Abstract
Background:
Monogenic interferonopathies are thought to be mediated by type I interferon (IFN). For example, a gain-of-function mutation in STING (STING N153S) up-regulates type I IFN-stimulated genes (ISGs) and causes perivascular inflammatory lung disease in mice. The equivalent mutation in humans also causes lung disease, which is thought to require signaling through the cGAS-STING pathway and subsequent activation of IFN regulatory factors (IRF) 3/7, type I IFN, and ISGs.
Objective:
We set out to define the roles of cGAS, IRF3, IRF7, the type I IFN receptor (IFNAR1), T cells, and B cells in spontaneous lung disease in STING N153S mice.
Methods:
STING N153S mice were crossed to animals lacking cGAS, IRF3/IRF7, IFNAR1, adaptive immunity, αβ T cells, and mature B cells. Mice were evaluated for spontaneous lung disease. Additionally, bone marrow chimeric mice were assessed for lung disease severity and survival.
Results:
Lung disease in STING N153S mice developed independently of cGAS, IRF3/IRF7, and IFNAR1. Bone marrow transplantation revealed that certain features of STING N153S-associated disease are intrinsic to the hematopoietic compartment. Rag1−/− STING N153S mice that lack adaptive immunity had no lung disease, and Tcrβ−/− STING N153S animals only developed mild disease. STING N153S led to a reduction in percent and number of naive and regulatory T cells, as well as an increased frequency of cytokine-producing effector T cells.
Conclusion:
Spontaneous lung disease in STING N153S mice develops independently of type I IFN signaling and cGAS. STING N153S relies primarily on T cells to promote lung disease in mice.
Keywords: STING, vasculopathy, interferonopathy, innate immunity, SAVI, cGAS
CAPSULE SUMMARY:
STING gain-of-function mutations cause lung disease, and type I interferon has been hypothesized to be required for disease. We report that STING-associated lung disease in mice relies on T cells but not type I interferon.
INTRODUCTION
Stimulator of Interferon Genes (STING) is an endoplasmic reticulum-associated sensor of cytosolic DNA.1, 2 Dimerization and subsequent activation of STING is triggered by cGAMP, a cyclic dinucleotide produced by cyclic GMP-AMP synthase (cGAS) upon detection of viral or aberrant host DNA3. Microbial cyclic dinucleotides also activate STING signaling4 and subsequent up-regulation of antiviral type I interferon (IFN)-stimulated genes (ISGs) through a pathway that includes IFN regulatory factors (IRFs) 3 and 7.5
STING signaling impacts both innate and adaptive immunity, and gain-of-function mutations in STING cause spontaneous autoimmune disease.6–10 However, the mechanisms of disease pathogenesis are not well understood, nor are the relative contributions of the innate and adaptive compartments. Autosomal dominant gain-of-function mutations in STING cause an autoinflammatory disease known as STING-associated vasculopathy with onset in infancy (SAVI), a pediatric disease characterized by ulcerative skin lesions, vasculopathy, lung disease, and premature death.8 Diseases caused by STING gain-of-function mutations have been regarded as interferonopathies,11 in part because ISGs are up-regulated in the patients’ peripheral blood mononuclear cells (PBMCs),8 but a contribution of type I IFN in disease pathogenesis has not been demonstrated. We previously generated heterozygous STING N153S mice that have a SAVI-associated mutation in STING.7 Lung disease in STING N153S mice is 100% penetrant and associated with premature death. Surprisingly, we found that IRF3 is not required for lung disease in STING N153S mice. Nevertheless, IRF3-independent disease does not exclude a role for type I IFN or ISGs in disease pathogenesis.7 Indeed, cGAS-STING signaling also can activate IRF7 to up-regulate production of type I IFN and ISGs.5
Beyond its role in the antiviral type I IFN response,12 STING signaling also modulates adaptive immunity. For example, STING agonists are currently in clinical development for treatment of cancer because of their ability to induce T cell priming via effects on dendritic cells and macrophages,13 and STING signaling contributes to apoptosis of T cells.9 Prior studies have suggested that STING gain-of-function mutations may exert type I IFN-independent effects on T cells. For example, we found that T cell cytopenia in STING N153S mice develops independently of IRF3,7 and a study of SAVI patient PBMCs similarly identified IRF3-independent effects on CD4+ T cell proliferation.14 In a second mouse model of SAVI (heterozygous STING V154M mice) lung disease was reported, but only in a subset of animals.6 In contrast, the STING N153S mice develop lung disease with 100% penetrance, making the STING N153S mouse model especially useful for studying the role of type I IFN and adaptive immunity in STING-associated lung disease.
To determine whether the type I IFN gene expression signature is required for STING-associated vasculopathy in mice, we crossed the STING N153S mice to cGAS−/−, Irf3−/−Irf7−/− double knockout, and Ifnar1−/− animals. We found that lung disease develops independently of cGAMP, type I IFN signaling, IRF3, and IRF7. By crossing STING N153S mice to Rag1−/− or Rag2−/−, Tcrβ−/−, and μMT−/− animals, we discovered that a combined absence of T cells and B cells completely prevents STING N153S-related lung disease. Moreover, mice lacking αβ T cells exhibited only very mild lung disease. In contrast, wild-type (WT) bone marrow transplantation (BMT) into STING N153S recipients did not protect from lethality or lung disease, suggesting a possible contribution of radio-resistant cells in disease pathogenesis. Our key discoveries of Ifnar1-independence and Rag1/2-dependence were validated in two independently generated and separately housed STING N153S mouse lines—one in St. Louis, Missouri (USA), and another in Dresden (Germany). Thus, a STING gain-of-function mutant relies on T cells but not type I IFN signaling to cause disease in mice.
METHODS
Ethics statement.
This study was performed in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the NIH. The protocols were approved by the institutional animal care and use committee at the Washington University School of Medicine (assurance number A3381-01) and the Landesdirektion Dresden, Germany. All efforts were made to minimize animal suffering.
Mice.
Heterozygous Tmem173N153S/wt mice (STING N153S mice) were previously described7 and crossed to congenic Ifnar1−/− mice15 or Irf3−/−Irf7−/− mice generated by crossing congenic Irf3−/− and Irf7−/− mice (kind gifts of T. Taniguchi (Tokyo, Japan) and provided generously by M. Diamond, Saint Louis, MO) and to Vav1-Cre16 (Vav-Cre) transgenic mice crossed to Rela fl/fl17 mice obtained from Jax (Stock numbers 008610 and 024342, respectively). STING N153S mice also were crossed to cGAS−/−,18 Rag1−/−,19 μMT−/−,20 and Tcrβ−/−,21 mice purchased from Jax (Stock numbers 026554, 002216, 002288, and 002118, respectively). Rag2−/− mice were a gift of Claudia Waskow, Dresden. The second heterozygous STING N153S knock-in mouse line in Dresden, Germany was independently generated using CRISPR/Cas9 with two distinct guide RNAs (sgRNA1; 5’-GTTAAATGTTGCCCACGGGC-3’; sgRNA2; 5’-CAGACTGCAGAGACTTCCGC-3’) and oligo donor (5’-GAGCTTGACTCCAGCGGAAGTCTCTGCAGTCTGTGAAGAAAAGAAGTTAAGT-GTTGCCCACGGGCTCGCCTGGTCATACTACATTGGGTACTTGCGGTTGA) in C57BL/6N mice purchased from Charles River (Sulzfeld, Germany). B6.CD45.1 (B6.SJL-Ptprca Pepcb/BoyJ) mice22 were housed at the Experimental Center, Technische Universität (TU) Dresden. B6.CD45.1/CD45.2 recipients were generated by crossing B6.CD45.1 mice to C57Bl/6 mice (Janvier).
Bone Marrow Chimeric Mice.
