Abstract
Background
High resolution genome-wide copy number analysis, routinely used in clinical diagnosis for several years, retrieves new and extremely rare copy number variations (CNVs) that provide novel candidate genes contributing to disease etiology. The aim of this work was to identify novel genetic causes of neurodevelopmental disease, inferred from CNVs detected by array comparative hybridization (aCGH), in a cohort of 325 Portuguese patients with intellectual disability (ID).
Results
We have detected CNVs in 30.1% of the patients, of which 5.2% corresponded to novel likely pathogenic CNVs. For these 11 rare CNVs (which encompass novel ID candidate genes), we identified those most likely to be relevant, and established genotype-phenotype correlations based on detailed clinical assessment. In the case of duplications, we performed expression analysis to assess the impact of the rearrangement. Interestingly, these novel candidate genes belong to known ID-related pathways. Within the 8% of patients with CNVs in known pathogenic loci, the majority had a clinical presentation fitting the phenotype(s) described in the literature, with a few interesting exceptions that are discussed.
Conclusions
Identification of such rare CNVs (some of which reported for the first time in ID patients/families) contributes to our understanding of the etiology of ID and for the ever-improving diagnosis of this group of patients.
Electronic supplementary material
The online version of this article (10.1186/s13023-019-1135-0) contains supplementary material, which is available to authorized users.
Keywords: CNVs, Neurodevelopment, Genotype-phenotype correlation, CUL4B overexpression
Background
Intellectual disability (ID) is one of the most common neurodevelopmental disorders (NDDs), affecting nearly 3% of the population worldwide. ID has a complex etiology resulting from the combination of environmental and genetic factors [1]. Relatively recent approaches to the identification of copy number variations (CNVs), have highlighted the relevance of rare de novo, and essentially private mutations that contribute to a significant proportion of the risk of NDDs, being presently an unavoidable element of diagnosis in the field of Neuropsychiatry, Neuropediatrics and Neurodevelopmental Pediatrics.
A substantial number of ID patients have CNVs resulting from deletions or duplications [2, 3]. The frequency of detection of chromosome abnormalities and/or genomic rearrangements in patients with NDDs by array comparative genomic hybridization (aCGH) depends mainly on the patient inclusion clinical criteria and on the microarray design; nevertheless, detection rates are usually higher in patients with ID/developmental delay (DD) that also present malformations or dysmorphic features and more severe cognitive impairment [2]. The characterization of these CNVs in different patient cohorts as well as in the general population is necessary to clarify their clinical relevance and establish adequate genotype-phenotype correlations [4].
We present the results obtained by studying 325 Portuguese patients with idiopathic ID using aCGH, in whom we found known and new candidate pathogenic CNVs. As expected, the great majority of the detected CNVs were rare and restricted to one patient/family; nevertheless, the efforts towards their characterization represent a step forward in order to clarify their clinical and molecular significance.
Results
Global data
From the 325 patients, 30.1% had at least one non-polymorphic CNV detected by aCGH (Part 1 of Additional file 1: Table S1): 8% had pathogenic CNVs, 5.2% had likely pathogenic CNVs and 16.9% had genomic variants of unknown significance (VOUS). The remaining 69.9% patients had only known polymorphic CNVs.
Pathogenic CNVs
The pathogenic CNVs detected were mainly de novo CNVs, including deletions at 1p36.23-p36.21, 2p13.1–13.3, 3q22.1-q23, 5p15.33-p15.32, 6q25.3, 7q11.23, 8p23.1, 11q24.2-q25, 12q24.21-q24.22, 16p11.2, 17q21.31, 22q11.21 and 22q13.3, as well as duplications at 1q21.1, 12q24.21, 9q34.13–34.3, 13q12.12-q34, 14q32.31-q32.33, 14q32.33, 15q11.2-q13.1, 16p13.11, 21q11.2-q22.11, Xp11.22 and Xq28 (see Table 1 for the list of all patients and findings). For most of these CNVs there are reports in the literature describing the phenotypic and genetic findings for similar patients, therefore only some particular cases are described in detail and discussed in Part 1 of Additional file 1, namely: (a) the interstitial deletion at 1p36.23-p36.21 found de novo in patient R1, of interest since interstitial deletions in this region are rarely described in association with NDDs; (b) the deletion at 3q22.1-q23 found de novo in patient R3, which reinforces the association of deletions affecting FOXL2 gene with blepharophimosis syndrome; (c) 7q11.23 deletions, detected in two non-related patients (C2 and R29), neither of whom presents the classical Williams-Beuren syndrome phenotype; (d) the 22q13.3 deletion found in patient C7, due to the incomplete overlap of the patient’s phenotype with that previously described for Phelan-McDermid syndrome; (e) the 9q34 duplications, detected in two non-related patients (C19 and R14): patient C19 has an intragenic EHMT1 duplication and a clinical presentation that overlaps the core phenotype of Kleefstra syndrome, commonly caused by deletions or point mutations affecting the EHMT1 gene; patient R14 has three de novo duplications at 9q34.13-q34.3 (affecting the whole EHMT1 gene), at 14q32.31-q32.33 and at 14q32.33, illustrating the difficulty to ascertain the specific role of each imbalance in complex rearrangements. We also included in this category CNVs occurring in risk-associated loci.
Table 1.
Clinical overview of RC patients for whom non-polymorphic CNVs vs likely benign and polymorphic CNVs were detected in the aCGH
| Pathogenic + Likely pathogenic (n = 23) | Polymorphic CNVs (n = 134) |
|---|---|
| Gender | Gender |
| Males 15 (65%) | Males 84 (63%) |
| Females 8 (35%) | Females 50 (37%) |
| ID | ID |
| Syndromic 19 (83%) | Syndromic 74 (55%) |
| Non-syndromic 4 (17%) | Non-syndromic 60 (45%) |
| Borderline 1 (4%) | Borderline 8 (6%) |
| Mild 15 (65%) | Mild 75 (56%) |
| Moderate 6 (26%) | Moderate 30 (22%) |
| Severe 0 (0%) | Severe 15 (11%) |
| Profound 1 (4%) | Profound 6 (4%) |
| History | History |
| Sporadic 11 (48%) | Sporadic 54 (40%) |
| Family history of ID 15 (65%) | Family history of ID 80 (60%) |
| Co-morbidities | Co-morbidities |
| Congenital anomalies 11 (48%) | Congenital anomalies 64 (48%) |
| Epilepsy 2 (9%) | Epilepsy 19 (14%) |
| Microcephaly 4 (17%) | Microcephaly 23 (17%) |
| Macrocephaly 1 (4%) | Macrocephaly 13 (10%) |
Likely pathogenic CNVs
Likely pathogenic CNVs were detected in 5.2% of patients in this study (Table 2; Figs. 1 and 2). They comprise candidate ID-causative loci located in 1q43-q44, 2q11.2-q12.2, 7q33, 10q26.3, 17p11.2 and 20q13.12-q13.13 (losses); 1p22.1-p21.3, 7q33, 9q33.2-q33.3, 9q34.3, Xq24 and Xq26.3 (gains) (Table 2). Patients with 1q43-q44, 7q33 and 10q26.3 CNVs have been described elsewhere in detail [5–7]; the patient with a 9q34.3 gain is described together with patient R14 in Part 1 of Additional file 1; therefore, we focus next on the remaining candidate loci.
Table 2.
