Abstract
Compartmentalization is vital for biological systems at multiple levels, including biochemical reactions in metabolism. Organelle-based compartments such as mitochondria and peroxisomes sequester the responsible enzymes and increase the efficiency of metabolism while simultaneously protecting the cell from dangerous intermediates, such as radical oxygen species. Recent studies show intracellular nucleotides, such as ATP and GTP, are heterogeneously distributed in cells with high concentrations at the lamellipodial and filopodial projections, or leading edge. However, the intracellular distribution of purine nucleotide enzymes remains unclear. Here, we report the enhanced localization of GTP-biosynthetic enzymes, including inosine monophosphate dehydrogenase (IMPDH isotype 1 and 2), GMP synthase (GMPS), guanylate kinase (GUK1) and nucleoside diphosphate kinase-A (NDPK-A) at the leading edge in renal cell carcinoma cells. They show significant co-localization at the membrane subdomain, and their co-localization pattern at the membrane is distinct from that of the cell body. While other purine nucleotide biosynthetic enzymes also show significant localization at the leading edge, their co-localization pattern with IMPDH is divergent. In contrast, a key glycolytic enzyme, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), predominantly localized in the cytoplasm. Mechanistically, we found that plasma membrane localization of IMPDH isozymes requires active actin polymerization. Our results demonstrate the formation of a discrete metabolic compartment for localized purine biosynthesis at the leading edge, which may promote localized nucleotide metabolism for cell migration and metastasis in cancers.
Keywords: Metabolic compartmentalization, IMPDH, GMPS, GUK1, de novo purine synthesis, salvage purine synthesis
1. Introduction
Compartmentalization is a fundamental element in evolution and is critical for many cellular processes, including biochemical reactions in metabolism [1]. Many metabolic processes are highly spatially regulated within cells. Well-studied examples are organelle-based compartments such as mitochondria, lysosomes, and peroxisomes. Such subcellular localization serves many functions including sequestration of metabolic reactions that if not confined would lead to harmful effects such as radical oxygen formation within peroxisomes [2]. Organelle-based compartmentalization enables cells to increase efficiency and thus total activity of sequestered metabolic pathways [3,4].
In addition to organelles, the cytosol of mammalian cells is an intricate, structured environment of non-membrane-bound multi-enzyme complexes and compartments, often referred to as metabolons [5–8]. Cytosolic compartmentalization is important for promoting enzyme activity through functions such as the reduction of intermediate diffusion rates, metabolic channeling of intermediates, enhanced activation of enzymes through physical association with other proteins, and decreased enzyme-metabolite inhibition [4,9,10]. A classic example is the Krebs TCA cycle metabolon consisting of a multi-enzyme complex containing six of the eight enzymes bound to the inner surface of the mitochondrial inner membrane [11]. This close association of enzymes increases the local concentration of metabolic intermediates promoting enhanced enzymatic activity [12–16].
The leading edge of polarized, motile cells is a complex example of multifaceted protein and enzyme compartmentalization. Spatially restricted synthesis of PI(3,4,5)P3 at the leading edge directs polarized protein localization to the membrane, leading to the establishment of signaling gradients through the activation of signaling pathways and RhoGTPases that promote actin polymerization and microtubule reorganization [17–19]. Thus, ATP at the leading edge is heavily utilized to fuel functions such as F-actin polymerization, phosphorylation-based signaling cascades, and the recycling of nucleotide diphosphates into nucleotide triphosphates [20–22]. GTP binding is critical for microtubule dynamic instability and RhoGTPase activation cycling, among others [18,23]. In line with their putative high demand at the leading edge, previous studies using biosensors for ATP and GTP in live cells demonstrate the heterogenous distribution and restricted membrane localization of triphosphate nucleotides [21,24,25]. While the subcellular localization of biosynthetic enzymes is a key determinant for producing such metabolic gradients, the spatiotemporal regulation of purine nucleotide metabolic enzymes remains largely unknown and is the focus of this study.
There are two conserved purine biosynthesis pathways. De novo purine synthesis from glucose is multi-step and energy consuming, while the salvage pathway is an energy efficient process in which nucleosides and nucleobases from blood and/or intracellular catabolic processes are recycled to produce ATP and GTP. De novo purine nucleotide biosynthesis culminates in the production of the intermediate metabolite IMP. IMPDH is the rate-limiting enzyme in GTP biosynthesis and can influence both ATP and GTP biosynthetic flux from IMP [26–29]. Our lab and others have reported the upregulation of the IMPDH2 isozyme during tumorigenesis and its importance in fueling ribosome biogenesis through regulation of GTP biosynthesis [30–32].
In the present study, we assessed the subcellular localization of GTP biosynthetic enzymes and several key enzymes involved in nucleotide and glucose metabolisms in two highly motile renal cell carcinoma cell lines, 786-o and Caki-1. Our data reveal previously unappreciated F-actin dependent compartmentalization of GTP biosynthetic enzymes at the leading edge. Furthermore, we found distinct localization to the leading edge region of de novo and salvage purine biosynthetic enzymes. These findings suggest the presence of a distinct compartment for nucleotide metabolism at the plasma membrane microdomain that may be mechanistically important for fueling cell motility and influencing metastasis in malignant tumors.
