Abstract
Control of translation initiation plays a critical role in the regulation of gene expression in all organisms, yet the mechanics of translation initiation in eukaryotic organisms are not well understood. Confounding studies of translation are the large number and overlapping functions of many initiation factors in cells, and a lack of cap-dependence in many in vitro systems. To shed light on intricate mechanisms that are often obscured in vivo, we use a fully reconstituted translation initiation system for analyzing RNA interactions with eukaryotic translation initiation factors and complexes from the model organism Saccharomyces cerevisiae. This system exhibits strong cap dependence, and dependence on translation factors varies with mRNA 5’ UTR sequences as expected from genome-wide studies of translation. Here we provide optimized protocols for purification and analysis of labeled and unlabeled mRNA recruitment factors on both the rate and factor dependence of mRNA recruitment to the translation preinitiation complex in response to RNA sequence- and structure-changes. In addition to providing streamlined and detailed protocols, we provide a new construct for purification of higher yields of fluorescently labeled and unlabeled full-length eIF4G.
Keywords: ribosome, mRNA translation initiation, eukaryotic initiation factors, eIF4F, eIF4B, eIF3
1. Introduction
During translation initiation in eukaryotes, the small ribosomal subunit is recruited to the 5’ end of a capped mRNA in complex with initiator Met-tRNA and several eukaryotic translation initiation factors (eIFs) [1]. This preinitiation complex (PIC) is then thought to scan each nucleotide along the 5’ UTR, moving in a 5’ to 3’ direction, until the tRNA base pairs with a start codon in good context. Upon start codon recognition, the Met-tRNA clashes with initiation factor eIF1 and a conformational change in the 40S takes place. These events surrounding start codon recognition have been well-described, but the events leading up to start codon recognition are less characterized.
While the molecular mechanisms of mRNA loading on PICs and subsequent movement are not well understood, it is clear that the eIF4 factors and eIF3 play critical roles in these steps [2]. There are four eIF4 factors: eIF4E, the major cytoplasmic cap-binding protein, complexes tightly in yeast with eIF4G, a large scaffolding protein, an interaction which is thought to recruit PICs to the cap; eIF4A is a DEAD-box RNA helicase that when bound to eIF4E and eIF4G forms the eIF4F trimer, which has moderate RNA unwinding activity in yeast; and eIF4B binds to the 40S subunit and promotes the activity of eIF4A/eIF4F. eIF3 promotes eIF2•GTP•Met-tRNA binding to the 40S to form the PIC as well as mRNA binding and scanning of this PIC. Each of these factors binds to RNA independently, and eIF4B and eIF3 are known to bind directly to the 40S subunit [3].
Recent genome-wide and biochemical studies of translation have shed light on the importance of both mRNA features and the specific complement of mRNA recruitment factors associated with an mRNA for determining which mRNAs are effectively translated in cells [3–8]. For instance, a number of mRNAs with long structured 5’ untranslated regions exhibit a strong requirement for eIF4B, while mRNAs with short 5’ untranslated regions and strong potential to form a closed loop (i.e. those messages that show enhanced interactions with eIF4E, eIF4G, and PABP) exhibit stronger dependence on eIF4G [9]. This suggests that eIF4B and eIF4G can use different mechanisms to stimulate translation, but given that both factors promote the activity of eIF4A [10–12], the molecular basis for differential activities is unclear. Further analysis of factor association with mRNAs, ribosome complexes, and other proteins in vitro could lend new insights into these mechanisms.
Previous work in the Lorsch lab established the first fully reconstituted system for kinetic analysis of cap-dependent structured mRNA recruitment to the translation preinitiation complex [8]. Here we provide detailed methods for recently optimized purification of the yeast translation initiation factors that stimulate capped mRNA recruitment to the translation preinitiation complex, and present protocols for measuring interactions between these factors, mRNAs, ribosomes, and functional PICs. To improve yields and quality of the difficult to purify full-length eIF4G, we present a new construct that is purified from E.coli as an intein-fusion protein free of nucleic acids and nucleases, and which can be fluorescently-labeled during the intein-cleavage process.
2. Materials and Methods
2.1. Specialized Equipment and Materials Required
Shaker (e.g. Thermo #SHKE8000–7) that will accommodate 6 Fernbach flasks (2800 ml with bottom baffles, Corning #4424–2XL) with incubation and refrigeration for temperatures from 16–37°C.
Type I Molecular Biology grade water, free of RNases and DNases for all buffers, columns, reactions. Milli-Q and Nanopure sell Type I purification systems.
Deionized or Distilled water for media preparation
Bottle-top filtration units for preparation of sterile RNase-free solutions (e.g. VWR #97066–204)
Floor centrifuge with rotor for spinning 1 L bottles
SS-34 or JA-20 rotor for spinning 34 ml Oak ridge tubes
Refrigerated Centrifuge (e.g. Eppendorf #5810R) for 50 ml conical tubes
Nitrogen mill (Spex Sample Prep #6875) with large canisters (Spex Sample Prep #6801)
French press with 35 ml pressure cell (Glen Mills)
FPLC: Biorad NGC, GE AKTA Pure or equivalent with a sample pump or large superloop for loading large volumes of cell lysate and running size exclusion columns. FPLC should be located in a chromatography refrigerator or cold room.
SDS PAGE mini-gel apparatus (e.g. Biorad #1658000) and power supply
Cooled vertical electrophoresis apparatus (e.g. Hoefer #SE260–10A-.75)
Refrigerated Circulator for native electrophoresis (e.g. Thermo #HAK1525108)
Circulating water bath for biochemical assays (e.g. Fisher #13-874-177)
10K Molecular weight cutoff (MWCO) dialysis cassettes (Thermo #66810)
10K MWCO protein concentrators (Millipore #UFC901024)
0.45 μm syringe filters (Millipore #SLHV033RB)
LB Powder (BD #214906)
YPD Powder (Teknova #Y5301)
Carbenicillin (Goldbio #C-103) *Note: Ampicillin may also be used, but is less stable than Carb, so plates must be made frequently and will give satellite colonies lacking plasmid of interest. Liquid media may allow minor unproductive growth of cells that have lost AmpR plasmids during long inductions.
Isopropyl-beta-D-thiogalactoside (IPTG; Goldbio #I2481)
Roche Complete EDTA-Free Protease inhibitor tablets (Roche #05056489001)
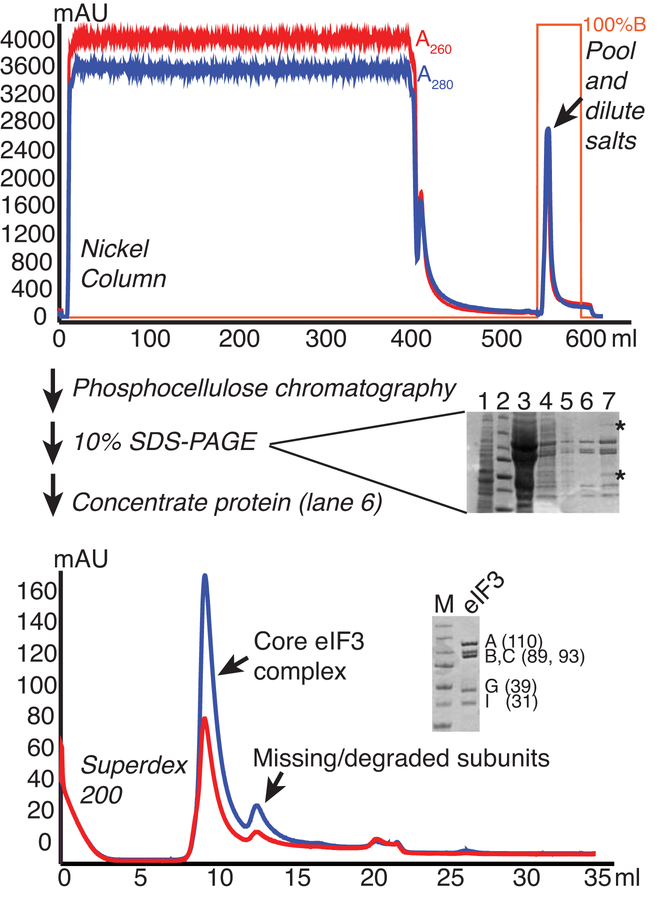
2.2. Preparation of His-tagged eIF3 from yeast (Figure 1)
Figure 1.
eIF3 purification from yeast. Lysate from cells overexpressing His-tagged eIF3B were fractionated on Nickel, Phosphocellulose (P11), and Superdex 200 columns to purify the core eIF3 complex. Chromatograms are shown for Nickel and Superdex columns, with A260 and A280 traces shown in red and blue, respectively and percentage buffer B (high imidazole) in orange. 10% SDS-PAGE gels stained with Coomassie are shown. The gel run following P11 (center) shows 1) clarified lysate, 2) MW marker, 3) P11 bound, 4) Ni eluate/P11 load, 5) P11 flowthrough, 6) P11-B200 eluate, 7) P11-B350 eluate. Asterisks mark contaminants that are consistently found in B350/1000 fractions. The larger contaminant is associated with RNase activity and the lower is a similar size to eIF3 ligands eIF5 and eIF2-gamma. The lower gel shows the final purified protein next to MW marker (Precision plus, Biorad: 250, 150, 100, 75, 50, 37, and 25 kDa bands), with the subunits and approximate MWs indicated.
