Abstract
The field of precision medicine – the possibility to accurately tailor pharmacological treatments to each specific patient – would be significantly advanced by the ability to rapidly, conveniently, and cost-effectively measure biomarkers directly at the point-of-care. Electrochemical aptamer-based (E-AB) sensors appear a promising approach to this end due to their low cost, ease of use, and good analytical performance in complex clinical samples. Thus motivated, we present here the development of an E-AB sensor for the measurement of the amino acid L-tryptophan, a diagnostic marker indicative of a number of metabolic and mental health disorders, in urine. The sensor employs a previously reported DNA aptamer able to recognize the complex formed between tryptophan and a rhodium-based amino-acid receptor. We adopted the aptamer to the EAB sensing platform by truncating it, causing it to undergo a binding-induced conformational change, modifying it with a redox-reporting methylene blue, and attaching it to an interrogating electrode. The resulting sensor is able to measure tryptophan concentrations in the micromolar range in minutes and readily discriminates between its target and other aromatic and non-aromatic amino acids. Using it we demonstrate the measurement of clinically relevant tryptophan levels in synthetic urine in a process requiring only a single dilution step. The speed and convenience with which this is achieved suggest that the E-AB platform could significantly improve the ease and frequency with which metabolic diseases are monitored.
Keywords: Amino acids detection, Electrochemical aptamer-based biosensor, DNA aptamer, Point of Care device, Metabolic diseases
Introduction
In recent years, the concept of precision medicine is inspiring the development of diagnostic technologies that can be implemented at the point of care or even employed in the home, thus improving the convenience and frequency with which health is monitored [1,2]. Towards this end, significant effort has been focused on the development of devices able to monitor drugs and biomarkers rapidly and conveniently [2,3]. Glucometers, for example, allow for the frequent measurement of glucose in finger-prick blood samples [4,5], thus dramatically improving the management of blood sugar [6]. To date, however, glucose remains one of only a small number of metabolites for which such “self-test” clinical measurements are possible [2,3]. Moreover, as is true with most of the single-step molecular measurement technologies reported to date, the glucose sensor is critically reliant on the chemical (in this case, enzymatic) reactivity of its target and thus is not generalizable to other analytes.
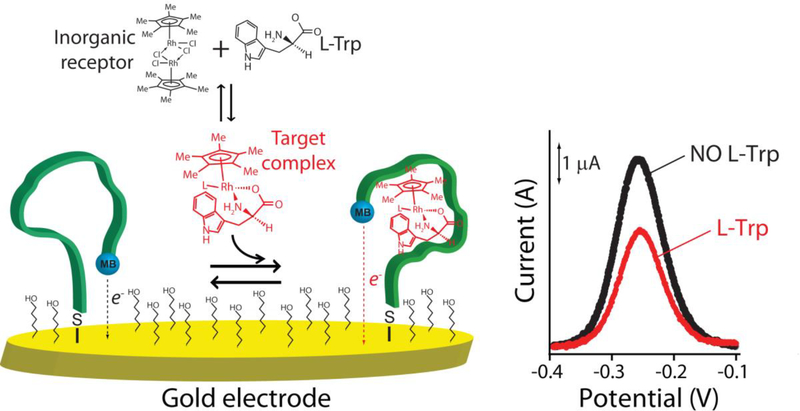
In response to the need for platforms supporting the rapid, convenient measurement of specific molecules irrespective of their chemical reactivity we are developing Electrochemical aptamer-based (E-AB) sensors, an approach that supports such measurements directly in complex clinical samples [7,8]. In this platform, an aptamer selected to specifically bind the desired target is attached to the surface of a gold electrode via a thiol group and modified with a redox reporter (here methylene blue) to support electrochemical signaling (Fig. 1, left). The binding of the target induces a conformational change in the aptamer, altering electron transfer from the redox reporter and, ultimately, resulting in a concentration-dependent change in current (Fig. 1, right). Because E-AB signaling is predicated on a binding-induced conformational change (and not simply the adsorption of the target to a surface) the platform is insensitive to the nonspecific binding of interferants and performs well even when challenged directly in, for example, undiluted serum or blood [7,8]. And because they are electrochemical, they support detection in small volumes using low-cost, hand-held electronics [7,8]. Finally, because E-AB sensors rely on the binding of aptamers, and not the chemical or enzymatic reactivity of the target, the platform is fairly general, allowing for the detection of targets ranging from proteins [9,10] to small molecules [11,12]. Given these attributes, the E-AB platform appears a promising approach to point-of-care molecular measurements [7,8]. Exploring this idea we describe here the rapid and convenient measurement of the aromatic amino acid L-tryptophan in urine using a newly developed E-AB sensor.