2. Materials and Methods
2.1. Materials
Primary antibodies: anti-IMPDH1 (WH0003614M1, dilution 1:200) antibody (Sigma-Aldrich); anti-IMPDH2 (ab131158, dilution 1:400) antibodies (Abcam); anti-GFP (11814460001, dilution 1:1000) antibody (Roche); anti-c-Myc (sc-789, dilution 1:400), anti-c-Myc (sc-40, dilution 1:400), anti-nm23-H1 (sc-465, dilution 1:400) antibodies (Santa Cruz Biotechnology); and anti-GAPDH (#5174, dilution 1:400) antibody (Cell Signaling Technology). Secondary antibodies: goat anti-mouse IgG Alexa 488 (#A-11029), goat anti-rabbit IgG Alexa 568 (#A-11011), and goat anti-rabbit IgG Alexa 488 (#A-11034) antibodies (Invitrogen); Alexa Fluor 546 phalloidin (#A-12380, dilution 1:200) probe (Invitrogen); and DAPI. Plasmids: PRPS1-Myc-DDK (#RC200698), GUK1-Myc-DDK (#RC202510), Atic-Myc-DDK (#MR219425), ADK-Myc-DDK (#RC200628), APRT-Myc-DDK (#RC216874), HPRT1-Myc-DDK (#RC200462), Pnp-Myc-DDK (#MR203940), Ampd1-Myc-DDK (#MR220907), Nt5c1a-Myc-DDK (#MR217369), GAPDH-Myc-DDK (#MR204932) were purchased from OriGene; FGAMS-EGFP (#99107), PAICS-EGFP (#99108), GFP-ATIC (#99110), PPAT-EGFP (#99105), ADSL-EGFP (#99109), ADSS-V5 (#98316) were purchased from Addgene. Rat GMPS (rGMPS) and mCherry were amplified using standard PCR procedures and combined into the pTRIPZ vector using the Gibson assembly cloning kit (New England Biolabs #E5510S).
2.2. Cell Culture
All cells were cultured in Dulbecco’s modified Eagle’s medium (HyClone #SH30243.01) supplemented with 10% Fetalplex (Gemini #100–602), 1% Penicillin-Streptomycin antibiotic solution (HyClone #SV30010), and 0.1% Normocin (InvivoGen #ant-nr-1) at 5% CO2 and 37°C. Lipofectamine 2000 (Invitrogen # 11668027) was used for transfection according to manufacturer’s instructions. Cells were plated at 0.75 × 105 cells/well (Caki-1) and 0.5 × 105 cells/well (786-o) in 24-well culture plates (Thermo Scientific Biolite) 24 h before transfection. Virus production and transduction were performed as previously described [32].
2.3. Immunofluorescence and Microscopy
Cells were plated on 0.01% poly-L-lysine-coated (Sigma-Aldrich #P1524) coverslips at 0.5 × 105 cells/well in 24-well culture plates 24 h pre-fixation. Cell tracer probe (Invitrogen #V12883) was used in live cells per manufacturer’s instructions. Cells were incubated for 15 min in 4% paraformaldehyde (Thermo Scientific #J19943-K2) at room temperature. Alexa fluor-conjugated wheat germ agglutinin (WGA) (Invitrogen #W-11262) was diluted to 5 μg/mL in PBS and cells were incubated for 10 min at room temperature, followed by three washes in PBS. Following fixation and/or WGA staining, cells were permeabilized using 0.2% Triton X-100 (ACROS Organics #21568–3500)/PBS for 10 min. Cells were blocked using 1% goat serum (Gibco #16210–064)/PBS for 15 min. Immunofluorescence staining was performed overnight at 4°C with primary antibodies diluted in 1% goat serum/PBS. Secondary antibodies were used at a 1:2000 dilution in 1% goat serum/PBS for 1 h at room temperature. Coverslips were mounted onto glass slides using Fluoromount-G (Southern Biotech #0100–01). Cells were imaged using a Keyence BZ-9000 microscope on 60x magnification. WGA-stained samples were imaged on a Zeiss LSM710 confocal microscope. Keyence images were deconvoluted using Keyence BZ-II Analyzer software.
2.4. Immunofluorescence Quantification
Microscopy images were quantified using ImageJ (National Institutes of Health), modified from previously described [33]. Briefly, 3 equal-length lines were drawn spanning the leading edge of cells. Using the “plot profile” function, raw data points depicting the fluorescence intensity along the length of the line were saved. For co-stained images, lines were copied to identical locations in each fluorescence channel image. Raw data points were normalized to the highest intensity, compiled, averaged, and plotted using GraphPad Prism 8. For leading edge versus cell body correlation analysis, a similar strategy was used, with the line draw function drawn free-hand at either the leading edge or inside the body of the cell. Correlation analysis was performed between the fluorescence channels using GraphPad Prism 8. The ratio of fluorescence intensity at the leading edge was quantified using the “freehand selections” function of ImageJ, followed by measurement of the fluorescence intensity and subsequent analysis using GraphPad Prism 8 and Microsoft Excel. All image quantification was performed on single z-stack slices.
