Abstract
De novo variants represent a significant cause of neurodevelopmental delay and intellectual disability. A genetic basis can be identified in only half of individuals who have neurodevelopmental disorders (NDDs); this indicates that additional causes need to be elucidated. We compared the frequency of de novo variants in patient-parent trios with (n = 2,030) versus without (n = 2,755) NDDs. We identified de novo variants in TAOK1 (thousand and one [TAO] amino acid kinase 1), which encodes the serine/threonine-protein kinase TAO1, in three individuals with NDDs but not in persons who did not have NDDs. Through further screening and the use of GeneMatcher, five additional individuals with NDDs were found to have de novo variants. All eight variants were absent from gnomAD (Genome Aggregation Database). The variant carriers shared a non-specific phenotype of developmental delay, and six individuals had additional muscular hypotonia. We established a fibroblast line of one mutation carrier, and we demonstrated that reduced mRNA levels of TAOK1 could be increased upon cycloheximide treatment. These results indicate nonsense-mediated mRNA decay. Further, there was neither detectable phosphorylated TAO1 kinase nor phosphorylated tau in these cells, and mitochondrial morphology was altered. Knockdown of the ortholog gene Tao1 (Tao, CG14217) in Drosophila resulted in delayed early development. The majority of the Tao1-knockdown flies did not survive beyond the third instar larval stage. When compared to control flies, Tao1 knockdown flies revealed changed morphology of the ventral nerve cord and the neuromuscular junctions as well as a decreased number of endings (boutons). Furthermore, mitochondria in mutant flies showed altered distribution and decreased size in axons of motor neurons. Thus, we provide compelling evidence that de novo variants in TAOK1 cause NDDs.
Keywords: TAO kinase 1, neurodevelopmental disorders, de novo variants, fly model
Main Text
Approximately 2%–5% of children are born with major congenital malformations,1 which are often accompanied by neurodevelopmental disorders (NDDs) with variable severity and different behavioral abnormalities. Of note, NDDs often arise from pathogenic de novo variants in genes critical for brain development.2, 3 As a result of the enormous genetic heterogeneity of these disorders, next-generation sequencing approaches have been effective means of diagnosing individuals with NDDs, but the diagnostic yield is about 50% at best.4 The molecular diagnosis provides an approach to shifting from a more phenotype-driven management of the symptoms to a more refined treatment based on genotype.5
To further elucidate the genetic spectrum of NDDs, we analyzed exome sequencing data from 4,785 patient-parent trios that were referred to Centogene AG (Rostock, Germany) for diagnostic exome sequencing in the period between January 2014 and June 2017. Most of the individuals with NDDs originated from the Middle East (64%) and from Europe (21%). Clinical information was provided by the referring physicians. These trios included 2,030 individuals with some form of NDD (as defined by the Human Phenotype Ontology [HPO] nomenclature6, 7: global developmental delay, seizures, microcephaly, macrocephaly, motor delay, delayed speech and language development, or intellectual disability). The remaining 2,755 trios were comprised of persons with other diseases. Written informed consent was obtained from affected individuals and/or guardians after the benefits and risks of clinical exome sequencing testing were explained to them. This study was approved by the Ethical Commission of the faculty of Medicine of the University of Rostock, Germany (registry no. A2015-0102). The samples were processed in Centogene’s laboratory (see Supplemental Data). Sequencing data were filtered for good quality (mean sequencing depth of >100×) and variants were only considered if the sequencing depth was >20×, the variant allele fraction was >20% of called reads, and the quality Phred score was >220. We performed Sanger sequencing validations for all variants with quality Phred scores <300 to rule out false-positive variants, as previously described.8
The frequency of de novo variants was compared between the 2,030 NDD and the 2,755 non-NDD trios in the 3,230 genes with a pLI score (the probability of being loss-of-function [LoF] intolerant) of ≥0.9, which indicated a high intolerance to LoF variants.9 The gene with the highest difference in the occurrence of de novo changes was TAOK1 (thousand and one amino acid [TAO] kinase 1, MIM: 610266, pLI = 1.00), which encodes the serine/threonine-protein kinase TAO1, which is highly expressed in the brain.10 We detected three de novo changes in TAOK1 among the NDD trios (individuals 1–3) but none in the non-NDD individuals (p = 0.08, Fisher′s exact test). The de novo variants included two missense (c.50A>G [p.Glu17Gly], c.892A>G [p.Lys298Glu]), and one nonsense change (c.2341G>T [p.Glu781∗])(GenBank: NM_020791.2).
In a next step, we specifically searched for TAOK1 de novo variants in 1,719 NDD patient-parent trios that were more recently (between July 2017 and February 2019) referred to Centogene AG for diagnostic exome sequencing. This revealed two additional carriers (individuals 4 and 5) of a missense (c.914A>C [p.Asp305Ala]) and a nonsense (c.1630C>T [p.Gln544∗]) variant.
Finally, three additional individuals with de novo variants in TAOK1 were identified through GeneMatcher11 (individual 6, c.332C>T [p.Ser111Phe]; individual 7, c.2366_2367insC [p.Leu790Phefs∗3]; and individual 8, c.2488G>T [p.Glu830∗]) (Table 1, Figure 1, Supplemental Data). These individuals were sequenced in a diagnostic setting at the University of Leipzig (individual 6); at Telemark Hospital Trust, Norway (individual 7); or at the Technical University of Dresden, Germany (individual 8) according to standard procedures.
Table 1.
List of Variants Identified in TAOK1 in This Study
| Individual | Chr | Position (GRCh37/hg19) | Ref | Alt | cDNA (GenBank: NM_020791.2) | Protein (GenBank: NP_065842.1) | Country of Origin | Sex | CADD score12 (v1.4) | De Novo | GnomAD | ACMG Scoring38 | Pathogenicity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Initial Exome Screening | |||||||||||||
| 1 | 17 | 27778616 | A | G | c.50A>G | p.Glu17Gly | SA | F | 23.2 | yes | not reported | PS2, PM2 | likely path |
| 2 | 17 | 27822638 | A | G | c.892A>G | p.Lys298Glu | Israel | M | 27 | yes | not reported | PS2, PM2 | likely path |
| 3 | 17 | 27857617 | G | T | c.2341G>T | p.Glu781∗ | SA | M | 42 | yes | not reported | PVS1, PS2, PM2 | path |
| Additional Carriers (Centogene) | |||||||||||||
| 4 | 17 | 27822660 | A | C | c.914A>C | p.Asp305Ala | Israel | F | 27.6 | yes | not reported | PS2, PM2 | likely path |
| 5 | 17 | 27837936 | C | T | c.1630C>T | p.Gln544∗ | Finland | M | 38 | yes | not reported | PVS1, PS2, PM2 | path |
| Additional Carriers (GeneMatcher) | |||||||||||||
| 6 | 17 | 27804704 | C | T | c.332C>T | p.Ser111Phe | Germany | F | 26.8 | yes | not reported | PS2, PM2 | likely path |
| 7 | 17 | 27861140 | G | GC | c.2366_2367insC | p.Leu790Phefs∗3 | Norway | M | 35 | yes | not reported | PVS1, PS2, PM2 | path |
| 8 | 17 | 27861262 | G | T | c.2488G>T | p.Glu830∗ | Germany | M | 46 | yes | not reported | PVS1, PS2, PM2 | path |
Abbreviations are as follows: Chr—chromosome, Ref—reference allele, Alt—alternate allele, De novo—parents available to confirm de novo status,
SA—Saudi Arabia, likely path—likely pathogenic, and path—pathogenic.
