Abstract
We report biallelic missense and frameshift pathogenic variants in the gene encoding human nucleoporin NUP214 causing acute febrile encephalopathy. Clinical symptoms include neurodevelopmental regression, seizures, myoclonic jerks, progressive microcephaly, and cerebellar atrophy. NUP214 and NUP88 protein levels were reduced in primary skin fibroblasts derived from affected individuals, while the total number and density of nuclear pore complexes remained normal. Nuclear transport assays exhibited defects in the classical protein import and mRNA export pathways in affected cells. Direct surface imaging of fibroblast nuclei by scanning electron microscopy revealed a large increase in the presence of central particles (known as “plugs”) in the nuclear pore channels of affected cells. This observation suggests that large transport cargoes may be delayed in passage through the nuclear pore channel, affecting its selective barrier function. Exposure of fibroblasts from affected individuals to heat shock resulted in a marked delay in their stress response, followed by a surge in apoptotic cell death. This suggests a mechanistic link between decreased cell survival in cell culture and severe fever-induced brain damage in affected individuals. Our study provides evidence by direct imaging at the single nuclear pore level of functional changes linked to a human disease.
keywords: febrile encephalopathy, nucleoporins, NUP214, NUP88, nuclear pore complex, central channel particles, neurodegeneration
Introduction
The hallmark of eukaryotic cells is a membrane-bound nucleus that physically separates the major steps in the gene expression pathway. Nuclear pore complexes (NPCs) are massive gateways embedded within the double membranes of the nuclear envelope that control molecular traffic into and out of the nucleus.1, 2 NPCs are composed of multiple copies of ∼30 different proteins called nucleoporins (Nups), which are generally conserved at the structural level between different eukaryotic organisms.3, 4 Only a small number of inherited human diseases have been reported to be caused by pathogenic variants in Nup-coding genes,5, 6, 7, 8, 9, 10, 11 but these form part of a broader group termed nuclear envelopathies.12 Nuclear envelopathies are caused by pathogenic variants in the genes encoding nuclear lamins and their close interactors, inner and outer nuclear membrane proteins and NPC components. Surprisingly, although many of these proteins are ubiquitously expressed, most of the disease phenotypes are highly tissue specific with a particular emphasis on muscle and neuronal cells.12, 13, 14
The cytoplasmic facade of the NPC is characterized by distinct architectural features including a cytoplasmic ring and eight free-ended proteinaceous filaments that protrude from it.1, 15 This distinctive structural element contains a subset of asymmetrically localized Nups that are not present in the nuclear ring and basket on the other side of the NPC.3, 16, 17, 18 NUP214 (MIM: 114350), localized to the base of the cytoplasmic filaments, plays a critical role in CRM1-mediated nuclear export and interacts with the essential DEAD box protein DBP5, which is critical for mRNA export.19, 20, 21, 22 NUP214, together with its close partner NUP88 (MIM: 602552), are thought to anchor the cytoplasmic filaments containing NUP358 to the cytoplasmic ring.16
Here we describe a rare neurodegenerative disease involving severe damage to the central nervous system and associated with two types of pathogenic variants in human NUP214. Affected individuals presented with febrile encephalopathy. Although the major affected tissues remain out of reach, we used primary skin fibroblasts derived from affected individuals to determine a set of distinct phenotypic changes at the single-cell level. These range from changes in the response to apoptotic signals to defects in nuclear transport and a dramatic increase in the presence of large particles, known as “central plugs,” in the NPC channel.
Material and Methods
Affected Individuals
Informed consent was obtained from all the parents for research analysis of clinical exomes and for sharing MRI images and skin biopsy after IRB review at Johns Hopkins University and Hadassah-Hebrew University Medical Center.
Exome Analysis
After informed consent, exome analysis was pursued on DNA extracted from whole blood of individual III-3 and of III-12 in family A (Figure 1A). Exonic sequences from DNA were enriched with the SureSelect Human All Exon 50 Mb V4 and V5 Kits for III-3 and III-12, respectively (Agilent Technologies). Sequences were generated on a HiSeq2500 (Illumina) as 60-bp and 125-bp (for III-3 and III-12, respectively) paired-end runs. Read alignment and variant calling were performed with DNAnexus using default parameters with the human genome assembly hg19 (GRCh37) as reference. Exome analysis of the proband (III-3) yielded 126.3 million mapped reads with a mean coverage of 129× and of III-12 yielded 49.2 million reads with mean coverage of 83×. Following alignment to the reference genome (hg19) and variant calling, variants were filtered out if the total read depth was less than 8 × and if they were off-target (>8 bp from splice junction), were synonymous, were predicted benign by Mutation Taster, or had minor allele frequency (MAF) > 0.01 in the ExAC database (Exome Aggregation Consortium, Cambridge, MA). For family B (Figure 1A), quad exome analysis was performed by GeneDx, using their standard protocol.23 Research analysis was performed by the BHCMG using PhenoDB and candidate genes were entered into GeneMatcher.24
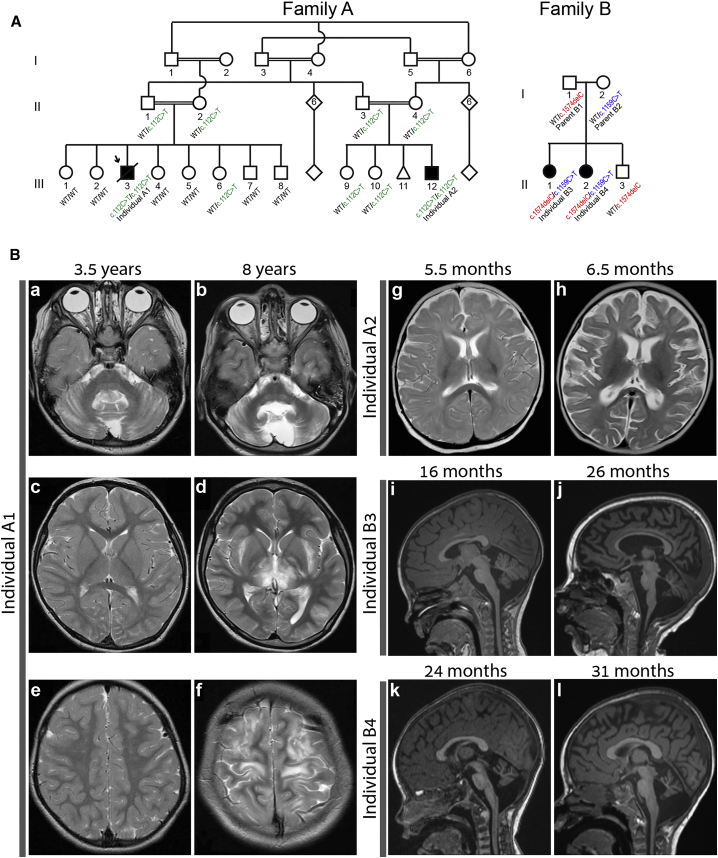
Figure 1.
Genetic Segregation of the NUP214 Variants and Brain Magnetic Resonance Imaging (MRI) of the Affected Individuals
(A) Pedigrees of family A, indicating genotypes of the individuals who were available for testing as regards the c.112C>T (p.Arg38Cys) variant, and family B, indicating siblings compound heterozygous for the c.1574delC and c.1159C>T variants. Shapes shaded in black indicate affected individuals. Multiple loops of consanguinity can be appreciated in family A.
(B) Axial T-2 weighted MRI for family A. (a–f) Individual A1: family A, individual III-3. Progressive cerebellar atrophy and interval changes in basal ganglia, internal capsula, subcortical white matter, and cerebral cortex with hyperintense foci at the age of 3.5 years (a, c, e) and 8 years (b, d, f). (g and h) Individual A2: family A, individual III-12. Occipital edema at age 5.5 months (g) and progressive atrophic changes with dilatation of the CSF spaces between the ages of 5.5 months (g) and 6.5 months (h). (i–l) Sagittal T-1 weighted MRI for family B. (i and j) Individual B3: family B, individual II-1. Progressive supratentorial and infratentorial parenchymal volume loss with ex vacuo dilatation of the entire ventricular system at 16 months (i) and 26 months (j). (k and l) Individual B4: family B, individual II-2. Progressive supra- and infratentorial atrophy similar to sibling with progression from 24 months (k) to 31 months (l).
Segregation Analysis
Amplicons containing the potential pathogenic variant in NUP214 were amplified by conventional PCR of genomic DNA from available family members. PCR products were purified and analyzed by Sanger di-deoxy nucleotide sequencing.
Sequence Analysis and Comparative Modeling
NUP214 orthologs were sampled from mammalia. The crystallized structure of the NUP214 N-terminal domain was available under the PDB: 3fhc.pdb.22 The retrieved sequences, including the proposed crystallized structures (GenBank: NP_005076.3), were aligned using ClustalW. PyMOL was used to generate a 3D model of the human NUP214_R38C mutant (N-terminal domain) protein according to established protocols.25 The obtained 3D comparative model was energetically minimized by using the biochemical/computational tools of Chimera. PyMOL was used for manual inspection of the investigated 3D models and for generating figures.25
Cell Culture
Primary fibroblast cell cultures were established from skin biopsies from three affected individuals and three unrelated unaffected control subjects, as well as the two unaffected parents from family B, according to IRB guidelines. Cultures were routinely propagated in high-glucose Dulbecco’s Modified Eagle Medium supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 U/mL penicillin, 100 mg/mL streptomycin, and 1 mM sodium pyruvate (Biological Industries). All of the experiments were performed with control and affected individual fibroblasts from identical or highly similar passage numbers and in all cases the passage number did not exceed 15. For the experiments testing the effect of transcription or translation inhibitors on the prevalence of central NPC channel particles in cells from an affected individual, cycloheximide and actinomycin D (Sigma-Aldrich) were dissolved in dimethyl sulfoxide (DMSO) and diluted at 1:1,000 into the growth medium. Cells were pre-treated with 5 μg/mL cycloheximide, 1 μg/mL actinomycin D, or DMSO only (mock treatment) for 24 h before the exposure of nuclei for FESEM imaging.