3. Results
3.1. IMPDH localizes at the leading edge in invasive renal cell carcinoma
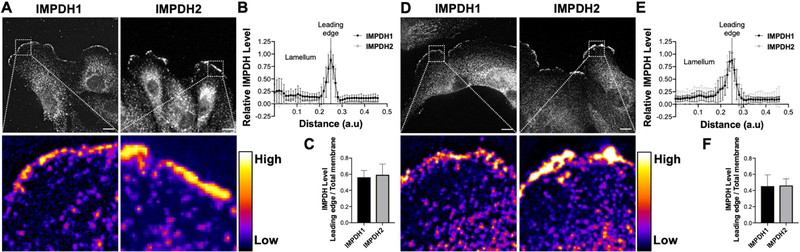
In previous reports, IMPDH1 and IMPDH2 are primarily localized in the cytoplasm in HeLa, CHO and MCF7 cells [34–36]. Consistent with these reports, we found that IMPDH1 and IMPDH2 are localized throughout the cell body in U87MG, a glioblastoma cell line which migrates in an actin polymerization-independent manner [37] (Supplemental Figure 1A). Interestingly, we found that IMPDHs significantly localized to the membrane at the leading edge in Caki-1 and 786-o cells, two highly motile renal cell carcinoma cell lines protruding prominent lamellipodia (Figure 1A, 1D; Supplemental Figure 1B). Analysis of the fluorescence intensity of IMPDH1 and IMPDH2 immunofluorescence staining at the leading edge compared to the lamellum, the flat section posterior to the leading edge lamellipodia, revealed a nearly 4-fold accumulation of IMPDH protein at the leading edge (Figure 1B, 1E). The leading edge localization of IMPDH1 and IMPDH2 accounts for approximately 55% and 45% of the total membrane localization in Caki-1 and 786-o cells, respectively (Figure 1C, 1F). In striking contrast to HeLa, CHO, MCF7 and U87MG cells, the results show robust membrane localization, confined at the leading edge, of IMPDH1 and IMPDH2 in RCC cells.
Figure 1: IMPDH1 and IMPDH2 localize to the leading edge of RCC cells.
Immunofluorescence staining of IMPDH1 (left) and IMPDH2 (right) in Caki-1 (A) and 786-o (D) cells. Insets of leading edge in pseudocolor shown below. Intensity measurements of immunofluorescence staining at leading edge of Caki-1 (B) and 786-o (E) cells. Ratio of fluorescence intensity at leading edge membrane compared to total cell membrane for Caki-1 (C) and 786-o (F) cells. Scale bars indicate 10 μm.
3.2. GTP biosynthetic enzymes localize to the leading edge
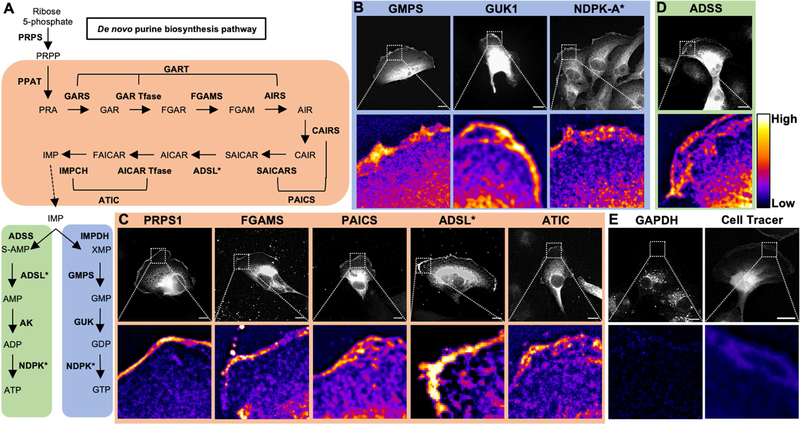
We next assessed the localization of the downstream enzymes of IMPDH (Figure 2). Similar to IMPDH, GMPS, the second step in GTP biosynthesis which converts XMP to GMP, showed distinct localization at the leading edge, with a nearly 2-fold increase in fluorescence signal at the leading edge compared to the lamellum (Figure 2B). Interestingly, both IMPDH and GMPS localized in discrete sub-membrane clusters at the leading edge, rather than localizing homogenously. Additionally, GUK1, the enzyme converting GMP to GDP, and NDPK-A, the final enzyme to produce GTP from GDP, localize at the leading edge with nearly 4-fold increases in fluorescence signal compared to cytosol (Figure 2B). These results reveal that all GTP biosynthetic enzymes localize to the leading edge in RCC cells.
Figure 2: Nucleotide metabolic enzymes localize at the lamellipodial membrane.
