreover, we found that conjugation of α-N-acetylgalactosamine (GalNAc) with two lncRNA-directed siRNAs allows for direct liver cell targeting of this lncRNA and functional enhanced uptake of cholesterol. Collectively, these data suggest that targeting the BM450697 lncRNA regulator of LDLR may result in a more specific, long-term, targeted approach to regulating cholesterol in the blood.
Keywords: LDL, LDLR, lncRNA, cholesterol, transcriptional gene silencing, Galnac, siRNA, hypercholesterolemia
Introduction
Hypercholesterolemia is a condition characterized by high levels of cholesterol in the blood and is a major factor that correlates with heart disease. Specifically, apolipoprotein-B (ApoB)-containing lipoproteins have been established as the causative agents in the development of atherosclerotic cardiovascular disease (ASCVD).1 Low-density lipoprotein (LDL) is a member of the ApoB lipoproteins,1 and reducing LDL levels in the blood may lead to a decreased risk of developing ASCVD. LDL blood levels are regulated by the LDL receptor (LDLR) in the liver, where active cycling of cholesterol from the blood occurs.2, 3 Recent findings have suggested that long non-coding RNAs (lncRNAs) play an increasingly important role in cholesterol and lipid homeostasis. lncRNAs are non-coding transcripts that are over 200 nt in length and have been shown to modulate transcription or translation in a myriad of ways.4, 5 For example, lncRNAs have been shown to modulate transcription through the recruitment of chromatin remodelling factors6, 7, 8 or by acting as mRNA decoys to sponge away9 or disrupt transcription factors or transcriptional machinery from binding to promoter sites10 or as enhancer RNAs to modulate transcription.11, 12 In addition, lncRNAs have been shown to act as miRNA sponges, modulate splicing events, or alter activation states of proteins that lead to different transcriptional outcomes.4, 13, 14, 15
The modulation of LDL cholesterol occurs via a multi-step pathway.16, 17 One branch of this pathway controls the transcription and recycling of the LDLR, whereas the other branch modulates cholesterol biogenesis, and the cross-talk between the pathways maintains intracellular cholesterol levels.18, 19, 20 Several lncRNAs have been found to participate in the modulation of cholesterol. For example, the lncRNAs liver-expressed LXR-induced sequence (LeXis)21 and macrophage-expressed LXR-induced sequence (MeXis)22 have been found to be integral in cholesterol biogenesis through modulation of the liver X receptor (LXR) pathway in mouse hepatocytes and macrophages, respectively, and both may contribute to the development of atherosclerosis.21, 22 Further, overexpression of LeXis, using an adeno-associated virus (AAV) vector in mice, was found to significantly decrease cholesterol levels, suggesting that lncRNAs may be useful candidates for cholesterol maintenance.23 Matsui et al.24 found that small RNAs direct transcriptional gene activation (TGA) of LDLRs, indicating that a lncRNA is actively involved in LDLR regulation. This lncRNA (expressed sequence tag [EST] BM450697) is notably antisense (AS) to the LDLR promoter and is discordantly regulated relative to the LDLR, suggesting that this RNA may be an endogenous transcript involved in the direct regulation of LDLR transcription. Further, Matsui et al.24 determined that this lncRNA may be a useful therapeutic target for cholesterol modulation.
To this end, we sought to determine the molecular functioning of the lncRNA BM450697 to modulate LDLR mRNA levels and to identify candidate transcriptional-gene-silencing (TGS)-mediated small interfering RNAs (siRNAs) that may be therapeutically useful in modulating LDL levels. In addition, we used α-N-acetylgalactosamine (GalNAc)-conjugated siRNAs as a delivery tool to directly target the liver for this siRNA-mediated effect. To determine mechanistically the involvement of the lncRNA BM450697 in LDLR regulation, we both overexpressed and repressed BM450697 expression. Notably, we found that BM450697 is an active transcriptional modulator of LDLR and that two siRNAs were observed to repress BM450697 and induce LDLR expression.
Results
BM450697 Is a lncRNA That Is Located in Both the Nuclear and Cytoplasmic Fractions and Is Enriched at the Promoter of the LDLR Gene in the Form of DNA:RNA Hybrids
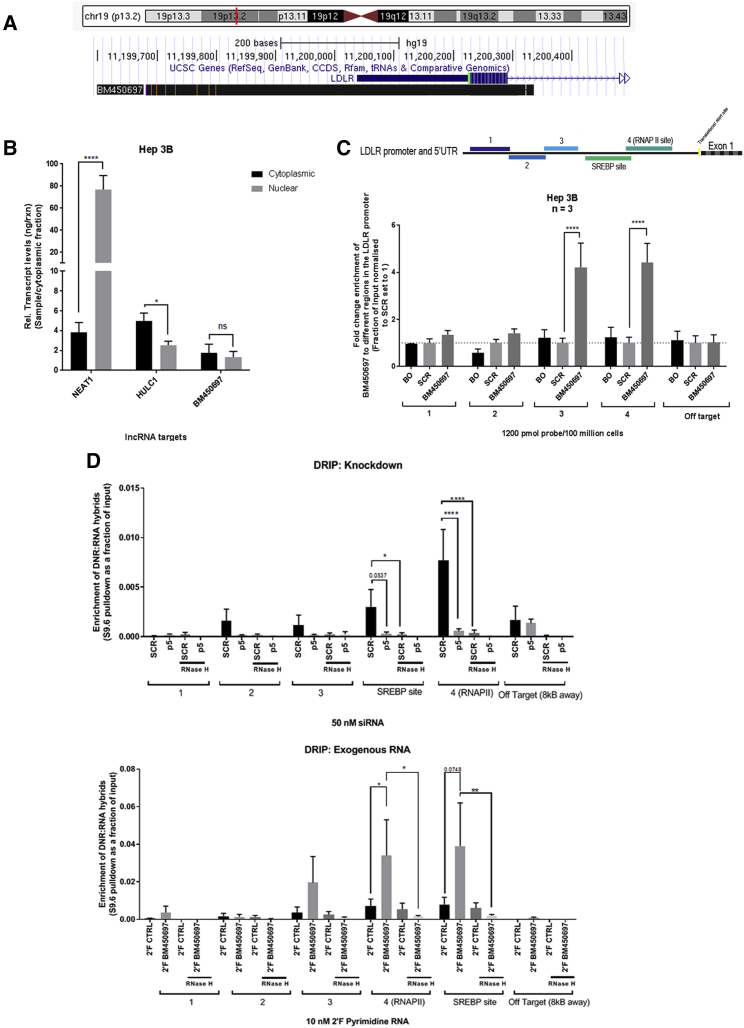
BM450697 is an AS RNA that overlaps the promoter of the LDLR gene according to the University of California, Santa Cruz (UCSC), Genome Browser (Figure 1A). According to the Annolnc database, BM450697 appears to be conserved in primates, but poorly conserved in vertebrates.25 Previous studies have observed that AS lncRNAs are functional in controlling the transcription of some gene promoters.5, 26 Recent studies with the LDLR gene observed an AS lncRNA BM450697 (Figure 1A) that may be functional in RNA directed activation of LDLR.24 To interrogate this notion further, we assessed both Hep 3B and Hep G2 cell lines for the expression of BM450697 by directional RT and qPCR (Figure S1A). Indeed, we observed that both Hep G2 and Hep 3B liver cells express BM450697 (Figure S1A), which is AS to the LDLR promoter (Figure 1A). In addition, we find using a standard curve method27 that BM450697 has a low level of expression (around 45 copies/ng) of RNA (Figure S1B) in Hep 3B cells. To further characterize this transcript, we confirmed that BM450697 is mostly polyadenylated, using poly deoxythymine (dT) magnetic beads to separate the RNA species (Figure S1C). Cytoplasmic fractionation showed that BM450697 was localized in both the cytoplasmic and nuclear fractions, using the nuclear lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) and the predominantly cytoplasmic lncRNA highly upregulated in liver cancer 1 (HULC1) as positive controls for each fraction (Figure 1B).
Figure 1.
BM450697 lncRNA Characterization in Hepatic Carcinoma Cell Lines
(A) A schematic diagram from the UCSC Genome Browser showing the position of BM450697 relative to the LDLR gene. (B) BM450697 appears to be equally present in the nuclear and cytoplasmic fractions. Hep 3B cells (10 million) were separated into nuclear and cytoplasmic fractions, using subcellular fractionation for RNA. HULC1 transcripts were used as positive controls for cytoplasmic expression, and NEAT1 was used as a positive control for nuclear expression. (C) BM450697 is enriched at the promoter site of LDLR in Hep 3B cells. Hep 3B cells (100 million) were harvested and incubated with either 5′ biotinylated ASOs toward BM450697 or scrambled controls, overnight at 37°C. Thereafter, a ChIRP assay was performed, the resultant genomic DNA was isolated, and subsequent qPCR was performed to determine the fold enrichment of BM450697 at the different promoter sites. BO, beads only; SCR, scrambled. (D) BM450697 formed DNA:RNA hybrids over the promoter region of the LDLR gene. Genomic DNA was isolated from either knockdown of BM450697 or exogenously added 2′ fluorinated BM450697 in Hep 3B cells after 72 h. Thereafter, 5 μg DNA was used per immunoprecipitation, in DNA samples treated with or without RNase H. Student’s t test was used in (B), comparing cytoplasmic and nuclear fractions for each gene amplified. Two-way ANOVA with the post hoc Tukey’s test was used in (C) and (D). *p < 0.05, **p < 0.01, and ***p < 0.0001.
