Abstract
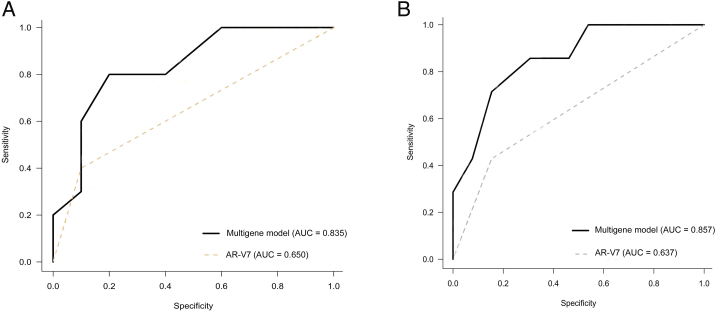
While circulating tumor cell (CTC)–based detection of AR-V7 has been demonstrated to predict patient response to second-generation androgen receptor therapies, the rarity of AR-V7 expression in metastatic castrate-resistant prostate cancer (mCRPC) suggests that other drivers of resistance exist. We sought to use a multiplex gene expression platform to interrogate CTCs and identify potential markers of resistance to abiraterone and enzalutamide. 37 patients with mCRPC initiating treatment with enzalutamide (n = 16) or abiraterone (n = 21) were prospectively enrolled for CTC collection and gene expression analysis using a panel of 89 prostate cancer–related genes. Gene expression from CTCs was correlated with PSA response and radioclinical progression-free survival (PFS) using Kaplan-Meier and Cox regression analyses. Twenty patients (54%) had detectable CTCs. At a median follow-up of 11.3 months, increased expression of the following genes was significantly associated with shorter PSA PFS and radioclinical PFS: AR, AR-V7, PSA, PSCA, TSPAN8, NKX3.1, and WNT5B. Additionally, high SPINK1 expression was associated with increased PFS. A predictive model including all eight genes gave an area under the curve (AUC) of 0.84 for PSA PFS and 0.86 for radioclinical PFS. In comparison, the AR-V7 only model resulted in AUC values of 0.65 and 0.64.These data demonstrate that clinically relevant information regarding gene expression can be obtained from whole blood using a CTC-based approach. Multigene classifiers in this setting may allow for the development of noninvasive predictive biomarkers to guide clinical management.
Abbreviations: ARSI, androgen receptor signaling inhibitors; AUC, area under the curve; CTC, circulating tumor cell; mCRPC, metastatic castrate-resistant prostate cancer; EpCAM, epithelial cell adhesion molecule; EMT, epithelial mesenchymal transition; HR, hazard ratio; IQR, interquartile range; IRB, institutional review board; LHRH, luteinizing hormone-releasing hormone; PFS, progression free survival
Introduction
Treatment options for patients with metastatic castration-resistant prostate cancer (mCRPC) include androgen receptor signaling inhibitors (ARSI), such as abiraterone or enzalutamide, and taxane-based chemotherapy [1], [2], [3], [4]. However, optimal treatment algorithms remain controversial, and there is substantial heterogeneity in treatment response [5]. For example, 30%-40% of patients with mCRPC do not respond to ARSI treatment or develop resistance within a brief period of time [1], [2], [6]. Predictive biomarkers that can help align an individual's tumor biology with an appropriate treatment remain an area of great clinical need.
While there has been marked progress in the development of tissue-based approaches that integrate precision genomics into clinical workflows [7], it is not practical or feasible to perform repeat biopsies with each new treatment decision over time, limiting the clinical utility of these discoveries. As a result, the development of noninvasive liquid markers to recapitulate and/or augment tissue-based information remains a priority, and CTC-based AR-V7 expression is the first such marker to accurately predict ARSI response in patients with mCRPC [6]. There is a critical need to identify additional markers, however, as AR-V7–positive patients account for only a small percentage of ARSI nonresponders and some patients expressing AR-V7 do respond to ARSI treatment [8], [9].
Here, we hypothesized that a CTC-based gene expression platform could be utilized to identify potential molecular markers of response and resistance to ARSI therapy. Through prospective enrollment of mCRPC patients undergoing treatment with abiraterone or enzalutamide, we sought to better understand the degree to which clinically relevant tumor profiles could be identified using blood-based CTC isolation strategies.
Patients and Methods
Patient Cohorts
Thirty-seven patients with mCRPC initiating treatment with abiraterone or enzalutamide and 27 normal controls were prospectively enrolled between January 2015 and September 2016. Genetic predictors of ARSI resistance were examined using a CTC-based mRNA expression assay. All patients consented to an IRB-approved protocol that permits blood sampling and tracking of clinical data.
CTC Isolation and Gene Expression
The CTC isolation and mRNA recovery methods are described in our previous study [10]. Approximately 5 ml of whole blood was drawn into 10-ml EDTA-containing Vacutainer tubes with a cell preservative. All blood samples were obtained before or within 4 weeks after initiation of ARSI therapy. CTCs were positively selected using anti-EpCAM antibody-conjugated Dynabeads (16,203; Thermo Fisher Scientific), which are antibody-coated magnetic beads that isolate cells that express epithelial cell adhesion molecule. Cells were washed and lysed to collect mRNA, which was captured using Oligo(dT)25 mRNA Dynabeads. Reverse transcription was performed to obtain cDNA, and multiplex qPCR was then performed to generate to target gene preamplified library. Real-time qPCR was then utilized to evaluate a panel of 89 genes, plus 3 internal controls (Supplementary Table 1).
CTC Characterization and Normalization of Gene Expression
To classify samples as positive or negative for CTCs, we previously developed an epithelial expression-based model using blood samples from 27 healthy controls with or without exogenous spike-ins of prostate cancer cell lines comprised of eight markers: CD326, CDH1, CDH2, DSG2, EGFR, KRT8, KRT18, and KRT19 (Supplementary Methods).
