Abstract
Upon activation, naive T cells give rise to a heterogeneous cell population of effector and memory T cells that mediate antigen clearance and provide long-lived protection from rechallenge. Many of the transcriptional regulators that direct the differentiation of naive T cells to acquire the range of phenotypic and functional characteristics of effector and memory T cells have been described. However, the active programs that maintain the specific subsets of memory T cells are less clear. Here, we discuss recent studies that suggest effector and memory CD8+ T cells exist in cellular ‘states’ with inherent plasticity. Further, we consider the newly identified role of active transcriptional and epigenetic programming in maintaining the identity of the distinct subsets within the memory population.
Introduction
Essential for the elimination of intracellular pathogens and malignant cells, CD8+ T cells are an important component of the adaptive immune response. Upon activation, antigen-specific CD8+ T cells proliferate and differentiate into a heterogenous population of effector cells that provide protection through cytolytic activity and the secretion of inflammatory cytokines [1]. A portion of the effector cell population has been considered to be terminally-differentiated—providing immediate, acute function, then undergoing apoptosis at the resolution of the infection or shortly thereafter. In contrast, other cells are programmed for long-term survival after the contraction of the effector population to afford durable immunological protection [1]. Heterogeneity in phenotype, function, location, and trafficking ability is also observed within the long-lived memory population [2]. Subsets within the effector or memory CD8+ T cell populations have largely been considered to be cellular ‘fates’ with fixed differentiation paths. However, recent studies suggest that CD8+ T effector and memory populations consist of cells in ‘states’ that require reinforcement by active regulatory programs which, when lost, reveal significant ‘plasticity’ among the distinct subsets. Understanding the functional heterogeneity that exists within the effector and memory T cell population and their corresponding differentiation paths and maintenance programs will allow for efficient design of T cell-based vaccines and adoptive therapies targeting emerging infectious diseases and cancers.
CD8+ T cell heterogeneity
The population of cells with effector function at the peak of infection exhibits substantial phenotypic and functional diversity [3,4], and considerable effort has been made to define cellular phenotypes that predict the ‘fate’ of an effector T cell following resolution of the infection. Expression of KLRG1 and CD127 can be used to delineate the memory potential of effector CD8+ T cells [5–8]. In transfer experiments, CD8+ T cells with high expression of CD127 and low expression of KLRG1 (KLRG1loCD127hi) were found to have a significantly greater capacity to survive following infection compared to the KLRG1hiCD127lo counterparts and exhibited stem-like properties such as multipotency and the capacity for proliferation and self-renewal [5,9,10]. Although both express cytokines and cytolytic molecules, KLRG1loCD127hi CD8+ T cells define a pool of memory precursors (MP) while the KLRG1hiCD127lo subset represents terminal effector (TE) cells that are more likely to die following the resolution of infection. This demarcation is by no means precise as further heterogeneity exists within the TE and MP cell populations [6], and TE cells do persist following infection at memory time points [11–13]. Furthermore, KLRG1 and CD127 are not necessary or sufficient to drive generation of the TE or MP CD8+ T cell populations, respectively [8,14]. Early expression of additional molecules has also been employed to predict the differentiation path of effector CD8+ T cells. Expression of the transcriptional regulator Id3 or TCF1 and reduced levels of IL-2Rα bias an effector CD8+ T cell to a longer-lived memory T cell state [15–18].
The memory population that persists after pathogen clearance is also comprised of cells with a range of phenotypes. Several approaches have been applied to categorize these cells based on phenotype and function, and at least 5 subsets have been identified (Table 1). Traditionally, the circulating CD8+ memory T cells have been divided into two broad subsets, effector memory (TEM) and central memory (TCM), based on anatomical location, expression of cell-surface molecules and effector function [19]. TEM lacking CD62L and CCR7 were originally described to continually recirculate through non-lymphoid tissues and blood surveying for reinfection. With the ability to elicit direct effector function, TEM would be poised to provide immediate protection should reinfection occur [19,20]. CD62LhiCCR7hi TCM are a long-lived subset that can traffic to secondary lymph nodes, have the ability to self-renew, and exhibit a high proliferation capacity upon reactivation [19,21–24]. Recently, surface expression of the chemokine receptor, CX3CR1, was used to refine this classification [25,26]. While classically defined TEM and TCM exhibit high or no CX3CR1 expression, respectively, a novel CX3CR1int subset was recently identified and termed peripheral memory T cells (TPM) [26]. TPM show a superior steady-state self-renewal capacity and can proliferate in a recall response to comparable levels as the CX3CR1− TCM population [26], but have functional abilities such as IL-2 secretion and cytotoxicity intermediary between TCM and TEM. Interestingly, TPM were found to preferentially surveil the non-lymphoid tissues presumably using a unique pattern of migration from blood to tissue to lymph [26]. Within this scheme, the CX3CR1hi TEM population appeared primarily restricted to the intravascular space [25,26] and presumably the spleen. These data emphasize that the TEM population as defined in much of the literature likely contains subsets of cells with distinct trafficking patterns and functional characteristics and highlight the pitfalls associated with phenotypic-based classifications in this context. An additional non-circulating, resident-memory (TRM) subset has also been described that is likely the prominent memory T cell type residing in the non-lymphoid tissues without recirculating, providing the first-line of defense upon pathogen reinfection (reviewed in [27]). Finally, a memory T stem cell (TSCM) population has been described in both human and mouse [28–31]. In some contexts, TSCM are concealed within the CD44loCD62Lhi naive T cells pool but can be identified by their expression of stem cell antigen-1 (Sca-1), B-cell lymphoma protein-2 (Bcl-2), and CD122. Like stem cells, TSCM are multipotent able to generate TE, TEM and TCM subsets, and have the ability to self-renew [29].
Table 1.