Figure 1.
De novo Variants in TAOK1 Detected in Individuals with NDDs
(A) Photographs of individual 6 show dysmorphic features including microsomia with short proximal extremities, macrocephaly with a high forehead, low-set ears, and down-slanting eyelids. The asymmetry of the lips is caused by the surgical intervention to correct the cleft lip and is therefore a secondary phenomenon.
(B) Photographs of individual 8 show dysmorphic features including a long face, a high forehead, big ears, a long nose, down-slanting palpebral fissures, retrognathia, congenital flat feet, and hyperextensibility of the distal joints. The latter three features might be linked to the FBNI mutation and fit the diagnosis of Marfan syndrome.
(C) Schematic representation of the encoded TAO1 kinase with its 1,001 amino acids. The functional domains (protein kinase [amino acids 28–281] and coiled-coil motifs 1 [amino acids 458–651] and 2 [amino acids 754–877]) are highlighted in red and blue, respectively. Arrows indicate the locations of the detected variants (see Table 1 for further descriptions). GenBank: NM_020791.2; NP_065842.1.
All eight TAOK1 variants were in silico predicted to be pathogenic (CADD [Combined Annotation Dependent Depletion] score12 >20) and not reported in gnomAD (Genome Aggregation Database) (Table 1).
Phenotypically, all eight variant carriers had developmental delay affecting speech and language development and/or motor development. Furthermore, muscular hypotonia was present in six, and intellectual disability was present in four individuals. Abnormal facial shape was found in five variant carriers, three of whom also had macrocephaly. Several other abnormalities were reported only in one or two individuals, and it is still unclear whether these signs and symptoms belong to the phenotypic spectrum (for details, see Table S1). Only one of the individuals had seizures. Individual 8 carried an additional de novo variant in FBN1 (MIM: 134797). This person also had features of Marfan syndrome (MIM: 154700) including congenital club feet, retrognathia, high palate, and joint laxity. (Because of the relation between these features and the FBN1 variant, these findings have not been included in Table S1.)
The frequency of carriers of de novo variants among our 3,749 NDD trios was 0.13%; the carriers included three carriers of de novo missense variants and two carriers of truncating variants. In gnomAD, a total of 12 carriers of LoF variants, all heterozygous and spread over the gene, are listed among the about 140,000 sequenced individuals (0.009%). Of note, it is not confirmed that these variants are real nor is there information about whether they occurred de novo or about the phenotype of the carriers. The gnomAD consortium removed individuals known to be affected by severe pediatric disease from the database; however, it should be noted that some individuals with severe disease might still be included in the dataset, albeit most likely at a frequency equivalent to or lower than that seen in the general population, as stated at the gnomAD homepage. Comparing the frequency of truncating variants in our samples (0.05%) and in gnomAD (0.009%) reveals a significantly higher rate of truncating variants among the individuals with NDDs (p = 0.049, Fisher′s exact test) than among those without. In addition, there are 163 unique missense variants listed in gnomAD, and the Z score for missense variants (a measurement of the deviation of observed missense variants from expectations)13 in TAOK1 is 4.92, suggesting a pathogenic role for at least a subset of missense variants. At this stage, functional consequences cannot be confirmed for the missense variants located within or near the kinase domain, as found in individuals 1, 2, 4, or 6. However, the phenotypic overlap with other individuals and the de novo occurrence support pathogenicity.
For individual 7, RNA could be extracted from a fresh blood sample via a PAXgene tube sampling. We also included three healthy, mutation-negative controls. Total RNA was extracted with the PAXgene Blood RNA Kit (QIAGEN) and then reverse transcribed into cDNA through use of the Maxima First Strand cDNA Synthesis Kit (Thermo Scientific). First, we sequenced the TAOK1 cDNA, which confirmed the expression of the variant but indicated lower levels of the mutant allele than of the wild-type allele (Figure 2A). Second, we performed quantitative PCR with SYBR Green on the LightCycler 480 system (Roche Diagnostics) that revealed TAOK1 expression of only 20%–45% in individual 7 compared to the controls (Figure 2B) when normalizing to two different reference genes in the blood sample (ACTB, G6PD). Next, we established a fibroblast line from individual 7 (L-13908) and extracted RNA by using the QIAmp RNA Extraction Kit (QIAGEN). Oligo(dT) nucleotides from the Maxima First Strand cDNA Synthesis Kit (ThermoFisher) served as primers for synthesis the complementary DNA (cDNA) by use of reverse transcriptase (RT). Expression analyses of TAOK1 in fibroblasts revealed a level of 20%–30% TAOK1 in individual 7 compared to the controls. Because this person carries one wild-type allele, the decrease in expression levels is more pronounced than expected. It can be speculated that TAO1 kinase regulates its own expression through a feedback loop. Confirming this hypothesis will require investigations of the expression in additional mutation carriers and elucidation of a potential mechanism. In the fibroblast line from individual 7, the mutated mRNA could be stabilized by cycloheximide treatment (CHX, for 8 h at 100 μg/mL final concentration) indicating nonsense-mediated mRNA decay as an explanation for the reduced abundance of the mutant allele (Figures 2A and 2B).
Figure 2.
Characterization of the TAOK1 Variant c.2366_2367insC in Blood and a Fibroblast Line Derived from the Mutation Carrier
(A) Electropherograms of Sanger sequencing of genomic (gDNA) and complementary DNA (cDNA) of individual 7 in the reverse direction. The variant is shown on the genomic level (upper left panel) from a blood sample, and the inserted C (G on the reverse strand) is highlighted. The expression of the mutant allele is decreased in blood (lower left panel), and in the fibroblast line (upper right panel) compared to the wild-type allele, as shown by the lower height of the peaks using Sanger sequencing. The mutant allele could be stabilized upon treatment with cycloheximide (CHX, 100 μg/mL, 8 h) as illustrated in the lower right panel.
