Abstract
Extracellular vesicles (EVs) generated from redox active anticancer drugs are released into the extracellular environment. These EVs contain oxidized molecules and trigger inflammatory responses by macrophages. Using a mouse model of doxorubicin (DOX)-induced tissue injury, we previously found that the major sources of circulating EVs are from heart and liver, organs that are differentially affected by DOX. Here, we investigated the effects of EVs from cardiomyocytes and those from hepatocytes on macrophage activation. EVs from H9c2 rat cardiomyocytes (H9c2 EVs) and EVs from FL83b mouse hepatocytes (FL83 b EVs) have different levels of protein-bound 4-hydroxynonenal and thus different immunostimulatory effects on mouse RAW264.7 macrophages. H9c2 EVs but not FL83 b EVs induced both pro-inflammatory and anti-inflammatory macrophage activation, mediated by NFκB and Nrf-2 pathways, respectively. DOX enhanced the effects of H9c2 EVs but not FL83 b EVs. While EVs from DOX-treated H9c2 cells (H9c2 DOXEVs) suppressed mitochondrial respiration and increased glycolysis of macrophages, EVs from DOX-treated FL83b cells (FL83b DOXEVs) enhanced mitochondrial reserve capacity. Mechanistically, the different immunostimulatory functions of H9c2 EVs and FL83 b EVs are regulated, in part, by the redox status of the cytoplasmic thioredoxin 1 (Trx1) of macrophages. H9c2 DOXEVs lowered the level of reduced Trx1 in cytoplasm while FL83b DOXEVs did the opposite. Trx1 overexpression alleviated the effect of H9c2 DOXEVs on NFκB and Nrf-2 activation and prevented the upregulation of their target genes. Our findings identify EVs as a novel Trx1-mediated redox mediator of immune response, which greatly enhances our understanding of innate immune responses during cancer therapy.
Keywords: Extracellular vesicles, Macrophage activation, Thioredoxin 1, NFκB, Nrf-2
Abbreviations
- DAMPs
damage-associated molecular patterns
- DOX
doxorubicin
- ECAR
extracellular acidification rate
- EVs
extracellular vesicles
- FL83b NTEVs
Non-treated FL83b hepatocyte-derived EVs
- FL83b DOXEVs
DOX-treated FL83b-derived EVs
- 4HNE
4-hydroxynonenal
- H9c2 NTEVs
Non-treated H9c2 cardiomyocyte-derived EVs
- H9c2 DOXEVs
DOX-treated H9c2-derived EVs
- Hmox1
heme oxygenase 1
- IAA
iodoacetic acid
- Keap-1
Kelch-like ECH-associated protein-1
- l-NAME
N(G)-Nitro-l-arginine methyl ester
- NFκB
nuclear factor kappa-light-chain-enhancer of activated B cells
- Nos
nitric oxide synthase
- Nqo1
NAD(P)H Quinone Dehydrogenase 1
- Nrf-2
nuclear factor (erythroid-derived 2) like 2
- OCR
oxygen consumption rate
- Prdx1
peroxiredoxin 1
- ROS
reactive oxygen species
- Sod2
superoxide dismutase 2
- Trx1
thioredoxin-1
- Txnrd1
thioredoxin reductase 1
1. Introduction
Extracellular vesicles (EVs), small membrane-encapsulated structures that are released by most cell types as alternative pathways, maintain cellular homeostasis by removing toxic oxidized molecules. Accumulating evidence has revealed the important role of EVs in mediating intercellular communication in the presence of local and systemic inflammation that results from such oxidative stresses as aging, cancer, cardiovascular diseases and lung diseases [[1], [2], [3], [4], [5]]. EVs can suppress or activate immune response depending on the bioactive contents of the EVs. The immunostimulatory effects of EVs are influenced by the transfer of damage-associated molecular patterns (DAMPs) from damaged cells and activate immune cells [[6], [7], [8], [9]]. In contrast, the immunoinhibitory effects of EVs are associated with the transfer of immunomodulatory transcription factors, non-coding RNAs, and anti-inflammatory cytokines, most of which are released from cancer cells and stem cells [10].
Macrophages are mononuclear phagocytic cells that mediate an innate immune response by recognizing microbial and endogenous danger-signaling molecules [11]. Macrophages are activated in response to environmental stimuli and produce distinct sets of cytokines, as well as enhance or inhibit inflammation by changing cellular metabolism. In physiologic conditions, the balance of the pro- and anti-inflammatory activities of macrophages is strictly regulated. A balanced response is critical for containment or elimination of pathogens. Severe pro-inflammatory immune response leads to secondary cytotoxicity and tissue destruction, and excessive anti-inflammatory response leads to tissue fibrosis.
Doxorubicin (DOX) is one of the most effective chemotherapeutic agents for treating many aggressive malignancies [12]. However, its efficacy is limited by severe side effects to normal tissues that result primarily from its redox capacity [13]. DOX can cause both direct and indirect tissue injuries: the drug has a direct effect on target tissues as well as indirect effects that result from systemic inflammation initiated by an innate immune response. For example, the migration of circulating inflammatory cytokines into an organ such as the brain leads to secondary tissue damage known as chemotherapy-induced cognitive impairment or chemobrain [14]. This influence is different from effects caused by cellular penetration by the drug.
4-hydroxynonenal (4HNE) is the most toxic lipid peroxidation product generated during DOX-induced oxidative stress [15]. The effects of 4HNE are primarily due to covalent modification of amino acids that contain a nucleophilic functional group, such as lysine, histidine, and cysteine, leading to inactivation or functional change of the adducted proteins [16]. That 4HNE induces a toxic effect that leads to cardiac injury following DOX treatment is well-established. Adducts of 4HNE to the proteins involved in the mitochondrial electron transport chain in DOX-treated heart tissues are formed, which results in mitochondrial dysfunction [17]. Furthermore, 4HNE adduction to mitochondrial apoptotic inducing factor 2 (AIFm2) can change AIFm2 function from oxidoreductase to pro-apoptosis and mediate cardiomyocyte cell death [18]. Our previous study demonstrates that serum EVs of mice treated with DOX were 4HNE-adducted. Also, we identified heart and liver tissues as the major contributors to oxidized EVs [19]. However, the heart and liver are differentially susceptible to DOX toxicity. While cardiac injury is a well-known long-term side effect of DOX, liver injury is rarely observed in DOX-treated patients [20]. The reason for this might be that enrichment of antioxidant enzymes [21] and drug transporters [22] protect liver from oxidative damage. However, how EVs impact immune cells has never been investigated. Here, we hypothesize that the EVs released from cardiomyocytes and hepatocytes exposed to DOX may have differential effects on macrophage activation, which would lead to differences in secondary tissue injury.
Thioredoxin (Trx) is a small redox-sensitive protein that contains two redox-active cysteine residues—Cys-32 and Cys-35—in its active site. Trx maintains the reducing environment of the cells by a thiol-disulfide exchange [23] and a denitrosylation reaction [24]. Trx1 overexpression, as well as administration of recombinant Trx1, is protective against severe inflammatory diseases, including inflammatory bowel disease [25], pancreatitis [26], and sepsis [27]. However, the role of Trx1 in EVs-mediated innate immune response is unknown.
In macrophages, Trx1 regulates immune activation by interacting with several proteins. Depending on cellular localization, Trx1 has the dichotomous functions of regulating two opposing redox-sensitive immunomodulatory transcription factors: nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB) and nuclear factor (erythroid-derived 2) like 2 (Nrf-2). NFκB is responsible for pro-inflammatory cytokines, chemokines, and adhesion molecules, whereas Nrf-2 drives anti-inflammatory response by upregulating anti-oxidant gene expression [28,29] and also by directly interfering with pro-inflammatory cytokine gene transcription [30]. Cytosolic Trx1 prevents dissociation of IκB from NFκB [31] and also directly interacts with the Nrf-2 interacting protein, Kelch-like ECH-associated protein-1 (Keap-1), maintaining KEAP1 in a reduced state [32] and thus blocking NFκB and Nrf-2 nuclear translocation, respectively. However, should NFκB or Nrf-2 translocate into the nucleus, Trx1 can promote its transcriptional activity by maintaining it in a reduced state [33,34].
In this study, we tested the hypothesis that cardiomyocytes and hepatocytes release EVs with different oxidized protein levels in response to DOX treatment, as exemplified by 4HNE protein adduction, and that they have differential effects on macrophages. We found that Trx1 regulates EV-induced macrophage activation by controlling the redox-sensitive transcription factors NFκB and Nrf-2. The findings of this study provide an insight into the intercellular communication between oxidatively injured cells and macrophages, and suggest a new therapeutic intervention to prevent DOX-induced inflammation.
2. Materials and methods
2.1. Cell culture
H9c2 rat cardiomyocyte, FL83b mouse hepatocyte, RAW264.7 mouse macrophage, and NR8383 rat macrophage cell lines were obtained from American Type Culture Collection (Manassas, VA). H9c2 and FL83b are immortalized normal cells from heart and liver, respectively. H9c2 cells were maintained in DMEM low glucose supplemented with 10% FBS, 1% penicillin/streptomycin, 1 mM pyruvate, 25 mM HEPES, 2 mM l-glutamine, and 1% non-essential amino acids. FL83b cells were maintained in F12 medium supplemented with 10% FBS and 1% penicillin/streptomycin. RAW264.7 cells were maintained in DMEM high glucose supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. NR8383 cells were maintained in Ham's F12K medium supplemented with 15% heat inactivated FBS, 2 mM l-glutamine, and 1.5 g/L sodium bicarbonate. The absence of mycoplasma contamination was confirmed in all cell lines.
2.2. Cell viability test
The sensitivity of H9c2 and FL83b cells to DOX toxicity was measured by the MTT assay. This assay relies on electron transfer from NADH and NADPH to MTT, which is achieved by oxidoreductase enzyme activity in viable cells [35]. The final product is an insoluble purple colored formazan. In this experiment, H9c2 or FL83b cells were seeded in 96-well plates at the density of 1000 cells/well. The next day the cells were exposed to 0.5 μM of DOX for 24 h. Cells were then washed with PBS, and 30 μl of MTT solution (5 mg/ml in PBS) were added to each well and incubated for 1 h. Then, 70 μl of 50% DMSO and 50% SDS were added to solubilize the formazan crystals. Absorbance was determined at 540 nm by SpectraMax Plus 384 microplate reader (Molecular Devices, San Jose, CA). DOX cytotoxicity was expressed as the percent of viability in treated cells relative to the non-treated control.
2.3. EV isolation and quantification
H9c2 and FL83b were treated or not treated with 0.5 μM DOX for 1 h followed by washing three times with 10 ml PBS. The media were changed to an EVs-free growing medium and incubated for 48 h before the conditioned media were collected. The conditioned media were filtered through 0.8 μm syringe filters to exclude vesicles larger than 0.8 μm, which are typically the apoptotic bodies. The filtered conditioned media were stored at −80 °C. The frozen conditioned media were thawed at room temperature before starting the EV isolation process.