Fig. 1.
Electrochemical aptamer-based (E-AB) sensors generate a detectable electrochemical signal via a binding-induced conformational change in a redox-reporter-modified, electrode-bound aptamer. (Left) In the absence of the target, the aptamer is relatively unfolded and extended. Target binding induces a conformational change, leading in turn to a change in the electron transfer rate of the methylene blue reporter. (Right) Binding of the tryptophan /inorganic receptor complex produces decreases the electrochemical signal in a manner quantitatively related to the concentration of tryptophan in the solution.
Tryptophan is an essential amino acid in humans is necessary for protein synthesis but also plays key roles in many metabolic functions [13].For example, it is a precursor of the neurologically important molecules serotonin and melatonin [13]. In addition, the level of tryptophan in biological fluids has been proposed as a means of diagnosing and monitoring multiple diseases [14]. Among these are numerous inborn metabolic disorders (hypertryptophanemia, tryptophanuria [15,16] and Hartnup disease [17]), as well as emotional, cognitive and developmental disorders [18,19]. To treat these clinicians are increasingly turning to tailored dietary therapies or nutritional regimes [14,20]. The rapid and convenient measurement of urinary tryptophan levels could impact the efficacy of these treatments by supporting the rapid, frequent adjustment of therapy [20, 21]. Current methods for measuring this amino acid in clinical samples, however, which include chromatography and mass spectrometry, fail to fill this need because they required specialized personnel and expensive instrumentation and are slow (typically days) to return an answer [22]. Here, however, we describe an E-AB sensor able to measure clinical tryptophan levels rapidly and conveniently enough to support point-of-care and even in-home measurements.
Materials and Methods
Chemical reagents
Reagent-grade chemicals, including sodium hydroxide, sulfuric acid, 6-mercapto-1-hexanol, sodium chloride, ethanol, HEPES (4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid), tris(2-carboxyethyl)-phosphine (TCEP), hydrochloride tris[hydroxymethyl]- aminomethane hydrochloride, magnesium chloride, ethylenediaminetetraacetic acid (EDTA), potassium chloride, Pentamethylcyclopentadienyl rhodium(III) chloride dimer, L-tryptophan, L-phenylalanine, L-tyrosine, L-glutamine, L-histidine, L-arginine, L-alanine, sodium phosphate monobasic and artificial urine (Surine™ Negative Urine Control – composition: 95% water, with 9.3 g/L of urea, 1.87 g/L of chloride, 1.17 g/L of sodium, 0.750 g/L of potassium, 0.670 g/L of creatinine and other dissolved ions, organic and inorganic compounds [23]) (all from Sigma-Aldrich, St. Louis, Missouri, USA) were used as received.
HPLC purified oligonucleotide were purchased from Biosearch Technologies (Novato, CA, USA). The aptamer variant was modified with a thiol-C6-SS group at its 3’ end, and a methylene blue attached by a six-carbon linker to an amine at its 5’ end. The oligonucleotide was dissolved in buffer (100 mM Tris buffer, 10 mM MgCl2, pH 7.8) at a concentration of 100 μM and then aliquot and stored at −20 °C. The final concentration of the oligonucleotide was confirmed using a Beckman Coulter DU 800 UV-Vis Spectrophotometer (Männedorf, Switzerland) using a 100 μL quartz cuvette and measuring the relative absorbance at 260 nm.
