Abstract
The renin angiotensin system (RAS) of the brain produces a series of biologically active angiotensinogen derived peptides involved in physiological homeostasis and pathophysiology of disease. Despite significant research efforts to date, a comprehensive understanding of brain RAS physiology is lacking. A significant challenge has been the limited set of bioanalytical assays capable of detecting angiotensin (Ang) peptides at physiologically low concentrations (2–15 fmol/g of wet tissue) and sufficient chemical specificity for unambiguous molecular identifications. Additionally, a complex brain anatomy calls for microanalysis of specific tissue regions, thus further taxing sensitivity requirements for identification and quantification in studies of the RAS. To fill this technology gap, we here developed a microanalytical assay by coupling a laboratory-built capillary electrophoresis (CE) nanoelectrospray ionization (nanoESI) platform to a high-resolution mass spectrometer (HRMS). Using parallel reaction monitoring, we demonstrated that this technology achieved confident identification and quantification of the Ang peptides at approx. 10 amol to hundreds of zmol sensitivity. This microanalytical assay revealed differential Ang peptide profiles between tissues that were micro-sampled from the subfornical organ and the paraventricular nucleus of the hypothalamus, important brain regions involved in thirst and water homeostasis and neuroendocrine regulation to stress. Microanalytical CE-nanoESI-HRMS extends the analytical toolbox of neuroscience to help better understand the RAS.
Keywords: Capillary electrophoresis, Nano-liquid chromatography, High-resolution mass spectrometry, Parallel reaction monitoring, Angiotensin, Renin-Angiotensin system, Peptidomics, Mouse, Subfornical organ, Paraventricular nucleus
INTRODUCTION
The classical renin angiotensin system (RAS) plays a critical role in maintaining cardiovascular and fluid homeostasis. As illustrated in Figure 1A, the RAS produces angiotensin (Ang) peptides by enzymatic cleavages from angiotensinogen (Ang T) [1–3], which have many physiological roles involved in cardiovascular homeostasis (e.g.., fluid balance) and disease processes (e.g., hypertension) [4, 2, 3]. Ang II, a metabolic product from Ang I and principle effector peptide, has long been known for its role in renal and blood pressure regulation in peripheral organs while much less is understood regarding the role of Ang peptides in the brain [4–7]. For example, the brain RAS and Ang peptides have been implicated in broad types of neurobiological processes [8–11] including neuroendocrine and metabolic regulation, neuronal function, memory, cognition, emotional responses to stress, and cerebral blood flow regulation [9, 10, 12]. The receptor subtypes (AT1R, AT2R, AT4R, MasR) that modulate the physiological effects of Ang derived peptides are differently distributed in the brain [12], thus raising intriguing questions about their respective physiological and neurobiological roles [13, 2]. Furthermore, bioactive roles have been identified for other Ang peptides, including Ang III, Ang IV, and Ang 1–7 [14–16]. Despite these significant research efforts and progress to date, there is still much to be learned about brain RAS physiology and pathophysiology [17, 18].
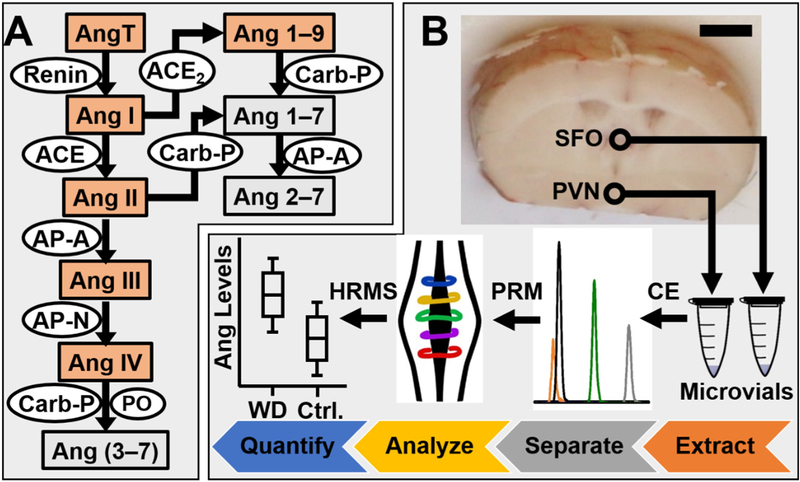
Figure 1.
Detection of angiotensin (Ang) peptides of the (A) renin-angiotensin system using (B) microanalytical high-resolution mass spectrometry (HRMS). Ang peptides are presented in boxes. Enzymes are inscribed in ovals. Peptides chosen for this study are highlighted in orange. The subfornical organ (SFO) and the paraventricular nucleus (PVN) were sampled by micropunch needles in water-deprived (WD) and control (Ctrl) mice (see circles). Ang peptides were microextracted, separated by a laboratory-built microanalytical capillary electrophoresis (CE) instrument, and detected using parallel reaction monitoring (PRM) HRMS. Key: Scale bar, 2 mm.
Currently, only a limited array of technologies is available for Ang peptides at physiologically relevant concentrations. Liquid-chromatography (LC) radioactive immunoassay (RIA) [19] is highly sensitive and quantitative, but lacks chemical specificity to unambiguously identify peptide signals [20], has low throughput (usually 1 antibody per peptide at a time), and can be costly for studies targeting multiple peptides (multiple antibodies needed). Other concerns pertain to antibody specificity [21]: The Ang peptides in the RAS only differ by one or two amino acids, which can lead to competitive interactions [22], thus requiring careful validation for each antibody. Additionally, the use of radioactive reagents calls for specialized expertise, hindering broader adoptability of LC-RIA from specialized laboratories.
Mass spectrometry (MS) has emerged as a powerful alternative for the development of assays with specificity to Ang peptides (reviewed in [23–25]). MS eliminates the need for antibodies and radioactive compounds and permits the detection of multiple Ang peptides in the same experiment, thus enhancing analytical throughput. Tandem MS augments chemical selectivity for detection and quantification by performing peptide-specific ion-molecule reactions, typically via collision-induced dissociation or higher-energy collisional dissociation (HCD) [26, 27]. Moreover, with the implementation of targeted ion detection, such as multiple/parallel reaction monitoring (MRM/PRM), tandem MS has emerged as a highly sensitive quantitative assay to characterize relative or absolute levels of Ang peptides in plasma and tissues [28–32]. For example, MRM on a triple-quadrupole mass spectrometer has yielded sub 10-amol levels of lower limit of detection [31]. MRM also permits the use of nonradioactive isotopically labeled peptides to facilitate relative or absolute quantification with ~10 amol sensitivity [33, 34], revealing endogenous angiotensin concentrations in brain hemispheres and the cerebellum [33]. These promising developments have enabled the confident identification and relative/absolute quantitation of Ang peptides from larger amounts of brain tissues. Furthermore, the recent development of table-top high-resolution MS (HRMS) instruments have provided exquisite chemical specificity for peptide identifications while maintaining sensitivity, promising to further the characterization of the RAS [26].