We used the chromatin immunoprecipitation by RNA pulldown (ChIRP) method to identify the loci associated with BM450697 (Figure 1C). Using several primer sets that tile along the LDLR promoter, we observed that BM450697 was enriched near the 3′ end of the LDLR promoter near the translational start site (Figure 1C), suggesting that the transcript directly interacts with the DNA of the LDLR promoter. In addition, we isolated the RNA fraction from the ChIRP assay to verify BM450697 pulldown with our AS oligonucleotides (Figure S1D). These results suggest that BM450697 has a functional role in the nucleus, by acting as a cis-regulatory lncRNA. The ChIRP data suggest that BM450697 forms DNA:RNA hybrids at the 3′ end of the LDLR promoter. In order to elucidate whether we had a true DNA:RNA hybrids, we performed DNA:RNA immunoprecipitation (DRIP) assays in Hep 3B cells with either exogenously added 2′ fluorinated (2′F) pyrimidine BM450697 RNA or under knockdown conditions with the siRNA p5 that targets BM450697 (see Figure 3). We found that BM450697 forms DNA:RNA hybrids near the 3′ end of the promoter (Figure 1D), with a significant enrichment around the transcription factor binding site of sterol regulatory element binding protein (SREBP) and near the transcriptional start site (primer set 4). Further, these hybrids were lost with either RNase H treatment or under knockdown conditions. Similarly, we found that exogenously added BM450697 increased enrichment of these hybrids at the 3′ end of the LDLR promoter (Figure 1D). These observations and those of others24 strongly support the notion that BM450697 is involved in transcriptional control of LDLR.
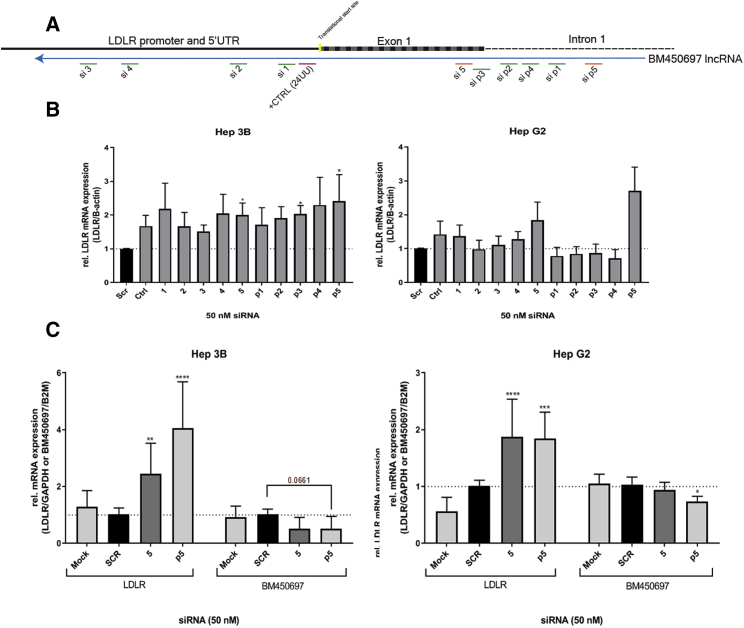
Figure 3.
siRNA Targeted Silencing of BM450697
(A) A schematic is shown depicting the tile of siRNAs developed for both the BM450697 transcript and its predicted promoter region. (B) siRNA screen targeting both BM450697 transcripts and predicted promoter in Hep G2 and Hep 3B cells. (C) siRNAs 4, 5, and p5 targeting of BM450697 resulted in activation of and a decrease in BM450697 in siRNA-treated Hep G2 and Hep 3B cells. (B and C) Data are representative of the mean ± SD of three independent experiments with triplicate treated conditions. The significance of differences between two groups was determined by non-parametric one-way ANOVA with the post hoc Dunn’s test (B) or by two-way ANOVA with the post hoc Tukey’s test or Student’s t test (C). *p < 0.05, **p < 0.01, and ***p < 0.0001.
BM450697 Modulates LDLR mRNA Levels through Transcriptional Interference
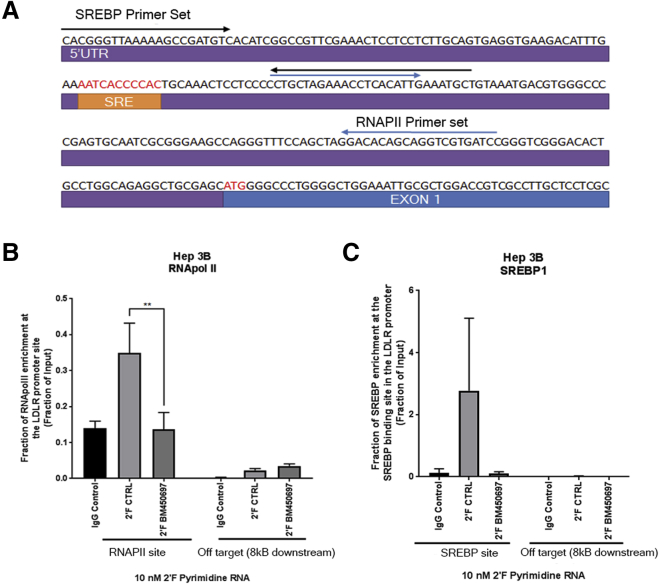
We sought to determine the mechanism by which BM450697 may regulate LDLR mRNA levels. Our ChIRP and DRIP assays suggested that BM450697 binds directly to the LDLR promoter and, based on our work and that of others,24 has a low level of expression. Thus, we postulated that BM450697 may act by interfering with transcription factor SREBP1a or RNA polymerase II (Pol II) association and/or binding at the LDLR promoter (Figure 2A). SREBP1a is one of the main transcription factors that drives LDLR gene expression in the cholesterol pathway;17thus, disruption of either SREBP1a or Pol II could result in a loss of LDLR transcription. We therefore performed chromatin immunoprecipitation (ChIP) assays with SREBP1a and Pol II (using the antibody toward the catalytically active phosphorylated serine 5 Pol II). We observed that when exogenous BM450697 RNA was transfected into Hep 3B cells, Pol II association at the LDLR promoter was significantly less compared with the control (scrambled, or a control lambda RNA; Figure 2B). Similarly, it appeared as if SREBP1a enrichment at the SREBP1 binding site was less than the control. However, this observation was not significant. Taken together, these data suggest that BM450697 decreases LDLR mRNA levels mechanistically by blocking interactions with Pol II and possibly SREBP1a at the promoter of LDLR.
Figure 2.
BM450697 Decreases LDLR mRNA Transcription through Displacement of Pol II and Possibly SREBP1a at the Promoter Site of LDLR
(A) A schematic of the LDLR promoter shows the position of the SREBP1 response element (SRE) and the primer sets used to amplify the select genomic regions for Pol II and SREBP1 binding. Hep 3B cells (2 million) were seeded and transfected the next day with either 10 nM 2′ fluorinated (2′F) pyrimidine BM450697 RNA or a lambda RNA control. Cells were cross-linked and processed for ChIP 48 h after transfection. (B) Pol II and (C) an SREBP1a ChIP. DNA was amplified with either the promoter-specific LDLR primer sets shown in (A) or with an off-target control primer set 8 kb upstream of the LDLR promoter site. Histograms are representative of the mean ± SEM of two independent experiments, performed in triplicate. One-way ANOVA with the post hoc Dunnett’s test was performed; **p < 0.01.
Repression of BM450697 by Small RNAs Induces the LDLR
Previous observations in studies of promoter regulatory AS lncRNAs suggest that they can be targeted and repressed by the action of siRNAs28 and AS oligonucleotides.5, 29 Rather than focus on the TGA of LDLR at the promoter site,24 we designed 10 siRNAs that could result in the TGS of the lncRNA, which could lead to an increase in LDLR expression, using a previously designed algorithm that generates siRNAs that target transcriptionally important regions.30 We interrogated the ability to target BM450697 with siRNAs tiled across the lncRNA (Figures 3A and S2). Relative to both scrambled controls and an siRNA control (CTRL, 24UU) from Matsui et al.24 we found that the siRNAs 5 and p5 appeared to induce LDLR expression (Figure 3B). Notably, the previously described LDLR activating siRNA (24UU, CTRL) and the siRNAs 5 and p5 all demonstrated activation of LDLR in both Hep G2 and Hep 3B cells (Figure 3B). Interestingly, p5 demonstrated the most reproducible repression of BM450697 in both Hep G2 and Hep 3B (Figures 3C and S3A) cells. We observed an ∼30% reduction in BM450697 expression levels compared with the scrambled control with both 5 and p5 siRNAs in Hep3B cells. In addition, p5 resulted in a significant decrease in BM450697 levels in Hep G2 cells (Figure 3C). Using intron-exon spanning primers, we observed that si5 and p5 increased nascent LDLR mRNA transcripts (Figure 3C). It has also been established that LDLR levels are regulated, in part, by a negative-feedback loop that increases PCSK9 levels.17 Thus, we wanted to determine whether knockdown of BM450697 increases PCSK9 mRNA levels. We found that siRNA 5 and p5 did not significantly increase PCSK9 mRNA levels (Figure S3B). Collectively, these data, juxtaposed with those presented in Figures 1 and 2 and observed by others,24 suggest that BM450697 is a transcriptional regulator of LDLR and can therefore serve as a convenient and potentially highly specific target to control LDLR expression via directed small RNAs.