2.2.1. Materials Required
SC-Leu-Ura plate and SC-Leu-Ura liquid media (SC-Leu-Ura dropout Powder, Yeast Nitrogen Base, Dextrose, and Agar can be purchased from Sunrise Scientific, BD, Teknova, and USB)
YPD media (6 baffled Fernbach flasks with 1.5 L each)
LPY87 strain (gift from A. Hinnebusch, NICHD, NIH, [13])
5 ml HisTrap HP column (GE #17524801)
Superdex 200 increase 10/300 column (GE #28990944)
Phosphocellulose resin (Sigma #C2258)
Econopac column (Biorad #7321010)
Low Imidazole Buffer (1L): 20 mM HEPES-KOH (pH 7.4), 350 mM KCl, 5 mM MgCl2, 10% (v/v) Glycerol, 20 mM Imidazole, 10 mM 2-Mercaptoethanol (BME)*
High Imidazole Buffer (0.5 L): 20 mM HEPES-KOH (pH 7.4), 350 mM KCl, 5 mM MgCl2, 10% (v/v) Glycerol, 250 mM Imidazole, 10 mM BME*
Phosphocellulose B100, B200, B350, and B1000 Buffers (Make 1 L B100, and 50 ml B200, B350, and B1000): 20 mM HEPES-KOH (pH 7.4), 100/200/350/1000 mM KCl, 0.1 mM EDTA, 10% (v/v) Glycerol, and 2 mM DTT*
-
Enzyme Storage Buffer: 20 mM HEPES-KOH (pH 7.4) 100 mM KOAc, 10% (v/v) Glycerol, and 2 mM DTT*
*Add BME and DTT to buffers just before use
Note: Buffers are kept at 4°C unless otherwise specified.
2.2.2. Cell growth
From a glycerol stock at −80°C, streak cells (LPY87) on an SC-Leu-Ura plate and grow at 30°C for 2–3 days. Do not place plate at 4°C.
Inoculate 50 ml SC-Leu-Ura using multiple isolated colonies, and grow at 30°C overnight.
Inoculate six flasks of 1.5 L YPD with 3 ml of overnight culture per flask and grow 18–24 hours at 30°C with shaking at 250 rpm. Note: Yields are similar when cells are grown in 9 L selective media.
Collect cells by pelleting for 10 minutes at 6500 × g at 4°C; resuspend each pellet in 10 ml Lysis Buffer and combine in a weighed empty bottle(s). Pellet cells again and weigh the pellet(s). Resuspend in 1/3 pellet volume of Lysis Buffer (Low Imidazole Buffer without protease inhibitors added, i.e. 45 g cells would require 15 ml buffer). Make a sieve in an ice bucket by poking small holes in aluminum foil lining the bucket with a pair of fine-tip forceps. Fill the ice bucket with liquid N2 and slowly drip the cell suspension into the liquid N2 with a serological pipette to form individual pellets. Use sieve to collect frozen droplets into a chilled beaker or bottle and store at −80°C. (The yeast can be stored at −80°C for years).
2.2.3. Cell lysis
Note: Blender lysis or microfluidizer lysis are also acceptable for this step, but a nitrogen mill imparts less variability in our hands.
Chill large nitrogen mill canister in dry ice.
Fill canister containing chilled mallet no more than 1/2 full. Efficiency of lysis decreases as canister is overfilled.
Cap canister just prior to placing in mill. Lyse cells using the following parameters: Cycles = 10, Precool = 15 minutes, Run = 1 minute, Cool = 2 minutes, Rate = 15.
Repeat steps 1–3 as needed to lyse cells. Store powdered lysate in a chilled bottle or beaker at −80°C. We have stored lysate for at least 2 months with no observed changes in activity of the subsequently purified protein.
2.2.4. Phosphocellulose column preparation
Column can be prepared ~2 days in advance or while preparing and running Nickel column. Combine 0.8 g phosphocellulose with 125 ml 0.5 M NaOH in a glass beaker with magnetic stir bar. Stir 5’.
Allow resin to settle for 5 minutes; Pour off NaOH and fines that did not settle. Some liquid will remain in the beaker.
Wash resin with ~500 ml H2O ~4–5X, allowing resin to settle for 5 minutes after each addition of water and discarding supernatant between washes until pH <11 (detected by pH paper).
Add 125 ml 0.5 M HCl and stir 5 minutes.
Allow resin to settle for 5 minutes; Pour off HCl and fines. Rinse with ~500 ml H2O ~4–5X until pH>4.
Pour resin into a 20 ml column; Pack column by washing with B100 Buffer until pH of buffer exiting column is 7.
Cap the column and store at 4°C until use. Check pH just prior to binding and wash with additional B100 Buffer if necessary.
2.2.5. Nickel column
Dissolve lysate powder in ~400 ml Low Imidazole Buffer with freshly added 10 mM BME, 1 Roche EDTA-Free complete tablet added per 50 ml buffer, and 100 μl of 5 mg/ml DNase I.
Clarify the lysate by centrifugation at 25,000 × g for 30 minutes at 4°C (or equivalent spin in 250 ml bottle). During this spin, equilibrate Nickel column. Nickel columns will give better yield if EDTA-stripped and Ni-regenerated according to the manufacturer’s instructions between preps.
Vacuum-filter low speed-supernatant through 2 layers of Whatman glass-fiber filters or use 5 μm syringe filters, then filter through a bottle-top 0.8 μm filter unit. Keep lysate on ice during filtration.
Load the lysate onto the nickel column at a flow rate of ~2 ml/minute. Wash the column with 100 ml Low Imidazole Buffer and elute with High Imidazole Buffer at 2–3 ml/minute (1–2 ml fractions). Nickel columns will give better yield if EDTA stripped and Ni-regenerated between preps.
Pool entire peak and dialyze against 2 L B100 Buffer for 2 hours to overnight.
2.2.6. Phosphocellulose column
Load dialyzed Nickel eluate onto a freshly prepared phosphocellulose column, equilibrated in B100 Buffer.
Wash the phosphocellulose column with 80 ml B100 Buffer, then 20 ml each of B200, B350, and B1000 Buffer, collecting 5 ml fractions. Analyze fractions by SDS-PAGE using 10% polyacrylamide gels; 15 μl/fraction for loading; 200 V, ~50 minutes. A 200–350 elution peak contains eIF3 and lacks RNases, and a 350–1000 elution peak contains eIF3 with major RNase contamination. Pool peak fractions and concentrate to ~0.3 ml in 10K MWCO concentrator.
2.2.7. Superdex 200
Filter retentate through a 0.45 μm syringe filter. Apply up to 0.25 ml to a Superdex 200 Increase 10/300 GL prepacked column from GE in B100 Buffer at 0.75 ml/minute. Collect 0.3 ml fractions.
Analyze fractions from each peak by 10% SDS-PAGE. Pool fractions enriched in stoichiometric eIF3 (the first peak that elutes from ~8–11 ml), determine concentration by Bradford assay using bovine serum albumin (BSA) to generate a standard curve, and concentrate protein if needed, using molecular weight of 361 kDa.
Test the eIF3 for RNase activity by incubating eIF3 with labeled RNA in reaction buffer and running on a denaturing gel, or by using an RNase alert kit according to manufacturer’s instructions (Ambion). Set up one reaction with excess protein over what would be used in an experiment to detect trace activity, and one with the amount that would typically be used in an experiment. Set up controls with RNase inhibitor added. Incubate at 26°C for at least as long as an experiment using the protein would run.
Flash-freeze protein in small aliquots (10–15 μl) in liquid N2 and store at −80°C. Typical yields are low: ~1 nmol protein/L cells.
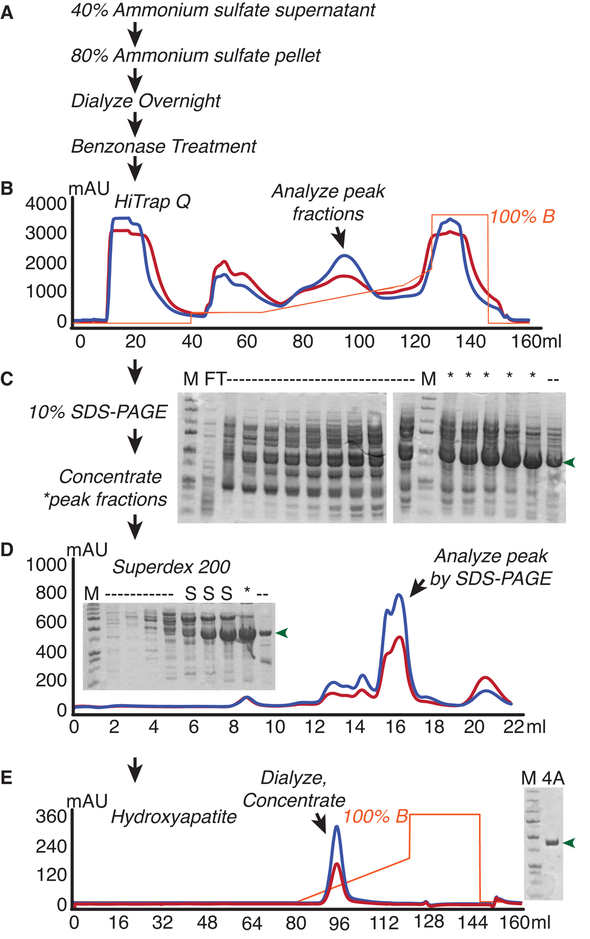
2.3. Preparation of nuclease-free yeast eIF4A from E. coli (Figure 2)
Figure 2.