However, new HRMS technologies are needed to extend studies on the RAS to small tissue samples, such as identified nuclei of the brain. The brain contains Ang peptides in limited concentrations (~2–15 fmol/g of each Ang peptide in wet tissue [22, 35]). These amounts become trace-limited in defined regulatory regions, such as the SFO and PVN, which are critical hypothalamic and forebrain regions for angiotensin mediated cardiovascular and thirst homeostasis [13, 2, 36]. Assuming an ~1 g/mL average tissue density [37] and uniform Ang peptide production across the brain, a standard 0.5 mm (diameter) × 1 mm (height) micropunch (~200 nL voxel) is estimated to encompass only ~400 zmol to ~3 amol of Ang peptides. These peptide amounts are challenging the sensitivity of even modern MS methodologies (see earlier). Therefore, to facilitate our understanding of the brain RAS, there is a high and yet unmet need for new analytical technologies that combine unambiguous identification, trace-level sensitivity in detection and quantification, and a capability for microanalysis in the brain using HRMS.
To fill this technology gap, we have developed a microanalytical HRMS approach with trace-sensitive capability for Ang peptides of the RAS. The approach builds on microanalytical CE electrospray ionization (ESI) platforms that we and others have custom-built and optimized for high sensitivity analysis of volume-limited specimens [38–40]. CE-ESI-HRMS has been used to analyze proteins [41–43] and metabolites [44–48] with zmol–amol sensitivity in single cells in Xenopus laevis embryos (reviewed in reference [49]), single neurons in Aplysia californica [50, 41], as well as limited populations of neurons [43, 51] and single neurons from the mouse brain [52]. CE with matrix-assisted laser desorption ionization MS has been extensively used for profiling neuropeptides [39, 40, 53–55]. In this study, we have hyphenated a laboratory-constructed microanalytical CE-ESI platform to a high-resolution ESI mass spectrometer (quadrupole-ion trap-orbitrap tribrid-analyzer) and developed a microanalytical methodology for Ang peptides. We demonstrate that, once optimized, this microanalytical assay enables the simultaneous identification and quantification of Ang peptides in <10 amol to hundreds of zmol sensitivity. As a proof-of-principle experiment, we have used this assay to profile Ang peptide production between the SFO and PVN in control and water-deprived mice, providing new data to study the RAS in the brain.
MATERIALS AND METHODS
Standards and Reagents.
All chemicals and reagents were purchased from Fisher Scientific (Hampton, NH). Method development/validation was performed using a mixture of reference standards containing each of the following peptides at 1×10−4 g/L concentration: Ang T, Ang I, Ang II, Ang III, Ang IV, and Ang 1–9. For nano-flow LC (nanoLC) MS, the peptide mixture was reconstituted in 2% acetonitrile in water containing 0.1% formic acid. For CE-MS, the peptide mixture was prepared in 75% acetonitrile in water containing 0.05% or 0.5% acetic acid (see optimization below). CE was performed in bare fused silica capillaries from Polymicro Technologies (Phoenix, AZ). CE-nanoESI emitters were fabricated by pulling borosilicate capillaries (0.75/1 mm inner/outer diameter, Sutter Instruments, Novato, CA) using a Flaming/Brown type capillary puller (model P-1000, Sutter Instruments).
Animal Care and Handling.
All protocols regarding the humane treatment of animals were approved by the Institutional Care and Use Committee of The George Washington University (IACUC no. A279). Adult male (3–4 months old) C57BL/6J mice were purchased from Jackson Laboratory (Bar Harbor, ME) and were housed in temperature- and humidity-controlled polyethylene cages on a 12-h light/dark cycle. All animals were supplied with water and food ad libitum leading up to the experiment. As previously described, water deprivation is a strong stimulus to induce peripheral and brain RAS stimulation and increased angiotensin levels [56–59]. Therefore, in our current study, one cohort of mice were water restricted for 24 h while the second cohort served as the control and had continued access to water ad libitum [59].
Tissue Preparation.
Following 24 h of either experimental condition (water-restricted vs. control), n = 3 male mice were sacrificed (biological replicates). For method optimization, whole brains were isolated, immediately frozen on dry ice, and kept in −80 °C until analysis. Using a standardized brain tissue punching tool (Braintree Scientific Inc., Braintree, MA), tissue punches of 0.5 mm internal diameter and 1 mm in depth were reproducibly obtained from the SFO and the PVN following coordinates specified by the Mouse Brain Atlas [60]: −0.22 mm caudal, 0 mm lateral to bregma, and 3.25 mm below the skull surface for the SFO; −0.7 mm caudal, ±0.25 mm lateral to bregma, and 4.7 mm below the skull surface for the PVN.
Peptide Extraction.
A protocol to extract peptides was optimized using whole mouse brains before applying it to tissue punches from the SFO and PVN. Whole mouse brains were transferred into a 2 mL microcentrifuge tube, where peptides were extracted in 300 μL of 75% acetonitrile in water containing 0.5% acetic acid or 1% formic acid, facilitated by 3 rounds of periodic sonication and vortex mixing. Tissue debris was separated by centrifugation at 15,000 × g for 5 min at 4 °C. The supernatant was collected into a pristine microcentrifuge tube, and its content was dried at +4 °C in a vacuum concentrator (Labconco Corp., Kansas City, MO). The optimized sample extraction method was scaled to the brain punches as follows: Peptides were extracted in 5 μL of optimized extraction solution (containing 1% formic acid, see below), vacuum-dried, resuspended in 2 μL of 75% acetonitrile in water with 0.5% acetic acid, followed by centrifugation at 10,000 × g for 5 min (+4 °C) to pellet potential cell debris. The resulting “samples” were analyzed by CE-HRMS.
NanoLC-nanoESI.
A commercial nanoLC instrument (Dionex Ultimate 3000 RSLCnano, ThermoFisher Scientific, Palo Alto, CA) was used to benchmark our CE platform. For nanoLC, 100 nL of a peptide mixture containing Ang I, Ang II, Ang 1–9, and Ang IV were loaded onto a trapping column (C18 Acclaim PepMap 100, 300 μm inner diameter, 50 mm length, 5 μm beads with 100 Å pore size, ThemoFischer Scientific) using the microliter pickup function at 10 μL/min. The peptides were separated on a C18 Acclaim PepMap RSCL analytical column (75 μm inner diameter, 250 mm in length, 3 μm beads with 100 Å pore size, ThermoFisher Scientific) using a 60-min linear gradient from 2% to 35% of acetonitrile (0.1% formic acid) at 300 nL/min. Molecules were charged by nanoelectrospray ionization (emitter with 5 μm aperture, New objective, Woburn, MA) at +2,500 V spray potential. The generated ions were detected by HRMS.
CE-nanoESI.
This study used the same CE platform as that described in references [41, 42]. The platform was constructed, optimized, operated, and validated as detailed elsewhere [50, 44, 41–43]. Samples of 300-nL to 1-μL volume were deposited into a sample-loading microvial, whence ~14–20 nL were hydrodynamically loaded into the CE separation capillary filled with 25% acetonitrile containing 1 M formic acid (pH 2.3). Electrophoretic separation was performed by applying +20,000 V to the capillary inlet against the grounded outlet, generating ~5.5 μA initial CE current. Compounds migrating through the capillary entered an electrokinetically pumped co-axial sheath nanoESI source [61] for ionization. The electrospray was generated by applying +1,500 V onto the sheath reservoir containing 10% acetonitrile with 0.05% acetic acid. The generated ions were detected by HRMS.
HRMS Detection.
