Abstract
We investigated the relationships between ruminal pH, gene expression in the rumen epithelium (RE), peripheral blood mononuclear cell subpopulations, and blood metabolites in Holstein calves during weaning transition. Calves (Weaning group, n=7) were assigned to one of two groups, and fed calf starter with forage (Forage group, n=3) or without forage (Starter group, n=4). Ruminal pH was measured continuously. Samples were collected at −1, 0, 1, and 3 weeks (blood and rumen fluid) or 3 weeks (rumen epithelium) after weaning. In the Weaning group, ruminal pH increased, and several blood metabolites increased (blood urea nitrogen [BUN], beta-hydroxybutyrate [BHB], and gamma-glutamyl transferase [GGT]) or decreased (total cholesterol [T-CHO] and phospholipid) after weaning. Ruminal pH was positively correlated with CD8+CD45R− cell populations and blood metabolites (BUN, glucose, and BHB) and negatively correlated with GGT activity. The 24 hr mean ruminal pH was higher in the Forage group during weaning transition, and toll-like receptor 4 mediated signaling pathway was activated in the Starter group at 3 weeks post-weaning. The number of CD8+CD45R− cells tended to be higher, and several blood metabolites (glucose, triglycerides, T-CHO, and phospholipid) were higher in the Forage group after weaning. Calves with higher ruminal pH also showed a greater energy metabolism status simultaneously with lesser hepatic disturbance enzymes in the peripheral blood. The results of our study indicate that serum GGT activity may be a plausible biomarker for predicting ruminal acidosis in Holstein calves during weaning transition.
Keywords: blood metabolites, peripheral blood mononuclear cell subpopulations, ruminal acidosis, toll-like receptor 4, weaning transition
During weaning transition, increased intake of calf starter to supply energy requirement causes ruminal acidosis, and this intake affects metabolic and immune responses in calves. For example, feeding only calf starter during weaning transition resulted in higher acidity in the rumen, and downregulation of genes encoding ketogenic enzymes (3-hydroxymethyl-3-methylglutaryl-CoA lyase, 3-hydroxybutyrate dehydrogenase type 1, and 3-hydroxybutyrate dehydrogenase type 2) and cholesterol biosynthesis (sterol regulatory element binding protein 2; SREBP2) was observed in the rumen epithelium (RE) [12]. Furthermore, weaning in calves is one of the biggest stressors affecting immune ability [10]. Previously, Hulbert et al. [8] reported that the concentrations of cortisol and tumor necrosis factor alpha (TNF-α) in peripheral blood stimulated by lipopolysaccharide (LPS) increased gradually until weaning, concurrent with a gradual reduction in the amount of milk replacer. Furthermore, stress levels and systemic inflammatory responses of calves are assessed by measuring the serum levels of cortisol, acute phase proteins (APPs), inflammatory cytokines, and leukocyte subsets, and these parameters are influenced by weaning stress in Holstein calves [10].
Past studies have examined the transcriptomic dynamics of the RE [6] and toll-like receptor (TLR) expression in the gastrointestinal tract [16] from Holstein calves, and TLR4 has been reported to recognize bacterial LPS and stimulate the production of cytokines like interleukin (IL)-6, IL-12, and TNF-α [22]. Specifically, a transcription factor, peroxisome proliferator-activated receptor alpha, was identified as an important regulator of molecular changes among calves fed commercial milk replacer only, milk plus orchardgrass hay, and milk plus calf starter [6]. Furthermore, the expression of ten bovine TLRs (TLR1-10) that regulate innate immune responses to commensal microflora were examined throughout the gastrointestinal tract in 3-week- and 6-month-old calves [16]. However, the relationships during weaning transition between rumen fermentation, rumen epithelial gene expression, and peripheral blood immune and metabolites are still unclear.
Therefore, this study aimed to investigate the effects of ruminal pH on gene expression in the RE and changes in immunological and metabolic parameters of peripheral blood in Holstein calves during weaning transition. We hypothesized that the acidic stimulus in the rumen caused by a low ruminal pH might alter peripheral blood mononuclear cell (PBMC) subpopulations, cytokine concentrations, and metabolites in the peripheral blood, as well as gene expression in the RE.
MATERIALS AND METHODS
In this study, we re-analyzed our previously published data regarding rumen fermentation parameters (pH and VFAs; [11, 12]) and differentially expressed genes in the RE examined by one-color microarray analysis for the upstream regulator analysis [12] to evaluate, during weaning transition, the relationships between ruminal pH, gene expression in the RE, and PBMC subpopulations and metabolites.
Animals and management
All animals were cared for according to protocols approved by Iwate University Laboratory Animal Care and Use Committee. Details of animal care have been reported previously [12]. Briefly, Holstein bull calves (Weaning group, n=7) were fed a commercial milk replacer (Meiji Feed, Kashima, Japan), calf starter (Meiji Feed), and mixed forage (orchard and timothy hay) until 6 weeks of age. Then, the calves were divided into forage provision and non-provision groups at 7 weeks of age, whereby forage was restricted in the forage non-provision group. All calves were weaned at 8 weeks of age. At 11 weeks of age (3 weeks after weaning), the calves that had adapted well to the experimental schedule were then selected from the forage provision (Forage group, n=3) and forage non-provision (Starter group, n=4) groups. The two groups were fed an identical amount of total dry matter from 8 weeks of age. The diet was supplied in two equal portions at 08:00 and 16:30 daily. The chemical compositions of the milk replacer, calf starter concentrate, and mixed forage fed to calves were reported previously [11, 12]. The amount of feeding was based on the Japanese Feeding Standards for Dairy Cattle.