Yeast eIF4A purification from E.coli. E.coli overexpressing yeast eIF4A were lysed and subjected to (A) Ammonium sulfate fractionation followed by Benzonase nuclease treatment to remove nucleic acids. eIF4A was then purified from lysates by B) Anion Exchange chromatography, D) Size Exclusion Chromatography, and E) Hydroxyapatite chromatography. Flowthrough (FT) and peak fractions were analyzed alongside marker (M) after each chromatographic step to monitor purity. Lanes with discarded fractions are noted as dashes, whereas desirable eIF4A fractions that were retained are denoted with asterisks, and shoulder fractions from the superdex column are denoted as “S”. eIF4A is indicated with green arrows.
2.3.1. Materials Required
BL21-CodonPlus (DE3)-RIL Competent Cells (Agilent #230245)
pET21c-TIF1 plasmid (Addgene #37230)
LB Carbenicillin agar plate
250 ml flask with 50 ml and 6 × 2.8L Fernbach flasks with 1.5L LB containing 50 μg/ml Carbenicillin and 34 μg/ml Chloramphenicol (LB Carb/Cm)
10% SDS PAGE materials and Coomassie stain
HiTrap Q HP column (5 ml, GE #17115401)
Superdex 200 increase 10/300 column (GE #28990944)
CHT Type II Hydroxyapatite column (Biorad #7324332)
Ammonium sulfate
0.45 μm PVDF syringe filters
2 M MgCl2
Benzonase nuclease (Sigma, ≥250 U/ml)
0.1M NaOH
Lysis Buffer: 20 mM HEPES-KOH (pH 7.4), 150 mM KCl, 0.1 mM EDTA, 10% (v/v) Glycerol, 2 mM DTT*, 1 Roche Complete EDTA-Free tablet/50ml*
Q50 Buffer: 20 mM HEPES-KOH (pH 7.4), 50 mM KCl, 0.1 mM EDTA, 10% (v/v) Glycerol, 2 mM DTT*
Q1000 Buffer: 20 mM HEPES-KOH (pH 7.4), 1 M KCl, 0.1 mM EDTA, 10% (v/v) Glycerol, 2 mM DTT*
Low Phosphate Buffer: 5 mM Potassium Phosphate, pH 6.8, 150 mM KCl, 10% (v/v) Glycerol, 2 mM DTT*
High Phosphate Buffer: 500 mM Potassium Phosphate, pH 6.8, 150 mM KCl, 10% (v/v) Glycerol, 2 mM DTT*
-
Enzyme Storage Buffer: 20 mM HEPES-KOH, pH 7.4, 250 mM KOAc, 10% (v/v) Glycerol, 2 mM DTT*
*Add Protease Inhibitors and/or DTT to buffers just before use
Note: Buffers are kept at 4°C unless otherwise specified.
2.3.2. Cell growth
Inoculate 2 × 5 ml LB Carb/Cm cultures with 10 isolated colonies from a fresh transformation of pET21c-eIF4A in BL21-CodonPlus(DE3) -RIL Competent Cells. Grow with shaking overnight at 37°C.
Inoculate two 1.5 L LB Carb/Cm cultures with the entire saturated overnight cultures from step 1.
When the cells reach an OD at 600 nm of 0.5 – 0.6, induce T7 transcription by adding 1 M IPTG to a final concentration of 1 mM. Shake the flasks for an additional 3 hours at 37°C. Pellet the cells in large bottles at 6500 × g for 5 minutes at 4°C. Wash cells by resuspending each pellet in 10 ml Lysis Buffer (without protease inhibitors) and pelleting the combined suspension in a 50 ml Falcon tube at 4000 × g for 10 minutes at 4°C. The pellet can be stored at −80°C for future use if needed.
2.3.3. Cell Lysis
Resuspend the cells in ≤25 ml of Lysis Buffer using a serological pipette, for a final volume ≤35 ml. Lyse cells twice through French press.
Clarify the lysate by spinning at 25,000 × g for 30 minutes at 4°C.
2.3.4. Ammonium sulfate fractionation
Decant the clarified low speed supernatant into a small beaker with stirring magnetic bar, and slowly add solid ammonium sulfate to make a 40% saturated solution to avoid disrupting the stir bar as the ammonium sulfate is dissolved upon addition. The amount to add is easily determined with an online calculator (e.g. http://www.encorbio.com/protocols/AM-SO4.htm). Stir at 4°C for ~20 minutes to 1 hour, then pellet at 20,000 × g at 4°C for 15 minutes.
Decant supernatant from 40% precipitation to a new beaker. Add additional solid ammonium sulfate to the supernatant to bring the solution to 80% saturation. Stir at 4°C for 20 minutes to 1 hour, then pellet at 20,000 × g at 4°C for 15 minutes.
Dissolve the pellet from the 80% cut in 8–10 ml Q50 Buffer (for a volume that can be accommodated in the dialysis cassette) and dialyze overnight in a 10K MWCO dialysis cassette against 2 L Q50 Buffer.
2.3.5. Anion exchange
Remove the fractionated lysate from dialysis to a 50 ml Falcon tube, add 10 μl Benzonase nuclease, gently mix, and incubate on ice for 1 hour.
While incubating, wash and equilibrate Q column with Q50 Buffer.
Filter the sample through a 0.45 μm filter and load onto Q column. Wash with 5 column volumes (cv) Buffer A, then 10% B over 5 cv (5 ml fractions), then a linear gradient of 10–35% B over 10 cv, then 35–50% over 2 cv, then wash with 100% B for 5 cv.
Combine 12 μl of each peak fraction to analyze with 4 μl 4X Laemmli buffer, heat at >90°C for 2–5 minutes, and run on 10% TGS gels beside a protein ladder until bromophenol blue dye reaches the bottom (~30 minutes at 250 V). While the gel is running, wash and equilibrate the Superdex 200 increase column with 2 cv of Low Phosphate Buffer.
2.3.6. Superdex 200
Pool the fractions from the indicated peak that contain large amounts of eIF4A and lack contaminants noted on the gel in Figure 2D.
Concentrate to 0.3 ml using a 15 ml 10K MWCO protein concentrator.
Filter the protein through a 0.45 μm syringe filter. Some sample loss is expected, and we do not force residual protein through to dry the filter, as drying of the filter with excessive force could denature protein. Load ≤0.25 ml protein onto 24 ml Superdex 200 increase column pumping at 0.75 ml/minute of Low Phosphate Buffer.
Run on an SDS-PAGE gel as described previously. Pool the peak fractions avoiding any ~75 kDa contaminants.
2.3.7. Hydroxyapatite column
Run the protein over a hydroxyapatite column, washing with 5–10 cv Low Phosphate Buffer and eluting with a linear gradient from 0–50% High Phosphate Buffer over 8 cv. Wash with 100% High Phosphate Buffer for 5 cv. Wash the column with water and store in 0.1 M NaOH.
Dialyze the combined peaks overnight in 2 L of Enzyme Storage Buffer. Check with SDS-PAGE.
Spin dialyzed fraction in a 10K MWCO concentrator at 5000 × g in a fixed angle, or 4000 × g in a swinging bucket rotor in 25-minute increments to concentrate. Between each concentration step, pipette the liquid up and down against the membrane to prevent the sample from precipitating on the concentrator. Aliquot in amounts that will be convenient for only a few experiments. We have seen no decrease in activity from ~5 or more freeze-thawing events, but generally try to limit the number of uses, as occasionally we have observed aggregation of protein in the wells of native gels when an aliquot has undergone extensive freezing and thawing. Flash freeze and store at −80°C. Test for the presence of RNase if necessary. Typical yields are very high: ~140 pmols/L cells.