(B) The quantification of TAOK1 mRNA levels by real-time PCR indicated highly reduced levels of TAOK1 in the affected individual (AI) compared to the control individuals (controls) in blood and fibroblasts. The expression in fibroblasts can be increased upon treatment with cycloheximide (CHX).
(C) Immunoblot analysis of total protein extract from fibroblasts of the affected individual (AI) and a control (control 1) with an antibody against phosphorylated TAO kinases TAO1, TAO2, and TAO3 indicates the absence of detectable, phosphorylated TAO1 kinase (p-TAOK1) in cells of the affected individual. Phosphorylation and abundance of TAO2 and 3 kinases (p-TAOK2/3) seemed to be unaltered. β-actin (ACTIN) was used as a loading control.
(D) Immunoblot analysis of total protein extract from the fibroblast line of the affected individual (AI) and two controls (control 1, control 2) with antibodies against total TAOK1 and phosphorylated tau protein (p-tau) indicating absence of both TAOK1 and p-tau in cells of the affected individual. β-actin (ACTN) was used as a loading control.
(E) Form factor as a measurement for mitochondrial interconnectivity was calculated in the fibroblast line of the affected individual (AI) and two controls (control 1, control 2) (10 cells each). Each dot represents the value in a single cell, and the mean and the interquartile range per individual are indicated.
We also performed immunoblotting with total protein extracts from the fibroblast line from individual 7 and two different control fibroblast lines. Antibodies used for immunoblotting were as follows: anti-TAOK3 (phospho S177) + TAOK2 (phospho S181) + TAOK1 (phospho S181) antibody (1:1000; Abcam), anti-TAOK1 (1:1000; Bethyl Laboratories), anti-p-tau (1:1000; Cell Signaling), and anti-β-actin (1:1,000,000; Sigma). This revealed a band for phosphorylated TAO1 kinase only in the control but not in individual 7 indicating loss of detectable phosphorylated TAO1 kinase in the mutated fibroblasts (Figure 2C). Total TAO1 kinase was also not evident in the cells of individual 7 (Figure 2D).
In Drosophila, Tao1 has been reported to affect overall brain volume at the mid-third-instar larval stage (L3).14 Furthermore, mutant Tao1 in Drosophila has been demonstrated to negatively regulate Par-1 as part of a signaling pathway in developing neurons, causing defects in the morphology of the central brain.15 Also, mammalian TAO1 kinase has been shown to phosphorylate and thereby activate PAR-1 (microtubule affinity-regulating kinase[MARK2]), leading in turn to phosphorylation of the mictotubule-stabilizing protein tau; this phosphorylation results in decreased tau affinity for microtubules.16 Thus, mammalian TAO1 kinase controls the extension of neurites in immature cultured neurons through its effects on the microtubule cytoskeleton.16, 17 Using patient-derived fibroblast cells from individual 7 and two different control fibroblast lines, we performed immunoblotting against phosphorylated tau protein and showed that there was no detectable phosphorylated tau (p-tau) protein in the mutant fibroblasts (Figure 2D). In accordance with previous observations,18 our results demonstrate that decreased TAO1 kinase protein levels effectively decrease tau phosphorylation in fibroblasts of a TAOK1 mutation carrier. Notably, higher p-tau levels have been shown to elongate the mitochondria as a result of delocalization of dynamin related protein 1 (DRP1), the fission protein for mitochondria.19 Having in mind the decreased levels of p-tau in the fibroblast culture of the affected individual, we speculated that reduced p-tau could result in shorter or smaller mitochondria. Thus, we analyzed the effect of decreased TAO1 kinase levels on the integrity of the mitochondrial network by calculating the form factor in fibroblasts of a TAOK1 mutation carrier and two unrelated healthy controls (Figure 2E). The form factor as a measure of the integrity of the mitochondrial network was determined as previously described.20 The form factor analysis revealed a reduction in the degree of mitochondrial branching and interconnectivity in fibroblasts from the affected individual compared to fibroblasts from healthy controls, implicating an impaired mitochondrial network.
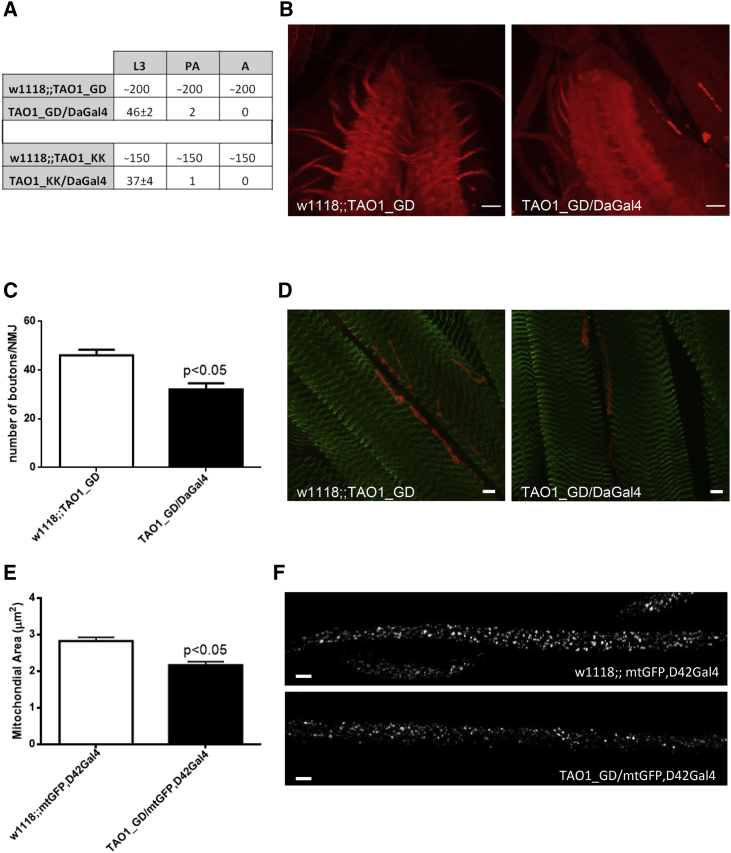
Four of the eight TAOK1 variants that we detected in our individuals result in null alleles as a result of premature protein truncation or decreased expression due to nonsense-mediated mRNA decay, as shown for the c.2366_2367insC variant. To further investigate the consequences of loss of TAO1 kinase and to confirm its role in neurodevelopment, we used two independent Drosophila melanogaster RNAi-mediated knockdown lines for Tao1 depletion: the TAO1_GD RNA interference line (17432) and the TAO1_KK RNA interference line (107645) (purchased from the Vienna Drosophila Resource Center). Tao1 was ubiquitously knocked down via the Daughterless Gal4 (DaGal4) driver, and w1118 (purchased from the Bloomington Drosophila Stock Center) was used as a control line. Drosophila Tao1 (Tao, CG14217) is the single representative of the TAO kinase subfamily21 that is highly similar to all three mammalian TAO kinase family proteins, TAO1, TAO2, and TAO3 (60%–70% identity in the protein kinase domain and 45% identity in the conserved C-terminal domain). All fly experiments were performed at least three times. Compared with controls, both TAO1_KK and TAO1_GD knockdown flies showed delayed development (Figure S1) and early death (discontinued development) between the L3 and the pharate adult (PA) stage (Figure 3A). Furthermore, we used immunostaining to investigate the structure of the