Memory T cell Subsets
| Memory Subset | Phenotypic Markers | Properties |
|---|---|---|
| Long-lived Effectors [11,12,32] | KLRG1hiCD127lo/int CD27loCD43lo |
High effector function Weak proliferative capacity |
| TEM [19,25,26] | CCR7loCD62Llo CX3CR1hiCD127hi |
Increased effector function Circulating - blood/spleen |
| TPM [25,26] | CD62Lhi/loCX3CR1int CD127hi |
High self-renewal capacity Increased proliferative capacity Intermediate effector function Circulating -tissues |
| TRM [20] | CD103hi/loCD69hi/lo CD127int |
Proliferative capacity Increased effector function Resident - tissues |
| TCM [19,25,26] | CCR7hiCD62Lhi CX3CR1loCD127hi |
Increased proliferative capacity Self-renewal ability Circulating - SLO |
| TSCM [28,31] | CD44loCD62LhiSca-1hi CD122hi, Bcl-2hi |
High self-renewal capacity Multipotent |
Note: CD127 and CX3CR1 are not necessarily conventional phenotypic marker of memory subsets but their differential expression has highlighted the heterogeneity in memory subsets that previously been under appreciated.
As noted above, T cells with the phenotype of TE T cells are often present for extended periods of time following infection (Long-lived effector cells) and contribute to CD8+ T cell recall responses. Specifically, a CD27loCD43lo T cell population with effector-like attributes, including high expression of KLRG1, T-bet, and granzyme B, but lo/intermediate levels of CD127, has been described [11,32]. Following a primary response, these cells eventually decline; however, boosting greatly enriches the frequency of this population. Despite weak proliferative ability, these cells provide cytolytic-dependent control of certain acute pathogens including Listeria monocytogenes and vaccinia virus [11]. CX3CR1 expression inversely correlates with the degree of effector T cell differentiation [25,26] so the long-lived effector cells likely are included in the CX3CR1hi classification of memory T cell subsets.
While the use of the defined populations introduced here proves useful in the discussion of the molecules involved in establishing and sustaining CD8+ memory T cell responses, it is increasingly apparent that subset heterogeneity is vast and can be flawed as functional and trafficking attributes often span phenotypic subgroups [33]. Thus, it is expected that our understanding of effector and memory T cell differentiation will continue to evolve reflecting perhaps instead a continuum of effector functions, recirculation patterns, and longevity/selfrenewal characteristics.
Transcriptional regulation of effector and memory CD8+ T cells
T cells responding to infection will integrate multiple diverse signals including those from antigen exposure, costimulation and the cytokine and tissue microenvironmental milieu that ultimately influence the final states of the progeny, directing the heterogeneity within the effector and memory T cell populations. Importantly, transcriptional programs are induced that instruct the commitment of a T cell to a particular effector or memory T cell subset (reviewed in [1,34]). Numerous transcription factors have been defined that instruct CD8+ T cell differentiation—formation of MP and TCM subsets require key transcriptional regulators such as Id3 [15,35], TCF1 [17,18], Eomesodermin [36,37], FOXO1 [38] and Bcl6 [39] whereas TE and TEM cell differentiation is supported by Id2 [40–42], Blimp1[43,44], T-bet [6] and Zeb2 [45,46]. However, the transcriptional regulation that is necessary to maintain the phenotypic and functional characteristics of the cellular subsets once established has not been studied as extensively.
Memory CD8+ T cell maintenance
Homeostasis of the persisting memory T cells is a key component of their ability to provide durable protection from reinfection. Memory T cells are maintained in an antigen-independent manner; however, their homeostasis is supported by the cytokines IL-7 and IL-15 [47]. IL-7 and IL-15 initiate the downstream JAK-STAT signaling cascade and subsequently activate transcription factors that promote homeostatic proliferation and survival of T cells by regulating expression of proliferation-associated, anti-apoptotic and pro-apoptoic genes [47]. Further, IL-7 has been shown to support the metabolic fitness of memory T cells. IL-7 induces expression of the glycerol channel aquaporin 9 in antigen-specific memory T cells allowing for uptake of glycerol for use in triglyceride synthesis and storage to meet the metabolic requirements of memory T cells [48]. Interestingly, human and mouse TRM have recently been shown to uniquely express high levels of fatty-acid binding proteins 4 and 5 (FABP4/5) that mediate lipid uptake and intracellular transport [49]. Skin TRM but not TCM deficient for both FABP4/5 were unable to take up exogenous free fatty acid and failed to persist suggesting that oxidative metabolism of exogenous FFAs is required for TRM to survive long-term in the peripheral tissues [49]. The purinergic receptor, P2RX7, has also recently been shown to be important for directing mitochondrial homeostasis and metabolic function in differentiating TCM and TRM populations in mice [50]. Importantly, mice transiently treated with a pharmacological inhibitor of P2RX7, a promising therapy to treat neuropathic pain, from day 40 to 50 following LCMV infection resulted in a loss of the established TCM population suggesting a necessary role in memory maintenance [50].
What additional transcription factors and downstream cellular processes these affect in memory T cells to assure longevity and robust recall responses remain to be fully appreciated. Importantly, the regulatory programs necessary to maintain the substantial diversity that exists within the effector and memory CD8+ T cell populations are only beginning to be defined. While precursors within the effector T cell population predetermined to give rise to the distinct memory subsets can be identified, plasticity between CD8+ T cell fates also has been demonstrated. Recent technological advances have allowed for the initiation of a more thorough examination of these important aspects of CD8+ T cell memory. Here we describe several studies that have utilized novel fate-mapping strategies and epigenetic profiling techniques to examine origin and subsetspecific long-term maintenance of CD8+ T cell memory.
Fate-mapping
Memory populations may be derived from a discrete group of memory precursors that arise from naive T cells following infection as early as the first division following T cell activation [51]. TCM and TEM have been suggested to chiefly differentiate from the KLRG1loCD127hi MP T cells and TRM from a KLRG1lo precursor [52–54]. In support of T cells having a predefined precursor-product relationship, Smith et al. showed that a T cell’s fate may be imprinted during development and be contingent at least in part on which time in life the precursor undergoes thymic maturation [55]. For this study, a new fate-mapping mouse model was established whereby a CD4 promoter-driven inducible cre recombinase could direct expression of red fluorescent protein TdTomato (RFP) in CD4+CD8+ double positive thymocytes upon tamoxifen treatment. In this way, a wave of newly developing CD8+ T cells were ‘timestamped’ and could be followed in the periphery [55]. Importantly, the authors found that T cells made during infancy exhibited more rapid effector-like qualities while T cells generated during adulthood more efficiently seeded the long-lived memory pool. The fate choice defined by the T cell’s developmental origin was governed by a unique transcriptional profile and chromatin landscape [55].