The aptamer sequence we employed was a truncated version of a previously reported aptamer against this target [22]:
5’- CGCGGTAGTCTTAACCTAAAGCGGTGTCA −3’
Sensor Fabrication
Electrode Polishing and Cleaning.
The sensors were fabricated using an established approach [24]. Briefly, E-AB sensors were fabricated on rod gold disk electrodes (3.0 mm diameter, BAS, West Lafayette, IN, USA). The disk electrodes were prepared by polishing on a microcloth pad soaked before with a 1 μm diamond suspension slurry (MetaDi, Buehler, Lake Bluff, IL, USA) and then with a 0.05 μm alumina powder aqueous suspension. Each polishing step is followed by sonication of the electrodes in a solution 1:1 water/ethanol for 5 min. The electrodes were then electrochemically cleaned following this procedure: a) The electrodes are placed in a 0.5 M NaOH solution and through cyclic voltammetry (CV) 1000–2000 scans are performed using a potential between −0.4 and −1.35 V versus Ag/AgCl at a scan rate of 2 V s−1; b) The electrodes were moved to a 0.5 M H2SO4 solution and using Chronoamperometry an oxidizing potential of 2 V was applied for 5 s. After a reducing potential of −0.35 V was then applied for 10 s; c) Using CV we cycled the electrodes rapidly (4 V s−1) in the same solution between −0.35 and 1.5 V for 10 scans followed by 2 cycles recorded at 0.1 V s−1 using the same potential window.
Electrode functionalization.
We first reduced the probe DNA (100 μM) by treating it for 1 h in a solution of 10 mM tris(2-carboxyethyl)-phosphine hydrochloride (TCEP) in the dark. This was then dissolved in “assembling buffer” (10 mM Na2HPO4 with 1 M NaCl and 1 mM MgCl2 at pH 7.3) at a final concentration of 500 nM. The electrochemically cleaned gold electrodes were then immersed in 200 μL of this solution for 1 h in the dark. Following this the electrode surface was rinsed with distilled water and incubated overnight at 4°C in assembling buffer containing 5 mM 6-mercaptohexanol, followed by a further rinse with distilled water before use. Of note, we have previously explored the use of other monolayers in sensors in this class and find that six-carbon-hydroxyl-terminated monolayers exhibit excellent performance [25].
Electrochemical Experiments
Electrochemical measurements were performed at room temperature using a CHI660D potentiostat with a CHI684 Multiplexer (CH Instruments, Austin, TX) and a standard three-electrode cell containing a platinum counter electrode and a Ag/AgCl (3 M KCl) reference electrode. Square Wave Voltammetry (SWV) was performed using a potential window of −0.1 to −0.4 V, a potential step of 0.001 V and 0.05 V amplitude.
Titration curves.
Experimental titration curves (Fig. 2b and 3) were performed in 10 mL of working buffer (20 mM HEPES, 1 M NaCl, 10 mM MgCl2, 5 mM KCl, pH 7.5) using three EAB sensors modified with the oligonucleotide probe and using a SWV frequency of 500 Hz. Initially, in absence of target and rhodium-based receptor, we performed a preliminary treatment by interrogating the electrodes with 60–120 scans until a stable current peaks were obtained. Once the sensor’s signal was stable, the desired rhodium-based receptor concentration was added to the solution to reach the final concentration of 100 μM. Then, increasing concentrations of the target was added and the sensors were interrogated after 10 min. Titration curves in Fig. 4 were performed using the same experimental approach but adding the appropriate volume of artificial urine spiked with the target (30 μL and 100 μL) to achieve dilution factors of 1:333 and 1:100.