ventral nerve cord (VNC), neuromuscular junctions (NMJs), and the NMJs’ endings (boutons). Standard dissection and immunostaining procedures were used for larval tissue analysis. For immunocytochemical analysis, larval fillets at the L3 stage were dissected in the physiological solution HL-322 then fixed for 20 min in 4% formaldehyde, washed in PBS, permeabilized with 0.4% Triton X-100 dissolved in PBS, and blocked for 1 h with 10% normal goat serum. Larval fillets were incubated overnight with the following primary antibodies: the synaptic marker anti-DLG (1:400; 4F3 anti-Disc large; Developmental Studies Hybridoma Bank) for labeling VNC and NMJs and anti-GABARABAP (1:100; Abcam) for defining muscular structures. The respective secondary antibodies were used: anti-rabbit Alexa Fluor® 488 (1:100; Thermo Fisher Scientific) and anti-mouse Alexa Fluor® 596 (1:100; Thermo Fisher Scientific). The images were taken on a confocal microscope (LSM710; Zeiss). Images of z stacks were examined with ImageJ (NIH software), and the number of boutons per muscle section was counted. At least 15 images from four different larvae were analyzed. Immunostaining of the larval VNC showed a reduction in size of VNC, less-defined structures of VNC, and less-developed axons originating in the VNC in Tao1 knockdown larvae (Figure 3B). Additionally, immunostaining of the NMJs revealed changed NMJ morphology and a significantly decreased number of boutons per muscle section in Tao1 knockdown larvae compared to control larvae (Figures 3C and 3D).
Figure 3.
Impaired Neurodevelopment in Drosophila with Knockdown of Tao1
(A) Early lethality in Tao1 knockdown flies. The table indicates the number of living flies in different stages (L3 — third instar larvae stage; PA — pharate adult stage; A — adult flies). Two Tao1 RNAi lines (TAO1_GD and TAO1_KK) were crossed with w1118 control line and the DaGal4 driver, which ubiquitously knocks down Tao1.
(B) Smaller-sized ventral nerve cord in Tao1 knockdown flies. Representative images of L3 control (w1118;;TAO1_GD) and L3 Tao1 knockdown (TAO1_GD/DaGal4) larval ventral nerve cord (VNC) are shown, indicating a reduction in the size of VNC and less-developed axons starting from VNC in Tao1 knockdown flies. Immunolabeling of the L3 larval VNC was done with the synaptic marker anti-DLG (red). Four animals per genotype were analyzed. The scale bar represents 20 μm.
(C) Significantly decreased amount of neuromuscular junction endings (boutons) in Tao1 knockdown flies. Quantification of boutons in control (w1118;;TAO1_GD) and Tao1 knockdown (TAO1_GD/DaGal4) larvae. p < 0.05 — significantly decreased number of boutons compared to those of the control; 15 images from four animals per genotype were analyzed. Error bars represent the SEM.
(D) Representative images of the neuromuscular junctions and boutons are shown, indicating changed morphology and a reduced number of boutons. Immunolabeling of L3 control (w1118;;TAO1_GD) and L3 Tao1 knockdown (TAO1_GD/DaGal4) larval neuromuscular junctions (NMJs) and their boutons with the synaptic marker anti-DLG (red) and anti-GABARAB (green) defined muscular structure. The scale bar represents 10 μm.
(E) Significantly decreased average mitochondrial size (μm2) in axons of Tao1 knockdown flies. The control line (w1118) and Tao1 RNAi line (TAO1_GD) were crossed with flies expressing mitochondrial-tagged GFP, predominantly in the motor neurons (mtGFP,D42Gal4), and quantification of axonal mitochondrial area in control (w1118;;mtGFP,D42Gal4) and Tao1 knockdown (TAO1_GD/mtGFP,D42Gal4) L3 larvae is shown. p < 0.05 — significantly decreased average mitochondrial area compared to the control; 10 images from three animals per genotype were analyzed. Error bars represent the SEM.
(F) Representative images of axonal mitochondria are shown, indicating disturbed mitochondrial distribution and reduced mitochondrial size. The scale bar represents 10 μm.
Given the changes in the mitochondrial network detected in fibroblasts from individual 7, we further analyzed the effect of decreased Tao1 levels on mitochondrial distribution and size in axons of Drosophila with knockdown of Tao1. Of note, dysmorphic mitochondria would result in decreased mitochondrial production of ATP, which is essential for the maintenance of axonal transport and function.23 We crossed control (w1118) and Tao1 RNAi lines (TAO1_GD) with flies expressing mitochondrial-tagged GFP (mtGFP) predominantly in motor neurons (mtGFP,D42Gal4) (purchased from the Bloomington stock center). mtGFP encodes the S65T spectral variant of GFP fused at the N terminus with the 31-amino-acid mitochondrial import sequence from human cytochrome C oxidase subunit VIII. Tao1 was ubiquitously knocked down via the D42Gal4 driver, which is predominantly expressed in motor neurons.24 Standard dissection and fixation procedures were used for the L3 stage larval tissue analysis. Quantification of mitochondrial area per axon (μm2) was counted from images of z stacks with ImageJ (NIH software). Differences were analyzed statistically using unpaired t tests with a Bonferroni–Dunn post hoc correction. We observed disturbed mitochondrial distribution and significantly decreased average mitochondrial size in axons of Tao1 knockdown flies compared to the control flies (Figures 3E and 3F), implicating impaired axonal transport and function. These data collectively demonstrate that the effect of knockdown of Tao1 on early development is characterized by reduction in the number of NMJ endings, the size of the VNC in flies, and the mitochondrial size in axons, confirming that depletion of Tao1 results in neurodevelopmental impairment in Drosophila.