Recipient mice received a single dose of 9 Gy total body γ-irradiation (Yxlon Maxi Shot source, Hamburg, Germany). Donor cells were administered via i.v. injection into the retro-orbital sinus. Recipient mice received neomycin via drinking water (1.17 g/l) for three weeks after γ-irradiation. Each Tmem173N153S/wt (STING N153S) or Tmem173wt/wt (WT) mouse was reconstituted with 2 × 106 B6.CD45.1 whole bone marrow cells. Each B6.CD45.1/CD45.2 recipient received 50,000 lin− c-kithi cells purified from bone marrow of either STING N153S or WT donor mice. Recipient peripheral blood (PB) T-lymphocytes (CD3+), B-lymphocytes (CD19+) and neutrophils (CD11b+ Gr-1hi) were analyzed for their donor origin and Sca-1 expression using a LSRII flow cytometer (Becton-Dickinson, Heidelberg, Germany).
Cell Preparations.
For isolation of lin− c-kithi donor cells, whole bone marrow cells were hematopoietic lineage-depleted using anti-biotin microbeads (Miltenyi) according to the manufacturer’s protocol and sorted on a BD FACS ARIA III flow cytometer. Peripheral blood was drawn by retrobulbar puncture directly into EDTA-coated tubes (Sarstedt, Nuembrecht, Germany) and analyzed directly on a Sysmex XT-2000i Vet analyzer (Sysmex, Norderstedt, Germany). For chimerism analysis, blood was subjected to erythrocyte lysis, immuno-stained and analyzed by flow cytometry. Lung cell suspensions were prepared by digestion of tissue with 0.5 mg/ml Collagenase IV and 25 μg/ml DNase I (both Sigma-Aldrich, Taufkirchen, Germany) for 15 minutes and rubbing through a 100 μm cell strainer. After erythrocyte lysis in NH4Cl-buffer, cells were filtered through a 30 μm mesh.
Flow Cytometry.
Cells were incubated with antibodies in PBS / 2% FCS for 30 min, washed twice with PBS / 2% FCS and analyzed on a BD FACS LSR II flow cytometer (BD Biosciences, Heidelberg, Germany). Data were analyzed using FlowJo V9 software (Tree Star, Ashland, OR) and gates were set according to Fluorescence-Minus-One (FMO) controls. For a detailed overview of antibody clones, refer to Table E1.
Table E1.
Antibodies used in flow cytometry experiments.
| Epitope | Clone | Manufacturer |
|---|---|---|
| CD117 | 2B8 | eBioscience |
| CD11b | M1/70 | eBioscience, Biolegend |
| CD19 | eBio1D3 | 4 eBioscience, Biolegend |
| CD3e | 145–2C11, eBio500A2 | eBioscience, Biolegend |
| CD4 | GK1.5 | eBioscience, Biolegend |
| CD45.1 | A20 | eBioscience, Biolegend |
| CD45.2 | 104 | eBioscience, Biolegend |
| CD45R (B220) | RA3–6B2 | eBioscience, Biolegend |
| CD8a | 53–6.7 | eBioscience, Biolegend |
| Ly6C | AL-21 | Biolegend |
| Ly6G | 1A8 | Biolegend |
| NK1.1 | PK136 | eBioscience, Biolegend |
| Sca-1 (Ly-6A/E) | D7 | eBioscience, Biolegend |
| Ter119 | TER-119 | eBioscience |
| CD44 | IM7 | eBioscience, Biolegend |
| CD25 | PC61.5 | eBioscience, Biolegend |
| F4/80 | BM8 | eBioscience, Biolegend |
Histological Staining and Analysis.
Spleen and lung tissue were isolated from mice, fixed in 4% formaldehyde for 24 h at 4°C, and suspended in 70% ethanol before being embedded in paraffin. Tissue sections (thickness of 3 μm) were stained with hematoxylin and eosin. Whole slides were scanned by Axioscan Z1 and ZEN software (both Zeiss, Jena, Germany). Quantification of lung tissue was done with ImageJ. Lung lesions were encircled by a blinded histologist, who quantitated the number of pixels within lung lesions divided by the total number of pixels in the lung tissue, excluding large airway spaces.
Gene Expression Analysis.
RNA was isolated from tissue homogenates using RNeasy Mini Kit according to manufacturer’s instructions (Qiagen). Quantitative Real Time PCR Assays were carried out using Gotaq® qPCR MasterMix with SYBR green fluorescence (Promega A6001) or TaqMan RNA-to-Ct 1-Step kit (Applied Biosystems). Primers in SYBR green assays were generated from online Primer Bank database, https://pga.mgh.harvard/edu/primerbank/ (Table E2). Taqman assays used primers and probes purchased from Integrated DNA Technologies. ΔΔCt values were calculated and normalized with respect to each housekeeping gene.
Table E2.
Primers used in qRT-PCR experiments.
| Primer | PrimerBank ID | Sequence |
|---|---|---|
| cxcl10-for | 10946576a1 | CCAAGTGCTGCCGTCATTTTC |
| cxcl10-rev | 10946576a1 | GGCTCGCAGGGATGATTTCAA |
| ifi44-for | 19527086a1 | AACTGACTGCTCGCAATAATGT |
| ifi44-rev | 19527086a1 | GTAACACAGCAATGCCTCTTGT |
| hprt-for | 7305155a1 | TCAGTCAACGGGGGACATAAA |
| hprt-rev | 7305155a1 | GGGGCTGTACTGCTTAACCAG |
| rpl13a-for | 334688867c2 | AGCCTACCAGAAAGTTTGCTTAC |
| rpl13a-rev | 334688867c2 | GCTTCTTCTTCCGATAGTGCATC |
| eef2-for | 237858599c1 | CCGACTCCCTTGTGTGCAA |
| eef2-rev | 237858599c1 | AGTTCAGGTCGTTCTCAGAGAG |
Multiplex Cytokine Assay.
Lungs were harvested and homogenized in 500 μl of PBS. Cytokine and chemokine levels were measured using Bio-Plex Pro Mouse Cytokine Group I Panel 23-Plex Assay Kit (Bio-Rad) on a Luminex platform. Twenty-three cytokines and chemokines including IL-1α, IL-1β, IL-6, IL-10, IL-12(p40), IL-12(p70), IFN-γ, MCP-1, MIP-Iα, MIP-1β, RANTES (CCL5), and TNF-α were examined.
Western blotting.
Fifty μl of lung homogenate in PBS was mixed with 50 μl 2X RIPA buffer (CST 9806S) with protease inhibitor (Thermo Fisher 78430) and phosphatase inhibitor (Thermo Fisher 88667). Fifteen μl of lysate were fractionated by 10% SDS-PAGE (Bio-Rad) and transferred to polyvinylidene fluoride membranes (EMD Millipore). Membranes were incubated in blocking buffer (5% non-fat milk in TBS-T buffer) for 1 h. The proteins p65 and IκBα were detected with 1:1000 rabbit anti-p65 (CST D14E12) and mouse anti-IκBα antibody (CST L35A5), respectively, in blocking buffer at 4° C overnight. Membranes were washed 3 times in TBS-T and then incubated with goat anti-rabbit IgG (Invitrogen 31460) or goat anti-mouse IgG (Sigma A8924) horseradish-peroxidase-conjugated secondary antibody in blocking buffer for 1 h. All Western blots were developed using Pierce™ ECL Substrate (Thermo Fisher Scientific) and acquired with a ChemiDoc™ Touch Imaging System (Bio-Rad).
RESULTS
STING N153S causes lung disease independently of type I IFN signaling, IRF3, IRF7, and cGAS.
