Abstract
Background
Vancomycin is a prevalent cause of the severe hypersensitivity syndrome drug reaction with eosinophilia and systemic symptoms (DRESS) which leads to significant morbidity and mortality and commonly occurs in the setting of combination antibiotic therapy which impacts future treatment choices. Variations in human leukocyte antigen (HLA) class I in particular have been associated with serious T-cell mediated adverse drug reactions which has led to preventive screening strategies for some drugs.
Objective
To determine if variation in the HLA region is associated with vancomycin-induced DRESS.
Methods
Probable vancomycin DRESS cases were matched 1:2 with tolerant controls based on sex, race, and age using BioVU, Vanderbilt’s deidentified electronic health record database. Associations between DRESS and carriage of HLA class I and II alleles were assessed by conditional logistic regression. An extended sample set from BioVU was utilized to conduct a time-to-event analysis of those exposed to vancomycin with and without the identified HLA risk allele.
Results
Twenty-three individuals met inclusion criteria for vancomycin-associated DRESS. 19/23 (82.6%) cases carried HLA-A*32:01 compared to 0/46 (0%) of the matched vancomycin tolerant controls (p=1×10−8) and 6.3% of the BioVU population (n=54,249) (p=2×10−16). Time-to-event analysis of DRESS development during vancomycin treatment among the HLA-A*32:01 positive group indicated that 19.2% developed DRESS and did so within four weeks.
Conclusions
HLA-A*32:01 is strongly associated with vancomycin DRESS in a population of predominantly European ancestry. HLA-A*32:01 testing could improve antibiotic safety, help implicate vancomycin as the causal drug and preserve future treatment options with co-administered antibiotics.
Clinical Implications
HLA-A*32:01 testing to help preempt and implicate vancomycin as the causal drug for DRESS could improve the safety and efficacy of antibiotic treatment.
Keywords: vancomycin, drug reaction with eosinophilia and systemic symptoms, human leukocyte antigen, antibiotic allergy, delayed hypersensitivity, T-cell hypersensitivity
Capsule Summary
In a population of predominantly European ancestry, we show that carriage of the HLA-A*32:01 allele is strongly associated with the development of vancomycin DRESS. HLA-A*32:01 testing could increase the safety of vancomycin treatment.
Graphical Abstract
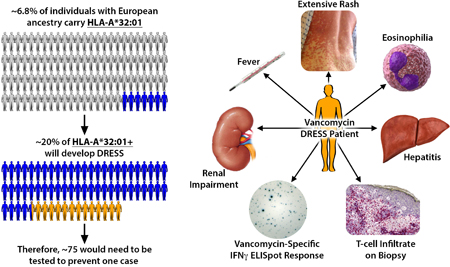
INTRODUCTION
Vancomycin is a widely used antibiotic of global importance for the treatment of serious, deep-seated, antibiotic-resistant Gram-positive infections which frequently require prolonged treatment courses. Worldwide the use of vancomycin is increasing because of the increasing incidence of methicillin-resistant Staphylococcus aureus infections. Vancomycin is associated with infusional pruritus and rash (“red man syndrome”) which is managed by slow infusion and anti-histamines. However, vancomycin is also a very common cause of a life-threatening delayed T-cell mediated reaction known as drug reaction with eosinophilia and systemic symptoms (DRESS) and has been implicated in up to 40% of antibiotic-related cases1,2. DRESS, otherwise known as drug-induced hypersensitivity syndrome, typically develops 2–8 weeks after drug initiation and presents with features including fever, a widespread rash, facial edema, white cell abnormalities, and involvement of internal organs such as the liver, kidneys, heart and lungs3. The mortality of DRESS is 1–10% and long-term morbidity such as autoimmune disease has been described up to 4 years following acute disease4,5. When DRESS develops in the setting of combination antibiotics and other co-administered drugs, all treatment is stopped and future exposure to all concurrently-dosed drugs is contraindicated due to the associated risks of morbidity and mortality if DRESS reoccurs and the inability to implicate any one drug on clinical grounds alone. The ability to more definitively diagnose DRESS associated with vancomycin may allow patients not only to avoid the current and future risk of vancomycin exposure but also to continue or resume therapy, particularly with other falsely implicated antibiotics.
METHODS
Vancomycin DRESS cases
Retrospective patients were detected using Vanderbilt’s BioVU repository, a deidentified electronic health record (EHR) database linked to a DNA biobank in operation since February 7, 2007. Prospective patients with potential vancomycin DRESS were enrolled to confirm genetic findings from the BioVU analysis using vancomycin-specific immunological studies to support clinical diagnoses. Patients were prospectively recruited between 2010 and 2018 through drug allergy clinics and inpatient facilities at participating institutions (Vanderbilt University Medical Center in Nashville (Tennessee, USA), Austin Health, Peter MacCallum Cancer Centre, and Alfred Health in Melbourne (Victoria, Australia), Fiona Stanley Hospital and Royal Perth Hospital in Perth (Western Australia, Australia)). Institutional review board (IRB) approvals were in place for the BioVU study and for all sites contributing to the prospective study. All aspects of the study including the collection and storage of DNA, plasma, peripheral blood mononuclear cells (PBMCs) and skin were IRB-approved and all patients provided written or electronic informed consent. Saliva and blood were routinely collected from prospective patients, processed and stored as repositories of DNA, PBMCs and plasma. Patients >17 years of age who were diagnosed with DRESS with vancomycin identified as a primary implicated drug, a corresponding Naranjo adverse drug reaction score of ≥5 (probable adverse drug reaction), a RegiSCAR score of ≥4 (probable DRESS) and available DNA or genotyping were included in the study6,7. Any potential duplicate cases between the prospective patients and the BioVU cohort were eliminated by an observer blinded to identifiable patient information.
Vancomycin tolerant controls
Controls from the BioVU genotyped cohort (n=54,249) were defined as individuals who tolerated intravenous vancomycin for greater than 5 weeks and had at least five vancomycin therapeutic trough levels over the treatment period recorded in the Vanderbilt EHR. 297/54,249 individuals were prescribed at least 5 weeks of vancomycin treatment. Using this subset, controls were matched 2:1 with cases on sex, race and age within five years. Vancomycin tolerance and length of treatment was verified by manual review of the EHR by a reviewer blinded to the HLA results. Additional controls for vancomycin Enzyme-Linked ImmunoSpot (ELISpot) assays were recruited from our Vanderbilt IRB-approved studies to investigate drug responses in individuals with a broad range of immune-mediated adverse drug reactions and healthy volunteers.