EVs were isolated by an affinity-based membrane method using exoEasy maxi kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. All types of EVs, including exosomes, microvesicles, and apoptotic bodies, were bound to the spin column and eluted to obtain EV products. The conditioned media were thawed at room temperature, and 32 ml of the media were then mixed 1:1 with the binding buffer XBP and applied onto the affinity spin column allowing EVs to bind to the membrane. After centrifugation at 500×g for 1 min, the flow-through was discarded. Washing buffer XWP was added to the column and centrifuged at 5000×g for 5 min to wash off contaminating proteins and residual buffer. Buffer XE was then applied onto the membrane and centrifuged at 500×g for 5 min to elute EVs from the column. The elution was applied to the column again and centrifuged at 5000×g for 5 min. Since XE buffer contains primarily inorganic salts that are toxic to cells, buffer exchanges with PBS were performed using 3 kDa Amicon Ultra-15 Centrifugal Filter units (Millipore Sigma, Burlington, MA). The amount of EVs was evaluated by bicinchoninic acid assay (Thermo Fisher Scientific, Waltham, MA). The EVs were stored at −80 °C until used.
2.4. 4HNE adducted protein measurement
EVs from non-treated H9c2 (H9c2 NTEVs), DOX-treated H9c2 (H9c2 DOXEVs), non-treated FL83b (FL83b NTEVs), and DOX-treated FL83b (FL83b DOXEVs) cells were lysed with radioimmunoprecipitation assay (RIPA) buffer and mixed with an equal volume of Laemmli buffer. Equivalent protein samples (250 ng) were then loaded on nitrocellulose membrane using Bio-Dot SF microfiltration apparatus (Bio-Rad). The membranes were left to air dry overnight. The next day, the membranes were blocked with blocking buffer for 2 h followed by incubation with anti-4HNE antibody (1:5000 dilution) for 1 h on an orbital shaker. After washing with TBST three times, the membranes were incubated with IRDye® 800CW donkey anti-rabbit (1:15,000 dilution) for 1 h. Then, the membranes were washed and left to air dry. The 4HNE-adducted protein bands were visualized using an Odyssey scanner (LI-COR).
2.5. Multiplex Cytokines Array
For macrophage activation, RAW264.7 cells were seeded at a density of 5 × 105 cells/well in a 6 well-plate overnight and incubated with FBS-free media supplemented with 20 μg of H9c2 NTEVs, H9c2 DOXEVs, FL83b NTEVs, FL83b DOXEVs, or an equal volume of PBS as the vehicle for 24 h. The conditioned media were then collected and were frozen at −80 °C until used. V-PLEX mouse cytokine immunoassay kit (Meso Scale Diagnostics, Rockville, MD) was used to measure TNF alpha, IL-1 beta, IL-6, and IL-10. Mouse TGF-beta 1 DuoSet ELISA (R&D Systems, Minneapolis, MN) was used to measure TGF- beta 1. All assays were performed according to manufacturers’ protocols. All standards and samples were measured in duplicate or triplicate.
2.6. Nitric oxide (•NO) measurement
RAW264.7 cells were treated with FBS-free DMEM without phenol red and supplemented with 20 μg of H9c2 NTEVs, H9c2 DOXEVs, FL83b NTEVs, FL83b DOXEVs, or an equal volume of the vehicle control. The conditioned media were collected at 24 h post-treatment. Since •NO is unstable and completely oxidized in biological fluid, we measured the accumulation of the stable oxidized products of •NO —nitrate and nitrite— in the conditioned media using Nitrate/Nitrite Colorimetric Assay Kit (Cayman, Ann Arbor, MI). Briefly, the conditioned media were incubated with nitrate reductase to convert nitrate to nitrite. Then, Griess reagents were added to react with total nitrite and generate a deep purple azo compound. The change in color was evaluated by reading the absorbance at 540 nm using a microplate reader (Molecular Devices, San Jose, CA). The concentration of nitrate/nitrite in samples was calculated from a standard curve generated by using the known concentration of nitrate.
2.7. Mitochondrial and glycolysis stress test
RAW264.7 cells were seeded into a 96-well plate (Seahorse Bioscience, North Billerica, MA) at a density of 20,000 cells/well for both the mitochondrial and glycolysis stress tests. The next day, the cells were treated with 20 μg/ml of EVs or an equal volume of PBS for 24 h prior to the test. For the mitochondrial stress test, oxygen consumption rate (OCR) was determined using the Seahorse XF-96 analyzer (Seahorse Bioscience). OCR was measured at baseline (Basal OCR) followed by the sequential addition of oligomycin (1 μM) to inhibit ATP synthase, FCCP (1 μM) uncoupler to permeabilize the inner mitochondrial membrane and promote maximum electron flow, and rotenone (1 μM)/antimycin (1 μM) to inhibit complex I and complex III, respectively [36]. ATP-linked OCR was calculated by subtracting the OCR after oligomycin treatment from the basal OCR. The spare respiratory capacity was calculated by subtracting the basal OCR from the maximal OCR after FCCP treatment. For the glycolysis stress test, the extracellular acidification rate (ECAR) was determined by the Seahorse XF-96 analyzer. ECAR was measured at baseline followed by the sequential addition of glucose (10 mM), Oligomycin (1 μM), and 2-deoxyglucose (50 mM) [37]. Basal glycolysis was calculated by subtracting the ECAR measurement post-2-deoxyglucose addition from the ECAR measurement post-glucose addition. The glycolytic reserve capacity was calculated by subtracting the post-oligomycin measurement from the post-glucose measurement.
2.8. Western blot analysis
Western blot analysis was done in reducing condition. EV suspension in PBS (0.5 μg/μl) was lysed with RIPA buffer containing protease inhibitor and sonicated for 3 × 30 s using a water sonicator. For western blots of macrophage proteins, cells were lysed by incubating with RIPA buffer containing protease inhibitor on ice for 30 min. Protein concentrations were determined using bicinchoninic acid assay. SDS polyacrylamide gels (8–15%) were loaded with 50 μg of lysed EVs or 40 μg of whole cell lysates and electrophoresis was performed at 100 V for 2 h. The proteins were transferred onto nitrocellulose membranes. After blocking with Odyssey blocking buffer (LI-COR Biosciences, Lincoln, NE) for 1 h, the membranes were incubated overnight with primary antibodies at 4 °C. The following primary antibodies were used: anti-Alix, anti-CD81, anti-HDAC1, anti-Lamin A/C (Santa Cruz Biotechnology, Dallas, TX), anti-beta actin (Sigma-Aldrich, St. Louis, MO), anti-MnSOD (Upstate Biotechnology, Waltham, MA), anti-Trx1 (AdipoGen Life Sciences, San Diego, CA). The membranes were washed three times with TBST buffer and were incubated with IRDye 800 CW donkey anti-goat for anti-Alix; donkey anti-rabbit for anti-CD81, anti-MnSOD, and anti-Trx1; goat anti-mouse for beta-actin, anti-HDAC1, and anti-Lamin A/C for 1 h at room temperature. After washing three times, the membranes were air dried and the protein bands were visualized using the Odyssey scanner (LI-COR).
2.9. NFκB and Nrf-2 DNA binding activity
RAW264.7 cells were seeded at a density of 5 × 105 cells/well in a 6 well-plate overnight and incubated with FBS-free media supplemented with 20 μg of H9c2 NTEVs, H9c2 DOXEVs, FL83b NTEVs, FL83b DOXEVs, or an equal volume of PBS as the vehicle, or Vitamin E (d-α-Tocopherol succinate, St. Louis, MO) for 24 h. To measure NFκB and Nrf-2 transcription factor activity, DNA binding ELISA was performed using TransAM NF-κB and Nrf-2 kits (Active Motif, Carlsbad, CA), according to the manufacturer's protocol. Briefly, nuclei from RAW264.7 cells were extracted by the nuclear extract kit (Active Motif). The nuclear extract was added onto the oligonucleotide-coated well and incubated for 1 h. After the washing step, the primary antibody for NF-κB (p50) or Nrf-2 was added and incubated for 1 h. Then, the plates were washed and incubated with horseradish peroxidase-conjugated secondary antibody for 1 h. The color was developed by the developing solution and the absorbance was read at 450 nm.
2.10. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
RNA was isolated from RAW264.7 cells treated with vehicle or EVs using the MagNA Pure Compact RNA Isolation kit (Roche, Risch-Rotkreuz, Switzerland). RNA integrity and quantification were evaluated using the Agilent 2100 Bioanalyzer System (Santa Clara, CA). The mRNAs were reverse transcribed to cDNA using the first strand cDNA Synthesis Kit (Roche). The Nanodrop Spectrophotometer (Thermo Fisher Scientific) was used to measure cDNA concentration. Quantitative real-time PCR was performed using a LightCycler 480 Real-Time PCR System (Roche) with gene-specific primers. All primers were purchased from Invitrogen. Primer sequences for mouse and rat genes were as follows:
Mouse Tnf 5′-CCTGTAGCCCACGTCGTAGC-3’ (Forward) and.
5′-AGCAATGACTCCAAAGTAGACC-3’ (Reverse)
Mouse Il1b 5′-GCACTACAGGCTCCGAGATGAAC-3’ (Forward) and.
5′-TTGTCGTTGCTTGGTTCTCCTTGT-3’ (Reverse);
Mouse Hmox1 5′-GGTGATGGCTTCCTTGTACC-3′ (Forward) and.
5′-GGTGATGGCTTCCTTGTACC-3′ (Reverse)
Mouse Prdx1 5′-ACACCCAAGAAACAAGGAGGATT-3’ (Forward) and.
5′-CAACGGGAAGATCGTTTATTGTTA-3’ (Reverse);
Mouse Nqo1 5′-TTCTGTGGCTTCCAGGTCTT-3’ (Forward) and.
5′-AGGCTGCTTGGAGCAAAATA-3’ (Reverse)
Mouse Sod2 5′-ATGTTGTGTCGGGCGGCG-3’ (Forward) and.
5′-AGGTAGTAAGCGTGCTCCCACACG-3’ (Reverse);
Mouse Txnrd1 5′-GGCCAACAAAATCGGTGAACACATGGAAG-3’ (Forward) and.
5′-CGCCAGCAACACTGTGTTAAATTCGCCCT-3’ (Reverse)
Mouse Trx1 5′-ATGGTGAAGCTGATCGAGAGC-3′ (Forward) and.