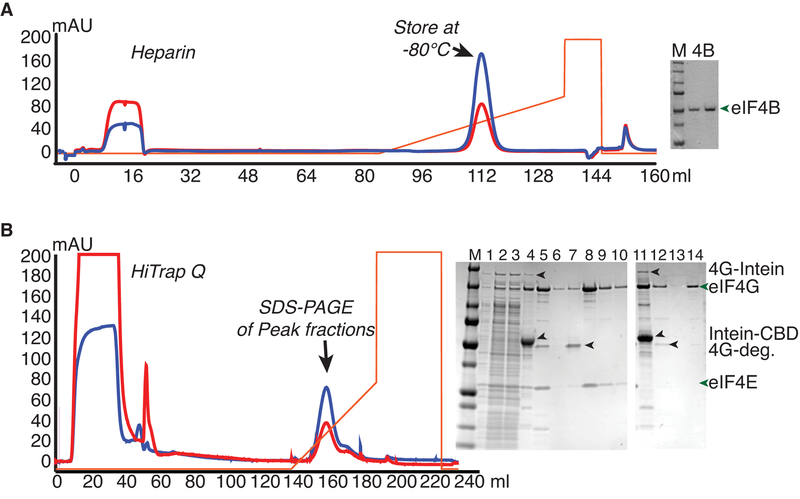
2.4. Preparation of yeast eIF4B from E. coli (Figure 3)
Figure 3.
Purification of Intein-Fusions of yeast eIF4B and eIF4G from E.coli. A. eIF4B was expressed as an intein-chitin-binding domain (CBD) fusion protein, and purified by running over a chitin column and eluting by intein cleavage with DTT buffer. Chromatogram shows eluate fractionation over a Heparin column. Gel shows Precision plus marker (M; Biorad) next to two dilutions of purified eIF4B. B. eIF4G fused to the intein-CBD was purified by binding to and washing on chitin resin followed by on-column nuclease digestion with micrococcal nuclease. Further washing was followed by DTT-induced intein-cleavage to elute eIF4G (complexed with eIF4E or without eIF4E). Chromatogram of anion exchange (Q) chromatography of eIF4G•eIF4E chitin eluate, from which full-length eIF4G eluted after degradation products. SDS-PAGE analysis shows separation of eIF4G•eIF4E (1–10) or eIF4G (11–13) from contaminants, with following loading order: M - Marker, 1) Lysate, 2–3) Low Speed supernatant, 4) Chitin Beads, 5) Q load, 6) Q flowthrough, 7–10) fractions from peaks 1 and 2, 11) Chitin Beads, 12) Q load, 13) Q flowthrough, and 14) major eIF4G peak fraction.
2.4.1. Materials Required
Plasmid pTYB2-TIF3 (Addgene #37226)
BL21-CodonPlus (DE3)-RIL Competent Cells (Agilent #230245)
250 ml flask with 50 ml and 6 × 2.8 L Fernbach flasks with 1.5 L LB Carb/Cm
1 M IPTG
Roche Complete EDTA-Free Protease inhibitor tablet
- Chitin Column Buffers (Make 1 L each and store at 4°C):
- Intein Lysis Buffer: 20 mM HEPES-KOH (pH 7.4), 500 mM KCl, 1 mM EDTA (pH 8)
- Intein Wash Buffer: 20 mM HEPES-KOH (pH 7.4), 1 M KCl, 1 mM EDTA (pH 8) and 0.1% Triton X-100.
- Intein Cleavage Buffer: 20 mM HEPES-KOH (pH 8), 500 mM KCl and 1 mM EDTA (pH 8)
Econopac column (Bio-Rad #7321010)
Chitin Resin (New England Biolabs #S6651L)
0.45 μm syringe filters
HiTrap Heparin HP column (GE #17040701)
- Heparin Column Buffers:
- Low Salt Buffer: 20 mM HEPES-KOH (pH 7.4), 100 mM KCl, 10% (v/v) Glycerol, 2 mM DTT*.
- High Salt Buffer: 20 mM HEPES-KOH (pH 7.4), 1 M KCl, 10% (v/v) Glycerol, 2 mM DTT*.
-
Enzyme Storage Buffer: 20 mM HEPES-KOH (pH 7.4), 100 mM KOAc, 10% (v/v) Glycerol, 2 mM DTT*.
*Add DTT to buffers just before use
Note: Buffers are kept at 4°C unless otherwise specified.
10% SDS-PAGE materials
10K MWCO protein concentrators
2.4.2. Cell Growth
Transform BL21/DE3-CodonPlus-RIL competent cells with pTYB2-TIF3 and plate on LB media containing 50 μg/ml carbenicillin. Incubate the plates overnight at 37°C.
Inoculate 50 ml LB Carb/Cm with 10 isolated colonies. Grow the culture with shaking at 250 rpm overnight at 37°C.
Inoculate 6 × 1.5 L flasks of LB Carb/Cm with 6 ml overnight culture each. Grow the culture with shaking at 250 rpm at 37°C.
When each culture reaches mid-log phase (OD600 ~ 0.6), pack each flask in ice in a large tray for 0.5 to 1 hours and allow the incubator to cool to 16°C. Induce the cells by adding 1 M IPTG to final concentration of 0.5 mM. Shake at 250 rpm overnight (~20 hours) at 16°C.
Collect the cells by pelleting the cultures at 6500 × g for 5 minutes. Wash combined cells with water, immediately pellet the cells again and dump the supernatant. If using French press, freeze pellet or proceed. For nitrogen mill, weigh the pellet to determine its volume, resuspend in 1/3 of the pellet’s volume of Lysis Buffer. Flash-freeze cell droplets as in 2.2.2 and store at −80°C.
2.4.3. Cell Lysis and chitin column
Lyse the cells in a Nitrogen mill or using French press (as in step 2.3.3.) For mill, transfer lysate to a prechilled 250 ml bottle. Resuspend the lysate in 80 ml Lysis Buffer with 1 protease inhibitor tablet per 50 ml.
Clarify the lysate by spinning at 25,000 × g for 30 minutes at 4°C.
Add 6 ml pre-equilibrated chitin resin to bottle with the clarified supernatant. Rock at 4°C for 30 minutes to allow protein to bind.
Pellet chitin beads at low speed (3000 × g) for 2 minutes.
Wash beads with 50 ml Wash Buffer.
Decant supernatant and resuspend beads in 20 ml Wash Buffer and pour into Econopac column. Allow column to drain to the bed surface.
Wash column with 100 ml Intein Wash Buffer (4 × 25 ml washes).
Run 25 ml of Intein Cleavage Buffer over the column. Drain to the bed surface. For labeled purification, proceed to section 2.4.5.
Add 8 ml of Intein Cleavage Buffer supplemented with 50 mM DTT to the column. Cap the bottom of the column when ~2 ml buffer remains above the chitin bed. Pipette slurry up and down with a 1 ml pipette tip to thoroughly mix. Cover the top and wrap the bottom cap with parafilm, and incubate at room temperature overnight (~15–20 hours) for intein cleavage.
Collect 7–8 ml eluate in a 50 ml falcon tube, using Intein Cleavage Buffer to wash the column. Check by Bradford using BSA as a protein standard or a nanodrop at 280 nm to ensure a protein peak has eluted. Avoid excessive rinsing as this will result in intein contamination. Dilute eluate 5-fold with no-salt buffer to bring concentration to 100 mM KCl.
2.4.4. Heparin column
Filter diluted eluate through a 0.45 μm syringe filter. Run eluate over a 5 ml Heparin column equilibrated in Low Salt Buffer. Wash the column with 10 cv Low Salt Buffer, then elute with a linear gradient from 0–50% High Salt over 10 cv, then with 5 cv 100%.
Dialyze combined peak overnight in 2 L of Enzyme Storage Buffer. Analyze the purity of the protein using SDS-PAGE. If the protein has contaminating intein domain, this can be removed by running the protein over chitin beads equilibrated in Storage Buffer.
Check the concentration by a Bradford assay using BSA as a protein standard. If concentration is necessary, use 10K MWCO concentrators, and check the concentration frequently so that the concentration won’t exceed 50 μM. We observe precipitation at higher concentrations. Aliquot in small amounts to avoid excessive freeze-thawing. Flash freeze in liquid nitrogen and store at −80°C. Typical yields are ~25 nmols/L cells.
2.4.5. Labeling eIF4B during purification by expressed protein ligation
After washing the chitin column with Intein Cleavage Buffer, drip 3 ml Intein Cleavage Buffer containing 200 mM MESNA (sodium 2-mercaptoethanesulfonate) through the column.
Add 0.5 ml 1 mM Cys-Lys fluorophore in 200 mM MESNA to the column, and allow to enter the column. Cap the column bottom before fluorophore drips out, and mix by pipetting slowly with a 1 ml pipette tip. Parafilm the column top and around the bottom cap, and wrap the column carefully in foil, incubating overnight for elution and labeling. Addition of MESNA allows transthioesterification of the protein, promoting efficient ligation of the fluorescent dipeptide to the purified protein during intein cleavage/elution [14].
Proceed with step 9 of 2.4.3. Keep the sample tubes covered with foil and avoid excessive exposure to fluorescent lighting if possible before and after the Heparin purification, and wrap the dialysis buffer beaker in foil for overnight dialysis to avoid photobleaching.
