Abstract
Introduction
Accurate diagnosis of maturity-onset diabetes of the young (MODY) is required in order to select appropriate treatment options and to assess prognosis. The aim of this study was to explore potential clinical indicators that could be used to differentiate MODY2, MODY3, and type 1 diabetes (T1D) in young subjects.
Methods
Twelve patients with MODY3 and 29 patients with MODY2 were characterized and compared to 26 patients with T1D. These three groups were matched for age and gender. Clinical profiles of the 67 patients were collected. Receiver operating characteristic (ROC) curves were used to identify the optimal cutoff values of clinical indicators.
Results
Compared to patients with T1D, subjects with MODY3 had higher fasting C-peptide levels (1.34 ± 1.51 vs. 0.29 ± 0.22 ng/mL; P < 0.001) and lower high-sensitivity C-reactive protein (hsCRP) levels (0.18 ± 0.15 vs. 1.22 ± 1.49 mg/L, P = 0.004); patients with MODY2 had lower hsCRP (0.37 ± 0.39 vs. 1.22 ± 1.49 mg/L; P = 0.003), total cholesterol (4.12 ± 0.68 vs. 4.61 ± 0.81 mmol/L, P = 0.034), and low-density lipoprotein cholesterol (LDL-C) (2.24 ± 0.68 vs. 2.67 ± 0.79 ng/L, P = 0.002) levels and higher fasting C-peptide levels (0.96 ± 0.42 vs. 0.29 ± 0.22 ng/mL, P = 0.002). The ROC-derived hsCRP values for discriminating MODY2 from T1D, MODY3 from T1D, and MODY3 from MODY2 were 0.675, 0.833, and 0.763, respectively. The ROC-derived fasting C-peptide levels for discriminating MODY2 from T1D and MODY3 from T1D were 0.951 and 0.975, respectively. The ROC-derived total cholesterol and LDL-C values for discriminating MODY2 from T1D were 0.670 and 0.662, respectively; the ROC-derived triglyceride value for discriminating MODY3 from MODY2 was 0.756. Additionally, a combination of indicators permitted better discrimination of MODY subtypes than any single parameter.
Conclusion
Our findings suggest that fasting C-peptide, hsCRP, and lipid levels permit good discrimination among MODY2, MODY3, and T1D. These clinical indicators could be used as markers of MODY2 and MODY3 in young patients with diabetes.
Electronic Supplementary Material
The online version of this article (10.1007/s13300-019-0647-x) contains supplementary material, which is available to authorized users.
Keywords: C-peptide, hsCRP, Lipid, MODY, Type 1 diabetes
Introduction
Maturity-onset diabetes of the young (MODY) is a group of heterogeneous monogenic disorders characterized by early-onset autosomal-dominant inheritance and dysfunction of pancreatic beta cells [1]. Currently, 14 causative genes involved in beta-cell development or insulin secretion have been identified for MODY [1, 2]. The two most common subtypes, MODY2 and MODY3, which are caused by mutations in a gene encoding glucokinase and mutations in a gene responsible for hepatocyte nuclear factor 1-alpha (HNF1A), respectively, account for approximately 90% of these patients [1]. Individuals with MODY2 present with mild fasting hyperglycemia, rarely suffer from chronic complications, and do not require antidiabetic agent treatment. Patients with HNF1A mutations manifest with progressive beta-cell dysfunction and a high risk of diabetes-related micro- and macrovascular complications, and these cases are particularly sensitive to sulfonylurea [1, 3]. Thus, precise diagnosis promotes optimal glycemic control. However, it is still challenging for clinicians to differentiate MODY2 and MODY3 from other forms of diabetes due to their overlapping clinical features. Additionally, molecular genetic testing is too expensive and is not available in many areas. Consequently, cost-effective and practical biomarkers that can help to identify patients with a high probability of GCK/HNF1A mutations are needed. Our previous study (as well as other studies) indicated that high-sensitivity C-reactive protein (hsCRP) is significantly lower in MODY3 patients than in those with young-onset type 2 diabetes (T2D) and other subtypes of diabetes [4, 5]. Laboratory characteristics that help to distinguish MODY2 from other subtypes of diabetes include a small increase in glucose levels according to a oral glucose tolerance test (OGTT) at 120 min, as 95% of MODY2 patients will show glucose levels < 83 mg/dL at 120 min [6]. MODY and type 1 diabetes (T1D) patients are relatively young at disease onset, and most are non-obese. Therefore, some MODY2 and MODY3 patients are misdiagnosed with T1D. Nevertheless, studies defining the clinical spectrum are still scarce in comparison to those focused on T1D in Chinese patients. Hence, we aimed to analyze the clinical features of subjects with MODY2 and MODY3. In addition, the present study evaluated the feasibility of using clinical features as diagnostic markers for differentiating HNF1A/GCK mutation carriers from T1D patients.