5′-GGCATATTCAGTAATAGAGGC-3′ (Reverse)
Mouse Actb 5′-TGGAATCCTGTGGCATCCATGAAAC-3’ (Forward)
5′-TAAAACGCAGCTCAGTAACAGTCCG-3’ (Reverse)
Rat Il1b 5′- GCAATGGTCGGGACATAGTT-3’ (Forward)
5′- AGACCTGACTTGGCAGAGGA-3’ (Reverse)
Rat Nos2: 5′-TAGTCAACTACAAGCCCCACG-3’ (Forward)
5′-GTGAGGAACTGGGGGAAACC-3′ (Reverse)
Rat Il10 5′GTTGCCAAGCCTTGTCAGAAA-3’ (Forward)
5′- TTTCTGGGCCATGGTTCTCT-3’ (Reverse)
Rat Actb 5′-AGGCATCCTCACCCTGAAGTA-3’ (Forward)
5′- CACACGCAGCTCATTGTAGA-3’ (Reverse).
The PCR cycle for initial denaturation was 10 min at 95 °C. This was followed by 40 cycles of 15 s at 95 °C, 30 s at 60 °C, and 30 s at 72 °C. The gene expression levels were calculated by the 2−ΔΔCt method and the data represent fold change relative to vehicle control.
2.11. Trx1 redox western
Reduced and oxidized forms of Trx1 in nuclear and cytoplasmic portions of RAW264.7 cells treated with EVs were detected as described previously [[38], [39], [40]]. After incubation of EVs for 24 h, the cells were washed with ice-cold PBS and lysed with hypotonic solution supplemented with protease inhibitor and 50 mM iodoacetic acid (IAA). The lysate was centrifuged at 16,100 g for 2 min and the supernatant was collected as the cytoplasmic fraction. The nuclear pellets were lysed with G-lysis buffer supplemented with protease inhibitor and IAA. Iodoacetic acid (IAA) was used to modify the thiol group, resulting in one negative charge addition per one modification. Microspin G-25 columns (GE Healthcare, Chicago, IL) were used to remove excessive IAA. Fifty μg of derivatized samples were loaded per lane. Trx1 redox forms were separated by charge using native polyacrylamide gel electrophoresis. The more negative charge of derivatized Trx1, which indicates the more reduced Trx1, appear at the lower bands relative to the more oxidized form [40]. Redox forms of Trx1 were detected using anti-Trx1 antibody (AdipoGen Life Sciences) as a primary antibody and IRDye® 800CW donkey anti-Rabbit as a secondary antibody. The protein bands were visualized using an Odyssey scanner (LI-COR, Lincoln, NE). The band intensities were assessed using Image Studio 5. x software (LI-COR, Lincoln, NE).
2.12. Trx activity measurement
Trx activity was measured based on the ability to reduce eosin-labeled insulin disulfide, as determined by using a commercial kit (Cayman Chemical, Ann Arbor, MI) [41]. In brief, EVs-treated or vehicle-treated RAW264.7 cells were washed with ice-cold PBS and collected by cell scraper. After centrifugation at 1000 g for 10 min, the supernatant was discarded, and the pellets were sonicated in 0.2 ml TE buffer and centrifuged at 10,000 g for 20 min at 4 °C. Next, 20 μg of cell lysate were pre-incubated in an assay buffer containing thioredoxin reductase and NADPH for 30 min at 37 °C. Then, eosin-labeled insulin was added and incubated for 5 min at room temperature. Fluorescence intensity was recorded with excitation/emission spectra of 520/545 nm every 5 min for 1 h. The reduction rate of eosin-labeled insulin was calculated based on the rate of fluorescence gained per time in the linear range, and the quantity of active Trx was determined using the standard curve for known amount of Trx.
2.13. Trx1 overexpression
Open reading frame expression clone of mouse Trx1 with EF1a promoter in pReceiver-LV233 lentiviral vector was purchased from GeneCopoeia (Rockville, MD). Empty control vector for pReceiver-LV233 was used as a negative control for Trx1 overexpression. Lentiviral production was performed in HEK293T cells, and RAW264.7 cell transduction was done according to the manufacturer's protocol. Transduced cells with Trx1 expression vector were selected in 3.0 μg/ml puromycin (Takara Bio USA, Mountain View, CA) containing media.
2.14. Statistical analysis
Results are presented as the mean ± standard deviation (SD) or standard error of the mean (SEM), as indicated in the figure legend. Analysis of variance (ANOVA) was used for comparison of study endpoints including 4HNE slot blot, Western blot, and Trx1 redox western, EVs, and other parameters in the macrophages from cardiomyocytes and hepatocytes. Pairwise comparisons were performed using the ANOVA model for vehicle versus treated and differences between H9c2 NTEVs vs H9c2 DOXEVs, FL83b NTEVs vs FL83b DOXEVs, H9c2 NTEVs vs FL83b NTEVs, and H9c2 DOXEVs vs FL83b DOXEVs. Adjusted p-values due to multiple testings were calculated using the Holm's method. A value of P < 0.05 was considered significant. The SAS system version 9.4 was used for data analysis. Excel and GraphPad Prism were used for graphing.
3. Results
3.1. DOX-treated cardiomyocytes—but not hepatocytes—release EVs with higher levels of protein-bound 4HNE
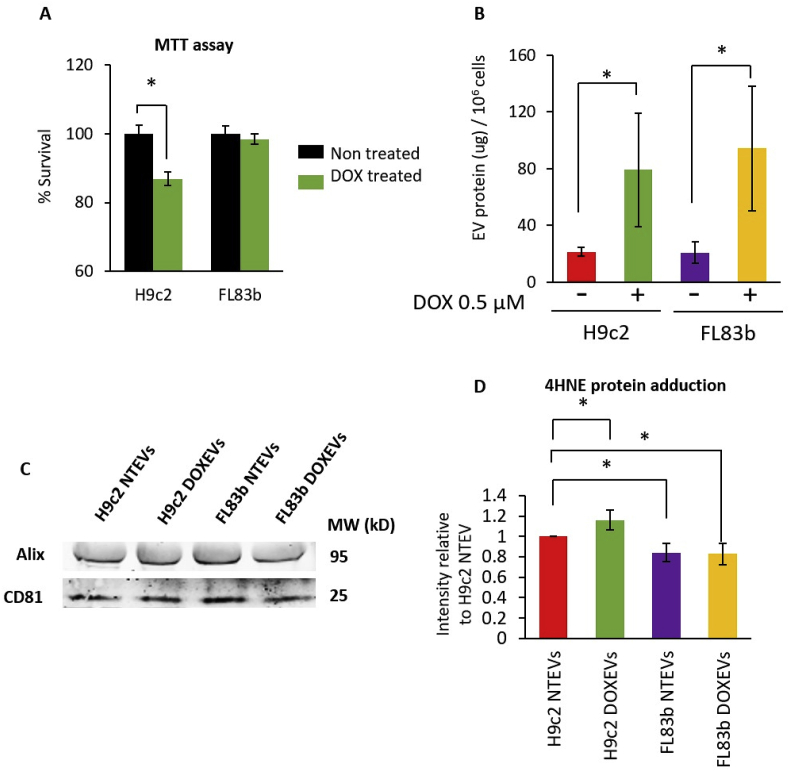
Cardiomyocytes are a major target of DOX in vivo. The results of measuring cell viability after 24 h of DOX treatment show that the viability of H9c2 cells decreased by 13% while FL83b cell viability was similar to that of the non-treated control. These data confirm that H9c2 cells are more sensitive to DOX than FL83b cells are (Fig. 1A). The quantity of EVs, as indicated by the level of EVs protein, reveals that H9c2 and FL83b cells release equal amounts of EVs per equal number of cells under normal conditions. DOX caused at least a 4-fold increase in EVs released from both H9c2 and FL83b cells (Fig. 1B). The presence of EVs, following isolation by the ExoEasy maxi kit, was validated by Western blot to detect EVs markers. EVs from non-treated and DOX-treated H9c2 and FL83b cells contained EVs markers, including Alix (a protein involved in EV biogenesis) and CD81 (a tetraspanin protein) (Fig. 1C).
Fig. 1.
Comparison of DOX toxicity, the levels of EVs and the levels of 4HNE protein adduction in the EVs released from cardiomyocytes and hepatocytes
(A) Percentage of surviving H9c2 cells and FL83b cells after 0.5 μM DOX treatment for 24 h (green bars) compared to non-treated control (black bars). (B) Amount of EVs released from one million cells of non-treated H9c2, DOX-treated H9c2, non-treated FL83b, or DOX-treated FL83b cells. (C) Western blots of EVs markers: Alix and CD81 in EVs derived from non-treated H9c2 (H9c2 NTEVs), DOX-treated H9c2 (H9c2 DOXEVs), non-treated FL83b (FL83b NTEVs), or DOX-treated FL83b (FL83b DOXEVs). (D) The levels of 4HNE protein adduction in H9c2 NTEVs, H9c2 DOXEVs, FL83b NTEVs, or FL83b DOXEVs. Data represent the mean of band intensities relative to H9c2 NTEVs ±SD of 5 independent experiments * p < 0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Measurement of 4HNE adduction in equal amounts of EVs protein reveals that EVs from H9c2 cells had more protein-bound 4HNE than EVs from FL83b cells. Moreover, comparisons between DOX-treated EVs (DOXEVs) and non-treated EVs (NTEVs) show that the H9c2 DOXEVs contained more 4HNE adducted proteins than the H9c2 NTEVs did. However, there was no difference between FL83b DOXEVs and FL83b NTEVs (Fig. 1D). These data suggest that DOX generates a higher level of oxidized proteins in cardiomyocyte than in hepatocyte EVs. To confirm that DOX is responsible for the increased 4HNE in EVs, increasing doses of DOX were used to treat H9c2 cells. The results show a dose-related increase in 4HNE in EVs derived from the DOX-treated H9c2 cells (Supplementary Fig. 1).
