Abstract
Tlymphocyte development and differentiation is a multi-step process that begins in the thymus and completed in the periphery. Sequential development of thymocytes is dependent on T cell receptor (TCR) signaling and an array of transcription factors. In this study we show that special 00AT-rich binding protein 1 (SATB1), a T lineage-enriched chromatin organizer and regulator, is induced in response to TCR signaling during early thymocyte development. SATB1 expression profile coincides with T lineage commitment and upregulation of SATB1 correlates with positive selection of thymocytes. CD4 thymocytes exhibit a characteristic bimodal expression pattern that corresponds to immature and mature CD4 thymocytes. We also demonstrate that GATA3, the key transcriptional regulator of αβ T cells positively regulates SATB1 expression in thymocytes suggesting an important role for SATB1 during T cell development.
Keywords: T cell receptor, SATB1, GATA-3, Signaling
1. Introduction
The T lineage developmental program begins with the migration of the hematopoietic progenitors into the thymus. Thymic environment provides the necessary signals for the commitment of early progenitors towards T lineage, which then undergo an orderly process of differentiation (Starr et al., 2003). Maturing thymocytes acquire CD4 and/or CD8 coreceptors in a developmentally relevant manner. Early thymic precursors that lack either of the coreceptors are known as CD4– CD8– double negative (DN) thymocytes. This population of cells is further subdivided into four stages based on the expression of CD44 and CD25: CD44 + CD25– (DN1), CD44+ CD25+ (DN2), CD44– CD25+ (DN3) and CD44– CD25– (DN4), which bear a precursor product relationship (Lind et al., 2001 ). DN3 thymocytes initiate the rearrangement of T cell receptor (TCR)-P chain, which is expressed together with a non-rearranging pre-T-a chain and receive pre-TCR signals to progress to CD4+ CD8 +double positive (DP) stage. At the DP stage TCR-α chain locus undergoes rearrangement and the mature receptor is expressed on the cell surface. TCR- α β+ DP thymocytes finally mature into CD4+ CD8– or CD4– CD8+ single positive (SP) thymocytes (Germain, 2002; Singer et al., 2008).
The fate of developing thymocytes is determined at multiple checkpoints. The first checkpoint is the β-selection checkpoint experienced by DN3 thymocytes that directs development to the DP stage. This checkpoint ensures the maturation of sufficient number of TCR-β expressing thymocytes that are available to rearrange and express the TCR- α chain allowing sufficient numbers of TCR- α β expressing thymocytes to audition for selection (Singer et al., 2008). At the second checkpoint, DP thymocytes undergo positive and negative selection in response to TCR interacting with peptide-bound MHC molecules on the antigen presenting cells (Zerrahn et al., 1997). TCR signaling is critical for appropriate positive and negative selection of thymocytes to ensure appropriate tolerance and immune function both through proper positive and negative selection processes as well as the development of natural regulatory T cells (nTregs) (Starr et al., 2003). While the sequence of events during T cell development is well defined, the molecular mechanisms including the role of transcription factors are just beginning to emerge (Rothenberg et al., 2008). The role of Special AT-rich binding protein 1 (SATB1) has been studied during T cell activation (Pavan Kumar et al., 2006) and differentiation (Notani et al., 2010). SATB1 is a global regulator and chromatin organizer (Cai et al., 2003; Galande et al., 2007). In absence of SATB1 function thymocyte development is blocked at the DP stage. SATB1 null DP thymocytes ectopically express IL-2Rα and IL-7R and undergo activation induced cell death (Alvarez et al., 2000). However, the molecular mechanisms governing the role of SATB1 in T cell development remain to be determined.
As a first step towards unraveling the role of SATB1 in T cell development and function, we monitored the expression profile of SATB1 in various thymocyte subsets and observed that SATB1 is differentially expressed. Our results further indicate that induction of SATB1 is dependent on TCR signaling and might in turn regulate the development of CD4 and CD8 thymocytes. Finally, we show that the transcription factor GATA-3 directly regulates SATB1 expression in developing thymocytes by binding to its upstream regulatory region. Collectively, this study provides mechanistic insights into the regulation of SATB1 during early T cell development.
2. Materials and methods
2.1. Mice
Age matched C57BL/6 control mice were used in all experiments. All the mice were bred and maintained according to the regulations and guidelines of National Institute of Aging, Baltimore, USA and National Centre for Cell Science (NCCS), Pune, India. The animals and procedures used in the experiments were in compliance with guidelines from the regulatory bodies of both the institutes.
2.2. Antibodies
The following fluorescent conjugated antibodies were obtained from either BD biosciences or eBioscience – CD4 (GK1.5), CD8 (53–6.7), CD24, CD69 (H1.2F3), Qa-2, FoxP3, TCR-β (H57–597), CD44 (1M7), CD25, anti-lL-4 and anti- IFN-γ. Anti-SATB1 antibody used for ChlP and immunostaining was raised in rabbit and purified using immunoaffinity chromatography using standard procedures. F1TC conjugated anti-Rabbit IgG was obtained from Boehringer Ingelheim. Anti-tubulin (T6557) and anti-β-actin (A2228) antibodies were procured from Sigma.
2.3. Cell culture
CD4+ T cells were isolated from spleen and lymph nodes by negative selection using CD4 isoaltion kit (BD Biosciences). The cells were cultured with anti-CD3 (1 μg/ml) and anti-CD28 (1 μg/ml). CD4+ T cells were treated with LY294002 (5 μM) (Calbiochem). Briefly, the cells were treated with LY294002 at indicated concentrations or vehicle control DMSO for 6 h. At the end of the time point cells were harvested and analyzed by immunoblotting.
2.4. RNA isolation and cDNA synthesis
Total RNA was isolated using RNAeasy isolation kit (Qiagen). Then total RNA (500 ng) was reverse transcribed using Dynamo cDNA synthesis Kit (Finnzymes) and oligo dT primer. Quantitative real-time PCRs were performed using SYBR green PCR master mix (Applied Biosystems), with annealing and extension of primers at 60°C. Fold changes were calculated using the formula; Fold change = 2–(dd Ct).
2.5. Immunoblotting
Cell lysates were prepared using R1PA lysis buffer (Tris pH 7.4, NP40 1%, Na deoxycholate 0.25%, NaCl 150mM, EDTA 1 mM) and immunoblotting was performed as described previously (Pavan Kumar et al., 2006).
2.6. Immunostaining and FACS analysis
The cells were fixed using 2% paraformaldehyde (Sigma-Aldrich) and permeabilized using 0.1% Triton-X-100 (Sigma) followed by staining with anti-SATB1 antibody. Secondary antibody used was conjugated with Alexafluor 594. DNA counter staining was performed using DAP1. Cells were visualized under confocal microscope (Carl Zeiss). Image analysis was performed using Zen 2011 software (Carl Zeiss).