Methods
This study included 26 subjects with T1D, 29 subjects with MODY2, and 12 subjects with MODY3 who were matched in age and gender and were of Chinese origin. T1D was diagnosed according to the criteria of the American Diabetes Association [7] and the positivity of at least two persistent [including glutamic acid decarboxylase antibody (GAD), islet cell antibody (ICA), and protein tyrosine phosphatase antibody (IA2)]. MODY2 and MODY3 were diagnosed based on the following criteria: (1) a family history of diabetes with an autosomal-dominant mode of inheritance in at least two generations; (2) an age at onset of diabetes of ≤ 45 years; (3) absence of autoantibodies; (4) non-obesity (BMI < 28 kg/m2), and positive genetic testing results [8]. The patients included in our study were aged 6–45 and were consecutively recruited from the outpatient clinic of the endocrinology department at Peking Union Medical College Hospital (PUMCH), Beijing, China, between January 2017 and December 2018. All procedures followed were in accordance with the ethical standards of the Peking Union Medical College Hospital Ethics Committee and with the 1964 Declaration of Helsinki, as revised in 2013. Written consent was obtained from all participants or from responsible family members.
The exons of the genes associated with MODY2 (GCK) and MODY3 (HNF1A) were screened using primer sequences designed using Premier 5 software (see Table S1 in the Electronic supplementary material, ESM). The NCBI BLAST database was used to identify variants through alignment with the reference sequences NM_000162 (GCK) and NM_000545.6 (HNF1A). All variants found in our study were considered pathogenic based on prediction analysis, and were cosegregated with clinical phenotypes of MODY within families.
Clinical indices including height and weight were recorded for each participant. Venous blood samples were collected after an overnight (≥ 10 h) fast. None of the participants were suffering from an illness (fever, infection, etc.) in the days before the study. Fasting insulin and C-peptide as well as 2-h postprandial insulin and C-peptide were measured by chemiluminescent analysis. The patients were told to have regular meals without too much sugar and fat. Glycated hemoglobin A1c (HbA1c) was measured using a dedicated high-performance liquid chromatography system. GAD and IA2 were determined by ELISA (SIMENS ADVIA Centaur XP, Germany), and ICA was measured using indirect immunofluorescence. The plasma concentrations of high-sensitive C-reactive protein (hs-CRP), total cholesterol, high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), triglyceride, and glycated albumin were assayed using an automatic biochemical analyzer (AU5800; Beckman Coulter, USA).
Statistical Analysis
All statistical analyses were performed using the Statistical Package for Social Sciences (SPSS 21.0 for Windows, Chicago, IL, USA). Skewed distributions were natural-logarithmically transformed prior to analysis. ANOVA with Bonferroni’s post hoc test was utilized for continuous outcomes, and parameter values were expressed as the mean ± standard deviation (SD). Logistic regression models were applied to analyze the associations of metabolic traits with different diabetic subtypes after controlling for gender and age. Odds ratios (ORs) and 95% confidence intervals (CIs) were evaluated. P < 0.05 was considered to indicate a statistically significant difference. Receiver operating characteristic (ROC) curves were used to determine the optimal cutoff values of metabolic traits to apply in order to distinguish among T1D, MODY2, and MODY3 patients. The optimal cutoff points were defined based on the maximum Youden index calculated as sensitivity + specificity − 1 [9].
Results
Clinical and Laboratory Characteristics of Patients with MODY2 and MODY3
Among the 67 diabetic participants in this study, there were 26 T1D, 29 MODY2, and 12 MODY3 patients. The basic features of the subjects are summarized in Table 1. The mean ages of the T1D, MODY2, and MODY3 patients were 22.88 ± 5.85, 23.29 ± 11.54, and 21.75 ± 8.74 years, respectively. The three groups were matched in age and gender, and showed comparable heights and weights.
Table 1.