Purine nucleotide metabolism pathway. De novo enzymes (orange), ATP biosynthetic branch (green), and GTP biosynthetic branch (blue) (A). Fluorescence imaging for GTP biosynthetic enzymes GMPS (mCherry-tagged, 786-o), GUK1 (Myc, Caki-1), and NDPK-A (antibody, Caki-1) (B). Immunofluorescence staining for de novo enzymes PRPS1 (Myc, Caki-1), FGAMS (GFP, Caki-1), PAICS (GFP, Caki-1), ADSL (GFP, 786-o), and ATIC (GFP, Caki-1) (C). Insets of leading edge in pseudocolor shown below. Scale bars indicate 10 μm. Asterisk (*) indicates enzymes involved in multiple pathways. PRPS: phosphoribosyl pyrophosphate synthetase. PPAT: phosphoribosyl pyrophosphate amidotransferase. GART: phosphoribosylglycinamide formyltransferase. FGAMS: phosphoribosyl formylglycinamidine synthase. PAICS: phosphoribosyl aminoimidazole succinocarboxamide synthetase. ADSL: adenylosuccinate lyase. ATIC: 5-amino-4-imidazolecarboxamide ribonucleotide transformylase/IMP cyclohydrolase. ADSS: adenylosuccinate synthase. AK: adenylate kinase. IMPDH: inosine-5′-monophosphate dehydrogenase. GMPS: GMP synthase. GUK1: guanylate kinase 1. NDPK: nucleoside-diphosphate kinase. GAPDH: glyceraldehyde 3-phosphate dehydrogenase.
3.3. Nucleotide metabolic enzymes localize to the leading edge
Next, we tested the localization of key enzymes involved in the de novo and salvage purine and the ATP synthesis pathways. Phosphoribosyl pyrophosphate synthetase 1 (PRPS1), phosphoribosyl formylglycinamidine synthase (FGAMS), phosphoribosyl aminoimidazole succinocarboxamide synthetase (PAICS), adenylosuccinate lyase (ADSL), and 5-amino-4-imidazolecarboxamide ribonucleotide transformylase/IMP cyclohydrolase (ATIC) showed significant localization to the leading edge (Figure 2C). ATP biosynthetic enzymes ADSL (also involved in de novo synthesis) and adenylosuccinate synthase (ADSS) showed accumulation within the lamellipodium (Figure 2D). Similarly, the purine salvage enzymes localized at the leading edge (Supplemental Figure 2). In contrast, the glycolytic enzyme GAPDH exhibited negligible localization at the leading edge (Figure 2E; Supplemental Figure 3). Furthermore, the protein-dye adducts formed during treatment with the Cell Tracer dye accumulated less at the leading edge than all enzymes tested (Figure 2E). Taken together, these results show that enzymes from the entire purine nucleotide biosynthetic pathway localize to the leading edge in highly motile RCC cells.
3.4. Metabolic enzymes are compartmentalized at the leading edge
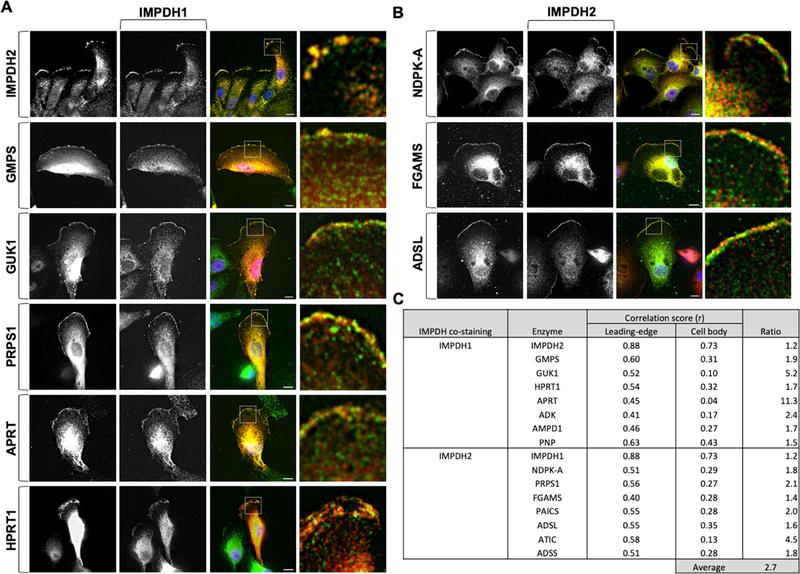
To determine if they were forming metabolons or distinct sub-clusters at the membrane, colocalization of the enzymes was assessed using quantitative co-localization analysis (Supplemental Figure 4). We found the correlation of the localization between IMPDH1 and IMPDH2 was highest at the leading edge, compared to the body of the cell, in both cell lines tested (Figure 3A, C). GMPS, GUK1, and NDPK-A were highly co-localized with IMPDH1 and IMPDH2, respectively, at the leading edge; however, the co-localization within the cell body was significantly lower (Figure 3). Similarly, all enzymes tested in Fig. 2 exhibited a high degree of co-localization with IMPDH1 or IMPDH2. Importantly, we found that the co-localization scores at the leading edge were approximately 2.7-fold higher than those in the general cell body (Figure 3; Supplemental Figure 5, 6). These data suggest that the leading edge increases the clustering formation of GTP biosynthetic and purine nucleotide biosynthetic enzymes.
Figure 3: Purine nucleotide enzymes compartmentalize at the leading edge.