Antibodies
Commercially obtained antibodies included anti-nuclear pore complex proteins mAb414 (MMS-120P; Covance), anti-NUP214 (ab-70497, Abcam), anti-importin β (ab-36775, Abcam), anti-NUP88 (ab-79785, Abcam), anti-tubulin (sc-8035; Santa Cruz Biotechnology), and anti-cleaved caspase 3 (Asp175; #9661, Cell Signaling). Secondary antibodies for immunofluorescent staining were TRITC goat anti-rabbit (Jackson ImmunoResearch) and Alexa Fluor 488-donkey anti-mouse IgG (Invitrogen). Affinity purified anti-NUP133 polyclonal antibody has been previously described.26
Western Immunoblotting
To prepare total cell lysates, fibroblasts were grown in 10 cm tissue culture dishes to ∼70% confluence. Prior to lysis, cells were washed three times in ice-cold PBS and lysed in 1 mL of RIPA-SDS lysis buffer supplemented with a protease inhibitor cocktail (Pierce-88665; Thermo Fisher Scientific), on ice. Lysates were passed three times through an 18-gauge needle and centrifuged for 5 min at 20,000 × g 4°C, to precipitate debris. Supernatants were mixed with SDS-PAGE loading buffer. SDS-PAGE and immunoblotting were performed using PVDF membranes (Millipore) and standard techniques. Quantification of band intensities was performed with the Gels sub-menu of ImageJ, using importin β or α-tubulin as general loading controls and normalizing specific bands of interest to the samples from control fibroblasts.
Fluorescence Microscopy
For indirect immunofluorescence, cells were grown on poly-lysine-coated coverslips for 24 h, fixed with 3.7% paraformaldehyde for 30 min, permeabilized with PBS/0.1% Triton X-100 for 3 min on ice, and blocked 30 min with PBS/5% fetal bovine serum, followed by the application of primary and secondary antibodies according to the manufacturer’s instructions. Coverslips were mounted in Fluoromount-G (SouthernBiotech). Images were acquired on (1) a fully motorized wide-field Zeiss AxioObserver Z1 driven by ZEN software (Figures 3 and 5A); (2) an Olympus BX61TRF motorized microscope, equipped with a DP70 digital camera and driven by DP Controller and Manager software (Olympus; Figures 4A and 5B); or (3) a Zeiss upright Axio Imager.M2 with ApoTome 2.0 equipped with an Orca Flash 4.0 V3 digital CMOS camera (Hamamatsu; Figures 7 and 8). Quantification of the average staining intensity per nucleus or per cell was performed by ZEN or ImageJ software. The draw spline contour or polygon selection tool were used to delineate areas of interest and measure intensity mean values. For measuring anti-Nup staining intensity or nuclear import, images were acquired after focus correction for the mid-plane of the nucleus and 5 × 5 tile merging, with background subtraction performed after delineation of cell or nuclear boundaries. For both mRNA export and cleaved caspase measurements, background subtraction was performed on nearby areas outside cells. In mRNA export experiments, both cytoplasmic and nuclear compartments were defined and normalization was to the cytoplasmic/nuclear ratio in control cells not expressing GFP. In rescue experiments measuring cleaved caspase 3, the cellular signal was normalized to control fibroblasts transfected with an empty vector. To analyze NPC density by light microscopy, tangential views of mAb 414 staining of the nuclear surface were used to measure fluorescent puncta per unit area.
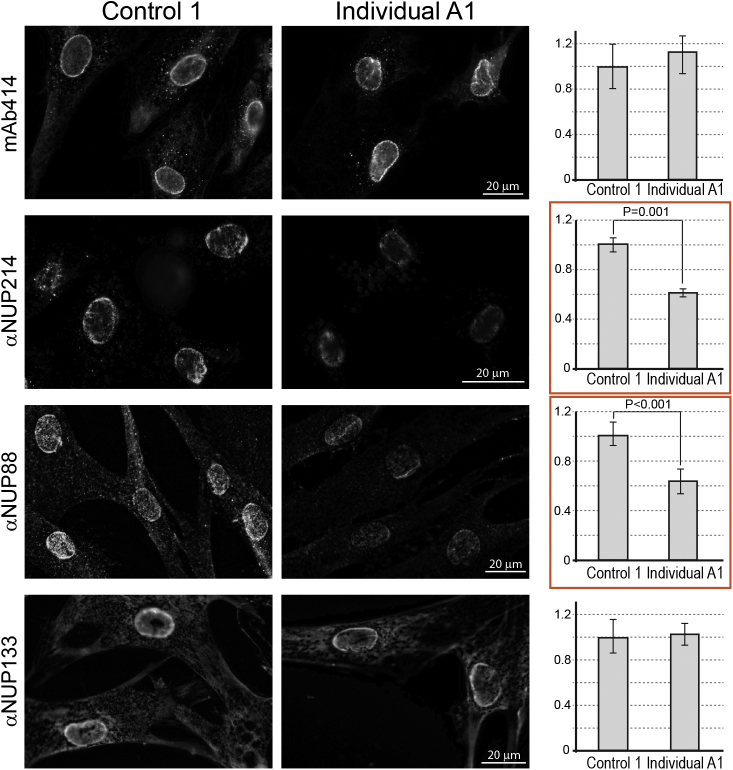
Figure 3.
Immunofluorescent Staining of Nucleoporins in Control and Affected Fibroblasts
Primary fibroblasts derived from affected individual A1 and an unrelated control individual were analyzed by indirect immunofluorescence with focus correction for the mid-plane of the nucleus and images captured as 5×5 tiles and merged into wider fields to allow quantitative analysis. Representative images of staining with several Nup-specific antibodies are shown on the left. Quantitative summaries of the average staining intensity per cell, derived from multiple fields and >100 cells in each category, are shown on the right. NUP214 and NUP88 staining intensity was reduced in the affected individual’s fibroblasts both at the nuclear rim and in the total cellular signal. Bars indicate SEM. See Material and Methods for further details.
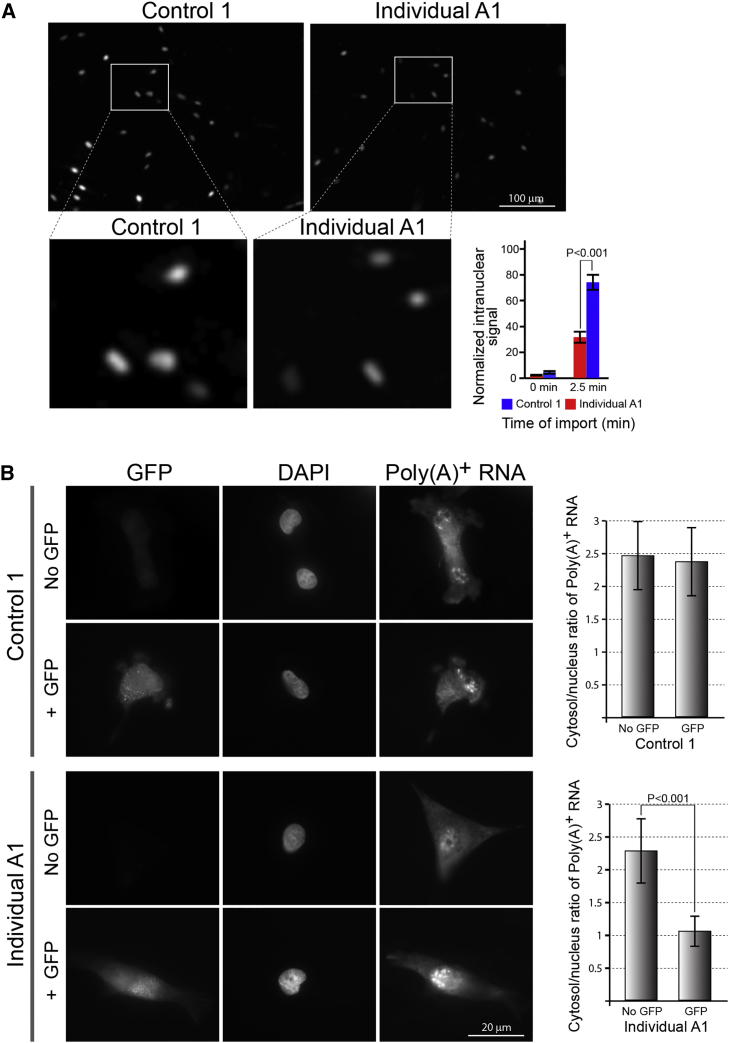
Figure 5.
Functional Analysis of Specific Nuclear Import and Export Pathways in Affected Fibroblasts
(A) Primary fibroblasts derived from individual A1 and an unrelated control individual were permeabilized by digitonin and then supplemented with cytosol, an ATP regenerating system, and a TRITC-NLS-BSA import substrate to follow import in the classical pathway. Nuclear accumulation of the fluorescent substrate at different time points was monitored by fluorescence microscopy, as in Figure 3. The upper panel shows large fields stitched together from 5×5 individual tiles at the 2.5 min time point. Specific areas are enlarged in the lower panel. A quantitative summary (bottom right) is shown for 3 independent experiments, each derived from multiple stitched fields acquired at time 0 and 2.5 min. The combined data and each of the individual experiments all show a significant delay in the accumulation of the substrate in the affected cell nuclei in comparison to control cells. Bars indicate SEM.
(B) Overexpression of GFP by transient 24 h transfection causes an increased accumulation of mRNA in the nuclei of affected cells. In situ hybridization to poly(A)+ was performed on fixed cells and compared between GFP-expressing and non-expressing cells in control and affected individual fibroblasts. Representative images are shown on the left and quantification of the cytoplasmic/nuclear ratio of poly(A)+ RNA in >30 individual cells in each group is shown on the right. Bars indicate SEM. See Material and Methods for further details and Figure S3 for a quantitative comparison of GFP expression levels in control and affected individual A1 cells.