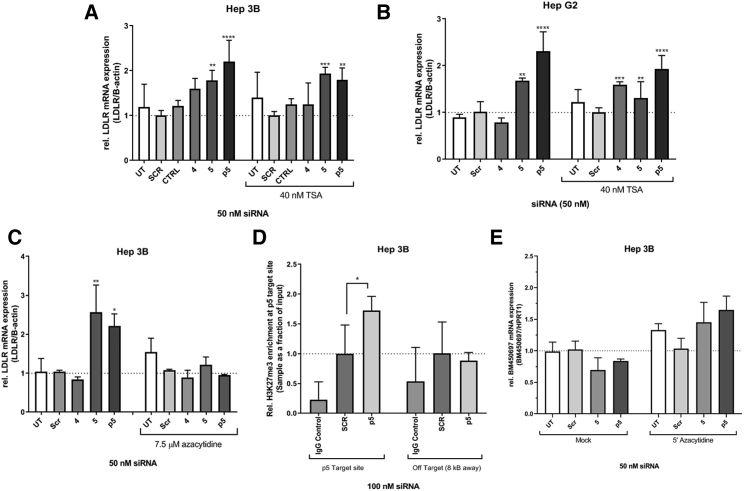
siRNA p5 May Function to Transcriptionally Control BM450697 Expression
We next wanted to elucidate whether any of these siRNAs that target BM450697 and result in the increase of LDLR mRNA expression levels are functional, by initiating TGS26 of BM450697. Using the ZENBU browser (http://fantom.gsc.riken.jp/zenbu) and the data obtained from Matsui et al.,24 we found that several of our target siRNAs were within potentially important transcriptional regions for BM450697 expression (Figure S2). Thus, we performed both trichostatin A (TSA) and 5′ azacytidine (5′ aza) experiments after siRNA treatment. TSA inhibits multiple histone deacetylases (HDACs), and whereas 5′ aza inhibits DNA methyltransferase activity, both histone and DNA-methylation-associated proteins have been thought to be involved in the TGS pathway.26 Interestingly, siRNA 5 and p5, were found to be resistant to TSA treatment (Figures 4A and 4B), whereas the siRNA-mediated effects on LDLR mRNA levels were lost when treated with 5′ aza in Hep 3B cells (Figure 4C). To confirm the 5′ aza findings, we performed a ChIP assay to determine whether there was an enrichment of the methylation marker H3K27me3 at the siRNA target site (Figure 4D and S5). We observed a significant increase in H3K27me3 at the p5 target site in the p5-transfected condition after 72 h, whereas no increase in H3K27me3 was observed at the LDLR promoter site (Figure S4). Further, when assessing the effects of 5′ aza on BM450697 mRNA expression, both si-5 and p5 silencing of the lncRNA were abrogated in the presence of 5′ aza (Figure 4E). Taken together, our data suggest that we may be able to direct TGS to the promoter region for BM450697 (Figure 4C), which controls LDLR expression and, ultimately, LDL uptake by the liver.
Figure 4.
siRNA p5 Functions in a Transcription-Silencing Manner
Hep 3B or Hep G2 cells were transfected with 50 nM siRNAs and co-incubated with DMSO or either (A and B) 40 nM trichostatin A (TSA) or (C and E) 7.5 μM 5′ azacytidine. (A and C) Hep 3B and (B) Hep G2 cells were harvested 72 h later and assessed for LDLR expression by qRT-PCR. (D) Hep 3B cells were transfected with 100 nM p5 and processed for ChIP with H3K27me3 or its mouse IgG control. DNA elutes were amplified with primers toward the target region of p5 or with an off-target primer set, located 8 kb upstream of the LDLR promoter in the LDLR gene. (E) Hep 3B treated with 7.5 μM 5′ azacytidine were harvested 72 h later and assessed for BM450697 expression by qRT-PCR. Data are the mean ± SD of three (A and B) or two (C–E) independent experiments with triplicate treated conditions. Two-way ANOVA (A–C) or one-way ANOVA (D) with the post hoc Tukey’s test was performed. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
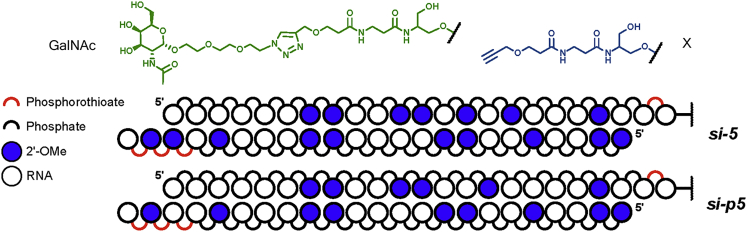
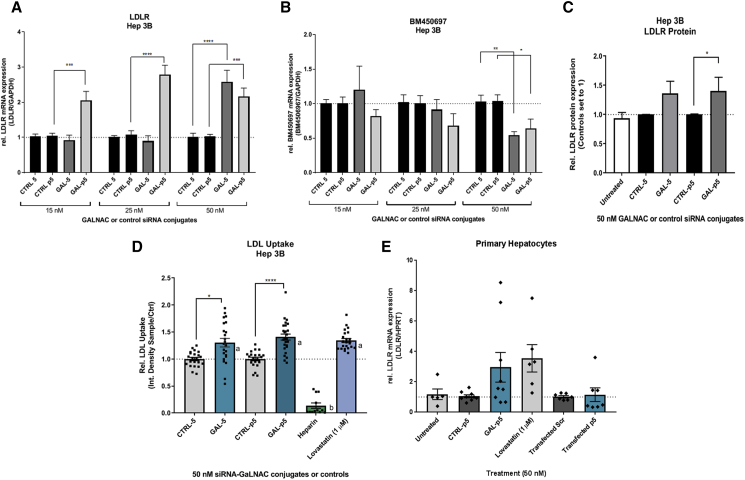
siRNA-GalNAc Conjugates Gal-5 and -p5 Functions to Transcriptionally Control BM450697 Expression and Subsequently LDL Uptake in Hep 3B Cells
All natural 5 and p5 siRNAs were converted into their corresponding therapeutic analogs by insertion of 2′-OMethyl (2′-OMe) on the sugar moiety and phosphorothioate (PS) linkages in the backbone. Furthermore, the sense (S) strands of each pair were modified with GalNAc on the 3′ end to yield the corresponding modified siRNA-GalNAc conjugates of 5 and p5, i.e., Gal-5 and Gal-p5 (Table 1), respectively. The unconjugated controls (CTRL-5 and CTRL-p5) contained a free alkyne on the 3′ end, as designated in Table 1. It has been well established in the literature that GalNAcs are readily recognized and taken up by the asialoglycoprotein receptor (ASGPR) in hepatocytes.31 Upon ligand binding, the ASGPRs are endocytosed, whereupon the receptor is recycled back to the surface of the cell, after which the therapeutic siRNA can exert its function through the RNA interference (RNAi) pathway or association with argonaute 1 (AGO1) complexes, in the cell.26, 31 Dosing of the siRNA conjugates showed that both Gal-5 and Gal-p5 had dose-dependent effects on LDLR mRNA and BM450697 RNA expression, with 50 nM showing significant effects for BM450697 repression and LDLR activation (Figures 5A and 5B). In addition, Gal-p5 appeared to be more potent than Gal-5, showing increased LDLR mRNA activation at 15 nM (Figure 5A). However, significant repression of BM450697 was observed only with 50 nM treatment with Gal-p5 (Figures 5A and 5B). It may be that a small reduction in the lncRNA is sufficient to elicit observable effects on LDLR mRNA expression, because of the low expression levels of this lncRNA in the cell (Figure S1B). In addition, we found a significant (∼1.4-fold) increase in LDLR protein levels, with 50 nM Gal-p5 in Hep 3B cells (Figure 5C), suggesting that this conjugate increases both LDLR mRNA and protein levels in Hep 3B cells. Upon validation of the siRNA-GalNAc conjugates, we next sought to determine whether these siRNA conjugates had any physiological effect on cholesterol uptake in the Hep 3B cell line. We observed that Gal-5 and Gal-p5 had significantly higher levels of uptake than their respective controls (unconjugated siRNAs of CTRL-5 and CTRL- p5; Figures 5D and S7). The increased LDL uptake (Figure 5D) in the Gal-5 and Gal-p5 conditions (1.3- and 1.4-fold, respectively) appear to mimic the observed increase in LDLR protein levels in Figure 5C, suggesting that the increase in uptake may be proportional to the increase in LDLR protein levels. We observed similar effects when transfecting Hep 3B cells with the natural siRNAs, 5 or p5, in Hep 3B cells (Figure S6). Furthermore, Gal-5 and -p5 had similar levels of LDL uptake compared to 1 μM Lovastatin (the positive control). Taken together, these data suggest that Gal-5 and Gal-p5 modulated LDL uptake in this cell line model.
Table 1.
Design of Candidate siRNAs-GalNAc Conjugates and Unconjugated Controls
S, sense; AS, antisense.
Figure 5.