Measurements and RE biopsies
Ruminal pH was measured continuously every 10 min throughout the experiment using a radio telemetry system (YCOW-S; DKK-TOA Yamagata, Yamagata, Japan), as reported previously [23]. The pH sensor was placed in the ventral sac of the rumen, from which rumen fluid samples were collected at 7, 8, 9, and 11 weeks of age (−1, 0, 1, and 3 weeks after weaning, respectively). Immediately after sampling, the rumen fluid samples were filtered through two layers of cheesecloth. For the VFA analysis, 2 ml of 25% HO3P in 3 N H2SO4 was added to 10 ml of rumen fluid. The total VFA and VFA components were separated and quantified by gas chromatography (Model 135, Hitachi, Tokyo, Japan) using a packed glass column (Thermon-3000, 3%) on a Shimalite TPA 60–80 mesh support (Shinwa Chemical Industries, Kyoto, Japan). To measure ruminal LPS activity, rumen fluid samples were centrifuged at 11,000 × g for 15 min at 4°C and assayed using a kinetic Limulus amebocyte lysate assay (Pyrochrome with Glucashield, Seikagaku Corp., Tokyo, Japan) [7]. The RE was biopsied from the ventral sac of the rumen at a site adjacent to the pH sensor at 3 weeks after weaning, as described previously [12].
Blood collection and biochemical analysis
Blood samples were collected from the jugular vein, simultaneously with the rumen fluid sample, using an evacuated serum-separating tube and a sodium-fluoride-containing tube (BD Vacutainer, Franklin Lakes, NJ, U.S.A.). The number of white blood cells was counted using a pocH-100iV Diff analyzer (Sysmex, Kobe, Japan) and the proportion of PBMCs was determined using the hemogram method with Diff-Quick staining. Serum and plasma were separated by centrifugation at 1,500 × g for 15 min at 4°C, and biochemical analysis was performed using an automated biochemistry analyzer (Accute, Toshiba, Tokyo, Japan).
Flow cytometry assay
Flow cytometry analysis was used to evaluate the PBMC subpopulations, as described previously [20]. Briefly, the PBMCs were labeled with a primary monoclonal antibody (CD3 [MM1A], CD4 [CACT138A], CD8 [BAT82A], CD14 [CAM36A], WC1 [ILA29], or CD21 [BAQ15A]; Washington State Monoclonal Antibody Center, Pullman, WA, U.S.A.). The CD3+, CD4+, and CD8+ T cell subsets were also labeled with CD45R (GC6A) monoclonal antibody. The labeled cells were also stained with specific purified IgG conjugated to phycoerythrin polyclonal secondary antibody (goat anti-mouse IgG1 RPE, AbD Serotec, Kidlington, Oxford, U.K.) or fluorescein isothiocyanate-conjugated antibody (goat anti-mouse IgG or IgM FITC, Southern Biotech, Birmingham, AL, U.S.A.). Flow cytometry analysis was performed using a FACScan analyzer (Becton Dickinson, Franklin Lakes, NJ, U.S.A.), equipped with a computer running the Cell Quest software (Becton Dickinson, Milan, Italy).
Enzyme linked immunosorbent assay for cytokines
Enzyme linked immunosorbent assays were performed to quantify the peripheral concentrations of inflammatory cytokines, as described previously [14]. The minimum detectable concentration of TNF-α was 150 pg/ml, and those of IL-1β, IL-6, and interferon (IFN)-γ were 30 pg/ml. The intra-assay coefficient of variation for TNF-α, IL-1β, IL-6, and IFN-γ was 5%.
RNA isolation from the RE
Total RNA was extracted from the RE of each calf using TRIzol Reagent and samples were further purified using an RNeasy RNA Clean-up Kit (QIAGEN, Valencia, CA, U.S.A.), as described previously [12]. RNA was quantified using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Thermo Fisher Scientific, Waltham, MA, U.S.A.) and RNA quality was assessed using a 2100 Bioanalyzer and RNA 6000 Nano LabChip kit (Agilent Technologies, Palo Alto, CA, U.S.A.); the RNA integrity value was 9.0 ± 0.7 (mean ± SD).
Microarray and upstream regulator gene analysis
Microarray and upstream regulator gene analysis were performed following a previous study [12]. Briefly, a customized bovine oligonucleotide microarray (Agilent Technologies) was used to detect differentially expressed genes (DEGs) in the RE using one-color microarray analysis as reported elsewhere [13]. Fluorescently labeled (Cy3) complementary RNA probes were hybridized, and the array was scanned using an Agilent Microarray Scanner (Agilent Technologies). Feature Extraction ver. 9.1 software (Agilent Technologies) was used to process the microarray images, align spots, and create raw numerical total spot intensity data. The microarray data were imported into GeneSpring 12.0 (Agilent Technologies), and normalization was performed (per-chip normalization). The Gene Expression Omnibus accession numbers are as follows: platform, GPL22091; samples, GSM2219049 to GSM2219055; and series, GSE83813 [12].
Lists of DEGs that corresponded to the raw estimated fold changes (FC; ≥2.0) were uploaded into the Ingenuity Pathway Analysis (IPA) software (www.ingenuity.com; Ingenuity Systems, Redwood City, CA, U.S.A.). The IPA knowledge base was used for DEG enrichment analysis, and top upstream regulator analysis and statistical calculations were performed to compare the two groups.
