Abstract
Bile acids (BAs) play an integral role in digestion through the absorption of nutrients, emulsification of fats and fat soluble vitamins, and maintenance of cholesterol levels. Metabolic disruption, diabetes, colorectal cancer and numerous other diseases have been linked with BA disruption, making improved BA analyses essential. To date, most BA measurements are performed using liquid chromatography separations in conjunction with mass spectrometry measurements (LC-MS). However, 15–40 minute LC gradients are often used for BA analyses and these may not even be sufficient for distinguishing all the important isomers present in the human body. Ion mobility spectrometry (IMS) is a promising tool for BA evaluations due to its ability to quickly separate isomeric molecules with subtle structural differences. In this study, we utilized drift tube IMS (DTIMS) coupled with MS to characterize 56 different unlabeled BA standards and 16 deuterated versions. In the DTIMS-MS analyses of the 12 isomer groups studied, BAs with smaller m/z values were easily separated in either their deprotonated or sodiated forms (or both). However, as the BAs grew in m/z value, they became more difficult to separate with two isomer groups being inseparable. Metal ions such as copper and zinc were then added to the overlapping BAs, and due to different binding sites, the resulting complexes were separable. Thus, the rapid separation capabilities of DTIMS-MS show great potential for the many needed BA analyses.
Keywords: Ion Mobility Spectrometry, Bile Acids, Collisional Cross Sections
Introduction
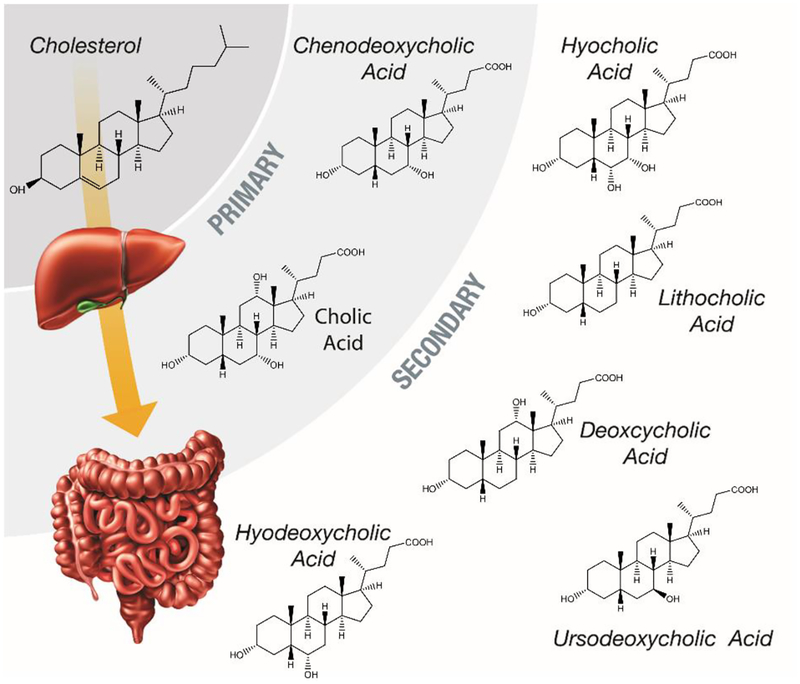
Bile acids (BAs) are components of the gastrointestinal (GI) tract that enable absorption of lipids, cholesterol, and fat-soluble vitamins, ultimately regulating the GI tract [1, 2]. BAs were first proposed as a potential tumor-promoting agent in 1939, due to their high concentrations in certain areas of the body [3]. In the last two decades, information about BA circulation during cholesterol metabolism (Figure 1) has led to new treatments for hypercholesterolemia [4, 5]. Results from these studies have also shown that BAs can be strongly cytotoxic and able to act as nuclear receptor ligands, detecting and controlling their own concentrations within the body [2, 6]. These properties have made it apparent that BAs are not only important in hepatic, biliary, and intestinal diseases, but many others [7–11]. Thus, gaining a better understanding of BA metabolic pathways is essential for determining their role in each process, especially in individuals that have disrupted BA metabolisms due to genetic predisposition, diet, and/or cancer.
Figure 1.
A schematic of the metabolic pathway that transforms cholesterol into primary and secondary bile acids through processing in the liver and the intestines. Structures of primary and secondary bile acids to illustrated to show their similarities.