Mouse thymocytes were acquired on FACS Cantoll (Becton Dickinson) and analysis was performed using Flowjo software (Tree star). Dead cells were excluded using forward and side scatter. Staining of intranuclear antigens such as SATB1 and FoxP3 was performed using Foxp3 nuclear staining kit as described by the manufacturer (eBiosciences).
2.7. Chromatin immunoprecipitation (ChIP)
ChlP was performed as described previously (Notani et al., 2010). Briefly, mouse thymocytes or CD4+ T cells were crosslinked by 1% (v/v) formaldehyde at room temperature for 10 min and neutralized with 125 mM glycine (final) followed by washing with PBS pH 7.4. Cells were sheared using Bioruptor (Diagenode) to obtain 200–500 base pair fragments. Chromatin was incubated with specific antibodies overnight at 4 °C and respective lgG types were used as isotype controls. Protein A/G bead cocktail was then added to pulldown the antibody-bound chromatin and was subjected to elution using sodium biocarbonate buffer containing SDS and DTT (Sigma-Aldrich). Eluted chromatin was de-crosslinked and proteins were removed by digesting with proteinase K. Purified immuno-precipitated chromatin was subjected to PCR amplification using specific primers. lnput chromatin was used as control.
2.8. ChlP-seq analysis
ChlP-seq analysis was performed using the data available from GEO database (GSE20898). The ChlPseq reads were aligned to the mm8 genome and peak calling on binding site was performed using MACS (Zhang et al., 2008).
3. Results
3.1. Differential expression of SATBl in subsets of mouse thymocytes
To monitor if SATB1 is differentially expressed in various subsets of thymocytes, we sorted mouse thymocytes based on the surface expression of CD4 and CD8 coreceptors into four distinct populations – DN, DP, CD4 SP and CD8 SP and analyzed for SATB1 expression by quantitative PCR. We observed that SATB1 is minimally expressed in DN population and highly expressed in DP populations; CD4 and CD8 thymocytes have intermediate expression (Fig. 1A). Further, we monitored the expression of SATB1 at protein level by flow cytometric analysis. Mouse thymocytes were stained using lineage specific antibodies and the lineage negative subset (Lin –ve) (Fig. S1A) was analyzed for expression of CD44 and CD25 antigens to distinguish them into DN1 to DN4 stages (Fig. S1B). We then monitored the expression of SATB1 in individual DN subsets and observed that SATB1 is minimally expressed in DN1 to DN3 subsets and induced in DN4 population (Fig. 1B). These data suggested that SATB1 expression might be induced in response pre-TCR signals and facilitates β-selection checkpoint.
Fig. 1.
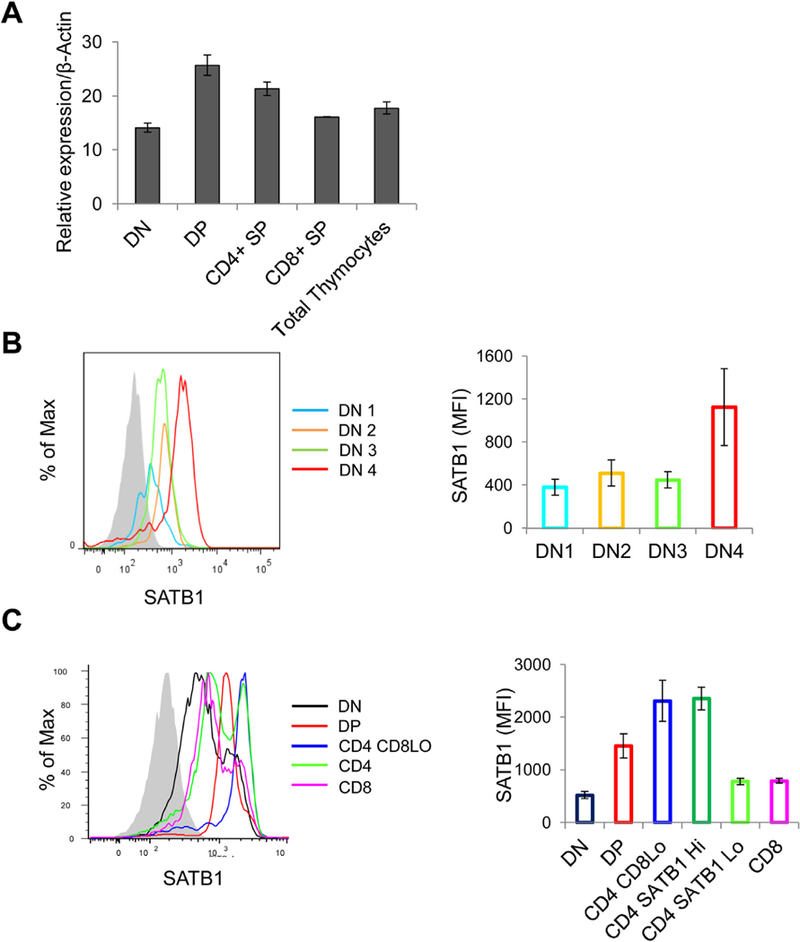
SATB1 expression is induced during thymocyte development. (A) Bar graph representing quantitative RT-PCR analysis of SATB1 expression in mouse thymocytes sorted based on anti-CD4 and anti-CD8 staining. (B) Mouse thymocytes were stained with lineage (Lin) specific factors along with CD44, CD25 and SATB1. The Lin –ve population is analyzed for CD44 and CD25 to differentiate into DN1-DN4 population and each subset is analyzed for SATB1 as represented in the histogram and bar graph. (C) SATB1 expression levels in various thymocyte subsets based on CD4 and CD8 co-receptors were quantified using flow cytometry analysis and corresponding histogram and bar graphs are plotted. Error bars represent standard deviation from three independent experiments.
Next, we monitored the expression of SATB1 in DP, CD4 CD8Lo, CD4 SP and CD8 SP thymcoyte populations and observed that expression of SATB1 in DP thymocytes was similar to the expression profile seen in DN4 thymocytes (Fig. 1B and C). However, CD4 + CD8Lo intermediate thymocytes exhibited relatively higher expression of SATB1 as compared to DP subset. lnterestingly, CD4 SP thymocytes exhibited bimodal distribution of SATB1 expression and we refer to the populations as SATB1Hi and SATB1L◦CD4 thymocytes (Fig. 1C). The CD8 thymocytes exhibited SATB1 expression profile similar to that of SATB1Lo subset of CD4 thymocytes. We then analyzed the expression pattern of SATB1 in thymocyte subsets based on TCRβ positive population and observed that the expression profile of SATB1 in TCRβHi thymocytes was similar to that of total thymocytes except in DP population (Fig. 2A–C). TCRβHi DP thymocytes exhibited higher expression in comparison to total DP thymocytes and similar to that of CD4 + CD8Lo intermediate thymocytes (Fig. S1C). These findings suggest that SATB1 is induced in response to TCR signaling in thymocytes.