General characteristics of the study subjects
| T1D (n = 26) | MODY 2 (n = 29) | MODY 3 (n = 12) | P | |
|---|---|---|---|---|
| Age (years) | 22.88 ± 5.85 | 23.29 ± 11.54 | 21.75 ± 8.74 | 0.889 |
| BMI (kg/m2) | 19.83 ± 3.18 | 19.47 ± 4.37 | 20.70 ± 1.65 | 0.623 |
| HbA1c% | 9.52 ± 2.99 | 6.42 ± 0.35* | 8.31 ± 2.62# | 1.18×10 −5 |
| Fasting glucose (mmol/L) | 9.58 ± 4.42 | 6.92 ± 0.58* | 8.26 ± 3.17 | 0.010 |
| 2-h glucose (mmol/L) | 15.57 ± 5.64 | 9.22 ± 1.78* | 13.46 ± 6.34# | 3.52×10 −5 |
| Fasting C-peptide (ng/mL) | 0.29 ± 0.22 | 0.96 ± 0.42* | 1.34 ± 1.51* | 1.88×10 −4 |
| 2-h C-peptide (ng/mL) | 0.62 ± 0.57 | 3.93 ± 2.02* | 2.42 ± 1.35*# | 1.91×10 −9 |
| hsCRP (mg/dL) | 1.22 ± 1.49 | 0.37 ± 0.39* | 0.18 ± 0.15* | 0.004 |
| Total cholesterol (mmol/L) | 4.61 ± 0.81 | 4.12 ± 0.68* | 4.41 ± 0.93 | 0.098 |
| Triglyceride (mmol/L) | 0.74 ± 0.32 | 0.65 ± 0.38 | 0.97 ± 0.43 | 0.051 |
| HDL-C (mmol/L) | 1.28 ± 0.53 | 1.47 ± 0.25 | 1.32 ± 0.39 | 0.227 |
| LDL-C (mmol/L) | 2.67 ± 0.79 | 2.24 ± 0.68 | 2.57 ± 0.86 | 0.125 |
Results are from one-way ANOVA; significant differences (P < 0.05) versus T1D are indicated by *, and significant differences (P < 0.05) versus MODY2 individuals are indicated by #
Values in bold are significant (P < 0.05) after Bonferroni correction
T1D type 1 diabetes, MODY 2 maturity-onset diabetes of the young type 2, MODY 3 maturity-onset diabetes of the young type 3, hs-CRP high-sensitivity C-reactive protein, LDL-C low-density lipoprotein cholesterol, HDL-C high-density lipoprotein cholesterol
Compared to subjects with T1D, those with MODY3 had higher fasting C-peptide levels (1.34 ± 1.51 vs. 0.29 ± 0.22 ng/mL; P < 0.001) and 2-h C-peptide (2.42 ± 1.35 vs. 0.62 ± 0.57 ng/mL; P = 0.002) and lower hsCRP levels (0.18 ± 0.15 vs. 1.22 ± 1.49 mg/L, P = 0.004).
Similarly, patients with MODY2 showed different clinical features to subjects with T1D; they had lower HbA1c levels (6.42 ± 0.35% vs. 9.52 ± 2.99%; P < 0.001), fasting glucose (6.92 ± 0.58 vs. 9.58 ± 4.42 mmol/L; P = 0.003), 2-h glucose (9.22 ± 1.78 vs. 15.57 ± 5.64 mmol/L, P < 0.001), hsCRP (0.37 ± 0.39 vs. 1.22 ± 1.49 mg/L; P = 0.003), total cholesterol (4.12 ± 0.68 vs. 4.61 ± 0.81 mmol/L, P = 0.034), and LDL-C levels (2.24 ± 0.68 vs. 2.67 ± 0.79 mmol/L, P = 0.050), but they had higher levels of fasting C-peptide (0.96 ± 0.42 vs. 0.29 ± 0.22 ng/mL, P = 0.002) and 2-h C-peptide (3.93 ± 2.02 vs. 0.62 ± 0.57 ng/mL, P < 0.001).
Potential of Clinical Indices to Discriminate Among MODY Subtypes and T1D
A separate multivariable logistic regression analysis was performed to examine the feasibility of using each variable as a MODY subtype predictor. As shown in Table 2, after adjusting for age and gender, decreased HbA1c (OR 0.163; 95% CI 0.04–0.61), fasting glucose (OR 0.69; 95% CI 0.52–0.91), 2-h glucose (OR 0.58; 95% CI 0.42–0.80), lnCRP (OR 0.48; 95% CI 0.25–0.94), total cholesterol (OR 0.37; 95% CI 0.16–0.89), and LDL-C (OR 0.36; 95% CI 0.14–0.92) levels and increased fasting C-peptide (OR 870; 95% CI 10.6–(7.2×104)) were independent predictors of MODY2 compared to T1D. Additionally, decreased lnCPR levels (OR 0.19; 95% CI 0.04–0.95) and increased fasting C-peptide (OR 5.7×104; 95% CI 1.9–(1.7×109)) and 2-h C-peptide (OR 17.6; 95% CI 2.0–152) levels were clinical markers of MODY3 versus T1D. Further, HbA1c (OR 2.94; 95% CI 1.15–7.56), 2-h glucose (OR 1.47; 95% CI 1.02–2.12), and triglyceride (OR 12.47; 95% CI 1.64–94.80) levels were positive predictors whereas hsCRP level (OR 0.21, 95% CI 0.05–0.86) was a negative predictor of MODY3 versus MODY2.