Statistical analysis
Ruminal pH, total VFA concentration, individual VFA proportions, ruminal LPS activity, PBMC subpopulations, and cytokine concentrations for the Weaning, Forage, and Starter groups at −1, 0, 1, and 3 weeks after weaning were evaluated using Prism software, version 8.01 (GraphPad Software, La Jolla, CA, U.S.A.). Distributions of variables were tested for normality using the Shapiro-Wilk test. Significant differences at the same week point were evaluated between the Forage and Starter groups using the unpaired Student’s t-test, while non-parametric data were analyzed using the Mann-Whitney test. One-way repeated-measures ANOVA, followed by Dunnett’s multiple comparison method, was used to determine within-group differences. Pearson’s correlation coefficient (r) and significance level (P-value) were used to determine the relationships between rumen parameters (24 hr mean ruminal pH, time with pH under 5.6 and 5.8, total VFA, proportions of individual VFAs, and LPS activity) and peripheral immune parameters or blood metabolites. The microarray data were normalized and an unpaired Student’s t-test was used to compare gene expression levels between the two groups using GeneSpring 12.0. Fold changes were calculated to identify the direction of changes in gene expression by comparing the Starter and Forage groups. Differences were considered significant at a cut-off of P<0.05, and a trend suggesting possible significance was determined at P<0.10.
RESULTS
Ruminal pH, VFA concentrations, and LPS activity
During the weaning transition period, calf body weight at −1, 0, 1, and 3 weeks after weaning (mean ± SE) was 81.1 ± 6.3, 85.9 ± 6.9, 93.6 ± 5.5, and 106.8 ± 6.8 kg, respectively, in the Weaning group, and the body weight increased significantly (P<0.05) at 0, 1, and 3 weeks within the group. Furthermore, body weight was not different between the Forage and Starter groups at 1 week before weaning, while it increased significantly (P<0.05) at 0, 1, and 3 weeks after weaning in the Forage group and at 1 and 3 weeks after weaning in the Starter group. No significant difference in total dry matter intake was observed at 3 weeks after weaning (P=0.336) between the two groups as reported previously [12].
In the Weaning group, the mean or minimum ruminal pH increased significantly (P<0.05) at 1 and 3 weeks or 3 weeks after weaning, respectively, when compared with those at 1 week before weaning (Table 1). Furthermore, the duration of ruminal pH <5.8 decreased significantly (P<0.05) at 3 weeks after weaning compared with 1 week before weaning (Table 1). No clinical symptoms, such as dehydration, diarrhea, fever, or a reduction in appetite, were observed in the Weaning group of calves.
Table 1. Changes in 24 hr mean ruminal pH, duration of time <5.6 and 5.8, total VFA, individual VFA proportions, acetic acid-to-propionic acid ratio, and LPS activity in weaning calves during weaning transition.
| Items | Weaning group (n=7) | SEM | ||||
|---|---|---|---|---|---|---|
| Week −1 | Week 0 | Week 1 | Week 3 | |||
| 24 hr mean ruminal pH | ||||||
| Minimum | 5.76 | 5.78 | 5.96A) | 6.16a) | 0.17 | |
| Mean | 5.36 | 5.38 | 5.70a) | 5.73a) | 0.10 | |
| Maximum | 6.42 | 6.23 | 6.43 | 6.58 | 0.24 | |
| Duration of ruminal pH | ||||||
| pH<5.6, min/d | 72.4 | 60.4 | 40.6 | 28.1 | 22.6 | |
| pH<5.8, min/d | 154.0 | 167.6 | 103.0 | 67.0a) | 66.6 | |
| Total VFA (mmol/dl) | 10.36 | 9.9 | 10.67 | 11.72 | 1.51 | |
| Individual VFA (%) | ||||||
| Acetic acid | 56.9 | 56.3 | 58.0 | 57.2 | 2.97 | |
| Propionic acid | 28.9 | 29.4 | 28.2 | 27.9 | 3.61 | |
| Butyric acid | 9.8 | 9.3 | 8.4 | 9.8 | 1.11 | |
| Others1) | 4.5 | 5.1 | 5.4 | 5.1 | 0.51 | |
| A/P ratio | 2.3 | 2.1 | 2.4 | 2.4 | 0.41 | |
| LPS (EU ×104/ml) | 13.0 | 14.0 | 14.8 | 16.0 | 7.71 | |
1) Volatile fatty acid (VFA) components that exclude acetate, propionate, and butyrate from total VFA. a, A) denotes significant difference (P<0.05 and 0.10, respectively) compared with the Week −1.
At 3 weeks after weaning, the mean (6.42 vs. 5.66) and maximum (6.98 vs 5.95) ruminal pH were significantly (P<0.05) higher in the Forage group, and the duration of ruminal pH <5.8 was significantly shorter (97.6 vs 1,050 min/d, P<0.05) in the same group compared with the Starter group [12]. The proportion of acetic acid (64.5 vs 51.7%) and the ruminal acetic acid-to-propionic acid ratio (3.39 vs 1.67) were significantly (P<0.05) higher in the Forage group relative to Weaning group, and a tendency toward a higher proportion of propionic acid (19.1 vs 34.5%, P=0.051) was observed in the Starter group relative to Weaning group [12]. The ruminal LPS activity in the Starter group was higher at 0 to 3 weeks after weaning, but no statistical significance (P>0.1) was identified between the Forage and Starter groups during weaning transition.
Microarray and upstream regulator analysis
In total, 1,024 DEGs were identified (446 upregulated and 577 downregulated; P<0.05) when comparing the Starter (n=4) and Forage (n=3) groups, of which 212 DEGs (P<0.05; FC≥2) could be mapped to a known molecular function in the top upstream regulator using IPA software. Among the genes encoding receptors, TLR4 (FC=1.61; P<0.05) was significantly upregulated in the Starter group compared with the Forage group, as reported previously [12].