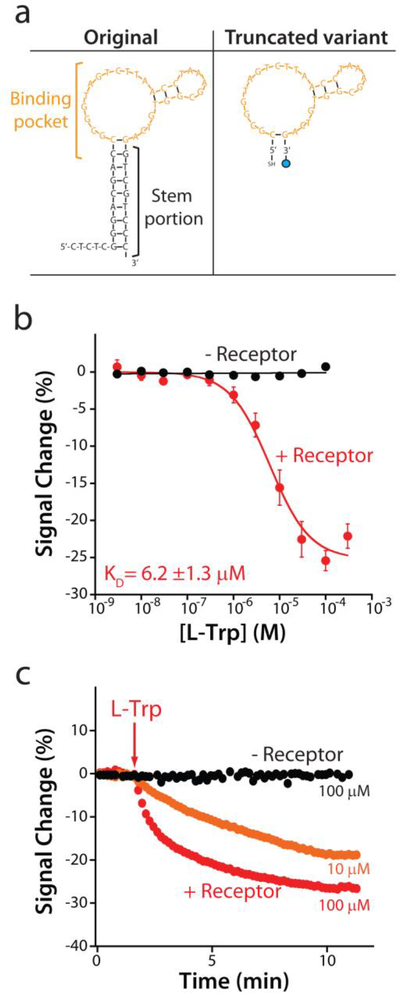
Fig. 2.
The re-engineered E-AB sensor responds sensitively and rapidly to its target. (a) To ensure that the aptamer undergoes a large-scale conformational change upon target binding we truncated the parent aptamer sequence to better populate the aptamer’s unfolded state in the absence of target. (b) A sensor fabricated using this truncated aptamer responds to increasing concentrations of tryptophan only in the presence (red curve) of the rhodium-based inorganic receptor, producing a well-defined Langmuir isotherm with a dissociation constant (KD) of 6.2 ± 1.3 μM. (c) The sensor rapidly responds to the addition of 10 or 100 μM tryptophan. The data and the error bars represent the average of three independently fabricated sensors.
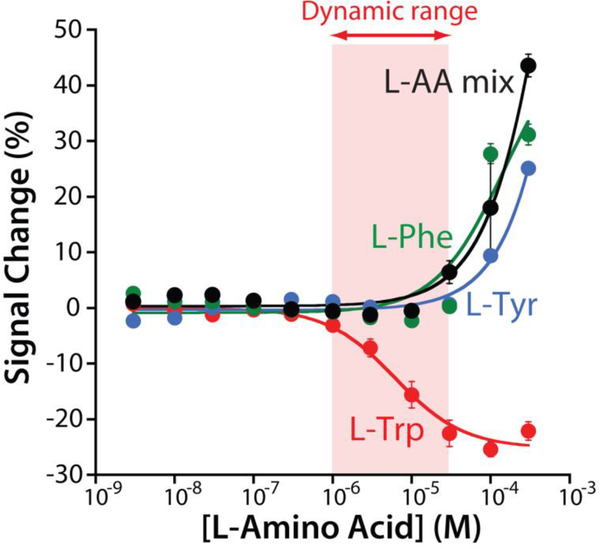
Fig. 3.
To test sensor specificity we challenged it against the other two aromatic amino acids (L-phenylalanine and L-tyrosine), as well as a mix of five amino acids (L-glutamine, L-histidine, L-proline, L-arginine, L-alanine). None of these potential interferents produces a measurable signal change below 30 μM, suggesting that none of them will interfere under realistic clinical conditions. The data and the error bars represent the average of three independently fabricated sensors.