List of pathogenic CNVs
| Patients | Gender | Alteration (Hg19) | Type | Size (Mb) | Genes | Key gene(s) involved | Associated syndrome | Phenotype overlap | Inheritance | Confirmation | Array platform | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R1 | Male | arr 1p36.23-p36.21(8,593,674-15,396,672)x1dn | del | 6.7 | 86 | ANGPTL7, CASZ1, MAD2L2, RERE | – | – | de novo | NP | 1 | – |
| R2 | Male | arr 2p13.1-p13.3(70,894,906-74,986,518)x1dnc | del | 4 | 62 | CYP26B1, EXOC6B | – | – | de novo | NP | 1 | Wen J, 2013 |
| R3 | Male | arr 3q22.1-q23(131,415,639-141,618,552)x1dn | del | 1.020 | 65 | FOXL2 | BPES | Yes (eye features) | de novo | NP | 1 | – |
| C1 | Male | arr 5p15.33-p15.32(204,849-5,014,883)x1 | del | 4.81 | 30 | TERT [CTNND2 not involved] | – | – | ND | NP | 2 | – |
| R4 | Male | arr 6q25.3(156,012,754-158,804,494)x1dnc | del | 2.6 | 14 | ARID1B | Coffin-Siris syndrome | Yes | de novo | NP | 1 | Santen GW, 2013 |
| C2 | Male | arr 7q11.23(72,721,760-74,140,846)x1 | del | 1.419 | 28 | BAZ1B, STX1A, WBSCR22, ELN | Williams-Beuren syndrome | Partially | ND | NP | 2 | – |
| R5 | Female | arr 8p23.1(7,039,276-12,485,558)x1dn | del | 5.5 | 70 | SOX7, GATA4 | 8p23.1 deletion syndrome | Yes (cardiac) | de novo | NP | 1 | – |
| C3 | Male | arr 11q24.2-q25(125,232,584-134,446,160)x1dn | del | 9.214 | 54 | KIRREL3, ETS1, FLI1, KCNJ1, KCNJ5, RICS | – | Partially | de novo | qPCR | 2 | – |
| R6 | Female | arr 12q24.21-q24.22(115,505,500-117,441,683)x1dnc | del | 0.2 | 10 | MED13L | – | Yes | de novo | qPCR | 1 | Adegbola A, 2015 |
| C4 | Male | arr 16p11.2(29,674,336-30,198,123)x1dn | del | 0.524 | 29 | KCTD13 | 16p11.2 deletion syndrome | – | de novo | NP | 2 | – |
| C5 | Male | arr 17q21.31(43,710,371-44,215,352)x1 | del | 0.505 | 8 | CRHR1, MAPT, STH, and part of the KIAA1267 (KANSL1) | 17q21.31 deletion syndrome (Koolen-De Vries syndrome) | – | ND | NP | 3 | – |
| C6 | Male | arr 22q11.21(18,894,835-21,505,417)x1 | del | 2.611 | 59 | TBX1 | 22q11 deletion syndrome | – | ND | NP | 2 | – |
| C7 | Male | arr 22q13.3(49,513,903-51,178,264)x1 | del | 1.664 | 39 | SHANK3 | 22q13.3 deletion syndrome (Phelan-McDermid syndrome) | Partially | ND | NP | 2 | – |
| C8 | Male | arr 1q21.1q21.2(146,106,723-147,830,830)x3dn | dup | 1.7 | 17 | HYDIN2, PRKAB2 | 1q21.1 duplication syndrome e | Partially | de novo | qPCR | 4 | – |
| R7 | Male | arr 1q21.1(145,883,119-148,828,690)x3pat | dup | 2.5 | 23 | HYDIN2, PRKAB2, GJA5 | 1q21.1 duplication syndrome e | Yes | paternal | NP | 1 | – |
| R8 | Male | arr 12q24.21(116,408,736-116,704,303)x3dnc | dup | 0.3 | 2 | MED13L | – | Yes | de novo | qPCR | 1 | Adegbola A, 2015 |
| C9 | Male | arr 13q12.12-q34(23,749,431-115,083,342)x2.15a | dup | 91.33 | ## | – | Trisomy 13 (mosaicism) | Yes | ND | Karyotyped | 2 | – |
| C10 | Female | arr 15q11.2-q13.1(22880274–29,331,964)x3mat | dup | 6.45 | 111 | CYF1P1, NIPA2, NIPA1, MKRN3, NDN, MAGEL2, SNURF/SNRPN, UBE3A GABRB3 | 15q11-q13 duplication syndromeb | Yes | maternal | NP | 2 | – |
| C11 | Female | arr 16p13.11(15,034,010-16,199,882)x3 | dup | 1.166 | 11 | NDE1 | 16p13.11 duplication syndrome e | – | ND | NP | 5 | – |
| R9 | Male | arr 16p13.11(15,421,671-16,443,968)x3mat | dup | 1 | 19 | NDE1 | 16p13.11 duplication syndrome e | Yes | maternal | NP | 1 | – |
| R10 | Male | arr 16p13.11(15,484,180-16,308,344)x3mat | dup | 0.8 | 9 | NDE1 | 16p13.11 duplication syndrome e | Yes | maternal | NP | 1 | – |
| C12 | Male | arr 21q11.2-q22.11(14,417,523-34,894,625)x3 | dup | 20.47 | 110 | DSCR1, DSCR2, DSCR3, DSCR4, APP | – | No | ND | NP | 2 | – |
| R11 | Male | arr Xp11.22(53,569,653-53,769,748)x2mat | dup | 0.2 | 3 | HUWE1 | – | Yes | maternal | qPCR | 1 | – |
| R12 | Male | arr Xq28(152,348,378-155,228,013)x2dn | dup | 2.8 | 78 | MECP2 | MECP2 duplication syndrome | Yes | de novo | NP | 1 | – |
| R13 | Male | arr Xq28(153,130,545-153,602,293)x2mat | dup | 0.5 | 16 | MECP2 | MECP2 duplication syndrome | Yes | maternal | NP | 1 | – |
| R14 | Male | arr 9q34.13-q34.3(135,767,911-141,153,431)x3dn | dup | 5.516 | 135 | EHMT1, RXRA, GRIN1, UAP1L1 | 9q34 duplication syndrome | Partially | de novo | NP | 1 | – |
| arr 14q32.31-q32.33(102,959,110-104,578,612)x3dn | dup | 1.620 | 22 | MARK3, KLC1, EIF5 | – | – | de novo | NP | 1 | – | ||
| arr 14q32.33(105,104,831-106,531,339)x3dn | dup | 1.427 | 24 | – | – | de novo | NP | 1 | – |
Patients R1 to R14: from research cohort; Patients C1 to C12: from clinical cohort; NP Not performed, ND Not determined; (a): mosaicism; (b) methylation status for SNRPN is normal (studied by MLPA); (c): Published in detail elsewhere; (d): karyotype revealed a balanced translocation between chromosomes 13 and 14, resulting in mosaic trisomy 13; (e): Other causes of disease were not excluded therefore the variant might not explain the total phenotypic presentation. Array platform 1: Agilent 180 K; 2: KaryoArray®v3.0 (Agilent 8x60k); 3: Affymetrix CytoScan HD array; 4: Affymetrix CytoScan 750 K; 5: Agilent Whole Genome 244 K
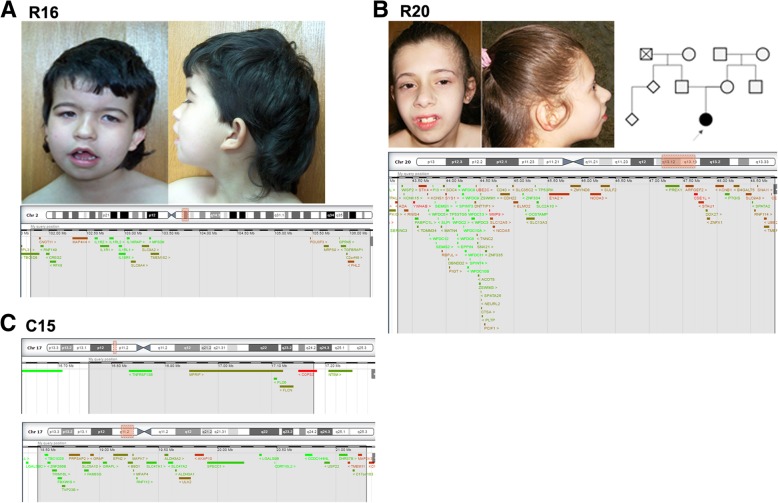
Fig. 1.