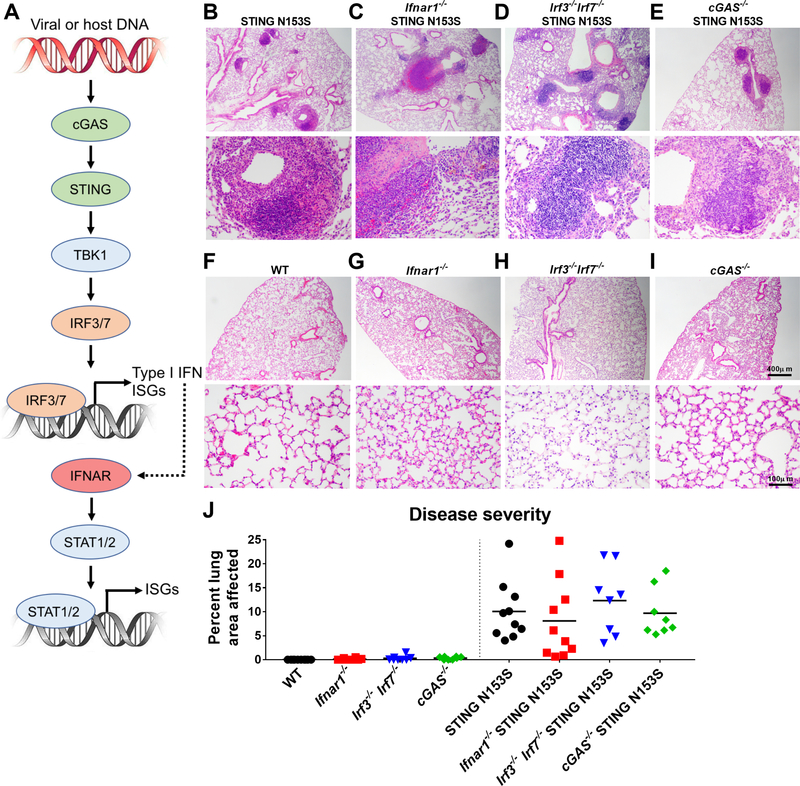
We anticipated that components of the STING and type I IFN signaling pathways would be required for lung disease pathogenesis in heterozygous STING N153S mice (Fig 1, A). To test this hypothesis, we examined histologically the lungs of STING N153S mice crossed to Ifnar1−/−, Irf3−/−Irf7−/− double knockout, and cGAS−/− animals (Fig 1, B-J). Although the STING N153S mutation causes lung disease in the absence of IRF3,7 we reasoned that STING may still mediate an interferonopathy by activating IRF7, which also triggers production of type I IFN and transcription of ISGs downstream of STING.5, 23 To address this question, we generated Ifnar1−/− STING N153S animals that lack the type I IFN receptor (IFNAR1) and discovered that the STING N153S mice develop lung disease independently of type I IFN signaling (Fig 1 B, C, and J). One hundred percent of Ifnar1−/− STING N153S mice had histologically appreciable lung disease, with no difference in the severity of lung disease in STING N153S and Ifnar1−/− STING N153S animals. These results suggest that type I IFN is not required for lung disease pathogenesis in mice. To determine whether IRF3 and IRF7 may cooperate to cause lung disease, we generated STING N153S mice lacking both of these genes (Irf3−/−Irf7−/− STING N153S mice) and found that these animals also exhibit severe lung pathology (Fig 1, D and J). Since the combined actions of IRF3 and IRF7 are required for upregulation of type I IFN downstream of STING,5 these results also are consistent with type I IFN-independence of lung disease pathogenesis in the STING N153S mice. Prior cell culture experiments using patient fibroblasts suggested that cGAMP may have a stronger effect on signaling via gain-of-function STING mutants than through WT STING.8 To determine whether cGAMP contributes to lung disease, we examined the lungs of cGAS−/− STING N153S mice and discovered that cGAS also is not required for STING-associated lung disease (Fig 1, E and J). All knockout littermate control animals that express only WT STING had histologically normal-appearing lungs (Fig 1, F-J). Thus, a STING N153S gain-of-function mutation causes lung disease in mice independently of cGAMP, IRF3, IRF7, and IFNAR1. This does not, however, exclude a role for type I IFN in SAVI in humans, although it does suggest that STING gain-of-function mutations can have type I IFN-independent effects.
FIG 1. STING N153S causes lung disease independently of type I IFN signaling, IRF3/7, and cGAS.
A, Diagram of the cGAS-STING signaling pathway and the relationships among cGAS, STING, IRFs, Type I IFN, and ISGs. B-I, Representative hematoxylin and eosin (H&E) images of lung sections from age-matched 9–21 week-old mice of the indicated genotypes, including littermate control mice. Upper panel scale bar = 400 μm. Lower panel scale bar = 100 μm. J, Quantitation of the affected lung area of mice of indicated genotype from n = 8–10 animals per genotype from two independent experiments. Histological analysis was performed by a blinded observer. Data in (J) were analyzed by Kruskal-Wallis test. P > 0.99 for the STING N153S group.
STING N153S-related T cell cytopenia and myeloid cell expansion occur independently of IFNAR1, IRF3, IRF7, and cGAS.
We previously found that SAVI-associated STING mutations cause T cell cytopenia and myeloid cell expansion independently of IRF3,7, 8 but this did not exclude a role for type I IFN and other components of the STING N153S signaling pathway. To determine whether type I IFN signaling, cGAS, or the combined activities of IRF3 and IRF7 contribute to leukocyte phenotypes in STING N153S mice, we performed flow cytometric analysis of peripheral blood leukocyte populations in STING N153S mice lacking IFNAR1, IRF3/IRF7, or cGAS, as well as the appropriate WT STING littermate animals. STING N153S had no effect on circulating B220+ B cell numbers (Fig 2, A), but consistently led to a reduction in CD4+ and CD8+ T cells (Fig 2, B and C). These effects on CD4+ and CD8+ T cell numbers occurred independently of IFNAR1, IRF3, and IRF7 (Fig 2, B and C). CD4+ but not CD8+ T cell cytopenia was independent of cGAS (Fig 2, C). These results demonstrate that IFNAR1 and IRFs downstream of STING are not required for T cell cytopenia and myeloid cell expansion in the STING N153S mice.
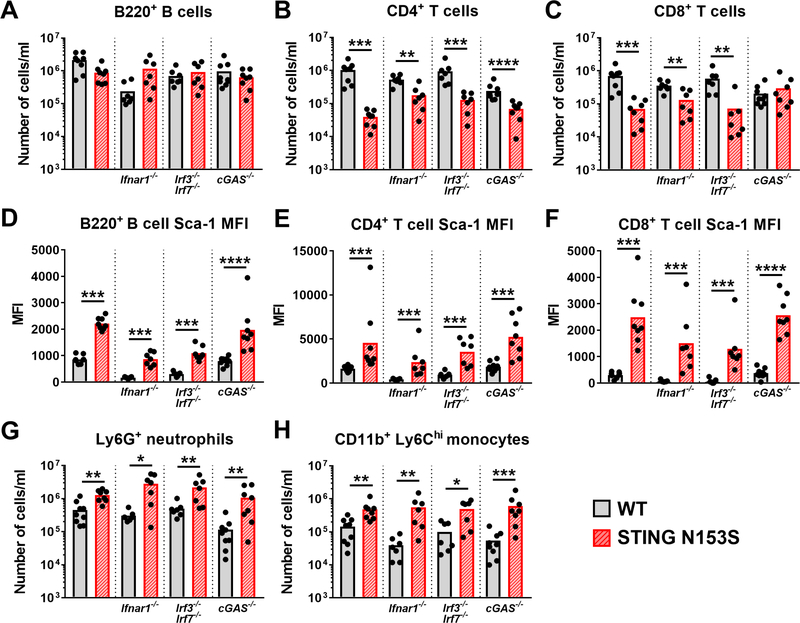
FIG 2. STING N153S-related T cell cytopenia and myeloid cell expansion occur independently of IFNAR1, IRF3, IRF7, and cGAS.
Peripheral blood was taken from age-matched STING N153S and WT littermate control mice for subsequent flow cytometric analysis of the indicated immune cell populations. A-C, Number of B220+ B cells (A), CD4+ T cells (B), and CD8+ T cells (C). D-F, Median fluorescence intensity (MFI) of Sca-1 expression on B220+ B cells (D), CD4+ T cells (E), and CD8+ T cells (F). G-H, Number of Ly6G+ neutrophils (G), and CD11b+ Ly6Chi inflammatory monocytes (H). Data in A-H represent the mean of n = 7–9 mice per indicated genotype pooled from two individual experiments. Results in A-H were analyzed by Mann-Whitney test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; **** P < 0.0001.