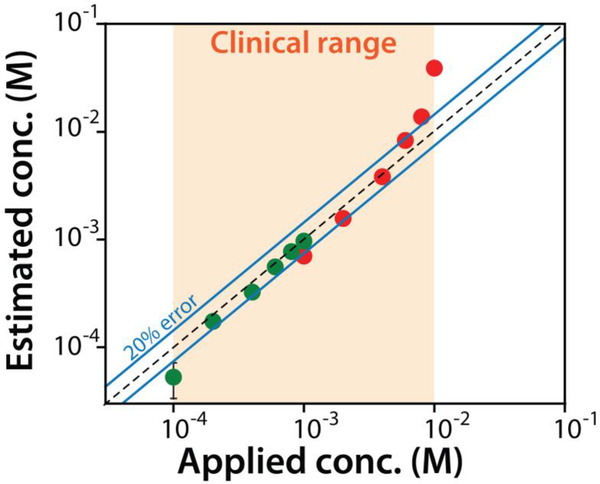
Fig. 4.
The sensor exhibits clinically relevant accuracy when challenged in synthetic urine. Using two different dilution factors of 1:333 (green) and 1:100 (red) we can detect across the entire clinically relevant range of tryptophan seen for patients with metabolic disorders. Shown are the known (spiked concentration prior to dilution) versus E-AB estimated concentrations (back calculated to correct for the dilution). The data and the error bars represent the average of three independently fabricated sensors.
The peak current was extracted for each sensor by subtracting the baseline current from the peak maxima in absence (C0) and for each concentrations of target (CTarget). We converted the signal current in signal change % (C%) using the following equation:
Where CTarget is the current peak in the presence of target and C0 is the current peak in the absence of target. The resultant data were fitted using a Langmuir equation (single-site binding mechanism) in Kaleidagraph (Synergy Software):
Where [X] is the target concentration, is the relative change in signal current in the presence of target, is the background current seen in the absence of the target, is the relative signal change seen at saturating target, and KD is the dissociation constant of the surface-bound aptamer.
Sensor equilibration time.
We determined the sensor’s equilibration time (Fig. 2c) using the above experimental approach and interrogating the sensor every 10 s in working buffer. After we achieved a stable current baseline, in presence (100 μM) or in absence of the rhodium-based receptor, we added to the solution the target and then we monitored the voltammetric signal for over 15 min. The observed signal change was fitted to a single exponential decay in Kaleidagraph (Synergy Software) to obtain the equilibration time constant of the sensor.
Results
We employed a DNA aptamer previously developed by the Stojanovic group [22], which displays high affinity and good specificity for a supramolecular complex between tryptophan and an amino-acid-binding rhodium-based inorganic receptor, pentamethylcyclopentadienyl rhodium(III) (Fig. 1). In order to adapt this into the E-AB platform we re-engineered the “original” sequence (Fig. 2a) via truncation so that, in absence of the target, the aptamer populates a non-binding, partially unfolded conformation [26]. We then modified this with a thiol group on its 5’end (to anchor the aptamer to a gold electrode) and a methylene blue redox reporter on its 3’ end. We employed this redox reporter due to its outstanding electrochemical reversibility (i.e., against repeated voltammetric scans) and stability when exposed to biological fluids [27]. The redox potential of the methylene blue is also far from the oxidation peak of tryptophan (above +0.5 V) [28], preventing interference.
When interrogated using square wave voltammetry (SWV) the re-engineered aptamer responds as expected to tryptophan (Fig. 1, left). For example, in the presence of the rhodium-based inorganic receptor (at 100 μM) and saturating target (100 μM) the sensor responds with a 25.2 ± 1.1% decrease in its electrochemical signal (Fig. 1, right). (Note: the error bars here and elsewhere in this paper represent standard deviations of multiple, interdependently fabricated devices.). This response is rapid; at 10 μM tryptophan the sensor responds with an equilibration time constant of 2.1 ± 0.1 min (Fig. 2c), a time scale reasonable for point of care applications. Titrating the sensor with increasing concentrations of tryptophan we obtain the expected Langmuir binding curve with a dissociation constant (KD) of 6.2 ± 1.3 μM (Fig. 2b, red curve). Titrations with other amino acids indicate that the sensor’s specificity is sufficient to support clinical measurements. Specifically, when challenge with the amino acids L-phenylalanine, L-tyrosine, or a solution formed by 5 different amino acids (L-glutamine, L-histidine, L-proline, L-arginine, L-alanine) (Fig. 3) we do not observe any significant signal change at concentrations below 30 μM, and thus none are expected to interfere with tryptophan under clinical sample conditions [15–17,22].