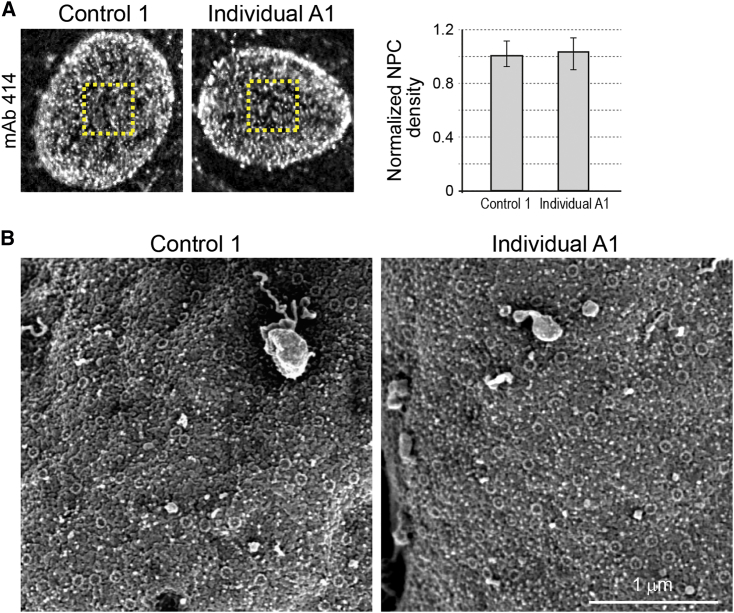
Figure 4.
A Comparison of NPC Density in Control and Affected Cells
(A) Primary fibroblasts derived from individual A1 and a control individual were analyzed by indirect immunofluorescence as in Figure 3. Tangential views of mAb414 staining (left) were used to estimate the average density of NPCs on the surface of nuclei from control and affected cells. Quantification of these signals from multiple images (right) suggests that there is no significant difference in the density of NPCs. Bars indicate SEM.
(B) Control and affected fibroblasts were subjected to hypotonic treatment and prepared for direct surface imaging of exposed nuclei by FESEM as described in Material and Methods. Representative flat areas from two nuclei demonstrate that NPC density is similar in both cell types. A quantitative summary of >60 different 1×1 μm flat areas from >30 nuclei in each category indicated a density of 12.8 ± 2.5 pores/μm2 in control cells and 12.1 ± 2 pores/μm2 in the affected individual’s cells.
Figure 7.
The Response to Heat Shock (HS) Stress in Affected Individual Fibroblasts
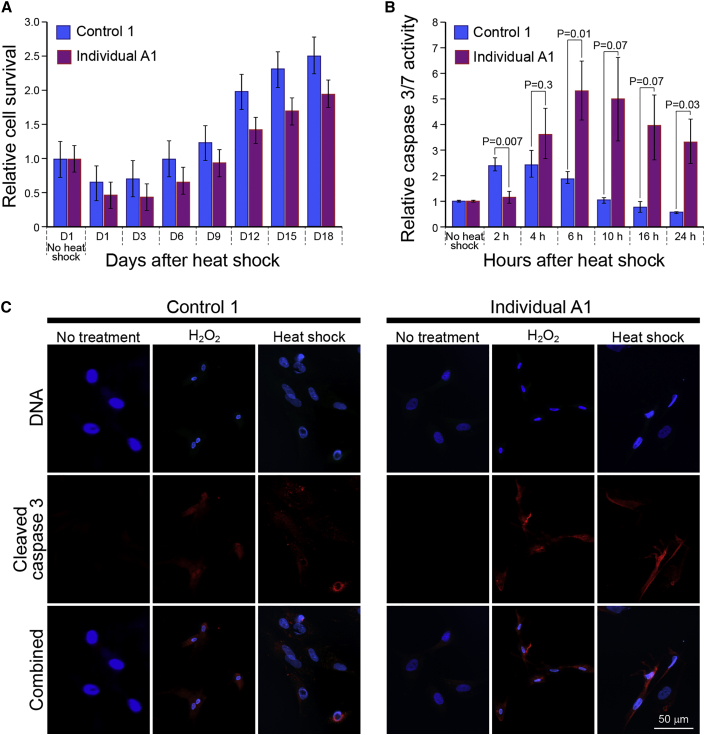
(A) Cell survival after HS stress over a duration of 18 days. Control and affected individual A1-derived primary fibroblasts were grown under identical conditions, subjected to HS of 43°C for 2 h and then returned to normal growth conditions at 37°C for 18 days. Cell viability was determined by Trypan blue staining. Bars represent mean ± SEM for three independent experiments.
(B) The apoptotic response to HS was determined by measuring cleaved caspase activity, following a 43°C, 2 h HS stress exposure, using the Caspase-Glo 3/7 Assay. Control and affected fibroblasts were compared. Bars represent mean ± SEM relative luminescence from three independent experiments, compared to untreated (no heat shock) controls.
(C) Immunostaining for cleaved caspase-3 in control and affected individual-derived fibroblasts after HS stress. Cells were subjected to 43°C, 2 h HS stress and returned to normal growth conditions at 37°C for 6 h, fixed, and processed for indirect immufluorescence with an anti-cleaved caspase-3 antibody. Overnight exposure to oxidative stress (2 mM H2O2) was used as a positive control for an apoptotic response in both cell types. DNA was stained with Hoechst 33258. Scale bar: 50 μm.
Figure 8.
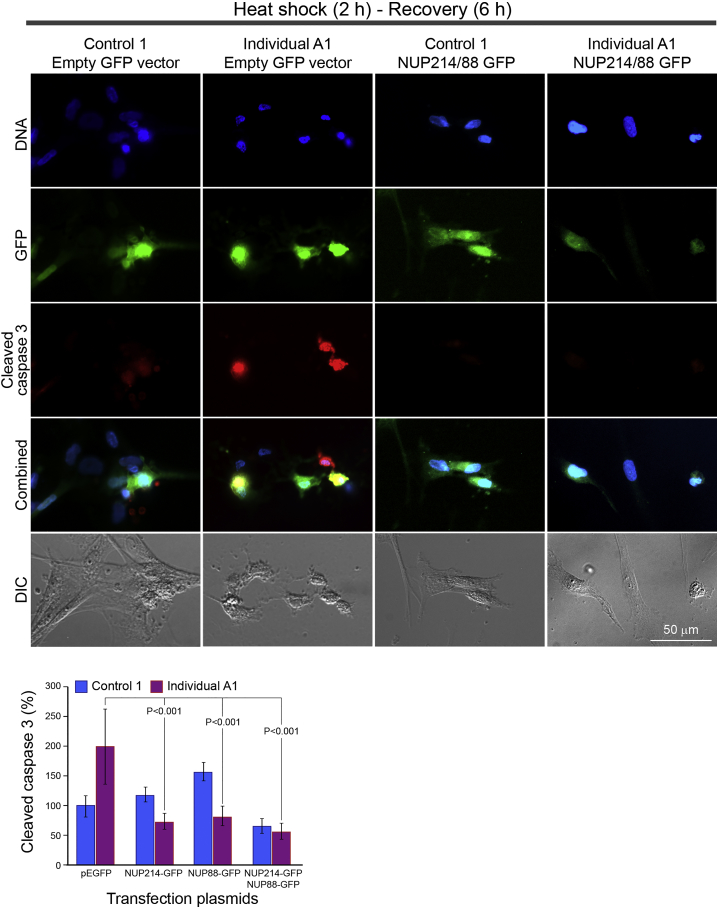
Rescue of the Stress Response Phenotype by Transient Transfection of Nup-Coding Constructs into Affected Fibroblasts
Control and affected individual A1-derived fibroblasts were transfected by electroporation with constructs encoding the full-length wild-type hNUP214-GFP, hNUP88-GFP, a combination of both constructs or an empty GFP-expressing vector as a control. HS exposure was performed 48 h after transfection, for 2 h 43°C, followed by a 6 h recovery period. Cells were then fixed and immunostained for cleaved caspase-3 as in Figure 7. Representative images from one experiment are shown for the double NUP214/NUP88 transfection and empty vector control in control and affected individual-derived fibroblasts. DNA was stained with Hoechst 33258. Scale bar: 50 μm. A quantitative summary of cleaved caspase-3 staining intensity in GFP-expressing cells measured in three independent experiments is shown at the bottom. Cell borders were delineated from DIC images by the ZEN software and anti-cleaved caspase-3 staining was measured per cell and normalized to control fibroblasts transfected with the empty GFP-expressing vector. Each bar represents at least 90 cells from 3 independent experiments, showing the mean intensity ± SEM.
Scanning Electron Microscopy
To expose fibroblast nuclei for direct surface imaging by FESEM, we used our previously published methods.27, 28 Briefly, cells were grown to 80% confluency, detached by trypsinization, subjected to hypotonic treatment without the addition of detergents, and passed through a 21-gauge needle. The cells were then gently centrifuged onto the surface of 5 × 5 mm2 silicon chips, fixed, and further processed for electron microscopy including dehydration through a graded series of ethanol solutions and critical-point drying using a K850 apparatus (Quorum Technologies). Samples were coated with a ∼1 nm thick layer of iridium using a Q150T turbo-pumped sputter coater (Quorum Technologies) and imaged on a Merlin scanning electron microscope (Zeiss) equipped with a secondary electron in-lens detector.
Nuclear Transport Assays
Nuclear import was assayed in digitonin-permeabilized fibroblasts supplemented with exogenous cytosol according to established protocols.29 Briefly, cells were grown for 24 h on 12 mm round coverslips placed in 24-well culture plates (50,000 cells per well) with 1 mL of growth medium. The coverslips were washed once in cold PBS, once in cold transport buffer (TB; 20 mM HEPES [pH 7.3], 110 mM potassium acetate, 2 mM magnesium acetate, 1 mM EDTA, 2 mM DTT, 1 mM PMFS, and 1 μg/mL each of aprotinin and leupeptin) and the cells were permeabilized in 50 μL TB supplemented with 0.005% digitonin (CAS 11024-24-1, Calbiochem) for 7 min on ice. Reaction mixtures (20 μL) contained Xenopus egg cytosol supplemented with an ATP regeneration system and TRITC-NLS-BSA as a cargo-reporter molecule.26 Alternatively, rabbit reticulocyte lysate (L4151, Promega) was used as an exogenous source of cytosol. Nuclear import was terminated at different time points by a brief wash in cold TB and fixation with 3.7% paraformaldehyde in PBS for 10 min. Quantification of the accumulated fluorescent signal is described above.