Sca-1 is a type I interferon-stimulated gene (ISG)24 that was up-regulated on the surface of STING N153S lymphocytes (Fig 2, D-F). Unexpectedly, we found that STING N153S causes up-regulation of Sca-1 in B cells and T cells independently of IFNAR1, IRF3, IRF7, and cGAS, suggesting type I IFN-independent up-regulation of Sca-1 by the STING N153S mutation (Fig 2, D-F). Neutrophil and inflammatory monocyte expansion in STING N153S also occurred independently of IFNAR1, IRF3, IRF7, and cGAS (Fig 2, G and H). Thus, consistent with the absence of any appreciable effect on spontaneous lung disease in STING N153S mice, these results demonstrate that IFNAR1, IRF3, IRF7, and cGAS are not required for these effects of STING N153S on hematopoietic cells.
Transplantation of WT bone marrow diminishes tissue ISG expression, but does not protect STING N153S mice from lung disease or lethality.
To begin to test the role of the hematopoietic compartment in disease pathogenesis, we generated bone marrow chimeric mice. Since STING gain-of-function mutants may intrinsically impact T cells and B cells,6, 7 we reasoned that transplantation of WT bone marrow into lethally irradiated adult (9–13 week-old) or juvenile (4-week-old) STING N153S recipients may ameliorate lung disease and prevent subsequent death. At multiple time points after transplantation, reconstitution efficiency of lymphocytes and neutrophils was measured and discovered to be more efficient in STING N153S mice than in WT recipient animals (Fig E1, A-F). In the lungs, STING N153S leukocytes were almost completely replaced by WT cells, and the diminutive STING N153S thymus remained small even after BMT (Fig E1, G and H). Additionally, we found that the type I IFN-stimulated gene Sca-1 is up-regulated in STING N153S T cells and B cells (Fig E1, I-L), but not in WT lymphocytes following transplantation into STING N153S recipients (Fig E1, M and N).
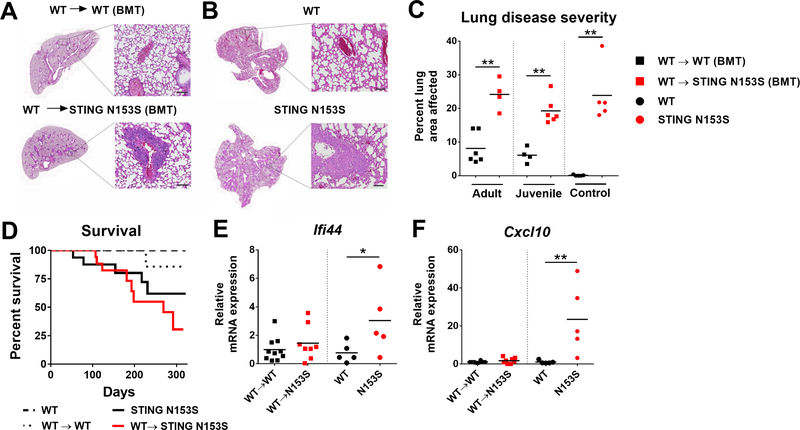
Despite efficient replacement of STING N153S leukocytes by WT cells, there was no amelioration of lung disease in these mice (Fig 3, A-C). Furthermore, in a 322-day survival study, STING N153S recipients of WT bone marrow still exhibited a high degree of lethality when compared with non-transplanted control animals (~70% lethality in STING N153S recipients compared with ~40% in non-transplanted STING N153S control animals, P > 0.05) (Fig 3, D). High expression of ISGs in the lungs of STING N153S mice was eliminated by transplantation of WT bone marrow (Fig 3, E-F). Similarly, elevated expression of ISGs in the STING N153S spleen and liver also was diminished after BMT (Fig E2, A-D). We previously found that STING N153S mice exhibit splenomegaly and aberrant splenic histopathology,7 which was ameliorated following BMT (Fig E2, E-K). Thus, despite the fact that transplantation of WT bone marrow results in diminished expression of ISGs, the STING N153S animals were not protected from lung disease and premature death.
FIG 3. Transplantation of WT bone marrow reduces ISG expression but does not protect STING N153S mice from lung disease or lethality.
A-B, Representative images of H&E-stained lung sections of adult (9–13 week-old) mice 32 weeks after bone marrow transplantation (BMT). Scale bar = 100 μm in high magnification image. C, Histological scoring of lung disease severity of adult (9–13 week-old) or juvenile (4–6 week-old) WT and STING N153S recipients of transplanted WT bone marrow, and age-matched, nonirradiated WT and STING N153S control animals. D, Survival analysis of age- and gender-matched WT, STING N153S and bone marrow chimeric mice. Animals were monitored daily for 322 days including 216 days after BMT. n = 11–17 mice per group. P > 0.05 by log-rank test for STING N153S versus transplanted STING N153S animals. E-F, Expression of ISGs Ifi44 (E) Cxcl10 (F) in the lungs of indicated mice as measured by qRT-PCR. qRT-PCR data are reported as the fold change relative to WT. n = 5–10 mice per group. Data in (C) and (E-F) were analyzed by Mann-Whitney test. *, P < 0.05; **, P < 0.01.
Transplantation of STING N153S hematopoietic stem/progenitor cells (HSPCs) into WT recipients causes weight loss and increased expression of ISGs in the lung and spleen.
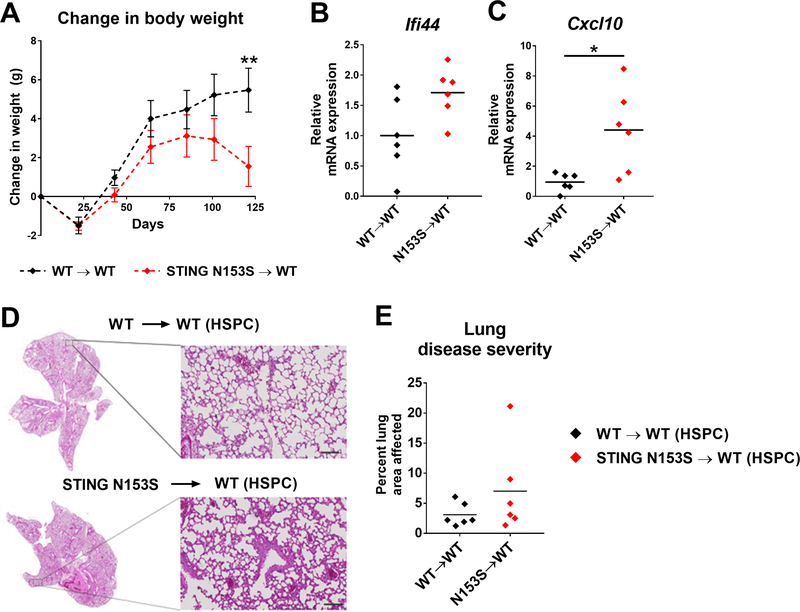
Our previous studies indicated that the lungs of STING N153S mice contain a mixture of T cells, B cells, and myeloid cells.7 We reasoned that residual radio-resistant cells (e.g., a subset of radio-resistant T cells) may be sufficient to cause lung disease in STING N153S recipients of WT bone marrow. To test this hypothesis, we transplanted equal numbers of Lin− c-Kithi HSPCs isolated from the bone marrow of either STING N153S or WT mice into lethally irradiated WT recipient animals. We found that after ~100 days, WT recipients of STING N153S HSPCs began to lose weight (Fig 4, A). Consistent with our previously published mixed bone marrow chimera studies,7 reconstitution of STING N153S lymphocytes was highly inefficient, with a ~15-fold difference in reconstitution of STING N153S donor CD3+ T cells compared to WT donor CD3+ T cells in WT recipient mice (Fig E3, A-D). Transplantation of STING N153S HSPCs was sufficient to up-regulate expression of certain ISGs (Ifi44 and Cxcl10) in the spleen (Fig E3, E-K) and Cxcl10 in the lung (Fig 4, B and C). Thus, STING N153S expression, even in a subset of leukocytes, is sufficient to up-regulate certain ISGs and to cause weight loss in mice. Histological evaluation of the lungs 120 days after transplantation revealed no significant difference in severity of lung disease between WT recipients of WT HSPCs and WT recipients of STING N153S HSPCs (Fig 4, D and E). These results suggest that STING N153S expression in the hematopoietic compartment is sufficient to up-regulate ISGs and cause weight loss, but without causing significant lung disease in the first ~3 months after HSPC transplantation. Since there was no difference between the groups, no definitive conclusion could be made in terms of whether STING N153S hematopoietic cells are sufficient to cause lung disease (Fig 4, F). Reconstitution of STING N153S T cells was highly inefficient (Fig E3, A and B), likely as a consequence of impaired capacity of STING N153S thymic progenitors to compete against radio-resistant WT progenitors.7 Thus, STING N153S T cells are sufficient to cause weight loss but not lung disease in WT recipient animals.