Table 2.
Potentials of different variables to discriminate between the MODY subtypes and T1D
| Variables | T1D vs. MODY 2 | T1D vs. MODY 3 | MODY 2 vs. MODY 3 | |||
|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |
| HbA1c% | 0.16 (0.04–0.61) | 0.007 | 0.81 (0.60–1.09) | 0.169 | 2.94 (1.15–7.56) | 0.025 |
| Fasting glucose (mmol/L) | 0.69 (0.52–0.91) | 0.009 | 0.94 (0.76–1.15) | 0.526 | 1.57 (0.97–2.54) | 0.069 |
| 2-h glucose (mmol/L) | 0.58 (0.42–0.80) | 0.001 | 0.94 (0.82–1.08) | 0.375 | 1.47 (1.02–2.12) | 0.039 |
| ln(fasting C-peptide) (ng/mL) | 870 (10.6–(7.2×104)) | 0.002 | 5.7×104 (1.9–(1.7×109)) | 0.037 | 2.31 (0.50–10.80) | 0.287 |
| ln(2-h C-peptide) (ng/mL) | 2.7×104 (0.5–(1.4×109)) | 0.065 | 17.6 (2.0–152) | 0.009 | 0.19 (0.03–1.13) | 0.067 |
| lnCRP (mg/dL) | 0.48 (0.25–0.94) | 0.033 | 0.19 (0.04–0.95) | 0.043 | 0.21 (0.05–0.86) | 0.031 |
| Total cholesterol (mmol/L) | 0.37 (0.16–0.89) | 0.025 | 0.81 (0.32–2.04) | 0.650 | 1.70 (0.65–4.45) | 0.277 |
| Triglyceride (mmol/L) | 0.30 (0.04–2.08) | 0.224 | 8.14 (0.86–77.50) | 0.068 | 12.47 (1.64–94.80) | 0.015 |
| HDL-C (mmol/L) | 3.85 (0.73–20.48) | 0.113 | 1.48 (0.28–7.71) | 0.641 | 0.14 (0.01–2.04) | 0.149 |
| LDL-C (mmol/L) | 0.36 (0.14–0.92) | 0.032 | 0.91 (0.36–2.35) | 0.852 | 1.98 (0.70–5.64) | 0.200 |
The OR (95% CI) and P values shown are from logistic regression and have been adjusted for age and gender
Values in bold are significant at P < 0.05
T1D type 1 diabetes, MODY 2 maturity-onset diabetes of the young type 2, MODY 3 maturity-onset diabetes of the young type 3, hs-CRP high-sensitivity C-reactive protein, LDL-C low-density lipoprotein cholesterol, HDL-C high-density lipoprotein cholesterol
ROC Curve Analysis
ROC curve analysis was used to evaluate the performance of the indicators as diagnostic markers for discriminating among different diabetic subtypes in young patients.
Diagnostic Value of hsCRP
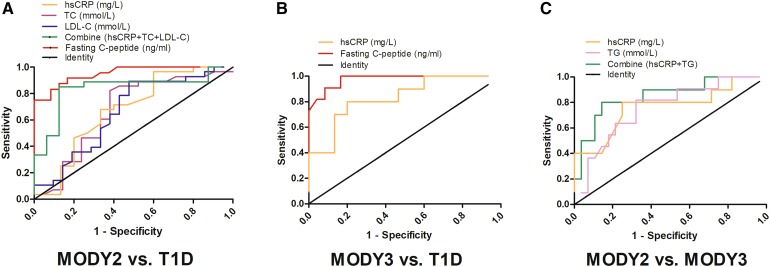
The results showed that the hsCRP level allowed good discrimination between MODY2 and T1D, with an AUC of 0.675. A cutoff hsCRP value of < 0.280 mg/L was identified which gave 66.7% sensitivity and 67.9% specificity. When discriminating MODY3 from T1D, hsCRP exhibited an AUC of 0.833, and a cutoff of < 0.200 mg/L gave 80.0% sensitivity and 80.0% specificity. For MODY3 vs. MODY2, the hsCRP level gave an AUC of 0.763, and a cutoff value of < 0.185 mg/L yielded 75.0% sensitivity and 80.0% specificity (Fig. 1).
Fig. 1.