Chemically Modified siRNA-GalNAc Conjugates Gal-5 and -p5 Significantly Increase LDLR mRNA Expression in Hep 3B and Primary Hepatocytes, Decrease the lncRNA BM450697, and Increase LDL uptake in Hep 3B Cells
Hep 3B cells were seeded at a concentration of 100,000 cells/well. siRNA conjugates targeted toward the lncRNA (Gal-5 and Gal-p5) or their corresponding unconjugated controls (CTRL-5 or CTRL-p5) were added to the cells at increasing concentrations (15, 25, and 50 nM). After 48 h, the cells were harvested for RNA and reverse transcribed with either random hexamers (A) or a strand-specific primer (B). (A) Relative LDLR mRNA expression levels and (B) relative BM450697 mRNA expression levels. Each siRNA-GalNAc conjugate was normalized to its respective unconjugated control with each concentration set to 1. Data are the mean ± SEM of three independent experiments, performed in triplicate. (C) An LDLR ELISA was used to determine LDLR protein levels with 50 nM treated with galNAc-siRNA. (A) A two-way ANOVA with a post hoc Tukey test was used; ***p < 0.001, ****p < 0.0001. (B) An unpaired Student’s t test (*p < 0.05, **p < 0.01) and (C) a non-parametric one-way ANOVA with a post hoc rank test (*p < 0.05) were performed. (D) siRNA conjugates (50 nM) were added to Hep 3B cells, after which the medium was replaced with serum-free EMEM 48 h later. As controls, Hep 3B cells were treated with 1 μM Lovastatin (replaced daily) and 100 nM heparin (30 min before the uptake assay). The cells were processed for LDL uptake 24 h after medium change, using an LDL uptake assay kit. Images were captured with the Nikon ECLPSE TS100 fluorescence light microscope, and data were analyzed using ImageJ. version 1.50i. Data represent the relative integrated density in three independent experiments, with three fields of view obtained for each experiment performed in triplicate. Data are the mean ± SEM. A non-parametric one-way ANOVA, with the post hoc Dunn’s test was used for comparisons between two conditions; ***p < 0.001 and ****p < 0.0001. When assessing the effects of Gal-5 and Gal-p5 to Lovastatin and heparin alone, a and b denote statistical significance; p < 0.001. (E) Primary hepatocytes were seeded at a concentration of 375,000 cells/well. Gal-p5 or its unconjugated control CTRL-p5 were added directly to the cells. Natural siRNAs (scrambled or p5) were transfected with RNAiMAX, as a control. After 48 h, the cells were harvested for RNA and 50 ng RNA was used in a one-step quantitative real-time PCR reaction. Relative LDLR mRNA expression levels Gal-5 or p5 was normalized to their respective unconjugated (CTRL-p5) or scrambled (Scr) control set to 1. Histograms are representative of the mean ± SEM of three independent experiments, performed in duplicate or triplicate.
siRNA-GalNAc Conjugate Gal-p5 Increases LDLR mRNA Expression in Primary Hepatocytes
Bolstered by our findings that Gal-p5 appeared to increase LDLR mRNA levels, decrease BM450697 RNA levels, and increase LDL uptake, we wanted to determine if some of these effects were evident in primary human hepatocytes. We observed that direct addition of the Gal-p5 conjugate lead to an ∼2.9-fold increase in LDLR mRNA expression, more so than the increase observed with the transfected natural siRNA p5 (Figure 5E) after 48 h. However, further optimizations are needed to improve the transfection efficiency in primary hepatocytes, as increased cytotoxicity related to the lipofectamine-based reagent may have masked the results after 48 h.32 Although these observations were not statistically significant, the trend suggests that the increase with Gal-p5 was similar to that with the positive control, 1 μM Lovastatin (Figure 5E). These data suggest that Gal-p5 is amenable to uptake in primary human hepatocytes and may increase LDLR mRNA levels ex vivo.
Discussion
Our understanding of gene regulation by RNAs has been rapidly expanding. lncRNAs represent a class of RNAs that exert gene modulatory effects in both the nucleus and cytoplasm. Nuclear effects include recruitment of chromatin remodelling factors to either repress or de-repress gene expression, working as enhancer RNAs, through transcriptional interference at the promoter site, or acting as a scaffold for regulatory proteins at target sites in the DNA.33 BM450697 is a lncRNA described by Matsui et al.24 It is a 1450 nt transcript that runs AS to the LDLR promoter. According to the coding potential calculator (CPC2), BM450697 has a coding potential of 0.16, classifying this RNA as non-coding.34
Based on our ChIRP and DRIP findings, this lncRNA is associated with the promoter of the LDLR gene in the form of DNA:RNA hybrids. We also determined that BM450697 is most likely to work through transcriptional interference, by preventing either transcriptional factor binding or Pol II binding to the promoter. Several studies have observed the effects of cis-acting lncRNAs in mediating transcriptional effects through interference. Martianov et al.35 observed that a non-coding transcript, upstream of the dihydrofolate reductase (DHFR) gene, interacts directly with the promoter region to form stable DNA:RNA triple-helix hybrids, displacing Pol II and the basal transcription initiation complex from the DHFR promoter. They also observed that this non-coding transcript directly interacts with the general transcription factor IIB, to mediate this repressive function on DHFR mRNA transcription. Martens et al.10 observed that the ncRNA SRG1 in Saccharomyces cerevisiae represses D-3-phosphoglycerate dehydrogenase 1 (SER3) through SRG1 transcription through the SER3 promoter, which leads to a displacement of transcriptional activators and Pol II binding to the SER3 gene. Interestingly, Latos et al.36 found that the lncRNA Airn causes silencing, not through its lncRNA product, but rather through the transcription of the lncRNA itself, which leads to the displacement of Pol II at the promoter of the Igf2r gene. The data reported here demonstrated that exogenously added BM450697 RNA led to a significant decrease in Pol II at the LDLR promoter. In addition, our ChIRP results suggest that lncRNA BM45069 is associated directly with the 3′ end of the LDLR promoter, and our DRIP assay confirmed the presence of an DNA:RNA hybrid. It thus appears that our mechanism is most likely to occur through an DNA:RNA triple helix or hybrid at the LDLR promoter that interferes with Pol II binding, rather than by transcriptional interference through lncRNA transcription, which would only occur in cis.
Having established and expanded upon the role of BM450697 in LDLR mRNA transcription, we next determined its suitability as a therapeutic target. Targeting LDLR expression may offer a treatment for type II familial hypercholesterolemia (FH)16 and ASCVD.1 Currently, statins represent the first-line strategy, targeting FH by increasing LDLR levels. However, 2%–12% of those with FH are unable to tolerate high-intensity statins because of the adverse effects associated with statin use.37, 38 Thus, an approach to directly targeting LDLR mRNA transcription may offer a suitable alternative to those unable to use statin therapy. Matsui et al.24 described that repressing BM450697 leads to a concomitant increase in LDLR expression, which we also observed. We found that siRNAs 5 and p5 appeared to be the most robust targets for LDLR expression and BM450697 silencing, and both did not significantly increase the expression in PCSK9 mRNA levels. Further, we observed that p5 repressed BM450697 RNA through a mechanism that may involve methylation related to the loss of the p5 effect with 5′ aza on both LDLR and BM450697 mRNA levels and an increased recruitment of H3K27me3 to the genomic target site of p5. The finding that siRNAs can mediate TGS through heterochromatin modification has been shown in several studies in yeast39, 40 and in mammalian cells, either through AGO1 recruitment41, 42 or through DNA methyltransferase 3A (DNMT3A) association at the target methylation site.26, 43 Taken together, the observations presented here suggest that p5 may be useful as an siRNA therapeutic that may mediate long-term silencing events of lncRNA BM450697 that results in stable activation of LDLR.
We converted siRNA 5 and p5 into therapeutically relevant constructs primed for liver targeting (Gal-5 and -p5), by using the liver-specific GalNAc ligand as a targeting appendage attached to the S strands of chemically modified versions of 5 and p5, to collectively, improve resistance toward nuclease activity, without increasing hepatotoxicity.44 The GalNAc moiety is recognized with high affinity by the ASGPR,45 exhibiting a turnover rate of approximately 15 min in the Hep G2 cell line.46 As such, the GalNAc conjugation of siRNAs has become an increasingly popular conjugation for liver-directed targets.31, 45 Gal-5 and Gal-p5 led to an increase in LDLR mRNA expression, while concomitantly decreasing BM450697 RNA expression, suggesting that these siRNAs-GalNAc conjugates exert their function in the cell and are delivered without the aid of a transfection reagent. Gal-p5 also significantly increased LDLR protein levels. Gal-5 and -p5 also increased LDL uptake to a similar extent, as 1 μM Lovastatin and Gal-p5 appeared to increase LDLR mRNA levels in primary hepatocytes. Collectively, the data presented here suggest that targeting the lncRNA BM450697 with GalNAc-conjugated siRNAs may offer a useful therapeutic approach to LDL maintenance (see Figure 6). Conjugation of GalNAc to siRNAs is a viable strategy for delivery of siRNAs into the liver. A completed phase II clinical trial using a PCSK9-targeted siRNA-GalNAc conjugate found that a single dose of 300 mg/person led to sustained low levels of LDL in patients for 180 days.47 Repression of PCSK9 results in the increased retention of LDLR on the cell surface because of the loss of PCSK9-targeted degradation of LDLR.18 Zimmermann et al.48 used a GalNAc-conjugated therapeutic siRNA, Revusiran, in a phase I clinical trial and found that this subcutaneously administered conjugate was well tolerated and reduced expression of its target transthyretin (TTR) by approximately 90%, with a multiple-dose strategy. The researchers found that the conjugate was well tolerated in humans and demonstrated robust and efficient knockdown of its target gene in vivo. However, Fitzgerald et al.47 reported several mild-to-moderate side effects in 11%–15% of the population treated with the PCSK9 inhibitor. Our strategy to target a lncRNA with a discreet expression profile and effect, may offer an alternative strategy to reduce off-target effects that may result from targeting coding genes. However, this remains to be determined. Future studies would need to include a humanized-liver mouse or primate model to validate our findings in an in vivo setting, as BM450697 appears, like other lncRNAs,29 to be conserved to humans and primates.25 In addition, the use of humanized mouse models has been shown to better recapitulate the effects of human hepatocyte function.49, 50 Further, we would need to validate the efficacy, LDL uptake and dosing strategy, and toxicity of our siRNA-GalNAc conjugate (Gal-p5) compared to other commercially available cholesterol-lowering strategies in different hepatic cell lines, as well as in primary human hepatocytes, to determine its true value as a therapeutic agent.