Nuclear export of mRNA was monitored by in situ hybridization to poly(A)+ on fixed cells. Fibroblasts were grown on 12 mm round coverslips, as above, to ∼80% confluency, transfected with a pEGFP-C1 empty plasmid, and grown for a further 24 h. The cells were then fixed with 3.7% paraformaldehyde in PBS for 20 min on ice, washed in cold PBS, and permeabilized with 0.5% Triton X-100 in PBS for 10 min on ice. Following washes with cold 2× SSC (300 mM NaCl, 3 mM sodium citrate) containing RNase inhibitor (RNasin, Promega), the cells were blocked for 1 h at 37°C in 2× SSC containing 1% BSA, 10% dextran sulfate, 50% formamide, 10 mg/mL tRNA (Roche), and 1 mM RNasin. Hybridization was overnight at 37°C with the same hybridization buffer supplemented with 1 ng/mL oligo-dT-Cy3 probe (Syntezza Bioscience). GFP-expressing and non-expressing cells were compared in both categories (control and affected fibroblasts) and the cytoplasmic/nuclear ratio of the signal was quantified in > 30 cells in each group, as described above.
Plasmids and Transfection
Plasmids encoding the full-length wild-type hNUP214 and hNUP88 with C-terminally fused GFP reporters cloned in pEGFP-C1 were a kind gift from Birthe Fahrenkrog (Université Libre de Bruxelles, Belgium) and Ralph Kehlenbach (Georg-August-University of Gottingen, Germany). The same pEGFP-C1 vector, optimized for high expression levels in human cells of an intron-less mRNA encoding GFP, was used in the mRNA export assay. Transient cell transfections were performed by electroporation, using the human dermal fibroblast nucleofector kit and the Nucleofector 2b Device (Lonza) according to the manufacturer’s instructions.
Cellular Stress and Apoptosis
To expose cells to heat shock stress, fibroblasts were first grown at 37°C in dishes, 24-well culture plates, or 96-well ELISA microplates, as appropriate for the different assays. The growth medium was exchanged with pre-warmed medium and the cells were moved into a 43°C incubator calibrated by direct measurements inside growth media, using a traceable digital thermometer (Fisher Scientific), to ensure the correct temperature and length of exposure. Cell viability was determined by Trypan blue staining (Biological Industries). To provide a positive control for the induction of apoptosis, 2 mM H2O2 was added to the growth medium for 6–8 h and then replaced by fresh medium for overnight growth at 37°C. To measure cleaved caspase activity, the Caspase-Glo 3/7 assay (Promega) was used according to the manufacturer’s instructions and luminescence was measured on an Infinite M1000 microplate reader (Tecan). For rescue experiments, cells were electroporated as described above and incubated at 37°C for 48 h, moved to 43°C for 2 h, and returned to 37°C for 6 h recovery before being fixed and processed for indirect immunofluorescence with anti-cleaved caspase 3.
Statistical Analysis
Except where otherwise indicated, data are presented as mean ± SEM, and the p values were determined by two-tailed Student’s t tests. p values < 0.05 were considered to be statistically significant. For the quantitative analysis in Figures 6 and S4, the occurrence of central channel particles was scored blindly.
Figure 6.
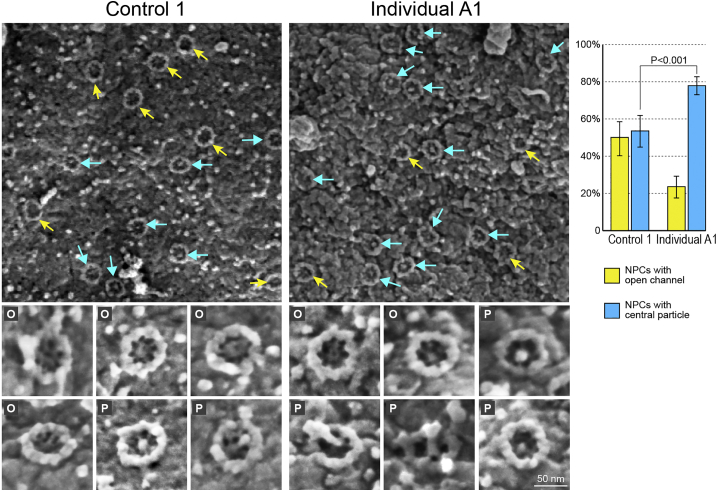
Direct Surface Imaging and Analysis of “Central Plugs” in NPC Channels
Nuclei from control and affected individual-derived fibroblasts were exposed by hypotonic treatment and prepared for FESEM imaging as in Figure 4B. Two representative flat areas of nuclear envelopes with NPCs are shown on the left and a gallery of enlarged, individual NPC images from both categories is shown below. Individual NPCs were scored as having either an open central channel (yellow arrows, O in gallery images) or a channel occupied by a recognizable particle (blue arrows, P in gallery images). A quantitative summary is shown for >1,000 NPCs from >30 nuclei, in each category, analyzed from multiple randomly chosen flat areas of 1×1 μm. Individual A1-derived nuclei are characterized by a larger fraction of NPCs containing a particle in their central channel. Bars indicate SEM. See Figure S4 for an extended analysis of nuclei from unaffected individuals and three affected individuals from both families.
Results
Clinical Summary
Our propositus was an undiagnosed male (family A, III-3 in Figure 1A and Table 1), the third of eight children born to first-cousin parents of Palestinian descent, who presented with ataxia, mental retardation, and intractable epilepsy and died at 11 years of presumed septic shock. The proband (henceforward called individual A1) exhibited minor developmental delay from infancy. At 21 months of age, and subsequently on several occasions, he suffered deterioration in association with febrile illnesses. These episodes caused transient encephalopathy and ataxia that was only partially reversible. Psychomotor development stagnated and regressed after peaking at age 3 when he was able to run and speak in sentences. Intractable epilepsy manifested around 6 years of age and included short non-provoked generalized tonic-clonic seizures and myoclonic jerks. Interictal EEG revealed bilateral nonsynchronous spikes. The major deterioration occurred abruptly during a febrile episode caused by influenza A at age 7 years. By age 11 years, he had lost voluntary mobility, was fed by gastrostomy, and could not verbalize although he seemed to understand simple commands. Examination revealed microcephaly (head circumference 48 cm at 7 years, −3 SD), normal weight (30 kg at 11 years, 5th–10th percentile), and severe truncal ataxia with appendicular spasticity requiring propped sitting. Brain magnetic resonance imaging (MRI) at age 3.5 years showed cerebellar atrophy (Figure 1B; a, c, and e) and repeat MRI at age 8 years, after the severe encephalopathic episode, demonstrated further atrophy along with symmetrical T2-prolongation in the thalami, hippocampi, and brainstem (Figure 1B; b, d, and f). Extensive metabolic investigations and muscle biopsy were normal, prompting evaluation by exome sequencing.
Table 1.
Clinical Features of Affected Individuals
| Family A Individual III-3 (A1) | Family A Individual III-12 (A2) | Family B Individual II-1 (B3) | Family B Individual II-2 (B4) | |
|---|---|---|---|---|
| Age at last examination | 11 years (exitus) | 7.5 months | 6 years 7 months | 3 years 3 months |
| Gender | M | M | F | F |
| Parental consanguinity | + | + | − | − |
| Developmental delay | + | + | + | + |
| Developmental regression | + | + | + | + |
| Febrile-induced regression | + | + | + | + |
| First episode of regression | 21 months | 5.5 months | 15 months | 30 months |
| Epilepsy | + | − | + | + |
| Progressive microcephaly | + | + | + | + |
| Hypotonia | − | + | + | + |
| Appendicular spasticity | + | − | − | − |
| Ataxia | + | NR | + | + |
| Myoclonic jerks | − | + | + | + |
| Cerebellar atrophy | + | + | + | + |
| Basal ganglia involvement | + | − | − | − |
| Hyponatremia upon acute presentation | + | + | + | + |
| NUP214 variant | c.112C>T (p.Arg38Cys) [hom] | c.112C>T (p.Arg38Cys) [hom] | c.1159C>T (p.Pro387Ser); c.1574delC (p.Pro525fs) | c.1159C>T (p.Pro387Ser); c.1574delC (p.Pro525fs) |
Abbreviations: F, female; M, male; NR, not relevant.
The male cousin of the proband (family A, III-12 in Figure 1A, henceforward called individual A2) presented at 5.5 months with partially reversible encephalopathy and developmental regression after a febrile illness. Developmental milestones were age appropriate preceding the illness; he was communicative, had a social smile, and rolled over. Notably, microcephaly with a closed anterior fontanelle was noted upon initial exam (40 cm, below 3rd percentile). Encephalopathy, axial hypotonia alternating with episodes of increased tone, tongue thrusting, and later constant myoclonic jerks of the distal extremities and episodes of downward gaze deviation were evident though epileptiform activity was not present on EEG. Infectious workup identified HHV6 in the CSF and blood by polymerase chain reaction (PCR), which may have triggered the infection-induced neurodegeneration. Brain MRI at 5.5 months showed cortical edema in the occipital region. Subsequent MRI at 6.5 months showed supratentorial and infratentorial (not shown) atrophy and widening of the CSF spaces (Figure 1B; g and h). Two months after the initial episode, he had right esotropia, abnormal movements of the hands had significantly decreased, and he could roll over independently.
The third and fourth affected individuals (family B, II-1 and II-2; individuals B3 and B4 in Figure 1A and Table 1) were sisters born to non-consanguineous parents of Northern European (non-Finnish) descent. The older sister (individual B3) had nystagmus at 2 months of age that self-resolved after 1 month and mild hypotonia, but she was otherwise meeting milestones appropriately and was walking at 13 months. At 15 months of age, she developed a fever that led to a rapid neurological decline, seizures, and abnormal movements. Initial brain MRI at age 16 months showed cerebellar atrophy (Figure 1B; i). She was treated with IVIG and steroids for presumed autoimmune encephalitis. During a prolonged admission, she self-extubated and suffered hypoxic injury due to the inability to re-intubate her. Subsequent MRI showed extensive parenchymal loss and further cerebellar atrophy (Figure 1B; j). She required tracheostomy and gastrostomy placement and continued to have a hyperkinetic movement disorder, epilepsy, and hyponatremia requiring sodium supplementation. At 5 years of age, during an RSV infection, she went into status epilepticus, requiring a pentobarbital-induced coma, ketamine infusion, and ketogenic diet.