Human leukocyte antigen (HLA) typing
High resolution four-digit HLA A B C DP DR DQ typing was performed using either sequence-based typing on 454FLX or Illumina Miseq8,9 or imputed from SNP data from HumanExome BeadChip and GWAS platforms by Expanded Multi-Ethnic Genotyping Array (MEGAEX, Illumina), HumanOmni-Quad, HumanOmni5-Quad and Human660W-Quad using SNP2HLA as previously described10. Imputation for HLA-A*32:01 using SNP2HLA has a reported accuracy of 99.46%10. Associations between DRESS and carriage of HLA-A/B/C/DRB1/DQA1/DQB1/DPB1 alleles at the 4-digit level were assessed by conditional logistic regression to accommodate the matching. Analyses were carried out in R version 3.4.3. (R Core Team (2017)). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/).
Enzyme-Linked ImmunoSpot (ELISpot) assays
Overnight IFN-γ ELISpot assays were performed in triplicate (Mabtech Kit 3420–2H) as previously described11–13 using negative (unstimulated) and positive (anti-CD3 Mabtech antibody and/or Staphylococcal enterotoxin B (SEB)) controls. PBMCs plated at 200,000 cells per well were incubated with pharmacy stock vancomycin and other implicated drugs at concentrations representative of peak serum concentrations (Cmax) as well as 10-fold higher and 10-fold lower than Cmax. As supported by consensus in the literature, a positive response was defined as >50 spot forming units (SFU)/million cells after background removal11–13. Figures representing ELISpot results were generated using GraphPad Prism 7.0a Macintosh Version, GraphPad Software, La Jolla California USA, www.graphpad.com.
Time-to-Event Analysis of Vancomycin-Exposed BioVU Cohort
In the BioVU cohort of 54,249 patients with available genotyping, we identified 137 patients that were HLA-A*32:01 positive and 1,672 who did not carry HLA-A*32:01 and for whom at least two weeks of intravenous vancomycin treatment was intended. 137 of the 1,672 HLA-A*32:01 negative individuals were randomly selected to serve as an equal-sized control group. The deidentified EHRs of the 274 patients in both sub-cohorts were reviewed during the period of vancomycin exposure to determine patient sex, race, age, longest treatment period, development of an adverse drug reaction (ADR) and specifically, the development of possible DRESS. Since DRESS is an immune-mediated reaction and vancomycin is renally cleared, patient immunosuppression, chronic renal failure and end stage renal failure with dialysis were documented as potential covariates. ADR latency, defined as the length of time from initiation of vancomycin to symptom onset, as well as any concurrent antimicrobials were documented. Analyses were carried out by Fisher’s exact tests, logistic and Cox regression as appropriate in R version 3.4.3.
Skin Testing and Histopathology
Intradermal skin testing (IDT) with 0.05, 0.5, 5 and 50 mg/mL of sterile pharmacy grade vancomycin was performed with readings at 20 minutes, 24 and 48 hours on two subjects: patient 18, a prospectively enrolled patient who had experienced probable vancomycin DRESS 6.5 months earlier and C50, an HLA-A*32:01 positive, vancomycin-naïve healthy control. For patient 18, histopathology was examined from the acute DRESS reaction and from a biopsy of the positive 5 mg/ml vancomycin delayed IDT. Formalin-fixed, paraffin-embedded skin biopsies were sectioned at 5 µm intervals. Slides were deparaffinized and stained with hematoxylin and eosin (H&E). For the immunohistochemistry (IHC), slides were placed on the Leica Bond Max IHC stainer and deparaffinized. Slides were incubated with anti-FOXP3 (Cat.14–4777-82, eBioscience, Inc.) for one hour at a 1:100 dilution, Ready-To-Use anti-CD4 (PA0427, Leica) for one hour, or Ready-To-Use anti-CD8 (MM39–10, StatLab) for 15 mins. The Bond Polymer Refine detection system was used for visualization. A dermatopathologist scored all slides.
Molecular Docking of Vancomycin with HLA-A*32:01
Sequences of HLA-A*32:01 and HLA-A*29:02 were obtained from the HLA/IGMT database (http://www.ebi.ac.uk/ipd/imgt/hla/allele.html). An atomic homology model for HLA-A*32:01 was generated with SWISS-MODELLER14 based on the most closely related crystal structure, PDB 6EI2, 92 % identical. To generate a peptide/HLA-A*32:01 complex model, the peptide from the crystal structure of 6EI2 was positioned into the antigen binding cleft of the HLA-A*32:01 model using SSM in the COOT program package, then mutated to RLYGKSLYSF, a peptide eluted from HLA-A*32:0115. The peptide/HLA-A*32:01 complex model was then geometry minimized using PHENIX16.
Vancomycin was docked into the HLA-A*32:01 model with AutoDock Vina17. The scoring grid dimensions were 40 × 40 × 40 Å, centered on a site corresponding to the Cα of the fifth peptide amino acid (P5). Vancomycin was docked with exhaustiveness set to 40. The top nine scoring orientations were output and compared. PyMol was used to generate molecular graphics (The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC.).
RESULTS
Baseline demographics
Twenty-three individuals were identified that met inclusion criteria for clinically diagnosed vancomycin-associated DRESS in our study including 15 prospectively recruited patients (7 from Vanderbilt University Medical Center in Nashville, 5 from Melbourne, and 3 from Perth) and 8 retrospectively ascertained individuals from Vanderbilt’s BioVU repository (Tables 1 and S1). The patient cohort was primarily of European ancestry and included 9 women and 14 men from 17 to 76 years of age who developed DRESS between 2004 and 2018. Only one patient was identified by a blinded observer that overlapped between the Vanderbilt retrospectively identified BioVU cohort and the prospectively collected patients and the duplicate was eliminated. All patients had Naranjo adverse drug reaction scores of 8 to 10 (probable or definite adverse drug reaction) and RegiSCAR scores of 4 to 7 (probable or definite DRESS). 21/23 (91%) patients were being treated with other antibiotics concurrently with vancomycin. The median latency period from vancomycin initiation to the first symptoms of DRESS was 21 days (mean, 22.9 days; range, 14 – 50 days) (Table 1). Age, race and sex matching was successful (Table S2) and indications for vancomycin treatment were similar between cases and controls (Tables 1 and S3). Similar to the DRESS cases who had a median vancomycin trough of 16 µg/mL (mean, 17.2 µg/mL; range, 3 – 44 µg/mL; n = 116), the tolerant controls had a median vancomycin trough of 18 µg/mL (mean, 19.6 µg/mL; range, 2 – 86 µg/mL; n = 644).