Tip: If one wishes to purify both labeled and unlabeled protein from the same lysate, a portion of the bound chitin resin can be removed from the washed column to a second column for labeling after step 8 of 2.4.3. We often remove 0.5 ml of washed resin slurry for labeling in a smaller Polyprep column, since we use much less labeled than unlabeled protein.
2.5. Preparation of full-length untagged eIF4G from E.coli
2.5.1. Materials Required
Plasmid pSW149 (Addgene #122248)
BL21-CodonPlus (DE3)-RIL Competent Cells (Agilent #230245)
250 ml flask with 50 ml and 6 × 2.8 L Fernbach flasks with 1.5 L LB Carb/Cm
1 M IPTG
DNase I
Roche Complete EDTA-Free Protease inhibitor tablets
Micrococcal Nuclease (NEB #M0247S)
- Chitin Column Buffers (Make 1 L each and store at 4°C):
- eIF4G-Intein Lysis Buffer: 20 mM HEPES-KOH (pH 7.4), 500 mM KCl, 1 mM EDTA (pH 7.4)
- MNase Buffer: 20 mM HEPES-KOH (pH 7.4), 100 mM KCl, 2 mM CaCl2
- eIF4G-Intein Cleavage Buffer: 20 mM HEPES-KOH (pH 8), 250 mM KCl, 1 mM EDTA (pH 8)
Chitin Resin
- Q Column Buffers:
- No salt Buffer: 20 mM HEPES-KOH (pH 7.4), 10% (v/v) Glycerol, 2 mM DTT*
- Q100 Buffer: 20 mM HEPES-KOH (pH 7.4), 100 mM KCl, 10% (v/v) Glycerol, 2 mM DTT*
- Q1000 Buffer: 20 mM HEPES-KOH (pH 7.4), 1 M KCl, 10% (v/v) Glycerol, 2 mM DTT*
-
eIF4G Storage Buffer: 20 mM HEPES-KOH (pH 7.4), 250 mM KOAc, 10% (v/v) Glycerol, 2 mM DTT*
*Add DTT to buffers just before use
Note: Buffers are kept at 4°C unless otherwise specified.
Econopac column
0.45 μm syringe filter
5 ml HiTrap Q HP column
10% SDS-PAGE materials
10K MWCO Protein Concentrators
2.5.2. Cell Growth
Transform BL21-CodonPlus (DE3)-RIL cells with pSW149 and plate on LB Carb. Incubate overnight at 37°C.
Inoculate 50 ml LB Carb/Cm with 10 isolated colonies. Grow the culture with shaking at 250 rpm overnight at 37°C.
Inoculate 6 × 1.5 L flasks of LB Carb/Cm with 6 ml overnight culture each. Grow the culture with shaking at 250 rpm at 37°C.
When the culture reaches mid-log phase (OD600 0.5–0.6), add IPTG for 0.5 mM final concentration. Shake at 250 rpm for two hours at 37°C.
Collect cells by pelleting at 6500 × g for 5 minutes at 4°C. Wash the combined cells with water and then eIF4G-Intein Lysis Buffer, immediately pellet again and dump the supernatant. Weigh the cell pellet to determine its volume, and resuspend in 1/3 of the pellet volume of eIF4G-Intein Lysis Buffer. Flash-freeze cell drops in liquid nitrogen. Store cells at −80°C.
2.5.3. Cell Lysis
Lyse cells as in section 2.2.3.
Store lysate at −80°C until use.
2.5.4. Purification by Chitin column
Resuspend lysate powder in 50 ml eIF4G-Intein Lysis Buffer containing 1 protease inhibitor tablet. After lysate is thawed and resuspended, add 50 μl Triton X-100, 250 μl 1 M MgCl2, and 100 μl 5 mg/ml DNase I. Incubate on ice 30 minutes or until the lysate is less viscous.
Clarify the lysate by spinning at 25,000 × g for 30 minutes at 4°C.
Transfer clarified supernatant to a 50 ml conical tube with 8 ml chitin slurry equilibrated in Lysis Buffer. Rock tube at 4°C for 30 minutes.
Pellet chitin beads at ≤3000 × g for 2 minutes at 4°C and discard supernatant.
Wash beads by suspending in 50 ml eIF4G-Intein Lysis Buffer and repelleting.
Resuspend resin in 20 ml Intein Lysis Buffer and pour into column.
Wash the column with 100 ml Intein Lysis Buffer (4°C), then 25 ml of MNase Buffer (room temperature), and drain to bed surface.
Prewarm 3 ml MNase Buffer to 37°C and then add Micrococcal Nuclease to 3 U/μl. Drip nuclease solution through the column, cap both ends, and incubate in a 37°C incubator for 30 minutes.
Wash column with 5 ml Lysis Buffer with 5 mM EGTA and incubate 5 minutes at room temperature to stop the reaction.
Wash the column with 50 ml Intein Lysis buffer (4°C), then 25 ml Intein Cleavage Buffer (4°C).
Add 8 ml Intein Cleavage Buffer containing 50 mM DTT. Cap the column when ~2 ml Intein Cleavage Buffer remains above column bed. Mix well to distribute the DTT and incubate the column at 4°C for 24 hours. As with eIF4B in section 2.4.5, labeled eIF4G can be generated by addition of MESNA and CL-fluorescein/TAMRA/etc. peptide in place of DTT. Collect eluate, washing out protein with eIF4G-Intein Lysis Buffer.
2.5.5. Anion Exchange Column
Dilute chitin eluate to 100 mM KCl using No Salt Buffer, and filter through a 0.45 μm syringe filter.
Load filtered eluate onto a Q column equilibrated in Q100 Buffer and wash with 10 cv Q100. Elute with a linear gradient from 0–50% Q1000 over 10 cv, then wash with 5 cv Q1000.
Dialyze combined peak overnight in 2 L eIF4G Storage Buffer at 4°C. Analyze the purity of the protein using SDS-PAGE.
Check the concentration by Bradford assay using BSA as a protein standard. If concentration is needed, use a 10 kDa protein concentrator, stopping every 20 minutes to pipette the liquid up and down to prevent sticking to the concentrator. Aliquot in small amounts to minimize freeze-thawing. Flash freeze in liquid nitrogen and store at −80°C. Typical yields are ~2 nmols/L.
2.5.6. Purification of recombinant untagged eIF4G complexed with eIF4E
Coexpression of eIF4E stabilizes eIF4G, so for purposes in which WT eIF4E will be present in stoichiometric amounts, we prefer the copurified complex. The protocol for purification of the eIF4G•eIF4E complex is the same as that for isolated eIF4G in section 2.5, with the following modifications:
Cells are transformed with both the pSW149 plasmid expressing yeast eIF4G1 fused to the intein-chitin-binding domain, and the pLAMP-eIF4E plasmid (KanR, obtained from J. Lorsch lab, NICHD, NIH). Colonies are selected on LB with 50 μg/ml Carbenicillin and 50 μg/ml Kanamycin, and then grown in liquid media containing these antibiotics as well as 34 μg/ml Chloramphenicol.
Optimal expression of eIF4G complexed with eIF4E is achieved by induction overnight at 16°C, as outlined for eIF4B overexpression in Section 2.4.2. Cells should be lysed in the Nitrogen mill to prevent degradation.
Typical yields are ~5 nmols/L.
2.6. Preparation of capped RNA with and without a poly(A) tail
2.6.1. Materials Required
Plasmid DNA (e.g. pSW104, Addgene #122295)
Restriction enzyme and buffer
Gel Purification Kit (Qiagen or similar)
5X Transcription buffer: 400 mM HEPES-KOH (pH 7.5), 120 mM MgCl2, 10 mM Spermidine, and 200 mM DTT
100 mM NTPs: 25 mM of each, ATP, CTP, GTP, UTP (Affymetrix)
RiboLock RNase inhibitor (Thermo #EO0381)
T7 RNA Polymerase: His-tagged T7 RNAP can be purified on a Nickel column [15], or this can be purchased. The optimal concentration to use must be determined in small test reactions.
STOP dye: 0.05% xylene cyanol, 0.05% bromophenol blue, 0.5 M EDTA (pH 8), and formamide to final volume.
TURBO DNase (Thermo #AM2238)
Vaccinia Capping Enzyme Kit (NEB #M2080)
Alpha-32P-GTP (Perkin Elmer #BLU006H), optional
RNeasy mini kit (Qiagen #74104)
2.6.2. Preparation of plasmid DNA
We clone mRNA sequences containing known 5’ and 3’ untranslated regions 3’ of a T7 promoter with restriction sites before and after a 3’ 82 adenosine tail into a pSP65A plasmid (gift from Alan Jacobson). Plasmids encoding poly(A) mRNA must be maintained in cells containing the F’ episome or the poly(A) sequence will undergo random shortening. The plasmid encoding RPL41A mRNA and an 83 nucleotide tail is called pSW104. Cutting pSW104 with BamHI prior to transcription yields untailed mRNA, while cutting with HindIII gives mRNA with an 82A tail. The transcription buffer used here is optimized for higher concentrations of NTPs needed for production of longer RNAs and those with a poly(A). Small test reactions should be performed, varying the amounts of template, RNAP, NTPs, and Magnesium as needed.