Peptide ions that were generated by nanoLC-nanoESI and CE-nanoESI were detected using the same quadrupole-linear ion trap-orbitrap tribrid-analyzer mass spectrometer (Orbitrap Fusion Lumos, ThermoFisher Scientific) executing PRM under identical settings. The MS1 acquisition parameters were as follows: mass analyzer, orbitrap; resolution, 120,000 FWHM; AGC target, 5×105 counts; and maximum injection time, 75 ms. Target precursor ions were isolated in the quadrupole analyzer with an isolation window of 1 Da and fragmented by HCD at 28–35% normalized collision energy (NCE) in nitrogen gas. Fragment ions were routed to the orbitrap analyzer for detection with the following settings: first mass, 100; resolution, 30,000 FWHM; AGC target, 1×105 counts; and maximum injection time, 60 ms.
Data Analysis.
Primary MS–MS/MS data were processed using Skyline version 4.1 [62]. The amino acid sequence of the Ang peptides was submitted to Skyline to predict (in silico) a list of theoretical fragments from each peptide (m/z values). Chromatographic peak boundaries were inspected manually and redefined as necessary. Parameters of peptide and transition integration were set as follows: filter, peptides; precursor charges, +1–4; ion charges, +1–3; ion types, b and y ions; instrument, min m/z 280 and max m/z 1,600; method match tolerance, m/z 0.01; full-scan, MS1 filtering in the orbitrap at 120,000 FWHM resolution (at m/z 400); MS/MS filtering acquisition, 50,000 FWHM resolution (at m/z 400); retention time filtering was set to only use transitions within 5 min of MS/MS identification. Skyline was used to generate chromatographic traces for each precursor ion and transition that matched the in-silico reference, which was followed by integration of under-the-curve peak areas that serve as a proxy for peptide quantification. Peptide quantities were reported as summed peak areas (SPA) of the precursor ions and transitions integrated areas. Peptides were accepted only if their monoisotopic peak was detected and more than 50% of the transitions were quantified.
Safety Considerations.
Standard safety procedures were followed during the handling of chemicals and biological samples. Fused silica capillaries and nanoESI emitters, which present a needle-stick hazard, were handled with care using gloves and protective eyewear. All electrically connective parts of the CE platform were earth-grounded or isolated in a Plexiglas enclosure equipped with a safety interlock enabled door to prevent users from exposure to electrical shock hazard.
RESULTS
Microanalytical MS for Ang Peptides.
We here developed a microanalytical framework to enable the ultrasensitive detection and quantification of Ang peptides in mouse brain nuclei critical to thirst regulation, specifically the SFO and PVN. Microscale analysis of these tissues presented several analytical challenges that were addressed by our study design (Fig. 1). As in all neuropeptidomics studies [23], the chemical integrity of the tissue must be preserved by sampling the brain with accuracy and speed, ideally in a frozen state and/or with the help of enzymatic deactivation. In this study, we used micro-punch needles to biopsy ~200 nL voxel from the SFO and PVN areas in the frozen brain (see Materials and Methods). Alternatively, limited amounts of materials may be obtained by other strategies ranging from micro-collection of bodily fluids (e.g., lumbar puncture or by in vivo dialysis probes) to dissection of cells, tissues, and organs. With tissues from our study estimated to only contain sub-to-low amol amounts of Ang peptides (refer to Introduction), trace-sensitive detection required optimization of the analytical workflow (see Fig. 1B). In a reversed order, we optimized the following methods using chemical standards or biological tissues: conditions of extraction and sample reconstitution (nanoLC vs. CE using tissues); choice of technology for analytical separation (using standards); conditions of mass spectrometric detection (using standards and tissues).
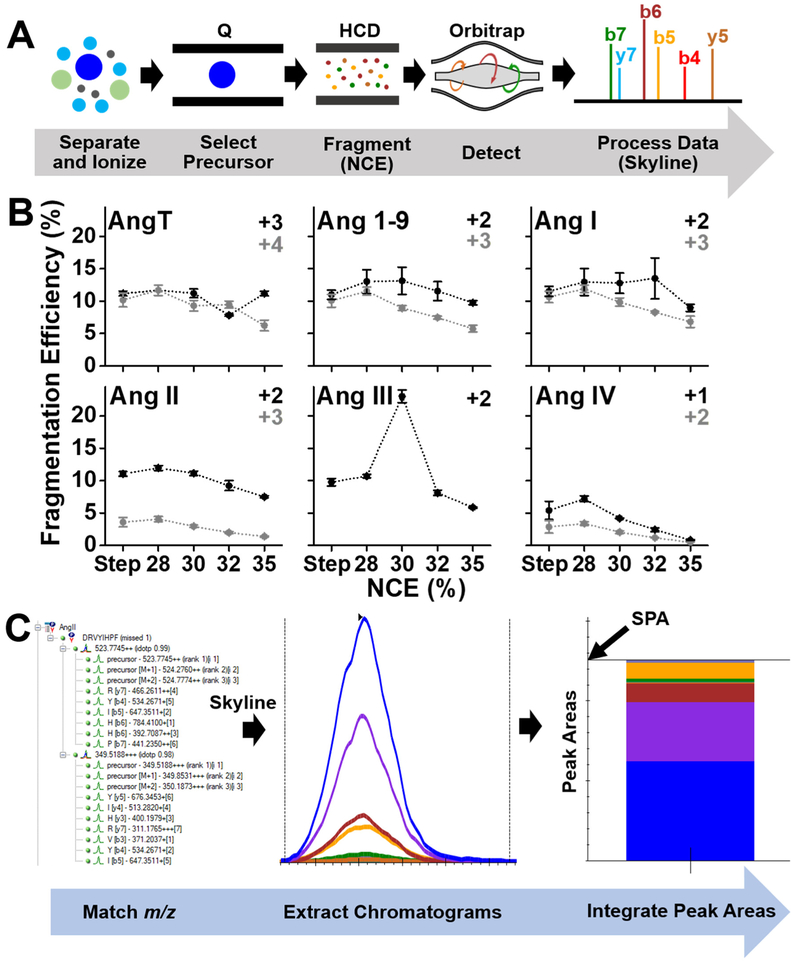
We designed an assay for the Ang peptides using PRM HRMS. The approach, presented in Figure 2, attains exquisite selectivity and quantification by monitoring m/z transitions specific to each peptide [26]. We used the Skyline software [62] to query specific m/z transition based on tandem MS data that we collected for each targeted Ang peptide standard (Fig. 1A). In principle, this data acquisition strategy also allows for the development of assays targeted for other peptides, including neuropeptides. The PRM assay from this work was applied on a late-generation quadrupole linear ion trap orbitrap mass spectrometer [26]; this approach is also adaptable to quadrupole orbitrap instruments outfitted with a collision cell. Peptide precursor ions were m/z-selected in the quadrupole, dissociated via HCD, and the resulting fragment ions were detected with high m/z accuracy (<10 ppm) in the orbitrap analyzer (Fig. 2A). To attain high data fidelity from biological samples, which may contain many isobaric signals (e.g., peptides and metabolites), data analysis in this study considered only peptides that i) exhibited the correct accurate m/z (<10 ppm), ii) produced fragment ions that matched theory (in silico reference) with co-elution with the precursor ion, and iii) gave signals that were quantified in at least 50% of the transitions. Detection efficiency was experimentally maximized for the dominant charge states (listed in Table 1) by optimizing the efficiency of fragmentation in the HCD cell (see Fig. 2B). Quantification was based on the total ion abundance of the fragment ions (m/z values) that matched theoretical prediction in Skyline (Fig. 2C). Peptide-specific chromatograms were extracted in the MS–MS/MS space, followed by the determination of summed peak area (SPA) for relative quantification of peptide abundance.