Table 1. Summary of case basic demographics, clinical characteristics, HLA risk allele carriage and DRESS history.
Legend: ID, patient identification; Age, age at time of vancomycin treatment; Latency, days between vancomycin initiation and symptoms development; HLA, human leukocyte antigen; Trough, last vancomycin trough level before development of hypersensitivity symptoms in µg/mL. F, female; M, male; W, Caucasian; W/H, Caucasian/Hispanic; B, African American; N/A, not available; MRSA, Methicillin-resistant Staphylococcus aureus; MRSE, Methicillin-resistant Staphylococcus epidermidis; AML, acute myeloid leukemia; E. coli, Escherichia coli.
| ID | Age | Sex | Race | Latency | RegiSCAR | Naranjo | HLA-A*32: 01 | Trough | Indication for vancomycin | Other implicated medications |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 56 | F | W | 18 | 5 | 8 | Positive | 27 | MRSA cervical spine post-operative wound infection | rifampin |
| 2 | 52 | M | W | 26 | 6 | 8 | Positive | 9 | Culture negative post-operative soft tissue infection of right foot | ceftriaxone, ciprofloxacin |
| 3 | 59 | M | W | 19 | 4 | 8 | Positive | 14 | Gram-positive cocci right chronic calcaneal osteomyelitis | ciprofloxacin |
| 4 | 66 | M | W | 21 | 4 | 8 | Positive | 3 | MRSA osteomyelitis | rifampin |
| 5 | 27 | M | W/H | 29 | 5 | 8 | Positive | 21 | Lumbar spine osteomyelitis | isoniazid, rifampin,ethambutol pyrazinamide |
| 6 | 33 | M | W | 28 | 5 | 8 | Positive | 14 | MRSE-infected mesh post bariatric surgery abdominal repai | None |
| 7 | 52 | M | W | 17 | 6 | 8 | Positive | 16 | Epidural abscess and osteomyelitis with Gram-positive bacteremia | ceftriaxone |
| 8 | 48 | M | B | 21 | 5 | 8 | Negative | 12 | Cellulitis post exploratory laparotomy and inguinal hernia repair | piperacillin-tazobactam |
| 9 | 53 | F | W | 23 | 7 | 8 | Positive | 7 | Culture negative soft tissue infection with underlying rib osteomyelitis | levofloxacin, fluconazole |
| 10 | 29 | M | W | 26 | 5 | 8 | Positive | 19 | Traumatic arm injury and possible osteomyelitis | trimethoprim/sulfa methoxazole, piperacillin-tazobactam, ciprofloxacin |
| 11 | 58 | M | W | 36 | 6 | 8 | Positive | 22 | Culture negative osteomyelitis | ciprofloxacin |
| 12 | 62 | M | W | 21 | 4 | 8 | Negative | 21 | Implantable cardioverter defibrillator pocket infection | None |
| 13 | 17 | M | W | 29 | 4 | 8 | Positive | 10 | MRSA bacteremia secondary to right pelvic myositis and phlebitis | ibuprofen, hydroxyzine |
| 14 | 51 | F | W | 14 | 6 | 8 | Positive | 23 | Enterococcus faecium bacteremia | ceftazidime |
| 15 | 57 | M | W | 15 | 5 | 8 | Negative | 24 | Culture negative febrile neutropenia and neutropenic colitis during AML treatment | meropenem |
| 16 | 24 | F | W | 14 | 6 | 10 | Positive | 16 | MRSE and Bacteroides fragilis post-operative wound infection following caesarean section and supracervical hysterectomy for uterine necrosis | azithromycin, clindamycin, gentamicin, piperacillin-tazobactam, amoxicillin,meropenem, metronidazole |
| 17 | 38 | M | W | 18 | 5 | 8 | Positive | 14 | MRSA right chest phlegmon, deep soft tissue infection, underlying osteomyelitis of 2nd rib with fracture | rifampin |
| 18 | 47 | F | W | 17 | 6 | 8 | Positive | 13 | MRSA and E. coli bacteremia w/ chest infiltrate | levofloxacin |
| 19 | 76 | M | W | 27 | 6 | 8 | Positive | 12 | Enterococcus bacteremia and endocarditis | gentamicin, benzylpenicillin |
| 20 | 61 | F | W | 50 | 5 | 8 | Positive | N/A | MRSA wound infection leading to hip prosthesis removal and placement of vancomycin spacer | ciprofloxacin |
| 21 | 71 | F | W | 28 | 5 | 8 | Negative | 16 | MRSA and E. coli hardware infection post knee replacement | ceftriaxone, rifampin |
| 22 | 40 | F | W | 15 | 4 | 8 | Positive | 17 | E. coli and Enterococcus faecalis urosepsis during pregnancy | ceftriaxone |
| 23 | 47 | F | W | 15 | 5 | 8 | Positive | 22 | Staphylococcus epidermis ventriculitis | ceftriaxone |
HLA Associations with DRESS
The HLA-A*32:01 allele was carried by 19/23 (86%) DRESS cases compared with 0/46 (0%) of the matched vancomycin tolerant controls (p=1×10−8, conditional logistic with Bonferroni multiple comparisons correction) (Tables 1, S3, S4 and S5 and Figure 1A). After adjusting for HLA-A*32:01 carriage, no other alleles were significant (p=0.13). In a larger BioVU cohort of DNA samples from 54,249 Vanderbilt patients, the HLA-A*32:01 allele carriage rate was 6.30% which matches the carriage rate in other cohorts of predominant European ancestry18,19. Carriage of HLA-A*32:01 in our BioVU cohort (n= 54,249) was more prevalent in European Americans (6.78%) than African American populations (2.78%).
Figure 1. HLA-A*32:01 is strongly associated with vancomycin DRESS.