Fig. 2.
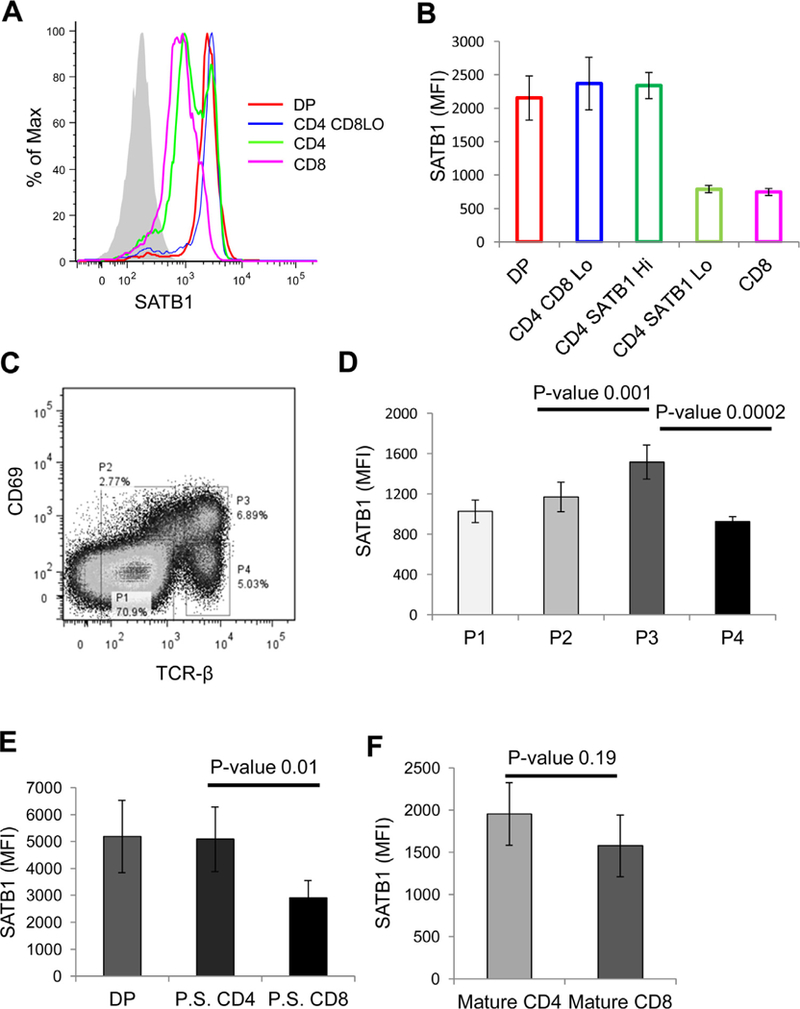
SATB1 expression is developmentally regulated during thymocyte development. (A) Mouse thymocytes were analyzed for SATB1 expression in TCR-βHI thymocyte subsets based on CD4 and CD8 as represented in the histogram, (B) represents the MFI values of SATB1 expression in thymocyte subsets as shown in (A). (C) Mouse thymocytes were stained with CD69 and TCR-β to distinguish them into developmental stages based on positive selection. Population 1 represents thymocytes that have not undergone positive selection. Population 2 represents thymocytes undergoing positive selection. Population 3 refers to thymocytes that have undergone positive selection and population 4 thymocytes represents mature thymocytes. (D) Bar graph represents SATB1 expression in each population represented in (C) and error bars represent standard deviation calculated from n = 5. (E) SATB1 expression in DP, CD4 and CD8 subpopulations of thymocytes that have undergone positive selection (population 3). (F) Bar graph denotes SATB1 expression in CD4 and CD8 thymocytes of mature subset (population 4). P-values were calculated using T-test (n = 5).
3.2. TCR signaling regulates SATB1 expression during thymocyte development
To test whether SATB1 expression is regulated in TCR-dependent manner in developing thymocytes, we determined its expression in thymocyte populations that are distinguished based on TCRβ and CD69, an early activation marker (Bendelac et al., 1992) (Fig. 2C). Thymocytes that exhibit intermediate expression of TCRβ and are negative for CD69 expression represents thymocytes that have not received a TCR signal (population 1), those that have intermediate expression of TCR and express CD69 represent the population undergoing thymic selection (population 2). Thymocytes that exhibit higher expression of TCRβ and downregulated CD69 expression on cell surface have successfully undergone positive selection (population 3). Population 4 corresponds to those cells that exhibit high TCRβ expression and downregulated CD69 expression and are the mature thymocytes (Fig. 2C). We observed that SATB1 is upregulated in populations 2 and 3 that have received TCR signals and express CD69, an early activation marker. Mature thymocytes that have diminished CD69 expression exhibit down-regulation of SATB1 (Fig. 2D). Expression level of SATB1 in positively selected thymocytes (population 3) was similar in DP and CD4 SP population of thymocytes and lower in CD8 SP thymocytes (Fig. 2D) indicating that SATB1 expression is regulated by TCR signaling. Finally, we observed that SATB1 was downregulated in mature CD4 SP and CD8SP subsets and the expression levels were comparable (Fig. 2F). Together, these data suggest that SATB1 expression is directly regulated by TCR signaling.
3.3. SATB1 is downregulated in mature CD4+ SP thymocytes
To better understand the significance of bimodal expression of SATB1 we compared the expression of SATB1 in CD4 SP mouse thymocytes with that of mature peripheral CD4+ T cells isolated from spleen and lymph nodes. We observed that peripheral CD4 + T cells do not exhibit dual populations as seen in thymocytes and the expression pattern in peripheral cells coincides with the SATB1Lo population of CD4+ thymocytes (Fig. 3 B). Further, we determined the expression pattern of SATB1 in CD69 positive and negative fractions (Fig. 3C) and observed that SATB1 expression is higher in CD69 +ve CD4 thymocytes and low in CD69–ve thymocytes (Fig. 3D and E).
Fig. 3.