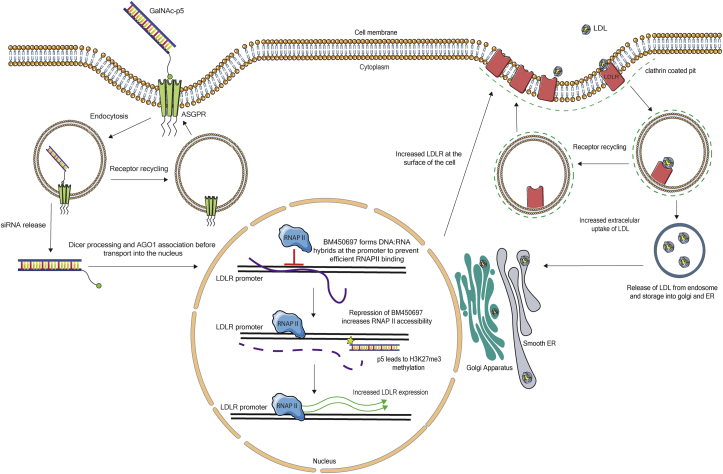
Figure 6.
Schematic Showing the Proposed Mechanism of BM450697 and the Proposed Effect of the Uptake of the siRNA-GalNAc Gal-p5 on BM450697 and LDLR Expression in the Cells
BM450697 expression results in a decrease in LDLR mRNA expression in hepatocytes by decreasing the accessibility of the Pol II binding site in the LDLR promoter. Upon GalNAc binding, the ASGPR is internalized into an endosome through endocytosis.31 Thereafter, the siRNA-GalNAc conjugate (Gal-p5) is processed and associates with AGO1.26 Once in the nucleus, the AGO1-associated complex exerts targeted gene silencing on BM450697, resulting in the loss of BM450697 through targeted TGS and H3k27me3 recruitment26 and an increase in LDLR mRNA expression. More LDLR protein is now processed and available in clathrin-coated pits located at the cell surface, allowing for an increase in LDL uptake and LDLR recycling to the surface of the cell.17, 20
The results of our study suggest that targeting of lncRNAs may result in positive therapeutic outcomes. Indeed, several reviews26, 44, 45 and studies in atherosclerosis and cholesterol maintenance12, 13, 14 have suggested that lncRNAs are viable targets that have robust and pronounced effects when targeted through RNAi-mediated strategies.21, 22, 23, 51, 52, 53 Invariably, there will be challenges in the delivery of RNAi-based therapeutics. However, the advent of the GalNAc-conjugated siRNA has shown great promise in liver-targeted diseases, with several GalNAc conjugates in phase I and II clinical trials.31 Our approach in targeting the TGS of BM450697 could be a promising strategy for long-term treatment of FH and those with hypercholesterolemia, where fewer doses of the siRNA-GalNAc conjugate are needed to maintain LDL levels in vivo.
Material and Methods
Cell Lines and Primary Hepatocytes
The human hepatocellular carcinoma cell lines Hep G2 and Hep 3B (ATCC, Manassas, VA, USA) were cultured in Eagle’s minimal essential medium (ATCC) supplemented with 10% fetal bovine serum (GeminiBio, Sacramento, CA, USA) at 37°C in a water-jacket incubator. The cells were mycoplasma free and were passaged twice weekly. Prepared and plateable primary human hepatocytes were purchased from Zen-Bio (Morrisville, NC, USA) and processed according to the manufacturer’s instructions, using human hepatocyte plating medium and human hepatocyte maintenance medium from the manufacturer (Zen-Bio). Primary hepatocytes were plated onto collagen-I-treated, 24-well plates purchased from Corning (Corning, NY, USA). The primary hepatocytes maintained their morphology for the duration of the study.
Antibodies, Plasmids, and All-Natural siRNAs
Pol II phosphorylated serine 5 C-terminal repeat domain (CTD; ab5131) and its respective rabbit IgG control antibody (ab171870), as well as mouse IgG controls (ab18413), were purchased from Abcam (Cambridge, UK). SREBP1 chip grade antibody (sc-13551X) was purchased from Santa Cruz (Dallas, TX, USA). S9.6 (MABE1095) was purchased from Merck Millipore (Burlington, MA, USA). A BM450697 expression vector was synthesized by GeneWiz (South Plainfield, NJ, USA) in a pcDNA3.1+ backbone. Ten all-natural siRNAs, designed to target the lncRNA BM450697 or its putative promoter region,30 were synthesized by Integrated Technologies (Skokie, IL, USA). Based on initial screening analysis, two siRNA pairs (5 and p5) were chosen for further investigation.
Design of Modified siRNA-GalNAc Conjugates
Therapeutic siRNA Gal-5 and Gal-p5 (Table 1) were designed to contain alternating 2′-OMe sugar modifications, along with PS linkages in the 3′ ends. Contrary to S strands bearing only a single PS linkage at the GalNAc-modified 3′ end, therapeutic AS strands were modified with three PS linkages at the 3′ end, to improve resistance toward 3′ exonuclease activity. Furthermore, AS strands were flanked at the 5 ends with two 3′-OMe modifications to improve stability toward 5 exonuclease, including alternating 2′-OMe motifs for protection against endonucleases.49
Synthesis of 2′-OH/2′-OMe PO/PS siRNAs
The corresponding base- and backbone-modified versions of siRNA 5 and p5 were designed to also contain a serinol alkyne modifier on the 3′ end of the S strands. Both pairs were synthesized in an N-dimethyltryptamine (DMT)-off mode on the Expedite 8909 (Perspective Biosystems, Framingham, MA, USA). RNA phosphoramidite reagents were purchased from Glen Research (Sterling, VA, USA); other reagents for the oligonucleotide synthesis were obtained from ABI Scientific (Sterling, VA, USA). The synthesis was carried out in a 1 μmol scale using 3′-alkyne-modifier serinol CPG and NittoPhase UnyLinker Solid Support.
After synthesis, the column was dried and emptied into a PCR-clean vial (RNase and DNase free), together with filters, and incubated with 1 mL methylamine (33% in ethanol) at 65°C for 2 h. The mixture was allowed to reach room temperature and kept at 4°C for an additional 30 min. The mixture was filtered, evaporated, and treated with 100 μL desilylation reagent (750 μL N-methyl-2-pyrrolidone, 375 μL triethylamine, and 500 μL triethylamine trihydrofluoride) on a thermomixer at 67°C for 3 h. Then, 10 μL quenching reagent (10 mL of 3 M NaOAc and 10 μL diethyl dicarbonate) was added, followed by the addition of 15 μL of 5 M NaClO4 and 10 μL of 3 M NaOAc. Cold acetone (1 mL) was added to precipitate the oligonucleotides. The vial was centrifuged at 1,000 rpm for 10 min and placed at 4°C for 30 min, and the supernatant was removed (washed three times). The pellet was dried on a speed-vac, redissolved in MilliQ, and subjected to ion exchange (IE) high-performance liquid chromatography (HPLC), using a Dionex UltiMate 3000 LC System equipped with a DNAPac PA-100 (9 × 250 mm; 0.5 M NaClO4, 5%–60% over 45 min; 0.25 M Tris-Cl [pH 8], 10% over 45 min; MilliQ water). Collected fractions were combined and subjected to desalting column chromatography (Illustra NAP-5; GE Healthcare), and the purity of each oligonucleotide 5X-S, 5-AS, p5X-S, and p5-AS was confirmed by the analytical IE HPLC (>90% purity). The identity of oligonucleotides was confirmed by MALDI-TOF mass spectrometry (MS; Bruker MicroTOF-Q II mass spectrometer) in positive mode, using 3-hydroxypicolinic acid matrix (10 mg/mL 3-hydroxypicolinic acid and 50 mM ammonium citrate in 70% aqueous acetonitrile). MS-calculated/MS-found values ([M+H]+) were 5-AS; 7394/7393, p5-AS; 7434/7432, 5X-S; 7930/7928, p5X-S; 7831/7830.
Synthesis of siRNA-GalNAc Conjugate S Strands
All reagents for the click reactions were obtained from Sigma-Aldrich (St. Louis, MO, USA) and were used as received. All reagents were prepared as fresh solutions in MilliQ water prior to setting up click reactions. Conjugation with α-GalNAc-PEG3-azide (Sigma-Aldrich, SMB00392) was carried out in an inert atmosphere (argon). A solution of an alkyne-modified oligonucleotide 5X-S or p5X-S (20 nmol, 12 μL) was added to 142 μL MilliQ water, after which α-GalNAc-PEG3-azide (200 nmol, 7.6 μL), aminoguanidine hydrochloride (50 mM, 4 μL), triethylammonium acetate (TEAA) buffer (1 M, pH 7.3, 20 μL), Cu(II) Tris((1-hydroxy-propyl-1H-1,2,3-triazol-4-yl)methyl)amine (THPTA; 10 mM, 10 μL), and ascorbic acid (50 mM, 4 μL) were added to get a total reaction volume of 200 μL. The reaction was stirred at room temperature for 12 h, after which the reaction mixture was subjected to gel filtration using a NAP-5 Sephadex column (Illustra; GE Healthcare, Pittsburgh, PA, USA). Purity was confirmed by the analytical IE HPLC at 20°C or 60°C above 90%). MS calculated/MS found values ([M+H]+): 5Gal-S, 8308/8306; and p5Gal-S, 8210/8208.