FIG 4. Transplantation of STING N153S hematopoietic stem/progenitor cells (HSPCs) into WT recipient mice causes weight loss and increased expression of Cxcl10 in the lung.
Lethally irradiated 10-week-old WT mice received either WT or STING N153S HSPCs isolated from 9–15 week-old mice and were monitored for 120 days. A, Weight change of WT recipient mice at 20-day intervals following HSPC transplantation. Data represent the mean ± SEM of n = 6 mice. B-C, Expression of ISGs (Ifi44 and Cxcl10) in the lungs of WT recipient mice 120 days after HSPC transplantation. D, Representative images of H&E-stained lung sections on day 120 after HSPC transplantation. Scale bar = 100 μm in high magnification image. E, Quantitation of the affected lung area of mice of indicated genotype from n = 6 animals per genotype. Data in (A) were analyzed by 2-way ANOVA. **, P < 0.01. Data in (B and C) and (F) were analyzed by Mann-Whitney. *, P < 0.05.
T cells promote lung disease pathogenesis in STING N153S mice.
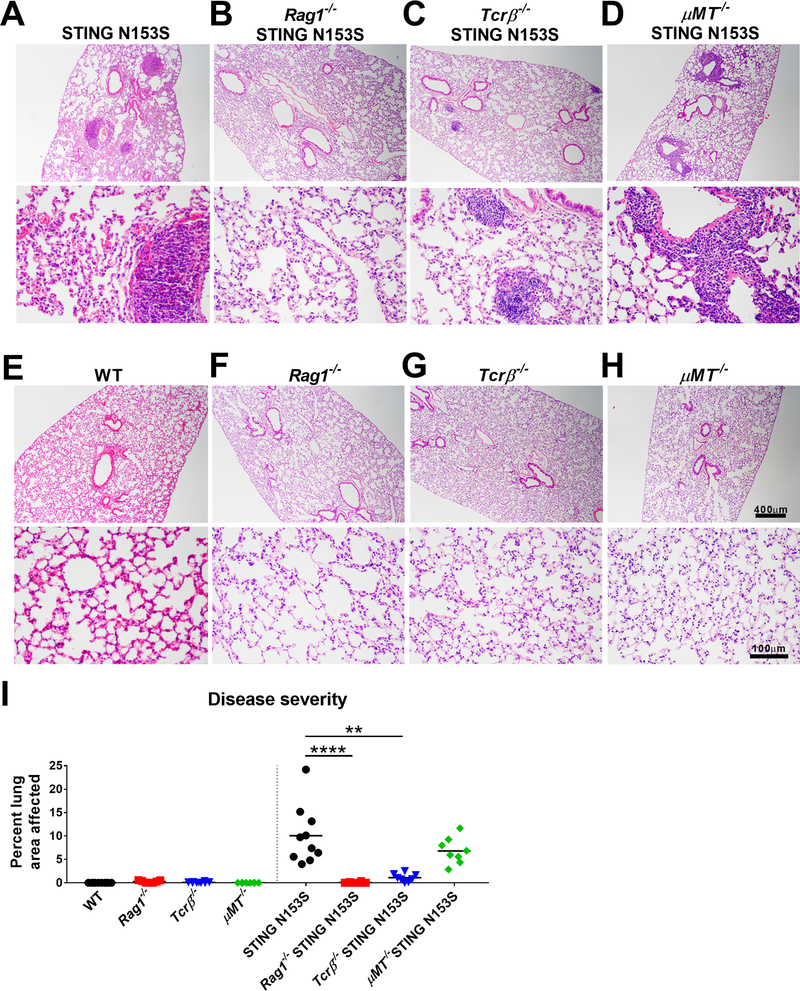
To more directly examine the role of T cells and B cells in spontaneous lung disease, we crossed our STING N153S mice to Rag1−/−, Tcrβ−/−, and μMT−/− animals. We found that 3-month-old Rag1−/− STING N153S mice, which lack T cells and mature B cells, exhibit no histological evidence of lung disease (Fig 5, A, B, and I), and that Tcrβ−/− STING N153S mice develop only very mild lung disease or no disease at all (Fig 5, C and I). Three-month-old Rag2−/− STING N153S mice also were protected from lung disease (data not shown). In contrast, μMT−/− STING N153S mice that lack mature B cells still have severe perivascular inflammation in the lung (Fig 5, D and I), indicating that mature B cells and antibodies are not required for STING N153S-induced lung pathology. None of the knockout littermate controls that express only WT STING had histologically appreciable lung disease (Fig 5, E-H).
FIG 5. T cells promote lung disease in STING N153S mice.
A-H, Representative images of H&E-stained lung sections from 9–21 week-old mice of indicated genotype. Images are representative of at least 4 tissue sections of n = 8–10 animals per genotype. Upper panel scale bar = 400 μm. Lower panel scale bar = 100 μm. I, Quantitation of the affected lung area of mice of indicated genotype from at least 4 tissue sections of n = 8–10 animals per genotype. Histological analysis was performed by a blinded observer. Data in (I) were analyzed by Kruskal-Wallis test. **, P < 0.01; ****, P < 0.0001.
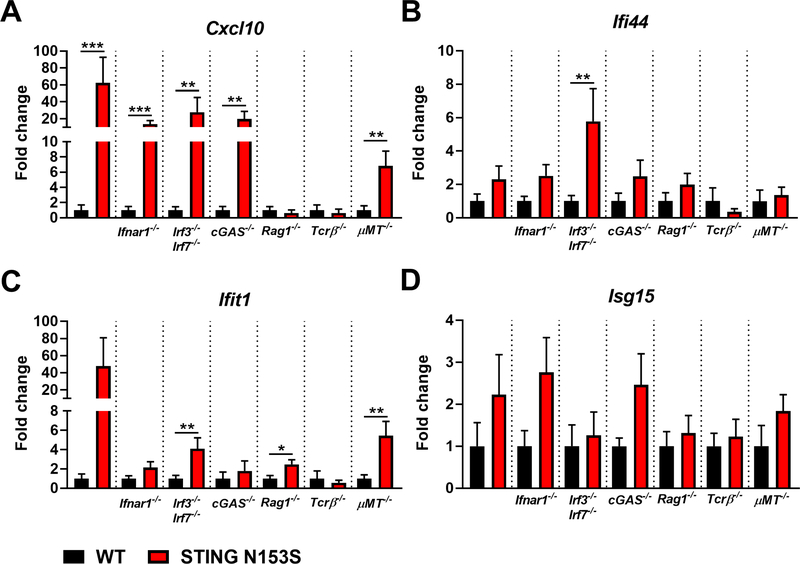
To examine the impact of innate and adaptive immune factors on ISG and cytokine expression in STING N153S mice, we measured ISG and cytokine levels in the lung homogenate from WT and STING N153S mice, as well as in corresponding WT and STING N153S animals lacking IFNAR1, IRF3/IRF7, cGAS, adaptive immunity (Rag1−/−), αβ T cells (Tcrβ−/−), or mature B cells (μMT−/−) (Fig 6 and 7). Up-regulation of Cxcl10 expression corresponded with the presence of lung disease, was not detected in Rag1−/− and Tcrβ−/− mice, and occurred in STING N153S mice lacking B cells, cGAS, IRF3, IRF7, or IFNAR1 (Fig 6, A). Thus up-regulation of Cxcl10 in the lungs of STING N153S mice requires T cells, but not B cells, type I IFN signaling, or IRFs that act downstream of STING. This effect is consistent with a previously established type I IFN-independent up-regulation of Cxcl10,25 but may also reflect infiltration of Cxcl10-expressing immune cells into the lungs of STING N153S mice. In contrast, expression levels of other ISGs (Ifi44, Ifit1, Isg15) did not correspond with the presence or absence of lung disease (Fig 6, B-D).