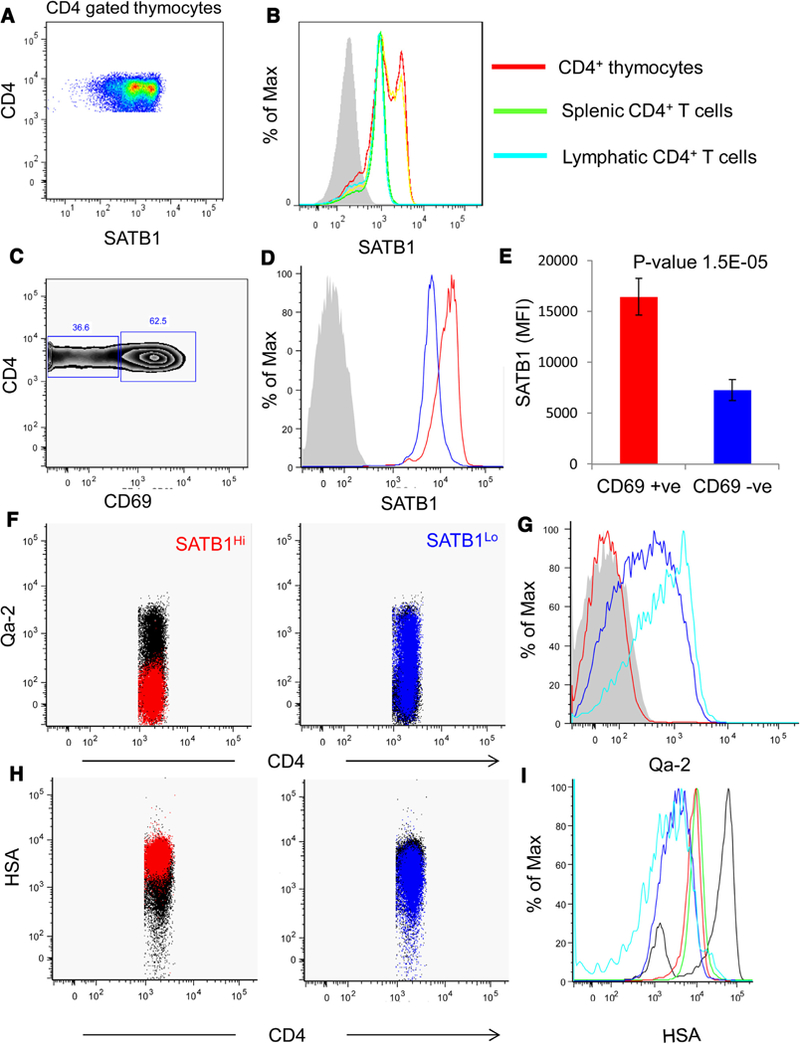
CD4+ thymocytes exhibit distinct bimodal pattern of SATB1 expression. (A) Flow cytometry data for gated CD4 + SP thymocytes analyzed for SATB1 expression and observed bimodal distribution (SATB1Hi and SATB1Lo). (B) Histogram comparing SATB1 expression in CD4 gated thymocytes of total (red), TCR-βHi CD4 dgated (yellow), splenic (green) and lymph node (cyan) CD4+ T cells. (C) Flow cytometryplot representing CD69 expression in CD4 gated thymocytes. (D) Histogram represents SATB1 expression inCD69 +ve (red) and CD69–ve (blue)thymocytes and corresponding MFI values of SATB1 are plotted in(F). Gated CD4 thymocytes were for Qa-2 expression and SATB1Hi (red) and SATB1Lo (blue) CD4 thymocytes were overlaid on the plots. (G) Histogram depicting the expression of Qa-2 in Satb1Hi (red), SATB1Lo (blue) and CD8 population (cyan). (H) Expression profile for HSA in CD4 thymocytes where SATB1Hi (red) and SATB1Lo(blue) are overlaid on the plot. (I) Histogram representing HSA expression in all major subsets –DN (black), DP (green), SATB1Hi (red), SATB1Lo (blue) and CD8 (cyan).
It has been shown that CD4 SP thymocytes are a heterogeneous population and remain functionally immature as demonstrated by high levels of CD24 expression (Scollay et al., 1984). CD4 SP thymocytes can also be distinguished from immature CD4 thymocytes by the expression of the non-classical class I antigen, Qa-2 (Vernachio et al., 1989). Immature HSAHi Qa-2–ve CD4 thymocytes are incompetent for TCR activation in contrast to mature HSALo Qa-2+ve thymocytes (Nikolic-Zugic and Bevan, 1990). Since maturation of SP thymocytes is associated with downregulation of HSA (Crispe and Bevan, 1987) and upregulation of Qa-2 expression (Ramsdell et al., 1991), we evaluated the expression of these surface markers on the SATB1Hi and SATB1L◦CD4 thymocyte populations. We observed that the expression of HSA in SATB1Hi CD4 thymocytes was similar to that of DP thymocytes. In contrast, the surface expression of HSA was downregulated in SATB1L◦CD4 thymocytes indicating the developmental progression of CD4 thymocytes from SATB1Hi to SATB1Lo compartment (Fig. 3H). We observed that SATB1Hi population was negative for the expression of Qa-2 while a significant fraction (>80%) of SATB1L◦CD4 thymocytes exhibited Qa-2 surface expression (Fig. 3F). CD5 expression on thymocytes is regulated by TCR signals and avidity of TCR-antigen interactions, such that intermediate levels of CD5 are observed in DP thymocytes and high levels of CD5 are observed in CD4SP and CD8SP thymocytes proportional to TCR signaling (Azzam et al., 1998). We observed that CD5 expression is elevated in SATB1Hi CD4+ thymocyte population compared to mature SATB1L◦CD4+ thymocytes (Fig. S2). These results suggest that SATB1 is downregulated in mature CD4+ SP thymocytes. Further, TCR-induced expression of SATB1 might play a role in the positive selection and maturation of thymocytes. We observed that TCR-βHi thymocytes are either positive or negative for SATB1 expression (Fig. S3A) and also observed that Foxp3+ CD4+ thymocytes are negative for SATB1 expression (Fig. S3B). Interestingly, we found elevated Foxp3 expression in subpopulations of DP, CD4+ CD8Lo, CD4+ SP thymocytes that were negative for SATB1 expression (Fig. S3B), suggesting that SATB1 and Foxp3 are expressed mutually exclusively during thymocyte development.