2.6.3. Restriction Digestion for Run-off Transcription
In a reaction volume of 1 ml, combine the following: purified plasmid DNA (1000 μg), restriction enzyme buffer (1X), restriction enzyme (1 unit per μg DNA, 1000 units in total), and water to bring the reaction to its final volume.
Incubate at 37°C for 4 hours to overnight to ensure complete digestion of the plasmid DNA.
Confirm complete digestion of the plasmid DNA on a 0.8% agarose gel. If undigested plasmid remains, cut plasmid can be isolated using a Qiagen or other similar gel purification kit. If digest appears complete when a large amount is loaded beside uncut, the digest does not require purification prior to transcription if an RNase inhibitor is added.
2.6.4. In vitro Transcription
In a reaction volume of 10 μl, combine the following: digested plasmid DNA (0.5–1 μg), Transcription buffer (1X), NTPs (20 mM, 5 mM of each NTP), RiboLock RNase inhibitor (1 U/μl), and T7 RNA polymerase (amount determined empirically).
Incubate the reaction for one hour at 37°C, and continue in an incubator at 30°C overnight to allow complete extension of transcripts.
Remove half the reaction to a tube containing 1 unit of Turbo DNase. Add 1 μl water to remaining reaction and incubate both samples at 37°C for 10 minutes to remove template from one tube.
Prepare a 10 cm × 7–8 cm, 6% polyacrylamide gel containing 1X TBE and 7M urea. Polymerize the gel with 10 μl of 10% APS and 1 μl of TEMED for every 1 ml of acrylamide solution.
Add 1 volume of STOP dye to each sample and heat at 95°C for 1 to 2 minutes prior to loading on the gel.
Run the gel at 200 V in 1X TBE until the xylene cyanol has traveled 3/4 of the way down the gel (approximately 1 hour).
Stain the gel with Methylene Blue for 20 minutes, destain with water if needed.
Evaluate the results and scale up the reaction using the conditions that gave the most full-length product without truncated transcripts. Adjust the volume to ≥1 ml for generating large amounts of RNA.
2.6.5. Purification and enzymatic capping of transcripts
Transcripts can be purified on 6% Urea PAGE if there are multiple species present, followed by Phenol:Chloroform extraction and ethanol precipitation [16]. With longer RNA species, however, the loss of RNA during gel purification is substantial, so it is preferable to optimize transcription conditions to generate a single RNA species that can be purified by Lithium Chloride precipitation [17]. If RNA is not capped before use, it is advisable to use an RNA cleanup kit, or further purify by use of a gel filtration column to remove contaminating nucleotides.
Add one volume of 7.5 M LiCl (3.75 M final), incubate at −20°C for at least 30 minutes, and pellet RNA for 15 minutes at 12,000 × g.
Wash pellet by adding 2.5 volumes 70% Ethanol, vortexing, and re-pelleting for two minutes at 12,000 × g.
Allow the pellet to dry in the open tube at room temperature for 5 minutes to one hour, and dissolve in water.
Following purification, a 5’ cap is enzymatically added using Vaccinia D1/D12 capping enzyme, either recombinantly purified [18], or purchased as a kit (NEB). Following capping, RNA is further cleaned up with an RNeasy or similar kit.
Efficiency of capping can be monitored by performing a 10 μl test reaction with a spike of 1 μl alpha-32P-GTP added, then running the test reaction on a gel beside the same amount of GTP in reaction buffer, and determining the fraction of GTP incorporated into full length RNA to calculate the fraction RNA capped. Alternatively, Primer extension can be performed with uncapped vs. capped RNA using a fluorescent or radiolabeled primer that anneals near the 5’ end and determining the fraction of RNA that is extended by one nucleotide following capping using a denaturing gel or capillary electrophoresis [8].
2.7. Monitoring mRNA recruitment factor interactions
Note: To measure binary interactions between two factors or between factors and ribosomes, use a labeled factor in place of labeled RNA, and titrate in the unlabeled binding partner.
Labeled factor or RNA interactions can also be monitored by running reactions on a native gel as in section 2.9 and visualizing the fluorescent RNA or protein on a fluorescent scanner. Annealing a fluorescent DNA oligo 16 or more nucleotides downstream of the start site allows the visualization of capped-mRNA on gels without use of radioactivity.
2.7.1. Materials Needed
10X Reconstitution Buffer: 300 mM HEPES-KOH (pH 7.4), 1 M KOAc (pH 7.6), 30 mM Mg(OAc)2, 20 mM DTT
Fluorescently-labeled RNA: We order 3’ fluorescein and TAMRA-labeled RNAs from IDT, Dharmacon, or Bio-synthesis. Our single-strand model is a 43-mer of 10 5’ and 3’ poly(UC) repeats flanking AUG, with a 3’ FAM.
Purified protein of interest
Enzyme Buffer: 20 mM HEPES-KOH (pH 7.4), 100 mM KOAc (pH 7.6), 10% (v/v) Glycerol, 2 mM DTT
Plate reader for measuring fluorescence polarization (e.g. Tecan Spark)
Corning NBS 384-well black plate (Corning #3820; we empirically determined that these plates are less susceptible to sticking than several others; Plates can be used multiple times by taping over used wells after each experiment and using open wells for subsequent experiments.)
Low retention tubes and tips are critical for accurate measurements with “sticky” RNA binding proteins such as eIF4B and eIF4G. We purchase posi-click tubes from Denville and low retention tips from Denville or VWR.
Kaleidagraph (Synergy software) or alternative fitting software (e.g. Graphpad Prism)
2.7.2. Protocol
Dilute the protein of interest in 0.6 ml Eppendorf tubes on ice using Enzyme Buffer. The dilutions to use depend on the KD of the interaction. If the KD is unknown, start with a wide-range, with final nanomolar concentrations of 0, 20, 40, 60, 80, 100, 200, 400, 600, 800, 1000, 1250, 1500, 2000, and 5000, and make 10X stocks of each (i.e. 0, 200, 400, etc.). Once the affinity is known, repeat the experiment with concentrations that cover the curve. Use 2.5 μl Enzyme Buffer for 0 protein reaction.
Make a master mix containing the buffer, RNA, and water for all of the reactions, so the final reaction volumes will be 25, and final concentrations in the reactions will be: 1X Recon Buffer, 30 nM fluorescently labeled RNA, and 1X protein. Using a master mix enhances reproducibility.
Aliquot 22.5 μl master mix for each reaction in 0.6 ml tubes at room temperature. Add 2.5 μl 10X protein to appropriate tube, and mix by pipetting 3 times. Reactions should incubate at room temperature or in a 26°C water bath for at least 10 minutes to ensure equilibrium is established prior to taking measurements.
Carefully pipette 10 μl of each reaction into two wells of the 384 well plate. Read fluorescence polarization for the fluorophore of interest. These technical replicates minimize noise, but biological replicates should be performed three times when optimal conditions are achieved.
Plot the anisotropy values (y) versus final protein concentration (x), and fit to a hyperbolic or quadratic model for binary interactions to determine KD [19]. A quadratic function is used when the concentration of the labeled component is close to the KD, generating an L-shaped curve instead of a hyperbola. An added correction factor should be added to either equation (i.e. m3 + equation in Kaleidagraph) so that the fitting software will adjust the y intercept based on the starting anisotropy value. Data can also be converted to fraction bound by subtracting the free RNA anisotropy and normalizing to the BMax.
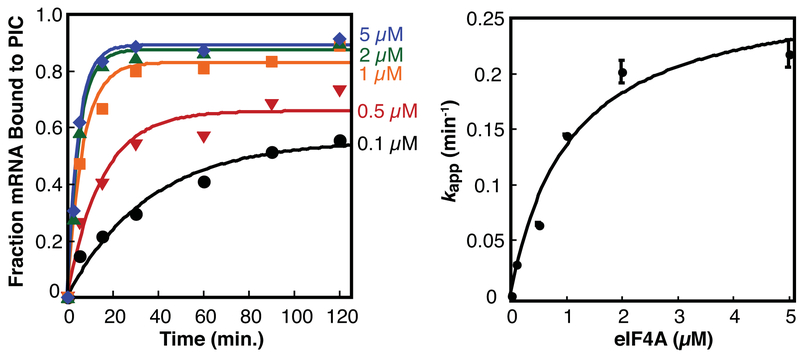
2.8. Monitoring factor dependence of mRNA recruitment to the ribosome (Figure 4)
Figure 4.
Kinetic stimulation and stabilization of mRNA binding to the translation preinitiation complex by eIF4A. The fraction of capped mRNA bound to the preinitiation complex was monitored by native gel shift at varying concentrations of eIF4A and plotted over time to derive the pseudo-first order apparent rate constant kapp. The kapp was plotted versus eIF4A concentration and fit to a hyperbolic function to determine the concentration of eIF4A that stimulates half-maximal rate, K1/2 or apparent affinity of eIF4A for the initiation complex.