Use of ROC curves to examine the abilities of various clinical indices to discriminate between different subtypes of diabetes. a MODY2 vs. T1D; the area under the curve is 0.951 for fasting C-peptide, 0.675 for hsCRP, 0.670 for total cholesterol, 0.662 for LDL-C, and 0.833 for the combination of hsCRP, total cholesterol, and LDL-C. b MODY3 vs. T1D; the area under the curve is 0.833 for hsCRP and 0.975 for fasting C-peptide. c MODY2 vs. MODY3; the area under the curve is 0.763 for hsCRP, 0.756 for triglyceride, and 0.85 for the combination of hsCRP and triglyceride
Diagnostic Value of Fasting C-Peptide
The results showed that the fasting C-peptide level permitted good discrimination between MODY2 and T1D, with an AUC of 0.951; a cutoff fasting C-peptide value of > 0.580 ng/mL yielded 87.5% sensitivity and 87.5% specificity. When this clinical parameter was used to discriminate MODY3 from T1D, we obtained an AUC of 0.975 and a cutoff fasting C-peptide value of > 0.485 ng/mL, which provided 100% sensitivity and 83.3% specificity. However, the fasting C-peptide level could not be used to distinguish between the two subtypes of MODY (Fig. 1).
Diagnostic Value of Lipids
The results showed good discrimination between MODY2 and T1D using total cholesterol, with an AUC of 0.670 and a cutoff value of < 4.525 mmol/L, which allowed 61.9% sensitivity and 82.1% specificity. When discriminating between MODY2 and T1D using LDL-C, the AUC was 0.662 and the identified cutoff value was < 2.65 mmol/L, yielding 52.4% sensitivity and 89.3% specificity. Additionally, discriminating between MODY3 and MODY2 using the triglyceride level led to an AUC of 0.756 and a cutoff value of > 0.65 mmol/L, which permitted 81.8% sensitivity and 67.9% specificity (Fig. 1).
Diagnostic Value of Combining Metabolic Indices
Since hsCRP, fasting C-peptide, total cholesterol, and LDL-C were all independent predictors of the MODY2 phenotype, we used ROC analysis to investigate whether a combination of those indicators could be used to predict MODY subtypes more effectively than the individual indices. The ROC curve for the combination of hsCRP with fasting C-peptide, total cholesterol, and LDL-C resulted in the highest AUC (1.0) for discriminating MODY2, whereas the combination of hsCRP, total cholesterol, and LDL-C yielded an AUC of 0.833. The combination of hsCRP with fasting C-peptide provided an AUC of 1.0 for distinguishing MODY3 from T1D. The combination of hsCRP and triglyceride gave an AUC of 0.85 for differentiating MODY3 from MODY2. Thus, we found that combinations of biochemical indicators can be used as markers to distinguish MODY subtypes.
Discussion
MODY is a relatively rare monogenic form of diabetes and is often misdiagnosed as T1D or T2D [1, 10]. Obtaining a definite genetic diagnosis of MODY is very important for predicting the clinical course of diabetes, guiding the optimization of treatment, and evaluating the risk to relatives. Therefore, it is crucial to identify novel and inexpensive biomarkers that could be used to identify these patients. To better define the phenotypic characteristics of Chinese patients with MODY2 and MODY3, we compared the clinical indicators of patients carrying mutations in GCK/HNF1A with those of T1D subjects and found that fasting C-peptide, hsCRP, and lipid levels could be used as biomarkers to distinguish MODY2 and MODY3 from T1D. This study therefore provides useful information for clinical workers who wish to identify the most common subtypes of MODY (MODY2 and MODY3) in patients with early-onset diabetes (< 45 years old).
Serum C-Peptide Levels
Fasting C-peptide levels were significantly lower in T1D patients than in those diagnosed with MODY subtypes—the cutoffs were 0.580 ng/mL and 0.485 ng/mL, respectively, for MODY2 and MODY3. Decreased beta-cell function in T1D individuals can be explained; see [7]. MODY2 is caused by an inactivating mutation of GCK, which serves as a key regulating enzyme in glucose-stimulated insulin secretion and acts as a glucose sensor in pancreatic beta cells; however, this mutation does not impair the function of beta cells [11]. Thus, MODY2 is characterized by asymptomatic fasting hyperglycemia, which is present from birth and remains stable throughout life. Unlike MODY2, MODY3 is caused by mutations of HNF1A, a transcription factor, and leads to decreased insulin secretion and progressive beta-cell damage [12]. The lack of a difference in fasting C-peptide levels between the two subtypes of MODY in our study may be due to the young age of the subjects included.