Facial appearance of patients and schematic representation of the deletions. a Patient R16 facies, with low set posteriorly rotated ears, anteverted ears with simplified helix, temporal narrowing with prominent metopic suture, arched eyebrows, synophrys, bilateral epicanthal folds, bulbous nasal tip, thin upper lip, open mouth with downturned corners, micrognathia; pedigree and deleted region of chromosome 2 (highlighted in red in the chromosome scheme (above) and in grey in the genes’ portion (below), adapted from DECIPHER). b Patient R20 facies, with wide forehead, strabismus, high nasal bridge, wide base of nose, bulbous nasal tip, short and smooth philtrum, thin upper lip with effaced cupid’s bow, prominent central incisors and micrognathia; pedigree and deleted region of chromosome 20. c Patient C15: deleted region in chromosome 17
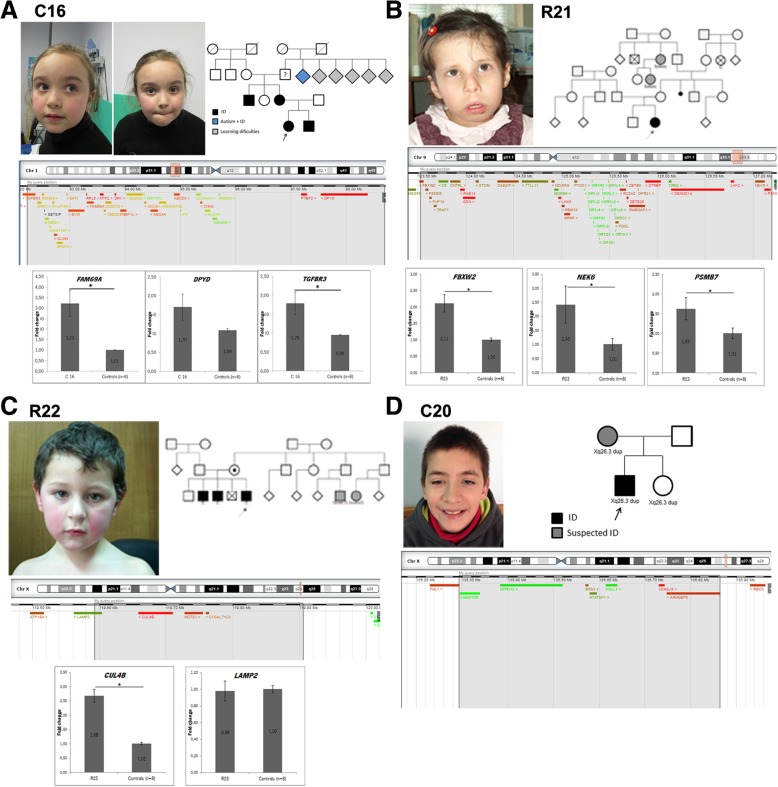
Fig. 2.
Overview of some patients with likely pathogenic duplications. a Patient C16 - facial appearance: mildly dysmorphic, with high forehead and frontal bossing, thick eyebrows and mildly anteverted nares; pedigree, schematic representation of the duplicated 1p region and expression pattern for genes FAM69A, DPYD and TGFBR3. b Patient R21 - facial appearance: large forehead, sparse lateral eyebrows, epicanthal folds, large nose, anteverted nares, long smooth philtrum, downturned corners of mouth and micrognathia; pedigree, schematic representation of the triplicated 9q region and expression pattern for genes FBXW2, NEK6 and PSMB7. c Patient R22 - facial appearance: mildly dysmorphic with large forehead and frontal central hair whorl; pedigree, schematic representation of the duplicated Xq region and expression pattern for CUL4B and LAMP2 genes. d Patient C20 - facial appearance: mildly dysmorphic patient with thick eyebrows, wide palpebral fissures and thin upper lip; pedigree and schematic representation of the duplicated Xq region. B2M and PPIB were used as housekeeping genes; * p < 0.05 (Student t-test)
2q11.2-q12.2 deletion
Patient R16 is a 17 year old girl with syndromic ID, cerebral ventricular enlargement, dysmorphic features and hirsutism. She carries a de novo 4.5 Mb deletion at 2q11.2-q12.2 affecting 26 genes, of which MAP4K4, FHL2, POU3F3 and CNOT11 have the highest haploinsufficiency score in DECIPHERr [8]. POU Class 3 Homeobox 3 (POU3F3) was previously reported deleted in a boy with ID and dysmorphic features (such as flat nose, prominent ears, large eyebrows and low hairline) [9], similar to those of our patient. This gene encodes a transcription factor present in post-mitotic cells and plays a role in neurogenesis and the correct destination of migratory neurons in the cerebral cortex in the mouse [10], thus standing out as a good candidate for the DD/ID in the patient.
17p11.2 deletions
Patient C15 is a 10 year old boy referred for consultation for DD, namely language and motor impairment, ataxia and some dysmorphic features, including hypertelorism, strabismus and low-set ears. It was not possible to reevaluate for IQ testing, but at the time of first evaluation he had no formal cognitive deficit (according with the GMDS score when he was 5 years old) and cerebral magnetic resonance imaging (MRI) showed no alterations. He has what appear to be two consecutive deletions at 17p11.2: a 420.6Kb deletion, that encompasses 5 genes, and a 2.77 Mb deletion that encompasses 36 genes. He has inherited them from his mother, who has confirmed learning difficulties, although she has completed the 6th grade. These deletions partially overlap the region involved in Smith-Magenis syndrome (SMS); however, the phenotype of the patient and mother is not similar to that of SMS, and the deletion does not affect the retinoic acid induced 1 (RAI1) gene, thought to cause most of the SMS core phenotype [11]. Among the genes affected by patient C15’s deletions, there are several others whose function could potentially contribute for his phenotype (detailed in Part 1 of Additional file 1).
20q13.12-q13.13 deletions
Patient R20 is a 16 year old girl with mild ID (IQ = 56), speech delay, MIC and facial dysmorphisms. Brain imaging studies revealed no structural alterations. She also has astigmatism and attention deficit hyperactivity disorder (ADHD). She carries a de novo 5.5 Mb deletion at 20q13.12-q13.13 encompassing 123 genes. Among these, the genes KCNB1, PIGT, CTSA, SLC2A10 and ARFGEF2 were associated with human disease (detailed in Part 1 of Additional file 1).
1p22.1p21.3 duplications
Patient C16 is a 7 year old girl with motor and speech delay, with a global DQ of 56.3 (GMDS). She carries a maternal 1p22.1p21.3 duplication of 6.461 Mb that affects 44 genes. Her mother has completed the 6th grade although with 2 in-grade retentions and always showing learning difficulties, especially in language skills. The girl has a 10 year old brother suspected of having cognitive deficit: he was not evaluated yet, but he is attending the 2nd grade and does not yet know how to read. There is also a positive history of learning difficulties on the maternal grandfather’s family side. The duplication affects several genes (Fig. 2a), including the FAM69A gene, which encodes a member of the FAM69 family of cysteine-rich type II transmembrane proteins. FAM69 proteins are thought to play a fundamental role in the endoplasmic reticulum, in addition to specialized roles in the vertebrate nervous system, according to a brain-specific or brain-including expression pattern [12]. Consistently, several FAM69 genes have been linked to neuropsychiatric disorders: C3ORF58 (DIA1) with autism [13]; CXORF36 (DIA1R) with X-linked ID [14] and FAM69A with schizophrenia and bipolar disease [15]. Even though the contribution of the excess of dosage for NDDS is still unknown, this gene can be considered a good candidate to explain the disease in the patient.