IMPDH1 (green) (A) and IMPDH2 (red) (B) co-localize with selected enzymes at the leading edge but not within the lamellum. Correlation scores of co-stained enzymes are an average 2.7-fold higher at the leading edge compared to the cell body (C). Insets shown at right. Scale bars indicate 10 μm.
3.5. Actin polymerization is required for IMPDH localization at the leading edge
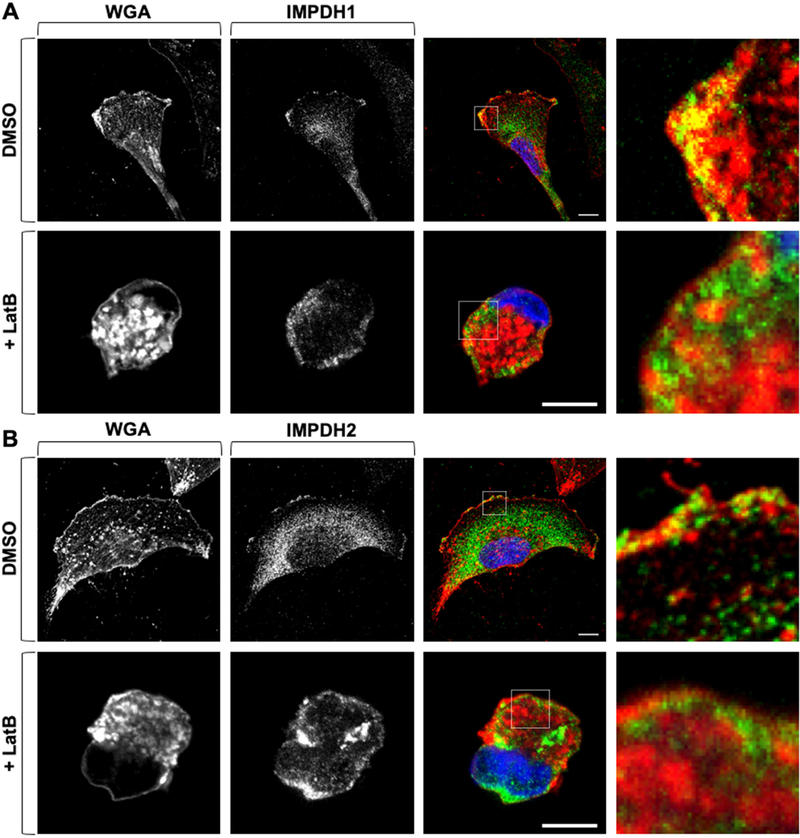
According to the amino acid sequence as well as previous studies, IMPDH has no membrane binding domain or lipidation motifs. To assess the mechanism of IMPDH localization at the leading edge, we focused on active actin reorganization in RCC cells since actin polymerization at the leading edge has been shown to promote the translocation of proteins, such as PI3K, and induce localized signaling [17,38–41]. Given the high association of metabolic enzymes to the leading edge, we next examined whether inhibition of actin polymerization could affect localization at the plasma membrane. Under normal conditions, IMPDH1 and IMPDH2 were co-merged with the fluorophore-conjugated wheat germ agglutinin (WGA), which binds to glycosylated membrane proteins (Figure 4 top row; Supplemental Figure 7). Strikingly, after treatment of latrunculin B (Lat B), an inhibitor of actin polymerization [42], the plasma membrane localization of IMPDH1 and IMPDH2 was diminished (Figure 4, bottom rows). Collectively, these data suggest that F-actin polymerization is functionally connected to the membrane localization of the IMPDH isozymes and may be involved in the localization of additional enzymes to the leading edge.
Figure 4: Actin polymerization is required for IMPDH localization to the membrane.
IMPDH1 (green) (A) and IMPDH2 (green) (B) co-localize with WGA (red) in DMSO-treated conditions (top row); 10 μM Lat B treatment for 2 h (bottom row) decreases co-localization. Confocal microscopy. Insets shown at right. Scale bars indicate 10 μm. WGA: wheat germ agglutinin.
4. Discussion
In the present study, we found the distinct localization of de novo and salvage nucleotide biosynthetic enzymes to the leading edge of highly motile RCC cells. All these enzymes portrayed a higher degree of co-localization with IMPDH1 or IMPDH2 at the membrane in comparison to the cell body, suggesting that the lamellipodia behaves as a microcompartment for spatiotemporal nucleotide biosynthesis. Together, our results show compartmentalization of the entire GTP biosynthetic pathway and purine biosynthetic pathway at the leading edge of RCC cells, pointing to site specific enhanced enzymatic reactions of nucleotide biosynthesis at the leading edge.
Previous reports demonstrate the importance of actin polymerization for promoting the translocation and activation of proteins at the membrane [17,41,43]. In the current work, we report the membrane localization of IMPDH isozymes is driven partially, if not all, by F-actin polymerization. Whether additional signaling dynamics are required for this translocation would provide significant insight into the mechanisms mediating translocation of these enzymes to the membrane.