Figure 2.
Development of a high-resolution mass spectrometry assay for Ang peptides. (A) Parallel reaction monitoring (PRM) selecting Ang peptides in the quadrupole (Q) for fragmentation via higher-energy collisional dissociation (HCD), followed by orbitrap detection of peptide-specific fragments (e.g., b- and y- ions listed). (B) Experimental optimization of fragmentation efficiency as a function of normalized collision energy (NCE) for HCD. Each data point was a separate analysis of the standard peptide mixture (1×10−4 g/L) using capillary electrophoresis (CE) high-resolution mass spectrometry (HRMS), analyzed in technical triplicate. Key: Error bars, 1 × S.E.M. (C) Data analysis in the Skyline software extracting specific precursor-fragment transitions for integration of under-the-curve peak areas to be used as a proxy for peptide quantification.
Table 1.
PRM-HRMS for microanalytical detection and quantification of angiotensin peptides from the renin-angiotensin system. The dominant charge state is highlighted in bold phase.
| Peptide | Peptide Sequence |
m/z (charge) | Calculated Lower Limit of Quant. |
Calculated Lower Limit of Det. |
||
|---|---|---|---|---|---|---|
| nM | amol | pM | amol | |||
| AngT | DRVYIHPFHLVIH |
412.2299 (+4); 549.3039 (+3) |
0.32 | 4.3 | 17 | 0.35 |
| Ang 1–9 | DRVYIHPFH |
395.2054 (+3); 592.3043 (+2) |
0.42 | 5.8 | 20 | 0.39 |
| Ang I | DRVYIHPFHL |
432.9001 (+3); 648.8463 (+2) |
0.77 | 11 | 30 | 0.60 |
| Ang II | DRVYIHPF | 349.5191 (+3); 523.7748 (+2) |
0.96 | 13 | 0.13 | 2.6 |
| Ang III | RVYIHPF | 466.2613 (+2) | 0.54 | 7.4 | 0.10 | 2.0 |
| Ang IV | VYIHPF |
388.2108 (+2); 775.4137 (+1) |
6.5 | 88 | 0.21 | 4.2 |
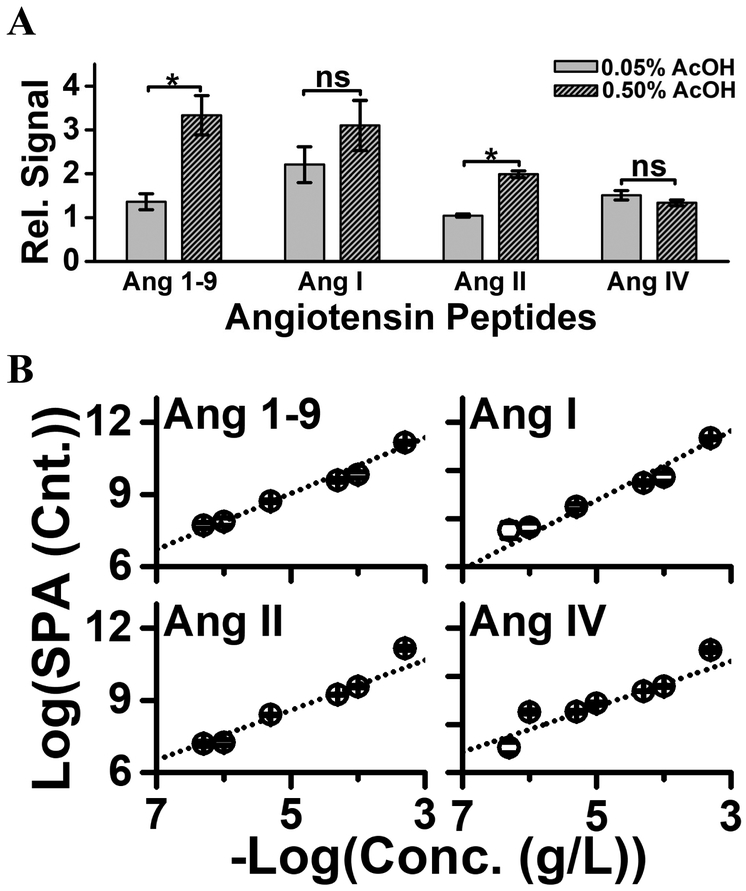
The PRM assay was configured to a microanalytical CE platform; this instrument has been described in detail elsewhere [41, 42]. The platform featured a sample-loading microvial, where ~300 nL of sample was deposited. Approximately 14 nL of the deposited sample was loaded into the CE capillary, followed by electrophoresis. For enhanced detection sensitivity, on-column enrichment was performed via field-amplified sample stacking (FASS) in the CE capillary. To lower the conductivity of the sample plug for a successful FASS [63], the background electrolyte was prepared to contain 25% acetonitrile with 1 M formic acid, whereas the sample (e.g., peptide standards or extracts from brain tissues) were reconstituted in aqueous 75% acetonitrile containing 0.05% (v/v) or 0.5% (v/v) acetic acid. As shown in Figure 3A, higher acid content significantly improved electrophoretic separation and sensitivity (Student’s t test, p < 0.05). The theoretical number of plates (N) was higher by ~40% for Ang 1–9 and ~25% for Ang I and Ang IV. Under optimal conditions, we determined a theoretical plane number (N) of ~933,547 plates/m, demonstrating remarkable separation efficiency. As a result, higher acid content led to similar or up to ~7-fold higher sensitivity for the peptides.
Figure 3.
Experimental characterization of analytical performance. (A) Sample reconstitution in higher acid content enhanced sensitivity by improving field-amplified sample stacking. Key: Error bars, 1 × S.E.M; *p < 0.05 (Student’s t-test). (B) The dynamic range of quantification was tested to be linear over a ~3–4 log-order range for the angiotensin standards, extrapolating to a lower limit of quantification at ~1–10 amol (see Table 1).
The analytical figures of merit were established. Calibration curves were measured based on the triplicate analysis (technical replicate) of a dilution series prepared from the reference Ang peptide mixture. The linear dynamic range of quantification was found to span 3–4-log orders of magnitude (Fig. 3B). The lower limit of quantification (LLOQ) was calculated in the 1–10 amol (0.3–1 nM) range for all the peptides, except for Ang IV (see Table 1). These results revealed comparable-to-improved sensitivity over studies that analyzed ~1–10 μL peptide extracts using nanoLC-HRMS [28, 29, 31, 33]. Compared to commercial nanoLC, microanalytical CE-nanoESI-HRMS has the advantage of inherent compatibility with limited samples: ~1,000-times smaller sample amounts were analyzed in this study than are typically analyzed by nanoLC. The calibration curves revealed low-amol sensitivity for most of the peptides using CE-nanoESI-HRMS. Based on the MS1 data, extrapolation to signal/noise = 3 predicts <10 amol to hundreds of zmol sensitivity for detection. Based on the triplicate analysis of each of the peptide standards, the average quantitative reproducibility was determined to be ~8% S.E.M technical error (same sample analyzed multiple times). Therefore, CE-nanoESI-HRMS combined microscale capability, sensitivity, and an ability for quantifying Ang peptides.