Since all BAs are originally derived from cholesterol as shown in Figure 1, they share a similar four ring steroid structure connected to a carbon side chain [12]. This unique amphipathic structure, makes BAs structurally appropriate for absorbing nutrients in the digestive system [13]. However, understanding and measuring the full repertoire of BAs can be exceptionally challenging due to their structural similarities. BAs are categorized into primary and secondary groups based on their processing in the liver and intestines [6, 14, 15]. The primary BAs, cholic acid (CA) and chenodeoxycholic acid (CDCA), are the end-products of cholesterol metabolism in the liver which can be conjugated (mainly with glycine or taurine, depending on animal species) to form taurocholic acid (TCA), glycocholic acid (GCA), taurochenodeoxycholic acid (TCDCA) and glycocheno-deoxycholic (GCDCA) acid as shown in Figure 2. This conjugation creates BAs that are impermeable to cell membranes, resulting in their high concentrations in bile and intestinal content. Conjugation also plays a pivotal role in fat digestion and absorption when the BAs reach the colon via the gall bladder, bile duct, and duodenum.
Figure 2.
A schematic of the primary BA structures (left) and examples of their glycine and taurine conjugated forms (right).
In the colon, deconjugation and dehydroxylation occur through bacterial enzymes, leading to the formation of dozens of distinct but structurally similar BAs [2]. For example, the enzymatic action of the bacterial microbiota converts CA and CDCA into secondary BAs by removing the hydroxyl group from the 7th carbon atom on the molecule forming deoxycholic acid (DCA) from CA, and lithocholic acid (LCA) from CDCA. The newly formed secondary BAs then pass into the portal vein and reach the liver, where they join new primary BAs and are reconjugated with glycine or taurine and stored in the gallbladder. However, other changes can occur to the BAs. For example, some BAs such as LCA, the most toxic substance produced in the body and a known carcinogen, enter the liver where they are sulfated or esterified to glucuronic acid and excreted. This recycling of BAs is known as enterohepatic circulation and can occur 10 times every day, forming additional BAs. The numerous resulting structurally similar BAs cause great difficulties in fully understanding BA metabolic pathways, and even though they have almost identical structures, they have quite different functions [16, 17]. Therefore, new analyses are needed to fully evaluate BAs responsible for pathogenic conditions.
While BAs can be analyzed directly with mass spectrometry (MS) based techniques, often in negative ionization mode due to their many hydroxyl group, the BA structural similarities and numerous isomers require LC separation times of between 20–45 minutes for their analyses [18–25]. LC gradients between 4 and 7 minutes have also been utilized but these are often for just a subset of the BAs [26], which makes new higher throughput and great coverage approaches greatly desirable. Ion mobility spectrometry coupled with mass spectrometry (IMS-MS) is an attractive approach due to its ability to separate isomeric structures on a millisecond time scale [27, 28]. In this work, we utilized drift tube IMS (DTIMS) coupled with MS (DTIMS-MS) to characterize the sodiated and deprotonated BA structures occurring in positive and negative mode electrospray ionization (ESI). Copper and zinc metal ions were also added to the BA isomers that were difficult to separate to evaluate if the metal bound complexes could be distinguished. The results of these analyses are detailed in this manuscript, showing the utility of DTIMS-MS for rapid BA analyses.
Materials and Methods
Chemicals, Solvents and Sample preparations:
72 BA standards (56 unlabeled and 16 deuterated) were characterized in this manuscript. The 56 unlabeled BA standards were purchased from Sigma-Aldrich (St. Louis, MO) and Steraloids (Newport, RI) and the 16 deuterium labeled BAs were purchased from C/D/N Isotopes Inc. (Point-Claire, Quebec Canada). Each BA standard was dissolved in high purity methanol purchased from Thermo Fisher Scientific (Waltham, MA) to a concentration of 5 mg/mL. Prior to analysis, each BA standard was further diluted with methanol to a final concentration of 0.5 μM.