A. 19/23 (83%) DRESS cases carried HLA-A*32:01 compared with 0/46 (0%) of the matched vancomycin tolerant controls (p=1×10–8, conditional logistic). If analyses are restricted to immunologically confirmed cases, then 11/12 (92%) vancomycin ELISpot positive patients carried HLA-A*32:01 compared with 0/24 (0%) of the BioVU matched controls (p=9×10–7, conditional logistic). HLA-A*32:01 carriage in all identified vancomycin DRESS cases and immunologically confirmed cases was also very significantly overrepresented compared to HLA-A*32:01 carriage in the entire BioVU cohort (6.3%) (p=2×10–16 and p=2=7×10–13 respectively, exact binomial tests). There was no significant difference in HLA-A*32:01 carriage between the vancomycin tolerant populations and the BioVU cohort (p=0.12 for all controls, p=0.40 for controls matched to immunologically confirmed cases, exact binomial tests). Additionally, there was no significant difference in HLA-A*32:01 carriage between the immunologically confirmed vancomycin DRESS cases and those that were not immunologically confirmed (p=0.32, Fisher’s exact test). All analyses shown included Bonferroni correction for multiple comparisons. B. IFN-γ release ELISpot results after 18-hour incubation with vancomycin at concentrations of 250 µg/mL (grey) or 500 µg/mL (black) using peripheral blood mononuclear cells from vancomycin DRESS patients. Controls included cells from vancomycin-naïve, HLA-A*32:01 positive healthy donors (n = 3) including the son of case patient 18 and the vancomycin skin test negative control C50, an HLA-A*32:01 positive individual tolerant of 4 weeks of vancomycin (n = 1), patients who had developed a non-DRESS immune-mediated adverse reaction to vancomycin (n = 5) and non-HLA matched healthy donors (n = 4). Means of the replicates are plotted. In patients with multiple blood draws at time points distant from the reaction, ELISpot results from the first blood draw are plotted. 12/14 (85.7%) DRESS cases had a positive vancomycin ELISpot compared to none of the controls (p=0.005 (DRESS vs. HLA-A-32:01 positive controls), p=0.002 (DRESS vs. non-DRESS ADRs), p=0.005 (DRESS vs. non-HLA matched healthy donors)). Positive results are those above the dotted line intersecting the y-axis at 50 SFU/million cells. Differences in proportion of positive responses between groups were assessed using Fisher’s exact tests. Patient and control PBMCs were also stimulated with vancomycin at concentrations of 5 µg/mL and 50 µg/mL and exhibited a dose-dependent response (data not shown). Legend: HLA, human leukocyte antigen; Vanc, vancomycin; DRESS, drug reaction with eosinophilia and systemic symptoms; SFU, spot-forming units; IM-ADR, immune-mediated adverse drug reaction.
IFN-γ ELISpot Responses in DRESS
IFN-γ ELISpot assays were performed on all prospectively enrolled cases for which cryopreserved PBMCs were available (14/15). PBMCs from 12/14 (86%) DRESS cases had a positive IFN-γ ELISpot response to vancomycin (Figure 1B). Analyses restricted to immunologically confirmed cases and matched controls, revealed that 11/12 (92%) IFN-γ ELISpot positive patients carried HLA-A*32:01 compared with 0/24 (0%) of the matched controls (p=9×10−7, conditional logistic) (Figure 1A). Three IFN-γ ELISpot positive patients had multiple blood draws at time points distant from the initial reaction with repeat positive results (Figure S1). In Patient 4, a positive IFN-γ ELISpot to vancomycin was demonstrated 9 years after the initial DRESS reaction. In samples with sufficient cell numbers, PBMCs were routinely tested against all other concurrently administered medications potentially implicated in DRESS development (Figure S2). Notably, patient 21, one of the two patients with a negative vancomycin IFN-γ ELISpot, is HLA-A*32:01 negative and demonstrated a reproducible positive IFN-γ ELISpot to rifampin stimulation leading us to conclude that her DRESS syndrome was associated with rifampin. Thirteen controls were tested concurrently with cases and none demonstrated a positive vancomycin IFN-γ ELISpot (Figure 1B and Table S6).
Time to DRESS Analysis of the Vancomycin-Exposed BioVU Cohort
While at least two weeks of vancomycin was intended in all patients, 22/137 (16%) HLA-A*32:01 positive and 18/137 (13%) HLA-A*32:01 negative patients completed <1 week of vancomycin therapy. Possible and definitive DRESS cases in the HLA-A*32:01 carriers occurred after one week to four weeks of vancomycin therapy and the estimated probability of developing DRESS was 19.2% at four weeks (Figures 2 and S3). The median time to DRESS symptoms after vancomycin initiation was 18 days in this cohort. In comparison, none of the 119 HLA-A*32:01 negative individuals who were exposed to at least one week of uninterrupted vancomycin treatment developed DRESS or symptoms suggestive of DRESS (p=6×10−5). Development of non-DRESS ADRs did not differ between risk allele positive and negative groups (p=0.35). Within the HLA-A*32:01 positive group, when considered jointly by logistic regression with DRESS as outcome, hemodialysis (p=0.03) and immunosuppression (p=0.04) were both protective factors against DRESS development. Among the DRESS cases, 2/13 (15%) had either hemodialysis or immunosuppression compared with 64/124 (52%) carrying HLA-A*32:01 who tolerated vancomycin (p=0.02). Notably, 18 HLA-A*32:01 positive individuals tolerated vancomycin for ≥5 weeks. This demonstrates that not all HLA-A*32:01 positive individuals will develop DRESS after prolonged vancomycin treatment.
Figure 2. Time-to-event analysis demonstrating that only HLA-A*32:01 positive patients developed DRESS.