For each new mRNA to be tested, controls must be run to map the PIC and ensure that mRNA bound to proteins doesn’t comigrate with the ribosome-bound complex. The appropriate control lacks 40S, but still contains all other components of the translation initiation complex to be analyzed. Comigration of RNA with labeled eIF1A is a strong indicator of mRNA recruited to a PIC.
A lack of dependence of the rate of mRNA recruitment on eIF4B or eIF4G concentration is indicative of a problem with the capping reaction, which is sometimes the result of inactive S-adenosyl-methionine [8].
2.8.1. Materials Required
10X Recon Buffer: 300 mM HEPES-KOH (pH 7.4), 1 M KOAc (pH 7.6), 30 mM Mg(OAc)2, 20 mM DTT
Purified eIF2, Methionylated initiator tRNA, 40S subunits, eIF1, eIF1A, and eIF5 [16, 20]
10 mM GDPNP with 10 mM Mg(OAc)2
100 mM ATP with 100 mM Mg(OAc)2
Purified eIF3 (Section 2.2)
Purified eIF4A (Section 2.3)
Purified eIF4B (Section 2.4)
Purified eIF4G•eIF4E or eIF4G only (Section 2.5)
Labeled capped mRNA (Section 2.6)
Native gel apparatus, plates, combs, and refrigerated circulator
10X THEM Buffer: 340 mM Tris Base, 570 mM HEPES, 1 mM EDTA, 25 mM MgCl2
40% 37.5:1 acrylamide: bisacrylamide solution
Native loading dye: 50% sucrose, 0.02% bromophenol blue
2.8.2. Reaction Setup
- Pour native gels. To measure a single K1/2 for factor or nucleotide titration, ~6–7 concentration points are needed at ~5–10 timepoints. When using gels with 15 wells, one gel is used for each timepoint, so K1/2 is measured for two proteins in an experiment.
- Wipe washed plates with ethanol, then SigmaCote. Distribute the SigmaCote by rubbing with a Kimwipe extensively. Assemble plates and spacers in a casting assembly
- Make a solution of 1X THEM and 4% 37.5:1 acrylamide (5 ml per gel.) Remove 0.5 ml to an Eppendorf tube and add 10 μl 10% APS, 1 μl TEMED. Pipette 0.4 ml into plates as a “plug” and allow to polymerize. Repeat for other gels.
- When plug is polymerized, add same ratio of APS and TEMED to remaining THEM-acrylamide mixture and pour gels using a serological pipette. Set combs, avoiding bubble formation, and allow to polymerize for at least 30 minutes.
- Carefully remove combs from gels and assemble gels using 1X THEM as running buffer. Use lane markers for easy loading.
- Set up racks with tubes containing 1 μl of native loading dye each.
- Make a master mix for the common components at room temperature, keeping stocks on ice if they will be returned to the freezer. 4 μl is sufficient loading volume for each timepoint for monitoring radiolabeled or fluorescent capped mRNA on a 15-well gel. Optimal reaction concentrations indicated in parentheses are for specific preparations of proteins. It is advisable to empirically determine the optimal concentration of each preparation of protein with a control mRNA, such as capped RPL41A, as we have observed slight changes in activity for independent preparations of eIF4A, eIF4G, and eIF3.
- Form ternary complex by incubating eIF2 (0.2 μM) with GDPNP (0.5 mM) at 26°C for 10 minutes, then add Met-tRNA (0.2 μM) and incubate an additional 5 minutes.
- To ternary complex, add 10X Recon Buffer (1X), water, ATP (5 mM), 40S subunits (30 nM), eIF1 (1 μM), eIF1A (1 μM), eIF5 (0.2 μM), eIF3 (0.3 μM), eIF4A (2 μM), eIF4B (0.3 μM), and eIF4E•eIF4G (50 nM). The component being analyzed should be omitted.
- Aliquot master mix (e.g. 40 μl), and add titrated protein of interest (e.g. 5 μl 10X stocks). Mix well, and incubate for at least 10 minutes at room temperature.
- Turn on native gel apparatus at 200 V, with the refrigerated circulator running at 18°C. The gels are loaded while running to quench the reactions. Otherwise, unlabeled capped mRNA can be used as a quench. Take care to not place hands in the open buffer tank.
- Add labeled capped mRNA (15 nM final from a 10X stock, e.g. 5 μl) to each tube, and mix by pipetting several times. Set up the reactions in a defined order, separated by a defined time interval (e.g. every 15 seconds), so that when loading gels, sample loading will be spaced by the same interval.
- ~15 seconds before the first timepoint, remove 4 μl of the first sample to a tube of native dye (from 2.8.2 1.), pipette twice and load onto the gel. Repeat for the rest of the concentration points. Repeat the loading of concentration points on additional gels for the additional timepoints.
- Run each gel for 1 hour at 200 V with cooling to 18°C. When fluorescent gels finish running, rinse and dry the outside of the plates and scan immediately. When radioactive gels finish, remove one plate (taking note of loading order), place a square of Whatman paper on top, and press gently to transfer gel to Whatman. Cover gel and paper with plastic wrap, taking care to wrap the back as well to avoid contamination. Wipe the surface of the plastic wrap to remove any bubbles and avoid contaminating the phosphor screen. Label gel and place at −20°C until all gels are finished running.
- Place gels in a phosphorimager cassette, wipe with a Kimwipe to remove any moisture, and place a phosphorimager screen on gels. Wrap the phosphorimager cassette in plastic (or store in a reusable plastic bag for smaller screens) to prevent moisture from accumulating on the screen and expose overnight at −20°C.
- Remove cassette from −20°C and allow to equilibrate for ~10 minutes. Wipe away moisture before unwrapping and opening cassette. Scan screen for radioactivity and quantify the fraction of mRNA located in the preinitiation complex band relative to the total mRNA signal in that lane using ImageQuant software. PIC bands for a new complex or mRNA should be verified using a no 40S control lane (all other components should be present in this control to distinguish PICs from multifactor complex), or by mapping complexes using fluorescently-labeled eIF1A, a stable component of the PIC.
- Plot fraction mRNA bound versus time for each reaction in Kaleidagraph, and fit to a single exponential equation to derive the apparent rate constant for each concentration point. Plot each apparent rate constant versus concentration and fit to a hyperbolic equation to determine the concentration of varied component at half-maximal rate (K1/2), and the maximal rate constant for that reaction.
3. Results and Discussion
To understand how mRNAs are recruited to the eukaryotic ribosome, an obvious and essential tool is a robust in vitro system of purified components, which can be coupled easily to studies performed in vivo. Here we add to previous work developing in vitro tools to study the yeast system [8, 16], by including optimized purification strategies for eIF3, eIF4A, and eIF4B, and a new full-length untagged eIF4G construct, as well as strategies for measuring interactions between RNA and components within these translation preinitiation complexes.
As previously described, eIF3 was expressed in yeast [8], but here a Nitrogen mill is employed for highly reproducible lysis [20]. Figure 1 shows the improved eIF3 purification scheme with representative chromatograms and gels. Lysate was loaded onto a Nickel column, washed, and eluted by adding High Imidazole Buffer. Use of a gradient is discouraged as this may fractionate the subunits. Yield and purity were marginally improved without affecting activity of eIF3 by performing phosphocellulose chromatography before size exclusion chromatography, which allowed for better separation of peaks on the latter. Phosphocellulose chromatography separated eIF3 from a number of contaminants in the flowthrough and into two elution pools: the first fractions eluting at lower salt concentrations were nearly pure, whereas the later fractions eluting at higher salt contained undesirable contaminants and high levels of RNases. A commercially available Superdex 200 size exclusion column was then used to both exchange the buffer and separate the full eIF3 core complex from smaller eIF3B-containing complexes.
Purification of eIF4A strictly requires the removal of nucleic acid, which often facilitates copurification of RNases. To effectively remove bound RNA and RNases, we treated the ammonium sulfate-fractionated overexpression lysate with Benzonase, a robust nuclease that targets both DNA and RNA, followed by a strong anion exchange column (Figure 2). A combination of isocratic followed by linear gradients were used to separate eIF4A from contaminants. The only peak showing higher 280 nm over 260 nm absorption was analyzed and those fractions lacked a doublet band near ~50 nucleotides and lower molecular weight contaminants. Those fractions containing substantial eIF4A and lacking contaminants were run on a Superdex 200 column, which separated a number of larger complexes from eIF4A, the largest peak, eluting after ~16 ml. Finally, a hydroxyapatite column removed residual RNase activity from the protein (data not shown). The purification method yielded ~5 mg of nuclease-free eIF4A per liter of cultured cells.
Purification of eIF4B and eIF4G rely on fusion of an intein-chitin-binding domain to each protein (Figure 3). For eIF4B, the fusion protein was purified on a chitin column, then a heparin column, similarly to previously published protocols for purification of eIF1 and eIF1A. It should be noted that full-length eIF4B is “sticky” and precipitated out at some threshold above 50 μM when concentrating. When working with eIF4B, low retention tips, tubes, and microplates led to less retention of protein over time (data not shown).