Drift tube IMS Analysis and Collision Cross Section (CCS) Measurements:
An Agilent 6560 IMS-QTOF MS (Agilent Technologies, Santa Clara) equipped with the gas kit upgrade was utilized for all of the drift tube IMS (DTIMS) measurements in this study [29, 30]. The gas kit contains a flow controller to automatically regulate the drift tube pressure and baratrons so drift gases other than nitrogen can be used. BAs were ionized using ESI in both positive and negative mode. For the DTIMS measurements, the ions were passed through the inlet glass capillary, focused by a high pressure ion funnel, and accumulated in an ion funnel trap. Ions were then pulsed into the 78.24 cm long IMS drift tube filled with ~ 3.95 torr of nitrogen gas, where they travelled under the influence of a weak electric field (10–20 V/cm). Ions exiting the drift tube were refocused by a rear ion funnel prior to QTOF MS detection and their arrival times (tA) were recorded and CCS values calculated [31]. All CCS values were measured using seven stepped electric field voltages to obtain the most accurate values and each sample was analyzed in both positive and negative ionization modes. CCS values are listed in Table S1 in the Electronic Supplementary Material (ESM). The detailed instrumental settings are given in the ESM, Tables S2–S4, and follow those previously published in an interlaboratory study [32] and a drift tube IMS CCS database study [33]. Each sample was measured in triplicate and relative standard deviations (RSD) were obtained for all of the analytes. The Agilent IM-MS Browser software was utilized for data processing and all stepped field CCS calculations.
Results and Discussion
To evaluate the structural complexity of BA isomers, 72 different BA standards were evaluated with DTIMS-MS. The standards included primary and secondary BAs, 12 isomer groups, and unlabeled and deuterated versions. To evaluate when and if separation was possible with DTIMS, both positive and negative ionization were utilized and the resulting sodiated and deprotonated forms of each standard were studied. All triplicate measured CCS values are noted in Table S1 (see ESM) along with CAS numbers for all molecules so the exact structure of each BA can be looked up in PubChem (https://pubchem.ncbi.nlm.nih.gov/) if desired. In our analyses, initially the results for the unlabeled and deuterated versions were compared. Of the 72 standards studied here, 56 BAs were unlabeled, while 16 BAs were in a deuterated form having either four or five deuterium substitutions. As expected, similar CCS values (<0.3% difference) were observed for the unlabeled and labeled forms in both positive and negative modes. Since <0.3% difference is the experimental error of the instrument used in this study, no structural changes were observed upon deuteration of the BAs. This information is important to understand since deuterated BAs can be spiked into complex mixtures in metabolomics studies for absolute quantitation of endogenous BAs. The identical CCS values therefore add confidence to the identification of the BAs in complex mixtures.
The role of conjugation is also of interest in the structural evaluation of BAs. Of the 56 unlabeled BAs analyzed in this study, 35 were unconjugated, while 9 where conjugated with glycine and 12 with taurine. To understand whether conjugation affects BA structure, the DTIMS CCS and m/z trends were evaluated for the whole collection of unlabeled BA standards. As shown in Figure 3, significant differences were observed in the positive and negative ion modes. In negative ion mode, the deprotonated forms of the unconjugated and conjugated BAs had very similar CCS values ranging from ~198 Å2 to 215 Å2 (Figure 3A). While the unconjugated BAs displayed a wide CCS distribution that spanned the whole CCS range, the glycine and taurine conjugates each had very narrow ranges with glycine conjugates only spanning 3 Å2 and taurine conjugates occurring over 6 Å2. However, in positive ion mode (Figure 3B), the range of CCS values for the sodiated complexes increased for all forms from ~188 Å2 to 222 Å2. Additionally, the range for each different type of BA also increased: unconjugated forms extended over 25 Å2, glycine conjugates spanned 8 Å2, and taurine conjugates occurred over 10 Å2. The extended CCS distribution illustrated the effect of both the sodium binding location and BA structural flexibility as the hydroxyl groups moved to coordinate with the sodium ion. These structural alterations thus resulted in either compaction or extension of the BAs as shown by the CCS values change. For example, the sodiated form of litocholenic acid (LCLA) was much smaller than its deprotonated form (Figure 4). Multiple forms of some sodiated BAs were also observed as seen by the small peaks in Figure 4 for the sodiated LCLA. Interestingly, sodium binding also induced multiple conformers in the BAs as shown by the three peaks for the sodiated form of LCLA. However, a single conformation often dominated the sodiated complexes (>80% of all forms), so only the dominate CCS conformer values are noted in Table S1 (see ESM).