Clinical relevant urinary tryptophan ranges in healthy individuals range from 20 to 70 μM. In individuals suffering from a number of inborn metabolic disorders, this rises to 0.1 to 10 mM [15–17]. The useful dynamic range of our sensor (defined here as the range from 10% to 90% of the maximum signal change), is 0.7 μM to 40 μM, rendering it both lower than and narrower than clinical range seen in the metabolic disorders alone, thus necessitating that we perform sample dilutions (Fig. 4). Specifically, using a dilution of 1:100 we can measure urinary concentrations from 0.1 mM to 1 mM (corresponding to concentrations of 1 to 10 μM in the test solution) and using 1:333 we can measure urinary tryptophan over the range 1 mM to 10 mM (corresponding to concentrations of 3 to 30 μM in the test solution). Using this approach to characterize synthetic urine spiked with known tryptophan concentrations we find that the sensor accurately measures its target with a relevant accuracy of ±20% from 0.2 to 8 mM (Fig. 4).
Discussion
Here we describe an E-AB sensor that can rapidly (<10 min) and conveniently (simply a dilution with buffer containing the inorganic receptor) measure urinary tryptophan in urine. The sensor can measure the entire clinical range (from 0.1 to 10 mM) achieving an accuracy of 20% across the majority of this range (from 0.2 to 8 mM). Finally, the sensor also efficiently rejects interference by other amino acids that might be present in the clinical samples.
The ability for patients to self-test for metabolite levels in the urine would provide immediate information regarding their metabolic and health status. This, in turn, would open new paths to individualized, patient-centered healthcare. To this end, the miniaturazibility of E-AB sensors and potential cost-effectiveness of E-AB sensors are also significant advantages. For example, exploiting screen-printed electrodes [29], it could be possible to adapt E-AB sensors on disposable strips read by a small portable potentiostat. At the same time, the development of a user-friendly interface could help patients converting the analytical signal into useful clinical data, for example, creating a specific application for the mobile phone. The resultant convenient measurement of tryptophan levels could support the tailored diets and treatments in a patient specific manner, circumventing the issue presented by individual differences in metabolism and nutrient absorption and correcting for them in real-time. In particular this could significantly impact the health of patients unable to care for themselves, such as children and patient with mental health disorders, whose intake of tryptophan could thus be easily and rapidly monitored.
Acknowledgements
The authors wish to thank Prof. Milan Stojanovic for advice and input. This work was funded in part by a grant from Aptatek Biosciences. The authors of this work have no financial interest in Aptakek Biosciences. This work was also funded by grant EB022015 from the National Institutes of Health.
Biographies

Andrea Idili is a postdoctoral researcher at the Institute of Collaborative Biotechnologies (ICB), University of California Santa Barbara. He received his PhD degree in Analytical Chemistry under the supervision of Prof. Francesco Ricci at University of Rome Tor Vergata in 2016. His current research focuses on the design and characterization of electrochemical aptamer-based (E-AB) sensors supporting the real-time, continuous monitoring of diagnostically relevant molecules both in vitro and in vivo.

Julian Gerson is a PhD student advised by Dr Tod Kippin in the Neuroscience and Behavior division of the department of Psychological and Brain Sciences at the University of California Santa Barbara. The general theme of his research focuses on the underlying neurobiology and behaviors connected with substance abuse. Currently, his specific research focuses on developing and employing E-AB biosensors to detect small molecules associated with drugs of abuse in both in vivo and in vitro settings.