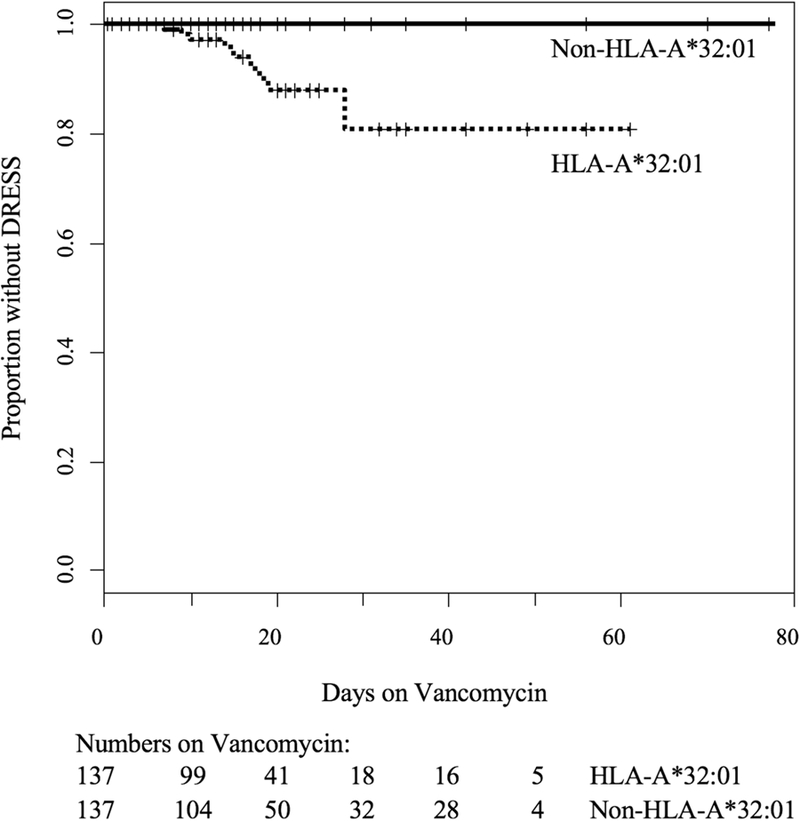
Kaplan-Meier estimates were used to determine time to DRESS or possible DRESS development during vancomycin treatment stratified by carriage of HLA-A*32:01. Cases of DRESS occurred in HLA-A*32:01 positive subjects between 1 and 4 weeks of vancomycin therapy but not in HLA-A*32:01 negative subjects. The estimated risk of DRESS prior to 4 weeks of treatment was 19.2% in those carrying the HLA-A*32:01 allele.
Skin testing, oral rechallenge, and skin histology results
Vancomycin intradermal testing (IDT) produced strong immediate histamine responses at 20 minutes in both HLA-A*32:01 positive individuals who were tested, but only the patient with a history of DRESS developed a delayed positive IDT with dermal induration and erythema at vancomycin concentrations of 0.5, 5 and 50 mg/mL recorded 24 and 48 hours after drug placement (Figure 3A and 3B). In addition, DRESS patient 18 had negative immediate testing, delayed IDT and oral challenge to levofloxacin which had been co-administered with vancomycin. H&E staining from skin biopsies obtained from patient 18 from the acute DRESS reaction and the 5 mg/ml vancomycin positive IDT skin test showed the papillary dermal edema, epidermal spongiosis and dense lymphocytic infiltrate classically seen in DRESS histology (Figure 3C and 3D). Immunohistochemistry of these same biopsies showed no appreciable difference in the distribution of CD4 and CD8 positive cells in the dermal infiltrate between the acute and skin test biopsies. The acute biopsy did, however, demonstrate a substantially higher number of intraepidermal CD8+ T cells when compared to the skin test biopsy. Conversely, dermal FOXP3+ T regulatory cells were present in the skin test biopsy but absent in the acute biopsy (Figure 3E).
Figure 3. Vancomycin skin testing, acute DRESS skin eruption and skin biopsy histology.

A and B. Vancomycin intradermal testing (IDT) results in patient 18 approximately 6.5 months after developing vancomycin DRESS and control C50, an HLA-A*32:01 positive, vancomycin-naïve healthy donor. IDT was performed on the volar forearm of the skin with 0.02 mL of vancomycin at concentrations of 0.05, 0.5, 5 and 50 mg/mL. The positive histamine and negative saline controls worked as expected. Vancomycin produced a strong immediate histamine response at 20 minutes in both control C50 and patient 18, but only patient 18 with a history of HLA-A*32:01 positive DRESS developed a concentration dependent induration of the skin at 48 hours at the 0.5, 5 and 50 mg/ml concentrations. Additionally, patient 18 had negative IDT to levofloxacin (not shown) and was successfully rechallenged with levofloxacin, a drug that, at the time of reaction, was administered with vancomycin. C. A representative example of the skin eruption from patient 18 during acute vancomycin DRESS. She had a diffuse morbilliform exanthema with facial involvement and facial edema (not shown). D. Hematoxylin and eosin staining of punch biopsies of skin from patient 18. Acute DRESS histology from a skin biopsy taken three days following onset of symptoms (upper panel) and skin test histology from the 5 mg/ml vancomycin positive intradermal skin test at 48 hours (lower panels) demonstrate papillary dermal edema, epidermal spongiosis and a dense lymphocytic infiltrate. Rare eosinophils are present on both biopsies. The skin test histology mirrors the results from the acute biopsy. E. Immunohistochemistry of T-cell subsets from acute DRESS and positive skin test biopsies from patient 18. Healthy skin is shown for comparison (upper panel). There was no discernable difference in the distribution of CD4 and CD8 positive cells in the dermal infiltrate between the acute (middle panel) and skin test (lower panel) biopsies. The number of intraepidermal CD8 positive cells was substantially higher in the acute biopsy. There was no appreciable exocytosis of CD4 T cells in the acute biopsy or CD4 or CD8 T cells in the biopsy from vancomycin IDT. Notably, dermal FOXP3+, regulatory T cells were present in the skin test biopsy, but absent in the acute biopsy.
Molecular Docking of Vancomycin with HLA-A*32:01
We used molecular docking to estimate potential interactions between vancomycin and HLA-A*32:01. We generated a homology model of peptide/HLA-A*32:01 complex based on the most similar solved structure (PDB 6EI2, HLA-A68, 92 % identical) and used AutoDock Vina to predict binding orientations and scores. Vancomycin was not predicted to bind HLA-A*32:01 with high affinity when peptide occupied the antigen binding cleft (∆G=−7.3 kcal/mole) (RLYGKSLYSF, corresponding to a peptide eluted from HLA-A*32:01). However, vancomycin was predicted to bind the antigen binding cleft of HLA-A*32:01 with higher affinity in the absence of peptide, ∆G=−7.7 kcal/mole (Figure 4). These data suggest that vancomycin has the potential to bind within the antigen binding cleft of HLA-A*32:01 in the absence of peptides that conform to the canonical HLA-A*32:01 binding motif (9mer P1 K or R, PΩ F, I or L)20. Vancomycin was predicted to bind antigen binding cleft residues in HLA-A*32:01 that differ between closely related alleles not associated with vancomycin induced DRESS, such as HLA-A*29:02 (polymorphic differences shown in magenta in Figure 5.9). Since the on-target mechanism of action for vancomycin is binding D-Ala-D-Ala in the bacterial cell wall, we asked if vancomycin has the potential to bind consecutive alanine residues in HLA-A*32:01. Molecular docking suggests that vancomycin is not likely to bind consecutive alanine residues (L isomers) in HLA-A*32:01. The top scoring molecular docking orientation shows that the vancomycin atoms contacting D-Ala-D-Ala were predicted to bind HLA-A*32:01 in the central region of the cleft normally occupied by the central positions in peptide backbone (shown in cyan in Figure 4).