FIG 6. ISG expression in the lungs of STING N153S mice, WT STING littermates, and corresponding knockout animals lacking genes involved in innate and adaptive immunity.
A-D, Expression of ISGs (Ifi44, Cxcl10, Ifit1, Isg15) in the lungs of STING N153S mice crossed to Ifnar1−/−, Irf3−/− Irf7−/−, cGAS−/−, Rag1−/−, Tcrβ−/−, and μMT−/− animals and appropriate littermate control. Data represent the mean ± SEM of n = 8 mice per genotype from two independent experiments. ISG expression of indicated knockout animals and STING N153S littermates were analyzed by Mann-Whitney test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
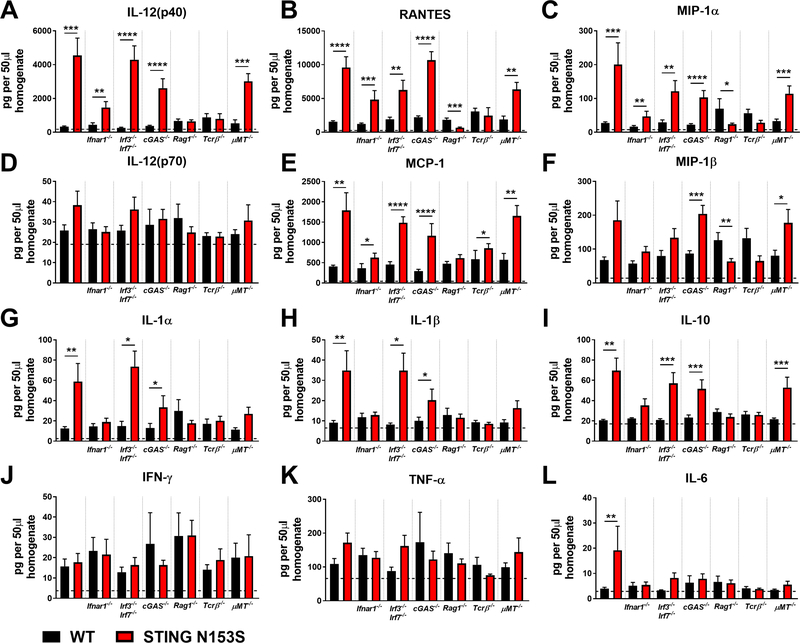
FIG 7. Chemokine and cytokine expression in the lungs of STING N153S mice, WT STING littermates, and corresponding knockout animals lacking genes contributing to innate or adaptive immunity.
A-L, Indicated chemokine and inflammatory cytokine expression in 50 μl of lung homogenate from STING N153S and WT littermate control animals, as well as STING N153S animals lacking Ifnar1−/−, Irf3−/− Irf7−/−, cGAS−/−, Rag1−/−, Tcrβ−/−, and μMT−/− animals, and the corresponding WT STING littermate controls. Data represent the mean ± SEM of n = 8 mice per genotype from two individual experiments. ISG expression of indicated knockout animals and STING N153S littermates were analyzed by Mann-Whitney test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Multiplex cytokine analysis of WT and STING N153S lung tissue revealed that IL-12(p40), RANTES, and MIP-1α were up-regulated in the lungs of STING N153S mice and genetic crosses that also had lung disease, including STING N153S mice lacking IFNAR1, IRF3, IRF7, or cGAS. Expression of other cytokines and chemokines did not consistently correlate with the presence or absence of lung disease (Fig 7, D-L). For example, IL-1α, IL-1β and IL-10 up-regulation was dependent on IFNAR1 but not on IRF3/IRF7 or cGAS, and did not consistently correlate with the presence of lung disease (Fig 7, G-I). We had previously speculated about a role for NF-κB in STING N153S-associated lung disease.7 However, cytokine analysis revealed that expression of certain pro-inflammatory cytokines (IFN-γ, TNF-α, IL-6) also did not correlate with the presence or absence of lung disease in the STING N153S mice, even in the absence of IFNAR1 or IRF3 and IRF7 (Fig. 7, J-L). Additionally, we made considerable efforts to generate STING N153S Vav-Cre Rela fl/fl mice. However, no STING N153S Vav-Cre Rela fl/fl mice were born from among 10 litters of Vav-Cre Rela fl/fl females crossed to STING N153S Rela fl/fl males, and no genetic linkage effect could explain the failure to generate STING N153S Vav-Cre Rela fl/fl animals. This suggests that STING N153S combined with Rela deletion in hematopoietic cells may result in embryonic lethality. To further examine a potential a role for NF-κB at the tissue and cellular level, we measured activation of p65 as well as IκBα expression in the lungs of WT and STING N153S mice, and we found that NF-κB activation was not increased in the lungs of STING N153S animals (Fig E4) or in mouse embryonic fibroblasts (data not shown). Thus, although we did not genetically determine whether NF-κB activation is required for disease, our data suggest that NF-κB may not play a major role in promoting lung disease pathogenesis.
In contrast to STING N153S mice and the corresponding genetic crosses that exhibited lung disease, STING N153S mice lacking αβ T cells did not express high levels of macrophage-derived cytokines or chemokines in the lung (Fig 7, A-F). This result suggests that although T cells initiate STING N153S-associated lung disease, there are likely to be combined effects of macrophages and T cells on lung disease pathogenesis.
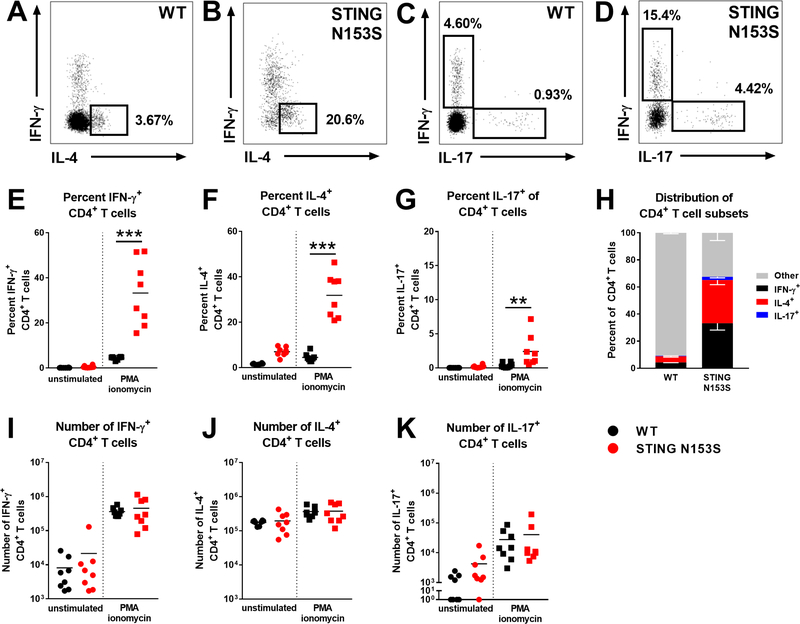
To better understand the impact of STING N153S on T cell subsets, we performed flow cytometric analysis of splenic T cell subsets and chemokine receptors. Expression of CXCR3, CX3CR1, and CCR4 was up-regulated on STING N153S CD4+ T cells, and CX3CR1 also was up-regulated on STING N153S CD8+ T cells (Fig E5). We reasoned that this may reflect increased effector cell differentiation of STING N153S T cells. Subsequent analysis of T cell subsets revealed a marked reduction in the number of naïve and effector memory CD4+ and CD8+ T cells in STING N153S animals, as well as a large increase in CD4+ and CD8+ central memory T cells. This corresponded with a trend toward diminished numbers of regulatory T cells (Tregs) in the spleens of STING N153S mice (Fig E6). We also examined splenic CD4+ T cell differentiation and discovered that the majority of STING N153S CD4+ T cells were either IFN-γ+ (Th1) or IL-4+ (Th2) cells (Fig 8). These findings are consistent with the notion that T cell dysregulation, perhaps intrinsically by STING or as a consequence of autoreactivity, may promote lung disease pathogenesis. Collectively, our results reveal that the STING N153S gain-of-function mutation promotes lung disease via effects on T cells, cytokines, and chemokines in this type I IFN-independent model of STING-mediated autoimmunity.