Gene expression was evaluated by qPCR in a 384-well format, and cycle threshold (Ct) values were normalized using the delta–delta Ct method (2(delta–delta Ct+ 1)) [11]. A total of 92 genes were assessed in each sample of 89 prostate cancer–related genes along with 3 internal control genes (ACTB, TUBA1B, and HMBS). Genes were selected based on a combination of potential relevance as a clinical biomarker and analytic performance characteristics, as previously described [10]. With a limited sample of 20 CTC-positive patients relative to a large number of candidate genes, a preliminary screening step was used to discard genes that could not differentiate between CTC-positive samples and those from 27 healthy controls (Supplementary Methods). In a secondary processing step, genes were categorized as “high” or “low” expression relative to a quantile selected from three fixed candidates to maximize the association with the primary clinical outcomes.
Clinical Outcomes
The primary outcomes for this study were (1) prostate-specific antigen (PSA) progression-free survival (PFS) and radioclinical PFS. PSA progression was defined using the Prostate Cancer Working Group 3 definition as a ≥25% increase in PSA levels above the nadir (and by ≫2 ng/ml), with confirmation ≥4 weeks later [12]. Radioclinical progression was defined as a ≫20% increase in the sum of the soft tissue lesion diameters during computed tomography, ≫2 new bone lesions on nuclear medicine bone scan, or symptomatic progression (pain aggravation or cancer-related complications) [12], [13]. Secondary outcomes included overall survival and best PSA response (maximum percentage decrease in PSA level from baseline).
Statistical Analysis
The association of each gene with PSA and radioclinical PFS was assessed using Kaplan-Meier analyses and the log-rank test, with P values adjusted using the Holm method to control the family-wise error rate across the three candidate quantiles for the gene-specific high/low cutoff values. These cutoff thresholds were selected for each gene to minimize the geometric mean of the P values from the PSA and radioclinical PFS analyses. Following selection of the split point, within the analysis of each outcome, we computed the Holm-adjusted P values for the selected genes. Genes nominally significant at the 5% level following this adjustment were considered as candidates for multigene models. In secondary analyses, Cox proportional-hazards regression was used to further evaluate the association of each gene with PSA and radioclinical PFS. Best PSA response for each gene was compared using Fisher's exact test. The utility of single and polygene scores for predicting 90-day PFS was measured using area under the receiver operating characteristic estimated from survival data [14]. Polygene scores were computed as simple sums of the number of component genes scored as 1 or “high”. The predictive performances of the polygene and single-gene models were compared. The association of the polygene score with PSA and radioclinical PFS was also assessed. All statistical analyses were performed using R software (version 3.3).
Results
Patient Characteristics
All patients enrolled continued to receive luteinizing hormone-releasing hormone (LHRH) agonist therapy while on secondary hormonal therapy, with 21 patients (56.8%) receiving abiraterone and 16 (43.2%) receiving enzalutamide. At the time of enrollment, the median baseline age was 72 years (interquartile range [IQR]: 67-79 years), and the median PSA level was 20.9 ng/ml (IQR: 11.6-96.8 ng/ml). Patients were followed for a median of 343 days (IQR: 142-640 days), and the majority of patients (54%) had received more than one previous treatment other than an LHRH agonist (Table 1). The baseline characteristics of the cohort are presented in Table 1.
Table 1.
The Baseline Demographic and Clinical Characteristics of Patients with Metastatic Castration-Resistant Prostate Cancer
| Variable | Overall (n = 37) |
CTC Positive (n = 20) |
CTC Negative (n = 17) |
P Value |
|---|---|---|---|---|
| Age, median, IQR, years | 72(67-79) | 73(64-78) | 72(68-80) | .437 |
| Race (%) | ||||
| Caucasian | 35 (94.6) | 18 (90.0) | 17 (100.0) | .407 |
| African American | 1 (2.7) | 1 (5.0) | 0 (0.0) | |
| Hispanic | 1 (2.7) | 1 (5.0) | 0 (0.0) | |
| PSA, median, IQR, ng/ml | 20.9 (11.6-96.8) | 65.8(17.4-359.3) | 14.4 (7.8-25.8) | .011 |
| No. of prior therapy (%) | ||||
| 0 | 17 (45.9) | 7 (35.0) | 10 (58.8) | .259 |
| 1 | 15 (40.5) | 9 (45.0) | 6 (35.3) | |
| 2 | 5 (13.5) | 4 (20.0) | 1 (5.9) | |
| Gleason score (%) | ||||
| ≤7 | 16 (43.2) | 8 (40) | 8 (47.0) | .803 |
| 8 | 3 (8.1) | 2 (10) | 1 (5.9) | |
| ≥9 | 17 (46.0) | 10 (50) | 7 (41.2) | |
| Unknown | 1 (2.7) | 0 (0.0) | 1 (5.9) | |
| ECOG (%) | ||||
| 0 | 21 (56.8) | 8 (40.0) | 13 (76.5) | .079 |
| 1 | 13 (35.1) | 10 (50.0) | 3 (17.6) | |
| 2 | 3 (8.1) | 2 (10) | 1 (5.9) | |
| Opioid analgesic (%) | ||||
| Yes | 26 (70.3) | 10 (50) | 16 (94.1) | .003 |
| No | 11 (29.7) | 10 (50) | 1 (5.9) | |
| Albumin, median, IQR, g/dl | 4.1(3.9-4.3) | 4.1(3.8-4.3) | 4.2(4.0-4.4) | .502 |
| Hemoglobin, median, IQR, g/dl | 12.8(11.7-13.9) | 12.6(10.6-13.2) | 12.8(12.5-14.6) | .825 |
| Alkaline phosphatase, Median, IQR, g/dl | 102.0(80.5-170.5) | 117.0(80.3-267.5) | 102.0(82.5-123.5) | ≪.001 |
| CTC probability | 0.95 (0.06-0.99) | 0.99(0.98-0.99) | 0.056(0.006-0.170) | ≪.001 |
| Extent of disease at baseline | ||||
| Bone metastasis | ||||
| Yes | 33 (89.2) | 19 (95.0) | 14 (82.4) | .217 |
| No | 4 (10.8) | 1 (5.0) | 3 (17.6) | |
| Nodal metastasis | ||||
| Yes | 24 (64.9) | 16 (80.0) | 8 (47.1) | .036 |
| No | 13 (35.1) | 4 (20.0) | 9 (52.9) | |
| Visceral metastasis (liver and/or lung) |
||||
| Yes | 10 (27.0) | 6 (30.0) | 4 (23.5) | .659 |
| No | 27 (73.0) | 14 (70.0) | 13 (76.5) |
P value: CTC+ vs. CTC−. ECOG, Eastern Cooperative Group.