Serum hsCRP Levels
The binding sites of the transcription factor HNF1A are harbored in the promoter of the hsCRP gene [13]. The inactivating mutations in HNF1A that are responsible for MODY3 have been shown to result in a significant reduction in hsCRP [14]. Our previous study [4] and a study from the UK [5] confirmed that the level of serum hsCRP was significantly lower in MODY3 patients than in T2D patients, and identified hsCRP cutoffs of 0.79 mg/L and 0.75 mg/L, respectively. In our study, we found that hsCRP levels were significantly lower in MODY3 than in T1D and MODY2 patients, in line with studies by McDonald [5] and Owen [15], and the relevant cutoffs were 0.200 mg/L and 0.185 mg/L, respectively. Additionally, we demonstrated that hsCRP levels were dramatically lower in MODY2 subjects than in T1D subjects, in accord with a study by McDonald [5], and the cutoff was 0.280 mg/L. However, Owen et al. [15] reached a different conclusion—that hsCRP levels were higher in MODY2 patients than in T1D patients, which may be due to racial/ethnic differences between the studies. Further large multicenter sample research may help to explain the results. Unlike MODY3, the mechanism for the association between MODY2 and hsCRP is still unclear, although the low inflammatory response in patients with MODY2 may partly explain this clinical manifestation.
Serum Lipid Levels
The hepatically expressed protein encoded by HNF1A is thought to bind to the genes responsible for lipid metabolism [16]. In humans, HNF1A defects are believed to increase the synthesis of bile acid [17] and influence the levels of total cholesterol, HDL-C, and LDL-C [18]. HNF1A-knockout mice (HNF1A−/−) were found to overexpress lipoprotein lipase (LPL) gene transcripts [19]. LPL encodes lipoprotein lipase, which is mostly found in the endothelial surfaces of capillaries in organs such as adipose tissues, liver, and skeletal muscle, where it hydrolyzes triglyceride-rich lipoprotein, including VLDL and chylomicrons [20]. This is why a previous study demonstrated that triglyceride levels were lower in MODY3 than in T2D [21]. Lipid levels in MODY3 patients were found to be no different from those in T1D patients in our study and in a previous study by Fendler et al. [22]; this may be explained by small sample sizes. Nevertheless, we found that subjects with MODY2 had lower total cholesterol and LDL-C levels than T1D subjects, and we identified cutoffs of 4.525 mmol/L and 2.65 mmol/L, respectively. Previous studies have reported that overexpression of liver GCK could increase glycogen deposits, lower blood glucose levels, and increase triacylglycerol and free fatty acid levels [23]. Decreasing GCK activity may reduce the production of both glycogen and malonyl-CoA by lowering glycolytic flux [24]. Because it plays an important role in lipid metabolism, lowering the level of malonyl-CoA leads to weakened inhibition of carnitine palmitoyltransferase 1, thus increasing the oxidation of fatty acids [25]. This is why Spégel et al. [26] suggested that there were lower levels of free fatty acids and triglycerides in MODY2 patients than in healthy control subjects. However, our results did not agree with those reported by Fendler et al. [27], who suggested that HDL-C could be used as a diagnostic tool for the clinical differentiation of GCK-MODY from HNF1A-MODY and T1D. These conflicting results may be explained by small sample sizes and ethnic differences between the studies. Additionally, we found that triglyceride levels were lower (cutoff value: 0.65 mmol/L) in subjects with MODY2 than in those with MODY3, which was also reported by Mughal [28], indicating that metabolic control is more intact in MODY2 patients than in MODY3 subjects.
Importantly, we found that the combination of hsCRP, fasting C-peptide, and lipid levels yielded better discrimination of MODY2 and MODY3 from T1D than that obtained with any of those metabolic parameters individually. Our finding supports the hypothesis that one metabolic abnormality contribute only to an insufficient degree, while additive metabolic indicators have a more significant effect on the risk of diabetes.
Although we aimed to identify diagnostic markers that could be used to differentiate young HNF1A/GCK mutation carriers from young T1D patients for the first time in mainland China, some potential limitations should be noted. First, the small number of MODY2 and MODY3 subjects included in the study restricted our analysis of genotype and phenotype correlations, resulting in a large OR (95% CI) for some parameters. Second, our findings are based on data from Chinese diabetic patients, so they cannot be generalized to other racial populations without further replication. Additionally, information on patient lifestyle (e.g., diet and activity) was scarce. Finally, although we analyzed the most common biochemical indicators in our study, we are aware that many others have also been implicated as discriminatory indicators [29]. Nonetheless, the observations made in the present study should contribute to a better understanding of markers allowing MODY2 and MODY3 patients to be distinguished from T1D patients.
Conclusion
In summary, we found that fasting C-peptide, hsCRP, total cholesterol, and LDL-C levels can be used to discriminate between MODY2 and T1D; fasting C-peptide and lnCRP levels can be used to distinguish between MODY3 and T1D; and hsCRP and triglyceride levels can be used to discriminate MODY2 from MODY3. Combinations of multiple biochemical indicators can also be used as markers to better distinguish between MODY subtypes. Our study may help to identify individuals at high risk of MODY2 or MODY3 among young patients with diabetes in China.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank the participants for their continuing participation in this research effort.