3.4. TCR signaling positively regulates SATB1 in peripheral CD4 + T cells
Peripheral CD4+ T cells differentiate into various T helper lineages in response to TCR signals, cytokine milieu and other ligands in the microenvironment. We wished to determine the expression levels of SATB1 in response to TCR signals in vitro. Mouse CD4+ T cells were activated in presence of plate-bound anti-CD3 or anti-CD28 or together with anti-CD3 plus anti-CD28. Immunostaining of SATB1 in CD4+ T cells activated in presence of anti-CD3 or anti-CD3 plus anti-CD28 revealed the characteristic cage-like nuclear substructure as seen in thymocytes (Cai et al., 2003; Galande et al., 2007) (Fig. 4A). In contrast, cells activated in the presence of only anti-CD28 exhibited poor induction of SATB1 expression (Fig. 4A and B). In activated CD4+ T cells highly compacted chromatin is seen as pockets of densely DAPI stained heterochromatic regions in the nucleus. SATB1 is localized exclusively in the lightly stained euchromatic regions and is virtually absent in the heterochromatic regions. In naïve CD4+ T cells SATB1 expression is almost undetectable and the prominent heterochromatic blobs as seen in activated nuclei are also not observed indicating that the gross chromatin organization could be very different (Fig. 4A). Immunoblot analysis indicated that SATB1 expression was induced within 2 h post anti-CD3 or anti-CD3/CD28 treatment and was sustained until 24 h of activation (Fig. 4B). In contrast, activation with only anti-CD28 antibody did not induce the expression of SATB1 in CD4+ T cells (Fig. 4B). Notably, kinetics of SATB1 expression is steep in cells activated by TCR (anti-CD3) alone in comparison to cells activated with TCR and co-stimulatory signals (anti-CD3 and anti-CD28). Further, CD4+ T cells cultured under control conditions do not exhibit significant induction of SATB1 expression (Fig. 4B and C). To confirm whether SATB1 is downstream effector of TCR signaling we monitored if SATB1 levels are affected upon perturbation of TCR signaling. Inhibition of TCR signaling in CD4+ T cells using PI3K chemical inhibitor LY294002 led to reduction in SATB1 expression when cultured for 6 h in presence of anti-CD3 and anti-CD28 (Fig. 4D and E). SATB1 is upregulated in CD4+ T cells cultured under activation signals and vehicle control (Fig. 4D and E). Together, these results suggested that SATB1 expression is induced in peripheral CD4 + T cells in response to TCR signaling and inhibition of TCR signaling suppresses SATB1 expression.
Fig.4.
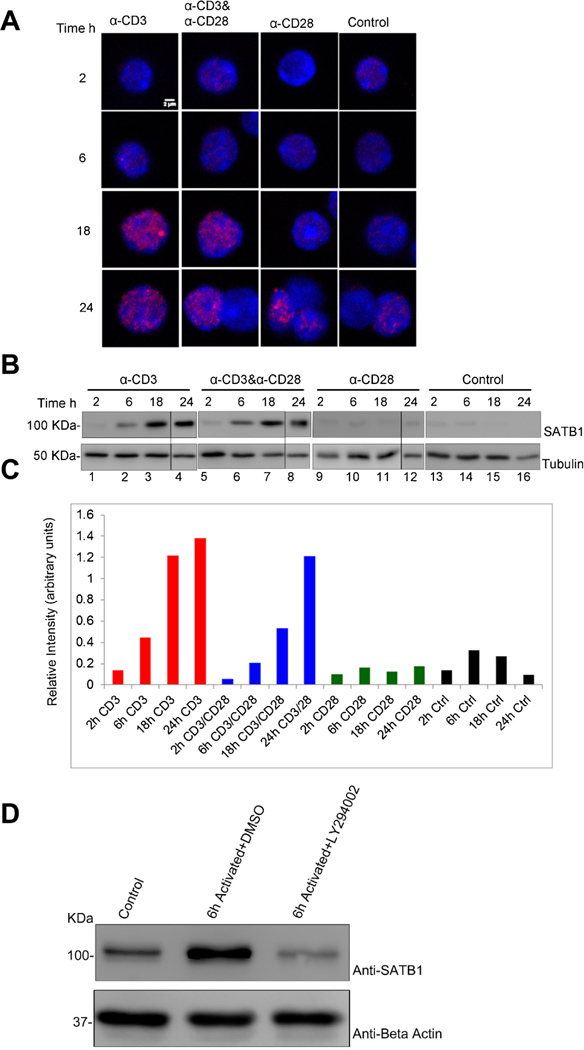
SATB1 is induced in peripheral CD4Tcells upon TCR signaling.(A) Immunofluorescence images of CD4T cells activated in presence of plate-boundanti-CD3,anti-CD28, anti-CD3/CD28 or control conditions (without antibody) for2h,6h,18h and 24h. Cells were stained with DAPI (blue) and SATB1 (red). (B) Immunoblot analysis for SATB1 expression in CD4+ T cells cultured under similar conditions as mentioned in (A). β-Actin blots served as loading control. (C) Bar graph represents densitometric analysis of immunoblots for SATB1 expression using Image-J software (NIH). (D) CD4+ T cells were treated with PI3K inhibitor (LY2940002) that affect downstream TCR signaling and analyzed for SATB1 expression by Western blot. (E) Bar graph represents densitometric analysis of immunoblots of three independent experiments including that from (D). Error bars represent standard deviation calculated from n = 3.
3.5. SATB1 expression correlates with Nur77 expression
Since SATB1 expression is regulated by TCR signaling, we next searched for markers to correlate these results with known molecules that are shown to be regulated by TCR signaling. Nur77 is one of the molecules that is upregulated early in response to TCR signaling (Liu et al., 1994). Furthermore, it has been shown that Nur77 expression levels are governed by TCR signal strength (Moran et al., 2011). Therefore we monitored the expression of Nur77 and correlated it with the expression of SATB1. Towards this, we compared the expression of SATB1 in total thymocytes to that of Nur77 positive thymocytes. We observed upregulated expression of SATB1 in Nur77+ve thymocytes compared to total thymocytes (Fig. 5A). Next, we compared the expression of SATB1 and Nur77 in individual thymocytes subsets. We observed that DP and CD4 thymocytes exhibit a difference in SATB1 expression in total and Nur77 +ve compartment, whereas in CD8 population we did not observe any significant difference in SATB1 expression between total CD8 and Nur77+ve compartment (Fig. 5B). This data suggests that SATB1 upregulation coincides with Nur77 expression during positive selection of DP thymocytes. We have shown that CD4 thymcoyes have bimodal expression of SATB1 (Fig. 3A and B). We therefore analyzed the expression of Nur77 in SATB1 Hi and Lo populations of CD4 thymocytes. Interestingly, we observed that Nur77 expression is higher in SATB1Hi poluation compared to SATB1Lo population (Fig. 5C). Taken together, this data suggests that similar to Nur77, SATB1 is also upregulated in response to TCR signaling.
Fig. 5.