Fate commitment and terminal-differentiation of T cells has been questioned and the notion that naive T cells give rise to an effector population where at least a portion of T cells maintain the flexibility to ‘dedifferentiate’ following resolution of the infection to seed the memory pool should be considered. Maintaining some degree of developmental plasticity would potentially drive functional heterogeneity within the memory T cell population. In support of this, a portion of TE cells can re-express CD127, suggesting that epigenetic remodeling and a loss of repressive marks from pro-memory genes can occur [6,12,13] . As well, memory lineage interconversion has been observed where CD62Llo TEM cells, considered to be more differentiated, converted to a CD62LhiCCR7hiCD27hi TCM phenotype in a proliferation-independent manner in the absence of antigen [21]. However, this observation was recently revised when memory T cells were subsetted based on CX3CR1 [25,26]. TEM with high expression of CX3CR1 were unable to adopt a CD62Lhi TCM phenotype when transferred to a new host. Yet, some CX3CR1int or CX3CR1− T cells could upregulate CD62L and join the TCM pool [26]. These findings support the idea that the phenotypic states among memory populations may not be fully stable and may be subject to additional regulation after the acute phase of infection.
In line with these findings that effector T cells do not necessarily have a fixed fate, we [12] and others [13] have examined the stability of ‘terminally-differentiated’ KLRG1hi effector CD8+ T cells and demonstrated plasticity within TE or long-lived effector T cells with the ability to convert into memory T cell populations in specific contexts. Using a KLRG1-cre reporter system that allows longitudinal tracking of KLRG1hi effector T cells throughout infection and memory development, T cells that lost KLRG1 expression during the late effector and contraction phase of infection—termed exKLRG1 T cells—were seen to make up 20–40% of the memory T cell population following Listeria monocytogenes or Vesicular stomatitis virus infection and persisted longer than the KLRGhi T cells that were observed at memory time points [13]. ExKLRG1 T cells preferentially differentiated from effector T cells that also expressed CD127, and reminiscent of KLRG1hiCD127hi double positive effector cells, expressed intermediate levels of Gzmb, T-bet, KI67, Bcl-2, Zeb2, Blimp1, and Bach2. ATAC-seq analysis revealed that exKLRG1 had open chromatin regions at both effector and memory-related gene loci. This seemingly allows the exKLRG1hi to efficiently generate most memory populations including TCM, TEM, TPM and TRM while preserving their effector-like past, permitting early participation in a secondary response [13]. Bach2, a transcriptional repressor shown to restrain terminal differentiation and promote memory formation in lymphocytes [56–58] appears to promote this developmental plasticity in CD8+ T cells and plays a role in the development of exKLRG1 memory cells from KLRG1hiCD127hi effector T cells in an AKT-mTOR-FOXO1 dependent manner [13].
We have further demonstrated the necessity for continued transcriptional regulation to sustain the differentiated state of a CD8+ T cell following acute viral infection [12]. The inhibitor of E protein transcription factors Id2, is expressed by effector CD8+ T cells to promote survival and terminal differentiation [40–42]. Notably, Id2 is also expressed by CD8+ T cells into the memory phase following infection to maintain the ‘effector-like’ KLRG1hi CD8+ T cell population. We used a tamoxifen-inducible knock-out mouse model in order to control the timing of Id2 deletion. Induced deletion of Id2 following resolution of lymphocytic choriomeningitis virus infection transformed the KLRG1hi long-lived TE and TEM subsets into a KLRG1lo population with the phenotype and gene expression program resembling that of the TCM memory subset [12]. Thus, constant Id2 regulation of E protein activity supports the persistence of an ‘effector-like’ memory population while preserving some degree of cellular plasticity that allows these cells to ‘dedifferentiate’ and reacquire expression of genes characteristic of the long-lived memory populations if inhibition of E proteins by Id2 is lost [12].
Conversely, continual reinforcement of the memory state has also been suggested to be necessary [59,60]. Induced deletion of the transcription factor FOXO1 in memory CD8+ T cells caused a reversion to a cell state reminiscent of terminally-differentiated effector T cells with increased KLRG1 and decreased CD62L expression in a proliferation independent manner. These cells also produced more granzyme B [59] and less IL-2 [60], and seemingly reverted away from the characteristic TCM memory state. As well, FOXO1-deleted memory T cells gradually declined in number as continuous FOXO1 expression was necessary to support the expression of pro-survival molecules and enable homeostatic turnover. Further, they had a greatly reduced re-expansion capacity, presumably due to a defect in proliferation or trafficking to secondary lymphoid organs where re-expansion takes place [60]. We have also demonstrated that the transcription factor Runx3 is important for the maintenance of TRM in nonlymphoid tissues [61]. Induced deletion of Runx3 in established small intestine TRM led to a significant loss of CD69+CD103+ cells likely due to an increased rate of apoptosis [27].
Taken together, these studies indicate that memory T cell ‘fates’ may not be absolute but rather an active process with regulatory networks working to uphold the memory cell ‘state’. This then raises the question of what signals are required to set up and sustain the reinforcement of memory cell states and can these be manipulated in the context of vaccination or immunopathology.
Epigenetic profiling
While several studies have worked to define the transcription programs necessary to uphold the subset-specific phenotypic and functional properties of memory CD8+ T cells, others have focused on the epigenetic modifications of histones and DNA that regulate the chromatin accessibility for those subset-defining transcription factors. DNA methylation is one such alteration that fixes genes in an ‘off’ position thus silencing expression. Histone modifications can promote repressive or permissive chromatin states, depending on the combination. For example, trimethylation of lysine 4 (H3K4me3) marks active promoters and is associated with gene expression while trimethylation of lysine 27 (H3K27me3) is associated with gene repression.