FIG 8. STING N153S splenic CD4+ T cells are mostly differentiated effector cells.
Splenocytes from 4-month-old WT and STING N153S mice were harvested and cultured. Some cells were treated with PMA/ionomycin for flow cytometric analysis of differentiated T cell subsets. A-D, Representative staining of intracellular IFN-γ, IL-4, and IL-17 in PMA/ionomycin-stimulated CD4+ T cells. E-G, Percent IFN-γ+ (E), IL-4+ (F), and IL-17+ (G) of total CD4+ T cells. H, Stacked bar graph of CD4+ differentiated T cell subset distribution from (E-G). IFN-γ− IL-4− IL-17− stimulated-CD4+ T cells were labeled “Other”. Data in (H) represent the mean ± SEM of populations from (E-G). I-L, Number of IFN-γ+ (I), IL-4+ (J), and IL-17+ (K) CD4+ T cells. Data in (E-G), (I-K) represent the mean of n = 8 mice per genotype from two independent experiments. Data were analyzed by Mann-Whitney test. *, P < 0.05; **, P < 0.01; ***, P < 0.005.
DISCUSSION
Although it has been speculated that STING gain-of-function mutations cause disease via type I IFN signaling, the hypothesis of type I IFN-mediated disease has never previously been confirmed in an animal model of the disease. We found that the STING N153S gain-of-function mutation does not require IRF3, IRF7, or type I IFN signaling to initiate lung disease in mice. Furthermore, we discovered that transplantation of WT bone marrow fails to prevent progression of lung disease and premature death in heterozygous STING N153S recipient animals. These results suggest that SAVI in humans might also develop independently of type I IFN and ISGs. Although this result is unexpected, it is important to underscore that our conclusions are bolstered by the fact that lung disease was found to be Ifnar1independent and Rag1/Rag2-dependent in two independently generated STING N153S mouse lines that were created in different laboratories and housed at separate institutions. More subtle effects of type I IFN on progression of lung disease or survival cannot be excluded based on our results. Although there may be species-specific effects of STING gain-of-function mutations, our study demonstrates that a SAVI-associated STING mutation can cause lung disease independently of type I IFN in mice.
Tcrβ−/− STING N153S mice that lack αβ T cells only have mild lung disease or no disease at all, and Rag1−/− STING N153S mice exhibit no histological sign of lung inflammation, suggesting that T cells play a major role in promoting lung disease in this animal model. Furthermore, we discovered that STING N153S promotes Th1, Th2, and Th17 effector cell differentiation in mice. These effects are likely type I IFN-independent. In support of this, Cerboni et. al. found that SAVI patient CD4+ T cells are impaired in proliferation because of IRF3-independent effects of another STING gain-of-function mutation.14 Whereas STING agonists can cause activation and recruitment of T cells through type I IFN-dependent priming by macrophages and dendritic cells,13 a tuberculosis vaccine that was adjuvanted by a STING agonist led type I IFN-independent immunity via effects on Th1 and Th17 cells.26 Thus, STING can exert both type I IFN-dependent and independent effects on T cells, although the precise molecular mechanisms remain incompletely defined.
JAK inhibitors have been partially effective in the treatment of SAVI.27, 28 One rationale for treatment of SAVI with JAK inhibitors is related to the idea that JAK/STAT signaling is initiated downstream of IFNAR1.29 Although treatment with the JAK inhibitor baricitinib diminished flares and stabilized interstitial lung disease in humans with SAVI, it did not eliminate the type I IFN gene expression signature.27 Additionally, JAK/STAT signaling also occurs downstream of an array of other cytokine receptors, suggesting that the effects of JAK inhibitors may occur via multiple pathways.30 Indeed, small molecular inhibitors have a variety of off-target effects, making it difficult to draw mechanistic conclusions because of a treatment response. Despite our animal model results suggesting that SAVI-associated lung disease may be initiated independently of type I IFN signaling and IRF3/7, we still cannot exclude a role for type I IFN in SAVI patients based on our mouse model findings. Indeed, the STING N153S mice are only a model and might not reflect what occurs in patients with SAVI. Our hope is that the results of our work may prompt further investigations into mechanisms of disease pathogenesis in patients, as well as the future identification of novel, highly effective therapies. If monoclonal antibodies against type I IFNs or IFNAR1 were discovered to ameliorate disease in SAVI patients, this would provide more specific and compelling evidence that type I IFN contributes to disease pathogenesis in humans.
Our multiplex cytokine and gene expression analysis indicated that certain cytokines and chemokines correlate with the presence of lung disease, including CXCL10, IL-12(p40), and RANTES. In contrast, levels of pro-inflammatory cytokines in the lung did not correlate with the presence of STING-associated lung disease. Indeed, levels of IL-1α, IL-1β, TNF-α, and IL-6 did not correspond with the presence or absence of disease. Consistent with this finding from the mouse model, drugs targeting each of these molecules were not reported to be effective in a small group of SAVI patients.8 Similarly, genetic elimination of mature B cells did not ameliorate disease in STING N153S mice, and B cell depletion with rituximab in a human patient with SAVI was not reported to be effective.8 We also considered potential involvement of the mTORC1 pathway based on our own previous work.7 However, we preliminarily found that up-regulation of this pathway was eliminated in STING N153S mice lacking IRF3, despite the fact that lung disease was still present,7 so we chose not to pursue further studies of mTORC1 in the STING N153S mice.
The fact that Rag1−/− STING N153S mice are protected from lung disease suggests that adaptive immunity might be required for initiation of lung disease pathogenesis. In contrast, BMT did not protect against spontaneous lung inflammation in STING N153S animals. This could be explained by a variety of possibilities. For example, once lung disease is initiated by STING N153S T cells at an early age, other cell types (e.g., lung endothelial cells or macrophages) may be sufficient to promote progression of lung disease as the animals age. Alternatively, a small subset of radio-resistant, autoreactive STING N153S T cells might persist and be sufficient to drive lung disease despite efficient replacement of recipient leukocytes.
The STING N153S mice develop lung disease with 100% penetrance, but another model of SAVI (STING V154M mutant mice) was recently reported to have lung disease with incomplete penetrance.6 Whether lung disease in STING V154M mice requires type I IFN was not tested, but these mice also exhibit lymphopenia that is independent of IFNAR1. Whereas the STING N153S mice have T cell cytopenia,7 the STING V154M mice have a more severe B cell deficiency, impaired T cell proliferation, and defective memory responses.6 Since we found that STING N153S promotes effector differentiation of T cells and T cell-driven lung disease in mice, the more profound defects in adaptive immunity reported in STING V154M animals may partially explain the incomplete penetrance of lung disease in that model. Alternatively, since STING also responds to bacterial cyclic dinucleotides,4 microbiome-associated differences may explain distinct phenotypes in these two animal models, but this remains to be tested.
Aside from activating the type I IFN response, the cGAS-STING pathway also affects cell senescence31 and plays a role in apoptosis.9, 32 Future studies of the STING N153S mice are likely to address these alternative functions of STING as potential mechanisms of disease pathogenesis, and experiments using precise tools (e.g., tissue-specific Cre-inducible STING N153S mice) will permit more complete identification of the immunological and cellular determinants of disease pathogenesis. If the immunological mechanisms are similar in humans and in mice with STING gain-of-function mutations, then SAVI patients may benefit from therapies that preferentially target T cells. Nevertheless, our findings from this preclinical model must be interpreted with caution, since mechanisms of disease pathogenesis may be distinct in different species.
Supplementary Material
KEY MESSAGES.
A gain-of-function STING mutation causes lung disease independently of cGAS, IRF3, IRF7, and IFNAR1, the type I IFN receptor.
WT bone marrow transplantation into STING N153S recipients failed to prevent lethality or progression of lung disease, suggesting a role for radio-resistant cells in disease pathogenesis.