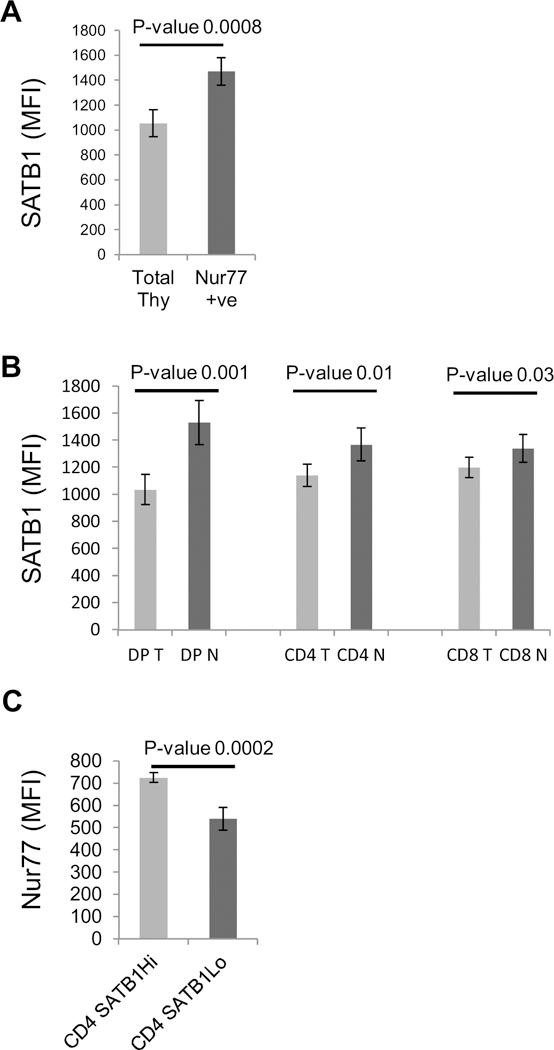
SATB1 expression correlates with that of Nur77 in developing thymocytes. A. Mouse thymocytes were stained with Nur77 and SATB1 and analysed for expression of SATB1 in total and Nur77+ve thymocytes. B. Expression pattern of SATB1 individual DP, CD4 and CD8 subsets of total (T) and Nur77+ve gated population (N). C. Nur77 expression in SATB1Hi and SATB1Lo populations of CD4 thymocytes. Error bars represent standard deviation and p values calculated using student’s T Test (n = 5).
3.6. Gata-3 directly regulates SATB1 expression
Thymocyte development from early precursors to mature cells and further differentiation into effector cells depends on array of transcription factors that are expressed temporally at distinct stages of development. Transcription factor GATA3 is indispensable for thymocyte development and overexpression of GATA3 in thymic precursors has been shown to initiate T lineage program (Rothenberg, 2011; Ho et al., 2009). Therefore, we used RNA-seq data available from the Gene Expression Omnibus (GEO) database using GATA3 KO mice (Wei et al., 2011) to investigate whether SATB1 expression is regulated by this core transcription factor during thymocyte development. GATA3 is known to play a critical role during differentiation of DN to CD3Lo, CD3Hi to CD4 and CD4 to Th2 phenotypes (Wei et al., 2011 ). Therefore, we tested whether GATA3 exerts a direct effect on SATB1 expression using the data from Wei et al. (Wei et al., 2011 ) (GEO database GSE20898). We compared the expression of SATB1 in control and GATA3 null mice thymocytes at CD3Hi stage of thymocytes (Fig. 6A) and observed that SATB1 expression is downregulated in GATA3 null mice in CD3Hi DP thymocytes. We further monitored if GATA3 directly regulates SATB1 expression by ChIP analysis and observed that GATA3 binds to Satb1 promoter in mouse thymocytes (Fig. 6B). Analysis of the ChIP-seq data sets for GATA3 available in the public database (GEO20898) using the peak identification software ‘MACS’ (Zhang et al., 2008; Wei et al., 2011) corroborated the findings from our ChIP analysis (Fig. 5C). The comparative occupancy analysis revealed that Satb1 promoter is bound by GATA3 in DN, DP and CD4 stages of thymocyte development (Fig. 6C). CD8 thymocytes from GATA3 KO mice served as negative control (Fig. 6C).
Fig. 6.
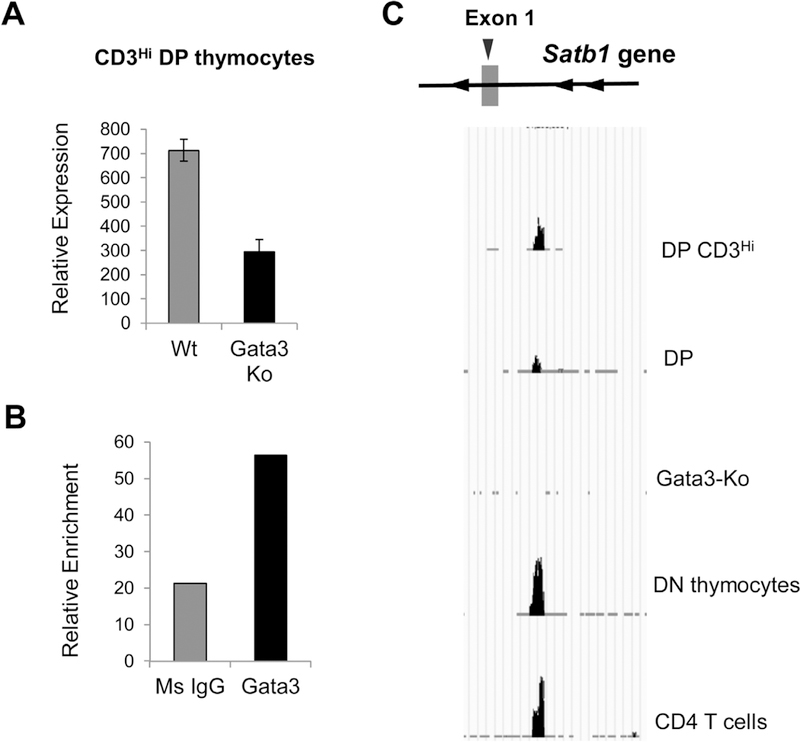
Transcriptional regulation of SATB1 by GATA3. (A) RNA-seq data from Wei et al. (Wei et al., 2011) analyzed for comparing relative expression pattern of SATB1 in CD3Hi thymocytes of control B6 and GATA3-KO mice. (B) Occupancy of GATA3 on Satb1 promoter monitored by ChIP analysis in total thymocytes. (C) Genome browser image showing the pattern of GATA3 occupancy on Satb1 promoter in each subset of mouse thymocytes. Thymocytes from GATA3-KO mice served as negative control.