Claudio Parolo is a postdoctoral researcher working at the Department of Chemistry and Biochemistry, University of California Santa Barbara. Throughout his scientific career he has worked on the development of biosensing devices that can positively impact diagnostics in developed and developing countries. In particular during his stay in the Plaxco group he worked on biosensors that harness the binding-induced changes in biomolecular dynamics to create easy-to-use electrochemical biosensors for point-of-care clinical applications.

Tod Kippin is a Professor at the University of California, Santa Barbara in Department of Psychological & Brain Sciences and a member of the Neuroscience Research Institute. His research program focuses on the neurobiology of substance abuse. A major aim of his group’s research is to evolve our ability to resolve the dynamic neurochemical perturbations produced by drugs of abuse through the implementation of novel biosensor techniques.

Kevin Plaxco is a Professor at the University of California, Santa Barbara, with shared appointments between the Department of Chemistry and Biochemistry, the Department of Mechanical Engineering, and the Biomolecular Science and Engineering Graduate Program. Dr. Plaxco’s research focus is on the physics of biomolecular folding and its many and varied engineering applications. A major aim of the group’s applied research is to harness the speed and specificity of folding in the development of sensors, adaptable surfaces, and smart materials.
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- 1.Schork NJ. Personalized medicine: time for one-person trials. Nature 2015;520:609–611. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed MU, Saaem I, Wu PC, Brown AS. Personalized diagnostics and biosensors: a review of the biology and technology needed for personalized medicine. Crit Rev Biotechnol. 2014;34:180–196. [DOI] [PubMed] [Google Scholar]
- 3.Nayak S, Blumenfeld NR, Laksanasopin T, Sia SK. Point-of-care diagnostics: recent developments in a connected age. Anal Chem. 2016;89:102–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoedemaekers CW, Gunnewiek JMK, Prinsen MA, Willems JL, Van der Hoeven JG. Accuracy of bedside glucose measurement from three glucometers in critically ill patients. Crit Care Med. 2008:36:3062–3066. [DOI] [PubMed] [Google Scholar]
- 5.Solnica B, Naskalski JW, Sieradzki J. Analytical performance of glucometers used for routine glucose self-monitoring of diabetic patients. Clin Chim Acta. 2003;331:29–35. [DOI] [PubMed] [Google Scholar]
- 6.Clarke SF, Foster JR. A history of blood glucose meters and their role in self-monitoring of diabetes mellitus. Bri J Biomed Sci. 2012;69:83–93. [PubMed] [Google Scholar]
- 7.Lubin AA, Plaxco KW. Folding-based electrochemical biosensors: the case for responsive nucleic acid architectures. Acc Chem Res. 2010;43:496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schoukroun-Barnes LR, Macazo FC, Gutierrez B, Lottermoser J, Liu J, White RJ. Reagentless, structure-switching, electrochemical aptamer-based sensors. Annu Rev Anal Chem. 2016;9:163–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao Y, Lubin AA, Heeger AJ, Plaxco KW. Label-free electronic detection of thrombin in blood serum by using an aptamer-based sensor. Angew Chem. 2005;117:5592–5595. [DOI] [PubMed] [Google Scholar]
- 10.Lai RY, Plaxco KW, Heeger AJ. Aptamer-based electrochemical detection of picomolar platelet-derived growth factor directly in blood serum. Anal Chem. 2007;79:229–233. [DOI] [PubMed] [Google Scholar]