CTC Isolation and Gene Analysis
Using a previously developed epithelial-based expression signature to assess for the presence of CTCs, 20 patients (54%) were classified as having detectable CTCs. A preliminary screening step was used to remove genes whose expression levels could not reliably distinguish between CTC-positive samples and those from healthy controls (Supplementary Figures S1 and S2). Based on this analysis, 41 genes (30 prostate cancer–related genes, 8 epithelial markers, and 3 internal controls) were excluded, and 51 genes were retained for survival analysis.
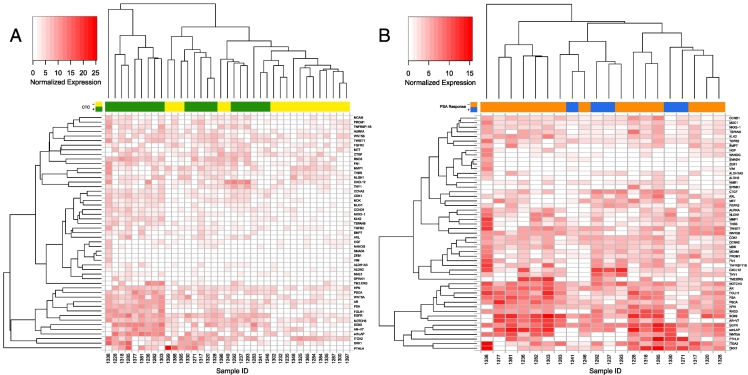
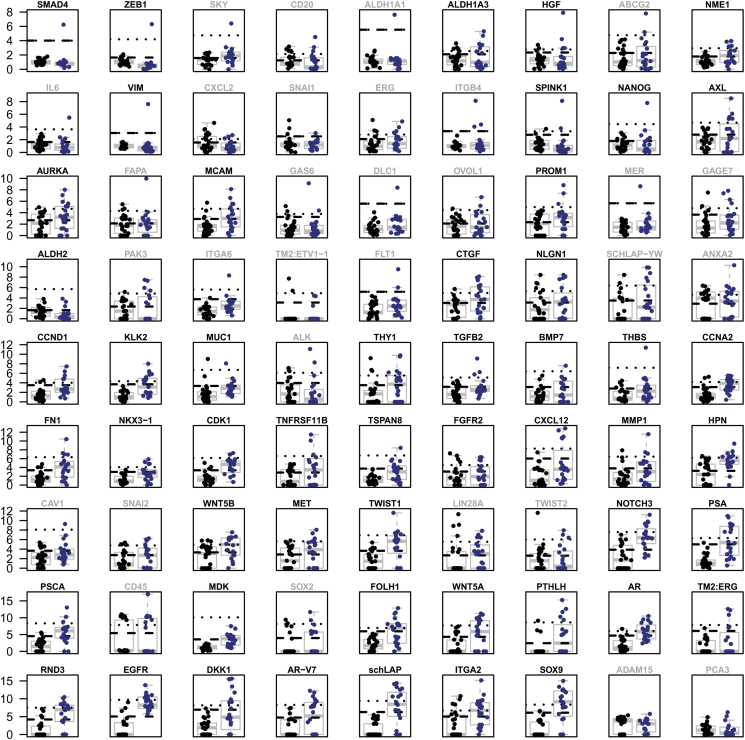
Compared to patients who were CTC-negative or ARSI responders, patients who were CTC-positive or ARSI nonresponders tended to express a number of prostate cancer–related genes (Figure 1, A and B). Continuous gene expression values were converted to categories of high and low expression for each gene based on association with survival in Kaplan-Meier analysis. As described in the methods, three potential thresholds were assessed for each gene, with cutoffs set at the 25th (A), 50th (B), and 75th (C) percentiles; i.e., for criteria A, the 15 samples with the lowest gene expression were grouped as “low” and the remaining 5 grouped as “high.” Threshold A was utilized for 18 genes, thrreshold B for 20 genes, and threshold C for 13 genes (Supplementary Tables S2 and S3).
Figure 1.
Integrative landscape analysis of gene signatures in metastatic castration-resistant prostate cancer with androgen receptor signaling inhibitor treatment. (A) A heat map representation of gene expression data from the CTC-positive samples (red) and the CTC-negative samples (blue). (B) Hierarchical clustering of gene expression in CTC-positive patients only. The 51 selected gene panels were enriched in the patients with a PSA response (≥50% decline in PSA level from baseline) (blue) compared to patients without a PSA response (red).