The younger sister (individual B4) presented to genetics clinic at 7 months with failure to thrive and hyponatremia but was meeting developmental milestones appropriately. By 24 months of age, she had motor and speech delay, ataxic gait, and occasional very mild head bobbing. She had acquired microcephaly. Brain MRI showed cerebellar hypoplasia at 24 months (Figure 1B; k). At 30 months of age, she had a first seizure in the setting of hyponatremia and RSV infection with low-grade fever (38.1°C). A few days later, she was admitted in status epilepticus in the setting of a febrile illness. During admission, she lost central tone and developed myoclonic jerks, persistent hyponatremia requiring chronic sodium supplementation, and recurrent seizures requiring multiple anti-convulsants. A repeat MRI during the admission showed stable cerebellar atrophy with supratentorial volume loss (Figure 1B; l). There was significant developmental regression during this hospitalization though she was able to regain some developmental skills, but at 44 months has not regained all skills that were present prior to hospitalization. Clinical exome sequencing done with her sister and parents as a quad was unrevealing.
Taken together, it became clear that the four affected individuals all suffered fever-induced, partially reversible acute encephalopathy and regression, progressive microcephaly, and brain atrophy. Onset of the first major developmental regression ranged from 5.5 months to 30 months, and viral triggers were identified in three of the four case subjects. The identification of different pathogens suggested that the trigger was the febrile episode itself (i.e., impaired cellular response to heat shock-induced nuclear damage) rather than a specific pathogen.
Identification of Biallelic Variants in NUP214 in Two Unrelated Families
To identify an underlying genetic diagnosis in family A, exome sequencing was pursued on affected individuals A1 and A2. The affected individuals shared two homozygous variants (Table S1). The variant in NUP214 (g.chr9:134002977C>T [hg19]; GenBank: NM_005085.3; c.112C>T [p.Arg38Cys]) segregated with the disease in available family members: both affected individuals were homozygous for the variant, whereas unaffected individuals were either homozygous for the wild-type allele or heterozygous (Figure 1A). This missense variant affects a highly conserved arginine residue, is predicted pathogenic by multiple bioinformatic algorithms, and has an allele frequency of 0.00006904 in the GnomAD database (Table S2), with no homozygotes. 120 population-matched control subjects were screened, and none were found to carry the p.Arg38Cys variant. Notably, within our exome database comprising 3,000 individuals, with ∼50% Palestinians, no other carriers were identified.
Family B had negative clinical exome sequencing, which included both affected individuals and their parents. The exome data were re-analyzed in a research context through the Baylor-Hopkins Center for Mendelian Genomics (BHCMG) to identify candidate genes and variants. Both sisters (individuals B3 and B4) were found to be compound heterozygous for a frameshift and a missense variant in NUP214 (g.chr9:134015962C>T [hg19]; GenBank: NM_005085.3; c.1159C>T [p.Pro387Ser] and g.chr9:134019946delC; GenBank: NM_005085.3; c.1574delC [p.Pro525Leufs∗6]). The c.1159C>T variant affects a highly conserved proline residue, is predicted to be pathogenic by multiple bioinformatics algorithms, and was observed only once in gnomAD in the heterozygous state. The c.1574delC variant leads to a frameshift in exon 12 of 36, resulting in a stop codon at amino acid 530 of 2,091 residues and is absent in gnomAD. Segregation analysis showed the c.1159C>T variant to be maternally inherited, and the c.1574delC to be paternally inherited. NUP214 was entered into GeneMatcher,30 a freely accessibly website designed to facilitate collaboration between clinicians and researchers with an interest in the same gene, and matched to family A above.
The NUP214 p.Arg38Cys and p.Pro387Ser Variants Are Predicted to Alter Protein Structure and Function
To examine the predicted consequence of the p.Arg38Cys (R38C) and of the p.Pro387Ser (P387S) variants on NUP214 structure, eight mammalian sequences with high homology to the human NUP214 were aligned (Figure S1A). The N-terminal domain of NUP214 orthologs is highly conserved, suggesting a functional role for this domain. In the generated 3D model of the wild-type NUP214 protein, R38 is located at the top portion of the crystallized domain and is involved in ionic/hydrophilic interactions with D154, D245, and H304 (Figure S1B). These residues are highly conserved and may form a gate at the top portion of NUP214. The replacement of R38 with cysteine is predicted to disturb this ion network and alter protein conformation (Figure S1B, top row). P387 is located at the bottom portion of the crystallized domain at the level of the aromatic residues Y48, W157, W248, W313, Y369, and W387 found at the base of the channel protruding toward the proposed gate (Figure S1B, bottom row). A similar arrangement of aromatic residues has been suggested to direct and stabilize structural changes during conformational transitions of membrane proteins.31 Thus, it may be speculated that the cited set of NUP214 aromatic residues participates in dynamic protein-protein interactions. Notably, proline residues located at the same level of the aromatic residues, i.e., P387 with P202 and P63 (not shown), may play a critical role in protein conformational changes through their kinking and hinging movements.32, 33 The disturbed ion network caused by R38C and the altered kinking motion due to the P387S replacement are predicted to change protein folding and interfere with protein-protein interactions with other nucleoporins or shuttling transport receptors.
Fibroblasts from Affected Individuals Exhibit a Decrease in NUP214 and NUP88 Levels
To check for potential effects of the p.Arg38Cys variant at the cellular level, we set out to test the total protein levels and sub-cellular localization of several nucleoporins in primary skin fibroblasts derived from affected individual A1. Total cell lysates were prepared from the primary fibroblasts and compared by immunoblotting to control fibroblast lysates from an unrelated, unaffected control individual. Representative immunoblots and quantitative summaries from multiple experiments are shown in Figure 2. Both NUP214 and its immediate neighbor and sub-complex partner protein NUP8816, 34 were consistently decreased in individual A1 cells to less than 50% of their levels in control fibroblasts. Note that the monoclonal antibody mAb414, which recognizes several FG-repeat Nups, provides an internal control for the specific decrease in NUP214 levels (Figure 2A; see Figure 2B for additional antibodies). Once affected individuals B3 and B4 in family B were identified, the immunoblot analysis was extended to include total cell lysates from additional control fibroblasts, two unaffected parents and the three available affected individuals (Figure S2). A comparison of NUP214 levels in total cell lysates between the three available affected individuals and three unrelated control subjects shows a reduction of about 50% in all of the affected fibroblasts (Figure S2).
Figure 2.
Immunoblot Analysis of Nucleoporin Levels in Fibroblasts Derived from an Affected Individual
Primary fibroblasts derived from affected individual A1 and an unrelated control individual were grown under identical conditions and total cell lysates were prepared for the analysis of protein content. Three separate preparations of cell lysates were compared by immunoblotting. Representative immunoblots are shown and a quantitative analysis of multiple blots from the three separate lysate preparations is shown in the bar charts (see Material and Methods; bars indicate SEM).
(A) Monoclonal antibody mAb414 (directed against five individual FG-repeat Nups, which are members of different NPC sub-complexes) demonstrates the specific effect of the NUP214 p.Arg38Cys variant.
(B) Additional protein specific Nup antibodies were used for confirmation and staining of NUP214, NUP88, NUP133 (a central scaffold Nup), and importin β, which was used as a loading control.
A consistent decrease in the levels of NUP214 and NUP88 was observed in all cases.
To test the intracellular localization of NUP214 and NUP88 within fibroblasts, we used indirect immunofluorescent staining on individual A1 cells and compared them to control fibroblasts. Wide-field fluorescence microscopy was deliberately chosen, to test whether specific Nups were mislocalized to the cytoplasm, or whether their levels were reduced in one or more of the cellular compartments. Algorithms for unified illumination, focus correction, and tiling and stitching enabled us to analyze large fields of cells and quantify the average cellular staining intensity for different Nups (Figure 3, bar charts on the right). All four of the primary anti-Nup antibodies used here generated the pronounced nuclear rim staining pattern typical of NPCs, with some background cytoplasmic staining. Both mAb414 (directed against multiple FG-repeat Nups) and anti-NUP133 (a central NPC scaffold component) showed no significant differences between control and affected fibroblasts. By contrast, nuclear rim staining by anti-NUP214 and anti-NUP88 was clearly diminished in the affected fibroblasts. Moreover, quantitative analysis showed that the total cellular signal for NUP214 and NUP88 was significantly reduced in the affected cells.
Taken together, our results suggest that the p.Arg38Cys missense variant destabilizes some of the interactions of NUP214 with its nearest neighbors at the NPC, resulting in a decrease in the overall steady-state levels of both NUP214 and NUP88, as well as their occurrence on the cytoplasmic side of affected cell NPCs. The levels of several other nucleoporins do not appear to be affected, as judged by immunoblotting and immunofluorescence (mAb414 and anti-NUP133 in Figures 2 and 3).
The Number and Density of NPCs Remain Unchanged in Affected Fibroblasts
The highly specific staining of monoclonal antibody mAb414 is often used as a first approximation for assessing the distribution of NPCs in the nuclear envelope by light microscopy.26, 35 Tangential views of mAb414 staining punctae on the nuclear surface show no difference between cells from control and affected individuals (Figure 4A), suggesting that the number and distribution of NPCs remain unchanged. To verify this conclusion, we used direct surface imaging of fibroblast nuclei by field emission scanning electron microscopy (FESEM)27 and compared large expanses of exposed nuclear envelopes between control and affected nuclei (Figure 4B). Individual NPCs were clearly detectable and a quantitative analysis indicated there was no significant difference in the average density of NPCs between control and affected cells (12.8 ± 2.5 versus 12.1 ± 2 pores/μm2). This conclusion correlates well with the previous results from immunoblots and immunofluorescence and hints at a partial occupancy of NUP214 and NUP88 on the cytoplasmic face of NPCs in the cells of affected individuals.
Disruption of Specific Nuclear Transport Pathways in Fibroblasts from Affected Individuals
To assess the functional consequences of a decreased occupancy of NUP214 and NUP88 in NPCs at the single-cell level, we first tested nuclear protein import. Nuclear import in the classical NLS (nuclear localization signal) importin α/β-mediated pathway was assayed in digitonin-permeabilized fibroblasts with a standard protein import substrate reporter (NLS-conjugated bovine serum albumin). A summary of three independent experiments, performed with two different sources of cytosolic extract (Xenopus egg cytosol and rabbit reticulocyte lysate), is shown in Figure 5A. In each individual experiment the affected cell nuclei accumulated significantly less import substrate compared to control nuclei at a short incubation time-point of 2.5 min. We note that at longer incubation times, the difference between control and affected fibroblasts was diminished and affected cells caught up with the controls. Thus, nuclear import in the classical pathway was significantly delayed, though not completely dysfunctional, in this widely used permeabilized cell assay.