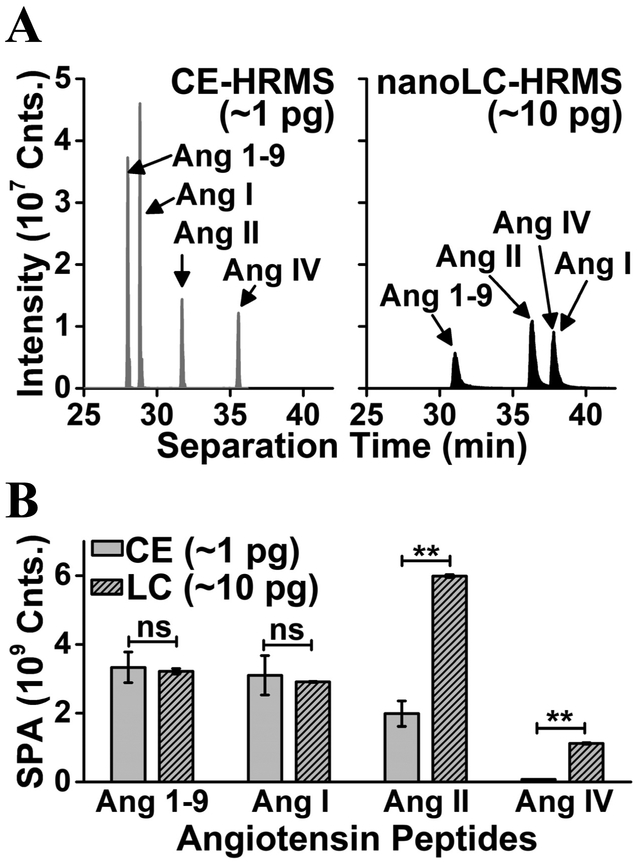
This Ang peptide assay was benchmarked against nanoLC PRM-HRMS (Fig. 4), which is the closest analytical technology. To enable direct comparison in accuracy, the assays were tested on the same mass spectrometer executing the same PRM method that was developed in this study (see earlier). The Ang peptide mixture was diluted to 100 μg/L/peptide concentration using solvents that were appropriate for the respective technologies. For nanoLC-PRM-HRMS, 100 nL (viz., ~100 ng/peptide) were analyzed over 60 min of separation with sample dilution using aqueous 2% acetonitrile (0.1% formic acid). For CE-PRM-HRMS, ~14 nL (viz., ~14 ng/peptide) were analyzed over ~40 min of separation with sample dilution in aqueous 75% acetonitrile (0.5% v/v acetic acid). Differences were evident in the metrics of analytical performance (Fig. 4A). All the peptides were resolved as sharp, symmetric peaks of high signal abundance using CE (peak capacity = ~83, peak symmetry = ~1.2). In contrast, two of the peptides were coeluted and all the peptides were detected as broadened, slightly tailing peaks of lower signal intensity using nanoLC (peak capacity = ~11, peak symmetry = ~2.3). Despite analyzing ~10-times less material, CE provided comparable ion signal intensities than nanoLC (compare Fig. 4B). Therefore, microanalytical CE provided a significant improvement over current nanoLC, raising unique potentials to detect and identify trace amounts of Ang peptides in the brain.
Figure 4.
Benchmarking microanalytical capillary electrophoresis (CE) against nano-flow liquid chromatography (nanoLC) for high-resolution mass spectrometry (HRMS). Analyzed sample volumes from the same standard angiotensin mixture (100 μg/L) were: 14 nL for CE and 100 nL for nanoLC. (A) Mass-selected ion traces comparing peptide separation during CE and nanoLC, revealing higher separation efficiency and detection sensitivity by electrophoresis. (B) Comparable summed peak areas (SPA) between the technologies suggest ~10-fold higher sensitivity by CE-HRMS. Key: Error bars, 1 × S.E.M; ns, not significant; **p ≤ 0.01 (Student’s t-test).
Angiotensin Peptides in the Brain.
As in most peptidomics studies [23], sensitive detection of Ang peptides required elimination/minimalization of their unwanted peptide degradation during sample handling as well as efficient extraction and recovery of these peptides from the brain tissues. Organic solvents containing high concentration of acids, such as formic or acetic acid, and rapid freezing or heating have been used to minimize enzymatic digestion due to denaturing proteases, allowing the efficient recovery of endogenous peptides from tissues and organisms [23, 64, 65]. To optimize recovery for CE-HRMS, we compared the efficiency of peptide detection from whole brain tissues upon extraction using 75% acetonitrile containing 0.5% acetic acid or 1% formic acid. The extracts were dried at 4 °C in a vacuum concentrator and reconstituted in 75% acetonitrile containing 0.5% acetic acid, which we previously found to facilitate separation and sensitivity for CE-HRMS (recall Fig. 3A). Compared to extraction in 0.5% acetic acid, SPA values obtained from 1% formic acid containing solution were higher by a factor of 5–8 for Ang 1–9, Ang I, Ang II, and Ang III and ~15–20 for Ang T and Ang IV. Superior extraction efficiency when using 1% formic acid prepared us to analyze tissues from the identified nuclei.
The optimized microassay was employed to profile Ang peptides in the SFO and PVN. As proof of concept, WD was used as model, which increases angiotensin levels [66, 56, 57]. Approximately 1 μL tissues were collected from the SFO and the PVN using micropunch needles (see Materials and Methods). Peptides were extracted from the collected tissues in 5 μL of 75% acetonitrile containing 1% formic acid (optimized for extraction, see earlier), dried, and reconstituted in 2 μL of 75% acetonitrile containing 0.5% acetic acid (optimized for electrophoresis, see earlier). An ~300 nL volume of the reconstituted extract was deposited onto the sample-loading microvial, whence ~20 nL were analyzed by CE-HRMS. By enabling the analysis of such low volumes of samples, CE-HRMS raised a potential to minimize peptide losses due to absorptive processes on the surfaces of vials and pipette tips, which would be difficult to circumvent for regular nanoLC requiring larger sample volumes for analysis.
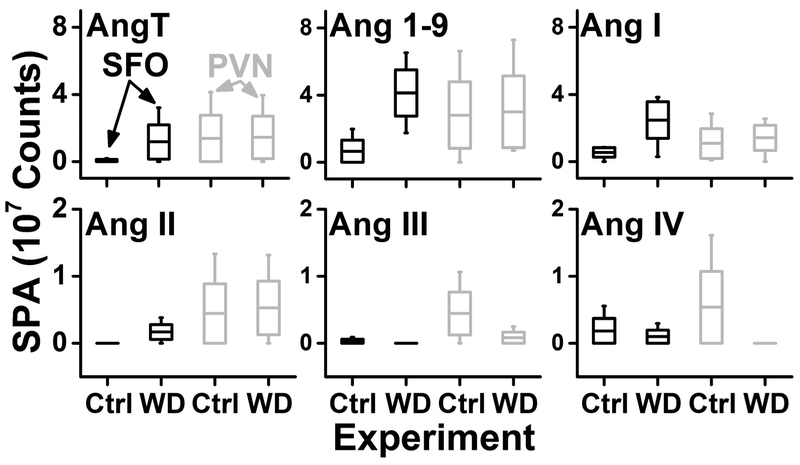
CE-nanoESI-HRMS revealed quantitative difference in Ang profiles. In Figure 5, the respective Ang levels are compared between the brain regions in the control and the experimental (WD). Following 24 h of WD, levels of AngT, Ang 1–9, Ang I, and Ang II increased in the SFO but not in the PVN. Conversely, the amounts of Ang III and Ang IV peptide fragments were unchanged or slightly decreased in the SFO post WD. These results may reflect increased Ang II receptor mediated function and reduced peptide degradation products following WD. On the other hand, WD caused marked decrease of Ang III and Ang IV concentrations in the PVN. This result combined with stable levels of AngT, Ang 1–9, Ang I, and Ang II suggest enhanced conversion from Ang II to Ang 1–7. Overall, these observations support the function of these brain nuclei, increasing brain Ang II production following water restriction and their overall importance in volumetric thirst regulation and fluid homeostasis [13, 2, 36, 67]. Future studies involving a higher number of biological replicates and the use of internal standards to enhance technical reproducibility are needed for further statistical analysis on the observed differences between the Ang profiles in our experimental groups. To our knowledge, these results provide, for the first time, the detection of biologically active Ang peptides in identified brain nuclei and may inform future experiments elucidating the role of the brain RAS in physiology and disease.