In silico analysis revealed the upstream regulator genes involved in the TLR4-mediated inflammatory signaling pathway. Among the genes identified as upstream regulators, interferon gamma (IFN-γ; z-score=3.251; P=5.60 × 10−23) was significantly activated. Among the genes involved in the TLR4-mediated inflammatory signaling pathway, TLR4 (z-score=2.150; P=6.54 × 10−11) and many downstream genes were significantly activated, including nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB; z-score=3.183; P=5.17 × 10−7), myeloid differentiation primary response 88 (MyD88; z-score=2.937; P=3.52 × 10−7), interferon regulatory factor 3 (IRF3; z-score=2.189; P=4.35 × 10−3), interferon regulatory factor 7 (IRF7; z-score=2.789; P=1.40 × 10−4), and interferon regulatory factor 8 (IRF8; z-score=2.583; P=1.14 × 10−4). In addition, signaling transducers and activators of transcription family of transcription factor 1 (STAT1; z-score=1.872; P=8.96 × 10−13), activator protein 1 (AP1; P=4.35 × 10−3), and toll-like receptor adaptor molecule 2 (TICAM2; P=2.04 × 10−2) were identified as significant upstream regulators based on their expression pattern. Figure 1 shows the activated TLR4-mediated signaling pathway caused by ruminal LPS, which includes the activation of key inflammatory regulator genes (NFκB, IRFs, STAT1, and AP1) and stimulation of proinflammatory cytokine production.
Fig. 1.
TLR4-mediated inflammatory signaling pathway in the rumen epithelium. Differentially expressed genes (P<0.05; fold change ≥2) identified by comparison of the Starter (n=4) and Forage (n=3) groups could be mapped to a known molecular function in the top upstream regulator using IPA software. z-Scores represent the activation or inhibition of genes as upstream regulators. Gray symbols denote significant activation (z-score ≥2) of genes, and white symbols denote no activation as an upstream regulator. IFNγ, interferon gamma; IFNγR, interferon gamma receptor; LPS, lipopolysaccharide; TLR4, toll-like receptor 4; MyD88, myeloid differentiation primary response 88; STAT1, signaling transducers and activators of transcription family of transcription factor 1; IRF3, interferon regulatory factor 3; NFκB, nuclear factor kappa-light-chain-enhancer of activated B cells; AP1, activator protein 1.
PBMC subpopulations and cytokine concentrations
No significant differences were identified in the PBMC subpopulations and cytokine concentrations in the Weaning group during weaning transition (Table 2). In contrast, the number of CD8+CD45R− T cells in the peripheral blood at 1 (237 vs. 83 count/µl) and 3 (176 vs. 72 count/µl) weeks after weaning tended to be higher (P=0.092 and 0.057, respectively) in the Forage group, whereas no significant differences between the Forage and Starter groups were seen in cytokine concentrations and the numbers of white blood cells, PBMCs, and T cell subsets. The population of CD14+ cells in the Starter group decreased significantly (P<0.05) at 0, 1, and 3 weeks after weaning (1,408, 1,159, and 1,028 count/µl, respectively) compared with 1 week before weaning (1,874 count/µl).
Table 2. Number of peripheral blood white blood cells (WBCs), mononuclear cells (PBMCs), and CD-positive T cells, and cytokine concentrations in weaning calves during weaning transition.
| Items | Weaning group (n=7) | SEM | ||||
|---|---|---|---|---|---|---|
| Week −1 | Week 0 | Week 1 | Week 3 | |||
| WBC (count/µl) | 11,129 | 11,529 | 10,429 | 9,314 | 806 | |
| PBMCs (count/µl) | 6,041 | 6,538 | 5,956 | 5,651 | 480 | |
| Subpopulation (count/µl) | ||||||
| CD3+CD45R− | 1,813 | 1,946 | 1,772 | 1,591 | 189 | |
| CD3+CD45R+ | 885 | 1,004 | 780 | 878 | 115 | |
| CD4+CD45R− | 318 | 388 | 352 | 274 | 40 | |
| CD4+CD45R+ | 312 | 406 | 304 | 330 | 67 | |
| CD8+CD45R− | 150 | 116 | 153 | 101 | 29 | |
| CD8+CD45R+ | 372 | 365 | 319 | 293 | 48 | |
| CD14+ | 1,394 | 1,477 | 1,349 | 1,161 | 186 | |
| WC1+ | 1,031 | 1,173 | 874 | 971 | 134 | |
| CD21+ | 1,136 | 973 | 1,269 | 1,177 | 151 | |
| Cytokine (ng/ml) | ||||||
| TNF-α | 575 | 529 | 706 | 408 | 165 | |
| IFN-γ | 359 | 463 | 485 | 363 | 54 | |
| IL-1β | 624 | 405 | 567 | 512 | 112 | |
| IL-6 | 2,518 | 2,026 | 1,851 | 1,879 | 232 | |
Biochemical analysis of peripheral blood
In the Weaning group, blood urea nitrogen (BUN) and BHB concentrations increased significantly (P<0.05) at 1 and 3 weeks, T-CHO and PL concentrations decreased significantly (P<0.05) at 1 and 3 weeks, and TP, AST, and alkaline phosphatase (ALP) concentrations increased significantly at 3 weeks after weaning compared with those at 1 week before weaning (Table 3). When comparing the Forage group with the Starter group, the concentrations of glucose (GLU; 107.1 vs. 94.8 mg/dl) and triglycerides (TG; 17.4 vs. 7.93 mg/dl) at 3 weeks post-weaning and total cholesterol (T-CHO; 72.8 vs. 60.2 and 62.9 vs. 43.4 mg/dl at 1 and 3 weeks post-weaning, respectively) and phospholipid (PL; 90.1 vs. 69.5 and 82.1 vs. 55.7 mg/dl at 1 and 3 weeks post-weaning, respectively) were significantly higher (P<0.05).