Next, we tested global mRNA export from fibroblast nuclei using in situ hybridization to poly(A)+ RNA in fixed cells. Initial observations suggested that there was no difference between fibroblasts derived from affected and unaffected individuals under normal conditions (data not shown); however, when we added a simple 24 h transfection with a plasmid overexpressing GFP, a clear difference emerged (Figure 5B). Presumably, the mRNA export machinery in transfected cells is challenged by a large excess of GFP-coding transcripts (see Figure S3 for a quantification of GFP expression levels in control and affected cells after transfection). This excess does not alter the cytoplasmic/nuclear ratio of poly(A)+ RNA in control fibroblasts (Figure 5B, top rows and bar chart), while in transfected fibroblasts from an affected individual the cytoplasmic/nuclear ratio dropped significantly (Figure 5B, bottom row and bar chart). The results demonstrate a second instance of decreased nuclear transport in fibroblasts derived from affected individual A1 and also hint that the full effect of biallelic NUP214 variants at the cellular level may only be appreciated by studying perturbations in homeostasis.
Nuclear Pore Channels in Affected Cells Contain More “Central Plugs”
To test the effects of the NUP214 p.Arg38Cys pathogenic variant at the single NPC level, we used high-resolution FESEM imaging and compared a large number of high-magnification images of NPCs from control and affected fibroblast nuclei (Figure 6). This analysis revealed a striking difference between the NPCs from the unaffected control and individual A1-derived cells: while in control cell nuclei roughly half of the NPCs had an open central channel, nearly 80% of the affected individual’s NPCs contained large particles in their channels. As shown in the gallery of individual NPC images at the bottom of Figure 6, these particles are not homogeneous and vary in size and in their orientation with respect to the central pore channel. Central channel particles, also known as “central plugs,” have long been thought to represent large cargo caught in transit through the NPC channel but have not yet been characterized by molecular means.15, 36 Thus, our observations suggest that large cargoes may be delayed in passage through NPC channels in the cells of affected individuals. The shift from a ∼1:1 to a ∼4:1 ratio in occupied versus open pore channels implies that the steady-state level of general transport in affected cell NPCs might be changed and likely relates to the changes we observed in specific import and export assays (Figures 5A and 5B).
We next sought to extend this analysis to additional primary fibroblasts, especially those derived from the two compound heterozygous individuals B3 and B4 (family B, II-1 and II-2 in Figure 1A). The extended FESEM analysis including three unrelated control subjects, the two unaffected parents from family B, and the three available affected individuals, is shown in Figure S4. Importantly, the results present a clear separation between the fibroblasts derived from unaffected individuals (three control subjects and two parents) and those derived from all three affected individuals, based on the prevalence of central channel particles (Figure S4). The “plugged” pore channel phenotype can therefore be considered to be a distinguishing feature of both types of the pathogenic variants of NUP214 identified in our study.
Finally, we tested the effect of pre-treatment with transcription and translation inhibitors on the prevalence of central channel particles in the cells derived from affected individual A1. As shown in Figure S5, the protein translation inhibitor cycloheximide had only a marginal effect, while the transcription inhibitor actinomycin D reduced the occurrence of “plugged” NPCs from 79% to 52% in individual A1 fibroblasts (Figure S5). This strongly suggests that messenger ribonucleoprotein complexes (mRNPs) account for most of the excess central channel particles observed in affected cell nuclei (see Discussion).
An Increased Sensitivity to Heat Shock Is Observed in Affected Fibroblasts
To further explore the effect of perturbations in homeostasis, we subjected control and affected fibroblasts to stress in the form of heat shock (HS). We first tested the response of these primary fibroblasts to different periods of time of exposure to a temperature of 43°C (Figure S6). The 2-hour exposure treatment was chosen for further investigation and cell survival was assessed at 72 h intervals from 1 to 18 days after this HS stress. As shown in Figure 7A, only 46% of individual A1 cells survived on average, 1 day after the treatment, compared to 63.8% in the unaffected control and a difference in the number of viable cells persisted in the cell cultures for up to 18 days after HS. Figure 7B shows a marked difference between control and affected cells in the apoptotic response to HS stress, as determined by a caspase-3/7 activity assay. Control fibroblasts responded more quickly and their caspase activity peaked between 2 and 4 h after HS. By contrast, the peak in caspase activity of individual A1-derived fibroblasts was only observed after 6 h, but it reached more than 2-fold higher levels compared to the control. Caspase-3/7 activity was quickly shut off in control fibroblasts and it was back to normal levels 10 h after HS, while high caspase activity persisted in the affected individual’s fibroblasts 24 h after HS stress (Figure 7B). Indirect immunofluorescent staining with anti-cleaved caspase-3 was performed in control and affected fibroblasts, 6 h after exposure to HS. As shown in Figure 7C, strong cytoplasmic staining of the cleaved caspase was detected in affected cells at this time point, confirming the results of the luminescent activity assay. Overnight exposure to strong oxidative stress (2 mM H2O2) was used as a positive control and caused a similar level of cleaved caspase staining in both control and affected individual’s cells (Figure 7C, middle columns). Thus, our results demonstrate a marked difference in the timing and efficiency of the cellular response to HS stress between primary fibroblasts derived from control and affected individuals. The end result is increased apoptotic cell death of the affected cells under the cell culture conditions of this assay.
The HS Response Phenotype Can Be Rescued by Transfection with Nup-Coding Constructs
To ask whether the sensitivity to HS stress is directly related to the homozygous p.Arg38Cys variant in NUP214, we designed specific rescue experiments in cell culture. Constructs encoding the full-length wild-type hNUP214 and hNUP88 with C-terminally fused GFP reporters were used for transient transfection of fibroblasts and followed by the 2 h HS-6 h recovery treatment scheme described above. We compared control and individual A1-derived fibroblasts transfected by an empty GFP-expressing vector, hNUP214-GFP, hNUP88-GFP, or a combined transfection with hNUP214-GFP and hNUP88-GFP. Immunostaining of cleaved caspase-3 was quantified in GFP-expressing cells and compared across all the experimental conditions, as summarized in the bar chart (Figure 8, bottom). The top part of Figure 8 shows representative images of the double NUP214/NUP88 transfection and empty vector control. The strongest anti-cleaved caspase-3 staining was observed in the affected individual’s cells transfected by the empty GFP-expressing vector, while the double NUP214/NUP88 transfection caused a significant decrease in this staining. Overall, as summarized in the bar chart, transfection with either of the wild-type hNUP214 or hNUP88 coding sequences was able to reduce the apoptotic response of affected individual fibroblasts while the double NUP214/NUP88 transfection had the strongest effect. We note that the overall transfection efficiency in these experiments was low and the relative expression levels or potential co-regulatory effects of the two nucleoporins could not be assessed. We conclude that the pathogenic variant-associated phenotype involving the apoptotic response to HS stress in fibroblasts can be reversed by overexpression of the wild-type coding sequences of human NUP214 and/or NUP88.
Discussion
Here we describe four individuals from two families, who presented with a distinct phenotype of febrile encephalopathy causing neurodevelopmental regression, seizures, myoclonic jerks, progressive microcephaly, and cerebellar atrophy with variable involvement of the basal ganglia. Whole-exome sequencing identified NUP214 as the candidate gene in both families. Affected individuals in family A were found to be homozygous for a missense variant, p.Arg38Cys, that segregated appropriately in the family. Affected individuals in family B were found to be compound heterozygous for a paternally inherited frameshift variant, p.Pro525Leufs∗6, and a maternally inherited missense variant, p.Pro387Ser.
Modeling of the N-terminal domain of wild-type versus mutant forms, p.Arg38Cys and p.Pro387Ser, of NUP214 suggested that the replacement of Arg38 by cysteine perturbs a local ion network that may behave as a gate for protein-protein interactions. The disturbed ion network is predicted to alter the protein structure, leading to incorrect folding and affecting local interactions. Notably, the p.Pro387Ser variant is spatially located among Y369 (3.5 Å distance) and W313 (3.9 Å distance) and would be predicted to seriously alter conformational changes and a putative interaction interface. These changes are proposed to impair interactions with neighboring Nups as well as the nuclear transport machinery. In particular, interactions with the NUP214 sub-complex partner NUP88 and docking onto the NPC scaffold might be impaired,16, 37 as well as the known interactions with CRM1 and DBP5.20, 21, 22
Collectively, functional studies of the NUP214 p.Arg38Cys variant in primary skin fibroblasts derived from affected individual A1 support the pathogenicity of this variant. First, immunoblot analysis showed that total cellular levels of NUP214, and its sub-complex partner NUP88, were significantly decreased in the affected fibroblasts compared to controls. This reduction reflected a decrease of at least 50% in the immunostaining intensity of these two nucleoporins in situ, at the nuclear envelope. Other nucleoporins tested by immunoblotting and immunofluorescence appeared unaffected, while the number and density of NPCs in affected individual cell nuclei remained normal as evident from direct FESEM imaging. These observations suggested that the p.Arg38Cys variant may affect nuclear transport through the NPC, rather than an overall change in its structure.
Defects in nuclear transport pathways were observed in standard functional assays, including a clear delay in classical NLS-mediated nuclear import and an accumulation of mRNA in affected cell nuclei. It is important to note that the complete inhibition of any major nuclear import or export pathway is expected to be lethal in all cell types. Thus, the changes in transport observed under normal growth conditions likely indicate that irreversible damage can occur under stress conditions or in particularly sensitive tissues such as the brain. The most striking cellular phenotype was obtained by high-resolution imaging of individual NPCs on the cytoplasmic face of the nuclear envelope of exposed fibroblast nuclei. FESEM imaging revealed a large increase in the presence of central particles in the pore channel of NPCs in cells derived from affected individuals compared to unaffected control subjects. This analysis was extended to include fibroblasts derived from affected individuals from both families, demonstrating that the “plugged” pore channel phenotype is a distinguishing feature of both the homozygous p.Arg38Cys pathogenic variant and the compound heterozygous p.Pro387Ser/p.Pro525Leufs∗6 condition. Thus, a distinct change observed in the nanometric scale, at the single-NPC level, is linked to human disease-causing variants. A logical interpretation of this finding is that large transport cargoes are delayed in passage through NPC channels in cells from affected individuals, causing a partial physical blockage or affecting the properties of the permeability barrier. Clearly, nuclear transport is still occurring in affected fibroblasts, but this represents a significant deviation from homeostasis. The strong effect of actinomycin D on central channel particles in affected individual cells implicates mRNPs as key molecular players in this novel disease phenotype. Together with the effect of excess GFP-coding transcripts on the export assay in affected cells and the known interactions of NUP214 with the DEAD box protein DBP5, this suggests that the mRNA export pathway is a major affected target of the pathogenic variants.