Figure 3.
The m/z versus CCS trend line for the 56 unlabeled BAs in their A) deprotonated [M-H]− and B) sodiated [M+Na]+ forms. The unconjugated, glycine conjugated and taurine conjugated BAs are noted with blue, orange and grey colors. The CCS measured in DTIMS with nitrogen gas are noted as DTCCSN2, with the unit of Å2.
Figure 4.
Arrival time distributions for the sodiated and deprotonated forms of lithocholenic acid (LCLA). Three peaks were observed for the sodiated LCLA complex, but the dominate conformer was much smaller than the deprotonated form.
The effect of different ion types was also investigated for the 12 isomer groups to understand if structural separation was possible and specific to deprotonation or sodiation. In the isomer groups, all but two groups could be distinguished in either positive or negative mode or a combination of both. For example, the isomer pair of 3-ketocholanic acid (3ketoCA) and LCLA with an exact mass of 374.2821 showed very similar arrival time distributions for their sodiated versions in positive mode and were inseparable (Figure 5A). However, in negative ion mode, the deprotonated form of 3ketoCA displayed a much shorter arrival time than LCLA providing separation at half height of the peak and suggesting differences in the BA conformations. The isomer pair 3a-hydroxy-7,12-diketocholanic acid (3aOH712diketoCA) and 3a-hydroxy-6,7-diketocholanic acid (3aOH67diketoCA) interestingly showed the opposite case (Figure 5B). In negative mode, the deprotonated structures for these two isomer showed by similar arrival times and were inseparable from each other. However, in positive mode, the sodiated forms displayed very different arrival times and were baseline separated from each other. These opposing trends suggest that isomer separation is specific to the group of BAs being studied and that an overall trend might not be established within the different isomer groups. However, this trend indicates that IMS-MS experiments for the analysis of the 56 BAs in the manuscript would have to be performed in both positive and negative mode unless only a subgroup of isomers was targeted. To eliminate this need, a short LC gradient could be coupled with the IMS-MS analyses if all the BAs could be distinguished either by LC elution time or IMS CCS in a single polarity. The multidimensional LC-IMS-MS evaluation would also provide even more confidence in the BA identifications due to the different resulting molecular characteristics that could be matched to a BA database.
Figure 5.
Isomeric arrival time distribution comparisons for A) 3-ketocholanic acid (black) and lithocholenic acid (red) and B) 3a-hydroxy-7,12-diketocholanic acid (black) and 3a-hydroxy-6,7-diketocholanic acid (red). The deprotonated spectra for each pair are shown on the left, while the sodiated spectra are shown on the left. Opposite separation trends were observed for the two different isomer pairs.
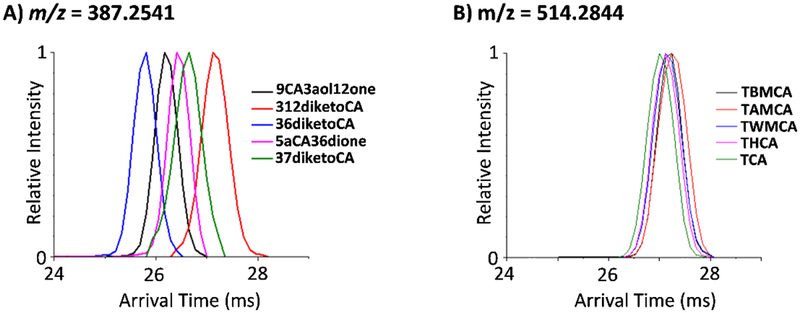
Upon further evaluation of the two isomer groups that could not be distinguished in either positive or negative ion mode or a combination of both, we noticed a trend. Of our 12 isomer groups studied, the 2 groups that could not be distinguished had some of the highest m/z values with exact masses of 449.3141 and 515.2917. In fact, the group with an exact mass of 515.2917 was the largest BA group analyzed in this work. This trend is shown in Figure 6 where the deprotonated BAs of 387.2541 m/z (Figure 6A) are compared to those at 514.2844 m/z (Figure 6B). The group at m/z = 387.2535 represented one of smaller mass studied and showed separation, while these at the largest m/z studied (m/z = 514.2844) showed very similar arrival time distributions and were inseparable from each other. Since we observed significant changes in BA structure upon sodium binding, we decided to assess whether the addition of metal ions could dramatically change the conformations of the larger BA isomers. Previous studies with other molecule types, such as glycans, have shown improved isomer separations with the use of copper and zinc, so we began our assays with these two metal cations [34–39]. To evaluate the effect of the metals, each