Identification of Genes Associated with Oncologic Endpoints
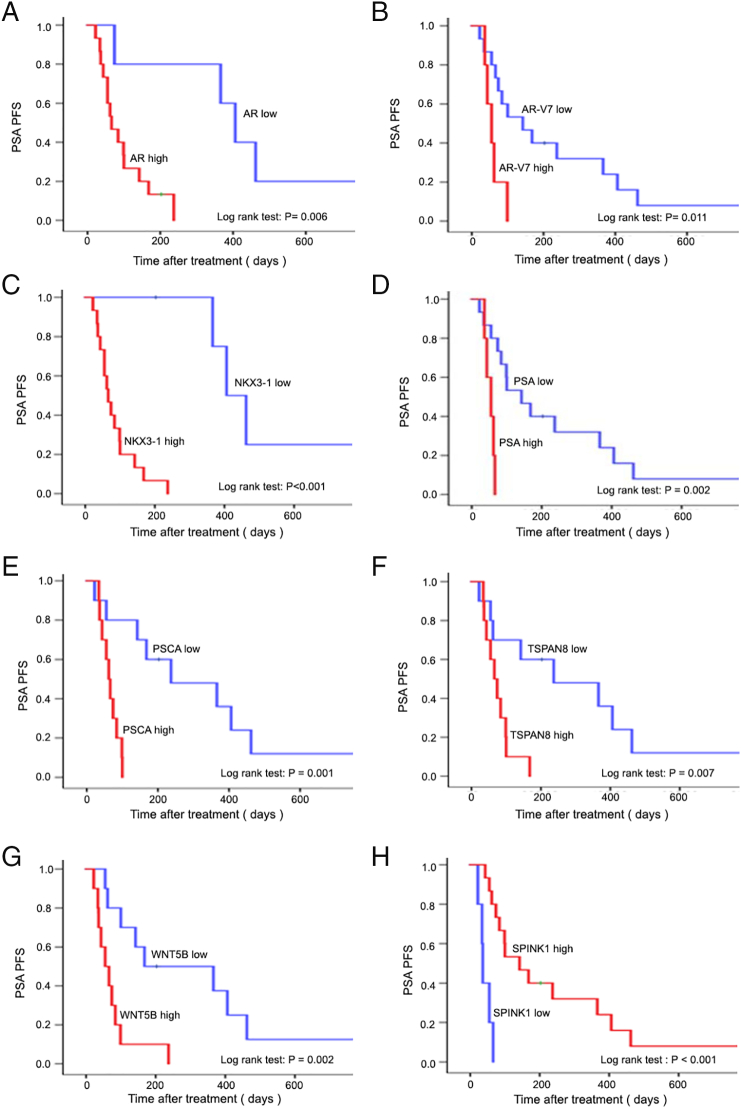
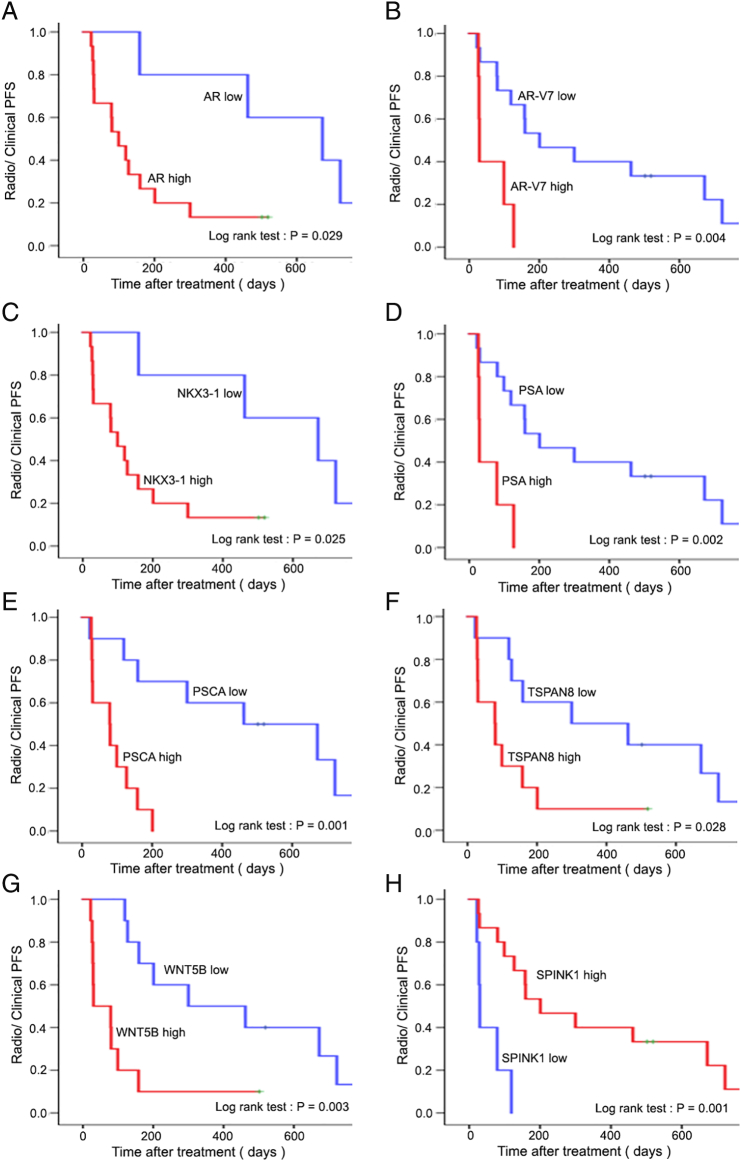
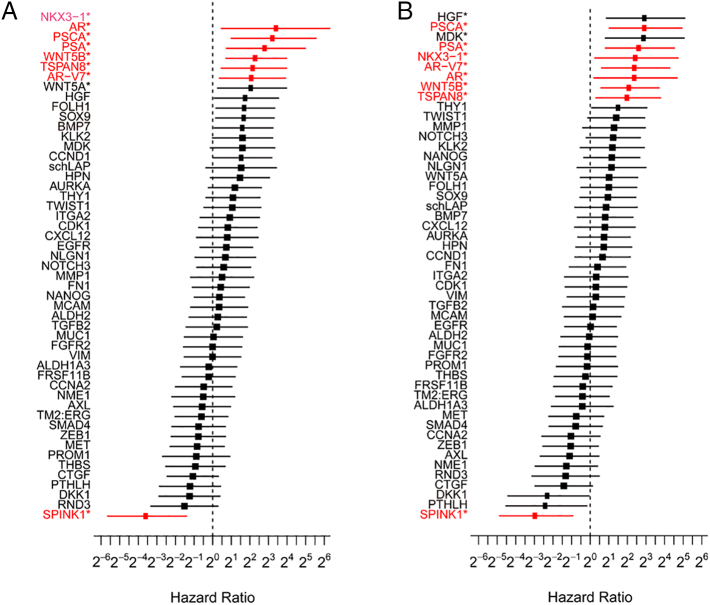
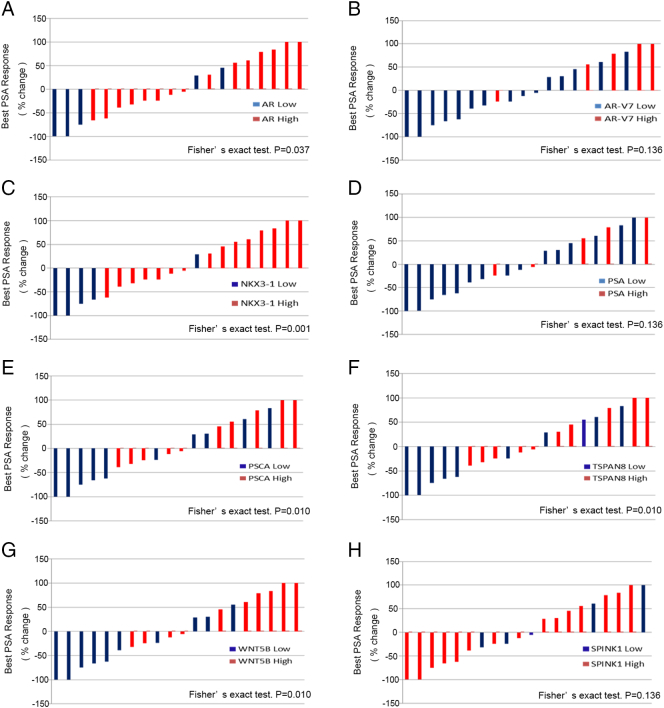
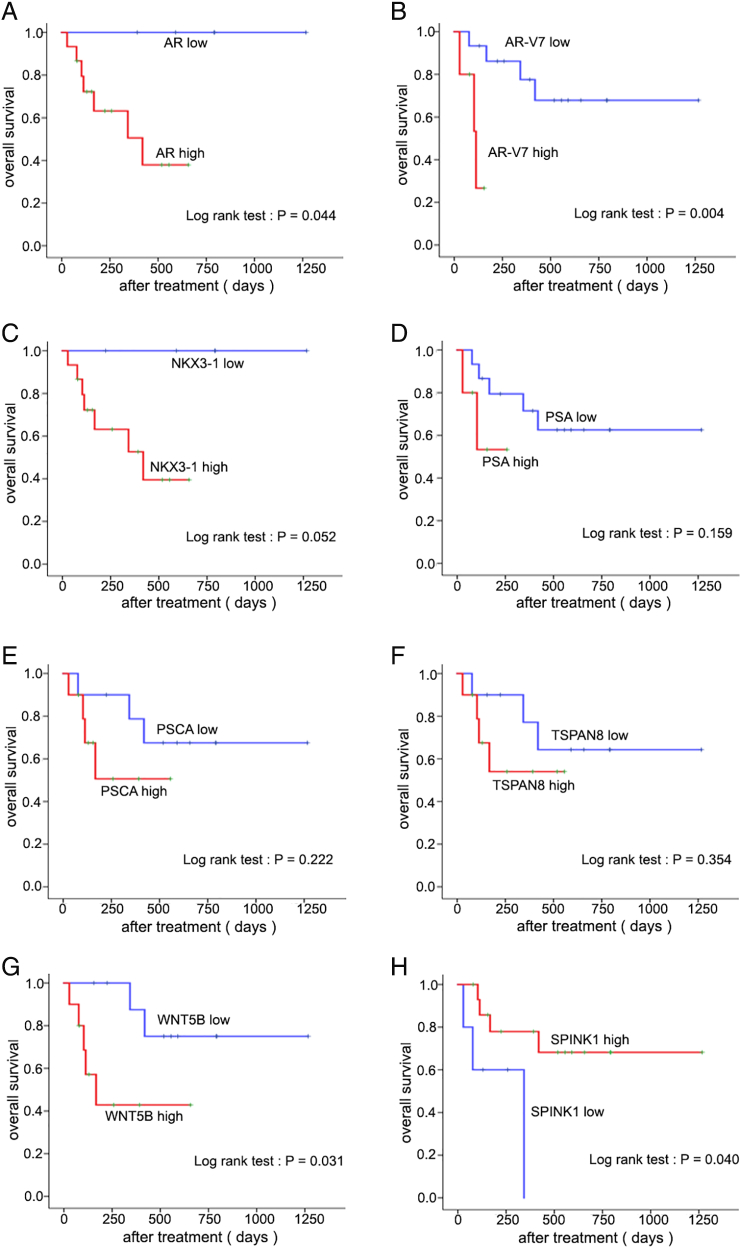
Among CTC-positive patients, the median times to PSA progression and radioclinical progression were 91 days (IQR: 55-228 days) and 142 days (IQR: 42-492 days), respectively. Among the 51 candidate genes, increased expression of AR, AR-V7, PSA, PSCA, TSPAN8, NKX3-1, and WNT5B was significantly associated with decreased PSA PFS (Figure 2) and radioclinical PFS (Figure 3), while SPINK1 was inversely associated with these outcomes. Univariate Cox regression also supported the association with both PSA and radioclinical PFS for each of these genes (Figure 4, A and B). Additionally, BMP7, FOLH1, SOX9, and WNT5a were nominally associated with PSA PFS, while THY1, PTHLH, MDK, and HGF were associated with radioclinical PFS (Supplementary Table S3). Waterfall plots displaying the maximum PSA response among CTC-positive patients and correlation with each of the candidate genes are shown in Supplementary Figure S3. The overall proportion of patients with a PSA response (≥50% decline from baseline) was 25% (5/20), and lack of response to ARSI treatment was significantly associated with high expression of AR, TSPAN8, PSCA, WNT5B, and NKX3-1. Finally, AR, AR-V7, WNT5B, and SPINK1 were all associated with overall survival in the Kaplan-Meier analyses for this secondary endpoint (Supplementary Figure S4).