Funding
This work was supported by grants from the National Key R&D Program of China (2017YFC1309603), the National Key Research and Development Program of China (2016YFA0101002), the National Natural Science Foundation of China (nos. 81170736, 81570715, 81870579), and the Medical Epigenetics Research Center, Chinese Academy of Medical Sciences (2017PT31036, 2018PT31021). The article processing charges were funded by the authors.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Disclosures
Junling Fu, Tong Wang, Jieying Liu, Xiaojing Wang, Qian Zhang, Ming Li, and Xinhua Xiao have nothing to disclose.
Compliance with Ethics Guidelines
All procedures followed were in accordance with the ethical standards of the Peking Union Medical College Hospital Ethics Committee and with the 1964 Declaration of Helsinki as revised in 2013. Informed consent was obtained from all individual participants included in the study.
Data Availability
The datasets obtained and/or analyzed during the current study are available from the corresponding author on reasonable request.
Open Access
This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Footnotes
Enhanced Digital Features
To view enhanced digital features for this article go to 10.6084/m9.figshare.8223536.
References
- 1.Anik A, Catli G, Abaci A, et al. Maturity-onset diabetes of the young (MODY): an update. J Pediatr Endocrinol Metab. 2015;28:251–263. doi: 10.1515/jpem-2014-0384. [DOI] [PubMed] [Google Scholar]
- 2.Prudente S, Jungtrakoon P, Marucci A, et al. Loss-of-function mutations in APPL1 in familial diabetes mellitus. Am J Hum Genet. 2015;97:177–185. doi: 10.1016/j.ajhg.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishay RH, Greenfield JR. A review of maturity onset diabetes of the young (MODY) and challenges in the management of glucokinase-MODY. Med J Austr. 2016;205:480–485. doi: 10.5694/mja16.00458. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Wang T, Yu M, et al. Screening of HNF1A and HNF4A mutation and clinical phenotype analysis in a large cohort of Chinese patients with maturity-onset diabetes of the young. Acta Diabetol. 2019;56:281–288. doi: 10.1007/s00592-018-1232-x. [DOI] [PubMed] [Google Scholar]
- 5.McDonald TJ, Shields BM, Lawry J, et al. High-sensitivity CRP discriminates HNF1A-MODY from other subtypes of diabetes. Diabetes Care. 2011;34:1860–1862. doi: 10.2337/dc11-0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDonald TJ, Ellard S. Maturity onset diabetes of the young: identification and diagnosis. Ann Clin Biochem. 2013;50:403–415. doi: 10.1177/0004563213483458. [DOI] [PubMed] [Google Scholar]
- 7.American Diabetes Association. Classification and diagnosis of diabetes: standards of medical care in diabetes 2019. Diabetes Care. 2019;42:S13–28. [DOI] [PubMed]
- 8.Kim SH. Maturity-onset diabetes of the young: what do clinicians need to know? Diabetes Metab J. 2015;39:468–477. doi: 10.4093/dmj.2015.39.6.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin J, Tian L. Joint confidence region estimation for area under ROC curve and Youden index. Stat Med. 2014;33:985–1000. doi: 10.1002/sim.5992. [DOI] [PubMed] [Google Scholar]
- 10.Pihoker C, Gilliam LK, Ellard S, et al. Prevalence, characteristics and clinical diagnosis of maturity onset diabetes of the young due to mutations in HNF1A, HNF4A, and glucokinase: results from the SEARCH for Diabetes in Youth. J Clin Endocrinol Metab. 2013;98:4055–4062. doi: 10.1210/jc.2013-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osbak KK, Colclough K, Saint-Martin C, et al. Update on mutations in glucokinase (GCK), which cause maturity-onset diabetes of the young, permanent neonatal diabetes, and hyperinsulinemic hypoglycemia. Hum Mutat. 2009;30:1512–1526. doi: 10.1002/humu.21110. [DOI] [PubMed] [Google Scholar]
- 12.Stride A, Ellard S, Clark P, et al. Beta-cell dysfunction, insulin sensitivity, and glycosuria precede diabetes in hepatocyte nuclear factor-1alpha mutation carriers. Diabetes Care. 2005;28:1751–1756. doi: 10.2337/diacare.28.7.1751. [DOI] [PubMed] [Google Scholar]
- 13.Nishikawa T, Hagihara K, Serada S, et al. Transcriptional complex formation of c-Fos, STAT3, and hepatocyte NF-1 alpha is essential for cytokine-driven C-reactive protein gene expression. J Immunol (Baltimore, Md: 1950) 2008;180:3492–3501. doi: 10.4049/jimmunol.180.5.3492. [DOI] [PubMed] [Google Scholar]