Full-length eIF4G, particularly in the absence of eIF4E, is not often used for in vitro studies of translation initiation, due to the difficulty of expressing and purifying sufficient yields of protein lacking nuclease activity and inhibitory degradation products [8]. To improve eIF4G and eIF4G•eIF4E purification methods and yields, the yeast eIF4G1 gene was cloned into pTYB2 for intein-fusion. Preliminary induction experiments indicated that optimal expression of the solitary yeast protein in E. coli was achieved after a two-hour IPTG induction at 37°C, while for the complex of eIF4G with eIF4E, higher soluble protein levels were yielded by inducing with IPTG at 16 degrees Celsius overnight. eIF4G requires micrococcal nuclease treatment to effectively remove RNase contaminants, but must first be concentrated for optimal nuclease activity without using prohibitively expensive volumes. However, full-length eIF4G (lacking intein) sticks to protein concentrators at the lower salt conditions required for micrococcal nuclease activity. Hence the nuclease digestion was performed while the intein-fusion protein was bound to the chitin column (Figure 3B). This method effectively concentrated a more soluble form of the protein without precipitation, allowed exchange into the proper Calcium-containing buffer with nuclease, and later allowed washing away of nuclease prior to elution. Following the chitin column, the eIF4G protein was fractionated on a Q column, separating full-length protein from a known inhibitory degradation product. The yield of eIF4G•eIF4E was increased by ~6–12-fold, whereas the yield of full-length eIF4G without eIF4E was increased by ~20-fold by these modifications [8]. Activity tests (not shown) indicated that the new eIF4G purification method gave protein that stimulated mRNA binding to the ribosome to the same degree as the published eIF4G purified with a GST-tag.
In order to generate high quality kinetic data, it is critical that the reagents used are of high quality. Common sources for loss of activity of reagents described here are through excessive freezing and thawing, and seemingly unavoidable loss of activity of S-adenosyl-methionine used for capping mRNA. It is recommended that proteins and RNAs be stored in very small aliquots at −80°C, and that the stocks for capping be made fresh every 6 months to 1 year. Nuclease contaminants in protein preparations may also lead to failure. These nucleases are often carried over by mutual association with RNA bound to a protein. Minor modifications introduced in the protocols here are present to avoid nucleases, such as treating with a nuclease to remove RNA during eIF4A and eIF4G preparation.
To understand how a particular mRNA sequence or structure alters translation initiation, it is useful to measure the concentration dependence of initiation factors on the rate [3, 6]. From these measurements, K1/2, the concentration of protein that stimulates half maximal rate, can also be determined, indicating how interactions of the titrated factor with the PIC and mRNP are affected by the RNA sequence of interest. In vitro assays described in sections 2.7 and 2.8 allow analysis of both RNA-binding to proteins, and capped mRNA-binding to the ribosome (mRNA recruitment). We provide protocols for fluorescence anisotropy measurements of RNA binding to single proteins in 384 well plates (Section 2.7). For mRNA recruitment measurements, we provide a basic protocol for monitoring rates of mRNA recruitment at varied concentrations of a reagent of choice by gel shift (Section 2.8), and an example of time courses of mRNA recruitment at different concentrations of eIF4A (Figure 4). Comparison of the ability of an RNA-binding protein to bind to a particular RNA sequence, and the ability of the protein to stimulate ribosome binding can provide insights into RNA sequence and structure-specific mechanisms of translational control.
Highlights.
Eukaryotic translation initiation factors bind RNA and the ribosome to affect changes in translation.
Capped mRNAs display strong dependence on the eIF4 factors for ribosome binding
Factor dependence of translation initiation can be kinetically determined
Optimized purification strategies allow larger studies of apparent affinities of translation initiation factors for the preinitiation complex
Acknowledgments
4. Funding: This work was supported by the National Institutes of Health (R00GM119173) and startup funds from the University at Buffalo College of Arts and Sciences.
5. Acknowledgements: The authors would like to thank Sarah Mitchell, Vaishnavi Rajogopal, Colin Aitken, and Jon Lorsch for useful discussions. Initial work optimizing this system was performed in the Lorsch lab.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- [1].Hinnebusch AG, Lorsch JR, The mechanism of eukaryotic translation initiation: new insights and challenges, Cold Spring Harb Perspect Biol 4(10) (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Mitchell SF, Walker SE, Rajagopal V, Aitken CE, Lorsch JR, Recruiting knotty partners: The roles of translation initiation factors in mRNA recruitment to the eukaryotic ribosome, Ribosomes: Structure, Function, and Dynamics (2011) 155–169. [Google Scholar]
- [3].Walker SE, Zhou F, Mitchell SF, Larson VS, Valasek L, Hinnebusch AG, Lorsch JR, Yeast eIF4B binds to the head of the 40S ribosomal subunit and promotes mRNA recruitment through its N-terminal and internal repeat domains, RNA 19(2) (2013) 191–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Costello J, Castelli LM, Rowe W, Kershaw CJ, Talavera D, Mohammad-Qureshi SS, Sims PFG, Grant CM, Pavitt GD, Hubbard SJ, Ashe MP, Global mRNA selection mechanisms for translation initiation, Genome Biol 16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Guenther UP, Weinberg DE, Zubradt MM, Tedeschi FA, Stawicki BN, Zagore LL, Brar GA, Licatalosi DD, Bartel DP, Weissman JS, Jankowsky E, The helicase Ded1p controls use of near-cognate translation initiation codons in 5’ UTRs, Nature 559(7712) (2018) 130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gupta N, Lorsch JR, Hinnebusch AG, Yeast Ded1 promotes 48S translation preinitiation complex assembly in an mRNA-specific and eIF4F-dependent manner, Elife 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Iwasaki S, Floor SN, Ingolia NT, Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor, Nature 534(7608) (2016) 558-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mitchell SF, Walker SE, Algire MA, Park EH, Hinnebusch AG, Lorsch JR, The 5 ‘−7-Methylguanosine Cap on Eukaryotic mRNAs Serves Both to Stimulate Canonical Translation Initiation and to Block an Alternative Pathway, Mol Cell 39(6) (2010) 950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sen ND, Zhou FJ, Harris MS, Ingolia NT, Hinnebusch AG, eIF4B stimulates translation of long mRNAs with structured 5 ‘ UTRs and low closed-loop potential but weak dependence on eIF4G, P Natl Acad Sci USA 113(38) (2016) 10464–10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Andreou AZ, Klostermeier D, eIF4B and eIF4G jointly stimulate eIF4A ATPase and unwinding activities by modulation of the eIF4A conformational cycle, J Mol Biol 426(1) (2014) 51–61. [DOI] [PubMed] [Google Scholar]
- [11].Ozes AR, Feoktistova K, Avanzino BC, Fraser CS, Duplex unwinding and ATPase activities of the DEAD-box helicase eIF4A are coupled by eIF4G and eIF4B, J Mol Biol 412(4) (2011) 674–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhou FJ, Walker SE, Mitchell SF, Lorsch JR, Hinnebusch AG, Identification and characterization of functionally critical, conserved motifs in the internal repeats and N-terminal domain of yeast translation initiation factor 4B (yeIF4B), J Biol Chem 289(17) (2014) 11860–11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Phan L, Zhang X, Asano K, Anderson J, Vornlocher HP, Greenberg JR, Qin J, Hinnebusch AG, Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5, Mol Cell Biol 18(8) (1998) 4935–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Schwarzer D, Cole PA, Protein semisynthesis and expressed protein ligation: chasing a protein’s tail, Curr Opin Chem Biol, 9 (2005) 561–569. [DOI] [PubMed] [Google Scholar]
- [15].Ichetovkin IE, Abramochkin G, Shrader TE, Substrate recognition by the leucyl/phenylalanyl-tRNA-protein transferase. Conservation within the enzyme family and localization to the trypsin-resistant domain, J Biol Chem, 272 (1997) 33009–33014. [DOI] [PubMed] [Google Scholar]
- [16].Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR, Reconstitution of yeast translation initiation, Method Enzymol 430 (2007) 111–145. [DOI] [PubMed] [Google Scholar]
- [17].Walker SE, Lorsch J, RNA Purification - Precipitation Methods, Laboratory Methods in Enzymology: RNA 530 (2013) 337–343. [DOI] [PubMed] [Google Scholar]
- [18].Cong PJ, Shuman S, Mutational Analysis of Messenger-RNA Capping Enzyme Identifies Amino-Acids Involved in GTP-Binding, Enzyme-Guanylate Formation, and GMP Transfer to RNA, Mol Cell Biol 15(11) (1995) 6222–6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Pollard TD, A guide to simple and informative binding assays, Mol Biol Cell, 21 (2010) 4061–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Munoz AM, Yourik P, Rajagopal V, Nanda JS, Lorsch JR, Walker SE, Active yeast ribosome preparation using monolithic anion exchange chromatography, RNA Biol 14(2) (2017) 188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]