Abstract
Persistent papillomatosis on footpads related to canine papillomavirus type 2 (CPV-2) infection has been described in dogs with immunocompromised condition. A 9-year-old, male French bulldog was presented with cauliflower-like nodules on the footpads of his left front leg. Histopathological examination revealed multiple finger-like projections of squamous epithelium with intranuclear inclusion bodies. Immunohistochemistry using an anti-bovine papillomavirus antibody demonstrated immunostaining in the keratinocytes. Partial genome DNA of CPV-2 was amplified from the lesion. Full genome sequence of CPV-2 in the subject showed 99.95% nucleotide identity with that of CPV-2 from the reference data. Two weeks after a biopsy, the skin lesion spontaneously regressed without any specific treatment. In non-immunocompromised dogs, CPV-2-related footpad papillomatosis could spontaneously resolve after a biopsy.
Keywords: canine papillomavirus type 2, footpad papillomatosis, spontaneous regression
Papillomaviruses cause proliferative disorders of epithelia in various mammals. In humans, high risk types of human papillomaviruses are known to be associated with development of cervical cancer. In dogs, six different clinical phenotypes have been associated with canine papillomavirus (CPV) infection: oral papillomatosis, venereal papillomatosis, exophytic cutaneous papillomas, inverted cutaneous papillomas, papillomas of the footpad, and canine pigmented viral plaque [4]. Currently, 20 types of canine papillomavirus have been reported according to The PapillomaVirus Episteme (PaVE), papillomavirus genomic information (https://pave.niaid.nih.gov/#home) [7]. CPV type 1 (CPV-1) is primarily involved in oral papillomatosis, which is a contagious disease found in young dogs; such papillomas usually last for about four weeks before regressing spontaneously [9]. Canine cutaneous papillomatosis has been associated with CPV-2, -6, and -7 [2]. These types of papillomas can usually occur on the face, ears, and feet in young dogs and occasionally occur in older dogs. CPV-2 was firstly isolated from a footpad papilloma of Golden retriever that had been in intensive training [9]. In contrast to oral papillomatosis caused by CPV-1, CPV-2-associated papillomas are more persistent and last for more than six months in dogs [9]. The CPV-2-associated skin lesions are usually associated with a clinical history of immunosuppression or with a stressful environmental setting [9]. Persistent footpad papillomas associated with CPV-2 have also been reported in dogs with X-linked severe combined immunodeficiency, and some of these dogs developed invasive and metastatic squamous cell carcinoma [1]. In this case report, we describe an old dog with CPV-2-related footpad papillomas affecting only one front paw, which showed spontaneous regression within a few weeks after skin biopsy.
A 9-year-old, male French bulldog presented with a one-month history of lameness due to developing nodular lesions on footpads of its left forelimb. While the dog was treated with oral antibiotics and prednisolone for a few weeks, the skin lesion was increasing in size, in addition to causing pain. Oral antibiotics and prednisolone were withdrawn about two weeks before initial visit to our hospital. On physical examination, cauliflower-like, exophytic and hyperkeratotic plaques with nodular lesions were recognized on the footpads and interdigital area of the paw (Fig. 1a). Some of the skin lesions were ulcerated and covered with dark red to yellowish crusts. Beside the skin lesions and lameness, the dog was otherwise healthy. From clinical presentation, we made a tentative diagnosis of papillomatosis. To confirm the diagnosis, 6 mm punch biopsy was performed from a part of nodular skin lesion under generalized anesthesia. Two weeks after skin biopsy, nodular skin lesions on the footpads of the limb almost completely disappeared without any specific treatment (Fig. 1b). Over the next 11 months, the dog remained systemically healthy and recurrence of skin lesion was not observed.
Fig. 1.
Gross appearance of the skin lesion on the left front paw. Cauliflower-like, exophytic nodular lesions were recognized on the footpads at the initial stage (a). Skin lesions almost completely diminished two weeks after biopsy (b).
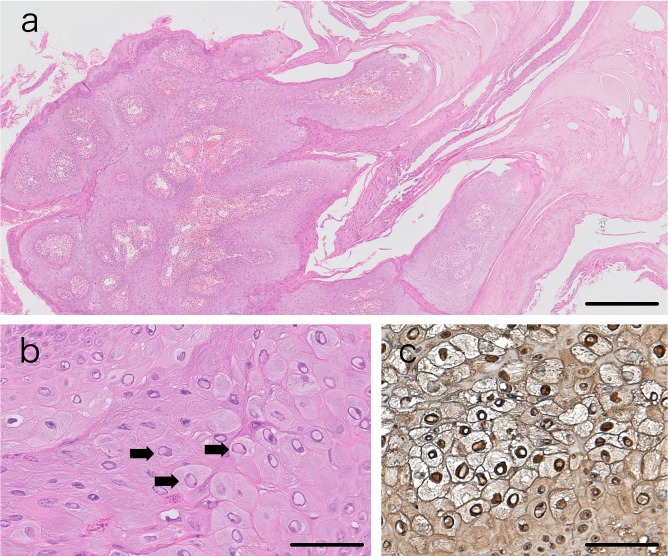
A half of the skin biopsy sample was fixed in 10% buffered neutral formalin and embedded in paraffin, then subjected to histopathological and immunohistochemical analyses. Hematoxylin and eosin staining revealed multiple finger-like exophytic proliferation of squamous epithelium with severe compact hyperkeratosis (Fig. 2a). The upper stratum spinosum or stratum granulosum was presented as clear cytoplasm with pyknotic nuclei and large keratohyalin granules. Pale basophilic intranuclear inclusion bodies were found in some of keratinocytes in the proliferative epidermis (Fig. 2b).