Figure 2.
Kaplan-Meier plots for PSA PFS according to expression of eight genes (A: AR, B: AR-V7, C: NKX3.1, D: PSA, E: PSCA, F: TSPAN8, G: WNT5B, H: SPINK1). The P value is calculated using the log-rank test.
Figure 3.
Kaplan-Meier plots for radiological and/or clinical PFS according to expression of eight genes (A: AR, B: AR-V7, C: NKX3.1, D: PSA, E: PSCA, F: TSPAN8, G: WNT5B, H: SPINK1). The P value is calculated using the log-rank test.
Figure 4.
Cox proportional-hazard analyses of the associations between individual gene expression and PSA PFS (A) or radioclinical PFS (B). The estimate for the hazard ratio of NKX3-1 in the Cox model for time to PSA progression diverged to infinity and has consequently been omitted from the plot. Gene names shown in red were nominally significant for both clinical outcomes.
Pilot Development of Multigene Model
The multigene model comprised of all candidate genes (AR, AR-V7, PSA, PSCA, TSPAN8, WNT5B, NKX3-1, and SPINK1) was assessed as an exploratory analysis and compared to a single-gene model for AR-V7 (Supplementary Figure S5). Receiver operating curves were constructed, and the AUCs for the multigene model showed increased accuracy compared to AR-V7 alone for PSA PFS (0.84 vs. 0.65) and radioclinical PFS (0.86 vs. 0.64). In addition, the multigene model score was significantly associated with decreased PSA PFS and radioclinical PFS (Supplementary Figure S6).
Discussion
Given the variable patterns of response to ARSI therapy in mCRPC, there is a critical need for predictive markers to guide precision-based therapeutic strategies. While AR-V7 provides an initial model for this approach, additional markers are needed [6], [9]. The present study lends additional support to the utility of AR-V7 in this setting, as all patients with high AR-V7 expression were ARSI nonresponders. However, 10 of 15 patients (66.7%) with low AR-V7 expression were also ARSI nonresponders, indicative of the variability of response and need to identify other detectable drivers of resistance. In this study, we demonstrate that multiple CTC-based biomarkers can be simultaneously evaluated and that there are several non–AR-V7 biomarkers that may be predictive of ARSI response.
Many of the genes identified in this biomarker development study—specifically, AR, AR-V7, NKX3-1, and PSA—are consistent with the known importance of AR signaling–driven resistance to ARSI treatment. For example, numerous prior studies have confirmed that amplification of the AR gene predicts response to ARSI therapy [15], [16]. Elevated AR transcription may also increase the generation of constitutively active truncated AR variants, as a CTC-based study revealed that AR-V7 amplification was directly proportional to AR levels and truncated AR splice variants were associated with ARSI resistance [6], [17]. NKX3-1 is an AR-regulated homeobox gene and well-known marker of AR signaling [18]. NKX3-1 has been shown to co-localize with AR and acts with other downstream pathways to promote cell survival in advanced prostate cancer [19]. Additionally, PSA transcripts, which are related to the AR signaling pathway, are known to be associated with time to ARSI treatment failure or death [20]. Fangfang et al. recently reported that detection of PSA transcripts in peripheral blood mononuclear cells could predict the time to ARSI treatment failure in patients with CRPC [21]. However, the present study is the first we are aware of to comprehensively and simultaneously assess AR signaling across multiple genes in this setting using a liquid-based approach.
In addition to the potential for AR signaling-based markers to serve as predictive markers, we identified markers of epithelial-mesenchymal transition (EMT) and “stemness” as having potential clinical relevance. Prostate stem cell antigen (PSCA) is a cell-surface 123–amino acid glycoprotein that is 30% identical to stem cell antigen type 2 (SCA-2) and was first identified in the LAPC-4 xenograft model of prostate cancer [22]. Elevated PSCA expression is correlated with higher tumor stage and progression to androgen independence, and one previous study has indicated its potential prognostic value as a CTC-based marker [23], [24]. The WNT pathway is also linked to treatment resistance in mCRPC [25], and WNT5 signaling can induce epithelial-to-mesenchymal transition in cancer, with mesenchymal transitioned cancer cells instigating the invasion of neighboring epithelial cancer cells through WNT5B secretion [26]. While each of these markers—and other candidates, TSPAN8 and SPINK1—needs validation, these data indicate that a liquid-based approach can provide clinically meaningful information surrounding the response to ARSI therapy.
The classification of patients as having high or low gene expressions is complicated, as the expression profiles vary for individual CTCs [25]. Technology for gene expression profiling based on CTCs is in its infancy, and at present, there are no robust reference sets against which to compare the relative expression of our samples. Instead, we used a data-driven approach to find relevant thresholds associated with PFS in order to demonstrate the potential utility of this technology. In doing so, we filtered out genes with expression patterns that could not distinguish between CTC-positive samples and normal controls, and optimized cutoff values for potentially useful genetic biomarkers. These analyses nominated a number of candidate genes consistent with potential ARSI resistance (AR, PSA, PSCA, TSPAN8, SPINK1, NKX3.1, WNT5B, and AR-V7), which may provide additional prognostic and predictive information beyond AR-V7 alone.