4. Discussion
We report that differential expression of SATB1 in various subsets of mouse thymocytes is contingent on TCR signaling. The early DN subsets in the thymus have potential to develop into alternate fates such as NK cells, dendritic cells, macrophages and γδ T cells (Rothenberg, 2012) and only those thymocytes that progress beyond DN3 stage are committed to the T lineage (Yui et al., 2010). Our data suggests that SATB1 is induced in thymocytes that are committed for T lineage and this phenomenon is independently documented by others (Satoh et al., 2013). The Immunological Genome Project (ImmGen) has identified various transcription regulators including SATB1 involved in αβ T cell development (Heng et al., 2008). We further gauged the expression of SATB1 during distinct developmental stages of thymocytes based on TCR-β and CD69 expression and observed that SATB1 is induced in cells that have undergone positive selection and later repressed in mature thymocytes. The strong correlation between SATB1 and Nur77 expression patterns indeed suggests the dependence of SATB1 expression on TCR signaling during positive selection of thymocytes.
Analysis of SATB1 expression in SP thymocytes revealed for the first time that CD4 thymocytes exhibit characteristic bimodal distribution of SATB1 expression, which we designated as SATB1Hi and SATB1Lo populations. Expression level of SATB1 in the CD8 SP thymocytes is similar to that of the SATB1Lo population. It is important to note here that in Satb1 null mice SP thymocytes fail to develop. The kinetic signaling model of CD4/CD8 lineage commitment proposes that MHC class II restricted CD4 thymocytes experience persistent TCR signaling whereas MHC class I restricted CD8 thymocytes undergo early cessation of TCR signals (Singer et al., 2008). Correspondingly, CD4 thymocytes that receive continuous and prolonged TCR signaling exhibit higher expression of SATB1 (SATB1Hi population) and CD8 thymocytes exhibit lower expression of SATB1. It is also noteworthy to mention that since cytosolic tail of CD4 is known to associate with higher levels of LCK (which initiates TCR signal transduction) compared to CD8 coreceptor (Wiest et al., 1993) and therefore TCR signaling in CD4+ T cells is stronger as compared to that in CD8+ T cells. Similarly, we observed that expression of SATB1 in CD69+ve CD4 thymocytes is higher compared to that of CD69+ve CD8+ thymocytes, corroborating the fact that TCR signal transduction dictates SATB1 expression. Previous studies have shown that CD4+ SP thymocytes are heterogeneous based on the expression of Qa-2 and HSA/CD24 (Ramsdell et al., 1991; Bhandoola et al., 2000). Qa-2 and HSA/CD24 markers distinguish mature from the immature populations. SATB1Hi thymocytes are negative for Qa-2 expression and positive for HSA/CD24 whereas SATB1Lo population is positive for Qa-2 expression and HSA/CD24 low suggesting that SATB1Hi thymocytes are immature CD4+ thymocytes and SATB1Lo population primarily represent mature CD4+ thymocytes. Since SATB1 expression is regulated by TCR signaling, it is natural to speculate that nTregs should exhibit even higher SATB1 expression compared to other conventional CD4 thymocytes, as nTregs originate from high affinity TCR engagement of thymocytes with self-peptide MHC (Itoh et al., 1999). Paradoxically, we observed that SATB1 expression is downregulated in nTregs compared to conventional CD4+ thymocytes and similar observations were reported by Beyer et al. (Beyer et al., 2011).
The genes necessary for the T cell development and function are regulated by transcription factors such as RBPJ, TCF1 and GATA3 and deletion of any of these key factors blocked thymocyte development (Rothenberg, 2012). We show that GATA3 transcriptionally regulates SATB1 expression and it was previously known that GATA3 itself is regulated in a TCR-dependent manner (Hernandez-Hoyos et al., 2003). Our results provide an experimental confirmation to a recent report analyzing the transcriptional profile of genes during the development of αβ T cells that has implicated that SATB1 expression is regulated by GATA3 during T cell development (Mingueneau et al., 2013).
Since an effective TCR signal transduction pathway involves signaling via the TCR and coreceptors such as CD28 (Acuto and Michel, 2003), we activated CD4+ T cells in time-dependent manner using only anti-CD3 or anti-CD28 or both. We observed that SATB1 expression is induced specifically with anti-TCR but not with costimulatory signals. We also observed by immunostaining that SATB1 is expressed only in TCR activated cells and localizes to the euchromatic regions and is almost absent in the DAPI intense heterochromatic regions in CD4+ T cell nuclei. Thus, these results suggest that SATB1 presumably functions as a global gene regulator by creating distinct nuclear compartments. Further, SATB1 is downregulated in CD4+ T cells when cultured with inhibitor of P13K, which acts as downstream mediator of TCR signal transduction pathway. These data provide further evidence that SATB1 is regulated by TCR signaling.
This study analyzes the expression of SATB1 during the ontogeny of thymocyte development and sheds light on the signaling mechanisms and the transcriptional regulators of SATB1 during thymocyte development. Intranuclear staining pattern of SATB1 in thymocyte and activated CD4 T cell suggests that SATB1 presumably demarcates the transcriptionally active and incactive regions of T cell nuclei. This study raises few pertinent questions to be addressed like, whether the waxing and waning of SATB1 levels has anything to do with strength of TCR signals. This is especially important due to the observations that expression levels of SATB1 vary in various subsets of thymocytes and correlation observed between SATB1 and Nur77. Since SATB1 is a transcriptional regulator it is important to understand the repertoire of genes that are turned on or silenced in T cells.
Supplementary Material
Acknowledgements
Work was supported by grants from the Centre of Excellence in Epigenetics program of the Department of Biotechnology, Government of India, Swarnajayanti Fellowship Award from the Department of Science and Technology, Government of India to SG and NIH-intramural research program to JMS. KG and RJ were supported by fellowships from the Council of Scientific and 1ndustrial Research, 1ndia. Authors wish to thank Dr. Krishanpal Karmodiya for help with Ch1P-seq analysis, Dr. Satyajit Rath for valuable suggestions, and staff at the experimental animal facility of NCCS.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.molimm.2016.07.005.