Rag1−/− and Tcrβ−/− STING N153S mice were protected from disease, demonstrating that T cells play a major role in promoting STING-associated lung disease in mice.
ACKNOWLEDGMENTS
The Miner laboratory is supported by NIH grant (K08 AR070918). The Rösen-Wolff laboratory is supported by the German Research Foundation (TRR237, B18). We acknowledge the Washington University Pulmonary Morphology Core for assistance with tissue processing and staining. We thank L. Schulze, C. Haase, T. Häring and K. Höhne for technical assistance and advice. We also appreciate the following core facilities for their support: Genome Engineering Facility, Transgenic Core (both MPI-CBG Dresden) and BioDip.
ABBREVIATIONS
- STING
Stimulator of interferon genes
- cGAS
Cyclic GMP-AMP synthase
- IFN
Interferon
- ISG
Interferon-stimulated genes
- IRF
Interferon regulatory factor
- SAVI
STING-associated vasculopathy with onset in infancy
- IFNAR1
Interferon alpha and beta receptor subunit 1
- PBMC
Peripheral blood mononuclear cells
- WT
Wild-type
- BMT
Bone marrow transplantation
- HSPC
Hematopoietic stem and progenitor cell
- Rag1
Recombination activating gene-1
- Tcrβ
T-cell receptor beta chain
- Sca-1
Stem cells antigen-1
- IFI44
Interferon-gamma-inducible protein 44
- CXCL10
C-X-C motif chemokine 10
- ISG15
Interferon-stimulated gene 15
- IFIT1
Interferon-induced protein with tetratricopeptide repeats 1
Footnotes
Conflict of interest statement: The authors have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Ishikawa H, Barber GN. STING an Endoplasmic Reticulum Adaptor that Facilitates Innate Immune Signaling. Nature 2008; 455:674–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 2009; 461:788–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013; 339:786–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature 2011; 478:515–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma S, DeOliveira RB, Kalantari P, Parroche P, Goutagny N, Jiang Z, et al. Innate immune recognition of an AT-rich stem-loop DNA motif in the Plasmodium falciparum genome. Immunity 2011; 35:194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouis D, Kirstetter P, Arbogast F, Lamon D, Delgado V, Jung S, et al. Severe combined immunodeficiency in stimulator of interferon genes (STING) V154M/wild-type mice. J Allergy Clin Immunol 2018. [DOI] [PubMed] [Google Scholar]
- 7.Warner JD, Irizarry-Caro RA, Bennion BG, Ai TL, Smith AM, Miner CA, et al. STING-associated vasculopathy develops independently of IRF3 in mice. J Exp Med 2017; 214:3279–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Jesus A, Marrero B, Yang D, Ramsey S, Sanchez GAM, et al. Activated STING in a Vascular and Pulmonary Syndrome. N Engl J Med 2014; 371:507–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gulen MF, Koch U, Haag SM, Schuler F, Apetoh L, Villunger A, et al. Signalling strength determines proapoptotic functions of STING. Nat Commun 2017; 8:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melki I, Rose Y, Uggenti C, Van Eyck L, Frémond M-L, Kitabayashi N, et al. Disease-associated mutations identify a novel region in human STING necessary for the control of type I interferon signaling. Journal of Allergy and Clinical Immunology 2017. [DOI] [PubMed] [Google Scholar]
- 11.Uggenti C, Lepelley A, Crow YJ. Self-Awareness: Nucleic Acid-Driven Inflammation and the Type I Interferonopathies. Annu Rev Immunol 2019. [DOI] [PubMed] [Google Scholar]
- 12.Ma Z, Damania B. The cGAS-STING Defense Pathway and Its Counteraction by Viruses. Cell Host Microbe 2016; 19:150–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corrales L, McWhirter SM, Dubensky TW Jr., Gajewski TF. The host STING pathway at the interface of cancer and immunity. J Clin Invest 2016; 126:2404–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerboni S, Jeremiah N, Gentili M, Gehrmann U, Conrad C, Stolzenberg M-C, et al. Intrinsic antiproliferative activity of the innate sensor STING in T lymphocytes. The Journal of Experimental Medicine 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang SY, Hertzog PJ, Holland KA, Sumarsono SH, Tymms MJ, Hamilton JA, et al. A null mutation in the gene encoding a type I interferon receptor component eliminates antiproliferative and antiviral responses to interferons alpha and beta and alters macrophage responses. Proc Natl Acad Sci U S A 1995; 92:11284–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Boer J, Williams A, Skavdis G, Harker N, Coles M, Tolaini M, et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol 2003; 33:314–25. [DOI] [PubMed] [Google Scholar]
- 17.Heise N, De Silva NS, Silva K, Carette A, Simonetti G, Pasparakis M, et al. Germinal center B cell maintenance and differentiation are controlled by distinct NF-kappaB transcription factor subunits. J Exp Med 2014; 211:2103–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 2014; 505:691–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 1992; 68:869–77. [DOI] [PubMed] [Google Scholar]
- 20.Kitamura D, Roes J, Kuhn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature 1991; 350:423–6. [DOI] [PubMed] [Google Scholar]
- 21.Mombaerts P, Clarke AR, Rudnicki MA, Iacomini J, Itohara S, Lafaille JJ, et al. Mutations in T-cell antigen receptor genes alpha and beta block thymocyte development at different stages. Nature 1992; 360:225–31. [DOI] [PubMed] [Google Scholar]
- 22.Morse HC. Genetic nomenclature for loci controlling surface antigens of mouse hemopoietic cells. J. Immunol. 1992; 149:3129–34. [PubMed] [Google Scholar]
- 23.Lazear HM, Lancaster A, Wilkins C, Suthar MS, Huang A, Vick SC, et al. IRF-3, IRF-5, and IRF-7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PLoS Pathog 2013; 9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Essers MA, Offner S, Blanco-Bose WE, Waibler Z, Kalinke U, Duchosal MA, et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature 2009; 458:904–8. [DOI] [PubMed] [Google Scholar]
- 25.Brownell J, Bruckner J, Wagoner J, Thomas E, Loo YM, Gale M Jr., et al. Direct, interferon-independent activation of the CXCL10 promoter by NF-kappaB and interferon regulatory factor 3 during hepatitis C virus infection. J Virol 2014; 88:1582–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Dis E, Sogi KM, Rae CS, Sivick KE, Surh NH, Leong ML, et al. STING-Activating Adjuvants Elicit a Th17 Immune Response and Protect against Mycobacterium tuberculosis Infection. Cell Rep 2018; 23:1435–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanchez GAM, Reinhardt A, Ramsey S, Wittkowski H, Hashkes PJ, Berkun Y, et al. JAK1/2 inhibition with baricitinib in the treatment of autoinflammatory interferonopathies. J Clin Invest 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fremond ML, Rodero MP, Jeremiah N, Belot A, Jeziorski E, Duffy D, et al. Efficacy of the Janus kinase 1/2 inhibitor ruxolitinib in the treatment of vasculopathy associated with TMEM173activating mutations in 3 children. J Allergy Clin Immunol 2016; 138:1752–5. [DOI] [PubMed] [Google Scholar]
- 29.Muller M, Briscoe J, Laxton C, Guschin D, Ziemiecki A, Silvennoinen O, et al. The protein tyrosine kinase JAK1 complements defects in interferon-alpha/beta and -gamma signal transduction. Nature 1993; 366:129–35. [DOI] [PubMed] [Google Scholar]
- 30.Larner AC, David M, Feldman GM, Igarashi K, Hackett RH, Webb DS, et al. Tyrosine phosphorylation of DNA binding proteins by multiple cytokines. Science 1993; 261:1730–3. [DOI] [PubMed] [Google Scholar]
- 31.Yang H, Wang H, Ren J, Chen Q, Chen ZJ. cGAS is essential for cellular senescence. Proceedings of the National Academy of Sciences 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abe T, Barber GN. Cytosolic-DNA-Mediated, STING-Dependent Proinflammatory Gene Induction Necessitates Canonical NF-κB Activation through TBK1. J Virol 2014; 88:5328–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.