We and others have reported the importance of IMPDH2 expression for fueling tumorigenesis [26,31,32,44,45]. De novo GTP biosynthesis has been implicated in cancer metastasis and in activation of RhoGTPases [46,47]. Future studies should clarify whether and how the concentrated biosynthesis of purine nucleotides regulate actin polymerization, microtubule dynamics, and RhoGTPase activation, which would uncover a potential contributing mechanism for cancer metastasis in patients.
Supplementary Material
Highlights.
Purine biosynthetic enzymes localize at leading edge of renal cell carcinoma cells
Co-localization of enzymes at leading edge is greater than in the cell body
Purine nucleotide biosynthetic enzymes compartmentalize at leading edge
Actin polymerization is required for leading edge localization of GTP enzymes
Acknowledgements
We thank the members of the Sasaki Lab, Drs. Carol Mercer, Maria Czyzyk-Krzeska, Susan Waltz, Satoshi Namekawa, Francis McCormack for feedback and sharing resources. We thank Ms. Emily Dobbs and Dr. Eric P. Smith for excellent editing. S. Kofuji is supported, in part, by Home for Innovative Researchers and Academic Knowledge Users (HIRAKU), JSPS KAKENHI Grant Number JP18K07233 and the Kanae Foundation. The work is supported in part by UC College of Medicine Research Innovation grant, MTP UC-Brain Tumor Center grant, MERF grant, Marlene Harris Ride Cincinnati grant, ABTA Discovery grant, B*Cured research grant, Ohio Cancer Research grant, R21NS100077, and R01NS089815 (A.T.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Diekmann Y, Pereira-Leal JB, Evolution of intracellular compartmentalization, Biochem. J 449 (2012) 319–331. doi: 10.1042/bj20120957. [DOI] [PubMed] [Google Scholar]
- [2].del Río LA, Sandalio LM, Palma JM, Bueno P, Corpas FJ, Metabolism of oxygen radicals in peroxisomes and cellular implications, Free Radic. Biol. Med 13 (1992) 557–580. doi: 10.1016/0891-5849(92)90150-F. [DOI] [PubMed] [Google Scholar]
- [3].Hinzpeter F, Gerland U, Tostevin F, Optimal Compartmentalization Strategies for Metabolic Microcompartments, Biophys. J 112 (2017) 767–779. doi: 10.1016/j.bpj.2016.11.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Alam MT, Olin-Sandoval V, Stincone A, Keller MA, Zelezniak A, Luisi BF, Ralser M, The self-inhibitory nature of metabolic networks and its alleviation through compartmentalization, Nat. Commun 8 (2017) 1–13. doi: 10.1038/ncomms16018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hudder A, Nathanson L, Deutscher MP, Organization of Mammalian Cytoplasm, Mol. Cell. Biol 23 (2003) 9318–9326. doi: 10.1128/mcb.23.24.9318-9326.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ovádi J, Saks V, On the origin of intracellular compartmentation and organized metabolic systems, Mol. Cell. Biochem 256 (2003) 5–12. doi: 10.1023/b:mcbi.0000009855.14648.2c. [DOI] [PubMed] [Google Scholar]
- [7].Moller BL, Dynamic Metabolons, 2Science. 330 (10AD) 1328–1329. [DOI] [PubMed] [Google Scholar]
- [8].Srere PA, The metabolon, Trends Biochem. Sci 10 (1985) 109–110. doi: 10.1016/0968-0004(85)90266-X. [DOI] [Google Scholar]
- [9].Gaertner FH, Unique catalytic properties of enzyme clusters, Trends Biochem. Sci 3 (1978) 63–65. doi: 10.1016/S0968-0004(78)94045-8. [DOI] [Google Scholar]
- [10].Al-Habori M, Microcompartmentation, metabolic channelling and carbohydrate metabolism, Int. J. Biochem. Cell Biol 27 (1995) 123–132. doi: 10.1016/1357-2725(94)00079-Q. [DOI] [PubMed] [Google Scholar]
- [11].Robinson JB, Inman L, Sumegi B, Srere PA, Further characterization of the Krebs tricarboxylic acid cycle metabolon., J. Biol. Chem 262 (1987) 1786–1790. [PubMed] [Google Scholar]
- [12].Vélot C, Mixon MB, Teige M, Srere PA, Model of a quinary structure between krebs TCA cycle enzymes: A model for the metabolon, Biochemistry. 36 (1997) 14271–14276. doi: 10.1021/bi972011j. [DOI] [PubMed] [Google Scholar]
- [13].Brangwynne CP, Echmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA, Germline P Granules Are Liquid Droplets That Localize by Controlled Dissolution/Condensation, Science (80-. ). 324 (2009) 1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- [14].Gall JG, Cajal Bodies : The First 100 Years, Annu. Rev. Cell Dev 16 (2000) 273–300. [DOI] [PubMed] [Google Scholar]