9q33.2-q33.3 triplication
Patient R21 is a 17 year old girl with mild ID (IQ = 53) and familial history of ID. During the neonatal period she presented seizures (flexion spasms and later generalized tonic-clonic), controlled with Phenobarbital, which was discontinued at 23 months; EEG initially showed lateral paroxystic activity, bilaterally, and a normal result at 6 months; brain MRI was normal. Additionally, she presented dysmorphic facial features (Fig. 2), a muscular ventricular septal defect that closed spontaneously, hypothyroidism, hypotonia, global DD, growth deceleration (height and weight around the 3rd centile after 12 months) with normal head size, around the 75th centile, delayed bone maturation (~ 3 years), growth hormone deficiency and short neck. She carries a 3.6 Mb de novo triplication at 9q33.2-q33.3 that affects 60 genes. Of those, only the CRB2 gene is associated with a human disease. Moreover, this triplication apparently disrupts the FBXW2 gene that encodes for an F-box protein. F-box proteins are one of the four subunits of ubiquitin protein ligases, called SCFs. SCF ligases bring ubiquitin conjugating enzymes to substrates that are specifically recruited by the different F-box proteins. Components of this complex, such as CUL4B, have been involved in ID pathogenesis [16]. Also included in the CNV are the LHX2 and LHX6 genes, both encoding transcription factors described to play roles in brain development [17, 18]. Additionally, LHX2 was also described to be involved in osteoclast differentiation and its overexpression inhibits skeletal muscle differentiation [19]. LHX6 is also known to play a role in cranial and tooth development [20], hence these genes could be of relevance to the cranioskeletal phenotype of the patient.
Based on the location within the triplication region and the expression levels described we selected the FBXW2, NEK6 and PSMB7 genes (detailed in Part 1 of Additional file 1) to study at the mRNA level in peripheral blood in the patient. The three genes had an increased expression when compared to controls (Fig. 2b). For NEK6 these findings are in accordance with the fact it is included inside the triplicated region. Regarding FBXW2 and PSMB7, we had hypothesized that their expression could be diminished since they are located at the breakpoints, which we concluded not to be the case. To the best of our knowledge no mutations in any of these three genes were reported in human NDDs, making their involvement in our patient’s symptomatology difficult to confirm at this stage.
Xq24 duplication
Patient R22 is a 14 year old boy with borderline IQ (IQ = 80) and a familial history of ID (two brothers and cousins with ID), an apparently benign cardiac arrhythmia, overweight (BMI 23.6 Kg/m2 P90), stereotypies and ADHD. He carries a 0.3 Mb maternally inherited duplication at Xq24 affecting four genes (CUL4B, LAMP2, C1GALT1C1, MCTS1), his mother being asymptomatic. Both point mutations and large deletions in the CUL4B gene are described as causative of X-linked ID and cerebral malformations [21, 22]. CUL4B is a scaffold protein member of the cullin family that works in the formation of protein complex that acts as an E3 ubiquitin ligase catalyzing the polyubiquitination of protein substrates. CUL4B was found to be responsible for TSC2 degradation in neocortical neurons positively regulating mTOR activity in those cells [23]. Additionally, CUL4B also targets WDR5 for ubiquitylation leading to its degradation in neurons nucleus, which causes impaired neurite outgrowth [24]. However, to our knowledge, there is only one 47.2 Mb duplication encompassing CUL4B (and other genes) described in a patient with ID [25], the present case being the first small, non-disruptive CUL4B duplication described in a patient with ID. CUL4B is entirely duplicated in the patient and its expression in peripheral blood cells is increased, leading to us to believe that the disorder in the patient is in fact driven by a dosage increase in CUL4B. The LAMP2 gene, located in the duplication breakpoint and encoding a protein with roles in autophagy/lysosomal function, does not present altered expression in the patient, suggesting that it may not be contributing to this phenotype (Fig. 2c).
Xq26.3 duplication
Patient C20 is a 17 year old boy referred to the consultation due to general DD. He carries a 570.1Kb duplication at Xq26.3 inherited from his mother, who has a suspicion of some cognitive impairment but for whom no formal intellectual assessment was possible. He has a global DQ of 57.1 (evaluated at the age of 10 years), scoring below the average in all GMDS sub-scales, namely on language and eye-hand co-ordination, and is described as a friendly boy. He has speech delay, dolichocephaly and several dysmorphisms, including micrognatia, syndactyly and clinodactyly. His younger sister (8 years old) also carries the duplication but has no ID and has a normal development for her age which, this being an X-linked gene, is not incompatible with the causality of disease. The duplication encompasses the several genes (Fig. 2d) including the ARHGEF6 gene. ARHGEF6 encodes for a protein that belongs to a family of cytoplasmic proteins which activate the Rho proteins by exchanging bound GDP for GTP. These Rho GTPases play a fundamental role in numerous cellular processes linked to the organization of the cytoskeleton, cell shape, and motility [26]. ARHGEF6 specifically has been implicated in the regulation of spine morphogenesis and loss of function (LoF) mutations have been found in patients with X-linked ID [27]. A 2.8 Mb duplication in Xq26.2-Xq26.3 has also been described in two brothers with ID and the ARHGEF6, PHF6, HPRT1 and SLC9A6 genes have been identified as potential contributors to those patients’ phenotype [28]. When compared to this publication, we can see that our patient’s duplication is smaller and affects only the ARHGEF6 gene; nevertheless, the phenotypic similarities between our patient and those described by Madrigal and colleagues (namely ID, dolichocephaly and facial dysmorphisms) suggest a determinant role for ARHGEF6 gene in phenotypes associated with Xq26 microduplications [28]. Expression data in the periphery for some of the genes involved in the duplication didn’t retrieve results that we could interpret.
CNVs of unknown significance
In the VOUS group, we included CNVs which did not encompass a known NDDs-related CNV region and for which (i) pathogenicity was not sufficiently supported by biological data, and/or (ii) similar copy number changes were described in control databases, and/or (iii) were inherited from a parent for whom the clinical presentation was not known. For 50% of these cases, inheritance from parents was not possible to determine due to parental sample unavailability, thus reducing our ability to interpret their clinical significance. A summary of the VOUS identified in this study is presented in Part 1 of Additional file 1: Table S2).
Discussion
This study of a cohort of ID patients in whom most common causes of disease had been excluded allowed us to find a reliable cause of disease in 8% of patients and to propose novel candidate ID loci in 5.2%. Making a stricter analysis and considering only the variants associated (or likely associated) with disease we can consider that this yield is comparable with several other similar studies, in which percentages ranging between 8.5 and 16% were achieved [29–31]. The CNVs classified as pathogenic often appear de novo and affect (in general) dozens of genes. Some difficulties arose when classifying several of these CNVs as, in some cases, although they occurred in known syndrome regions not all the patients carrying them presented the major clinical features established for that particular syndrome. In fact, even these well-established pathogenic CNVs can be associated with a broad and distinctive phenotypic presentation, as observed in patients C2 and R29, both with WBS associated deletions but not presenting the full-blown phenotype of this syndrome. In this perspective, we believe that the main contributions of this work are: (I) the reporting of new patients with CNVs in regions associated with identified syndromes but with different clinical presentations; (II) the reporting of novel candidate ID-causative loci at 2q11.2-q12.2 (del), 7q33 (del and dup), 10q26.3 (del), 17p11.2 (del), 20q13.12-q13.13 (del), 1p22.1-p21.3 (dup), 9q33.2-q33.3 (tri), 9q34.3 (dup), Xq24 (dup) and Xq26.3 (dup); (III) the study in patients with copy number gains of the mRNA expression in peripheral blood for genes located either inside the duplicated/triplicated regions and/or at the breakpoints, making it possible to determine if there is an actual effect of gene dosage at the transcription level. Many of the CNVs here detected by aCGH were rare and restricted to one patient/family, which made their contribution to the patient’s phenotype difficult to assess. Several of these have been therefore classified as VOUS and their clinical significance needs to be carefully addressed in future studies. Individually rare intermediate-size CNVs (frequency, ≤0.05%; ≥250 kb), and not necessarily assigned a priori as pathogenic, appear to be collectively common in unselected populations (10.5%), and have been associated with ID and negatively with educational attainment [4]; being so, even these should not be excluded as cause of disease but rather re-assessed in the face of accumulating information, in order to establish useful genotype-phenotype correlations. Nevertheless, one cannot exclude the possibility that some of these CNVs are unrelated to pathogenesis, namely in patients where no other genomic testing (such as whole-exome or whole-genome sequencing) was performed to rule out other causes, this being a potential limitation of this work.