Figure 4. Molecular docking prediction of vancomycin binding HLA-A*32:01.

Vancomycin is shown as sticks, white for carbon, blue for nitrogen, red for oxygen. Vancomycin atoms that mediate intermolecular contacts with D-Ala-D-Ala (in PDB 1FVM) are shown in cyan. A homology model of HLA-A*32:01 is shown in yellow as a ribbon diagram. Polymorphic positions that distinguish the associated HLA-A*32:01 allele from the closely related HLA-A*29:02 allele are shown in magenta. Intermolecular contacts between vancomycin and HLA-A*32:01 predicted by molecular docking are shown as black dashes.
DISCUSSION
The international implementation of routine pre-prescription screening for HLA-B*57:01 has eliminated abacavir hypersensitivity as a clinical entity and has paved the way for the translation of other HLA screening strategies for the prevention of drug hypersensitivity reactions into clinical practice8,9,21,22. Similar to the discovery of abacavir and HLA-B*57:01, our study highlights the utility of using large clinical databases and prospectively defined cases in combination with adjunctive immunological information to define genetic associations with a specific clinical phenotype8,9,23. Since vancomycin is frequently prescribed empirically in an urgent manner for acute life-threatening infections, and since DRESS typically takes 2 weeks to occur, unlike previous models that suggest HLA screening prior to intended prescription of a drug, use of HLA-A*32:01 typing may be more appropriate following initiation of vancomycin, when bacterial culture information is available, and in patients destined to receive longer or multiple treatment courses to identify those that could be at risk for vancomycin DRESS. This would be facilitated through the development of a single allele assay for HLA-A*32:01, similar to approaches developed for HLA-B*57:01 and HLA-B*15:02, which are now widely available through commercial laboratories with short turnaround times. Since certain HLA alleles are known to influence the natural history of some infections, one potential limitation of this study is that we were not able to match controls based on indication for vancomycin treatment. However, given that HLA-A*32:01 was not represented at all in our matched tolerant controls despite the good distribution of almost identical Gram positive and mixed infections and similar host risk factors and co-morbidities that led to the intent for prolonged vancomycin treatment in both vancomycin DRESS cases and tolerant controls, we feel confident that this finding is not a disease association.
Our time-to-event analysis suggests that the risk of DRESS approaches 20% at four weeks of therapy in those carrying HLA-A*32:01 (Figures 2 and S3). Based on this analysis and the prevalence of HLA-A*32:01 of approximately 6.8% in individuals of European ancestry, we can estimate that approximately 70 patients started on vancomycin would need to undergo HLA-A*32:01 testing to prevent or preempt one case of vancomycin DRESS, which is a favorable ratio compared with other well-defined HLA-drug associations where HLA testing is used in clinical practice3.
Ex vivo and in vivo diagnostic approaches such as IFNγ ELISpot assays and IDT warrant further study for their sensitivity, specificity and safety for vancomycin and concurrently administered medications24. Additionally, if these techniques were to become routine in the diagnosis of vancomycin DRESS, standardized IFNγ ELISpot assays would need to be commercially available for clinical laboratories. In patient 18, evidence of a localized DRESS reaction on histopathology from a positive IDT biopsy demonstrates that the immunopathology of the acute reaction can be recapitulated in the skin following disease recovery. Consistent with previous studies showing that the ratio of FOXP3+ T cells to overall CD3+ T cells in acute DRESS skin positively correlates with longer times from start of symptoms to skin biopsy, we observed an increase is FOXP3+ regulatory T cells in the dermis of recovery phase skin following intradermal vancomycin administration (Figure 3E)25. This also suggests that regulatory T cells may reside in the skin weeks to months following acute DRESS. While these immunohistopathologic results are compelling, they are from a single patient and require further study in additional patients with vancomycin DRESS.
Vancomycin has been associated with other ADRs including linear IgA bullous dermatosis, fixed drug eruption, acute generalized exanthematous pustulosis, and Stevens-Johnson syndrome/toxic epidermal necrolysis1,26. We have enrolled 10 individuals with non-DRESS vancomycin immune-mediated adverse drug reactions in our broader drug hypersensitivity studies. While the heterogeneity and small number of patients limits our ability to rule out other HLA associations with non-DRESS vancomycin-induced reactions, only 1/10 is HLA-A*32:01 positive. These HLA typing results suggest that this association is specific for vancomycin DRESS and that HLA screening would not prevent other vancomycin-induced delayed hypersensitivity reactions. Our study was powered to identify a strong association between HLA-A*32:01 and vancomycin DRESS in a population of primary European ancestry and we cannot generalize at this point to non-European ancestries where HLA associations with vancomycin DRESS will need to be independently studied.
Although the specific mechanism of vancomycin DRESS is unknown, our data may provide clues to the immunopathogenesis of this syndrome. The strong association with HLA-A*32:01 supports that vancomycin DRESS is an HLA Class I-restricted, CD8+ T-cell mediated process. For the HLA-B*57:01-restricted abacavir hypersensitivity reaction, immunologically-confirmed hypersensitivity can occur as early as 1.5 days of first dosing suggesting that a pre-existing memory T-cell response may be mechanistic9. In contrast, vancomycin DRESS in ours and other studies is characterized by a long latency period (median 21 days)27. Further, HLA-A*32:01 positive individuals who have not been exposed to vancomycin were observed to have negative responses to vancomycin by both in vivo (intradermal challenge) (n=1) and ex vivo (IFNγ ELISpot) assessments (n=4). These data might suggest that vancomycin DRESS pathogenesis is dependent upon a naïve T-cell response requiring CD4+ T-cell help. Vancomycin is a large glycopeptide and is excreted unchanged in the urine. Unlike abacavir which has been shown to alter the repertoire of self-peptides presented to T cells in HLA-B*57:01 positive individuals28,29, our model suggests that vancomycin may bind within the antigen binding cleft of HLA-A*32:01 in the absence of peptides that conform to the canonical HLA-A*32:01 binding motif (Figure 4).