Figure 5.
Quantitative comparison of Ang peptide amounts between the subfornical organ (SFO, data in black) and the paraventricular nucleus (PVN, data in grey) in control (Ctrl) and upon water deprivation (WD) of N = 3 male mice. Key: Boxes, 1 × S.E.M.; whiskers, 10–90 percentiles; mid-line, mean.
CONCLUSIONS
We have developed a microanalytical assay for Ang peptides based on a laboratory-built CE-nanoESI-HRMS platform. The technology attained exquisite detection specificity by integrating multiple orthogonal pieces of information for each peptide, namely accurate mass (<10 ppm error), time of separation (migration), and peptide-dependent fragmentation during PRM. By eliminating chemical/molecular probes, radioactive reagents, or antibodies, this approach aids robustness and can be readily adapted by other labs. The assay combined microanalytical capability for 10–20 nL of peptides extracted from a standard 0.5 mm (diameter) × 1 mm (height) micropunch (~200 nL voxel) with <10 amol to few hundreds of zmol sensitivity for detection and a ~4-log-oder linear dynamic range with an ~10-amol sensitivity for quantification. These performance metrics were adequate to profile endogenous Ang levels in the mouse, even in limited tissues that were collected from the SFO and PVN in this study. In proof of principle experiments, we demonstrated the ability of the assay to capture quantitative changes in Ang production upon WD. With a capability for label-free detection, our approach can be extended to other types of peptides and neuropeptides, brain regions, as well as other types of cells, tissues, organs and biological models. The approach presented here is readily adoptable for commercialized CE-MS instruments (e.g., CE 7100 from Agilent, Santa Clara, CA and CESI 8000 Plus from AB Scix, Ontario, Canada), thus facilitating broader adoption of the methodology presented in this study. Additionally, the microanalytical assay is compatible with the use of internal standards (e.g., isotopically labeled Ang peptides) to aid compound identification (e.g., migration time markers) and quantification (relative or absolute). With label-free detection, trace-level sensitivity for detection and quantification, and compatibility with microsampling (e.g., micropunch needles here), CE-nanoESI-HRMS extends the analytical toolbox of neuroscience to facilitate studies on the RAS.
ACKNOWLEDGEMENT
The reported work was partially supported by the Arnold and Mabel Beckman Foundation Beckman Young Investigator Grant (to P.N.) and the National Institutes of Health grants 1R35GM124755 (to P.N.) and R01HL137103 (to P.J.M.).
Footnotes
Conflict of Interest. The authors have no conflicts of interest.
REFERENCES
- 1.Vickers C, Hales P, Kaushik V, Dick L, Gavin J, Tang J et al. Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J Biol Chem. 2002;277(17):14838–43. doi: 10.1074/jbc.M200581200. [DOI] [PubMed] [Google Scholar]
- 2.Coble JP, Grobe JL, Johnson AK, Sigmund CD. Mechanisms of brain renin angiotensin system-induced drinking and blood pressure: importance of the subfornical organ. Am J Physiol-Regul Integr Comp Physiol. 2015;308(4):R238–R49. doi: 10.1152/ajpregu.00486.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forrester SJ, Booz GW, Sigmund CD, Coffman TM, Kawai T, Rizzo V et al. Angiotensin II signal transduction: an update on mechanisms of physiology and pathophysiology. Physiol Rev. 2018;98(3):1627–738. doi: 10.1152/physrev.00038.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mazzolai L, Pedrazzini T, Nicoud F, Gabbiani G, Brunner HR, Nussberger J. Increased cardiac angiotensin II levels induce right and left ventricular hypertrophy in normotensive mice. Hypertension. 2000;35(4):985–91. doi: 10.1161/01.Hyp.35.4.985. [DOI] [PubMed] [Google Scholar]
- 5.Weiss D, Kools JJ, Taylor WR. Angiotensin II-induced hypertension accelerates the development of atherosclerosis in ApoE-deficient mice. Circulation. 1999;100(18):474. doi: 10.1161/01.CIR.103.3.448. [DOI] [PubMed] [Google Scholar]
- 6.Nakagawa P, Sigmund CD. How Is the brain renin-angiotensin system regulated? Hypertension. 2017;70(1):10–8. doi: 10.1161/hypertensionaha.117.08550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uijl E, Ren LW, Danser AHJ. Angiotensin generation in the brain: a re-evaluation. Clin Sci. 2018;132(8):839–50. doi: 10.1042/cs20180236. [DOI] [PubMed] [Google Scholar]
- 8.Phillips MI. Angiotensin in brain. Neuroendocrinology. 1978;25(6):354–77. doi: 10.1159/000122756. [DOI] [PubMed] [Google Scholar]
- 9.McKinley MJ, Albiston AL, Allen AM, Mathai ML, May CN, McAllen RM et al. The brain renin-angiotensin system: location and physiological roles. Int J Biochem Cell Biol. 2003;35(6):901–18. doi: 10.1016/s1357-2725(02)00306-0. [DOI] [PubMed] [Google Scholar]
- 10.Grobe JL, Xu D, Sigmund CD. An intracellular renin-angiotensin system in neurons: Fact, hypothesis, or fantasy. Physiology. 2008;23(4):187–93. doi: 10.1152/physiol.00002.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright JW, Harding JW. Importance of the brain angiotensin system in Parkinson’s disease. Parkinsons Dis. 2012:860923. doi: 10.1155/2012/860923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Bohlen und Halbach O. Angiotensin IV in the central nervous system. Cell Tissue Res. 2003;311(1):1–9. doi: 10.1007/s00441-002-0655-3. [DOI] [PubMed] [Google Scholar]
- 13.Pyner S Neurochemistry of the paraventricular nucleus of the hypothalamus: Implications for cardiovascular regulation. J Chem Neuroanat. 2009;38(3):197–208. doi: 10.1016/j.jchemneu.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Jiang F, Yang JM, Zhang YT, Dong M, Wang SX, Zhang Q et al. Angiotensin-converting enzyme 2 and angiotensin 1–7: novel therapeutic targets. Nat Rev Cardiol. 2014;11(7):413–26. doi: 10.1038/nrcardio.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yugandhar VG, Clark MA. Angiotensin III: A physiological relevant peptide of the renin angiotensin system. Peptides. 2013;46:26–32. doi: 10.1016/j.peptides.2013.04.014. [DOI] [PubMed] [Google Scholar]