NDDs associated pathways: old and new genes
The likely pathogenic CNVs here proposed as novel candidate loci for ID encompass several genes that either were already associated with NDDs (like CUL4B) or are now proposed to have a role in ID and which can be grouped according to their function in several cellular aspects:
Transcriptional factors/cell cycle regulators/DNA repair proteins
Transcriptional regulation is an essential component of the neuronal differentiation programs and of the response to stimulation patterns underlying neuronal plasticity; genes involved in these pathways have been implicated in well-known NDDs, as is the case of FOXL2 [32], BAZ1B [33], and EBF3 [7]. This work revealed genes that appear to be good candidate loci for ID; of those, POU3F3, already described deleted in a patient with ID [9], stands as a strong candidate.
Chromatin modifiers/chromatin remodeling proteins
An excess of mutation genes encoding proteins involved in chromatin regulation have been described in NDDs [34]. EHMT1 and ARID1B belong to this category and are known to be associated with ID for many years. Here we describe two more patients with duplications affecting the EHMT1 gene, in one of which it was possible to show EHMT1 overexpression. ARID5A encodes for a protein belonging to the ARID family of proteins with important roles in development, tissue-specific gene expression and proliferation control [35].
Ubiquitin signaling
Ubiquitin-mediated degradation of proteins is a crucial mechanism for cell maintenance and viability [36]. Several genes belonging to this pathway are described to be associated with NDDs, as is the case of CUL4B [21], shown here to be duplicated in two patients. UBE2C encodes a key component of the ubiquitin proteasome system (UPS) that participates in cell cycle progression and checkpoint control [37]. The NEURL3 and CNOT4 genes also encode for proteins with E3 ubiquitin-protein ligase activity; as for FBXW2, it encodes for one of the four types of subunits of SCF ubiquitin-protein ligases. Neither of these genes has been linked, until now, with NDDs, but our findings reinforce the idea that genes encoding for proteins belonging to the UPS are possible new candidate genes for NDD phenotypes.
Cytoskeleton regulation and organization, cell shape and motility
Several NDDs are caused by mutations in genes regulating neuronal migration, which often encode for proteins involved in the function of the cytoskeleton [38]. TSC1, involved in microtubule-mediated protein transport due to unregulated mTOR signaling [39], and ARHGEF6, here described in different CNVs, have been previously associated with NDDs [39, 40]. B9D1 has been confirmed as a novel Meckel syndrome gene [41].
Intracellular vesicular trafficking and exocytosis
In this work we report a patient with a deletion encompassing ARFGEF2, previously described associated with epilepsy and ID (in the case of homozygous mutations) [42, 43]. The collection of patients presented herein also allowed the first description of EXOC6B gene haploinsufficiency in association with DD/ID (reported in detail in a dedicated publication) [44].
Signaling mediators/transducers/ receptor activity/transmembrane proteins
Disruption of synaptogenesis has been associated with ID and NDDs [45] and in this work we could identify CNVs in several genes associated with this pathway. SEMA4C gene encodes a transmembrane semaphorin which regulates axonal guidance in the developing nervous system [46]. Syntaxins, such as Syntaxin 1A, encoded by STX1A gene, are key molecules implicated in the docking of synaptic vesicles with the presynaptic plasma membrane [47]. Signaling processes are essential for proper cellular function and usually implicate enzymes, transmembrane proteins and voltage ion-channels whose disruption may be associated with disease [48]. Many of the genes described herein, including CACNA1C, GPR45, TNFRSF13B, FAM69A, AKT3 and CSE1L, are associated with these pathways, highlighting once again the crucial contribution of proper cellular signaling and synapse development and function for ID/DD.
Of notice, and although our attempts of establishing genotype-phenotype correlations was mostly focused on dosage impact of individual genes (e.g. haploinsufficiency/overexpression), CNVs may also lead to disease through other mechanisms, namely gene fusion generation [49] and impact on genome architecture, for example Topological Associated Domain disruption, with impact on the expression of genes located outside the affected regions [50].
Conclusion
The aCGH technology has for long been used in the research and clinical contexts allowing the delineation of many new microdeletion and microduplication syndromes. In the last decade a decrease in the rate at which new syndromes were described has been observed, most likely because the most frequent/recurrent CNVs were described in the early days of aCGH [51]. For the remaining and rarer (often “private”) forms, it is still important, however, to make an effort to share their clinical and genetic features as well as the CNV data, to support future diagnosis and establishment of genotype-phenotype correlations, as well as the identification of novel candidate genes for disease, as those advanced here.
Subjects and methods
Subjects
This work included the analysis of 325 ID patients (full IQ (FIQ) below 70 and borderline FIQ 70–80) of Portuguese origin (36.9% females, 63.1% males), of which 188 (mostly trios) were included in a research cohort (RC) and 137 were studied in the context of routine clinical genetics diagnostics (clinical cohort, CC), all being referenced as having NDDs (detailed description of inclusion criteria and clinical characterization provided in Part 1 of Additional file 1). For the RC we were able to obtain DNA for all the parents as well as a more extensive clinical description (see Table 3).
Table 3.