Table 3. Biochemical analysis of peripheral blood in weaning calves during weaning transition.
| Items | Weaning group (n=7) | SEM | |||
|---|---|---|---|---|---|
| Week −1 | Week 0 | Week 1 | Week 3 | ||
| TP (g/dl) | 6.25 | 6.58 | 6.49 | 6.80a) | 0.17 |
| ALB (g/dl) | 3.47 | 3.61 | 3.59 | 3.59 | 0.06 |
| BUN (mg/dl) | 14.0 | 17.4 | 19.6a) | 19.8a) | 1.35 |
| T-BIL (mg/dl) | 0.093 | 0.079 | 0.079 | 0.07 | 0.01 |
| GLU (mg/dl) | 93.4 | 93.4 | 89.9 | 100.1A) | 3.12 |
| TG (mg/dl) | 11.2 | 12.3 | 12.0 | 12.0 | 1.97 |
| T-CHO (mg/dl) | 100.3 | 89.7 | 65.6a) | 51.8a) | 5.19 |
| PL (mg/dl) | 109.0 | 96.5 | 78.3a) | 67.0a) | 5.58 |
| FFA (µEg/l) | 225.4 | 147.0 | 146.7 | 121.7A) | 30.1 |
| BHB (µmol/l) | 113.2 | 144.8 | 202.4a) | 283.7a) | 17.3 |
| Ca (mg/dl) | 10.6 | 10.6 | 10.7 | 10.9 | 0.1 |
| iP (mg/l) | 8.06 | 8.37 | 7.57 | 7.81 | 0.25 |
| AST (U/l) | 54.6 | 58.2 | 58.4 | 74.5a) | 3.95 |
| GGT (U/l) | 20.8 | 22.0 | 20.5 | 19.6 | 1.74 |
| ALP (U/l) | 634.3 | 671.5 | 646.4 | 757.7a) | 41.4 |
a, A) denotes significant difference (P<0.05 and 0.10, respectively) compared with Week −1.
Pearson correlation analyses
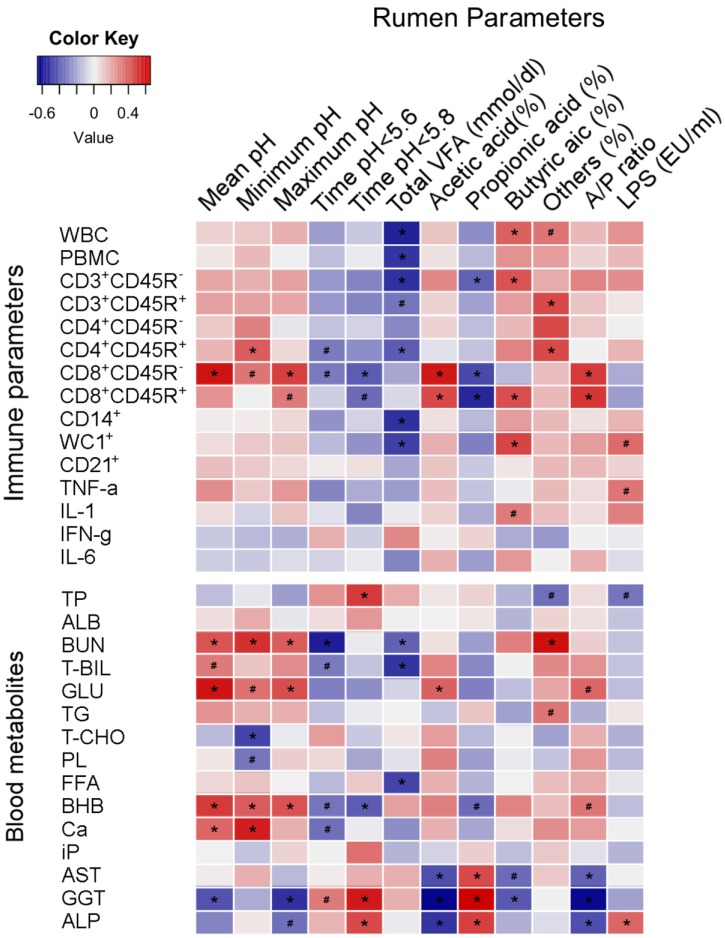
Analyses of Pearson’s correlation coefficients were performed between rumen fermentation parameters and peripheral immune parameters or blood metabolites in the Weaning group (Fig. 2). Among the immune parameters (P<0.05), the CD8+CD45R− T cell subset number was positively correlated (r=0.592) with 24 hr mean ruminal pH, and negatively correlated (r=−0.325) with the duration of ruminal pH<5.8. The CD8+CD45R− T cell subset number was also positively correlated with the proportion of acetic acid (r=0.582) and ruminal acetic acid-to-propionic acid ratio (r=0.500), and negatively correlated with the proportion of propionic acid (r=−0.450).
Fig. 2.
Correlation analyses between rumen parameters and peripheral immune parameters or blood metabolites. Cells are colored based on Pearson’s correlation coefficient analyses. Blue represents a negative correlation and red represents a positive correlation. *denotes significant (P<0.05) correlation.
Among the peripheral blood metabolites (P<0.05), the concentration of BUN was positively correlated (r=0.422) with 24 hr mean ruminal pH, and showed negative correlation (r=−0.566) with the duration of ruminal pH<5.6. The concentration of GLU was positively correlated with 24 hr mean ruminal pH (r=0.602) and proportion of acetic acid (r=0.384). Peripheral gamma-glutamyl transferase (GGT) activity was negatively (r=−0.432) correlated with 24 hr mean ruminal pH. Serum AST, GGT, and ALP activities were negatively correlated with the proportion of acetic acid (r=−0.433, −0.633, and −0.501, respectively) and ruminal acetic acid-to-propionic acid ratio (r=−0.386, −0.625, and −0.433, respectively), and were positively correlated with the proportion of butyric acid (r=0.455, 0.656, and 0.479, respectively).