Once activated, a naive T cell may undergo epigenetic programming that drives differentiation of effector cells and expression of genes that mediate pathogen clearance while genes associated with naive or resting cells are repressed. TE CD8+ T cells were found to epigenetically silence genes responsible for longevity and plasticity in part by the Polycomb Repressive Complex 2 (PRC2,) which catalyzes de novo repressive H3K27 trimethylation marks. Conversely, MP cells appeared to maintain permissive and active chromatin states at both MP- and TE-signature genes in a mechanism involving FOXO1 [62]. In a parallel study that used single-cell RNA sequencing to assess CD8+ T cells over the course of a viral infection, EZH2, a catalytic subunit of PRC2, was found to be highly expressed in TE cells after the first division following T cell activation [63]. TE cells were seen to gain repressive H3K27me3 marks compared to naive T cells and Ezh2 binding was observed at genes—many previously linked to memory—with reduced expression at day 4 relative to first division TE cells [63]. Furthermore, PRC2 deficiency affected the differentiating TE or ‘effector-like’ cells while minimally impacted the memory T cell maturation [62,63]. This argues that CD8+ TE cells restrict their fate while the MP population maintains multipotency to differentiate into memory cells that can turn-on effector function upon reactivation [62].
Two recent studies examined DNA methylation status to address the origin of human and mouse memory T cells and suggest that effector T cells dedifferentiate to seed the memory pool [64,65]. Youngblood et al. considered the observation that memory CD8+ T cells are hybrid in nature—able to elicit effector functions upon rechallenge but also have naive-associated properties such as pluripotency and secondary lymphoid homing ability [65]. The on-off-on pattern of expression for naive-associated genes during naive-to-effector-to-memory differentiation was analyzed through whole-genome bisulfite sequencing (WGBS) to reveal the DNA methylation status for CD8+ T cell subsets over the course of an infection [65]. Effector T cell subsets responding to infection were found to have increased regions of de novo DNA methylation at naive-associated loci in part by the action of the methyltransferase Dnmt3a, and canonical effector genes became demethylated [65]. Interestingly, the MP effector subset re-expressed naive-associated genes as they formed memory T cells, and this was associated with the erasure of the de novo DNA methylation that was acquired during the naive to effector T cell transition. Effector genes such as Gzmb and Prf1 remained unmethylated in memory T cells despite their reduced expression levels [65]. While previous work showed that Dmnt3a drives a de novo DNA methylation program specific to effector TE cells [66], this new work supports the idea that effector cells with memory potential dedifferentiate into memory T cells that are able to re-express naive-associated genes [65]. Similarly, Akondy et al. noted that the human memory population originated from effector T cells that had extensively proliferated during the initial two weeks following administration of the yellow fever vaccine [64]. Additionally, shared features of the effector and memory T cell epigenetic profiles were observed with notable similarities in chromatin accessibility and DNA methylation. While the transcriptional signature of memory T cells was more like naive than effector—with the exception of TCR, cytokine and toll-like receptor mediated signaling pathways—the memory T cells retained epigenetic memory of differentiation through an effector stage and appear to dedifferentiate from a subset of fate-permissive, activated CD8+ T cells [64].
Once established, memory CD8+ T cell populations are maintained through active transcriptional programs and presumably supported by epigenetic modifications. Abdelsamed et al. examined the acquired DNA methylation marks in human TSCM, TCM and TEM cells and how these are preserved during antigen-independent homeostasis [67]. WGS was performed on naive and memory T cells isolated from healthy donor blood. A unique methylation status was identified for each different memory populations; for instance, the CCR7 and CD62L regions where significantly methylated in the TEM population but remained predominately unmethylated in the TCM and TSCM subsets [67]. As well, the DMNT3A and TCF7 promoters became progressively enriched for methylation as the memory subset became more differentiated (ie. TSCM<TCM<TEM). However, loci of effector molecules were demethylated in all memory T cell subsets [67]. Following IL-7- and IL-15-driven in vitro homeostatic proliferation or adoptive transfer into transplant patients, the memory T cells maintained the effector-loci demethylation status [67]. Interestingly, in these experimental settings, the TCM and TSCM subsets differentiated towards a TEM phenotype and also increased methylation at the CCR7 and TCF7 loci [67]. This suggests that certain epigenetic marks can remain stable over time while others can be modified for adaptation to environmental changes.
Maintenance of other immune populations
The requirement for active reinforcement of cell ‘state’ by transcriptional networks is not unique to the CD8+ T cell population. In fact, heterogeneity and plasticity have been described among the CD4+ T cell lineages. It is suggested that CD4+ T cells undergo subset specification but retain the ability to dynamically regulate their cellular ‘state’ in response to changing conditions. Cytokine signaling mediating STAT transcription factors and subsequent activation or repression of subset-specific transcriptional programs is a major driver of plasticity [68]. As an example, regulatory T cells (Treg) can be induced by environmental cues to express T helper cell subset defining transcription factors presumably to support Treg homeostasis and better regulate immune responses [68]. Further, in several inflammatory settings, Treg can become unstable, lose Foxp3 expression and acquire an effector phenotype [69]. As well, induced deletion of Foxp3 in mature Tregs results in a loss of suppressive function and conversion to an effector T cell able to produce IL-2 and T helper type 1 cytokines [70]. Similarly, active regulation by transcription factors in innate lymphoid cells (ILCs) ensures subset-specific phenotypes and functional plasticity can occur as a result of cytokine stimuli likely to fine-tune the immune response [71]. While continual GATA-3 regulation [72,73] has been shown to support ILC homeostasis and T-bet, Rorγt, Notch transcription factors can mediate ILC subset conversion [71], additional transcriptional and epigenetic regulators reinforcing these ‘states’ have yet to be identified.