- 16.Ferrario CM, Trask AJ, Jessup JA. Advances in biochemical and functional roles of angiotensin-converting enzyme 2 and angiotensin-(1–7) in regulation of cardiovascular function. Am J Physiol-Heart Circul Physiol. 2005;289(6):H2281–H90. doi: 10.1152/ajpheart.00618.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakagawa P, Sigmund CD. How is the brain renin-angiotensinsSystem regulated? Hypertension. 2017;70(1):10–8. doi: 10.1161/hypertensionaha.117.08550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sigmund CD, Diz DI, Chappell MC. No brain renin-angiotensin system deja vu all over again? Hypertension. 2017;69(6):1007–10. doi: 10.1161/hypertensionaha.117.09167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chappell MC, Brosnihan KB, Diz DI, Ferrario CM. Identification of angiotensin-(1–7) in rat brain - evidence of differential processing of angiotensin peptides J Biol Chem. 1989;264(28):16518–23. [PubMed] [Google Scholar]
- 20.Desilva PE, Husain A, Smeby RR, Khairallah PA. Measurement of immunoactive angiotensin peptides in rat tissues - some pitfalls in angiotensin-II analysis. Anal Biochem. 1988;174(1):80–7. doi: 10.1016/0003-2697(88)90521-0. [DOI] [PubMed] [Google Scholar]
- 21.Herrera M, Sparks MA, Alfonso-Pecchio AR, Harrison-Bernard LM, Coffman TM. Response to lack of specificity of commercial antibodies leads to misidentification of angiotensin type-1 receptor protein. Hypertension. 2013;61(4):E32–E. doi: 10.1161/hypertensionaha.111.00982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chappell MC. Biochemical evaluation of the renin-angiotensin system: the good, bad, and absolute? Am J Physiol-Heart Circul Physiol. 2016;310(2):H137–H52. doi: 10.1152/ajpheart.00618.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Svensson M, Skold K, Nilsson A, Falth M, Nydahl K, Svenningsson P et al. Neuropeptidomics: MS applied to the discovery of novel peptides from the brain. Anal Chem. 2007;79(1):14–21. doi: 10.1021/ac071856q. [DOI] [PubMed] [Google Scholar]
- 24.Zestos AG, Kennedy RT. Microdialysis coupled with LC-MS/MS for in vivo neurochemical monitoring. Aaps J. 2017;19(5):1284–93. doi: 10.1208/s12248-017-0114-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scifo E, Calza G, Fuhrmann M, Soliymani R, Baumann M, Lalowski M. Recent advances in applying mass spectrometry and systems biology to determine brain dynamics. Expert Rev Proteomics. 2017;14(6):545–59. doi: 10.1080/14789450.2017.1335200. [DOI] [PubMed] [Google Scholar]
- 26.Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol Cell Proteomics. 2012;11(11):1475–88. doi: 10.1074/mcp.O112.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods. 2012;9(6):555–66. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 28.Cui L, Nithipatikom K, Campbell WB. Simultaneous analysis of angiotensin peptides by LCMS and LC-MS/MS: Metabolism by bovine adrenal endothelial cells. Anal Biochem. 2007;369(1):27–33. doi: 10.1016/j.ab.2007.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lortie M, Bark S, Blantz R, Hook V. Detecting low-abundance vasoactive peptides in plasma: Progress toward absolute quantitation using nano liquid chromatography-mass spectrometry. Anal Biochem. 2009;394(2):164–70. doi: 10.1016/j.ab.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Domenig O, Schwager C, van Oyen D, Poglitsch M. Ex vivo equilibrium analysis of the renin-angiotensin-system: Clinical implications for diagnosis and treatment of hypertension. Hypertension. 2014;64:A479. [Google Scholar]
- 31.Olkowicz M, Radulska A, Suraj J, Kij A, Walczak M, Chlopicki S et al. Development of a sensitive, accurate and robust liquid chromatography/mass spectrometric method for profiling of angiotensin peptides in plasma and its application for atherosclerotic mice. J Chrom A. 2015;1393:37–46. doi: 10.1016/j.chroma.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 32.Tan L, Yu ZR, Zhou XM, Xing D, Luo XY, Peng RF et al. Antibody-free ultra-high performance liquid chromatography/tandem mass spectrometry measurement of angiotensin I and II using magnetic epitope-imprinted polymers. J Chrom A. 2015;1411:69–76. doi: 10.1016/j.chroma.2015.07.114. [DOI] [PubMed] [Google Scholar]
- 33.van Thiel BS, Martini AG, te Riet L, Severs D, Uijl E, Garrelds IM et al. Brain renin-angiotensin system does it exist? Hypertension. 2017;69(6):1136–44. doi: 10.1161/hypertensionaha.116.08922. [DOI] [PubMed] [Google Scholar]
- 34.Pavo N, Goliasch G, Wurm R, Novak J, Strunk G, Gyongyosi M et al. Low- and high-renin heart failure phenotypes with clinical implications. Clin Chem. 2018;64(3):597–608. doi: 10.1373/clinchem.2017.278705. [DOI] [PubMed] [Google Scholar]
- 35.Hermann K, McDonald W, Unger T, Lang RE, Ganten D. Angiotensin biosythesis and concentrations in brain of normotensive and hypertensive rats. J Physiol Paris. 1984;79(6):471–80. [PubMed] [Google Scholar]
- 36.Bourassa EA, Speth RC. Water deprivation increases angiotensin-converting enzyme but not AT(1) receptor expression in brainstem and paraventricular nucleus of the hypothalamus of the rat. Brain Res. 2010;1319:83–91. doi: 10.1016/j.brainres.2009.12.079. [DOI] [PubMed] [Google Scholar]
- 37.Barber TW, Brockway JA, Higgins LS. The density of tissues in and about the head. Acta Neurol Scand. 1970;46(1):85–92. [DOI] [PubMed] [Google Scholar]
- 38.Lapainis T, Sweedler JV. Contributions of capillary electrophoresis to neuroscience. J Chrom A. 2008;1184(1–2):144–58. doi: 10.1016/j.chroma.2007.10.098. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Ma M, Chen R, Li L. Enhanced neuropeptide profiling via capillary electrophoresis off-line coupled with MALDI FTMS. Anal Chem. 2008;80(16):6168–77. doi: 10.1021/ac800382t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang XY, Chen RB, Wang JH, Metzler A, Tlusty M, Li LJ. Mass spectral charting of neuropeptidomic expression in the stomatogastric ganglion at multiple developmental stages of the lobster Homarus americanus. Acs Chem Neuro. 2012;3(6):439–50. doi: 10.1021/cn200107v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lombard-Banek C, Moody SA, Nemes P. Single-cell mass spectrometry for discovery proteomics: quantifying translational cell heterogeneity in the 16-cell frog (Xenopus) embryo. Angew Chem Int Ed. 2016;55(7):2454–8. doi: 10.1002/anie.201510411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lombard-Banek C, Reddy S, Moody SA, Nemes P. Label-free quantification of proteins in single embryonic cells with neural fate in the cleavage-stage frog (Xenopus laevis) embryo using capillary electrophoresis electrospray ionization high-resolution mass spectrometry (CE-ESI-HRMS). Mol Cell Proteomics. 2016;15(8):2756–68. doi: 10.1074/mcp.M115.057760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi SB, Zamarbide M, Manzini MC, Nemes P. Tapered-tip capillary electrophoresis nano-electrospray ionization nass spectrometry for ultrasensitive proteomics: The mouse cortex. J Am Soc Mass Spectrom. 2017;28(4):597–607. doi: 10.1007/s13361-016-1532-8. [DOI] [PubMed] [Google Scholar]