3.2. Differential effects of H9c2 EVs and FL83 b EVs on macrophage activation
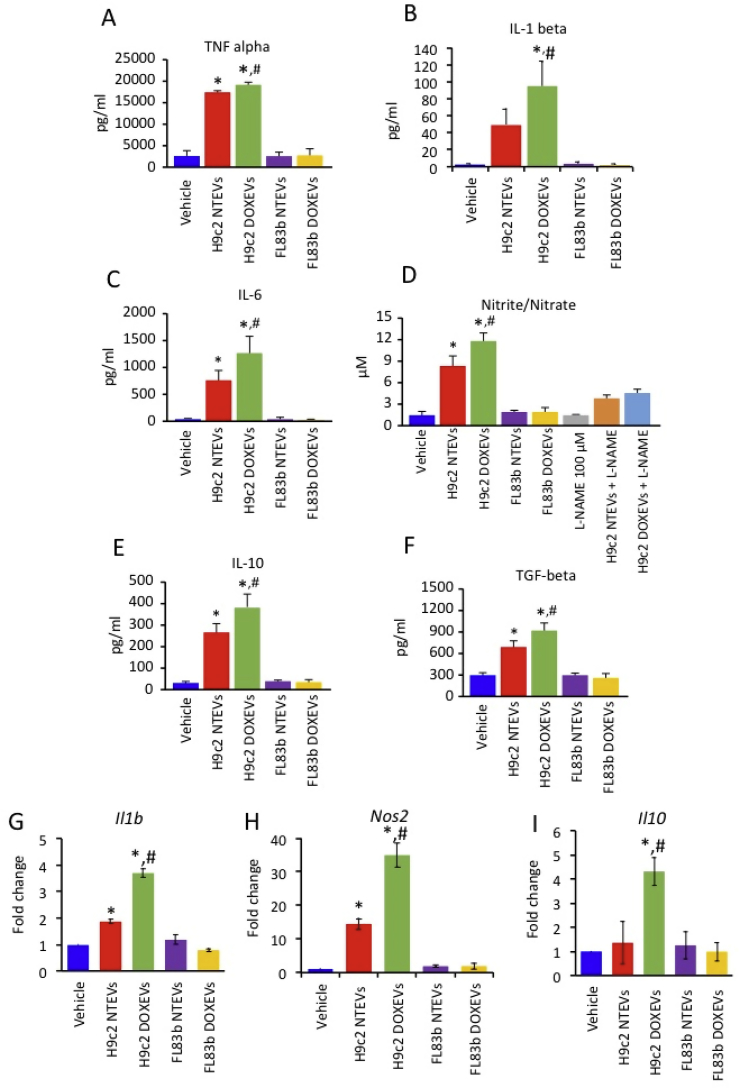
To investigate the effect of EVs on macrophage function, we determined the levels of pro- and anti-inflammatory cytokines, as well as •NO production, in conditioned media of RAW264.7 cells treated with H9c2 NTEVs, H9c2 DOXEVs, FL83b NTEVs, or FL83b DOXEVs for 24 h. The data show that the release of pro-inflammatory cytokines, including TNF alpha, IL-1 beta, IL-6, and •NO, significantly increased in H9c2 EVs-treated macrophages but not in FL83b EVs-treated macrophages (Fig. 2A and D). The •NO generation is specific to nitric oxide synthase (NOS) because it is suppressible by N(G)-Nitro-l-arginine methyl ester (l-NAME), which is a competitive inhibitor of NOS (Fig. 2D). Likewise, anti-inflammatory cytokines such as IL-10 and TGF beta also increased in response to H9c2 EVs but not FL83 b EV treatment (Fig. 2E and F). These data demonstrate that H9c2 EVs but not FL83 b EV caused both pro- and anti-inflammatory macrophage activation. DOX bolstered the immunostimulatory effects of H9c2 EVs but not FL83 b EV (Fig. 2A and F). The immunostimulatory effect of H9c2 EVs was not due to species differences between rat cardiomyocytes and mouse macrophages, as similar results were observed in pro- and anti-inflammatory gene expression in the NR8383 rat macrophage (Fig. 2G and I).
Fig. 2.
Cardiomyocyte-derived EVs but not hepatocyte-derived EVs promote both pro-inflammatory and anti-inflammatory cytokine release as well as nitric oxide production by macrophages. The effect was potentiated by DOX treatment
(A-C) Levels of pro-inflammatory cytokines TNF alpha, IL-1 beta, IL-6 in RAW264.7 cells treated for 24 h with vehicle, H9c2 NTEVs, H9c2 DOXEVs, FL83b NTEVs, or FL83b DOXEVs. (D) Level of Nitrite/Nitrate as a parameter of •NO production in RAW264.7 cells treated with vehicle, H9c2 NTEVs, H9c2 DOXEVs, FL83b NTEVs, or FL83b DOXEVs. l-NAME was used as a negative control as it is the inhibitor of •NO production. (E–F) Levels of anti-inflammatory cytokines; IL-10 and TGF beta in RAW264.7 cells treated for 24 h with vehicle, H9c2 NTEVs, H9c2 DOXEVs, FL83b NTEVs, or FL83b DOXEVs. Data represent the mean ± SD of 4 independent experiments, *p < 0.05 vs vehicle control, #p < 0.05 vs H9c2 NTEVs. (G–I) Gene expression of NR8383 rat macrophages after EVs treatment. Bar graphs represent fold change of Il1b(G), Nos2(H) and Il10(I) gene expression measured by RT-PCR of NR8383 rat macrophages treated with vehicle, H9c2 NTEVs, H9c2 DOXEVs, FL83b NTEVs, or FL83b DOXEVs. Data represent the mean ± SD of 3 independent experiments, *p < 0.05 vs vehicle, #p < 0.05 vs H9c2 NTEVs.
3.3. Differential effects of H9c2 EVs and FL83 b EV on macrophage metabolism
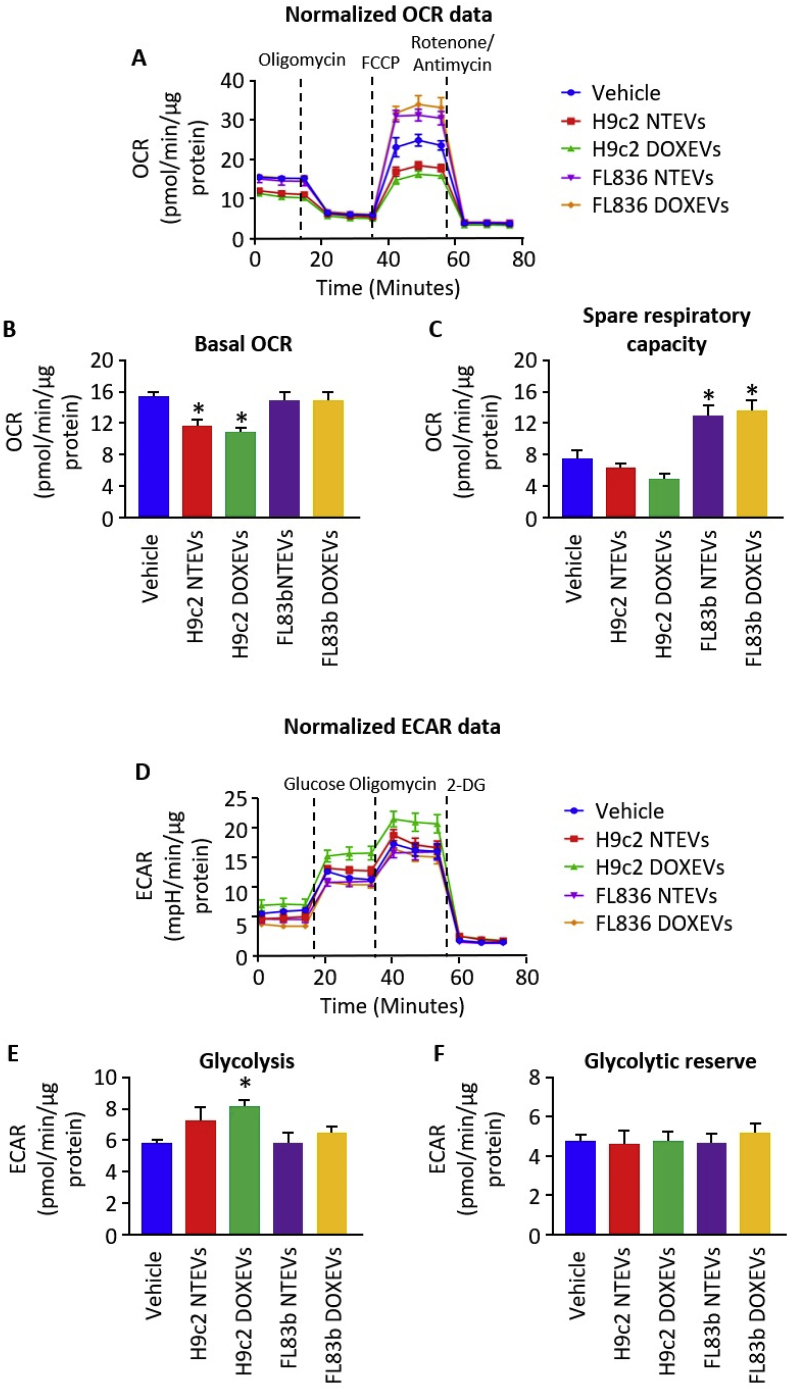
Pro- and anti-inflammatory macrophage functions are supported by the metabolic process used by macrophages to produce energy [42]. Pro-inflammatory macrophages rely more on glycolysis and less on mitochondrial respiration. The shift to glycolysis is suitable for rapid energy production during inflammation. In contrast, anti-inflammatory macrophages mainly utilize mitochondrial oxidative phosphorylation metabolism to make the ATP that supports cell survival and tissue repair.
Here, we observed that EVs induce the metabolic shifts in macrophages. The data from the mitochondrial stress and glycolytic stress tests show that EVs from DOX-treated H9c2 cells significantly suppressed basal oxygen consumption rate (OCR) (Fig. 3A and B) and enhanced glycolysis of macrophages (Fig. 3D and E), which supports the metabolic process expected for pro-inflammatory macrophage. EVs from non-treated H9c2 cells also suppressed basal OCR and slightly, but not significantly, increased glycolysis. EVs from non-treated and DOX-treated H9c2 cells had no effect on glycolytic reserve capacity (Fig. 3F). In contrast to H9c2 EVs, FL83b EVs did not have any effects on basal OCR, glycolysis, or glycolytic reserve (Fig. 3A and B, 3D-3F), but they did lead to higher mitochondrial spare respiratory capacity (Fig. 3C). These data suggest that EVs from hepatocytes did not cause either pro- or anti-inflammatory macrophage activation. However, their ability to enhance mitochondrial metabolism suggests that they may prime macrophages for the tissue repair process when conditions exist that increase the demand for ATP.
Fig. 3.
Differential effects of H9c2 EVs and FL83b EVs on mitochondrial respiration and glycolysis
(A) Kinetic graph of OCR at baseline and after oligomycin, FCCP, and rotenone/antimycin treatment of macrophages incubated with vehicle, H9c2 NTEVs, H9c2 DOXEVs, FL83b NTEVs, or FL83b DOXEVs. (B) Bar graph represents the average of basal OCR calculated from graph A. (C) Bar graph represents the average of Spare Respiratory Capacity calculated from graph A. (D) Kinetic graph of ECAR at baseline and after glucose, oligomycin, and 2-deoxyglucose (2-DG) treatment of macrophages incubated with vehicle, H9c2 NTEVs, H9c2 DOXEVs, FL83b NTEVs, or FL83b DOXEVs. (E) Bar graph represents the average of basal glycolysis calculated from graph D. (F) Bar graph represents the average of glycolytic reserve calculated from graph D. Data represent the mean ± SEM of 3 independent experiments, *p < 0.05 vs vehicle.