standard was injected without and with the metal solutions. As shown in Figure 7, the deprotonated and sodiated BA isomers with an exact mass of 449.3141 had similar IMS arrival time profiles and were inseparable (Figure 7, top panel). In contrast, when Cu2+ and Zn2+ bound to the BAs, reasonable separation of the isomers was attained (Figure 7, bottom panel). However, multiple peaks were also observed in these arrival time distributions, which is especially apparent for GUDCA. The changes in separation of the BAs indicates that the metal cations bind differently than sodium allowing additional resolution for the four BAs. The multiple peaks in the arrival time distributions however illustrate that Cu2+ and Zn2+ bind to multiple locations on the BAs, making the spectra more complex to analyze. Thus, if specific targeted analyses are desired, this may be one promising way of rapidly separating the BA isomers, but quantitation of the multiple complexed peaks (which may overlap with the other isomers) will be difficult. Another interesting observation in the Cu2+ and Zn2+ arrival time distributions was that GCDCA and GDCA switched conformational sizes in the different metal complexes, indicating that even Cu2+ and Zn2+ are binding in different ways. Therefore, other metal cations could be used with the BAs to possibly produce even better separations, but this study is beyond the scope of this manuscript. We did however try to complex Cu2+ and Zn2+ the largest BA isomer group in this study (having an exact mass of 515.2917). Unfortunately, no complexes were observed, so the effect of Cu2+ and Zn2+ addition could not be studied for this isomer group.
Figure 6.
Small deprotonated BAs illustrated greater IMS separations than larger BAs. The arrival time distributions for two representative isomer groups are shown in their deprotonated forms for A) m/z = 387.2541 and B) m/z = 514.2844. The group at m/z = 387.2541 represents one of the smaller isomer groups analyzed, while the group at m/z = 514.2844 was the largest isomer group characterized in this work.
Figure 7.
Metal ions enable better separations for the different BAs, but also induce multiple conformations. The IMS arrival time distributions for the BA isomers (GCDCA, GDCA, GUDCA and GHDCA) with the exact mass of 449.3141 are shown in their A) deprotonated, B) sodiated, C) copper complexed, and D) zinc complexed forms. The multiple peaks observed for the copper and zinc complexed forms illustrate the multiple binding locations for each cation on the BAs.
Conclusions
In this work, we applied DTIMS-MS to characterize BA standards with subtle structural differences. The analyses showed that the structure for BAs can change dramatically from positive to negative ion mode where sodiated and deprotonated ions are formed. In some cases, deprotonation enabled better isomeric separations, but the opposite trend was also observed. Ultimately, evaluation in both ion modes enabled the most confident identification for the entire set of BAs analyzed. Two of the isomer groups we studied were however indistinguishable in either their deprotonated or sodiated forms. In these cases, metal ions such as copper and zinc were added to the overlapping BA isomers and the metal complexation led to separation of the isomers. Our results suggest that IMS-MS is a powerful tool for rapidly identifying BAs and distinguishing them from their isomers. However, no specific IMS structural trends were observed in the study for the different isomer groups such as a certain hydroxyl position resulting in a smaller size or that sodiation would always separate the isomers. Standards will therefore be needed initially to identify the BAs in the assays and this work detailing the CCS values of 56 unlabeled BAs will serve as a powerful starting point for many IMS-MS-based BA analyses. We also believe that coupling short LC gradients with the IMS-MS assays would provide even more confidence in the BA identifications. These LC-IMS-MS analyses may even eliminate the need for both positive and negative mode evaluations if all of the BAs of interest can be separated in a single polarity either by their LC elution time or their IMS CCS value.