NDD is a highly heterogeneous disease group25 with a large number of biological pathways affected by pathogenic variants, reflecting the complexity of normal brain development. A microdeletion on chromosome 17, comprising TAOK1, had nominated TAOK1 as a potential candidate gene for developmental delay and microcephaly.26 The Decipher database reports four additional patients with large deletions that include the TAOK1 gene. These patients include one who has an intragenic deletion of exons 2–8 and a reported phenotype of microcephaly and seizures. We show here multiple sequence variants in persons with NDDs; all of these variants occurred de novo and thus support the possibility that these changes are pathogenic and that TAOK1 plays a role in NDDs. TAO1 kinase was shown to induce microtubule instability via activation of microtubule affinity-regulating kinase (MARK) and phosphorylation of microtubule-associated proteins, including tau, whose dissociation from microtubules results in microtubule disassembly.15, 27 Altered microtubule stability in maturing neurons could potentially lead to a perturbed neuronal network.28 Notably, previous studies have shown that pathogenic variants in a number of tubulin subunits, including TUBB2B (MIM: 612850), TUBB3 (MIM: 602661), and TUBG1 (MIM: 191135), cause brain malformations and NDDs.29, 30, 31, 32 These findings link different NDDs on the molecular level. Additionally, TAOK1 is activated during microtubule-dependent processes such as mitosis and neuritogenesis, and knockdown of TAOK1 expression inhibits these processes.27, 33 Furthermore, TAO1 kinase was previously implicated as an important regulator of checkpoint control because it caused the most penetrant checkpoint defect.34 Its dual function in regulating microtubule dynamics and mitotic progression contributes to correct congression of chromosomes, thereby protecting genomic stability in human cells. During neuronal development, an impaired congression of chromosomes during mitosis would lead to an altered proportion of symmetrical and asymmetrical progenitor cell division. This effect could be further enhanced by down-regulation of the G2/M checkpoint as a result of TAO1 kinase loss,35 leading to reduced apoptosis.36 Apoptosis is a necessary mechanism of programmed cell death during neuronal development and differentiation.37
Thus, through controlling the extension of neurites in immature cultured neurons,16, 17 through its role in the regulation of tau phosphorylation and dissociation from microtubules,16, 17, 32 and through congression of chromosomes during mitosis,34 TAO1 kinase plays a role in the establishment of neuronal polarity, neuronal differentiation, and early brain development. These molecular mechanisms could play an essential role in impaired brain development in TAOK1 mutation carriers. Accordingly, Tao1 kinase has been shown to affect the size of the larval brain in Drosophila,14 and mutations in Tao1 cause defects in the morphology of the central brain in flies.15
In conclusion, we provide strong genetic and functional support that LoF variants in TAOK1 are an additional cause of NDDs.
Declaration of Interests
G. Oprea, M. Werber, and A. Rolfs are employees and shareholders of Centogene AG. R. Affan is an employee and shareholder of Pronto Diagnostics. A. Rolfs is a founder of Centogene AG and a member of its scientific advisory board. N. Navot is a founder of Pronto Diagnostics and a member of its scientific advisory board. S. Kishore is a founder of his own startup, Mimamsia, and is the primary shareholder of this company (after his contribution to this manuscript and thus unrelated to this manuscript). The remaining authors declare no competing interests. None of the authors is member of a board or advisory committee or a paid consultant in relation to this study or holds any patent related to this work.
Acknowledgments
We would like to thank the participants and Eli Ormerod (Oslo University Hospital) for establishing the cell line from individual 7. The study was funded by the University of Lübeck. Exome sequencing was done as part of diagnostic work-up.
Published: June 20, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.05.005.
Web Resources
CADD score, https://cadd.gs.washington.edu/score
DECIPHER, https://decipher.sanger.ac.uk/search?q=TAOK1#consented-patients/results
GeneMatcher, https://genematcher.org/
GnomAD, https://gnomad.broadinstitute.org/gene/ENSG00000160551
OMIM, https://omim.org/
Transcript (GenBank: NP_065842), https://www.ncbi.nlm.nih.gov/nuccore/NM_020791
UniProt, https://www.uniprot.org/
Vienna Drosophila Resource Center, https://stockcenter.vdrc.at/control/main
Supplemental Data
References
- 1.Sheridan E., Wright J., Small N., Corry P.C., Oddie S., Whibley C., Petherick E.S., Malik T., Pawson N., McKinney P.A., Parslow R.C. Risk factors for congenital anomaly in a multiethnic birth cohort: an analysis of the Born in Bradford study. Lancet. 2013;382:1350–1359. doi: 10.1016/S0140-6736(13)61132-0. [DOI] [PubMed] [Google Scholar]; Sheridan, E., Wright, J., Small, N., Corry, P.C., Oddie, S., Whibley, C., Petherick, E.S., Malik, T., Pawson, N., McKinney, P.A., and Parslow, R.C. (2013). Risk factors for congenital anomaly in a multiethnic birth cohort: an analysis of the Born in Bradford study. Lancet 382, 1350-1359. [DOI] [PubMed]
- 2.Hoischen A., Krumm N., Eichler E.E. Prioritization of neurodevelopmental disease genes by discovery of new mutations. Nat. Neurosci. 2014;17:764–772. doi: 10.1038/nn.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hoischen, A., Krumm, N., and Eichler, E.E. (2014). Prioritization of neurodevelopmental disease genes by discovery of new mutations. Nat. Neurosci. 17, 764-772. [DOI] [PMC free article] [PubMed]