- 11.Rowe AA, Miller EA, Plaxco KW. Reagentless measurement of aminoglycoside antibiotics in blood serum via an electrochemical, ribonucleic acid aptamer-based biosensor. Anal Chem. 2010; 82:7090–7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker BR, Lai RY, Wood MS, Doctor EH, Heeger AJ, Plaxco KW. An electronic, aptamer-based small-molecule sensor for the rapid, label-free detection of cocaine in adulterated samples and biological fluids. J Am Chem Soc. 2006; 128:3138–3139. [DOI] [PubMed] [Google Scholar]
- 13.Palego L, Betti L, Rossi A, Giannaccini G. Tryptophan biochemistry: structural, nutritional, metabolic, and medical aspects in humans. Journal of amino acids. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kałużna-Czaplińska J, Gątarek P, Chirumbolo S, Chartrand MS, Bjørklund G. How important is tryptophan in human health? Crit Rev Food Sci Nutr. 2017; 1–17. [DOI] [PubMed] [Google Scholar]
- 15.Snedden W, Mellor CS, Martin JR. Familial hypertryptophanemia, tryptophanuria and indoleketonuria. Clin Chim Acta. 1983; 131:247–256. [DOI] [PubMed] [Google Scholar]
- 16.Martin JR, Mellor CS, Fraser FC. Familial hypertryptophanemia in two siblings. Clin Gen. 1995;47:180–183. [DOI] [PubMed] [Google Scholar]
- 17.Wilcken B, Yu JS, Brown DA. Natural history of Hartnup disease. Arch Dis Child. 1977;52:38–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindseth G, Helland B, Caspers J. The effects of dietary tryptophan on affective disorders. Arch Psychiatr Nurs. 2015;29:102–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kałuzna-Czaplinska J, Michalska M, Rynkowski J. Determination of tryptophan in urine of autistic and healthy children by gas chromatography/mass spectrometry. Med Sci Monit. 2010;16:CR488–CR492. [PubMed] [Google Scholar]
- 20.Camp KM, Lloyd-Puryear MA, Huntington KL. Nutritional treatment for inborn errors of metabolism: indications, regulations, and availability of medical foods and dietary supplements using phenylketonuria as an example. Mol Genet Metab. 2012;107:3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saudubray JM, Sedel F, Walter JH. Clinical approach to treatable inborn metabolic diseases: an introduction. J Inherit Metab Dis. 2006;29:261–274. [DOI] [PubMed] [Google Scholar]
- 22.Yang KA, Barbu M, Halim M, Pallavi P, Kim B, Kolpashchikov DM, Pecic S, Taylor S, Worgall TS, Stojanovic MN. Recognition and sensing of low-epitope targets via ternary complexes with oligonucleotides and synthetic receptors. Nat Chem.2014;6:1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Putnam DF. Composition and concentrative properties of human urine. McDonnell Douglas astronautics company; 1971. https://ntrs.nasa.gov/archive/nasa/casi.ntrs.nasa.gov/19710023044.pdf [Google Scholar]
- 24.Xiao Y, Lai RY, Plaxco KW. Preparation of electrode-immobilized, redox-modified oligonucleotides for electrochemical DNA and aptamer-based sensing. Nat. Protoc 2007; 2: 2875. [DOI] [PubMed] [Google Scholar]
- 25.Ricci F, Zari N, Caprio F, Recine S, Amine A, Moscone D, Palleschi G, Plaxco KW. Surface chemistry effects on the performance of an electrochemical DNA sensor. Bioelectrochemistry.2009;76:208–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White RJ, Rowe AA, Plaxco KW. Re-engineering aptamers to support reagentless, self-reporting electrochemical sensors. Analyst. 2010;135:589–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang D, Ricci F, White RJ, Plaxco KW. Survey of Redox-Active Moieties for Application in Multiplexed Electrochemical Biosensors. Anal. Chem 2016; 88:10452–10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Safavi A, Momeni S. Electrocatalytic oxidation of tryptophan at gold nanoparticle-modified carbon ionic liquid electrode. Electroanalysis. 2010; 22:2848–2855. [Google Scholar]
- 29.Grabowska I, Sharma N, Vasilescu A, Iancu M, Badea G, Boukherroub R., Ogale S, Szunerits S. Electrochemical Aptamer-Based Biosensors for the Detection of Cardiac Biomarkers. ACS omega. 2018;3:12010–12018. [DOI] [PMC free article] [PubMed] [Google Scholar]