DISCUSSION
This study investigated the effects of ruminal pH on gene expression in the RE and on peripheral immune function during weaning transition. Previously, it has been reported that hay intake might play an important role in mitigating ruminal acidosis during weaning transition [2, 11]. The details of changes in ruminal pH and VFAs during weaning transition [11] and at 3 weeks after weaning [12] have been previously described. For example, we reported significantly lower ruminal pH in calves fed only calf starter compared with calves fed calf starter with forage during weaning transition [11], and these calves showed a significantly longer duration of ruminal pH <5.8 at 3 weeks after weaning [12]. In addition, a significantly higher proportion of acetic acid and lower proportions of propionic and butyric acids in calves fed calf starter with forage were identified during weaning transition [11], and a significantly lower proportion of acetic acid and a tendency toward higher proportion of propionic acid were observed in the Starter group [12]. Thus, feeding calf starter without forage caused a severely lower ruminal pH, and the Starter group calves suffered from ruminal acidosis during weaning transition.
TLR4 recognizes bacterial LPS and activates the inflammatory response in bovine mammary epithelial cells [9] and the RE [18]. Moreover, TLR4 has been reported to modulate the inflammatory signaling pathway by inducing the expression of inflammatory regulator transcription factors, such as NFκB, IRFs, STAT1, and AP1, and to stimulate the production of cytokines [22]. Several studies reported that gene expression of TLR4 was significantly higher in subacute ruminal acidosis (SARA) positive cattle relative to SARA negative cattle, suggesting a negative correlation between ruminal pH and TLR4 gene expression in the RE [4]. In the present study, the expression of TLR4 was significantly upregulated in the Starter group, and TLR4 downstream genes (such as MyD88, NFκB, IRF3, IRF7, and IRF8) were identified as significantly activated upstream regulators, although only numerically higher ruminal LPS activity was identified in the Starter group. Therefore, we assumed that higher ruminal acidity rather than LPS activity may have induced activation of the TLR4 and TLR4-mediated signaling pathway in the RE of calves from the Starter group.
In the present study, although there was no significant changes in the Weaning group during weaning transition, the CD8+ T cell subset (CD8+CD45R−) tended to be lower in the Starter group, and CD14+ cells (LPS receptor; [1]) decreased in the Forage and Starter groups during weaning transition. Only a few previous reports have shown that an increase in bovine CD8+ cells are associated with immune stimulatory responses to various infections in cattle, such as bovine viral diarrhea virus [21], foot-and-mouth disease virus [5], and Mycobacterium bovis [15]. Therefore, these results suggest that the lower number of CD8+CD45R− cells after weaning in the Starter group might represent a reduction in peripheral cellular immunity. Meanwhile, Stefanska et al. [26] reported that peripheral blood mRNA abundance of CD14 was higher (P≤0.01) in SARA-risk and -positive herds compared to SARA-negative herds, and was negatively correlated (P≤0.01) with ruminal pH. In the present study, however, decrease in the number of CD14+ cells was found in both groups, which is not consistent with the previous study [26]. Therefore, it is more plausible that these changes were a physiological response during weaning transition, and significantly lower ruminal pH or numerically higher LPS activity in the Starter group might intensify the relevant changes in the present study. Collectively, higher ruminal acidosis or LPS activity in the Starter group might not be sufficiently severe enough for stimulating a systemic inflammatory response (no differences in serum cytokine levels in the two groups were found), and the immunostimulatory effect was limited to activating TLR4 and the TLR4-mediated signaling pathway in the RE.
In the Weaning group, AST activity and ALP concentration increased significantly at 3 weeks after weaning, and calves in the Starter group showed higher serum AST and GGT activities and ALP concentration, which are indicators of liver function in ruminants [3, 17, 24]. Cattle with hepatic toxicity induced by poultry litter consumption showed higher serum levels of GGT and ALP than healthy cattle [24], and the concentration of ALP increased in goats fed a high-grain diet [3]. Therefore, the higher levels of AST (68.8 vs. 78.7 IU/l), GGT (18.4 vs. 20.5 IU/l), and ALP (636.3 vs. 848.7 IU/l) observed at 3 weeks after weaning in the Starter group compared to Weaning group might imply decreased liver function caused by feeding a considerable amount of calf starter. Furthermore, disturbed liver function might induce lower glucose levels in the Starter group, which is consistent with hypoglycemia identified in an induced hepatic injury rat model [27]. Meanwhile, the Weaning group calves showed significant increases in T-CHO and PL at 3 weeks after weaning, and feeding calf starter without forage caused low peripheral TG, T-CHO, and PL concentrations in the present study. The rumen epithelial cholesterol biosynthesis genes controlled by SREBP were downregulated in dairy cattle fed a high-grain diet [25]. In addition, Xu et al. [28] reported that LPS derived from rumen induced the downregulation of sterol regulatory element binding transcription factor 1c and 2 (SREBF1c and SREBF2, respectively) in the liver, and was related to the significantly lower plasma concentrations of TG, FFA, and T-CHO in dairy cattle. Therefore, the higher ruminal acidity induced by feeding only calf starter caused the downregulation of SREBP1 and ketogenic genes in the RE, as reported previously [12]. This might result in the significantly lower peripheral concentrations of TG, T-CHO, and PL found in the Starter group compared with the Forage group.