- 3.Ku C.S., Polychronakos C., Tan E.K., Naidoo N., Pawitan Y., Roukos D.H., Mort M., Cooper D.N. A new paradigm emerges from the study of de novo mutations in the context of neurodevelopmental disease. Mol. Psychiatry. 2013;18:141–153. doi: 10.1038/mp.2012.58. [DOI] [PubMed] [Google Scholar]; Ku, C.S., Polychronakos, C., Tan, E.K., Naidoo, N., Pawitan, Y., Roukos, D.H., Mort, M., and Cooper, D.N. (2013). A new paradigm emerges from the study of de novo mutations in the context of neurodevelopmental disease. Mol. Psychiatry 18, 141-153. [DOI] [PubMed]
- 4.Wright C.F., McRae J.F., Clayton S., Gallone G., Aitken S., FitzGerald T.W., Jones P., Prigmore E., Rajan D., Lord J., DDD Study Making new genetic diagnoses with old data: iterative reanalysis and reporting from genome-wide data in 1,133 families with developmental disorders. Genet. Med. 2018;20:1216–1223. doi: 10.1038/gim.2017.246. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wright, C.F., McRae, J.F., Clayton, S., Gallone, G., Aitken, S., FitzGerald, T.W., Jones, P., Prigmore, E., Rajan, D., Lord, J., et al.; DDD Study (2018). Making new genetic diagnoses with old data: iterative reanalysis and reporting from genome-wide data in 1,133 families with developmental disorders. Genet. Med. 20, 1216-1223. [DOI] [PMC free article] [PubMed]
- 5.Aronson S.J., Rehm H.L. Building the foundation for genomics in precision medicine. Nature. 2015;526:336–342. doi: 10.1038/nature15816. [DOI] [PMC free article] [PubMed] [Google Scholar]; Aronson, S.J., and Rehm, H.L. (2015). Building the foundation for genomics in precision medicine. Nature 526, 336-342. [DOI] [PMC free article] [PubMed]
- 6.Robinson P.N., Köhler S., Bauer S., Seelow D., Horn D., Mundlos S. The Human Phenotype Ontology: A tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 2008;83:610–615. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; Robinson, P.N., Kohler, S., Bauer, S., Seelow, D., Horn, D., and Mundlos, S. (2008). The Human Phenotype Ontology: A tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 83, 610-615. [DOI] [PMC free article] [PubMed]
- 7.Groza T., Köhler S., Moldenhauer D., Vasilevsky N., Baynam G., Zemojtel T., Schriml L.M., Kibbe W.A., Schofield P.N., Beck T. The Human Phenotype Ontology: Semantic unification of common and rare disease. Am. J. Hum. Genet. 2015;97:111–124. doi: 10.1016/j.ajhg.2015.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Groza, T., Kohler, S., Moldenhauer, D., Vasilevsky, N., Baynam, G., Zemojtel, T., Schriml, L.M., Kibbe, W.A., Schofield, P.N., Beck, T., et al. (2015). The Human Phenotype Ontology: Semantic unification of common and rare disease. Am. J. Hum. Genet. 97, 111-124. [DOI] [PMC free article] [PubMed]
- 8.Trujillano D., Bertoli-Avella A.M., Kumar Kandaswamy K., Weiss M.E., Köster J., Marais A., Paknia O., Schröder R., Garcia-Aznar J.M., Werber M. Clinical exome sequencing: Results from 2819 samples reflecting 1000 families. Eur. J. Hum. Genet. 2017;25:176–182. doi: 10.1038/ejhg.2016.146. [DOI] [PMC free article] [PubMed] [Google Scholar]; Trujillano, D., Bertoli-Avella, A.M., Kumar Kandaswamy, K., Weiss, M.E., Koster, J., Marais, A., Paknia, O., Schroder, R., Garcia-Aznar, J.M., Werber, M., et al. (2017). Clinical exome sequencing: Results from 2819 samples reflecting 1000 families. Eur. J. Hum. Genet. 25, 176-182. [DOI] [PMC free article] [PubMed]
- 9.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lek, M., Karczewski, K.J., Minikel, E.V., Samocha, K.E., Banks, E., Fennell, T., O’Donnell-Luria, A.H., Ware, J.S., Hill, A.J., Cummings, B.B., et al.; Exome Aggregation Consortium (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285-291. [DOI] [PMC free article] [PubMed]
- 10.Hutchison M., Berman K.S., Cobb M.H. Isolation of TAO1, a protein kinase that activates MEKs in stress-activated protein kinase cascades. J. Biol. Chem. 1998;273:28625–28632. doi: 10.1074/jbc.273.44.28625. [DOI] [PubMed] [Google Scholar]; Hutchison, M., Berman, K.S., and Cobb, M.H. (1998). Isolation of TAO1, a protein kinase that activates MEKs in stress-activated protein kinase cascades. J. Biol. Chem. 273, 28625-28632. [DOI] [PubMed]
- 11.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sobreira, N., Schiettecatte, F., Valle, D., and Hamosh, A. (2015). GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 36, 928-930. [DOI] [PMC free article] [PubMed]
- 12.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kircher, M., Witten, D.M., Jain, P., O’Roak, B.J., Cooper, G.M., and Shendure, J. (2014). A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310-315. [DOI] [PMC free article] [PubMed]
- 13.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]; Samocha, K.E., Robinson, E.B., Sanders, S.J., Stevens, C., Sabo, A., McGrath, L.M., Kosmicki, J.A., Rehnstrom, K., Mallick, S., Kirby, A., et al. (2014). A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 46, 944-950. [DOI] [PMC free article] [PubMed]
- 14.Poon C.L., Mitchell K.A., Kondo S., Cheng L.Y., Harvey K.F. The hippo pathway regulates neuroblasts and brain size in Drosophila melanogaster. Curr. Biol. 2016;26:1034–1042. doi: 10.1016/j.cub.2016.02.009. [DOI] [PubMed] [Google Scholar]; Poon, C.L., Mitchell, K.A., Kondo, S., Cheng, L.Y., and Harvey, K.F. (2016). The hippo pathway regulates neuroblasts and brain size in Drosophila melanogaster. curr. biol. 26, 1034-1042. [DOI] [PubMed]
- 15.King I., Tsai L.T., Pflanz R., Voigt A., Lee S., Jäckle H., Lu B., Heberlein U. Drosophila tao controls mushroom body development and ethanol-stimulated behavior through par-1. J. Neurosci. 2011;31:1139–1148. doi: 10.1523/JNEUROSCI.4416-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]; King, I., Tsai, L.T., Pflanz, R., Voigt, A., Lee, S., Jackle, H., Lu, B., and Heberlein, U. (2011). Drosophila tao controls mushroom body development and ethanol-stimulated behavior through par-1. J. Neurosci. 31, 1139-1148. [DOI] [PMC free article] [PubMed]