List of likely pathogenic CNVs
| Patients | Gender | Alteration (Hg19) | Type | Size (Kb) | Genes | Relevant genes involved | Confirmation | Inheritance | DGV controls | DECIPHER | Array platform | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C13 | Male | arr 1q43-q44(240,043,427-249,233,096)x1dn f | del | 3.7 | 18 | AKT3 | qPCR | de novod | No | 250,152, 250,915 (smaller) | 1 | Lopes F, et al., 2019 |
| R15 | Female | arr 1q43-q44(243,552,007-243,738,675)x1dn f | del | 0.19 | 2 | AKT3 | qPCR | de novod | No | 252,432 (smaller) | 2 | Lopes F, et al., 2019 |
| C14 | Male | arr 1q43q44(243,592,147-243,749,968)x1pat f | del | 0.16 | 2 | AKT3 | qPCR | paternal | No | 252,432 (smaller) | 1 | Lopes F, et al., 2019 |
| R16 | Female | arr 2q11.2-q12.2(101,756,265-106,265,018)x1dn | del | 4500 | 24 | MAP4K4, FHL2, POU3F3, CNOT11 | qPCR | de novo | No | 251,756 | 2 | – |
| R17, R18e | Male, Female | arr 7q33(133,176,651-135,252,871)x1mat f | del | 2076 | 23 | AGBL3, CNOT4, CALD1, EXOC4 | qPCR | maternala | No | 256,036 | 2 | Lopes F, et al., 2018 |
| R19 | Female | arr 10q26.3(131,374,701-132,030,468)x1dn | del | 600 | 3 | EBF3 | qPCR | de novo | 3/6564b | No | 2 | Lopes F et al., 2017 |
| C15 | Male | arr 17p11.2(16,757,564-17,178,161)x1mat | del | 420 | 5 | COPS3 | NP | maternala | No | No | 3 | – |
| arr 17p11.2(18,478,816-21,255,056)x1mat | del | 2770 | 36 | EPN2, RNF112, ULK2, ALDH3A2, AKAP10, B9D1 | NP | maternala | No | 340,692 (smaller) | – | |||
| R20 | Female | arr 20q13.12-q13.13(43,283,820-48,850,844)x1dn | del | 5500 | 88 | KCNB1, PIGT, CTSA, SLC2A10, ARFGEF2 | NP | de novo | No | 309 | 2 | – |
| C16 | Female | arr 1p22.1p21.3(92,227,986-98,689,243)x3mat | dup | 6461 | 44 | FAM69A, TGFBR3, GLMN, EVI5, RPL5, MTF2, DR1, ABCA4, ABCD3, CNN3, PTBP2, DPYD | qPCR | maternala | No | 318,358 | 1 | – |
| C17,C18e | Male, Male | arr 7q33(134,598,205-134,815,177)x3mat f | dup | 216 | 2 | CALD1, AGBL3 | qPCR | maternala | No | No | 1 | Lopes F, et al., 2018 |
| R21 | Female | arr 9q33.2-q33.3(123,525,064-127,187,619)x4dn | tri | 3600 | 52 | CRB2, LHX2, LHX6, DENND1A, STRBP, RAB14, GSN, PSMB7, ZBTB26 | qPCR | de novo | No | No | 2 | – |
| C19 | Female | arr 9q34.3(140540819–140,659,057)×3mat | dup | 0.118 | 2 | EHMT1 | NP | maternal | 1/2504 (smaller) | No | 1 | – |
| R22, R23e | Male, Male | arr Xq24(119,592,606-119,904,981)x2mat | dupc | 300 | 4 | CUL4B, LAMP2, C1GALT1C1, MCTS1 | qPCR | maternal | No | No | 2 | – |
| C20 | Male | arr Xq26.3(135,293,144-135,863,290)x2mat | dup | 570 | 9 | ARHGEF6, CD40LG, BRS3, MAP7D3 | qPCR | maternal | No | No | 3 | – |
Patients R15 to R23: from research cohort; Patients C13 to C20: from clinical cohort; NP Not performed; (a): inherited from an affected parent; (b): doubt regarding the quality of the call in these controls; (c) duplication may disrupt gene if located in tandem; (d) paternity and maternity confirmed; (e): siblings; (f): family described elsewhere. Array platform 1: Affymetrix Cystoscan 750 K; 2: Agilent 180 K; 3: KaryoArray®v3.0 (Agilent 8x60k)
Methods
Genomic DNA was extracted from peripheral blood using either the Citogene® DNA isolation kit (Citomed, Portugal) manually or the QIAsymphony SP kit and apparatus. aCGH was performed using the following platforms Agilent 180 K (GPL15397); KaryoArray®v3.0 (Agilent 8x60k); Agilent Whole Genome 244 K (GPL10118); Affymetrix CytoScan HD (GPL1613) or CytoScan 750 K (GPL18637) (detailed description provided in Additional file 1).
Data analysis
CNVs detected were classified using criteria adapted from those previously described elsewhere [3, 52] as: pathogenic, likely pathogenic, CNVs of unknown clinical significance (VOUS) (detailed description in Part 2 of Additional file 1). For simplification of terminology throughout the text and in the tables, the term CNV is used for pathogenic and likely pathogenic variants, as well as VOUS. Polymorphic CNVs were not further considered in our analysis, except where specifically indicated (e.g. known risk loci, although relatively frequent, were considered pathogenic). All alteration are described in the tables as in the Decipher database (for example 12q24.21-q24). For CNV confirmation we performed qRT-PCR (7500-FAST Real Time PCR, Thermo Fisher Scientific, Waltham, MA, USA), using SDC4 and ZNF80 as reference genes (detailed description in Part 2 of Additional file 1; primers in Table S3). Total RNA was isolated from leukocytes using the QIAsymphony RNA Kit (QIAGEN GmbH, Germany), according to the manufacturer’s protocol. First-strand cDNA synthesized using SuperScript® III Reverse Transcriptase (RT) (Thermo Fisher Scientific, Waltham, MA, USA). Expression analysis was performed by quantitative real-time reverse transcription PCR (qRT-PCR) using Power SYBR Green® (Thermo Fisher Scientific, Waltham, MA, USA) (detailed description in Part 2 of Additional file 1; genes and primers listed in Table S4).
Additional file
Figure S1. Facial appearance of some patients carrying pathogenic variants. Figure S2. Clinical features of patients R14 and C19 and images of their CNVs. Table S1. Patients with altered aCGH results (i.e. with CNVs classified as non-polymorphic). Table S2. List of variants of unknown clinical significance (VOUS). Table S3. Primers used for quantitative PCR confirmation. Table S4. Primers used for expression studies. Table S5. OMIM entrance, haploinsufficiency score and constrain metrics for the selected genes in patient R16. Table S6. OMIM entrance, haploinsufficiency score and constrain metrics for the selected genes in patient C15. Table S7. OMIM entrance, haploinsufficiency score and constrain metrics for the selected genes in patient R20. Table S8. OMIM entrance, haploinsufficiency score and constrain metrics for the selected genes in patient C16. Table S9. OMIM entrance, haploinsufficiency score and constrain metrics for the selected genes in patient R21. Table S10. OMIM entrance, haploinsufficiency score and constrain metrics for the selected genes in patient C19. Table S11. OMIM entrance, haploinsufficiency score and constrain metrics for the selected genes in patients R22 and R23. Table S12. OMIM entrance, haploinsufficiency score and constrain metrics for the selected genes in patient C20. (DOC 11550 kb)
Acknowledgements
We would like to thank all the patients and their families for participation in this study and for allowing this publication. We would also like to acknowledge the DECIPHER Consortium, Database of Genomic Variants and OMIM since this study makes use of data generated and managed by these platforms.
Financial disclosure
FT, PR, PT and JPB authors are employed by company CGC Genetics. All other authors declare no financial competing interests.
Authors’ contributions
FL, FT, SS, SL and PR performed the molecular studies and analysed the molecular data. PE, JW and BY contributed to the molecular studies and to the analysis of molecular data. GS, MB, JS, FD, MR, JS, GO, MJS, TT, CM, CG, GB, AJ, FR, CM, SM, SL, EMC, MJC, AD, CN, CRM, DA, JD, SF, SF, SGS, SC, AS, MRL, JPB and AMF collected and analysed clinical data. FL, FT and PM drafted the paper. PM and PT obtained funding for this study. The study was performed under the direction of PM. All authors have agreed with and approved the final version.
Funding
This work has been funded by FEDER funds, through the Competitiveness Factors Operational Programme (COMPETE), and by National funds, through the Foundation for Science and Technology (FCT), under the scope of the projects: PIC/IC/83026/2007, PIC/IC/83013/2007 and POCI-01-0145-FEDER-007038. This work has also been funded by the project NORTE-01-0145-FEDER-000013, supported by the Northern Portugal Regional Operational Programme (NORTE 2020), under the Portugal 2020 Partnership Agreement, through the European Regional Development Fund (FEDER). FL was supported by Foundation for Science and Technology (FCT) through the fellowship SFRH/BD/90167/2012.
Availability of data and materials
All data generated or analysed during this study are included in this published article and in its supplementary information files.
Ethics approval and consent to participate
The enrollment of the patients and families was done by the referring doctor, clinical information was gathered in an anonymized database and written informed consent was obtained for all participants and/or their legal guardians for both study participation and publication of identifying information/images according to the Portuguese Data Protection Authority (CNPD). This study was approved by the ethics committee of Center for Medical Genetics Dr. Jacinto Magalhães, Porto Hospital Center and all research was performed in accordance with relevant guidelines/regulations.
Consent for publication
Informed consent was obtained for all participants and/or their legal guardians for publication of identifying information/images according to the Portuguese Data Protection Authority (CNPD).
Competing interests
FT, PR, PT and JPB authors are employed by company CGC Genetics. All other authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fátima Lopes and Fátima Torres contributed equally to this work.