Supplementary Material
Acknowledgements
The authors would like to thank Nathan Johnson for help with the figures. Portions of this research were supported by grants from the NIH National Institute of Environmental Health Sciences (R01 ES022190, R01 ES022186 and P42 ES027704), National Institute of General Medical Sciences (P41 GM103493), the PA Department of Health Tobacco CURE Program, and the Laboratory Directed Research and Development Program at Pacific Northwest National Laboratory. This research utilized capabilities developed by the Pan-omics program (funded by the U.S. Department of Energy Office of Biological and Environmental Research Genome Sciences Program). This work was performed in the W. R. Wiley Environmental Molecular Sciences Laboratory (EMSL), a DOE national scientific user facility at the Pacific Northwest National Laboratory (PNNL). PNNL is operated by Battelle for the DOE under contract DE-AC05–76RL0 1830.
Biographies

Xueyun Zheng is a research scientist in the Department of Chemistry at Texas A&M University. She has been working on instrument developments for mass spectrometry and ion mobility spectrometry, in addition to applications in biophysical analyses, metabolomics and lipidomics.

Francesca B. Smith is a graduate student in the Epidemiology program at Oregon State University. She has worked to develop ion mobility spectrometry assays to separate structurally similar molecules and isomers.

Noor A. Aly is a graduate student in the Toxicology program at Texas A&M University. Her current research focuses on the characterization of environmental chemicals coupling ion mobility spectrometry and mass spectrometry analyses.

Jingwei Cai is a scientist in Drug Metabolism and Pharmacokinetics Department at Genentech now. She received her molecular toxicology PhD in Dr. Andrew Patterson Lab at Penn State University in 2018. She has been studying the xenobiotics-gut microbiome-host interactions, especially physiological and metabolic role that gut microbiome play in host health and diseases with mass spectrometry and NMR-based metabolomics tools.

Richard D. Smith is a Battelle Fellow and Chief Scientist for the Biological Sciences Division at Pacific Northwest National Laboratory. His research has included the original development of the combinations of both supercritical fluid chromatography and capillary electrophoresis separations with mass spectrometry (MS), as well as high resolution capillary LC and high resolution MS for proteomics, and more recently Structures for Lossless Ion Manipulations (SLIM) for achieving much higher resolution ion mobility separations with MS.

Andrew D. Patterson is an Associate Professor of Molecular Toxicology at the Pennsylvania State University, University Park, PA and is the Scientific Director of Metabolomics. He and his students, postdocs, and collaborators focus on understanding the host-metabolite-microbiota communication network—specifically how the manipulation of gut microbiota by diet and/or xenobiotics impacts host metabolites (e.g., bile acids, short chain fatty acids), their metabolism, and how these co-metabolites interact with host nuclear/soluble receptors (e.g., farnesoid X receptor, aryl hydrocarbon receptor). The lab employs a variety of tools, including NMR- and mass spectrometry-based metabolomics, genomics, and conventional and gnotobiotic transgenic mice, to facilitate its study of these pathways and understand their impact on human health and disease.

Erin S. Baker is an Associate Professor in the Department of Chemistry at North Carolina State University. Her research group is utilizing informatics tools and multi-omic analyses to rapidly evaluate numerous samples in a short time period and connect this molecular information with phenotypic data. Her lab utilizes a variety of solid phase extraction techniques, chromatography methods, ion mobility spectrometry assays and mass spectrometry instruments to attain the molecular information needed for a better understanding of the environmental and biological systems being studied.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
The authors have no conflict of interest to declare.
No humans or animals were used in the studies in this manuscript.