Conclusion
Heterogeneity within lymphocyte populations is important for fighting a diverse array of infections (Figure 1). Maintaining developmental plasticity within memory T cell subpopulations allows for interpretation of environmental signals and affords the T cell compartment the capacity to evolve the most effective response. While recent work described above favors the re-examination of the notion that CD8+ T cells irreversibly commit to a particular cell ‘fate’ in the effector phase, it will be important to clearly understand the transcriptional and epigenetic programs in place that dictate stability or pliability of a memory T cell. While this review focuses on memory homeostasis, additional complexity emerges when considering the secondary effector population. For instance, memory T cells removed from nonlymphoid tissue can give rise to circulating memory populations in a secondary response [74]. Further, in a secondary nonlymphoid tissue infection TRM proliferate in situ producing secondary effector that maintain the TRM population [75,76]. Additional studies will be necessary to understand the plasticity in secondary effector T cell subsets and the subsequent heterogeneity that may develop in the memory populations. Importantly, with this knowledge will come the ability to therapeutically reprogram T cell populations in the form of T cell-based vaccines and treatments against infectious and inflammatory diseases as well as malignancies.
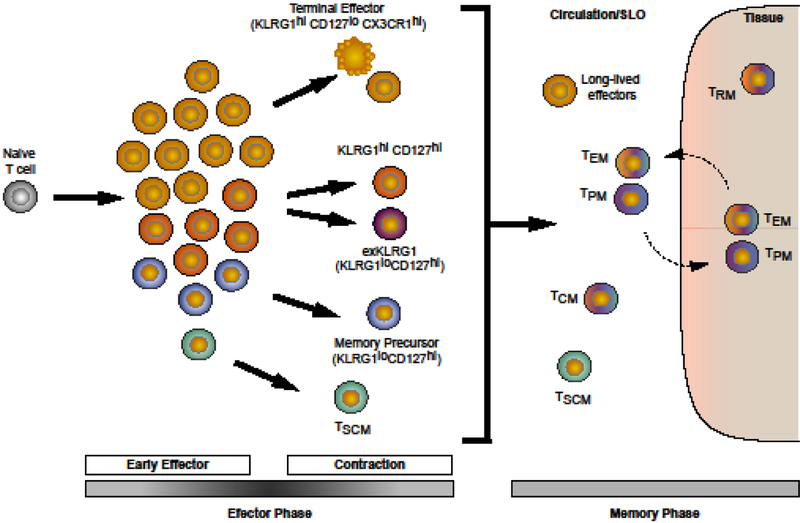
Figure 1. Heterogeneity within the effector and memory CD8+ T cell populations.
Activation of a naive T cell gives rise to phenotypically diverse effector T cells that contribute to the generation of the long-lived memory subsets. Memory T cells are represented with the corresponding effector precursor color that has a demonstrated role in that subset’s formation. Memory T cells retain the epigenetic modifications acquired during the effector phase as indicated by yellow nuclei. TEM = effector memory; TCM = central memory; TPM = peripheral memory; TRM = resident memory; TSCM = stem-cell memory.
Highlights.
The effector and memory CD8+ T cell populations exhibit significant heterogeneity
Effector and memory T cell subsets exist in ‘states’ and retain some degree of plasticity
Active transcriptional regulation maintains the identity of the memory T cell subsets
Epigenetic programming likely re-enforce memory T cell ‘states’
Acknowledgements
This review was supported by the US National Institutes of Health AI109976 and AI067545 to AWG. We thank Drs. Bingfei Yu, J. Justin Milner and Nadia Kurd for critical review of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chang JT, Wherry EJ, Goldrath AW: Molecular regulation of effector and memory T cell differentiation. Nat Immunol 2014, 15:1104–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamilton SE, Jameson SC: CD8 T cell memory: it takes all kinds. Front Immunol 2012, 3:353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM: Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity 2012, 36:142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arsenio J, Kakaradov B, Metz PJ, Kim SH, Yeo GW, Chang JT: Early specification of CD8+ T lymphocyte fates during adaptive immunity revealed by single-cell gene-expression analyses. Nat Immunol 2014, 15:365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R: Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol 2003, 4:1191–1198. [DOI] [PubMed] [Google Scholar]
- 6.Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM: Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity 2007, 27:281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, Ahmed R: Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J Exp Med 2008, 205:625–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grundemann C, Schwartzkopff S, Koschella M, Schweier O, Peters C, Voehringer D, Pircher H: The NK receptor KLRG1 is dispensable for virus-induced NK and CD8+ T-cell differentiation and function in vivo. Eur J Immunol 2010, 40:1303–1314. [DOI] [PubMed] [Google Scholar]
- 9.Huster KM, Busch V, Schiemann M, Linkemann K, Kerksiek KM, Wagner H, Busch DH: Selective expression of IL-7 receptor on memory T cells identifies early CD40L-dependent generation of distinct CD8+ memory T cell subsets. Proc Natl Acad Sci U S A 2004, 101:5610–5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schluns KS, Kieper WC, Jameson SC, Lefrancois L: Interleukin-7 mediates the homeostasis of naive and memory CD8 T cells in vivo. Nat Immunol 2000, 1:426–432. [DOI] [PubMed] [Google Scholar]
- 11.Olson JA, McDonald-Hyman C, Jameson SC, Hamilton SE: Effector-like CD8(+) T cells in the memory population mediate potent protective immunity. Immunity 2013, 38:1250–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Omilusik KD, Nadjsombati MS, Shaw LA, Yu B, Milner JJ, Goldrath AW: Sustained Id2 regulation of E proteins is required for terminal differentiation of effector CD8(+) T cells. J Exp Med 2018, 215:773–783. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This study with [13] is the first to suggest that CD8+ T cell terminal-differentiation is a cellular ‘state’ that must be actively maintained and is reversible. Id2/E-proteins were shown to transcriptionally and phenotypically sustain KLRG1hi effector and effector-memory T cells.