3.4. NFκB and Nrf-2 are activated by H9c2 EVs
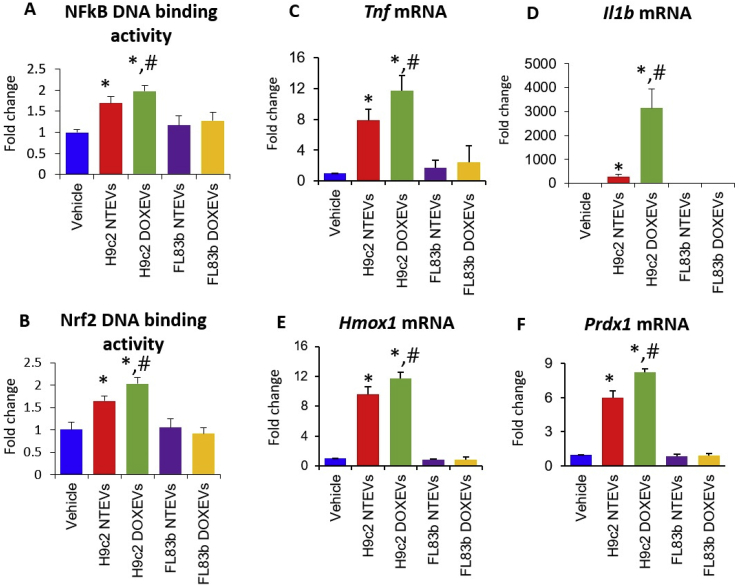
Because NFκB and Nrf-2 are sensitive to electrophiles and play an important role in macrophage activation, we hypothesized that H9c2 EVs, and especially H9c2 DOXEVs, which carry high levels of protein-bound 4HNE, might activate both NFκB and Nrf-2, leading to the observed pro- and anti-inflammatory macrophage activation.
By measuring the DNA binding activity of NFκB and Nrf2 in RAW264.7 macrophages treated with EVs, we found that H9c2 NTEVs significantly enhanced NFκB and Nrf-2 DNA binding activity. H9c2 DOXEVs slightly enhanced higher NFκB and Nrf-2 activation compared to H9c2 NTEVs (Fig. 4A and B). Interestingly, neither FL83b NTEVs nor FL83b DOXEVs significantly activated NFκB or Nrf-2 activity. The transcriptional activities of NFκB and Nrf-2 were confirmed by the expression of their target genes: TNF alpha (Tnf) and interleukin-1 beta (Il1b) for the former (Fig. 4C and D), and heme oxygenase 1 (Hmox-1) and peroxiredoxin 1 (Prdx1) for the latter (Fig. 4E and F).
Fig. 4.
Cardiomyocyte-derived EVs but not hepatocyte-derived EVs enhanced NFκB and Nrf-2 transcription factor activities. The effect was potentiated by DOX
(A) NFkB DNA binding activity and (B) Nrf2 DNA binding activity in macrophages treated with vehicle, H9c2 NTEVs, H9c2 DOXEVs, FL83b NTEVs, or FL83b DOXEVs. (C, D) Fold change of pro-inflammatory cytokines TNF alpha and IL-1 beta gene expression. (E, F) Fold change of anti-oxidant enzymes Hmox1 and Prdx1 gene expression after 24 h treatment of RAW264.7 cells with vehicle, H9c2 NTEVs, H9c2 DOXEVs, FL83b NTEVs, or FL83b DOXEVs. Data represent the mean ± SD of 3 independent experiments, *p < 0.05 vs vehicle, #p < 0.05 vs H9c2 NTEVs.
To verify that 4HNE adduction is responsible for immune activation of H9c2-derived EVs via NFκB and Nrf2 pathways, we co-treated RAW264.7 macrophages with 20 μg of H9c2 DOXEVs and various concentrations of vitamin E, a known lipid antioxidant that prevents polyunsaturated fatty acids from free radical attacks [43]. The results show that vitamin E inhibits NFκB and Nrf-2 responses in a dose-dependent manner with a greater inhibition effect on NF-kB than Nrf-2 at the same concentration (Supplementary Figs. 2A–2B).
To investigate why FL83b EVs, which also contain 4HNE-adducted proteins, did not activate NFκB or Nrf-2, we measured the mRNA levels of several antioxidants, including NAD(P)H Quinone Dehydrogenase 1 (Nqo1), superoxide dismutase 2 (Sod2), thioredoxin reductase 1 (Txnrd1), and Trx1, that might suppress the effect of FL83b EVs on macrophage activation (Supplementary Figs. 3A–3D). Interestingly, among these antioxidant genes, Trx1 is the gene that was upregulated by FL83b EVs, especially FL83b DOXEVs treatment (Supplementary Fig. 3D), suggesting that Trx1 may play an important role in preventing the activation of macrophages.
3.5. Trx1 function is critical for the differential effects of H9c2 EVs and FL83b EVs on macrophage activation
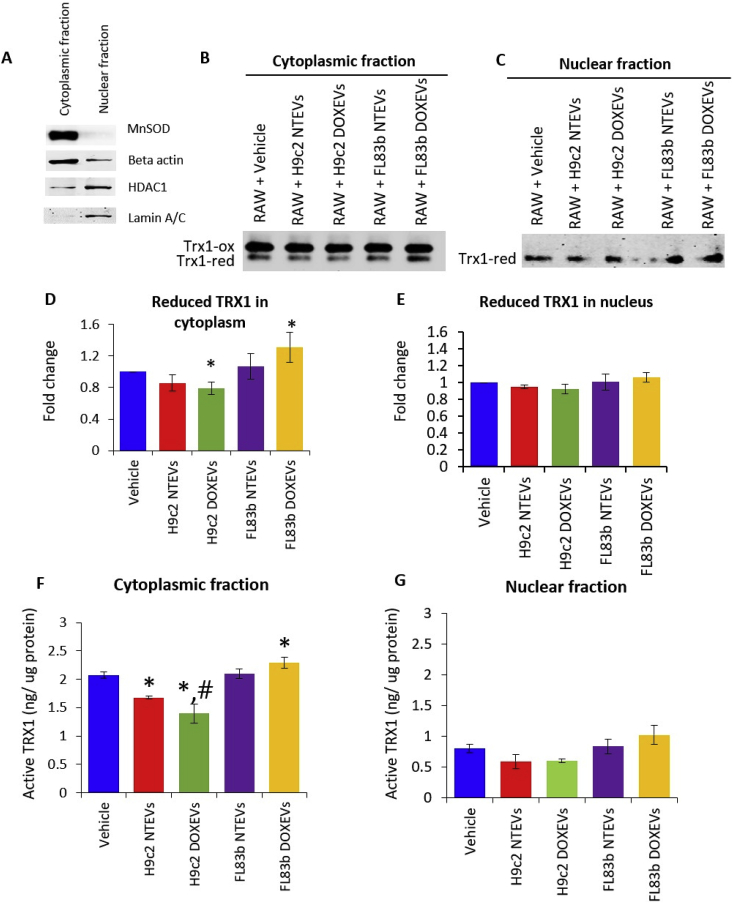
The redox status of Trx1 is important for its antioxidant activity and its immunoregulatory function. First, we evaluated the redox status of Trx1 in cytoplasmic and nuclear fraction of RAW264.7 macrophages in response to EVs treatment. The purity of cytoplasmic and nuclear fractions was verified by Western blot (Fig. 5A).
Fig. 5.
Differential effects of H9c2 EVs and FL83b EVs on reduced Trx1 and Trx1 activity
(A) Verification of cytoplasmic and nuclear fraction purification by Western blot of manganese superoxide dismutase (MnSOD) and beta actin for cytoplasmic fraction, as well as histone deacetylase 1 (HDAC1) and lamin A/C for nuclear fraction. (B and C) Redox western of Trx1 in cytoplasmic fraction (B) and nuclear fraction (C) of RAW264.7 treated with vehicle, H9c2 NTEVs, H9c2 DOXEVs, FL83b NTEVs, or FL83b DOXEVs. (D and E) Bar graph shows the average fold change (n = 3) of reduced Trx1 levels in cytoplasm (D) and nuclei (E). (F and G) Trx1 activity from equal amounts of protein in cytoplasm (F) and nuclei (G) of RAW264.7 treated with vehicle, H9c2 NTEVs, H9c2 DOXEVs, FL83b NTEVs, or FL83b DOXEVs, *p < 0.05 vs vehicle, #p < 0.05 vs H9c2 NTEVs.
Next, we determined the redox status of Trx1 by redox western analysis. The results show that H9c2 NTEVs-treated macrophages have lower levels of reduced Trx1 in the cytoplasm compared to the vehicle-treated control (though not statistically significant) and that treatment with H9c2 DOXEVs significantly lowered the reduced Trx1. In contrast, FL83b DOXEVs significantly increased the cytoplasmic levels of reduced Trx1. Macrophages treated with FL83b NTEVs also increased cytoplasmic reduced Trx1, but levels did not reach statistical significance (Fig. 5B and D). In the nuclear compartment, only the reduced form of Trx1 was detected and there was no significant change after EVs treatment (Fig. 5C and E). The changes in Trx1 activity in cytoplasm and nucleus are consistent with the levels of reduced Trx1 (Fig. 5F and G). These data suggest that EVs exert their effect on Trx1 redox state mainly in the cytoplasm of macrophages. The sharp contrast between the reduced Trx1 levels in cytoplasm of H9c2 DOXEVs-treated cells and FL83b DOXEVs-treated cells highlights the significance of Trx1 activity in macrophage activation.