References
- 1.Hofmann AF. The continuing importance of bile acids in liver and intestinal disease. Arch Intern Med. 1999;159(22):2647–58. [DOI] [PubMed] [Google Scholar]
- 2.Stamp D, Jenkins G. Chapter 1 An Overview of Bile-Acid Synthesis, Chemistry and Function. Bile Acids: Toxicology and Bioactivity: The Royal Society of Chemistry; 2008. p. 1–13. [Google Scholar]
- 3.Cook JW, Kennaway EI, Kennaway NM. Production of tumours in mice by deoxycholic acid. Nature. 1940;145:627-. [Google Scholar]
- 4.Princen HMG, Post SM, Twisk J. Regulation of bile acid biosynthesis. Curr Pharm Design. 1997;3(1):59–84. [Google Scholar]
- 5.Shneider BL, Fox VL, Schwarz KB, Watson CL, Ananthanarayanan M, Thevananther S, et al. Hepatic basolateral sodium-dependent-bile acid transporter expression in two unusual cases of hypercholanemia and in extrahepatic biliary atresia. Hepatology. 1997;25(5):1176–83. [DOI] [PubMed] [Google Scholar]
- 6.Li T, Chiang JY. Bile acid signaling in metabolic disease and drug therapy. Pharmacol Rev. 2014;66(4):948–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ajouz H, Mukherji D, Shamseddine A. Secondary bile acids: an underrecognized cause of colon cancer. World J Surg Oncol. 2014;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dvorak K, Payne CM, Chavarria M, Ramsey L, Dvorakova B, Bernstein H, et al. Bile acids in combination with low pH induce oxidative stress and oxidative DNA damage: relevance to the pathogenesis of Barrett’s oesophagus. Gut. 2007;56(6):763–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon MF, Mapstone NP, Neville PM, Moayyedi P, Axon ATR. Bile reflux gastritis and intestinal metaplasia at the cardia. Gut. 2002;51(3):351–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ross RK, Hartnett NM, Bernstein L, Henderson BE. Epidemiology of adenocarcinomas of the small intestine: is bile a small bowel carcinogen? Br J Cancer. 1991;63(1):143–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohtaki Y, Hida T, Hiramatsu K, Kanitani M, Ohshima T, Nomura M, et al. Deoxycholic acid as an endogenous risk factor for hepatocarcinogenesis and effects of gomisin A, a lignan component of Schizandra fruits. Anticancer Res. 1996;16(2):751–5. [PubMed] [Google Scholar]
- 12.Jenkins G, Hardie LJ, Royal Society of Chemistry (Great Britain) Bile acids: toxicology and bioactivity. Cambridge: SC Pub.; 2008. xi, 163 p. p. [Google Scholar]
- 13.Dawson PA. Biochemistry of Lipids, Lipoproteins and Membranes. Sixth ed McLeod NRaR, editor: Elsevier; 2016. [Google Scholar]
- 14.Jian-Shan Cai J HC. The Mechanism of Enterohepatic Circulation in the Formation of Gallstone Disease. The Journal of Membrane Biology. 2014;247:1067–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ajouz H, Mukherji D, Shamseddine A. Secondary bile acids: an underrecognized cause of colon cancer. World J Surg Oncol. 2014;12:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ajouz Hana D M, Shamseddine Alli. Secondary bile acids: an underrecognized cause of colon cancer 2014. [ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li T, Chiang JYL. Bile Acid Signaling in Metabolic Disease and Drug Therapy. Pharmacological Reviews. 2014;66(4):948–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Canaveras JC, Donato MT, Castell JV, Lahoz A. Targeted profiling of circulating and hepatic bile acids in human, mouse, and rat using a UPLC-MRM-MS-validated method. J Lipid Res. 2012;53(10):2231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wegner K, Just S, Gau L, Mueller H, Gerard P, Lepage P, et al. Rapid analysis of bile acids in different biological matrices using LC-ESI-MS/MS for the investigation of bile acid transformation by mammalian gut bacteria. Anal Bioanal Chem. 2017;409(5):1231–45. [DOI] [PubMed] [Google Scholar]
- 20.Humbert L, Maubert MA, Wolf C, Duboc H, Mahe M, Farabos D, et al. Bile acid profiling in human biological samples: comparison of extraction procedures and application to normal and cholestatic patients. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;899:135–45. [DOI] [PubMed] [Google Scholar]
- 21.Gnewuch C, Liebisch G, Langmann T, Dieplinger B, Mueller T, Haltmayer M, et al. Serum bile acid profiling reflects enterohepatic detoxification state and intestinal barrier function in inflammatory bowel disease. World J Gastroenterol. 2009;15(25):3134–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sayin SI, Wahlstrom A, Felin J, Jantti S, Marschall HU, Bamberg K, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17(2):225–35. [DOI] [PubMed] [Google Scholar]