References
- Acuto O, Michel F, 2003. CD28- mediated co-stimulation: a quantitative support for TCR signalling. Nat. Rev. Immunol. 3, 939–951. [DOI] [PubMed] [Google Scholar]
- Alvarez JD, Yasui DH, Niida H,Joh T, Loh DY, Kohwi-Shigematsu T, 2000. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 14,521–535. [PMC free article] [PubMed] [Google Scholar]
- Azzam HS, Grinberg A, Lui K, Shen H, Shores EW, Love PE, 1998. CD5 expression is developmentally regulated byT cell receptor(TCR) signals and TCR avidity. J. Exp. Med. 188, 2301–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendelac A, Matzinger P, Seder R, Paul W, Schwartz R, 1992. Activation events duringthymic selection.J. Exp. Med. 175, 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer M, Thabet Y, Muller RU, Sadlon T, Classen S, Lahl K, Basu S, Zhou X, Bailey-Bucktrout SL, Krebs W, Schonfeld EA, Bottcher J, Golovina T, Mayer CT, Hofmann A, Sommer D, Debey-Pascher S, Endl E, Limmer A, Hippen KL, Blazar BR, Balderas R, Quast T, Waha A, Mayer G, Famulok M, Knolle PA, Wickenhauser C, Kolanus W, Schermer B, Bluestone JA, Barry SC, Sparwasser T, Riley JL, Schultze JL, 2011. Repression ofthe genome organizer SATB1 in regulatory T cells is required for suppressive function and inhibition of effector differentiation. Nat. Immunol. 12, 898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandoola A, Dolnick B, Fayad N, Nussenzweig A, Singer A, 2000. Immature thymocytes undergoing receptor rearrangements are resistant to an Atm-dependent death pathway activated in mature T cells by double-stranded DNA breaks.J. Exp. Med. 192, 891–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Han HJ, Kohwi-Shigematsu T, 2003Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat. Genet. 34, 42–51. [DOI] [PubMed] [Google Scholar]
- Crispe IN, Bevan MJ, 1987. Expression and functional significance of the J11d marker on mouse thymocytes.J. Immunol. 138, 2013–2018. [PubMed] [Google Scholar]
- Galande S, Purbey PK, Notani D, Kumar PP, 2007. The third dimension of gene regulation: organization of dynamic chromatin loopscape by SATB1. Curr. Opin. Genet. Dev. 17, 408–414. [DOI] [PubMed] [Google Scholar]
- Germain RN, 2002. T-cell development and the CD4-CD8 lineage decision. Nat. Rev. Immunol. 2, 309–322. [DOI] [PubMed] [Google Scholar]
- Heng TS, Painter MW, Elpek K, Lukacs-Kornek V, Mauermann N, Turley SJ, Koller D, Kim FS, Wagers AJ, Asinovski N, 2008. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9,1091–1094. [DOI] [PubMed] [Google Scholar]
- Hernandez-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-lla J, 2003. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation Immunity 19, 83–94. [DOI] [PubMed] [Google Scholar]
- Ho IC, Tai TS, Pai SY, 2009. GATA3 and the T-cell lineage: essential functions before and afterT-helper-2-cell differentiation. Nat. Rev. Immunol. 9,125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Hemmer B, Martin R, Germain RN, 1999. Serial TCR engagement and down-modulation by peptide: MHC molecule ligands: relationship to the quality of individual TCR signaling events. J. Immunol. 162, 2073–2080. [PubMed] [Google Scholar]
- Lind EF, Prockop SE, Porritt HE, Petrie HT, 2001. Mapping precursor movement through the postnatal thymus reveals specific microenvironments supporting defined stages of early lymphoid development. J. Exp. Med. 194, 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z-G, Smith SW, McLaughlin KA, Schwartz LM, Osborne BA, Apoptotic signals delivered through the T-cell receptor of a T-cell hybrid require the immediate-early gene nur77 (1994). [DOI] [PubMed] [Google Scholar]
- Mingueneau M, Kreslavsky T, Gray D, Heng T, Cruse R, Ericson J, Bendall S, Spitzer MH, Nolan GP, Kobayashi K, 2013. The transcriptional landscape of [alpha][beta] T cell differentiation. Nat. Immunol. 14,619–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, Hogquist KA, 2011. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse.J. Exp. Med. 208,1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic-Zugic J, Bevan MJ, 1990. Functional and phenotypic delineation oftwo subsets of CD4 single positive cells in the thymus. Int. Immunol. 2, 135–141. [DOI] [PubMed] [Google Scholar]
- Notani D, Gottimukkala KP,Jayani RS, Limaye AS, Damle MV, Mehta S, Purbey PK, Joseph J, Galande S, 2010. Global regulator SATB1 recruits β-catenin and regulates TH2 differentiation in Wnt-dependent manner. PLoS Biol. 8,e1000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavan Kumar P, Purbey PK, Sinha CK, Notani D, Limaye A, Jayani RS, Galande S, 2006. Phosphorylation of SATB1 a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo. Mol. Cell 22, 231–243. [DOI] [PubMed] [Google Scholar]
- Ramsdell F, Jenkins M, Dinh Q, Fowlkes BJ, 1991. The majority of CD4 + 8-thymocytes are functionally immature. J. Immunol. 147,1779–1785. [PubMed] [Google Scholar]
- Rothenberg EV, Moore JE, Yui MA, 2008. Launching the T-cell-lineage developmental programme. Nat. Rev. Immunol. 8, 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV, 2011. T cell lineage commitment: identity and renunciation. J. Immunol. 186, 6649–6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV, 2012. Transcriptional drivers of the T-cell lineage program. Curr. Opin. Immunol. 24,132–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh Y, Yokota T, Sudo T, Kondo M, Lai A, Kincade PW, Kouro T, Iida R, Kokame K, Miyata T, 2013. The Satb1 protein directs hematopoietic stem cell differentiation toward lymphoid lineages. Immunity 38, 1105–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scollay R, Wilson A, Shortman K, 1984. Thymus cell migration: analysis of thymus emigrants with markers that distinguish medullary thymocytes from peripheral T cells. J. Immunol. 132,1089–1094. [PubMed] [Google Scholar]
- Singer A, Adoro S, Park JH, 2008. Lineage fate and intense debate: myths, models and mechanisms of CD4-versus CD8-lineage choice. Nat. Rev. Immunol. 8, 788–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr TK, Jameson SC, Hogquist KA, 2003. Positive and negative selection of T cells. Annu. Rev. Immunol. 21,139–176. [DOI] [PubMed] [Google Scholar]
- Vernachio J, Li M, Donnenberg AD, Soloski MJ, 1989. Qa-2 expression in the adult murine thymus. A unique marker for a mature thymic subset. J. Immunol. 142, 48–56. [PubMed] [Google Scholar]
- Wei G, Abraham BJ, Yagi R, Jothi R, Cui K, Sharma S, Narlikar L, Northrup DL, Tang Q, Paul WE, Zhu J, Zhao K, 2011. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity 35, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiest DL, Yuan L, Jefferson J, Benveniste P, Tsokos M, Klausner RD, Glimcher LH, Samelson LE, Singer A, 1993. Regulation of T cell receptor expression in immature CD4+CD8+ thymocytes by p56lck tyrosine kinase: basis for differential signaling by CD4 and CD8 in immature thymocytes expressing both coreceptor molecules. J. Exp. Med. 178, 1701–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui MA, Feng N, Rothenberg EV, 2010. Fine-scale staging of T cell lineage commitment in adult mouse thymus. J. Immunol. 185, 284–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerrahn J, Held W, Raulet DH, 1997. The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell 88, 627–636. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS, 2008. Model-based analysis of chIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