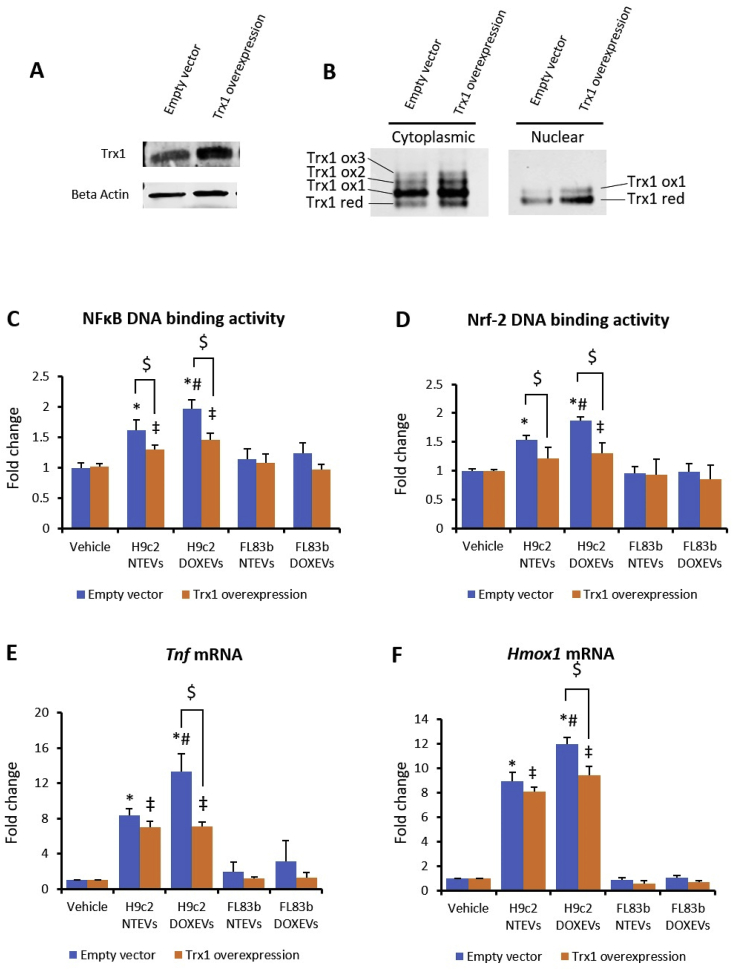
3.6. Trx1 at the center of the NFkB/Nrf-2 axis
To validate the role of Trx1 on EVs-mediated macrophage activation through NFκB and Nrf-2 pathways, we overexpressed mouse Trx1 gene in RAW264.7 cells. We then compared the DNA binding activity of NFκB and Nrf-2 24 h after EVs treatment. As shown in Fig. 6A, we confirmed that the level of Trx1 did increase in overexpressed cells. Trx1 redox western analysis in cytoplasmic and nuclear fraction shows that both reduced and oxidized Trx1 increased in Trx1 overexpressed cells. In nuclear fraction, reduced Trx1 was more predominant than oxidized Trx1 (Fig. 6B). Trx1 overexpression suppressed the effect of H9c2 NTEVs and H9c2 DOXEVs on NFκB and Nrf-2 activities, causing a reduction approximating that of the vehicle-treated control. The inhibitory effect of Trx1 was more prominent in H9c2 DOXEVs-treated cells, where NFκB and Nrf-2 activities decreased to the same levels as in the H9c2 NTEVs-treated cells (Fig. 6C and D). As a target gene of NFκB, Tnf in H9c2 DOXEVs-treated cells was significantly downregulated by Trx1. Tnf in H9c2 NTEVs-treated Trx1 overexpressed cells was slightly but not significantly lower than in the empty vector control. However, Trx1 overexpression significantly downregulated Tnf in H9c2 DOXEVs-treated cells (Fig. 6E). In H9c2 DOXEVs-treated cells, Hmox1, an Nrf-2 target gene, was also suppressed by Trx1 overexpression (Fig. 6F). These data support the central role of Trx1 in regulating both the NFκB and Nrf-2 pathways.
Fig. 6.
Trx1 overexpression suppressed the effect of H9c2 EVs on macrophage activation via NFκB and Nrf-2
(A) Western blot of Trx1 in RAW264.7 macrophage with Trx1 overexpression compared to empty vector. (B) Redox western of Trx1 in cytoplasmic and nuclear fraction of RAW264.7 macrophage with Trx1 overexpression compared to empty vector. (C and D) NFkB DNA binding activity (C) and Nrf2 DNA binding activity (D) in empty vector (blue) and Trx1 overexpressed RAW264.7 macrophages (orange) treated with vehicle, H9c2 NTEVs, H9c2 DOXEVs, FL83b NTEVs, or FL83b DOXEVs. (E and F) Fold change of pro-inflammatory cytokines Tnf(C) anti-oxidant Hmox1(D) gene expression after 24 h treatment of empty vector or Trx1 overexpressed RAW264.7 cells treated with vehicle, H9c2 NTEVs, H9c2 DOXEVs, FL83b NTEVs, or FL83b DOXEVs. Data represent the mean ± SD of 3 independent experiments, *p < 0.05 vs empty vector + vehicle, #p < 0.05 vs empty vector + H9c2 NTEVs, ‡p < 0.05 vs Trx1 overexpression + vehicle, $ p < 0.05 vs empty vector with the same treatment. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
In this study, we demonstrate that cardiomyocytes with a high sensitivity to DOX release EVs containing oxidized molecules that activate the pro- and anti-inflammatory responses of macrophages. In contrast, hepatocytes with a high tolerance to DOX release EVs that do not have immunological effects. This differential effect of EVs on macrophage functions is regulated by the redox state of Trx1, which influences the NFκB and Nrf-2 pathways.
DOX is a highly effective chemotherapeutic agent that is well-known to cause redox cycling in vivo. Our previous study demonstrates that heart and liver are two major normal tissue targets of DOX that release EVs into the circulation [19]. The results from this study of cardiomyocytes and hepatocytes are consistent with the animal data and supportive of the premise that cardiomyocytes and hepatocytes contribute to EVs released in response to DOX treatment. The higher levels of 4HNE-adducted proteins in H9c2-derived EVs, which increased after DOX treatment, might be due to cardiomyocytes being more susceptible to oxidative stress than hepatocytes are, since the activities of catalase, superoxide dismutase, and glutathione peroxidase in the heart are 150 times, 4 times, and 27 times lower than those in the liver, respectively [21,44].
Oxidized phospholipids are known to induce or inhibit inflammation. It has been shown that when macrophages are treated with oxidized 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine (OxPAPC), which is a major component of low-density lipoproteins, they are stimulated to express both pro- and anti-inflammatory phenotypes called Mox macrophages. Mox macrophages are a redox-regulatory phenotype with a high capacity to withstand oxidative stress by upregulating anti-oxidant enzymes through Nrf-2 activation [45]. Mox macrophages contribute to 30% of macrophages found in atherosclerotic plaque in a mouse model and play a key role in promoting chronic low-grade inflammation [46]. Since we observed that EVs from cardiomyocytes contain 4HNE, which is an oxidized lipid product, and because Nrf-2 activation also occurred, it is tempting to speculate that Mox may be the macrophage phenotype induced by cardiomyocyte EVs. However, extensive studies will be needed to substantiate the presence of Mox in immune responses. In our system, cardiomyocytes released oxidized EVs and stimulated an innate immune cell response without DOX treatment, which is unlikely to occur in vivo. This in vitro observation may be due, in part, to the cells being exposed to 21% O2 in the cell culture system, which is higher than the 4–14% O2 concentration in tissue. The higher concentration of oxygen caused oxidative stress to the cells at baseline [47]. By contrast, this O2 concentration is a negligible issue for hepatocytes since they contain high levels of antioxidant enzymes. A comparison of the non-treated and DOX-treated cells shows that DOX promoted release of immunostimulatory EVs from cardiomyocytes but not hepatocytes. This effect is not likely due to DOX, since the drug had been removed during the washing step. Our in vitro model mimics the clinical scenario where cardiotoxicity occurs later, after the cessation of DOX treatment [48]. A low-level but persistent inflammation, mediated by macrophage activation that is caused by cardiomyocyte-derived EVs, could lead to impaired cardiac function and systemic inflammation that can also affect indirect targets of DOX, such as brain. Although the hepatocytes-derived EVs contained fewer 4HNE-adducted proteins than the cardiomyocytes-derived EVs, there were some HNE-adducted proteins in the hepatocyte EVs. However, only the H9c2-derived EVs but not the FL83b-derived EVs activated macrophages. Therefore, the FL83b EVs may activate some genes, which prevents macrophage stimulation. Antioxidants have been shown to inhibit EV-associated 4HNE adduction. We have previously demonstrated that treatment with an antioxidant such as Mn(III) meso-tetrakis (N-(n-butoxyethyl)pyridium-2-yl)porphyrin, MnTnBuOE-2-PyP5+ (MnP) decreased 4HNE adduction in serum EVs in animals [19]. However, whether blocking 4HNE-adducted EVs alone is sufficient to block the activity of NF-kB and Nrf-2 would need to be investigated in depth as we found 4HNE-adducted proteins in the EVs. Thus, future investigation should focus on this and how HNE-adducted proteins contribute to EVs effects on the signaling pathways.
Metabolic reprogramming provides substrates needed for macrophage function. In our study, macrophages treated with cardiomyocyte DOXEVs suppressed mitochondrial oxidative phosphorylation (OXPHOS) and promoted glycolysis. This metabolic shift provides the environment required for antioxidant synthesis to maintain redox homeostasis [49]. In contrast to cardiomyocyte DOXEVs, hepatocyte DOXEVs did not have any effects on basal OXPHOS or glycolysis, but they did induce higher reserve capacity. Mitochondrial reserve capacity indicates the ability of mitochondria to generate ATP under high energy demand conditions, which is critical for the tissue repair process [50]. Our data suggest it is possible that, following DOX treatment, hepatocytes release EVs that provide biomolecules to help prepare macrophages for an anti-inflammatory response and thus help with the subsequent tissue repair. We propose that one of the key biomolecules is Trx1, because we found that Trx1 was the only major antioxidant enzyme that showed an increase in macrophages treated with hepatocyte EVs but a decrease in cardiomyocyte EVs. In heart tissue, Trx1 is known to enhance mitochondrial function by upregulation of genes that are involved in mitochondrial OXPHOS, including cytochrome c oxidase [51]. Trx1 also can help to maintain mitochondrial function by preserving the activity of the mTOR pathway [52]. In macrophages, Trx1 enhances the upregulation of IL-10 and CD206, the two genes commonly expressed in anti-inflammatory macrophages [53]. It is possible that, in macrophages that promote tissue healing, EVs-driven Trx1 expression may be a mechanism that protects the liver against indirect tissue damage by DOX.
Trx1 plays a major role in regulating NFκB and Nrf-2 activities, which depend on redox state and localization. In our model, the change in Trx1 redox state occurred predominantly in cytoplasm. Thus, our data suggest that cytosolic Trx1 is a key to EVs-mediated macrophage activation. However, we could not completely rule out the role of nuclear Trx1. We propose that the low level of the reduced Trx1 in cytoplasm caused by cardiomyocyte DOXEVs treatment leads to the low ability to sequester NFκB and Nrf-2 by IκB and Keap-1, respectively, resulting in the activation of these transcription factors. In contrast to cardiomyocyte EVs, hepatocyte EVs increase the level of reduced cytoplasmic Trx1 leading to a lack of macrophage activation (Fig. 7). Trx1 overexpression causes higher levels of Trx1 in both cytoplasm and in nucleus. Trx1 in cytoplasm inhibits both NFκB and Nrf-2 activation while Trx1 in nucleus promotes NFκB and Nrf-2 activities. In Trx1-overexpressed cells, increased cytoplasmic Trx1 might not be high enough to completely block NFκB and Nrf-2 nuclear translocation. Consequently, the translocated NFκB and Nrf-2 transcription activities will be enhanced by higher levels of nuclear Trx1. Therefore, the effect of H9c2 EVs is still significantly higher than vehicle and/or mouse hepatocytes.