- 13.Herndler-Brandstetter D, Ishigame H, Shinnakasu R, Plajer V, Stecher C, Zhao J, Lietzenmayer M, Kroehling L, Takumi A, Kometani K, et al. : KLRG1(+) Effector CD8(+) T Cells Lose KLRG1, Differentiate into All Memory T Cell Lineages, and Convey Enhanced Protective Immunity. Immunity 2018, 48:716–729 e718. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Using fate-mapping methods, the authors show that KLRG1hi terminally-differentiated effector CD8+ T cells can de-differentiate and contribute to memory formation. This opposes the notion that only KLRG1lo memory precursor effector T cells can generate memory.
- 14.Goldrath AW, Sivakumar PV, Glaccum M, Kennedy MK, Bevan MJ, Benoist C, Mathis D, Butz EA: Cytokine requirements for acute and Basal homeostatic proliferation of naive and memory CD8+ T cells. J Exp Med 2002, 195:1515–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang CY, Best JA, Knell J, Yang E, Sheridan AD, Jesionek AK, Li HS, Rivera RR, Lind KC, D’Cruz LM, et al. : The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol 2011, 12:1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalia V, Sarkar S, Subramaniam S, Haining WN, Smith KA, Ahmed R: Prolonged interleukin-2Ralpha expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity 2010, 32:91–103. [DOI] [PubMed] [Google Scholar]
- 17.Jeannet G, Boudousquie C, Gardiol N, Kang J, Huelsken J, Held W: Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc Natl Acad Sci U S A 2010, 107:9777–9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou X, Yu S, Zhao DM, Harty JT, Badovinac VP, Xue HH: Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity 2010, 33:229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A: Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 1999, 401:708–712. [DOI] [PubMed] [Google Scholar]
- 20.Masopust D, Vezys V, Marzo AL, Lefrancois L: Preferential localization of effector memory cells in nonlymphoid tissue. Science 2001, 291:2413–2417. [DOI] [PubMed] [Google Scholar]
- 21.Wherry EJ, Teichgraber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian UH, Ahmed R: Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol 2003, 4:225–234. [DOI] [PubMed] [Google Scholar]
- 22.von Andrian UH, Mempel TR: Homing and cellular traffic in lymph nodes. Nat Rev Immunol 2003, 3:867–878. [DOI] [PubMed] [Google Scholar]
- 23.Kaech SM, Wherry EJ: Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity 2007, 27:393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jameson SC, Masopust D: Diversity in T cell memory: an embarrassment of riches. Immunity 2009, 31:859–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bottcher JP, Beyer M, Meissner F, Abdullah Z, Sander J, Hochst B, Eickhoff S, Rieckmann JC, Russo C, Bauer T, et al. : Functional classification of memory CD8(+) T cells by CX3CR1 expression. Nat Commun 2015, 6:8306. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This study with [26] uses the surface molecule, CX3CR1, to categorize the heterogeneity observed in the effector and memory CD8+ T cell population.
- 26.Gerlach C, Moseman EA, Loughhead SM, Alvarez D, Zwijnenburg AJ, Waanders L, Garg R, de la Torre JC, von Andrian UH: The Chemokine Receptor CX3CR1 Defines Three Antigen-Experienced CD8 T Cell Subsets with Distinct Roles in Immune Surveillance and Homeostasis. Immunity 2016, 45:1270–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]; * A novel CX3CR1int memory subset is identified and termed peripheral memory (TPM). This study further suggests that instead of TEM cells, the TPM population is the dominant memory subset circulating through the peripheral tissues and the TEM are restricted to the blood.
- 27.Milner JJ, Goldrath AW: Transcriptional programming of tissue-resident memory CD8(+) T cells. Curr Opin Immunol 2018, 51:162–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gattinoni L, Zhong XS, Palmer DC, Ji Y, Hinrichs CS, Yu Z, Wrzesinski C, Boni A, Cassard L, Garvin LM, et al. : Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat Med 2009, 15:808–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Joe G, Hexner E, Zhu J, Emerson SG: Host-reactive CD8+ memory stem cells in graft-versus-host disease. Nat Med 2005, 11:1299–1305. [DOI] [PubMed] [Google Scholar]
- 30.Turtle CJ, Swanson HM, Fujii N, Estey EH, Riddell SR: A distinct subset of self-renewing human memory CD8+ T cells survives cytotoxic chemotherapy. Immunity 2009, 31:834–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gattinoni L, Lugli E, Ji Y, Pos Z, Paulos CM, Quigley MF, Almeida JR, Gostick E, Yu Z, Carpenito C, et al. : A human memory T cell subset with stem cell-like properties. Nat Med 2011, 17:1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hikono H, Kohlmeier JE, Takamura S, Wittmer ST, Roberts AD, Woodland DL: Activation phenotype, rather than central- or effector-memory phenotype, predicts the recall efficacy of memory CD8+ T cells. J Exp Med 2007, 204:1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jameson SC, Masopust D: Understanding Subset Diversity in T Cell Memory. Immunity 2018, 48:214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaech SM, Cui W: Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol 2012, 12:749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji Y, Pos Z, Rao M, Klebanoff CA, Yu Z, Sukumar M, Reger RN, Palmer DC, Borman ZA, Muranski P, et al. : Repression of the DNA-binding inhibitor Id3 by Blimp-1 limits the formation of memory CD8+ T cells. Nat Immunol 2011, 12:1230–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, et al. : Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science 2003, 302:1041–1043. [DOI] [PubMed] [Google Scholar]
- 37.Banerjee A, Gordon SM, Intlekofer AM, Paley MA, Mooney EC, Lindsten T, Wherry EJ, Reiner SL: Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J Immunol 2010, 185:4988–4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao RR, Li Q, Gubbels Bupp MR, Shrikant PA: Transcription factor Foxo1 represses T-bet-mediated effector functions and promotes memory CD8(+) T cell differentiation. Immunity 2012, 36:374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ichii H, Sakamoto A, Hatano M, Okada S, Toyama H, Taki S, Arima M, Kuroda Y, Tokuhisa T: Role for Bcl-6 in the generation and maintenance of memory CD8+ T cells. Nat Immunol 2002, 3:558–563. [DOI] [PubMed] [Google Scholar]