The present study has several additional limitations. First, the small sample size supports only a discovery-based approach, and prospective validation in larger cohorts is needed. Second, we relied on epithelial expression for enrichment and identification of CTCs, which likely misses some clinically relevant CTCs that have undergone EMT. However, the findings here suggest that this did not preclude identification of EMT/stem cell markers of progression. Third, drug resistance can occur through numerous mechanisms that may not be detected through RT-PCR, including translocation and mutation, and thus assessment of circulating cfDNA could augment the approach utilized here. Finally, we performed cell lysis immediately after cell enrichment, which precludes CTC enumeration and assessment of the level of CTC purity in the sample. However, the clinical utility of enumeration-based approaches remains unclear, and background leukocyte contamination is accounted for through normalization of gene expression to control samples [27].
Conclusion
We confirmed the potential for molecular analysis of enriched CTCs to help derive markers of treatment response and resistance in this prospective study of patients undergoing ASRI therapy. In addition to AR-V7, which is now a well-known marker in this setting, we identified multiple additional clinically relevant genes that can be detected through a simple blood draw. Although additional research is needed to validate these findings, CTC isolation and molecular characterization are an important and feasible avenue for biomarker discovery. Using methods such as the one described here, the ability to interrogate tumor expression via a simple blood draw offers substantial potential for enhancing precision-based treatment selection in mCRPC.
The following are the supplementary data related to this article.
Supplementary Figure S1.
Normalized gene expression values. The cycle threshold (Ct) values were normalized using the delta–delta Ct method, and bold gene signatures were retained for the survival analysis. Light gray gene signatures, which cannot distinguish between positive samples (blue) and negative controls (black), were omitted. Each dot represents one analyzed gene expression datum from a patient with metastatic castration-resistant prostate cancer or a healthy control. The dashed and dotted lines represent grouping thresholds determined by k-means clustering used to test association between sample source and grouping. The y-axis shows the normalized gene expression values.
Normalized gene expression values (y-axis) for internal control genes (ACTB, TUBA1B, and HMBS) and eight epithelial marker genes. Black present controls and blue present CTC-positive samples.
Supplementary Figure S3.
Waterfall plots displaying the best prostate-specific antigen (PSA) response to androgen receptor signaling inhibitors according to expression of eight genes. The best PSA response –50% change line indicates the threshold for defining a PSA response (≥50% decline in PSA level from baseline). (A:AR,B: AR-V7, C: NKX3.1, D: PSA, E: PSCA, F: TSPAN8, G: WNT5B, H: SPINK1).
Supplementary Figure S4.
Overall survival as a function of eight gene expressions (A: AR, B: AR-V7, C: NKX3.1, D: PSA, E: PSCA, F: TSPAN8, G: WNT5B, H: SPINK1). The P value is calculated using the log-rank test.
Supplementary Figure S5.
Receiver operating characteristic curves of the polygene model and individual model using AR-V7 for predicting PSA progression (A) and radioclinical progression (B) among patients with metastatic castration-resistant prostate cancer undergoing androgen receptor signaling inhibitor treatment.
PSA-PFS and radiological and/or clinical PFS as a function of multigene model score, categorized by median score. The P value is calculated using the log-rank test.
Acknowledgments
Acknowledgements
We would like to thank Amy Kasputis whose efforts as research coordinator made this work possible.
Conflicts of Interest
T. M. M.: Myriad Genetics (advisory board, research funding); GenomeDx (research funding). No potential conflicts of interest were disclosed by the other authors.
Funding
This work was supported by a Department of Defense Physician Research Training Award (T.M.M. W81XWH-14-1-0287), the Prostate Cancer Foundation (T.M.M., S.A.T.), Alfred A. Taubman Institute (T.M.M., S.A.T), the National Institutes of Health (J.H.; 1T32CA180984-01A1), and a grant from Research year of Inje University in 2016 (J.S.C.).
References
- 1.de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, Chi KN, Jones RJ, Goodman OB, Jr., Saad F. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364(21):1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, de Wit R, Mulders P, Chi KN, Shore ND. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367(13):1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 3.Beer TM, Armstrong AJ, Rathkopf DE, Loriot Y, Sternberg CN, Higano CS, Iversen P, Bhattacharya S, Carles J, Chowdhury S. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med. 2014;371(5):424–433. doi: 10.1056/NEJMoa1405095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrylak DP, Tangen CM, Hussain MH, Lara PN, Jr., Jones JA, Taplin ME, Burch PA, Berry D, Moinpour C, Kohli M. Docetaxel and estramustine compared with mitoxantrone and prednisone for advanced refractory prostate cancer. N Engl J Med. 2004;351(15):1513–1520. doi: 10.1056/NEJMoa041318. [DOI] [PubMed] [Google Scholar]
- 5.Lorente D, Mateo J, Perez-Lopez R, de Bono JS, Attard G. Sequencing of agents in castration-resistant prostate cancer. Lancet Oncol. 2015;16(6):279–292. doi: 10.1016/S1470-2045(15)70033-1. [DOI] [PubMed] [Google Scholar]
- 6.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, Chen Y, Mohammad TA, Chen Y, Fedor HL. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371(11):1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, and Attard G, et al (2015). Integrative clinical genomics of advanced prostate cancer. Cell 2015;161(5), 1215–1228. [DOI] [PMC free article] [PubMed]