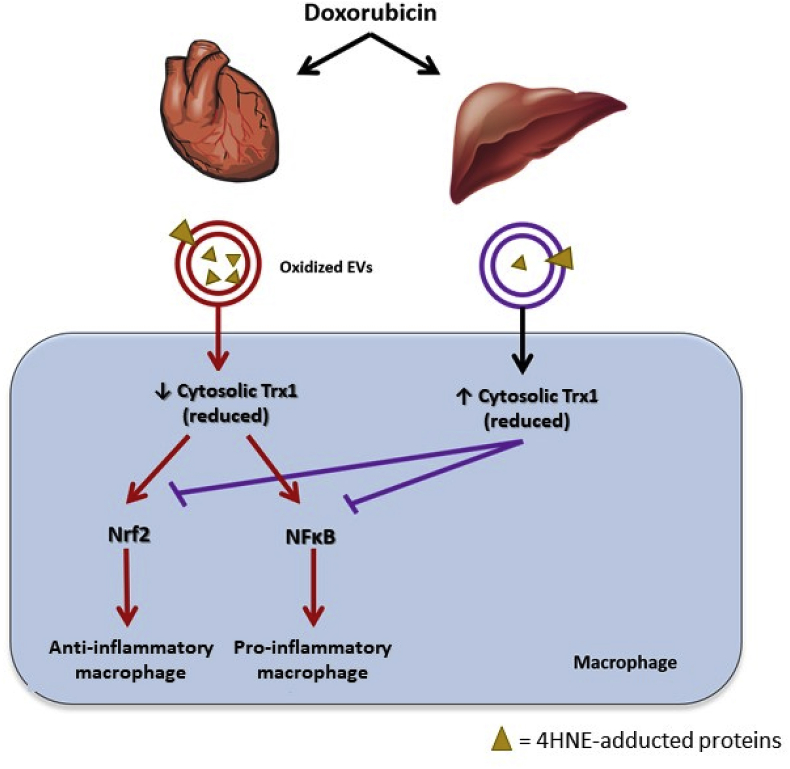
Fig. 7.
Proposed mechanistic model for the differential effects of normal tissue-derived EVs on macrophages.
Cardiomyocyte EVs induce pro- and anti-inflammatory macrophage activation by transferring 4HNE-adducted proteins to the macrophages, which in turn exacerbates oxidative stress in the macrophages by decreasing cytosolic reduced Trx1. As a result of reduced Trx1 exhaustion, NFκB is activated and pro-inflammatory cytokines are produced. Nrf-2 activation occurs as Keap-1 becomes more oxidized by the loss of the Trx1 that is required for it to maintain its reducing state. DOX aggravates the propagation of oxidative stress induced by cardiomyocyte EVs. In contrast, hepatocyte EVs upregulate cytosolic Trx1, which maintains macrophages in an inactivated status. DOX potentiates the effect of hepatocyte EVs on Trx1 induction, thus preventing macrophage activation after Dox treatment.
In conclusion, our data suggest that not all EVs released upon exposure to pro-oxidant conditions are detrimental. EVs released by cells that survive oxidative stress can induce macrophages to generate more redox regulatory proteins, such as Trx1, that help limit immune activation. Therefore, preventing the release of EVs by interfering with EVs biogenesis will not only block the ability of injured cells to remove toxic molecules, it also eliminates any beneficial effects of EVs released by surviving cells. Consequently, modulation of Trx1 function could be an effective approach to modulate the innate immune system to protect against the indirect effects of inflammatory mediators following cancer therapy.
Disclosures
The authors declare no financial conflicts of interest.
Financial support
This study was supported by the National Institutes of Health, RO1 CA217934, P30 CA177558, and P20 GM121327; and a Ministry of Science and Technology Royal Thai Government Scholarship to C. Yarana.
Acknowledgments
We thank Dr. Edward Kasarskis for his insights and helpful discussions, and Dr. William St. Clair for his informed advice. Furthermore, we would like to acknowledge Dr. Donald Cohen for permission to use the Odyssey instrument, Donna Wall for RNA integrity analysis, and Michael Alstott for assistance with the Seahorse Bioenergetics study. We acknowledge with gratitude the assistance of the Redox Metabolism Shared Resource Facility of the Markey Cancer Center, accompanied by a Cancer Center Support Grant from the National Cancer Institute (2P30 CA177588-01).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101237.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Benedikter B.J., Weseler A.R., Wouters E.F.M., Savelkoul P.H.M., Rohde G.G.U., Stassen F.R.M. Redox-dependent thiol modifications: implications for the release of extracellular vesicles. Cell. Mol. Life Sci. : CM. 2018;75:2321–2337. doi: 10.1007/s00018-018-2806-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robbins P.D. Extracellular vesicles and aging. Stem Cell Invest. 2017;4:98. doi: 10.21037/sci.2017.12.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Minciacchi V.R., Freeman M.R., Di Vizio D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin. Cell Dev. Biol. 2015;40:41–51. doi: 10.1016/j.semcdb.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loyer X., Vion A.C., Tedgui A., Boulanger C.M. Microvesicles as cell-cell messengers in cardiovascular diseases. Circ. Res. 2014;114:345–353. doi: 10.1161/CIRCRESAHA.113.300858. [DOI] [PubMed] [Google Scholar]
- 5.Fujita Y., Kosaka N., Araya J., Kuwano K., Ochiya T. Extracellular vesicles in lung microenvironment and pathogenesis. Trends Mol. Med. 2015;21:533–542. doi: 10.1016/j.molmed.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Heiserman J.P., Chen L., Kim B.S., Kim S.C., Tran A.L., Siebenborn N., Knowlton A.A. TLR4 mutation and HSP60-induced cell death in adult mouse cardiac myocytes. Cell Stress Chaperones. 2015;20:527–535. doi: 10.1007/s12192-015-0577-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mancek-Keber M., Frank-Bertoncelj M., Hafner-Bratkovic I., Smole A., Zorko M., Pirher N., Hayer S., Kralj-Iglic V., Rozman B., Ilc N., Horvat S., Jerala R. Toll-like receptor 4 senses oxidative stress mediated by the oxidation of phospholipids in extracellular vesicles. Sci. Signal. 2015;8 doi: 10.1126/scisignal.2005860. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Martinez I., Santoro N., Chen Y., Hoque R., Ouyang X., Caprio S., Shlomchik M.J., Coffman R.L., Candia A., Mehal W.Z. Hepatocyte mitochondrial DNA drives nonalcoholic steatohepatitis by activation of TLR9. J. Clin. Investig. 2016;126:859–864. doi: 10.1172/JCI83885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai Y., Xu M.J., Koritzinsky E.H., Zhou Z., Wang W., Cao H., Yuen P.S., Ross R.A., Star R.A., Liangpunsakul S., Gao B. Mitochondrial DNA-enriched microparticles promote acute-on-chronic alcoholic neutrophilia and hepatotoxicity. JCI insight. 2017;2 doi: 10.1172/jci.insight.92634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burrello J., Monticone S., Gai C., Gomez Y., Kholia S., Camussi G. Stem cell-derived extracellular vesicles and immune-modulation. Frontiers in cell and developmental biology. 2016;4:83. doi: 10.3389/fcell.2016.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X., Mosser D.M. Macrophage activation by endogenous danger signals. J. Pathol. 2008;214:161–178. doi: 10.1002/path.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meredith A.M., Dass C.R. Increasing role of the cancer chemotherapeutic doxorubicin in cellular metabolism. J. Pharm. Pharmacol. 2016;68:729–741. doi: 10.1111/jphp.12539. [DOI] [PubMed] [Google Scholar]
- 13.Zhu H., Sarkar S., Scott L., Danelisen I., Trush M.A., Jia Z., Li Y.R. Doxorubicin redox biology: redox cycling, topoisomerase inhibition, and oxidative stress. Reactive oxygen species. 2016;1:189–198. doi: 10.20455/ros.2016.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tangpong J., Cole M.P., Sultana R., Joshi G., Estus S., Vore M., St Clair W., Ratanachaiyavong S., St Clair D.K., Butterfield D.A. Adriamycin-induced, TNF-alpha-mediated central nervous system toxicity. Neurobiol. Dis. 2006;23:127–139. doi: 10.1016/j.nbd.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Chaiswing L., Cole M.P., St Clair D.K., Ittarat W., Szweda L.I., Oberley T.D. Oxidative damage precedes nitrative damage in adriamycin-induced cardiac mitochondrial injury. Toxicol. Pathol. 2004;32:536–547. doi: 10.1080/01926230490502601. [DOI] [PubMed] [Google Scholar]
- 16.Dalleau S., Baradat M., Gueraud F., Huc L. Cell death and diseases related to oxidative stress: 4-hydroxynonenal (HNE) in the balance. Cell Death Differ. 2013;20:1615–1630. doi: 10.1038/cdd.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Y., Miriyala S., Miao L., Mitov M., Schnell D., Dhar S.K., Cai J., Klein J.B., Sultana R., Butterfield D.A., Vore M., Batinic-Haberle I., Bondada S., St Clair D.K. Redox proteomic identification of HNE-bound mitochondrial proteins in cardiac tissues reveals a systemic effect on energy metabolism after doxorubicin treatment. Free Radical Biol. Med. 2014;72:55–65. doi: 10.1016/j.freeradbiomed.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miriyala S., Thippakorn C., Chaiswing L., Xu Y., Noel T., Tovmasyan A., Batinic-Haberle I., Vander Kooi C.W., Chi W., Latif A.A., Panchatcharam M., Prachayasittikul V., Butterfield D.A., Vore M., Moscow J., St Clair D.K. Novel role of 4-hydroxy-2-nonenal in AIFm2-mediated mitochondrial stress signaling. Free Radical Biol. Med. 2016;91:68–80. doi: 10.1016/j.freeradbiomed.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yarana C., Carroll D., Chen J., Chaiswing L., Zhao Y., Noel T., Alstott M., Bae Y., Dressler E.V., Moscow J.A., Butterfield D.A., Zhu H., St Clair D.K. Extracellular vesicles released by cardiomyocytes in a doxorubicin-induced cardiac injury mouse model contain protein biomarkers of early cardiac injury. Clin. Cancer Res. : an official journal of the American Association for Cancer Research. 2018;24:1644–1653. doi: 10.1158/1078-0432.CCR-17-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King P.D., Perry M.C. Hepatotoxicity of chemotherapy. Oncol. 2001;6:162–176. doi: 10.1634/theoncologist.6-2-162. [DOI] [PubMed] [Google Scholar]
- 21.Odom A.L., Hatwig C.A., Stanley J.S., Benson A.M. Biochemical determinants of Adriamycin toxicity in mouse liver, heart and intestine. Biochem. Pharmacol. 1992;43:831–836. doi: 10.1016/0006-2952(92)90250-m. [DOI] [PubMed] [Google Scholar]
- 22.Patel M., Taskar K.S., Zamek-Gliszczynski M.J. Importance of hepatic transporters in clinical disposition of drugs and their metabolites. J. Clin. Pharmacol. 2016;56(Suppl 7):S23–S39. doi: 10.1002/jcph.671. [DOI] [PubMed] [Google Scholar]
- 23.Prinz W.A., Aslund F., Holmgren A., Beckwith J. The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J. Biol. Chem. 1997;272:15661–15667. doi: 10.1074/jbc.272.25.15661. [DOI] [PubMed] [Google Scholar]