ChIRP Assay
The ChIRP assay was performed as described by Chu et al.54, 55 Briefly, 100 million cells were used per condition for the assay. The cells were harvested from 500 cm2 dishes in 1× PBS and cross-linked with 0.5% methanol-free formaldehyde (Sigma-Aldrich) for 10 min, before quenching the reaction with 0.125 mM glycine for 5 min at room temperature. Cells were washed once with ice-cold 1× PBS (Ca and Mg free; Thermo Fisher Scientific, Waltham, MA, USA) and resuspended in 10 mL 1× radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-Cl [pH 8.0], 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% SDS, and 40 mM NaCl) with 1× Halt Protease Inhibitor Cocktail and 0.02 vol SUPERase In RNase Inhibitor (Invitrogen). The cells were resuspended with a 26G needle, and lysed at 4°C for 45 min on a rotator, and centrifuged at 10,000 × g for 10 min at 4°C. The supernatant was removed, and the nuclei were resuspended in 1× micrococcal nuclease buffer (New England Biolabs). Two thousand units micrococcal nuclease (MNase) was added and the nuclei suspension was incubated at 37°C for 20 min. The MNase reaction was inhibited by using 0.1 vol 0.5 M EDTA. Nuclei were centrifuged at 10,000 × g for 10 min at 4°C and resuspended in 1 mL nuclear lysis buffer (1× PBS, 1% SDS, 1% NP-40, and 1% Na-deoxycholate with 1× Halt Protease Inhibitor and 0.02 vol SUPERase In RNase Inhibitor), resuspended with a 26G needle, and rotated for 30 min at 4°C. The nuclei were subsequently sonicated for 10 cycles (30 s on and 30 s off), to rupture the nuclei, and centrifuged at 10,000 × g for 10 min at 4°C, to remove the debris. The supernatant was collected, and 10% was collected as the input. The remainder of the nuclei lysate was used in the ChIRP assay.
Lysates were pre-cleared using Dynabeads MyOne Streptavidin C1 (Invitrogen) for 2 h at 4°C with rotation. Thereafter, biotinylated AS oligonucleotides (ASOs) toward BM450697 (Table S1) and scrambled biotinylated ASOs (Table S1) were used for each condition, to a final concentration of 1,200 pmol. Nuclei lysates were incubated in hybridization buffer (750 mM NaCl, 1% SDS, 50 mM Tris-Cl [pH 7.0], 1 mM EDTA, and 15% formamide) with 1× Halt Protease Inhibitor Cocktail and 0.02 vol SUPERase In RNase Inhibitor. Hybridization occurred overnight at 37°C. Dynabeads MyOne Streptavidin C1 (Invitrogen) were washed according to the manufacturer’s instructions and resuspended in nuclear lysis buffer. Then, 200 μL of the prepared magnetic beads was used for each ChIRP condition. Magnetic beads were incubated with the lysates for 30 min at 37°C with rotation. The beads were collected on a magnetic stand and washed five times with 2× saline sodium citrate (SSC) buffer (Invitrogen) with 0.5% SDS.
DNA was eluted off the beads, using 100 μg RNase A (Thermo Fisher Scientific) and 100 U RNase H (New England Biolabs) in 1 mL elution buffer (50 nM NaHCO3 and 1% SDS) at 37°C for 30 min with rotation. Thereafter, the elutes were collected, treated with proteinase K (New England Biolabs), and reverse cross-linked overnight at 65°C. DNA was isolated using the QIAGEN PCR purification kit.
The remainder of the reaction was used for RNA isolation with QIAzol (QIAGEN, Hilden, Germany), followed by a TURBO DNase treatment (Ambion, Invitrogen) for 30 min. Primer-specific RT was performed using the Quantitect Reverse Transcription kit (QIAGEN). qPCR was performed as previously described, using primers for the LDLR promoter (DNA) as well as primers specific for the lncRNA BM450697 (RNA; see Table S1 for the primer sets).
ChIP and DRIP Assay
Linearized BM450697 DNA or a lambda DNA control were transcribed using the Durascribe Synthesis Kit (Lucigen, Middleton, WI, USA). Thereafter, the DNA-free 2′F RNA was dephosphorylated with calf intestinal phosphatase (New England Biolabs) for 30 min at 37°C. The RNA was subsequently purified with the RNA MEGAClear Kit (Ambion, Invitrogen), according to the manufacturer’s instructions.
For the ChIP assay, 2 million Hep 3B cells were seeded onto 15 cm2 dishes. The next day, the cells were transfected with 10 nM BM450697 or the control lambda RNA or with 100 nM scrambled or p5 siRNA, and the media were replaced 24 h later. After 24 (RNA) or 48 (siRNA) h, the cells were cross-linked with 0.5% methanol-free formaldehyde for 10 min, before the reaction was quenched with 0.125 mM glycine for 5 min at room temperature. The cells were washed once with ice cold 1× PBS and resuspended in 1 mL 1× RIPA buffer with 1× Halt Protease Inhibitor Cocktail. They were resuspended with a 26G needle, lysed at 4°C for 45 min on a rotator, and centrifuged at 10,000 × g for 10 min at 4°C. The supernatant was removed, and the nuclei were resuspended in 1× micrococcal nuclease buffer (New England Biolabs). Four hundred units of micrococcal nuclease (MNase) was added, and the nuclei suspension was incubated at 37°C for 20 min. The MNase reaction was inhibited using 0.1 vol 0.5 M EDTA. The nuclei were centrifuged at 10,000 × g for 10 min at 4°C and resuspended in 1 mL nuclear lysis buffer (with 1× Halt Protease Inhibitor Cocktail), resuspended with a 26G needle, and rotated for 30 min at 4°C. The nuclei were subsequently sonicated for five cycles (30 s on and 30 s off ), to rupture the nuclei, and centrifuged at 10,000 × g for 10 min at 4°C, to remove debris. The supernatant was collected, and 10% was used as the input. MNase-digested lysates were incubated with 2 μg antibody overnight with rotation at 4°C. Thereafter, 20 μL magnetic protein A/G beads (Thermo Fisher Scientific) was added to the lysates for 2 h at 4°C with rotation. Using a magnetic rack, bound beads were washed four times with a high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-Cl [pH 8.1], and 500 mM NaCl), one time with LiCl washing buffer (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, and 10 mM Tris-Cl [pH 8.1]), and one time with 1× Tris-EDTA (TE) buffer (Ambion, Invitrogen). Samples were subsequently eluted in 100 μL elution buffer (1% SDS and 0.1 M NaHCO3) for 20 min at 68°C. The elutes were subjected to sequential RNase and proteinase K digestion for 1 h at 37°C, before reverse cross-linking overnight at 65°C. DNA was isolated by using a PCR purification kit (QIAGEN). To analyze the ChIP data, qPCR was performed with a standard curve in each run. The calculated concentrations were used to determine the enrichment of the protein at the target DNA site, as a fraction of input.
DRIP assays were performed according to Halász et al.56 Briefly, after treatment with either exogenously added 2′F control or 2′F BM450697 or with 100 nM scrambled or siRNA p5, cells were harvested for DNA using the Maxwell RSC Cultured Cells DNA Kit (Promega, Madison, WI, USA). Thereafter, cells were sonicated for 10 cycles of 30 s on and 30 s off, to shear the DNA to approximately 500 bp fragments. Thereafter, 5 μg DNA was used per condition, with half of the samples treated with RNase H (New England Biolabs) overnight at 37°C. RNase H-treated samples were purified using the DNA Clean and Concentrator kit (Zymo Research, Irvine, CA, USA). DNA samples were incubated with the S9.6 antibody, which specifically recognizes DNA:RNA hybrids (Merck Millipore), overnight at 4°C in IP buffer (50 mM HEPES/KOH [pH 7.5], 0.14 M NaCl, 5 mM EDTA, 1% Triton X-100, and 0.1% Na-deoxycholate), after which pre-blocked magnetic protein A/G beads were added for 2 h at 4°C. Samples were washed successively with 1 mL low-salt buffer (50 mM HEPES/KOH [pH 7.5], 0.14 M NaCl, 5 mM EDTA [pH 8], 1% Triton X-100, and 0.1% Na-deoxycholate), 1 mL high-salt buffer (50 mM HEPES/KOH [pH 7.5], 0.5 M NaCl, 5 mM EDTA [pH 8], 1% Triton X-100, and 0.1% Na-deoxycholate), 1 mL LiCl washing buffer (10 mM Tris-HCl [pH 8], 0.25 M LiCl, 0.5% NP-40, 0.5% Na-deoxycholate, and 1 mM EDTA [pH 8]), and two washes of 1 mL TE (100 mM Tris-HCl [pH 8] and 10 mM EDTA [pH 8]) at 4°C. Thereafter, samples were eluted with 100 μL elution buffer (50 mM Tris-HCl [pH 8], 10 mM EDTA, and 1% SDS) for 15 min at 65°C. After purification, the nucleic acids were cleaned with the DNA Clean and Concentrator kit (Zymo Research, Irvine, CA, USA) in 30 μL TE buffer. The recovered DNA was then analyzed by qPCR, as mentioned above.