References
- 1.Reichenberg A, Cederlöf M, McMillan A, Trzaskowski M, Kapra O, Fruchter E, et al. Discontinuity in the genetic and environmental causes of the intellectual disability spectrum. Proc Natl Acad Sci. 2015;113(4):1098–1103. doi: 10.1073/pnas.1508093112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coe BP, Girirajan S, Eichler EE. The genetic variability and commonality of neurodevelopmental disease. Am J Med Genet Part C Semin Med Genet. 2012;160 C(2):118–129. doi: 10.1002/ajmg.c.31327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86(5):749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mannik K, Magi R, Macé A, Cole B, Guyatt AL, Shihab HA, et al. Copy number variations and cognitive phenotypes in unselected populations. JAMA. 2015;313(20):2044–2054. doi: 10.1001/jama.2015.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopes F, Torres F, Soares G, van Karnebeek CD, Martins C, Antunes D, et al. The role of AKT3 copy number changes in brain abnormalities and neurodevelopmental disorders: four new cases and literature review. Front Genet. 2019;10(February):1–8. doi: 10.3389/fgene.2019.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopes F, Torres F, Lynch SA, Jorge A, Sousa S, Silva J, et al. The contribution of 7q33 copy number variations for intellectual disability. Neurogenetics. 2018;19(1):27–40. doi: 10.1007/s10048-017-0533-5. [DOI] [PubMed] [Google Scholar]
- 7.Lopes F, Soares G, Gonçalves-Rocha M, Pinto-Basto J, Maciel P. Whole gene deletion of EBF3 supporting Haploinsufficiency of this gene as a mechanism of neurodevelopmental disease. Front Genet. 2017;8(143):1–7. [DOI] [PMC free article] [PubMed]
- 8.Firth HV et al. Am J Hum Genet. 2009;84(4):524–33. [DOI] [PMC free article] [PubMed]
- 9.Dheedene A, Maes M, Vergult S, Menten B. A de novo POU3F3 deletion in a boy with intellectual disability and dysmorphic features. Mol Syndromol. 2014;5:32–35. doi: 10.1159/000356060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominguez MH, Ayoub AE, Rakic P. POU-III transcription factors (Brn1, Brn2, and Oct6) influence neurogenesis, molecular identity, and migratory destination of upper-layer cells of the cerebral cortex. Cereb Cortex. 2013;23:2632–2643. doi: 10.1093/cercor/bhs252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Potocki L, Bi W, Treadwell-deering D, Carvalho CMB, Eifert A, Friedman EM, et al. Characterization of Potocki-Lupski syndrome critical interval that can convey an autism phenotype. Am J Hum Genet. 2007;80(April):633–649. doi: 10.1086/512864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dudkiewicz M, Lenart A, Pawłowski K. A novel predicted calcium-regulated kinase family implicated in neurological disorders. PLoS One. 2013;8(6):1–10. doi: 10.1371/journal.pone.0066427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science (80- ) 2008;321(5886):218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thiselton DL, McDowall J, Brandau O, Ramser J, d’Esposito F, Bhattacharya SS, et al. An integrated, functionally annotated gene map of the DXS8026-ELK1 interval on human Xp11.3-Xp11.23: potential hotspot for neurogenetic disorders. Genomics. 2002;79(4):560–572. doi: 10.1006/geno.2002.6733. [DOI] [PubMed] [Google Scholar]
- 15.Wang KS, Liu XF, Aragam N. A genome-wide meta-analysis identifies novel loci associated with schizophrenia and bipolar disorder. Schizophr Res. 2010;124(1–3):192–199. doi: 10.1016/j.schres.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 16.Silfhout ATV, Nakagawa T, Bahi-Buisson N, Haas SA, Hu H, Bienek M, et al. Variants in CUL4B are associated with cerebral malformations. Hum Mutat. 2014;36(Umr 8104):106–117. doi: 10.1002/humu.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy A, Gonzalez-Gomez M, Pierani A, Meyer G, Tole S. Lhx2 regulates the development of the forebrain hem system. Cereb Cortex. 2014;24:1361–1372. doi: 10.1093/cercor/bhs421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flandin P, Zhao Y, Vogt D, Jeong J, Long J, Potter G, et al. Lhx6 and Lhx8 coordinately induce neuronal expression of Shh that controls the generation of interneuron progenitors. Neuron. 2011;70(5):939–950. doi: 10.1016/j.neuron.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JH, Youn BU, Kim K, Moon JB, Lee J, Nam K-I, et al. Lhx2 regulates bone remodeling in mice by modulating RANKL signaling in osteoclasts. Cell Death Differ. 2014;21:1613–1621. doi: 10.1038/cdd.2014.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z, Gutierrez D, Li X, Bidlack F, Cao H, Wang J, et al. The LIM homeodomain transcription factor LHX6: a transcriptional repressor that interacts with pituitary homeobox 2 (PITX2) to regulate odontogenesis. J Biol Chem. 2013;288(4):2485–2500. doi: 10.1074/jbc.M112.402933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarpey PS, Raymond FL, Meara SO, Edkins S, Teague J, Butler A, et al. Mutations in CUL4B, which encodes a ubiquitin E3 ligase subunit, cause an X-linked mental retardation syndrome associated with aggressive outbursts, seizures, relative macrocephaly, central obesity, hypogonadism, pes Cavus, and tremor. Am J Hum Genet. 2007;80:345–352. doi: 10.1086/511134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isidor B, Pichon O, Baron S, David A, Le Caignec C. Deletion of the CUL4B gene in a boy with mental retardation , minor facial anomalies , short stature. Am J Med Genet Part A. 2009;152A:175–180. doi: 10.1002/ajmg.a.33152. [DOI] [PubMed] [Google Scholar]
- 23.Wang HL, Chang NC, Weng YH, Yeh TH. XLID CUL4B mutants are defective in promoting TSC2 degradation and positively regulating mTOR signaling in neocortical neurons. Biochim Biophys Acta - Mol Basis Dis. 2013;1832(4):585–593. doi: 10.1016/j.bbadis.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Nakagawa T, Xiong Y. X-linked mental retardation gene CUL4B targets Ubiquitylation of H3K4 methyltransferase component WDR5 and regulates neuronal gene expression. Mol Cell. 2011;43(3):381–391. doi: 10.1016/j.molcel.2011.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin Z, Yu L, Geng J, Wang J, Jin X, Huang H. A novel 47.2 Mb duplication on chromosomal bands Xq21.1–25 associated with mental retardation. Gene. 2015;567(1):98–102. doi: 10.1016/j.gene.2015.04.083. [DOI] [PubMed] [Google Scholar]
- 26.Murali A, Rajalingam K. Small rho GTPases in the control of cell shape and mobility. Cell Mol Life Sci. 2014;71(9):1703–1721. doi: 10.1007/s00018-013-1519-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kutsche K, Yntema H, Brandt A, Jantke I, Nothwang HG, Orth U, et al. Mutations in ARHGEF6, encoding a guanine nucleotide exchange factor for rho GTPases, in patients with X-linked mental retardation. Nat Genet. 2000;26:247–250. doi: 10.1038/80002. [DOI] [PubMed] [Google Scholar]
- 28.Madrigal I, Fernández-Burriel M, Rodriguez-Revenga L, Cabrera JC, Martí M, Mur A, et al. Xq26.2-q26.3 microduplication in two brothers with intellectual disabilities: clinical and molecular characterization. J Hum Genet. 2010;55(12):822–826. doi: 10.1038/jhg.2010.119. [DOI] [PubMed] [Google Scholar]