- 16.Timm T., Matenia D., Li X.Y., Griesshaber B., Mandelkow E.M. Signaling from MARK to tau: regulation, cytoskeletal crosstalk, and pathological phosphorylation. Neurodegener. Dis. 2006;3:207–217. doi: 10.1159/000095258. [DOI] [PubMed] [Google Scholar]; Timm, T., Matenia, D., Li, X.Y., Griesshaber, B., and Mandelkow, E.M. (2006). Signaling from MARK to tau: regulation, cytoskeletal crosstalk, and pathological phosphorylation. Neurodegener. Dis. 3, 207-217. [DOI] [PubMed]
- 17.Biernat J., Wu Y.Z., Timm T., Zheng-Fischhöfer Q., Mandelkow E., Meijer L., Mandelkow E.M. Protein kinase MARK/PAR-1 is required for neurite outgrowth and establishment of neuronal polarity. Mol. Biol. Cell. 2002;13:4013–4028. doi: 10.1091/mbc.02-03-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]; Biernat, J., Wu, Y.Z., Timm, T., Zheng-Fischhofer, Q., Mandelkow, E., Meijer, L., and Mandelkow, E.M. (2002). Protein kinase MARK/PAR-1 is required for neurite outgrowth and establishment of neuronal polarity. Mol. Biol. Cell 13, 4013-4028. [DOI] [PMC free article] [PubMed]
- 18.Giacomini C., Koo C.Y., Yankova N., Tavares I.A., Wray S., Noble W., Hanger D.P., Morris J.D.H. A new TAO kinase inhibitor reduces tau phosphorylation at sites associated with neurodegeneration in human tauopathies. Acta Neuropathol. Commun. 2018;6:37. doi: 10.1186/s40478-018-0539-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Giacomini, C., Koo, C.Y., Yankova, N., Tavares, I.A., Wray, S., Noble, W., Hanger, D.P., and Morris, J.D.H. (2018). A new TAO kinase inhibitor reduces tau phosphorylation at sites associated with neurodegeneration in human tauopathies. Acta Neuropathol. Commun. 6, 37. [DOI] [PMC free article] [PubMed]
- 19.Perez M., Jara C., Quintanilla R.A. Contribution of Tau Pathology to Mitochondrial Impairment in Neurodegeneration. Front. Neurosci. 2018;12:441. doi: 10.3389/fnins.2018.00441. [DOI] [PMC free article] [PubMed] [Google Scholar]; Perez, M., Jara, C., and Quintanilla, R.A. (2018). Contribution of Tau Pathology to Mitochondrial Impairment in Neurodegeneration. Front. Neurosci. 12, 441. [DOI] [PMC free article] [PubMed]
- 20.Grünewald A., Voges L., Rakovic A., Kasten M., Vandebona H., Hemmelmann C., Lohmann K., Orolicki S., Ramirez A., Schapira A.H. Mutant Parkin impairs mitochondrial function and morphology in human fibroblasts. PLoS ONE. 2010;5:e12962. doi: 10.1371/journal.pone.0012962. [DOI] [PMC free article] [PubMed] [Google Scholar]; Grunewald, A., Voges, L., Rakovic, A., Kasten, M., Vandebona, H., Hemmelmann, C., Lohmann, K., Orolicki, S., Ramirez, A., Schapira, A.H., et al. (2010). Mutant Parkin impairs mitochondrial function and morphology in human fibroblasts. PLoS ONE 5, e12962. [DOI] [PMC free article] [PubMed]
- 21.Dan I., Watanabe N.M., Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]; Dan, I., Watanabe, N.M., and Kusumi, A. (2001). The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 11, 220-230. [DOI] [PubMed]
- 22.Uytterhoeven V., Kuenen S., Kasprowicz J., Miskiewicz K., Verstreken P. Loss of skywalker reveals synaptic endosomes as sorting stations for synaptic vesicle proteins. Cell. 2011;145:117–132. doi: 10.1016/j.cell.2011.02.039. [DOI] [PubMed] [Google Scholar]; Uytterhoeven, V., Kuenen, S., Kasprowicz, J., Miskiewicz, K., and Verstreken, P. (2011). Loss of skywalker reveals synaptic endosomes as sorting stations for synaptic vesicle proteins. Cell 145, 117-132. [DOI] [PubMed]
- 23.Ochs S., Hollingsworth D. Dependence of fast axoplasmic transport in nerve on oxidative metabolism. J. Neurochem. 1971;18:107–114. doi: 10.1111/j.1471-4159.1971.tb00172.x. [DOI] [PubMed] [Google Scholar]; Ochs, S., and Hollingsworth, D. (1971). Dependence of fast axoplasmic transport in nerve on oxidative metabolism. J. Neurochem. 18, 107-114. [DOI] [PubMed]
- 24.Sanyal S. Genomic mapping and expression patterns of C380, OK6 and D42 enhancer trap lines in the larval nervous system of Drosophila. Gene Expr. Patterns. 2009;9:371–380. doi: 10.1016/j.gep.2009.01.002. [DOI] [PubMed] [Google Scholar]; Sanyal, S. (2009). Genomic mapping and expression patterns of C380, OK6 and D42 enhancer trap lines in the larval nervous system of Drosophila. Gene Expr. Patterns 9, 371-380. [DOI] [PubMed]
- 25.Platzer K., Sticht H., Edwards S.L., Allen W., Angione K.M., Bonati M.T., Brasington C., Cho M.T., Demmer L.A., Falik-Zaccai T. De novo variants in MAPK8IP3 cause intellectual disability with variable brain anomalies. Am. J. Hum. Genet. 2019;104:203–212. doi: 10.1016/j.ajhg.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; Platzer, K., Sticht, H., Edwards, S.L., Allen, W., Angione, K.M., Bonati, M.T., Brasington, C., Cho, M.T., Demmer, L.A., Falik-Zaccai, T., et al. (2019). De novo variants in MAPK8IP3 cause intellectual disability with variable brain anomalies. Am. J. Hum. Genet. 104, 203-212. [DOI] [PMC free article] [PubMed]
- 26.Xie B., Fan X., Lei Y., Chen R., Wang J., Fu C., Yi S., Luo J., Zhang S., Yang Q. A novel de novo microdeletion at 17q11.2 adjacent to NF1 gene associated with developmental delay, short stature, microcephaly and dysmorphic features. Mol. Cytogenet. 2016;9:41. doi: 10.1186/s13039-016-0251-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xie, B., Fan, X., Lei, Y., Chen, R., Wang, J., Fu, C., Yi, S., Luo, J., Zhang, S., Yang, Q., et al. (2016). A novel de novo microdeletion at 17q11.2 adjacent to NF1 gene associated with developmental delay, short stature, microcephaly and dysmorphic features. Mol. Cytogenet. 9, 41. [DOI] [PMC free article] [PubMed]
- 27.Timm T., Li X.Y., Biernat J., Jiao J., Mandelkow E., Vandekerckhove J., Mandelkow E.M. MARKK, a Ste20-like kinase, activates the polarity-inducing kinase MARK/PAR-1. EMBO J. 2003;22:5090–5101. doi: 10.1093/emboj/cdg447. [DOI] [PMC free article] [PubMed] [Google Scholar]; Timm, T., Li, X.Y., Biernat, J., Jiao, J., Mandelkow, E., Vandekerckhove, J., and Mandelkow, E.M. (2003). MARKK, a Ste20-like kinase, activates the polarity-inducing kinase MARK/PAR-1. EMBO J. 22, 5090-5101. [DOI] [PMC free article] [PubMed]