Our results showing a difference in the degree and timing of the cellular response to heat shock between control and affected fibroblasts suggest a possible disease mechanism for the fever-induced neurodegeneration seen in affected individuals. Fibroblasts derived from affected individual A1 had a delayed, exaggerated, and prolonged cellular response to heat shock compared to control fibroblasts. Additionally, affected cell survival was decreased and led to a permanent lag in relative survival compared to control cells. These results suggest that changes in nucleocytoplasmic transport resulting from abnormal functioning of NUP214 affect the timing and efficiency of the cellular response to heat shock stress, eventually leading to greater apoptotic cell death. In particular, it appears that the affected individual’s cells are unable to quickly shut off the apoptotic response in the recovery from heat shock. We hypothesize that the neurons of affected individuals with pathogenic variants in NUP214 are more susceptible to apoptosis in the setting of a febrile illness. This in turn leads to the clinical manifestations of developmental regression, progressive microcephaly, and cerebellar atrophy. Although the identification of different viral pathogens in three of the affected individuals points to the febrile episode itself as the trigger, an alternative (but not mutually exclusive) explanation might be that viral infections compound the effect of compromised nucleocytoplasmic trafficking caused by the genetic variants.
Recently, a neurodegenerative disease of infantile onset, affecting mainly the central white matter, accompanied by fever-induced systemic cytokine storm (hypomyelinating leukodystrophy-13 [MIM: 616881]), was reported in association with a homozygous missense variant in HIKESHI/C11ORF73.38 Interestingly, the encoded transport factor Hikeshi interacts with nucleoporins via its N-terminal domain and regulates the import of heat-shock proteins from the Hsp70 family into the nucleus under stress conditions,39, 40 suggesting a potential pathomechanistic link between these diseases.
NUP214 has previously been noted for its involvement in chromosomal translocations leading to acute myeloid leukemia and other hematopoietic malignancies.41, 42 The mouse ortholog of human NUP214 was the first nucleoporin gene to be disrupted in higher eukaryotes, with the knockout proving to be non-viable and the embryos dying in utero after the depletion of the maternal protein pool.43 Recent work on recessive variants unmasked by chromosomal deletions has pointed to a single base-pair deletion in human NUP214 as a candidate disease-causing pathogenic variant in the case of a neurodevelopmental disorder.44 Another recent report45 describes a similar clinical phenotype to the one in our study, consisting of severe encephalopathy and microcephaly and associated with a different biallelic variant in NUP214. An intriguing new study by Bonin et al. identifies biallelic variants in human NUP88 as a novel cause of lethal fetal akinesia deformation sequence (FADS [MIM: 208150]) in two families.46 The authors present a zebrafish model in which the disruption of nup88 causes similar developmental defects to those observed in affected human fetuses.46
As mentioned above, only a small number of inherited human diseases have been linked to nucleoporins and these affect different tissues: two types of nephrotic syndrome (type 11 [MIM: 616730] and type 13 [MIM: 616893]),6, 8 triple A syndrome (MIM: 231550) consisting of adrenal insufficiency, achalasia, and alacrima,7 XX gonadal dysgenesis (ovarian dysgenesis 6 [MIM: 618078]),10 as well as atrial fibrillation and early sudden cardiac death (atrial fibrillation 15 [MIM: 615770]).11 Two central nervous system disorders have been linked to pathogenic variants in the genes encoding NUP358/RanBP2 (MIM: 601181) and NUP62 (MIM: 605815). NUP358/RanBP2 has been reported to cause autosomal-dominant susceptibility to infection-induced, acute encephalopathy 3 (MIM: 608033). Affected individuals present with infection-induced encephalopathy characterized by seizures, changes in mental status including coma, and gait abnormalities, as well as brain MRI findings of T2 signal abnormalities most commonly in the pons, thalami, and brainstem.47 Infantile striatonigral degeneration (MIM: 271930) is caused by biallelic variants in NUP62 and results in developmental arrest during late infancy, choreoathetosis, dysphagia, pendular nystagmus, and severe basal ganglia atrophy on MRI.5, 48 In some cases, the developmental arrest and neurodegeneration occurred in the setting of a febrile illness.
The most clinically striking phenotypic feature in the four affected individuals we describe is the acute encephalopathy that occurs in the setting of a febrile illness. The differential diagnosis for an individual with this clinical presentation includes autoimmune encephalopathy, meningoencephalitis, inborn errors of metabolism, and immunodeficiency. Thus far, the clinical manifestations of the affected individuals described here are limited to the central nervous system, which can be a factor that differentiates this disorder from other causes of acute encephalopathy. For instance, inborn errors of metabolism may have liver and/or kidney involvement, and in the setting of immunodeficiency, infections may be present in other organ systems. Here we propose that germline variants in the genes encoding nucleoporins may be a new category of disease genes that cause infection- and/or fever-induced encephalopathy.
The combined insight from the clinical and experimental aspects of the current study will guide future attempts to decipher the molecular mechanisms underlying nucleoporin-associated disorders, as well as a comparison to other neurodegenerative diseases that have recently been linked to NPCs and the nuclear transport machinery.49, 50, 51, 52
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank the affected individuals and their families for contributing samples and enthusiasm for this work. We thank David Valle and Gary Steele for their assistance with segregation analysis in family B. This work was supported by a research grant from the Israel Science Foundation (958/15) to A. Harel and a grant from the Baylor-Hopkins Center for Mendelian Genomics (1U54HG006542) to A. Hamosh and N. Sobreira.
Published: June 6, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.05.003.
Web Resources
ClustalW, https://www.genome.jp/tools-bin/clustalw
ExAC Browser, http://exac.broadinstitute.org/
GeneMatcher, https://genematcher.org/
gnomAD Browser, https://gnomad.broadinstitute.org/
MutationTaster, http://www.mutationtaster.org/
OMIM, https://www.omim.org/
Supplemental Data
References
- 1.Fahrenkrog B., Köser J., Aebi U. The nuclear pore complex: a jack of all trades? Trends Biochem. Sci. 2004;29:175–182. doi: 10.1016/j.tibs.2004.02.006. [DOI] [PubMed] [Google Scholar]; Fahrenkrog, B., Koser, J., and Aebi, U. (2004). The nuclear pore complex: a jack of all trades? Trends Biochem. Sci. 29, 175-182. [DOI] [PubMed]
- 2.Hetzer M.W., Wente S.R. Border control at the nucleus: biogenesis and organization of the nuclear membrane and pore complexes. Dev. Cell. 2009;17:606–616. doi: 10.1016/j.devcel.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hetzer, M.W., and Wente, S.R. (2009). Border control at the nucleus: biogenesis and organization of the nuclear membrane and pore complexes. Dev. Cell 17, 606-616. [DOI] [PMC free article] [PubMed]
- 3.Bilokapic S., Schwartz T.U. 3D ultrastructure of the nuclear pore complex. Curr. Opin. Cell Biol. 2012;24:86–91. doi: 10.1016/j.ceb.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bilokapic, S., and Schwartz, T.U. (2012). 3D ultrastructure of the nuclear pore complex. Curr. Opin. Cell Biol. 24, 86-91. [DOI] [PMC free article] [PubMed]
- 4.Hoelz A., Debler E.W., Blobel G. The structure of the nuclear pore complex. Annu. Rev. Biochem. 2011;80:613–643. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]; Hoelz, A., Debler, E.W., and Blobel, G. (2011). The structure of the nuclear pore complex. Annu. Rev. Biochem. 80, 613-643. [DOI] [PubMed]
- 5.Basel-Vanagaite L., Muncher L., Straussberg R., Pasmanik-Chor M., Yahav M., Rainshtein L., Walsh C.A., Magal N., Taub E., Drasinover V. Mutated nup62 causes autosomal recessive infantile bilateral striatal necrosis. Ann. Neurol. 2006;60:214–222. doi: 10.1002/ana.20902. [DOI] [PubMed] [Google Scholar]; Basel-Vanagaite, L., Muncher, L., Straussberg, R., Pasmanik-Chor, M., Yahav, M., Rainshtein, L., Walsh, C.A., Magal, N., Taub, E., Drasinover, V., et al. (2006). Mutated nup62 causes autosomal recessive infantile bilateral striatal necrosis. Ann. Neurol. 60, 214-222. [DOI] [PubMed]
- 6.Braun D.A., Sadowski C.E., Kohl S., Lovric S., Astrinidis S.A., Pabst W.L., Gee H.Y., Ashraf S., Lawson J.A., Shril S. Mutations in nuclear pore genes NUP93, NUP205 and XPO5 cause steroid-resistant nephrotic syndrome. Nat. Genet. 2016;48:457–465. doi: 10.1038/ng.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]; Braun, D.A., Sadowski, C.E., Kohl, S., Lovric, S., Astrinidis, S.A., Pabst, W.L., Gee, H.Y., Ashraf, S., Lawson, J.A., Shril, S., et al. (2016). Mutations in nuclear pore genes NUP93, NUP205 and XPO5 cause steroid-resistant nephrotic syndrome. Nat. Genet. 48, 457-465. [DOI] [PMC free article] [PubMed]