Interestingly, in the present study, analyses of Pearson’s correlation coefficients in the Weaning group revealed significant correlations between rumen parameters (ruminal pH, total VFA, and VFA components) and immune parameters (CD8+CD45R− T cells) or blood metabolites (BUN, GLU, BHB, and GGT). Previously, Nasrollahi et al. [17] suggested that blood metabolite concentrations may help to predict susceptibility of cows to SARA, and serum AST activity was negatively correlated with ruminal pH in dairy cows. In the present study, GGT activity was negatively correlated with 24 hr mean ruminal pH, and was lower in the Starter group during weaning transition, although we could not find any correlation between serum AST activity and ruminal pH. Collectively, serum GGT activity level together with the levels or concentrations of peripheral blood CD8+CD45R− T cells, BUN, GLU, and BHB may be valuable indicators of the occurrence of ruminal acidosis and its consequences for peripheral blood immune and metabolic statuses in Holstein calves during weaning transition. Meanwhile, Pollock et al. [19] reported that normal variation in the level of nutrition and weaning affects peripheral blood cellular immune responses in calves. In the present study, CD8+CD45R+ and CD8+CD45R− T cell numbers were positively correlated with the proportion of acetic acid. In addition, the proportion of acetic acid was also positively correlated with peripheral blood GLU level, which was significantly higher in the Forage group at 3 weeks after weaning. Therefore, these results support that a higher ruminal proportion of acetic acid improves peripheral blood energy status, and may subsequently enhance the peripheral blood cellular immune responses. However, to the best of our knowledge, the basis of the relationships between VFA components and immune parameters remains largely unstudied in cattle. Therefore, further studies are required to explore all the relevant relationships, and our results may suggest a new inspiration to the related future studies in Holstein calves or cattle.
In conclusion, we found that feeding only calf starter caused a low pH in the rumen and resulted in significant upregulation of TLR4 in the RE. Consequently, activation of the TLR4 signaling pathway was identified based on activated upstream regulator genes (TLR4, NFκB, MyD88, and IRFs), whereas activation did not markedly affect the systemic inflammatory response. Blood metabolite analysis indicated that feeding only calf starter induced lower cholesterol biosynthesis in the RE, and calves fed calf starter with forage showed higher energy status and lower AST, GGT, and ALP levels in the peripheral blood. Moreover, a significant negative correlation was found between 24 hr mean ruminal pH and serum GGT activity, which supports that serum GGT level could be an indicator of SARA severity in Holstein calves during weaning transition.
Acknowledgments
This research was supported financially by KAKENHI Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (No. 26292156).
REFERENCES
- 1.Berthon P., Hopkins J.1996. Ruminant cluster CD14. Vet. Immunol. Immunopathol. 52: 245–248. doi: 10.1016/0165-2427(96)05568-7 [DOI] [PubMed] [Google Scholar]
- 2.Castells L., Bach A., Aris A., Terré M.2013. Effects of forage provision to young calves on rumen fermentation and development of the gastrointestinal tract. J. Dairy Sci. 96: 5226–5236. doi: 10.3168/jds.2012-6419 [DOI] [PubMed] [Google Scholar]
- 3.Chang G., Zhang K., Xu T., Jin D., Seyfert H. M., Shen X., Zhuang S.2015. Feeding a high-grain diet reduces the percentage of LPS clearance and enhances immune gene expression in goat liver. BMC Vet. Res. 11: 67. doi: 10.1186/s12917-015-0376-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y., Oba M., Guan L. L.2012. Variation of bacterial communities and expression of Toll-like receptor genes in the rumen of steers differing in susceptibility to subacute ruminal acidosis. Vet. Microbiol. 159: 451–459. doi: 10.1016/j.vetmic.2012.04.032 [DOI] [PubMed] [Google Scholar]
- 5.Childerstone A. J., Cedillo-Baron L., Foster-Cuevas M., Parkhouse R. M.1999. Demonstration of bovine CD8+ T-cell responses to foot-and-mouth disease virus. J. Gen. Virol. 80: 663–669. doi: 10.1099/0022-1317-80-3-663 [DOI] [PubMed] [Google Scholar]
- 6.Connor E. E., Baldwin R. L., 6th, Li C. J., Li R. W., Chung H.2013. Gene expression in bovine rumen epithelium during weaning identifies molecular regulators of rumen development and growth. Funct. Integr. Genomics 13: 133–142. doi: 10.1007/s10142-012-0308-x [DOI] [PubMed] [Google Scholar]
- 7.Hirabayashi H., Kawashima K., Okimura T., Tateno A., Suzuki A., Asakuma S., Isobe N., Obitsu T., Kushibiki S., Sugino T.2017. Effect of nutrient levels during the far-off period on postpartum productivity in dairy cows. Anim. Sci. J. 88: 1162–1170. doi: 10.1111/asj.12743 [DOI] [PubMed] [Google Scholar]
- 8.Hulbert L. E., Cobb C. J., Carroll J. A., Ballou M. A.2011. Effects of changing milk replacer feedings from twice to once daily on Holstein calf innate immune responses before and after weaning. J. Dairy Sci. 94: 2557–2565. doi: 10.3168/jds.2010-3980 [DOI] [PubMed] [Google Scholar]
- 9.Ibeagha-Awemu E. M., Lee J. W., Ibeagha A. E., Bannerman D. D., Paape M. J., Zhao X.2008. Bacterial lipopolysaccharide induces increased expression of toll-like receptor (TLR) 4 and downstream TLR signaling molecules in bovine mammary epithelial cells. Vet. Res. 39: 11. doi: 10.1051/vetres:2007047 [DOI] [PubMed] [Google Scholar]