- 8.Scher HI, Graf RP, Schreiber NA, McLaughlin B, Lu D, Louw J, Danila DC, Dugan L, Johnson A, Heller G. Nuclear-specific AR-V7 protein localization is necessary to guide treatment selection in metastatic castration-resistant prostate cancer. Eur Urol. 2017;71(6):874–882. doi: 10.1016/j.eururo.2016.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernemann C, Schnoeller TJ, Luedeke M, Steinestel K, Boegemann M, Schrader AJ, Steinestel J. Expression of AR-V7 in circulating tumour cells does not preclude response to next generation androgen deprivation therapy in patients with castration resistant prostate cancer. Eur Urol. 2017;71(1):1–3. doi: 10.1016/j.eururo.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 10.Singhal U, Wang Y, Henderson J, Niknafs YS, Qiao Y, Gursky A, Zaslavsky A, Chung JS, Smith DC, Karnes RJ. Multigene profiling of CTCs in mCRPC identifies a clinically relevant prognostic signature. Mol Cancer Res. 2018;16(4):643–654. doi: 10.1158/1541-7786.MCR-17-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 12.Scher HI, Morris MJ, Stadler WM, Higano C, Basch E, Fizazi K, Antonarakis ES, Beer TM, Carducci MA, Chi KN. Trial design and objectives for castration-resistant prostate cancer: updated recommendations from the Prostate Cancer Clinical Trials Working Group 3. J Clin Oncol. 2016;34(12):1402–1418. doi: 10.1200/JCO.2015.64.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada J Natl Cancer Inst. 2000;92(3):205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 14.Heagerty PJ, Lumley T, Pepe MS. Time dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56(2):337–344. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 15.Taplin M-E, Bubley GJ, Shuster TD, Frantz ME, Spooner AE, Ogata GK, Keer HN, Balk SP. Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer. N Engl J Med. 1995;332(21):1393–1398. doi: 10.1056/NEJM199505253322101. [DOI] [PubMed] [Google Scholar]
- 16.Azad AA, Volik SV, Wyatt AW, Haegert A, Le Bihan S, Bell RH, Anderson SA, McConeghy B, Shukin R, Bazov J. Androgen receptor gene aberrations in circulating cell-free DNA: biomarkers of therapeutic resistance in castration-resistant prostate cancer. Clin Cancer Res. 2015;21(10):2315–2324. doi: 10.1158/1078-0432.CCR-14-2666. [DOI] [PubMed] [Google Scholar]
- 17.Scher HI, Lu D, Schreiber NA, Louw J, Graf RP, Vargas HA, Johnson A, Jendrisak A, Bambury R, Danila D. Association of AR-V7 on circulating tumor cells as a treatment-specific biomarker with outcomes and survival in castration-resistant prostate cancer. JAMA Oncol. 2016;2(11):1441–1449. doi: 10.1001/jamaoncol.2016.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erbaykent-Tepedelen B, Ozmen B, Varisli L, Gonen-Korkmaz C, Debelec-Butuner B, Muhammed Syed H, Yilmazer-Cakmak O, Korkmaz KS. NKX3.1 contributes to S phase entry and regulates DNA damage response (DDR) in prostate cancer cell lines. Biochem Biophys Res Commun. 2011;414(1):123–128. doi: 10.1016/j.bbrc.2011.09.035. [DOI] [PubMed] [Google Scholar]
- 19.Tan PY, Chang CW, Chng KR, Wansa KD, Sung WK, Cheung E. Integration of regulatory networks by NKX3-1 promotes androgen-dependent prostate cancer survival. Mol Cell Biol. 2012;32(2):399–414. doi: 10.1128/MCB.05958-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kantoff PW, Halabi S, Farmer DA, Hayes DF, Vogelzang NA, Small EJ. Prognostic significance of reverse transcriptase polymerase chain reaction for prostate-specific antigen in men with hormone-refractory prostate cancer. J Clin Oncol. 2001;19(12):3025–3028. doi: 10.1200/JCO.2001.19.12.3025. [DOI] [PubMed] [Google Scholar]
- 21.Qu F, Xie W, Nakabayashi M, Zhang H, Jeong SH, Wang X, Komura K, Sweeney CJ, Sartor O, Lee GM. Association of AR-V7 and prostate-specific antigen RNA levels in blood with efficacy of abiraterone acetate and enzalutamide treatment in men with prostate cancer. Clin Cancer Res. 2017;23(3):726–734. doi: 10.1158/1078-0432.CCR-16-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reiter RE, Gu Z, Watabe T, Thomas G, Szigeti K, Davis E, Wahl M, Nisitani S, Yamashiro J, Le Beau MM. Prostate stem cell antigen: a cell surface marker overexpressed in prostate cancer. Proc Natl Acad Sci. 1998;95(4):1735–1740. doi: 10.1073/pnas.95.4.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu Z, Thomas G, Yamashiro J, Shintaku IP, Dorey F, Raitano A, Witte ON, Said JW, Loda M, Reiter RE. Prostate stem cell antigen (PSCA) expression increases with high Gleason score, advanced stage and bone metastasis in prostate cancer. Oncogene. 2000;19(10):1288–1296. doi: 10.1038/sj.onc.1203426. [DOI] [PubMed] [Google Scholar]
- 24.Hara N, Kasahara T, Kawasaki T, Bilim V, Obara K, Takahashi K, Tomita Y. Reverse transcription-polymerase chain reaction detection of prostate specific antigen, prostate-specific membrane antigen, and prostate stem cell antigen in one milliliter of peripheral blood: value for the staging of prostate cancer. Clin Cancer Res. 2002;8(6):1794–1799. [PubMed] [Google Scholar]
- 25.Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, Desai R, Fox DB, Brannigan BW, Trautwein J. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349(6254):1351–1356. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato S, Hayakawa Y, Sakurai H, Saiki I, Yokoyama S. (2014). Mesenchymal-transitioned cancer cells instigate the invasion of epithelial cancer cells through secretion of WNT3 and WNT5B. Cancer Sci. 2014;105(3):281–289. doi: 10.1111/cas.12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smerage JB, Barlow WE, Hortobagyi GN, Winer EP, Leyland-Jones B, Srkalovic G, Tejwani S, Schott AF, O'Rourke MA, Lew DL. Circulating tumor cells and response to chemotherapy in metastatic breast cancer: SWOG S0500. J Clin Oncol. 2014;32(31):3483–3489. doi: 10.1200/JCO.2014.56.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Normalized gene expression values (y-axis) for internal control genes (ACTB, TUBA1B, and HMBS) and eight epithelial marker genes. Black present controls and blue present CTC-positive samples.
PSA-PFS and radiological and/or clinical PFS as a function of multigene model score, categorized by median score. The P value is calculated using the log-rank test.