- 44.Onjiko RM, Moody SA, Nemes P. Single-cell mass spectrometry reveals small molecules that affect cell fates in the 16-cell embryo. Proc Natl Acad Sci U S A. 2015;112(21):6545–50. doi: 10.1073/pnas.1423682112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Onjiko RM, Morris SE, Moody SA, Nemes P. Single-cell mass spectrometry with multi-solvent extraction identifies metabolic differences between left and right blastomeres in the 8-cell frog (Xenopus) embryo. Analyst. 2016;141(12):3648–56. doi: 10.1039/c6an00200e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Onjiko RM, Plotnick DO, Moody SA, Nemes P. Metabolic comparison of dorsal versus ventral cells directly in the live 8-cell frog embryo by microprobe single-cell CE-ESI-MS. Anal Methods. 2017;9(34):4964–70. doi: 10.1039/c7ay00834a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Onjiko RM, Portero EP, Moody SA, Nemes P. In situ microprobe single-cell capillary electrophoresis mass spectrometry: Metabolic reorganization in single differentiating cells in the live vertebrate (Xenopus laevis) embryo. Anal Chem. 2017;89(13):7069–76. doi: 10.1021/acs.analchem.7b00880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Onjiko RM, Portero EP, Moody SA, Nemes P. Microprobe capillary electrophoresis mass spectrometry for single-cell metabolomics in live frog (Xenopus laevis) embryos. J Vis Exp. 2017(130):e56956. doi: 10.3791/56956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lombard-Banek C, Portero EP, Onjiko RM, Nemes P. New-generation mass spectrometry expands the toolbox of cell and developmental biology. Genesis. 2017;55(1–2). doi: 10.1002/dvg.23012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nemes P, Rubakhin SS, Aerts JT, Sweedler JV. Qualitative and quantitative metabolomic investigation of single neurons by capillary electrophoresis electrospray ionization mass spectrometry. Nat Protoc. 2013;8(4):783–99. doi: 10.1038/nprot.2013.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi SB, Lombard-Banek C, Munoz-Llancao P, Manzini MC, Nemes P. Enhanced peptide detection toward single-neuron proteomics by reversed-phase fractionation capillary electrophoresis mass sspectrometry. J Am Soc Mass Spectrom. 2018;29(5):913–22. doi: 10.1007/s13361-017-1838-1. [DOI] [PubMed] [Google Scholar]
- 52.Aerts JT, Louis KR, Crandall SR, Govindaiah G, Cox CL, Sweedler JV. Patch clamp electrophysiology and capillary electrophoresis-mass spectrometry metabolomics for single cell characterization. Anal Chem. 2014;86(6):3203–8. doi: 10.1021/ac500168d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parkin MC, Wei H, O’Callaghan JP, Kennedy RT. Sample-dependent effects on the neuropeptidome detected in rat brain tissue preparations by capillary liquid chromatography with tandem mass spectrometry. Anal Chem. 2005;77(19):6331–8. doi: 10.1021/ac050712d. [DOI] [PubMed] [Google Scholar]
- 54.Rubakhin SS, Page JS, Monroe BR, Sweedler JV. Analysis of cellular release using capillary electrophoresis and matrix assisted laser desorption/ionization-time of flight-mass spectrometry. Electrophoresis. 2001;22(17):3752–8. doi:. [DOI] [PubMed] [Google Scholar]
- 55.Zhang ZC, Jia CX, Li LJ. Neuropeptide analysis with liquid chromatography-capillary electrophoresis-mass spectrometric imaging. J Sep Sci. 2012;35(14):1779–84. doi: 10.1002/jssc.201200051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Culman J, Hohle S, Qadri F, Edling O, Blume A, Lebrun C et al. Angiotensin as neuromodulator/neurotransmitter in central control of body-fluid and elctrolyte homeostasis. Clin Exp Hypertens. 1995;17(1–2):281–93. doi: 10.3109/10641969509087071. [DOI] [PubMed] [Google Scholar]
- 57.Reis LC, Saad WA, Camargo LAA, Menani JV, Silveira JEN, Saad WA. Inhibitory effect of DUP-753 on the drinking responses of rats to central administration of noradrenaline and angiotensin II and to dehydration. Braz J Med Biol Res. 1996;29(4):507–10. [PubMed] [Google Scholar]
- 58.Abdelaal AE, Mercer PF, Mogenson GJ. Plasma angiotensin II levels and water intake following beta-adrenergic stimulation, hypovolemia, cellular dehydration and water deprivation. Pharmacol Biochem Behav. 1976;4(3):317–21. doi: 10.1016/0091-3057(76)90248-3. [DOI] [PubMed] [Google Scholar]
- 59.Bekkevold CM, Robertson KL, Reinhard MK, Battles AH, Rowland NE. Dehydration parameters and standards for laboratory mice. J Am Assoc Lab Anim Sci. 2013;52(3):233–9. [PMC free article] [PubMed] [Google Scholar]
- 60.Paxinos G, Franklin KBJ, Franklin KBJ. The mouse brain in stereotaxic coordinates. 2nd ed. San Diego: Academic Press; 2001. [Google Scholar]
- 61.Sun LL, Zhu GJ, Zhao YM, Yan XJ, Mou S, Dovichi NJ. Ultrasensitive and fast bottom-up analysis of femtogram amounts of complex proteome digests. Angew Chem Int Ed. 2013;52(51):13661–4. doi: 10.1002/anie.201308139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–8. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang CX, Thormann W. Head-column field-amplified sample stacking in binary system capillary electrophoresis: A robust approach providing over 1000-fold sensitivity enhancement. Anal Chem. 1996;68(15):2523–32. doi: 10.1021/ac951250e. [DOI] [PubMed] [Google Scholar]
- 64.Reiher W, Shirras C, Kahnt J, Baumeister S, Isaac RE, Wegener C. Peptidomics and peptide hormone processing in the drosophila midgut. J Proteome Res. 2011;10(4):1881–92. doi: 10.1021/pr101116g. [DOI] [PubMed] [Google Scholar]
- 65.Jia CX, Lietz CB, Ye H, Hui LM, Yu Q, Yoo S et al. A multi-scale strategy for discovery of novel endogenous neuropeptides in the crustacean nervous system. J Proteomics. 2013;91:1–12. doi: 10.1016/j.jprot.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abdelaal AE, Mercer PF, Mogenson GJ. Plasma angiotensin II-levels and water intake following beta-adrenergic stimulation, hypovolemia, cellular dehydration and water deprivation. Pharmacol Biochem Behav. 1976;4(3):317–21. doi: 10.1016/0091-3057(76)90248-3. [DOI] [PubMed] [Google Scholar]
- 67.Thornton SN. Thirst and hydration: Physiology and consequences of dysfunction. Physiol Behav. 2010;100(1):15–21. doi: 10.1016/j.physbeh.2010.02.026. [DOI] [PubMed] [Google Scholar]