- 28.Chakraborti S., Natarajan K., Curiel J., Janke C., Liu J. The emerging role of the tubulin code: From the tubulin molecule to neuronal function and disease. Cytoskeleton (Hoboken) 2016;73:521–550. doi: 10.1002/cm.21290. [DOI] [PubMed] [Google Scholar]; Chakraborti, S., Natarajan, K., Curiel, J., Janke, C., and Liu, J. (2016). The emerging role of the tubulin code: From the tubulin molecule to neuronal function and disease. Cytoskeleton (Hoboken) 73, 521-550. [DOI] [PubMed]
- 29.Jaglin X.H., Poirier K., Saillour Y., Buhler E., Tian G., Bahi-Buisson N., Fallet-Bianco C., Phan-Dinh-Tuy F., Kong X.P., Bomont P. Mutations in the beta-tubulin gene TUBB2B result in asymmetrical polymicrogyria. Nat. Genet. 2009;41:746–752. doi: 10.1038/ng.380. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jaglin, X.H., Poirier, K., Saillour, Y., Buhler, E., Tian, G., Bahi-Buisson, N., Fallet-Bianco, C., Phan-Dinh-Tuy, F., Kong, X.P., Bomont, P., et al. (2009). Mutations in the beta-tubulin gene TUBB2B result in asymmetrical polymicrogyria. Nat. Genet. 41, 746-752. [DOI] [PMC free article] [PubMed]
- 30.Poirier K., Lebrun N., Broix L., Tian G., Saillour Y., Boscheron C., Parrini E., Valence S., Pierre B.S., Oger M. Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat. Genet. 2013;45:639–647. doi: 10.1038/ng.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]; Poirier, K., Lebrun, N., Broix, L., Tian, G., Saillour, Y., Boscheron, C., Parrini, E., Valence, S., Pierre, B.S., Oger, M., et al. (2013). Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat. Genet. 45, 639-647. [DOI] [PMC free article] [PubMed]
- 31.Poirier K., Saillour Y., Bahi-Buisson N., Jaglin X.H., Fallet-Bianco C., Nabbout R., Castelnau-Ptakhine L., Roubertie A., Attie-Bitach T., Desguerre I. Mutations in the neuronal ß-tubulin subunit TUBB3 result in malformation of cortical development and neuronal migration defects. Hum. Mol. Genet. 2010;19:4462–4473. doi: 10.1093/hmg/ddq377. [DOI] [PMC free article] [PubMed] [Google Scholar]; Poirier, K., Saillour, Y., Bahi-Buisson, N., Jaglin, X.H., Fallet-Bianco, C., Nabbout, R., Castelnau-Ptakhine, L., Roubertie, A., Attie-Bitach, T., Desguerre, I., et al. (2010). Mutations in the neuronal ß-tubulin subunit TUBB3 result in malformation of cortical development and neuronal migration defects. Hum. Mol. Genet. 19, 4462-4473. [DOI] [PMC free article] [PubMed]
- 32.Breuss M., Keays D.A. Microtubules and neurodevelopmental disease: the movers and the makers. Adv. Exp. Med. Biol. 2014;800:75–96. doi: 10.1007/978-94-007-7687-6_5. [DOI] [PubMed] [Google Scholar]; Breuss, M., and Keays, D.A. (2014). Microtubules and neurodevelopmental disease: the movers and the makers. Adv. Exp. Med. Biol. 800, 75-96. [DOI] [PubMed]
- 33.Wojtala R.L., Tavares I.A., Morton P.E., Valderrama F., Thomas N.S., Morris J.D. Prostate-derived sterile 20-like kinases (PSKs/TAOKs) are activated in mitosis and contribute to mitotic cell rounding and spindle positioning. J. Biol. Chem. 2011;286:30161–30170. doi: 10.1074/jbc.M111.228320. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wojtala, R.L., Tavares, I.A., Morton, P.E., Valderrama, F., Thomas, N.S., and Morris, J.D. (2011). Prostate-derived sterile 20-like kinases (PSKs/TAOKs) are activated in mitosis and contribute to mitotic cell rounding and spindle positioning. J. Biol. Chem. 286, 30161-30170. [DOI] [PMC free article] [PubMed]
- 34.Draviam V.M., Stegmeier F., Nalepa G., Sowa M.E., Chen J., Liang A., Hannon G.J., Sorger P.K., Harper J.W., Elledge S.J. A functional genomic screen identifies a role for TAO1 kinase in spindle-checkpoint signalling. Nat. Cell Biol. 2007;9:556–564. doi: 10.1038/ncb1569. [DOI] [PubMed] [Google Scholar]; Draviam, V.M., Stegmeier, F., Nalepa, G., Sowa, M.E., Chen, J., Liang, A., Hannon, G.J., Sorger, P.K., Harper, J.W., and Elledge, S.J. (2007). A functional genomic screen identifies a role for TAO1 kinase in spindle-checkpoint signalling. Nat. Cell Biol. 9, 556-564. [DOI] [PubMed]
- 35.Raman M., Earnest S., Zhang K., Zhao Y., Cobb M.H. TAO kinases mediate activation of p38 in response to DNA damage. EMBO J. 2007;26:2005–2014. doi: 10.1038/sj.emboj.7601668. [DOI] [PMC free article] [PubMed] [Google Scholar]; Raman, M., Earnest, S., Zhang, K., Zhao, Y., and Cobb, M.H. (2007). TAO kinases mediate activation of p38 in response to DNA damage. EMBO J. 26, 2005-2014. [DOI] [PMC free article] [PubMed]
- 36.Wu M.F., Wang S.G. Human TAO kinase 1 induces apoptosis in SH-SY5Y cells. Cell Biol. Int. 2008;32:151–156. doi: 10.1016/j.cellbi.2007.08.006. [DOI] [PubMed] [Google Scholar]; Wu, M.F., and Wang, S.G. (2008). Human TAO kinase 1 induces apoptosis in SH-SY5Y cells. Cell Biol. Int. 32, 151-156. [DOI] [PubMed]
- 37.Pinto-Teixeira F., Konstantinides N., Desplan C. Programmed cell death acts at different stages of Drosophila neurodevelopment to shape the central nervous system. FEBS Lett. 2016;590:2435–2453. doi: 10.1002/1873-3468.12298. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pinto-Teixeira, F., Konstantinides, N., and Desplan, C. (2016). Programmed cell death acts at different stages of Drosophila neurodevelopment to shape the central nervous system. FEBS Lett. 590, 2435-2453. [DOI] [PMC free article] [PubMed]
- 38.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]; Richards, S., Aziz, N., Bale, S., Bick, D., Das, S., Gastier-Foster, J., Grody, W.W., Hegde, M., Lyon, E., Spector, E., et al.; ACMG Laboratory Quality Assurance Committee (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405-424. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.