Fig. 2.
Histopathological and immunohistopathological findings of the papillomatous lesions. In hematoxylin and eosin sections, multiple finger-like exophytic proliferation of squamous epithelium with compact hyperkeratosis (a), and basophilic intranuclear inclusion bodies in proliferative keratinocytes (arrows) (b) were found. In immunohistochemistry, intranuclear inclusion bodies were positive for bovine papillomavirus-1 (c). Scale bar represents 500 µm in (a), and 50 µm in (b) and (c).
To further confirm the association of these findings with CPV, a standard immunoperoxidase technique was used for the immunohistochemistry using rabbit polyclonal anti-bovine papillomavirus antibody (B508, Dako, Carpinteria, CA, U.S.A., dilution 1:200). Deparaffinized sections were treated with citric acid buffer (pH 6) for 30 min in a microwave oven before incubation for 1 hr with primary antibody. Peroxidase-conjugated anti-rabbit IgG (Dako, dilution 1:200) was used as the secondary antibody. The reaction products were visualized by 3,3′-diaminobenzidine tetrahydrochloride (Dako; Liquid DAB + Substrate Chromogen System). The papillomavirus antigen was detected in the nuclei of the keratinocyte lesion (Fig. 2c).
To detect the papillomavirus genome in the skin lesion, PCR analysis was performed. DNA was extracted from a half of the biopsy sample using QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). Consensus primer set, MY09/11, was used for detection of papillomavirus genome (Table S1, Supporting information) [8]. The reaction mixture was prepared according to the manufacturer’s instructions (KOD FX Neo (Toyobo, Osaka, Japan). PCR analysis revealing a 450-bp product was amplified (Fig. 3). The amplicon was sequenced with BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific, Waltham, MA, U.S.A.) and 3130xl Genetic Analyzer (Thermo Fisher Scientific). The sequence of the amplicon corresponded to CPV-2 in the database of National Center for Biotechnology Information (Accession Number, NC_006564). To characterize the genes of CPV-2, full genome sequencing of CPV was also performed. Three primer sets were designed to amplify the full genome of CPV-2 (Table S1). The primers for sequencing are also listed in Table S1. The PCR and sequencing were conducted as described above. The full genome sequence of CPV-2 was deposited in the DNA Data Bank of Japan (Accession Number, LC363559). Blast analysis of the full genome sequence showed 99.95% nucleotide identity with the sequence, NC_006564, and there were four mutations including two missense mutations on E1 and E4 (Table 1). Mutations were analyzed by motif analysis using the Genetyx version 11 (Genetyx, Osaka, Japan) and secondary structure analysis was performed using Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index). In silico analysis revealed that these mutations could not be associated any motifs.
Fig. 3.

Detection of the canine papillomavirus (CPV) genome DNA in the lesion. A 450-bp band was detected by PCR using MY09/11 primer set. bp: base pairs, M: DNA Ladder Marker, S: the sample derived from the lesion.
Table 1. Genetic information of canine papillomavirus type-2 (CPV-2) genome in this study compared with the reference genome.
| Position (nt)a) | Reference genomeb) | CPV-2 in this study | Open reading frame | Mutation |
|---|---|---|---|---|
| 1919 | A | G | E1 | Silent mutation |
| 1977 | C | G | E1 | Missense mutation (His395Asp) |
| 2946 | T | C | E2, E4 | E2: Silent mutation E4: Missense mutation (Phe12Ser) |
| 4621 | - | T | NCRc) | Base insertion |
a) nt, Nucleotides. b) Accession No.: NC_006564. c) NCR, Non-coding region.