Currently, the use of HLA testing in clinical practice has been limited to pre-prescription screening strategies. This discovery of a strong association between HLA-A*32:01 and one of the most serious immunologically-mediated reactions associated with a commonly used antibiotic, vancomycin, raises the possibility that HLA testing could be used as a diagnostic risk stratification tool after initiation of vancomycin treatment but prior to development of vancomycin DRESS. Patients with complex and life-threatening infections commonly receive vancomycin dosed concurrently with beta-lactams or fluoroquinolone antibiotics as was noted in 21/23 (91%) of our cases. This often leads to patients with DRESS being labeled as allergic to all of these antibiotic classes, which significantly restricts current and future treatment options. In those found to be HLA-A*32:01 positive, vancomycin treatment could either be rationalized where therapeutically appropriate or continued under close clinical observation and laboratory monitoring with discontinuation of vancomycin at the first sign of early DRESS symptoms. Alternatively, for those who develop possible vancomycin-induced DRESS or who have a known history suggestive of vancomycin DRESS, HLA-A*32:01 testing could be combined with adjunctive testing such as IFNγ ELISpot to vancomycin and other co-administered drugs to improve drug causality assessment. These strategies have the immediate potential to improve patient care by improving drug safety, increasing short-term drug efficacy and reducing future constriction of antibiotic choices.
Supplementary Material
ACKNOWLEDGEMENTS
Thank you to the staff of the Division of Infectious Diseases at Vanderbilt University Medical Center for supporting this study, to Dr. Caroline A. Nelson, Dr. Ar Kar Aung and the Vanderbilt University Medical Center Department of Dermatology for helping with patient recruitment, to Dr. David E. Elder for providing skin histology slides, to Dr. Sharon Albers for assisting with skin biopsies and to Dr. Kaija Strautins, Cindy Hager, Louise Barnett, Lilanka Fernando, Rama Gangula, Dana King and Patricia Correia for laboratory assistance. Thanks to Dr. Michael Derrick and Lincoln Shade for their initial help in setting up the BioVU immunologically-mediated adverse drug reaction protocol and to Dr. Jason Karnes for his help with the VESPA cohort HLA project. Special thanks to the patients and volunteers who participated in this study.
Some of the dataset(s) used for the analyses described were obtained from Vanderbilt University Medical Center’s Synthetic Derivative and BioVU which are supported by numerous sources: institutional funding, private agencies, and federal grants. These include the NIH funded Shared Instrumentation Grant S10RR025141; and CTSA grants UL1TR002243, UL1TR000445, and UL1RR024975. Genomic data are also supported by investigator-led projects that include U01HG004798, R01NS032830, RC2GM092618, P50GM115305, U01HG006378, U19HL065962, R01HD074711; and additional funding sources listed at https://victr.vanderbilt.edu/pub/biovu/. We additionally acknowledge the Vanderbilt Translational Pathology Shared Resource supported by NCI/NIH Cancer Center Support Grant 5P30 CA68485–19.
Financial Support: This work was supported by the National Institutes of Health (F30 AI131780, P50 GM115305, T32 GM7347, P30 AI110527, R34 AI136815, R21 AI139021, T32 AI007474, K12 HL143956 and R01 AI134648), the National Centre for Infections in Cancer, the Austin Medical Research Foundation, and the National Health and Medical Research Council of Australia.
Abbreviations
- DRESS
Drug Reaction with Eosinophilia and Systemic Symptoms
- HLA
Human Leukocyte Antigen
- EHR
Electronic Health Record
- IRB
Institutional Review Board
- PBMC
Peripheral Blood Mononuclear Cell
- ELISpot
Enzyme-Linked ImmunoSpot
- SEB
Staphylococcal Enterotoxin B
- SFU
Spot Forming Units
- Cmax
maximum serum concentration
- ADR
Adverse Drug Reaction
- IDT
Intradermal Testing
- H&E
Hematoxylin and Eosin
- IHC
Immunohistochemistry
- MRSA
Methicillin-resistant Staphylococcus aureus
- MRSE
Methicillin-resistant Staphylococcus epidermidis
- AML
Acute Myeloid Leukemia
- E. coli
Escherichia coli
- ALL
Acute Lymphoblastic Leukemia
- CLL
Chronic Lymphoblastic Leukemia
- MSSA
Methicillin-sensitive Staphylococcus aureus
- HIV/AIDS
Human Immunodeficiency Virus/Acquired Immune Deficiency Syndrome
- IM-ADR
Immune-Mediated Adverse Drug Reaction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest: The authors have no conflicts of interest relevant to this manuscript. However, EP receives consulting fees from Biocryst. EP and SM receive royalties from UpToDate and have equity in IIID Pty Ltd that holds a patent for HLA-B*57:01 testing for abacavir hypersensitivity. MR serves as a consultant for Merck, Processa Pharma and aTyr Pharma and is a Deputy Editor for JAMA Dermatology.
Contributor Information
Katherine C Konvinse, Email: katherine.c.konvinse@vanderbilt.edu.
Jason A Trubiano, Email: jason.trubiano@austin.org.au.
Rebecca Pavlos, Email: rebecca.pavlos@telethonkids.org.au.
Ian James, Email: i.james@murdoch.edu.au.
Christian M Shaffer, Email: christian.m.shaffer@vumc.org.
Cosmin A Bejan, Email: adi.bejan@vumc.org.
Ryan J Schutte, Email: rschutte@pathology.ufl.edu.
David A Ostrov, Email: ostroda@pathology.ufl.edu.
Mark A Pilkinton, Email: mark.a.pilkinton@vumc.org.
Misha Rosenbach, Email: misha.rosenbach@uphs.upenn.edu.
Jeffrey P Zwerner, Email: jeffrey.p.zwerner@vumc.org.
Kristina B Williams, Email: kristina.b.williams@vumc.org.
Jack Bourke, Email: jack.bourke@health.wa.gov.au.
Patricia Martinez, Email: patricia.martinez@health.wa.gov.au.
Francois Rwandamuriye, Email: f.rwandamuriya@iiid.murdoch.edu.au.
Abha Chopra, Email: a.chopra@iiid.murdoch.edu.au.
Mark Watson, Email: m.watson@iiid.murdoch.edu.au.
Alec J Redwood, Email: alec.redwood@uwa.edu.au.
Katie D White, Email: katie.d.white@vumc.org.
Simon A Mallal, Email: s.mallal@vumc.org.
Elizabeth J Phillips, Email: elizabeth.j.phillips@vumc.org.