- [15].An S, Kumar R, Sheets ED, Benkovic SJ, Reversible compartmentalization of de novo purine biosynthetic complexes in living cells., Science. 320 (2008) 103–6. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- [16].Scalettar BA, Abney JR, Hackenbrock CR, Dynamics, structure, and function are coupled in the mitochondrial matrix., Proc. Natl. Acad. Sci 88 (2006) 8057–8061. doi: 10.1073/pnas.88.18.8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sasaki AT, Chun C, Takeda K, Firtel RA, Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement, J. Cell Biol 167 (2004) 505–518. doi: 10.1083/jcb.200406177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Raftopoulou M, Hall A, Cell migration: Rho GTPases lead the way, Dev. Biol 265 (2004) 23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- [19].Ciobanasu C, Faivre B, Le Clainche C, Actin Dynamics Associated with Focal Adhesions, Int. J. Cell Biol 2012 (2012) 1–9. doi: 10.1155/2012/941292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bernstein BW, Bamburg JR, Actin-ATP hydrolysis is a major energy drain for neurons, J. Neurosci 23 (2003) 1–6. papers2://publication/uuid/783C669C-C01A-48E1-8934-EB2A59D1ECE7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Suzuki R, Hotta K, Oka K, Spatiotemporal quantification of subcellular ATP levels in a single HeLa cell during changes in morphology, Sci. Rep 5 (2015) 1–9. doi: 10.1038/srep16874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Cunniff B, McKenzie AJ, Heintz NH, Howe AK, AMPK activity regulates trafficking of mitochondria to the leading edge during cell migration and matrix invasion, Mol. Biol. Cell 27 (2016) 2662–2674. doi: 10.1091/mbc.E16-05-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Etienne-Manneville S, Microtubules in Cell Migration, Annu. Rev. Cell Dev. Biol 29 (2013) 471–499. doi: 10.1146/annurev-cellbio-101011-155711. [DOI] [PubMed] [Google Scholar]
- [24].Bianchi-Smiraglia A, Rana MS, Foley CE, Paul LM, Lipchick BC, Moparthy S, Moparthy K, Fink EE, Bagati A, Hurley E, Affronti HC, V Bakin A, Kandel ES, Smiraglia DJ, Feltri ML, Sousa R, Nikiforov MA, Internally ratiometric fluorescent sensors for evaluation of intracellular GTP levels and distribution, Nat. Methods (2017). doi: 10.1038/nmeth.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Imamura H, Huynh Nhat KP, Togawa H, Saito K, Iino R, Kato-Yamada Y, Nagai T, Noji H, Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators, Proc Natl Acad Sci U S A. 106 (2009) 15651–15656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zou J, Han Z, Zhou L, Cai C, Luo H, Huang Y, Liang Y, He H, Jiang F, Wang C, Zhong W, Elevated expression of IMPDH2 is associated with progression of kidney and bladder cancer, Med. Oncol 32 (2015) 373. doi: 10.1007/s12032-014-0373-1. [DOI] [PubMed] [Google Scholar]
- [27].Hedstrom L, IMP Dehydrogenase : Structure , Mechanism , and Inhibition, (2009) 2903–2928. [DOI] [PMC free article] [PubMed]
- [28].JACKSON RC, WEBER G, MORRIS HP, IMP dehydrogenase, an enzyme linked with proliferation and malignancy, Nature. 256 (1975) 331–333. 10.1038/256331a0. [DOI] [PubMed] [Google Scholar]
- [29].Collart FR, Chubb CB, Mirkin BL, Huberman E, Increased Inosine-5′-phosphate Dehydrogenase Gene Expression in Solid Tumor Tissues and Tumor Cell Lines, Cancer Res. 52 (1992) 5826–5828. http://cancerres.aacrjournals.org/content/52/20/5826.abstract. [DOI] [PubMed] [Google Scholar]
- [30].Valvezan AJ, Turner M, Belaid A, Lam HC, Miller SK, McNamara MC, Baglini C, Housden BE, Perrimon N, Kwiatkowski DJ, Asara JM, Henske EP, Manning BD, mTORC1 Couples Nucleotide Synthesis to Nucleotide Demand Resulting in a Targetable Metabolic Vulnerability, Cancer Cell. 0 (2017) 1–15. doi: 10.1016/j.ccell.2017.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Huang F, Ni M, Chalishazar MD, Huffman KE, Kim J, Cai L, Shi X, Cai F, Zacharias LG, Ireland AS, Li K, Gu W, Kaushik AK, Liu X, Gazdar AF, Oliver TG, Minna JD, Hu Z, DeBerardinis RJ, Inosine Monophosphate Dehydrogenase Dependence in a Subset of Small Cell Lung Cancers, Cell Metab. (2018) 1–14. doi: 10.1016/j.cmet.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kofuji S, Hirayama A, Eberhardt AO, Kawaguchi R, Sugiura Y, Sampetrean O, Ikeda Y, Warren M, Sakamoto N, Kitahara S, Yoshino H, Yamashita D, Sumita K, Wolfe K, Lange L, Ikeda S, Shimada H, Minami N, Malhotra A, Morioka S, Ban Y, Asano M, Flanary VL, Ramkissoon A, Chow LML, Kiyokawa J, Mashimo T, Lucey G, Mareninov S, Ozawa T, Onishi N, Okomura K, Terakawa J, Daikoku T, Wise-Draper T, Majd N, Kofuji K, Sasaki M, Mori M, Kanemura Y, Smith EP, Anastasiou D, Wakimoto H, Holland EC, Yong WH, Horbinski C, Nakano I, DeBarardinis RJ, Bachoo RM, Mischel PS, Yasui W, Suematsu M, Saya H, Soga T, Grummt I, Bierhoff H, Sasaki AT, IMP dehydrogenase-2 drives aberrant nucleolar activity and structure in glioblastoma, Nat Cell Biol. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Osmani N, Peglion F, Chavrier P, Etienne-Manneville S, Cdc42 localization and cell polarity depend on membrane traffic, J. Cell Biol 191 (2010) 1261–1269. doi: 10.1083/jcb.201003091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Thomas EC, Gunter JH, Webster JA, Schieber NL, Oorschot V, Parton RG, Whitehead JP, Different Characteristics and Nucleotide Binding Properties of Inosine Monophosphate Dehydrogenase (IMPDH) Isoforms, PLoS One. 7 (2012). doi: 10.1371/journal.pone.0051096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Calise SJ, Carcamo WC, Krueger C, Yin JD, Purich DL, Chan EKL, Glutamine deprivation initiates reversible assembly of mammalian rods and rings, Cell. Mol. Life Sci 71 (2014) 2963–2973. doi: 10.1007/s00018-014-1567-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ji Y, Gu J, Makhov AM, Griffith JD, Mitchell BS, Regulation of the interaction of inosine monophosphate dehydrogenase with mycophenolic acid by GTP, J. Biol. Chem 281 (2006) 206–212. doi: 10.1074/jbc.M507056200. [DOI] [PubMed] [Google Scholar]
- [37].Panopoulos A, Howell M, Fotedar R, Margolis RL, Glioblastoma motility occurs in the absence of actin polymer, Mol. Biol. Cell 22 (2011) 2212–2220. doi: 10.1091/mbc.e10-10-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yamaguchi H, Condeelis J, Regulation of the actin cytoskeleton in cancer cell migration and invasion, Biochim. Biophys. Acta - Mol. Cell Res 1773 (2007) 642–652. doi: 10.1016/j.bbamcr.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sandilands E, Cans C, Fincham VJ, Brunton VG, Mellor H, Prendergast GC, Norman JC, Superti-Furga G, Frame MC, RhoB and actin polymerization coordinate Src activation with endosome-mediated delivery to the membrane, Dev. Cell 7 (2004) 855–869. doi: 10.1016/j.devcel.2004.09.019. [DOI] [PubMed] [Google Scholar]
- [40].Lambrechts A, Gevaert K, Cossart P, Vandekerckhove J, Van Troys M, Listeria comet tails: the actin-based motility machinery at work, Trends Cell Biol. 18 (2008) 220–227. doi: 10.1016/j.tcb.2008.03.001. [DOI] [PubMed] [Google Scholar]
- [41].Dandekar SN, Park JS, Peng GE, Onuffer JJ, Lim WA, Weiner OD, Actin dynamics rapidly reset chemoattractant receptor sensitivity following adaptation in neutrophils, Philos. Trans. R. Soc. B Biol. Sci 368 (2013) 1–11. doi: 10.1098/rstb.2013.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Coue M, Brenner SL, Spector I, Korn ED, Inhibition of actin polymerization by latrunculin A, FEBS Lett. 213 (1987) 316–318. [DOI] [PubMed] [Google Scholar]
- [43].Sasaki AT, Janetopoulos C, Lee S, Charest PG, Takeda K, Sundheimer LW, Meili R, Devreotes PN, Firtel RA, G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility, J. Cell Biol 178 (2007) 185–191. doi: 10.1083/jcb.200611138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Fellenberg J, Kunz P, Sahr H, Depeweg D, Overexpression of Inosine 5 9 - Monophosphate Dehydrogenase Type II Mediates Chemoresistance to Human Osteosarcoma Cells, PLoS One. 5 (2010). doi: 10.1371/journal.pone.0012179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhou L, Xia D, Zhu J, Chen Y, Chen G, Mo R, Zeng Y, Dai Q, He H, Liang Y, Jiang F, Zhong W, Enhanced expression of IMPDH2 promotes metastasis and advanced tumor progression in patients with prostate cancer, Clin. Transl. Oncol 16 (2014) 906–913. doi: 10.1007/s12094-014-1167-9. [DOI] [PubMed] [Google Scholar]
- [46].Bianchi-Smiraglia A, Wawrzyniak JA, Bagati A, Marvin EK, Ackroyd J, Moparthy S, Bshara W, Fink EE, Foley CE, Morozevich GE, Berman AE, Shewach DS, Nikiforov MA, Pharmacological targeting of guanosine monophosphate synthase suppresses melanoma cell invasion and tumorigenicity, Cell Death Differ. 22 (2015) 1858–1864. doi: 10.1038/cdd.2015.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Bianchi-Smiraglia A, Bagati A, Fink EE, Moparthy S, Wawrzyniak JA, Marvin EK, Battaglia S, Jowdy P, Kolesnikova M, Microphthalmia-associated transcription factor suppresses invasion by reducing intracellular GTP pools, Oncogene. (2016) 1–13. doi: 10.1038/onc.2016.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.