RNA, cDNA, and Quantitative Real-Time PCR
Cells were transfected with siRNAs (1–5 and p1–p5) using Lipofectamine RNAiMAX (Invitrogen) in Opti-MEM (GIBCO, Invitrogen). Briefly, the diluted siRNAs in Opti-MEM were added to RNAiMAX in Opti-MEM and incubated at room temperature for 20 min. Hep 3B cells were plated at a density of 75,000 cells/24-well plate and transfected the next day with the duplex siRNAs, using 1.5 μL RNAiMAX per condition. Hep G2 cells were reverse transfected at a density of 75,000 cells/24-well plate with 50 nM siRNA, using 2.4 μL RNAiMAX per condition. The media were replaced the following day, and the cells were harvested 72 h after transfection. In testing the modified siRNA-GalNAc conjugates (CTRL-5, CTRL-p5, Gal-5, and Gal-p5), the conjugates were added directly to the wells of either Hep 3B cells or primary hepatocytes, in the absence of any transfection reagent. Cells were harvested 48 h after transfection. RNA was isolated by using the SimplyRNA kit for RNA isolation (Promega), according to the manufacturer’s instructions. cDNA was synthesized by reverse transcribing 250 ng total RNA using the Quantitect Reverse Transcription kit, which includes a genomic DNA-wipeout step (QIAGEN). Quantitative real-time PCR was performed with 2× Fast Universal qPCR Mix (Kapa Biosystems, Wilmington, MA, USA), according to the manufacturer’s instructions, on a Roche LightCycler 96 real-time PCR system (Roche, Basel, Switzerland). Thermal cycling parameters started with 3 min at 95°C, followed by 40 cycles of 95°C for 3 s and 60°C for 30 s. Specificity of the PCR products was verified by melting-curve analysis (primers used in this study can be found in Table S1). To amplify BM450697, primer-specific RT was employed, with a primer complementary to the 3′ end of the lncRNA (see Table S1). Thereafter, touchdown qPCR was performed with the BM450697 primer set, downstream of the RT-specific primer. Thermal cycling parameters started with 3 min at 95°C, with touchdown PCR from 70°C to 60°C for 10 cycles, followed by 40 cycles of 95°C for 3 s and 60°C for 30 s.
For primary hepatocytes LDLR and HPRT1 were amplified, using the Luna Universal One-Step qRT-PCR kit (New England Biolabs). Briefly, 50 ng RNA was added to each amplification reaction, with the following thermal cycling parameters: 10 min at 55°C for RT and 1 min initial denaturation at 95°C, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s.
All data were analyzed by the 2−ΔΔCt method, calibrated to a reference gene, and normalized to the scrambled-control- or control-treated sample set to 1.
Poly(dT) Enrichment and Depletion
Ten million Hep 3B cells were used per experiment, and poly(dT) RNA was selected for use with the Oligo(dT)25 Dynabeads magnetic capture kit, according to the manufacturer’s instructions (Thermo Fisher Scientific). The negative fraction was used as the poly(dT)-depleted fraction. One microgram of RNA was converted to cDNA in either BM450697-specific RT primer or with random hexamers, using the Quantitect Reverse Transcription kit (QIAGEN). PCR was performed as described above for BM450697 amplification. The PCR products were separated on a 1.5% 1× Tris-acetate-EDTA (TAE) agarose gel at 100 V for 40 min and visualized using a Bio-Rad EZ Imager (Bio-Rad, Hercules, CA, USA).
Subcellular Fractionation
Ten million Hep 3B cells were used per experiment, in duplicate, according to the fractionation protocol for RNA by Gagnon et al.57 DNase-treated, total isolated RNA was converted to cDNA using the Quantitect Reverse Transcription kit (QIAGEN) with random hexamers or an RT-specific primer. In order to assess the quality of the fractionation, qPCR was performed, amplifying known nuclear (NEAT1) or cytoplasmic (HULC1) lncRNAs. BM450697 was amplified as described above (see Table S1 for primers). Relative amounts of each transcript were determined with a standard curve with Hep 3B genomic DNA and described as nanograms per reaction. The PCR products were separated on a 1.5% 1× TAE agarose gel at 100 V for 40 min with a Bio-Rad EZ Imager (Bio-Rad).
TSA and 5′ Aza Experiments
Hep 3B and Hep G2 cells were seeded at a density of 100,000 cells/12-well plate, and transfected as previously described. Trichostatin A (Merck Millipore) was added to Hep 3B cells at a final concentration of 40 nM. The cells were harvested 72 h later for RNA, as previously described. 5′ Aza (Sigma-Aldrich, St. Louis, MO) was added daily to a final concentration of 7.5 μM, and the cells were harvested at 72 h for total RNA isolation.
LDL Uptake Assay
Hep 3B cells were seeded at a density of 75,000 cells/24-well plate. The next day, modified conjugates (CTRL5, CTRL-p5, Gal-5, or Gal-p5) were added directly to each well, whereas unmodified siRNAs (scrambled, 5, or p5) were transfected using 1.5 μL RNAiMAX per condition. Media were replaced at 48 h with serum-free EMEM. The LDL uptake assay was performed according to the manufacturer’s instructions, using BODIPY-LDL at 2.5 μg/mL LDL per well (Molecular Probes, Invitrogen) the next day. Images were acquired with a Nikon ECLPSE TS100 fluorescence light microscope. Images were acquired using Nikon NIS Elements, and data were analyzed, using ImageJ, version 1.50i (Wayne Rasband; NIH, Bethesda, MD, USA). Lovastatin (Insolution Lovastatin, sodium salt; CalBiochem, Merck Millipore) at 1 μM was used as a positive control, with the medium replaced and fresh Lovastatin added each day. Heparin at 100 μg/mL was used as the negative control to prevent LDL uptake and was added 30 min prior to the assay (Thermo Fisher Scientific).
LDLR ELISA
Hep 3B cells were seeded at a density of 75,000 cells/24-well plate. The next day, modified conjugates (CTRL5, CTRL-p5, Gal-5, or Gal-p5) were added directly to each well, and 72 h later, total protein was harvested using RIPA buffer. LDLR protein levels were determined with a human LDLR Quantikine ELISA kit (R&D Systems, Minneapolis, MN, USA), according to the manufacturer’s instructions. Absorbance was measured on a Promega GloMax Explorer microplate reader (Promega).
Statistical Analysis
All experiments were performed in triplicate, with at least two biological experiments performed per data set. Graphing and statistical analyses were performed using Prism for Windows, version 7.03 (GraphPad Software, La Jolla, CA, USA). For experiments with multiple comparisons, one- or two-way ANOVA (parametric or non-parametric) was performed, with a post hoc Tukey’s, Dunnett’s, or non-parametric Dunn’s test. Where only two groups were compared, the two-tailed unpaired Student’s t-test was performed. Details of the graphical representation, statistics used, and n values for each figure are described in each legend.
Author Contributions
R.M.R. and K.V.M. conceived of and designed the experiments; R.M.R. conducted the experiments; A.H.H, S.S, M.T., and K.V. designed and synthesized the siRNA-GalNAc conjugates; and R.M.R. and K.V.M. wrote the main manuscript and prepared the tables and figures. All authors reviewed the manuscript.
Conflicts of Interest
R.M.R. and K.V.M. have patent PCT/US2018/016727. The remaining authors declare no competing interests.
Acknowledgments
This project was supported by NIH NIAID grant P01 AI099783-01 and NIH grant DK104681-02 to K.V.M. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We would like to thank the mass spectrometry core at City of Hope.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.05.024.