- 40.Cannarile MA, Lind NA, Rivera R, Sheridan AD, Camfield KA, Wu BB, Cheung KP, Ding Z, Goldrath AW: Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nat Immunol 2006, 7:1317–1325. [DOI] [PubMed] [Google Scholar]
- 41.Knell J, Best JA, Lind NA, Yang E, D’Cruz LM, Goldrath AW: Id2 influences differentiation of killer cell lectin-like receptor G1(hi) short-lived CD8+ effector T cells. J Immunol 2013, 190:1501–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masson F, Minnich M, Olshansky M, Bilic I, Mount AM, Kallies A, Speed TP, Busslinger M, Nutt SL, Belz GT: Id2-mediated inhibition of E2A represses memory CD8+ T cell differentiation. J Immunol 2013, 190:4585–4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rutishauser RL, Martins GA, Kalachikov S, Chandele A, Parish IA, Meffre E, Jacob J, Calame K, Kaech SM: Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity 2009, 31:296–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kallies A, Xin A, Belz GT, Nutt SL: Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity 2009, 31:283–295. [DOI] [PubMed] [Google Scholar]
- 45.Dominguez CX, Amezquita RA, Guan T, Marshall HD, Joshi NS, Kleinstein SH, Kaech SM: The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J Exp Med 2015, 212:2041–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Omilusik KD, Best JA, Yu B, Goossens S, Weidemann A, Nguyen JV, Seuntjens E, Stryjewska A, Zweier C, Roychoudhuri R, et al. : Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J Exp Med 2015, 212:2027–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abdelsamed HA, Zebley CC, Youngblood B: Epigenetic Maintenance of Acquired Gene Expression Programs during Memory CD8 T Cell Homeostasis. Front Immunol 2018, 9:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cui G, Staron MM, Gray SM, Ho PC, Amezquita RA, Wu J, Kaech SM: IL-7-Induced Glycerol Transport and TAG Synthesis Promotes Memory CD8+ T Cell Longevity. Cell 2015, 161:750–761. [DOI] [PMC free article] [PubMed] [Google Scholar]; * With [49] and [50], this work demonstrates that CD8+ memory T cells have unique metabolic requirements that support long-term persistence.
- 49.Pan Y, Tian T, Park CO, Lofftus SY, Mei S, Liu X, Luo C, O’Malley JT, Gehad A, Teague JE, et al. : Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature 2017, 543:252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]; * The authors show that TRM need to take-up and metabolize free fatty acids to ensure their long-term survival. This is the first study to provide evidence for subset-specific metabolic adaptations within the memory T cell population.
- 50.Borges da Silva H, Beura LK, Wang H, Hanse EA, Gore R, Scott MC, Walsh DA, Block KE, Fonseca R, Yan Y, et al. : The purinergic receptor P2RX7 directs metabolic fitness of long-lived memory CD8(+) T cells. Nature 2018, 559:264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This study identifies a role for the purinergic receptor, P2RX7, in directing and maintaining the metabolic fitness of TCM and TRM populations in mice.
- 51.Chang JT, Palanivel VR, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, Longworth SA, Vinup KE, Mrass P, Oliaro J, et al. : Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science 2007, 315:1687–1691. [DOI] [PubMed] [Google Scholar]
- 52.Mackay LK, Rahimpour A, Ma JZ, Collins N, Stock AT, Hafon ML, Vega-Ramos J, Lauzurica P, Mueller SN, Stefanovic T, et al. : The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat Immunol 2013, 14:1294–1301. [DOI] [PubMed] [Google Scholar]
- 53.Obar JJ, Lefrancois L: Early signals during CD8 T cell priming regulate the generation of central memory cells. J Immunol 2010, 185:263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheridan BS, Pham QM, Lee YT, Cauley LS, Puddington L, Lefrancois L: Oral infection drives a distinct population of intestinal resident memory CD8(+) T cells with enhanced protective function. Immunity 2014, 40:747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith NL, Patel RK, Reynaldi A, Grenier JK, Wang J, Watson NB, Nzingha K, Yee Mon KJ, Peng SA, Grimson A, et al. : Developmental Origin Governs CD8(+) T Cell Fate Decisions during Infection. Cell 2018, 174:117–130 e114. [DOI] [PubMed] [Google Scholar]; ** This study uses a fate-mapping technology to demonstrate the naive T cell pool itself is heterogenous in nature. Naive T cell sub-populations that are transcriptionally, epigenetically and phenotypically distinct differentially develop based on the time they undergo thymic development. Fetal-derived CD8+ T cells represent a more effector-like population while adult-derived CD8+ T cells have enhanced capacity generate long-lived memory.
- 56.Roychoudhuri R, Clever D, Li P, Wakabayashi Y, Quinn KM, Klebanoff CA, Ji Y, Sukumar M, Eil RL, Yu Z, et al. : BACH2 regulates CD8(+) T cell differentiation by controlling access of AP-1 factors to enhancers. Nat Immunol 2016, 17:851–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shinnakasu R, Inoue T, Kometani K, Moriyama S, Adachi Y, Nakayama M, Takahashi Y, Fukuyama H, Okada T, Kurosaki T: Regulated selection of germinal-center cells into the memory B cell compartment. Nat Immunol 2016, 17:861–869. [DOI] [PubMed] [Google Scholar]
- 58.Hu G, Chen J: A genome-wide regulatory network identifies key transcription factors for memory CD8(+) T-cell development. Nat Commun 2013, 4:2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Delpoux A, Michelini RH, Verma S, Lai CY, Omilusik KD, Utzschneider DT, Redwood AJ, Goldrath AW, Benedict CA, Hedrick SM: Continuous activity of Foxo1 is required to prevent anergy and maintain the memory state of CD8(+) T cells. J Exp Med 2018, 215:575–594. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This study with [60] demonstrate that active transcriptional maintenance is required to uphold the memory T cell ‘state’. The transcription factor FOXO1 is required to prevent anergy and support memory T cell longevity and self-renewal.