- 14.Thanabalasingham G, Shah N, Vaxillaire M, et al. A large multi-centre European study validates high-sensitivity C-reactive protein (hsCRP) as a clinical biomarker for the diagnosis of diabetes subtypes. Diabetologia. 2011;54:2801–2810. doi: 10.1007/s00125-011-2261-y. [DOI] [PubMed] [Google Scholar]
- 15.Owen KR, Thanabalasingham G, James TJ, et al. Assessment of high-sensitivity C-reactive protein levels as diagnostic discriminator of maturity-onset diabetes of the young due to HNF1A mutations. Diabetes Care. 2010;33:1919–1924. doi: 10.2337/dc10-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon EB, Lee HS, Shim YS, et al. The changes of subtypes in pediatric diabetes and their clinical and laboratory characteristics over the last 20 years. PLoS One. 2016;21:81–85. doi: 10.6065/apem.2016.21.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ekholm E, Nilsson R, Groop L, et al. Alterations in bile acid synthesis in carriers of hepatocyte nuclear factor 1alpha mutations. J Intern Med. 2013;274:263–272. doi: 10.1111/joim.12082. [DOI] [PubMed] [Google Scholar]
- 18.Babaya N, Ikegami H, Fujisawa T, et al. Association of I27L polymorphism of hepatocyte nuclear factor-1 alpha gene with high-density lipoprotein cholesterol level. J Clin Endocrinol Metab. 2003;88:2548–2551. doi: 10.1210/jc.2002-021891. [DOI] [PubMed] [Google Scholar]
- 19.Shih DQ, Bussen M, Sehayek E, et al. Hepatocyte nuclear factor-1alpha is an essential regulator of bile acid and plasma cholesterol metabolism. Nat Genet. 2001;27:375–382. doi: 10.1038/86871. [DOI] [PubMed] [Google Scholar]
- 20.Verges B. Lipid disorders in type 1 diabetes. Diabetes Metab. 2009;35:353–360. doi: 10.1016/j.diabet.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 21.St-Jean M, Boudreau F, Carpentier AC, et al. HNF1alpha defect influences post-prandial lipid regulation. PLoS One. 2017;12:e0177110. doi: 10.1371/journal.pone.0177110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fendler W, Rizzo M, Borowiec M, et al. Less but better: cardioprotective lipid profile of patients with GCK-MODY despite lower HDL cholesterol level. Acta Diabetol. 2014;51:625–632. doi: 10.1007/s00592-014-0567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Doherty RM, Lehman DL, Telemaque-Potts S, et al. Metabolic impact of glucokinase overexpression in liver: lowering of blood glucose in fed rats is accompanied by hyperlipidemia. Diabetes. 1999;48:2022–2027. doi: 10.2337/diabetes.48.10.2022. [DOI] [PubMed] [Google Scholar]
- 24.Velho G, Petersen KF, Perseghin G, et al. Impaired hepatic glycogen synthesis in glucokinase-deficient (MODY-2) subjects. J Clin Investig. 1996;98:1755–1761. doi: 10.1172/JCI118974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foster DW. Malonyl-CoA: the regulator of fatty acid synthesis and oxidation. J Clin Investig. 2012;122:1958–1959. doi: 10.1172/JCI63967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spegel P, Ekholm E, Tuomi T, et al. Metabolite profiling reveals normal metabolic control in carriers of mutations in the glucokinase gene (MODY2) Diabetes. 2013;62:653–661. doi: 10.2337/db12-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fendler W, Borowiec M, Antosik K, et al. HDL cholesterol as a diagnostic tool for clinical differentiation of GCK-MODY from HNF1A-MODY and type 1 diabetes in children and young adults. Clin Endocrinol. 2011;75:321–327. doi: 10.1111/j.1365-2265.2011.04052.x. [DOI] [PubMed] [Google Scholar]
- 28.Giuffrida FMA, Moises RS, Weinert LS, et al. Maturity-onset diabetes of the young (MODY) in Brazil: establishment of a national registry and appraisal of available genetic and clinical data. Diabetes Res Clin Pract. 2017;123:134–142. doi: 10.1016/j.diabres.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 29.Mughal S. A., Park R., Nowak N., Gloyn A. L., Karpe F., Matile H., Malecki M. T., McCarthy M. I., Stoffel M., Owen K. R. Apolipoprotein M can discriminate HNF1A-MODY from Type 1 diabetes. Diabetic Medicine. 2013;30(2):246–250. doi: 10.1111/dme.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets obtained and/or analyzed during the current study are available from the corresponding author on reasonable request.