Supplemental Information
References
- 1.Borén J., Williams K.J. The central role of arterial retention of cholesterol-rich apolipoprotein-B-containing lipoproteins in the pathogenesis of atherosclerosis: a triumph of simplicity. Curr. Opin. Lipidol. 2016;27:473–483. doi: 10.1097/MOL.0000000000000330. [DOI] [PubMed] [Google Scholar]
- 2.Wójcik C. Incorporation of PCSK9 inhibitors into prevention of atherosclerotic cardiovascular disease. Postgrad. Med. 2017;129:801–810. doi: 10.1080/00325481.2017.1376570. [DOI] [PubMed] [Google Scholar]
- 3.Brown M.S., Goldstein J.L. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 4.Ma L., Bajic V.B., Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris K.V., Mattick J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B.R., Protacio A., Flynn R.A., Gupta R.A. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Ruscio A., Ebralidze A.K., Benoukraf T., Amabile G., Goff L.A., Terragni J., Figueroa M.E., De Figueiredo Pontes L.L., Alberich-Jorda M., Zhang P. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Y., Rowley M.J., Böhmdorfer G., Wierzbicki A.T. A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Mol. Cell. 2013;49:298–309. doi: 10.1016/j.molcel.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang W.C., Fu W.M., Wong C.W., Wang Y., Wang W.M., Hu G.X., Zhang L., Xiao L.J., Wan D.C., Zhang J.F., Waye M.M. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513–22525. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martens J.A., Laprade L., Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 11.Ørom U.A., Derrien T., Beringer M., Gumireddy K., Gardini A., Bussotti G., Lai F., Zytnicki M., Notredame C., Huang Q. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim T.-K., Hemberg M., Gray J.M., Costa A.M., Bear D.M., Wu J., Harmin D.A., Laptewicz M., Barbara-Haley K., Kuersten S. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kung J.T.Y., Colognori D., Lee J.T. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L.L. Linking Long Noncoding RNA Localization and Function. Trends Biochem. Sci. 2016;41:761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Liu G., Zheng X., Xu Y., Lu J., Chen J., Huang X. Long non-coding RNAs expression profile in HepG2 cells reveals the potential role of long non-coding RNAs in the cholesterol metabolism. Chin. Med. J. (Engl.) 2015;128:91–97. doi: 10.4103/0366-6999.147824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouhairie V.E., Goldberg A.C. Familial hypercholesterolemia. Cardiol. Clin. 2015;33:169–179. doi: 10.1016/j.ccl.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horton J.D., Goldstein J.L., Brown M.S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levy E., Ben Djoudi Ouadda A., Spahis S., Sane A.T., Garofalo C., Grenier É., Emonnot L., Yara S., Couture P., Beaulieu J.F. PCSK9 plays a significant role in cholesterol homeostasis and lipid transport in intestinal epithelial cells. Atherosclerosis. 2013;227:297–306. doi: 10.1016/j.atherosclerosis.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 19.Attie A.D., Seidah N.G. Dual regulation of the LDL receptor--some clarity and new questions. Cell Metab. 2005;1:290–292. doi: 10.1016/j.cmet.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 20.Moore K.J., Rayner K.J., Suárez Y., Fernández-Hernando C. microRNAs and cholesterol metabolism. Trends Endocrinol. Metab. 2010;21:699–706. doi: 10.1016/j.tem.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sallam T., Jones M.C., Gilliland T., Zhang L., Wu X., Eskin A., Sandhu J., Casero D., Vallim T.Q., Hong C. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature. 2016;534:124–128. doi: 10.1038/nature17674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sallam T., Jones M., Thomas B.J., Wu X., Gilliland T., Qian K., Eskin A., Casero D., Zhang Z., Sandhu J. Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nat. Med. 2018;24:304–312. doi: 10.1038/nm.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tontonoz P., Wu X., Jones M., Zhang Z., Salisbury D., Sallam T. Long Noncoding RNA Facilitated Gene Therapy Reduces Atherosclerosis in a Murine Model of Familial Hypercholesterolemia. Circulation. 2017;136:776–778. doi: 10.1161/CIRCULATIONAHA.117.029002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsui M., Sakurai F., Elbashir S., Foster D.J., Manoharan M., Corey D.R. Activation of LDL receptor expression by small RNAs complementary to a noncoding transcript that overlaps the LDLR promoter. Chem. Biol. 2010;17:1344–1355. doi: 10.1016/j.chembiol.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou M., Tang X., Tian F., Shi F., Liu F., Gao G. AnnoLnc: a web server for systematically annotating novel human lncRNAs. BMC Genomics. 2016;17:931. doi: 10.1186/s12864-016-3287-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinberg M.S., Morris K.V. Transcriptional gene silencing in humans. Nucleic Acids Res. 2016;44:6505–6517. doi: 10.1093/nar/gkw139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whelan J.A., Russell N.B., Whelan M.A. A method for the absolute quantification of cDNA using real-time PCR. J. Immunol. Methods. 2003;278:261–269. doi: 10.1016/s0022-1759(03)00223-0. [DOI] [PubMed] [Google Scholar]
- 28.Morris K.V., Santoso S., Turner A.M., Pastori C., Hawkins P.G. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnsson P., Ackley A., Vidarsdottir L., Lui W.O., Corcoran M., Grandér D., Morris K.V. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat. Struct. Mol. Biol. 2013;20:440–446. doi: 10.1038/nsmb.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ackley A., Lenox A., Stapleton K., Knowling S., Lu T., Sabir K.S., Vogt P.K., Morris K.V. An Algorithm for Generating Small RNAs Capable of Epigenetically Modulating Transcriptional Gene Silencing and Activation in Human Cells. Mol. Ther. Nucleic Acids. 2013;2:e104. doi: 10.1038/mtna.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Y. Preclinical and Clinical Advances of GalNAc-Decorated Nucleic Acid Therapeutics. Mol. Ther. Nucleic Acids. 2017;6:116–132. doi: 10.1016/j.omtn.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park J.-S., Surendran S., Kamendulis L.M., Morral N. Comparative nucleic acid transfection efficacy in primary hepatocytes for gene silencing and functional studies. BMC Res. Notes. 2011;4:8. doi: 10.1186/1756-0500-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen H., Du G., Song X., Li L. Non-coding Transcripts from Enhancers: New Insights into Enhancer Activity and Gene Expression Regulation. Genomics Proteomics Bioinformatics. 2017;15:201–207. doi: 10.1016/j.gpb.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang Y.J., Yang D.C., Kong L., Hou M., Meng Y.Q., Wei L., Gao G. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017;45(W1):W12–W16. doi: 10.1093/nar/gkx428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martianov I., Ramadass A., Serra Barros A., Chow N., Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 36.Latos P.A., Pauler F.M., Koerner M.V., Şenergin H.B., Hudson Q.J., Stocsits R.R., Allhoff W., Stricker S.H., Klement R.M., Warczok K.E. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 37.Li B., Hao P.-P., Zhang Y., Yin R.-H., Kong Q.-Z., Cai X.-J., Zhao Z., Qi J.-N., Li Y., Xiao J. Efficacy and safety of proprotein convertase subtilisin/kexin type 9 monoclonal antibody in adults with familial hypercholesterolemia. Oncotarget. 2017;8:30455–30463. doi: 10.18632/oncotarget.10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waters D.D., Hsue P.Y., Bangalore S. PCSK9 Inhibitors for Statin Intolerance? JAMA. 2016;315:1571–1572. doi: 10.1001/jama.2016.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noma K., Sugiyama T., Cam H., Verdel A., Zofall M., Jia S., Moazed D., Grewal S.I.S. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat. Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- 40.Verdel A., Jia S., Gerber S., Sugiyama T., Gygi S., Grewal S.I.S., Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim D.H., Villeneuve L.M., Morris K.V., Rossi J.J. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 42.Janowski B.A., Huffman K.E., Schwartz J.C., Ram R., Nordsell R., Shames D.S., Minna J.D., Corey D.R. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat. Struct. Mol. Biol. 2006;13:787–792. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- 43.Weinberg M.S., Villeneuve L.M., Ehsani A., Amarzguioui M., Aagaard L., Chen Z.X., Riggs A.D., Rossi J.J., Morris K.V. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA. 2006;12:256–262. doi: 10.1261/rna.2235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Janas M.M., Schlegel M.K., Harbison C.E., Yilmaz V.O., Jiang Y., Parmar R., Zlatev I., Castoreno A., Xu H., Shulga-Morskaya S. Selection of GalNAc-conjugated siRNAs with limited off-target-driven rat hepatotoxicity. Nat. Commun. 2018;9:723. doi: 10.1038/s41467-018-02989-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willoughby J.L.S., Chan A., Sehgal A., Butler J.S., Nair J.K., Racie T., Shulga-Morskaya S., Nguyen T., Qian K., Yucius K. Evaluation of GalNAc-siRNA Conjugate Activity in Pre-clinical Animal Models with Reduced Asialoglycoprotein Receptor Expression. Mol. Ther. 2018;26:105–114. doi: 10.1016/j.ymthe.2017.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartz A.L., Fridovich S.E., Lodish H.F. Kinetics of internalization and recycling of the asialoglycoprotein receptor in a hepatoma cell line. J. Biol. Chem. 1982;257:4230–4237. [PubMed] [Google Scholar]
- 47.Fitzgerald K., White S., Borodovsky A., Bettencourt B.R., Strahs A., Clausen V., Wijngaard P., Horton J.D., Taubel J., Brooks A. A Highly Durable RNAi Therapeutic Inhibitor of PCSK9. N. Engl. J. Med. 2017;376:41–51. doi: 10.1056/NEJMoa1609243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zimmermann T.S., Karsten V., Chan A., Chiesa J., Boyce M., Bettencourt B.R., Hutabarat R., Nochur S., Vaishnaw A., Gollob J. Clinical Proof of Concept for a Novel Hepatocyte-Targeting GalNAc-siRNA Conjugate. Mol. Ther. 2017;25:71–78. doi: 10.1016/j.ymthe.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strom S.C., Davila J., Grompe M. Chimeric mice with humanized liver: tools for the study of drug metabolism, excretion, and toxicity. Methods Mol. Biol. 2010;640:491–509. doi: 10.1007/978-1-60761-688-7_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang J., Wang Y., Zhou T., Wong L.Y., Tian X.Y., Hong X., Lai W.H., Au K.W., Wei R., Liu Y. Generation of Human Liver Chimeric Mice with Hepatocytes from Familial Hypercholesterolemia Induced Pluripotent Stem Cells. Stem Cell Reports. 2017;8:605–618. doi: 10.1016/j.stemcr.2017.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fernández-Ruiz I. Atherosclerosis: A new role for lncRNAs in atherosclerosis. Nat. Rev. Cardiol. 2018;15:195. doi: 10.1038/nrcardio.2018.18. [DOI] [PubMed] [Google Scholar]
- 52.Kumar M.M., Goyal R. LncRNA as a Therapeutic Target for Angiogenesis. Curr. Top. Med. Chem. 2017;17:1750–1757. doi: 10.2174/156802661766616111644744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simion V., Haemmig S., Feinberg M.W. LncRNAs in vascular biology and disease. Vascul. Pharmacol. 2019;114:145–156. doi: 10.1016/j.vph.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chu C., Quinn J., Chang H.Y. Chromatin isolation by RNA purification (ChIRP) J. Vis. Exp. 2012;61:e3912. doi: 10.3791/3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chu C., Zhang Q.C., da Rocha S.T., Flynn R.A., Bharadwaj M., Calabrese J.M., Magnuson T., Heard E., Chang H.Y. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Halász L., Karányi Z., Boros-Oláh B., Kuik-Rózsa T., Sipos É., Nagy É., Mosolygó-L Á., Mázló A., Rajnavölgyi É., Halmos G., Székvölgyi L. RNA-DNA hybrid (R-loop) immunoprecipitation mapping: an analytical workflow to evaluate inherent biases. Genome Res. 2017;27:1063–1073. doi: 10.1101/gr.219394.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gagnon K.T., Li L., Janowski B.A., Corey D.R. Analysis of nuclear RNA interference in human cells by subcellular fractionation and Argonaute loading. Nat. Protoc. 2014;9:2045–2060. doi: 10.1038/nprot.2014.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.