REFERENCES
- 1.Lin YF, Yang CH, Sindy H, Lin JY, Rosaline Hui CY, Tsai YC, et al. Severe cutaneous adverse reactions related to systemic antibiotics. Clin Infect Dis 2014;58(10):1377–85. [DOI] [PubMed] [Google Scholar]
- 2.Blumenthal KG, Peter JG, Trubiano JA, Phillips EJ. Antibiotic allergy. Lancet 2018. [DOI] [PMC free article] [PubMed]
- 3.Pavlos R, Mallal S, Phillips E. HLA and pharmacogenetics of drug hypersensitivity. Pharmacogenomics 2012;13(11):1285–306. [DOI] [PubMed] [Google Scholar]
- 4.Aota N, Shiohara T. Viral connection between drug rashes and autoimmune diseases: how autoimmune responses are generated after resolution of drug rashes. Autoimmun Rev 2009;8(6):488–94. [DOI] [PubMed] [Google Scholar]
- 5.Ghislain PD, Roujeau JC. Treatment of severe drug reactions: Stevens-Johnson syndrome, toxic epidermal necrolysis and hypersensitivity syndrome. Dermatol Online J 2002;8(1):5. [PubMed] [Google Scholar]
- 6.Kardaun SH, Sekula P, Valeyrie-Allanore L, Liss Y, Chu CY, Creamer D, et al. Drug reaction with eosinophilia and systemic symptoms (DRESS): an original multisystem adverse drug reaction. Results from the prospective RegiSCAR study. Br J Dermatol 2013;169(5):1071–80. [DOI] [PubMed] [Google Scholar]
- 7.Naranjo CA, Busto U, Sellers EM, Sandor P, Ruiz I, Roberts EA, et al. A method for estimating the probability of adverse drug reactions. Clin Pharmacol Ther 1981;30(2):239–45. [DOI] [PubMed] [Google Scholar]
- 8.Mallal S, Nolan D, Witt C, Masel G, Martin AM, Moore C, et al. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet 2002;359(9308):727–32. [DOI] [PubMed] [Google Scholar]
- 9.Mallal S, Phillips E, Carosi G, Molina JM, Workman C, Tomazic J, et al. HLA-B*5701 screening for hypersensitivity to abacavir. N Engl J Med 2008;358(6):568–79. [DOI] [PubMed] [Google Scholar]
- 10.Karnes JH, Shaffer CM, Bastarache L, Gaudieri S, Glazer AM, Steiner HE, et al. Comparison of HLA allelic imputation programs. PLoS One 2017;12(2):e0172444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keane NM, Pavlos RK, McKinnon E, Lucas A, Rive C, Blyth CC, et al. HLA Class I restricted CD8+ and Class II restricted CD4+ T cells are implicated in the pathogenesis of nevirapine hypersensitivity. AIDS 2014;28(13):1891–901. [DOI] [PubMed] [Google Scholar]
- 12.Keane NM, Roberts SG, Almeida CA, Krishnan T, Chopra A, Demaine E, et al. High-avidity, high-IFNgamma-producing CD8 T-cell responses following immune selection during HIV-1 infection. Immunol Cell Biol 2012;90(2):224–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trubiano JA, Strautins K, Redwood AJ, Pavlos R, Konvinse KC, Aung AK, et al. The Combined Utility of Ex Vivo IFN-gamma Release Enzyme-Linked ImmunoSpot Assay and In Vivo Skin Testing in Patients with Antibiotic-Associated Severe Cutaneous Adverse Reactions. J Allergy Clin Immunol Pract 2018;6(4):1287–96 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc 2009;4(1):1–13. [DOI] [PubMed] [Google Scholar]
- 15.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 2010;66(Pt 4):486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 2010;66(Pt 2):213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31(2):455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gragert L, Madbouly A, Freeman J, Maiers M. Six-locus high resolution HLA haplotype frequencies derived from mixed-resolution DNA typing for the entire US donor registry. Hum Immunol 2013;74(10):1313–20. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez-Galarza FF, Takeshita LY, Santos EJ, Kempson F, Maia MH, da Silva AL, et al. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res 2015;43(Database issue):D784–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rapin N, Hoof I, Lund O, Nielsen M. MHC motif viewer. Immunogenetics 2008;60(12):759–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung WH, Hung SI, Hong HS, Hsih MS, Yang LC, Ho HC, et al. Medical genetics: a marker for Stevens-Johnson syndrome. Nature 2004;428(6982):486. [DOI] [PubMed] [Google Scholar]
- 22.Hetherington S, Hughes AR, Mosteller M, Shortino D, Baker KL, Spreen W, et al. Genetic variations in HLA-B region and hypersensitivity reactions to abacavir. Lancet 2002;359(9312):1121–2. [DOI] [PubMed] [Google Scholar]
- 23.Martin AM, Nolan D, Gaudieri S, Almeida CA, Nolan R, James I, et al. Predisposition to abacavir hypersensitivity conferred by HLA-B*5701 and a haplotypic Hsp70-Hom variant. Proc Natl Acad Sci U S A 2004;101(12):4180–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Empedrad R, Darter AL, Earl HS, Gruchalla RS. Nonirritating intradermal skin test concentrations for commonly prescribed antibiotics. J Allergy Clin Immunol 2003;112(3):629–30. [DOI] [PubMed] [Google Scholar]
- 25.Morito H, Ogawa K, Fukumoto T, Kobayashi N, Morii T, Kasai T, et al. Increased ratio of FoxP3+ regulatory T cells/CD3+ T cells in skin lesions in drug-induced hypersensitivity syndrome/drug rash with eosinophilia and systemic symptoms. Clin Exp Dermatol 2014;39(3):284–91. [DOI] [PubMed] [Google Scholar]
- 26.Baden LA, Apovian C, Imber MJ, Dover JS. Vancomycin-induced linear IgA bullous dermatosis. Arch Dermatol 1988;124(8):1186–8. [PubMed] [Google Scholar]
- 27.Minhas JS, Wickner PG, Long AA, Banerji A, Blumenthal KG. Immune-mediated reactions to vancomycin: A systematic case review and analysis. Ann Allergy Asthma Immunol 2016;116(6):544–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Illing PT, Vivian JP, Dudek NL, Kostenko L, Chen Z, Bharadwaj M, et al. Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature 2012;486(7404):554–8. [DOI] [PubMed] [Google Scholar]
- 29.Ostrov DA, Grant BJ, Pompeu YA, Sidney J, Harndahl M, Southwood S, et al. Drug hypersensitivity caused by alteration of the MHC-presented self-peptide repertoire. Proc Natl Acad Sci U S A 2012;109(25):9959–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