- 60.Utzschneider DT, Delpoux A, Wieland D, Huang X, Lai CY, Hofmann M, Thimme R, Hedrick SM: Active Maintenance of T Cell Memory in Acute and Chronic Viral Infection Depends on Continuous Expression of FOXO1. Cell Rep 2018, 22:3454–3467. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This work provides evidence that continuous FOXO1 programming is required to functionally maintain memory T cell populations following acute and chronic infection. This observation was extended to human memory CD8+ T cells.
- 61.Milner JJ, Toma C, Yu B, Zhang K, Omilusik K, Phan AT, Wang D, Getzler AJ, Nguyen T, Crotty S, et al. : Runx3 programs CD8(+) T cell residency in non-lymphoid tissues and tumours. Nature 2017, 552:253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gray SM, Amezquita RA, Guan T, Kleinstein SH, Kaech SM: Polycomb Repressive Complex 2-Mediated Chromatin Repression Guides Effector CD8(+) T Cell Terminal Differentiation and Loss of Multipotency. Immunity 2017, 46:596–608. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Together with [63], these groups argue that the fate of the terminal effector T cell subset is restricted due to epigenetic modifications that silence pro-memory genes and restrict memory potential. This is achieved in part by the action of Polycomb Repressive Complex 2 (PRC2) that catalyzes repressive de novo H3K27 trimethylation marks.
- 63.Kakaradov B, Arsenio J, Widjaja CE, He Z, Aigner S, Metz PJ, Yu B, Wehrens EJ, Lopez J, Kim SH, et al. : Early transcriptional and epigenetic regulation of CD8(+) T cell differentiation revealed by single-cell RNA sequencing. Nat Immunol 2017, 18:422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Using single-cell RNA sequencing methods, the authors identify EZH2, a catalytic subunit of PRC2, to be highly expressed in terminally-differentiated effector T cells. In parallel to [62], they demonstrate a role for PRC2 in silencing memory-associated genes and contributing to the terminal differentiation of effector T cells.
- 64.Akondy RS, Fitch M, Edupuganti S, Yang S, Kissick HT, Li KW, Youngblood BA, Abdelsamed HA, McGuire DJ, Cohen KW, et al. : Origin and differentiation of human memory CD8 T cells after vaccination. Nature 2017, 552:362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This study, [65], and [67] examine DNA methylation status to trace the origin of mouse and human memory T cells. The data suggests that memory cells ‘dedifferentiate’ from effector cells and retain epigenetic marks from the effector stage.
- 65.Youngblood B, Hale JS, Kissick HT, Ahn E, Xu X, Wieland A, Araki K, West EE, Ghoneim HE, Fan Y, et al. : Effector CD8 T cells dedifferentiate into long-lived memory cells. Nature 2017, 552:404–409. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This work further shows that the methyltransferase Dnmt3a is responsible in part for the de novo DNA methylation marks at naive-associated loci during the effector phase and that effector cells with memory potential can erase such marks in order to re-express naive-associated genes.
- 66.Ladle BH, Li KP, Phillips MJ, Pucsek AB, Haile A, Powell JD, Jaffee EM, Hildeman DA, Gamper CJ: De novo DNA methylation by DNA methyltransferase 3a controls early effector CD8+ T-cell fate decisions following activation. Proc Natl Acad Sci U S A 2016, 113:10631–10636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abdelsamed HA, Moustaki A, Fan Y, Dogra P, Ghoneim HE, Zebley CC, Triplett BM, Sekaly RP, Youngblood B: Human memory CD8 T cell effector potential is epigenetically preserved during in vivo homeostasis. J Exp Med 2017, 214:1593–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This study shows that DNA methylation status is preserved during homeostatic proliferation of human memory T cells. However, DNA methylation changes occur when memory T cells phenotypically convert from TCM and TSCM subset to the more differentiated TEM population.
- 68.DuPage M, Bluestone JA: Harnessing the plasticity of CD4(+) T cells to treat immunemediated disease. Nat Rev Immunol 2016, 16:149–163. [DOI] [PubMed] [Google Scholar]
- 69.Sawant DV, Vignali DA: Once a Treg, always a Treg? Immunol Rev 2014, 259:173–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Williams LM, Rudensky AY: Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat Immunol 2007, 8:277–284. [DOI] [PubMed] [Google Scholar]
- 71.Colonna M: Innate Lymphoid Cells: Diversity, Plasticity, and Unique Functions in Immunity. Immunity 2018, 48:1104–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yagi R, Zhong C, Northrup DL, Yu F, Bouladoux N, Spencer S, Hu G, Barron L, Sharma S, Nakayama T, et al. : The transcription factor GATA3 is critical for the development of all IL-7Ralpha-expressing innate lymphoid cells. Immunity 2014, 40:378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhong C, Cui K, Wilhelm C, Hu G, Mao K, Belkaid Y, Zhao K, Zhu J: Group 3 innate lymphoid cells continuously require the transcription factor GATA-3 after commitment. Nat Immunol 2016, 17:169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Masopust D, Vezys V, Wherry EJ, Barber DL, Ahmed R: Cutting edge: gut microenvironment promotes differentiation of a unique memory CD8 T cell population. J Immunol 2006, 176:2079–2083. [DOI] [PubMed] [Google Scholar]
- 75.Beura LK, Mitchell JS, Thompson EA, Schenkel JM, Mohammed J, Wijeyesinghe S, Fonseca R, Burbach BJ, Hickman HD, Vezys V, et al. : Intravital mucosal imaging of CD8(+) resident memory T cells shows tissue-autonomous recall responses that amplify secondary memory. Nat Immunol 2018, 19:173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Park SL, Zaid A, Hor JL, Christo SN, Prier JE, Davies B, Alexandre YO, Gregory JL, Russell TA, Gebhardt T, et al. : Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses. Nat Immunol 2018, 19:183–191. [DOI] [PubMed] [Google Scholar]