- 29.Rosenberg C, Knijnenburg J, Bakker E, Vianna-Morgante AM, Sloos W, Otto PA, et al. Array-CGH detection of micro rearrangements in mentally retarded individuals: clinical significance of imbalances present both in affected children and normal parents. J Med Genet. 2006;43(2):180–186. doi: 10.1136/jmg.2005.032268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu X, Shaw CA, Patel A, Li J, Cooper ML, Wells WR, et al. Clinical implementation of chromosomal microarray analysis: summary of 2513 postnatal cases. PLoS One. 2007;2(3):e327. doi: 10.1371/journal.pone.0000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sagoo GS, Butterworth AS, Sanderson S, Shaw-Smith C, Higgins JPT, Burton H. Array CGH in patients with learning disability (mental retardation) and congenital anomalies: updated systematic review and meta-analysis of 19 studies and 13,926 subjects. Genet Med Off J Am Coll Med Genet. 2009;11(3):139–146. doi: 10.1097/GIM.0b013e318194ee8f. [DOI] [PubMed] [Google Scholar]
- 32.Verdin H, De Baere E. FOXL2 impairment in human disease. Horm Res Paediatr. 2012;77:2–11. doi: 10.1159/000335236. [DOI] [PubMed] [Google Scholar]
- 33.Lalli MA, Jang J, Park JHC, Wang Y, Guzman E, Zhou H, et al. Haploinsufficiency of BAZ1B contributes to Williams syndrome through transcriptional dysregulation of neurodevelopmental pathways. Hum Mol Genet. 2016;25(7):1294–1306. doi: 10.1093/hmg/ddw010. [DOI] [PubMed] [Google Scholar]
- 34.Lasalle JM. Autism genes keep turning up chromatin. OA Autism. 2013;1(2):14. doi: 10.13172/2052-7810-1-2-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patsialou A, Wilsker D, Moran E. DNA-binding properties of ARID family proteins. Nucleic Acids Res. 2005;33(1):66–80. doi: 10.1093/nar/gki145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clague MJ, Coulson JM, Urbé S. Cellular functions of the DUBs. J Cell Sci. 2012;1251 Clagu(Pt 2):277–286. doi: 10.1242/jcs.090985. [DOI] [PubMed] [Google Scholar]
- 37.Hao Z, Zhang H, Cowell J. Ubiquitin-conjugating enzyme UBE2C: molecular biology, role in tumorigenesis, and potential as a biomarker. Tumor Biol. 2012;33(3):723–730. doi: 10.1007/s13277-011-0291-1. [DOI] [PubMed] [Google Scholar]
- 38.Stouffer MA, Golden JA, Francis F. Neuronal migration disorders: focus on the cytoskeleton and epilepsy. Neurobiol Dis. 2016;92(Pt A):18–45. doi: 10.1016/j.nbd.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Curatolo P, Bombardieri R, Jozwiak S. Tuberous sclerosis. Lancet. 2008;372(9639):657–668. doi: 10.1016/S0140-6736(08)61279-9. [DOI] [PubMed] [Google Scholar]
- 40.Nodé-Langlois R, Muller D, Boda B. Sequential implication of the mental retardation proteins ARHGEF6 and PAK3 in spine morphogenesis. J Cell Sci. 2006;119:4986–4993. doi: 10.1242/jcs.03273. [DOI] [PubMed] [Google Scholar]
- 41.Hopp K, Heyer CM, Hommerding CJ, Henke SA, Sundsbak JL, Patel S, et al. B9D1 is revealed as a novel Meckel syndrome ( MKS ) gene by targeted exon-enriched next-generation sequencing and deletion analysis. Hum Mol Genet. 2011;20(13):2524–2534. doi: 10.1093/hmg/ddr151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yilmaz S, Gokben S, Serdaroglu G, Eraslan C, Mancini GM, Tekin H, et al. The expanding phenotypic spectrum of ARFGEF2 gene mutation: cardiomyopathy and movement disorder. Brain and Development. 2016;38(1):124–127. doi: 10.1016/j.braindev.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 43.Banne E, Atawneh O, Henneke M, Brockmann K, Gärtner J, Elpeleg O, et al. West syndrome, microcephaly, grey matter heterotopia and hypoplasia of corpus callosum due to a novel ARFGEF2 mutation. J Med Genet. 2013;50(11):772–775. doi: 10.1136/jmedgenet-2013-101752. [DOI] [PubMed] [Google Scholar]
- 44.Wen J, Lopes F, Soares G, Farrell SA, Nelson C, Qiao Y, et al. Phenotypic and functional consequences of haploinsufficiency of genes from exocyst and retinoic acid pathway due to a recurrent microdeletion of 2p13.2. Orphanet J Rare Dis. 2013;8:100. doi: 10.1186/1750-1172-8-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshida T, Yasumura M, Uemura T, Lee S-JSJ, Ra M, Taguchi R, et al. IL-1 receptor accessory protein-like 1 associated with mental retardation and autism mediates synapse formation by trans-synaptic interaction with protein tyrosine phosphatase δ. J Neurosci. 2011;31(38):13485–13499. doi: 10.1523/JNEUROSCI.2136-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu H, Wang X, Liu S, Wu Y, Zhao T, Chen X, et al. Sema4C participates in myogenic differentiation in vivo and in vitro through the p38 MAPK pathway. Eur J Cell Biol. 2007;86(6):331–344. doi: 10.1016/j.ejcb.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 47.Quick MW. The role of SNARE proteins in trafficking and function of neurotransmitter transporters. Handb Exp Pharmacol. 2006;175:181–196. doi: 10.1007/3-540-29784-7_9. [DOI] [PubMed] [Google Scholar]
- 48.van Bokhoven H. Genetic and epigenetic networks in intellectual disabilities. Annu Rev Genet. 2011;45(1):81–104. doi: 10.1146/annurev-genet-110410-132512. [DOI] [PubMed] [Google Scholar]
- 49.Rippey C, Walsh T, Gulsuner S, Brodsky M, Nord AS, Gasperini M, et al. Formation of chimeric genes by copy-number variation as a mutational mechanism in schizophrenia. Am J Hum Genet. 2013;93(4):697–710. doi: 10.1016/j.ajhg.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spielmann M, Lupiáñez DG, Mundlos S. Structural variation in the 3D genome. Nat Rev Genet. 2018;19(7):453–467. doi: 10.1038/s41576-018-0007-0. [DOI] [PubMed] [Google Scholar]
- 51.van Ravenswaaij-Arts CMA, Kleefstra T. Emerging microdeletion and microduplication syndromes; the counseling paradigm. Eur J Med Genet. 2009;52(2–3):75–76. doi: 10.1016/j.ejmg.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 52.Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med. 2011;13(7):680–685. doi: 10.1097/GIM.0b013e3182217a3a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Facial appearance of some patients carrying pathogenic variants. Figure S2. Clinical features of patients R14 and C19 and images of their CNVs. Table S1. Patients with altered aCGH results (i.e. with CNVs classified as non-polymorphic). Table S2. List of variants of unknown clinical significance (VOUS). Table S3. Primers used for quantitative PCR confirmation. Table S4. Primers used for expression studies. Table S5. OMIM entrance, haploinsufficiency score and constrain metrics for the selected genes in patient R16. Table S6. OMIM entrance, haploinsufficiency score and constrain metrics for the selected genes in patient C15. Table S7. OMIM entrance, haploinsufficiency score and constrain metrics for the selected genes in patient R20. Table S8. OMIM entrance, haploinsufficiency score and constrain metrics for the selected genes in patient C16. Table S9. OMIM entrance, haploinsufficiency score and constrain metrics for the selected genes in patient R21. Table S10. OMIM entrance, haploinsufficiency score and constrain metrics for the selected genes in patient C19. Table S11. OMIM entrance, haploinsufficiency score and constrain metrics for the selected genes in patients R22 and R23. Table S12. OMIM entrance, haploinsufficiency score and constrain metrics for the selected genes in patient C20. (DOC 11550 kb)
Data Availability Statement
All data generated or analysed during this study are included in this published article and in its supplementary information files.