- 24.Sengupta R., Holmgren A. The role of thioredoxin in the regulation of cellular processes by S-nitrosylation. Biochim. Biophys. Acta. 2012;1820:689–700. doi: 10.1016/j.bbagen.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 25.Tamaki H., Nakamura H., Nishio A., Nakase H., Ueno S., Uza N., Kido M., Inoue S., Mikami S., Asada M., Kiriya K., Kitamura H., Ohashi S., Fukui T., Kawasaki K., Matsuura M., Ishii Y., Okazaki K., Yodoi J., Chiba T. Human thioredoxin-1 ameliorates experimental murine colitis in association with suppressed macrophage inhibitory factor production. Gastroenterology. 2006;131:1110–1121. doi: 10.1053/j.gastro.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 26.Ohashi S., Nishio A., Nakamura H., Kido M., Ueno S., Uza N., Inoue S., Kitamura H., Kiriya K., Asada M., Tamaki H., Matsuura M., Kawasaki K., Fukui T., Watanabe N., Nakase H., Yodoi J., Okazaki K., Chiba T. Protective roles of redox-active protein thioredoxin-1 for severe acute pancreatitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2006;290:G772–G781. doi: 10.1152/ajpgi.00425.2005. [DOI] [PubMed] [Google Scholar]
- 27.Chen G., Li X., Huang M., Li M., Zhou X., Li Y., Bai J. Thioredoxin-1 increases survival in sepsis by inflammatory response through suppressing endoplasmic reticulum stress. Shock. 2016;46:67–74. doi: 10.1097/SHK.0000000000000570. [DOI] [PubMed] [Google Scholar]
- 28.Thimmulappa R.K., Scollick C., Traore K., Yates M., Trush M.A., Liby K.T., Sporn M.B., Yamamoto M., Kensler T.W., Biswal S. Nrf2-dependent protection from LPS induced inflammatory response and mortality by CDDO-Imidazolide. Biochem. Biophys. Res. Commun. 2006;351:883–889. doi: 10.1016/j.bbrc.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thimmulappa R.K., Lee H., Rangasamy T., Reddy S.P., Yamamoto M., Kensler T.W., Biswal S. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J. Clin. Investig. 2006;116:984–995. doi: 10.1172/JCI25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi E.H., Suzuki T., Funayama R., Nagashima T., Hayashi M., Sekine H., Tanaka N., Moriguchi T., Motohashi H., Nakayama K., Yamamoto M. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 2016;7:11624. doi: 10.1038/ncomms11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayashi T., Ueno Y., Okamoto T. Oxidoreductive regulation of nuclear factor kappa B. Involvement of a cellular reducing catalyst thioredoxin. J. Biol. Chem. 1993;268:11380–11388. [PubMed] [Google Scholar]
- 32.Xu Y., Fang F., Miriyala S., Crooks P.A., Oberley T.D., Chaiswing L., Noel T., Holley A.K., Zhao Y., Kiningham K.K., Clair D.K., Clair W.H. KEAP1 is a redox sensitive target that arbitrates the opposing radiosensitive effects of parthenolide in normal and cancer cells. Cancer Res. 2013;73:4406–4417. doi: 10.1158/0008-5472.CAN-12-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirota K., Murata M., Sachi Y., Nakamura H., Takeuchi J., Mori K., Yodoi J. Distinct roles of thioredoxin in the cytoplasm and in the nucleus. A two-step mechanism of redox regulation of transcription factor NF-kappaB. J. Biol. Chem. 1999;274:27891–27897. doi: 10.1074/jbc.274.39.27891. [DOI] [PubMed] [Google Scholar]
- 34.Lukosz M., Jakob S., Buchner N., Zschauer T.C., Altschmied J., Haendeler J. Nuclear redox signaling. Antioxidants Redox Signal. 2010;12:713–742. doi: 10.1089/ars.2009.2609. [DOI] [PubMed] [Google Scholar]
- 35.Berridge M.V., Tan A.S. Characterization of the cellular reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT): subcellular localization, substrate dependence, and involvement of mitochondrial electron transport in MTT reduction. Arch. Biochem. Biophys. 1993;303:474–482. doi: 10.1006/abbi.1993.1311. [DOI] [PubMed] [Google Scholar]
- 36.Divakaruni A.S., Paradyse A., Ferrick D.A., Murphy A.N., Jastroch M. Analysis and interpretation of microplate-based oxygen consumption and pH data. Methods Enzymol. 2014;547:309–354. doi: 10.1016/B978-0-12-801415-8.00016-3. [DOI] [PubMed] [Google Scholar]
- 37.TeSlaa T., Teitell M.A. Techniques to monitor glycolysis. Methods Enzymol. 2014;542:91–114. doi: 10.1016/B978-0-12-416618-9.00005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Halvey P.J., Watson W.H., Hansen J.M., Go Y.M., Samali A., Jones D.P. Compartmental oxidation of thiol-disulphide redox couples during epidermal growth factor signalling. Biochem. J. 2005;386:215–219. doi: 10.1042/BJ20041829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Go Y.M., Ziegler T.R., Johnson J.M., Gu L., Hansen J.M., Jones D.P. Selective protection of nuclear thioredoxin-1 and glutathione redox systems against oxidation during glucose and glutamine deficiency in human colonic epithelial cells. Free Radical Biol. Med. 2007;42:363–370. doi: 10.1016/j.freeradbiomed.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watson W.H., Pohl J., Montfort W.R., Stuchlik O., Reed M.S., Powis G., Jones D.P. Redox potential of human thioredoxin 1 and identification of a second dithiol/disulfide motif. J. Biol. Chem. 2003;278:33408–33415. doi: 10.1074/jbc.M211107200. [DOI] [PubMed] [Google Scholar]
- 41.Montano S.J., Lu J., Gustafsson T.N., Holmgren A. Activity assays of mammalian thioredoxin and thioredoxin reductase: fluorescent disulfide substrates, mechanisms, and use with tissue samples. Anal. Biochem. 2014;449:139–146. doi: 10.1016/j.ab.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 42.Langston P.K., Shibata M., Horng T. Metabolism supports macrophage activation. Front. Immunol. 2017;8:61. doi: 10.3389/fimmu.2017.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X., Quinn P.J. The location and function of vitamin E in membranes (review) Mol. Membr. Biol. 2000;17:143–156. doi: 10.1080/09687680010000311. [DOI] [PubMed] [Google Scholar]
- 44.Doroshow J.H., Locker G.Y., Myers C.E. Enzymatic defenses of the mouse heart against reactive oxygen metabolites: alterations produced by doxorubicin. J. Clin. Investig. 1980;65:128–135. doi: 10.1172/JCI109642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kadl A., Meher A.K., Sharma P.R., Lee M.Y., Doran A.C., Johnstone S.R., Elliott M.R., Gruber F., Han J., Chen W., Kensler T., Ravichandran K.S., Isakson B.E., Wamhoff B.R., Leitinger N. Identification of a novel macrophage phenotype that develops in response to atherogenic phospholipids via Nrf2. Circ. Res. 2010;107:737–746. doi: 10.1161/CIRCRESAHA.109.215715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shalhoub J., Falck-Hansen M.A., Davies A.H., Monaco C. Innate immunity and monocyte-macrophage activation in atherosclerosis. J. Inflamm. 2011;8:9. doi: 10.1186/1476-9255-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jagannathan L., Cuddapah S., Costa M. Oxidative stress under ambient and physiological oxygen tension in tissue culture. Current pharmacology reports. 2016;2:64–72. doi: 10.1007/s40495-016-0050-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abu-Khalaf M.M., Juneja V., Chung G.G., DiGiovanna M.P., Sipples R., McGurk M., Zelterman D., Haffty B., Reiss M., Wackers F.J., Lee F.A., Burtness B.A. Long-term assessment of cardiac function after dose-dense and -intense sequential doxorubicin (A), paclitaxel (T), and cyclophosphamide (C) as adjuvant therapy for high risk breast cancer. Breast Canc. Res. Treat. 2007;104:341–349. doi: 10.1007/s10549-006-9413-7. [DOI] [PubMed] [Google Scholar]
- 49.Serbulea V., Upchurch C.M., Ahern K.W., Bories G., Voigt P., DeWeese D.E., Meher A.K., Harris T.E., Leitinger N. Macrophages sensing oxidized DAMPs reprogram their metabolism to support redox homeostasis and inflammation through a TLR2-Syk-ceramide dependent mechanism. Molecular metabolism. 2018;7:23–34. doi: 10.1016/j.molmet.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van den Bossche J., Baardman J., Otto N.A., van der Velden S., Neele A.E., van den Berg S.M., Luque-Martin R., Chen H.J., Boshuizen M.C., Ahmed M., Hoeksema M.A., de Vos A.F., de Winther M.P. Mitochondrial dysfunction prevents repolarization of inflammatory macrophages. Cell Rep. 2016;17:684–696. doi: 10.1016/j.celrep.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 51.Ago T., Yeh I., Yamamoto M., Schinke-Braun M., Brown J.A., Tian B., Sadoshima J. Thioredoxin 1 upregulates mitochondrial proteins related to oxidative phosphorylation and TCA cycle in the heart. Antioxidants Redox Signal. 2006;8:1635–1650. doi: 10.1089/ars.2006.8.1635. [DOI] [PubMed] [Google Scholar]
- 52.Oka S.I., Hirata T., Suzuki W., Naito D., Chen Y., Chin A., Yaginuma H., Saito T., Nagarajan N., Zhai P., Bhat S., Schesing K., Shao D., Hirabayashi Y., Yodoi J., Sciarretta S., Sadoshima J. Thioredoxin-1 maintains mechanistic target of rapamycin (mTOR) function during oxidative stress in cardiomyocytes. J. Biol. Chem. 2017;292:18988–19000. doi: 10.1074/jbc.M117.807735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.El Hadri K., Mahmood D.F., Couchie D., Jguirim-Souissi I., Genze F., Diderot V., Syrovets T., Lunov O., Simmet T., Rouis M. Thioredoxin-1 promotes anti-inflammatory macrophages of the M2 phenotype and antagonizes atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2012;32:1445–1452. doi: 10.1161/ATVBAHA.112.249334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.