- 23.Burkard I, von Eckardstein A, Rentsch KM. Differentiated quantification of human bile acids in serum by high-performance liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;826(1–2):147–59. [DOI] [PubMed] [Google Scholar]
- 24.Steiner C, von Eckardstein A, Rentsch KM. Quantification of the 15 major human bile acids and their precursor 7alpha-hydroxy-4-cholesten-3-one in serum by liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2010;878(28):2870–80. [DOI] [PubMed] [Google Scholar]
- 25.Sarafian MH, Lewis MR, Pechlivanis A, Ralphs S, McPhail MJ, Patel VC, et al. Bile acid profiling and quantification in biofluids using ultra-performance liquid chromatography tandem mass spectrometry. Anal Chem. 2015;87(19):9662–70. [DOI] [PubMed] [Google Scholar]
- 26.Scherer M, Gnewuch C, Schmitz G, Liebisch G. Rapid quantification of bile acids and their conjugates in serum by liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877(30):3920–5. [DOI] [PubMed] [Google Scholar]
- 27.Wyttenbach T, Bowers M. Gas-Phase Conformations: The Ion Mobility/Ion Chromatography Method In: Schalley C, editor. Modern Mass Spectrometry. Topics in Current Chemistry. 225: Springer; Berlin Heidelberg; 2003. p. 207–32. [Google Scholar]
- 28.Lanucara F, Holman SW, Gray CJ, Eyers CE. The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics. Nat Chem. 2014;6(4):281–94. [DOI] [PubMed] [Google Scholar]
- 29.May JC, Goodwin CR, Lareau NM, Leaptrot KL, Morris CB, Kurulugama RT, et al. Conformational Ordering of Biomolecules in the Gas Phase: Nitrogen Collision Cross Sections Measured on a Prototype High Resolution Drift Tube Ion Mobility-Mass Spectrometer. Anal Chem. 2014;86(4):2107–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ibrahim YM, Baker ES, Danielson Iii WF, Norheim RV, Prior DC, Anderson GA, et al. Development of a new ion mobility time-of-flight mass spectrometer. Int J Mass Spectrom. 2015;377:655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mason EA, McDaniel EW. Kinetic Theory of Mobility and Diffusion: Sections 5.1 – 5.2. Transport Properties of Ions in Gases: Wiley-VCH Verlag GmbH & Co. KGaA; 2005. p. 137–93. [Google Scholar]
- 32.Stow SM, Causon TJ, Zheng X, Kurulugama RT, Mairinger T, May JC, et al. An Interlaboratory Evaluation of Drift Tube Ion Mobility - Mass Spectrometry Collision Cross Section Measurements. Anal Chem. 2017;89(17):9048–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng X, Aly NA, Zhou Y, Dupuis KT, Bilbao A, Paurus V, et al. A structural examination and collision cross section database for over 500 metabolites and xenobiotics using drift tube ion mobility spectrometry. Chemical Science. 2017;8(11):7724–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Y, Dodds ED. Ion Mobility Studies of Carbohydrates as Group I Adducts: Isomer Specific Collisional Cross Section Dependence on Metal Ion Radius. Analytical Chemistry. 2013;85(20):9728–35. [DOI] [PubMed] [Google Scholar]
- 35.Huang Y, Dodds ED. Discrimination of Isomeric Carbohydrates as the Electron Transfer Products of Group II Cation Adducts by Ion Mobility Spectrometry and Tandem Mass Spectrometry. Analytical Chemistry. 2015;87(11):5664–8. [DOI] [PubMed] [Google Scholar]
- 36.Huang Y, Dodds ED. Ion-neutral collisional cross sections of carbohydrate isomers as divalent cation adducts and their electron transfer products. Analyst. 2015;140(20):6912–21. [DOI] [PubMed] [Google Scholar]
- 37.Gaye MM, Nagy G, Clemmer DE, Pohl NLB. Multidimensional Analysis of 16 Glucose Isomers by Ion Mobility Spectrometry. Analytical Chemistry. 2016;88(4):2335–44. [DOI] [PubMed] [Google Scholar]
- 38.Huang Y, Dodds ED. Ion-neutral collisional cross sections of carbohydrate isomers as divalent cation adducts and their electron transfer products. The Analyst. 2015;140(20):6912–21. [DOI] [PubMed] [Google Scholar]
- 39.Zheng X, Zhang X, Schocker NS, Renslow RS, Orton DJ, Khamsi J, et al. Enhancing glycan isomer separations with metal ions and positive and negative polarity ion mobility spectrometry-mass spectrometry analyses. Anal Bioanal Chem. 2017;409(2):467–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.