The present case showed exophytic papillomatous skin lesion only on the footpads and interdigital area of the front paw of an old dog, and CPV-2 was identified from the skin lesion. Morphological and histopathological findings of the skin lesion in the dog were similar to those of the previously reported cases of CPV-2 related persistent footpad papillomas [1, 9]. However, the dog showed clinical remission two weeks after biopsy without any specific treatment. The prognosis of this CPV-2 case was similar to that of other reported CPV-1 cases [6, 9]. Nevertheless, there are differences in genes between CPV-1 and CPV-2 [9]. For example, E5 open reading frame, which codes for an important host-cell transformation factor and a strong oncoprotein, is detected in CPV-2 but not in CPV-1 [5, 9]. While reports about CPV-associated disorders have been insufficient to conclude virulence of CPV, clinical phenotype, and prognosis are thought to depend on the type of CPV infections, namely genetic characteristics [5]. Compared with the reference full genome of CPV-2, the sequence of CPV-2 isolated from the canine from our study demonstrated two missense mutations, which were clearly not functional by in silico analysis and no mutation was detected in the E5 gene. Because there are only one complete (accession number: NC_006564) and two partial genome sequences of CPV-2 in NCBI [3, 9], detailed genetic analysis cannot be performed in our study. CPV-2 infected dogs with persistent skin lesions or having squamous cell carcinoma reported in the previous studies were living in stressful environmental setting or had X-linked severe combined immunodeficiency [1, 9]. While clinical diagnostic examinations such as blood test, urinalysis and diagnostic imaging test were not performed, immunocompromised condition was not found through its clinical history and physical examination. From these findings, we observed that clinical severity and prognosis of canine papillomatosis could depend not only on the CPV type, but also on host immunological condition of dogs infected by CPV-2.
A recent study revealed that 40 dogs with oral papillomatosis caused by CPV1 achieved clinical remission without any specific treatment after biopsy [6]. The same study showed that the healing process could be related to antibody response, and that antibody titers peaked around the time of clinically remission. In our study, although detailed immunological evaluation was not performed, it is possible that the biopsy after withdrawal of oral prednisolone triggered an active immune response against CPV-2 and contributed to clinical remission in this case.
In conclusion, we described a dog with CPV2-related footpad papillomatosis. It is possible that such papillomas spontaneously resolve after a biopsy procedure in non-immunocompromised dogs.
Supplementary Material
REFERENCES
- 1.Goldschmidt M. H., Kennedy J. S., Kennedy D. R., Yuan H., Holt D. E., Casal M. L., Traas A. M., Mauldin E. A., Moore P. F., Henthorn P. S., Hartnett B. J., Weinberg K. I., Schlegel R., Felsburg P. J.2006. Severe papillomavirus infection progressing to metastatic squamous cell carcinoma in bone marrow-transplanted X-linked SCID dogs. J. Virol. 80: 6621–6628. doi: 10.1128/JVI.02571-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lange C. E., Favrot C.2011. Canine papillomaviruses. Vet. Clin. North Am. Small Anim. Pract. 41: 1183–1195. doi: 10.1016/j.cvsm.2011.08.003 [DOI] [PubMed] [Google Scholar]
- 3.Lange C. E., Tobler K., Brandes K., Breithardt K., Ordeix L., Von Bomhard W., Favrot C.2010. Canine inverted papillomas associated with DNA of four different papillomaviruses. Vet. Dermatol. 21: 287–291. doi: 10.1111/j.1365-3164.2009.00817.x [DOI] [PubMed] [Google Scholar]
- 4.Miller W. H., Griffin C. E., Campbell K. L.2013. Neoplastic and non-neoplastic tumors. pp. 774–843. In: Muller & Kirk’s Small Animal Dermatology, 7th ed. (Miller, W. H., Griffin, C. E. and Campbell, K. L. eds.), Elsiver, St. Louis. [Google Scholar]
- 5.Rector A., Van Ranst M.2013. Animal papillomaviruses. Virology 445: 213–223. doi: 10.1016/j.virol.2013.05.007 [DOI] [PubMed] [Google Scholar]
- 6.Sancak A., Favrot C., Geisseler M. D., Müller M., Lange C. E.2015. Antibody titres against canine papillomavirus 1 peak around clinical regression in naturally occurring oral papillomatosis. Vet. Dermatol. 26: 57–59, e19–e20. doi: 10.1111/vde.12189 [DOI] [PubMed] [Google Scholar]
- 7.Van Doorslaer K., Li Z., Xirasagar S., Maes P., Kaminsky D., Liou D., Sun Q., Kaur R., Huyen Y., McBride A. A.2017. The Papillomavirus Episteme: a major update to the papillomavirus sequence database. Nucleic Acids Res. 45D1: D499–D506. doi: 10.1093/nar/gkw879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waropastrakul S., Munday J. S., French A. F.2012. Infrequent detection of papillomaviral DNA within canine cutaneous squamous cell carcinomas, haemangiosarcomas and healthy skin on the ventrum of dogs. Vet. Dermatol. 23: 197–e41. doi: 10.1111/j.1365-3164.2012.01043.x [DOI] [PubMed] [Google Scholar]
- 9.Yuan H., Ghim S., Newsome J., Apolinario T., Olcese V., Martin M., Delius H., Felsburg P., Jenson B., Schlegel R.2007. An epidermotropic canine papillomavirus with malignant potential contains an E5 gene and establishes a unique genus. Virology 359: 28–36. doi: 10.1016/j.virol.2006.08.029 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.