- 10.Kim M. H., Yang J. Y., Upadhaya S. D., Lee H. J., Yun C. H., Ha J. K.2011. The stress of weaning influences serum levels of acute-phase proteins, iron-binding proteins, inflammatory cytokines, cortisol, and leukocyte subsets in Holstein calves. J. Vet. Sci. 12: 151–157. doi: 10.4142/jvs.2011.12.2.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim Y. H., Nagata R., Ohtani N., Ichijo T., Ikuta K., Sato S.2016a. Effects of dietary forage and calf starter diet on ruminal pH and bacteria in Holstein calves during weaning transition. Front. Microbiol. 7: 1575. doi: 10.3389/fmicb.2016.01575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim Y. H., Toji N., Kizaki K., Kushibiki S., Ichijo T., Sato S.2016b. Effects of dietary forage and calf starter on ruminal pH and transcriptomic adaptation of the rumen epithelium in Holstein calves during the weaning transition. Physiol. Genomics 48: 803–809. doi: 10.1152/physiolgenomics.00086.2016 [DOI] [PubMed] [Google Scholar]
- 13.Kizaki K., Shichijo-Kizaki A., Furusawa T., Takahashi T., Hosoe M., Hashizume K.2013. Differential neutrophil gene expression in early bovine pregnancy. Reprod. Biol. Endocrinol. 11: 6. doi: 10.1186/1477-7827-11-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kushibiki S., Shingu H., Komatsu T., Itoh F., Kasuya E., Aso H.2006. Hodate, K. Effect of recombinant bovine tumor necrosis factor-α on hormone release in lactating cows. Anim. Sci. J. 77: 603–612. doi: 10.1111/j.1740-0929.2006.00392.x [DOI] [Google Scholar]
- 15.Liébana E., Girvin R. M., Welsh M., Neill S. D., Pollock J. M.1999. Generation of CD8(+) T-cell responses to Mycobacterium bovis and mycobacterial antigen in experimental bovine tuberculosis. Infect. Immun. 67: 1034–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malmuthuge N., Li M., Fries P., Griebel P. J., Guan L. L.2012. Regional and age dependent changes in gene expression of Toll-like receptors and key antimicrobial defence molecules throughout the gastrointestinal tract of dairy calves. Vet. Immunol. Immunopathol. 146: 18–26. doi: 10.1016/j.vetimm.2012.01.010 [DOI] [PubMed] [Google Scholar]
- 17.Nasrollahi S. M., Zali A., Ghorbani G. R., Kahyani A., Beauchemin K. A.2019. Short communication: Blood metabolites, body reserves, and feed efficiency of high-producing dairy cows that varied in ruminal pH when fed a high-concentrate diet. J. Dairy Sci. 102: 672–677. doi: 10.3168/jds.2018-15022 [DOI] [PubMed] [Google Scholar]
- 18.Plaizier J. C., Khafipour E., Li S., Gozho G. N., Krause D. O.2012. Subacute ruminal acidosis (SARA), endotoxins and health consequences. Anim. Feed Sci. Technol. 172: 9–21. doi: 10.1016/j.anifeedsci.2011.12.004 [DOI] [Google Scholar]
- 19.Pollock J. M., Rowan T. G., Dixon J. B., Carter S. D.1994. Level of nutrition and age at weaning: effects on humoral immunity in young calves. Br. J. Nutr. 71: 239–248. doi: 10.1079/BJN19940130 [DOI] [PubMed] [Google Scholar]
- 20.Qadis A. Q., Goya S., Yatsu M., Yoshida Y. U., Ichijo T., Sato S.2014. Effects of a bacteria-based probiotic on subpopulations of peripheral leukocytes and their cytokine mRNA expression in calves. J. Vet. Med. Sci. 76: 189–195. doi: 10.1292/jvms.13-0370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhodes S. G., Cocksedge J. M., Collins R. A., Morrison W. I.1999. Differential cytokine responses of CD4+ and CD8+ T cells in response to bovine viral diarrhoea virus in cattle. J. Gen. Virol. 80: 1673–1679. doi: 10.1099/0022-1317-80-7-1673 [DOI] [PubMed] [Google Scholar]
- 22.Roy A., Srivastava M., Saqib U., Liu D., Faisal S. M., Sugathan S., Bishnoi S., Baig M. S.2016. Potential therapeutic targets for inflammation in toll-like receptor 4 (TLR4)-mediated signaling pathways. Int. Immunopharmacol. 40: 79–89. doi: 10.1016/j.intimp.2016.08.026 [DOI] [PubMed] [Google Scholar]
- 23.Sato S., Mizuguchi H., Ito K., Ikuta K., Kimura A., Okada K.2012. Technical note: development and testing of a radio transmission pH measurement system for continuous monitoring of ruminal pH in cows. Prev. Vet. Med. 103: 274–279. doi: 10.1016/j.prevetmed.2011.09.004 [DOI] [PubMed] [Google Scholar]
- 24.Silanikove N., Tiomkin D.1992. Toxicity induced by poultry litter consumption: effect on measurements reflecting liver function in beef cows. Anim. Sci. 54: 203–209. [Google Scholar]
- 25.Steele M. A., Vandervoort G., AlZahal O., Hook S. E., Matthews J. C., McBride B. W.2011. Rumen epithelial adaptation to high-grain diets involves the coordinated regulation of genes involved in cholesterol homeostasis. Physiol. Genomics 43: 308–316. doi: 10.1152/physiolgenomics.00117.2010 [DOI] [PubMed] [Google Scholar]
- 26.Stefanska B., Człapa W., Pruszynska-Oszmałek E., Szczepankiewicz D., Fievez V., Komisarek J., Stajek K., Nowak W.2018. Subacute ruminal acidosis affects fermentation and endotoxin concentration in the rumen and relative expression of the CD14/TLR4/MD2 genes involved in lipopolysaccharide systemic immune response in dairy cows. J. Dairy Sci. 101: 1297–1310. doi: 10.3168/jds.2017-12896 [DOI] [PubMed] [Google Scholar]
- 27.Toshina Y., Dote T., Usuda K., Shimizu H., Tominaga M., Kono K.2004. Hepatic injury and gluconeogenesis after subcutaneous injection of monochloroacetic acid in rats. Environ. Health Prev. Med. 9: 58–62. doi: 10.1007/BF02897933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu T., Tao H., Chang G., Zhang K., Xu L., Shen X.2015. Lipopolysaccharide derived from the rumen down-regulates stearoyl-CoA desaturase 1 expression and alters fatty acid composition in the liver of dairy cows fed a high-concentrate diet. BMC Vet. Res. 11: 52. doi: 10.1186/s12917-015-0360-6 [DOI] [PMC free article] [PubMed] [Google Scholar]