- 7.Cronshaw J.M., Matunis M.J. The nuclear pore complex protein ALADIN is mislocalized in triple A syndrome. Proc. Natl. Acad. Sci. USA. 2003;100:5823–5827. doi: 10.1073/pnas.1031047100. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cronshaw, J.M., and Matunis, M.J. (2003). The nuclear pore complex protein ALADIN is mislocalized in triple A syndrome. Proc. Natl. Acad. Sci. USA 100, 5823-5827. [DOI] [PMC free article] [PubMed]
- 8.Miyake N., Tsukaguchi H., Koshimizu E., Shono A., Matsunaga S., Shiina M., Mimura Y., Imamura S., Hirose T., Okudela K. Biallelic Mutations in Nuclear Pore Complex Subunit NUP107 Cause Early-Childhood-Onset Steroid-Resistant Nephrotic Syndrome. Am. J. Hum. Genet. 2015;97:555–566. doi: 10.1016/j.ajhg.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; Miyake, N., Tsukaguchi, H., Koshimizu, E., Shono, A., Matsunaga, S., Shiina, M., Mimura, Y., Imamura, S., Hirose, T., Okudela, K., et al. (2015). Biallelic Mutations in Nuclear Pore Complex Subunit NUP107 Cause Early-Childhood-Onset Steroid-Resistant Nephrotic Syndrome. Am. J. Hum. Genet. 97, 555-566. [DOI] [PMC free article] [PubMed]
- 9.Neilson D.E., Adams M.D., Orr C.M., Schelling D.K., Eiben R.M., Kerr D.S., Anderson J., Bassuk A.G., Bye A.M., Childs A.M. Infection-triggered familial or recurrent cases of acute necrotizing encephalopathy caused by mutations in a component of the nuclear pore, RANBP2. Am. J. Hum. Genet. 2009;84:44–51. doi: 10.1016/j.ajhg.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; Neilson, D.E., Adams, M.D., Orr, C.M., Schelling, D.K., Eiben, R.M., Kerr, D.S., Anderson, J., Bassuk, A.G., Bye, A.M., Childs, A.M., et al. (2009). Infection-triggered familial or recurrent cases of acute necrotizing encephalopathy caused by mutations in a component of the nuclear pore, RANBP2. Am. J. Hum. Genet. 84, 44-51. [DOI] [PMC free article] [PubMed]
- 10.Weinberg-Shukron A., Renbaum P., Kalifa R., Zeligson S., Ben-Neriah Z., Dreifuss A., Abu-Rayyan A., Maatuk N., Fardian N., Rekler D. A mutation in the nucleoporin-107 gene causes XX gonadal dysgenesis. J. Clin. Invest. 2015;125:4295–4304. doi: 10.1172/JCI83553. [DOI] [PMC free article] [PubMed] [Google Scholar]; Weinberg-Shukron, A., Renbaum, P., Kalifa, R., Zeligson, S., Ben-Neriah, Z., Dreifuss, A., Abu-Rayyan, A., Maatuk, N., Fardian, N., Rekler, D., et al. (2015). A mutation in the nucleoporin-107 gene causes XX gonadal dysgenesis. J. Clin. Invest. 125, 4295-4304. [DOI] [PMC free article] [PubMed]
- 11.Zhang X., Chen S., Yoo S., Chakrabarti S., Zhang T., Ke T., Oberti C., Yong S.L., Fang F., Li L. Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell. 2008;135:1017–1027. doi: 10.1016/j.cell.2008.10.022. [DOI] [PubMed] [Google Scholar]; Zhang, X., Chen, S., Yoo, S., Chakrabarti, S., Zhang, T., Ke, T., Oberti, C., Yong, S.L., Fang, F., Li, L., et al. (2008). Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell 135, 1017-1027. [DOI] [PubMed]
- 12.Worman H.J., Dauer W.T. The nuclear envelope: an intriguing focal point for neurogenetic disease. Neurotherapeutics. 2014;11:764–772. doi: 10.1007/s13311-014-0296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Worman, H.J., and Dauer, W.T. (2014). The nuclear envelope: an intriguing focal point for neurogenetic disease. Neurotherapeutics 11, 764-772. [DOI] [PMC free article] [PubMed]
- 13.Chi Y.H., Chen Z.J., Jeang K.T. The nuclear envelopathies and human diseases. J. Biomed. Sci. 2009;16:96. doi: 10.1186/1423-0127-16-96. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chi, Y.H., Chen, Z.J., and Jeang, K.T. (2009). The nuclear envelopathies and human diseases. J. Biomed. Sci. 16, 96. [DOI] [PMC free article] [PubMed]
- 14.Schreiber K.H., Kennedy B.K. When lamins go bad: nuclear structure and disease. Cell. 2013;152:1365–1375. doi: 10.1016/j.cell.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; Schreiber, K.H., and Kennedy, B.K. (2013). When lamins go bad: nuclear structure and disease. Cell 152, 1365-1375. [DOI] [PMC free article] [PubMed]
- 15.Beck M., Förster F., Ecke M., Plitzko J.M., Melchior F., Gerisch G., Baumeister W., Medalia O. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306:1387–1390. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]; Beck, M., Forster, F., Ecke, M., Plitzko, J.M., Melchior, F., Gerisch, G., Baumeister, W., and Medalia, O. (2004). Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science 306, 1387-1390. [DOI] [PubMed]
- 16.Bernad R., van der Velde H., Fornerod M., Pickersgill H. Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export. Mol. Cell. Biol. 2004;24:2373–2384. doi: 10.1128/MCB.24.6.2373-2384.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bernad, R., van der Velde, H., Fornerod, M., and Pickersgill, H. (2004). Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export. Mol. Cell. Biol. 24, 2373-2384. [DOI] [PMC free article] [PubMed]
- 17.Hamada M., Haeger A., Jeganathan K.B., van Ree J.H., Malureanu L., Wälde S., Joseph J., Kehlenbach R.H., van Deursen J.M. Ran-dependent docking of importin-beta to RanBP2/Nup358 filaments is essential for protein import and cell viability. J. Cell Biol. 2011;194:597–612. doi: 10.1083/jcb.201102018. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hamada, M., Haeger, A., Jeganathan, K.B., van Ree, J.H., Malureanu, L., Walde, S., Joseph, J., Kehlenbach, R.H., and van Deursen, J.M. (2011). Ran-dependent docking of importin-beta to RanBP2/Nup358 filaments is essential for protein import and cell viability. J. Cell Biol. 194, 597-612. [DOI] [PMC free article] [PubMed]
- 18.Wälde S., Thakar K., Hutten S., Spillner C., Nath A., Rothbauer U., Wiemann S., Kehlenbach R.H. The nucleoporin Nup358/RanBP2 promotes nuclear import in a cargo- and transport receptor-specific manner. Traffic. 2012;13:218–233. doi: 10.1111/j.1600-0854.2011.01302.x. [DOI] [PubMed] [Google Scholar]; Walde, S., Thakar, K., Hutten, S., Spillner, C., Nath, A., Rothbauer, U., Wiemann, S., and Kehlenbach, R.H. (2012). The nucleoporin Nup358/RanBP2 promotes nuclear import in a cargo- and transport receptor-specific manner. Traffic 13, 218-233. [DOI] [PubMed]
- 19.Hodge C.A., Tran E.J., Noble K.N., Alcazar-Roman A.R., Ben-Yishay R., Scarcelli J.J., Folkmann A.W., Shav-Tal Y., Wente S.R., Cole C.N. The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1. Genes Dev. 2011;25:1052–1064. doi: 10.1101/gad.2041611. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hodge, C.A., Tran, E.J., Noble, K.N., Alcazar-Roman, A.R., Ben-Yishay, R., Scarcelli, J.J., Folkmann, A.W., Shav-Tal, Y., Wente, S.R., and Cole, C.N. (2011). The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1. Genes Dev. 25, 1052-1064. [DOI] [PMC free article] [PubMed]
- 20.Hutten S., Kehlenbach R.H. Nup214 is required for CRM1-dependent nuclear protein export in vivo. Mol. Cell. Biol. 2006;26:6772–6785. doi: 10.1128/MCB.00342-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hutten, S., and Kehlenbach, R.H. (2006). Nup214 is required for CRM1-dependent nuclear protein export in vivo. Mol. Cell. Biol. 26, 6772-6785. [DOI] [PMC free article] [PubMed]
- 21.Napetschnig J., Kassube S.A., Debler E.W., Wong R.W., Blobel G., Hoelz A. Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19. Proc. Natl. Acad. Sci. USA. 2009;106:3089–3094. doi: 10.1073/pnas.0813267106. [DOI] [PMC free article] [PubMed] [Google Scholar]; Napetschnig, J., Kassube, S.A., Debler, E.W., Wong, R.W., Blobel, G., and Hoelz, A. (2009). Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19. Proc. Natl. Acad. Sci. USA 106, 3089-3094. [DOI] [PMC free article] [PubMed]
- 22.von Moeller H., Basquin C., Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat. Struct. Mol. Biol. 2009;16:247–254. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]; von Moeller, H., Basquin, C., and Conti, E. (2009). The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat. Struct. Mol. Biol. 16, 247-254. [DOI] [PubMed]
- 23.Retterer K., Juusola J., Cho M.T., Vitazka P., Millan F., Gibellini F., Vertino-Bell A., Smaoui N., Neidich J., Monaghan K.G. Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 2016;18:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]; Retterer, K., Juusola, J., Cho, M.T., Vitazka, P., Millan, F., Gibellini, F., Vertino-Bell, A., Smaoui, N., Neidich, J., Monaghan, K.G., et al. (2016). Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 18, 696-704. [DOI] [PubMed]
- 24.Sobreira N., Schiettecatte F., Boehm C., Valle D., Hamosh A. New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum. Mutat. 2015;36:425–431. doi: 10.1002/humu.22769. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sobreira, N., Schiettecatte, F., Boehm, C., Valle, D., and Hamosh, A. (2015). New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum. Mutat. 36, 425-431. [DOI] [PMC free article] [PubMed]
- 25.Pierri C.L., Parisi G., Porcelli V. Computational approaches for protein function prediction: a combined strategy from multiple sequence alignment to molecular docking-based virtual screening. Biochim. Biophys. Acta. 2010;1804:1695–1712. doi: 10.1016/j.bbapap.2010.04.008. [DOI] [PubMed] [Google Scholar]; Pierri, C.L., Parisi, G., and Porcelli, V. (2010). Computational approaches for protein function prediction: a combined strategy from multiple sequence alignment to molecular docking-based virtual screening. Biochim. Biophys. Acta 1804, 1695-1712. [DOI] [PubMed]
- 26.Harel A., Orjalo A.V., Vincent T., Lachish-Zalait A., Vasu S., Shah S., Zimmerman E., Elbaum M., Forbes D.J. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell. 2003;11:853–864. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]; Harel, A., Orjalo, A.V., Vincent, T., Lachish-Zalait, A., Vasu, S., Shah, S., Zimmerman, E., Elbaum, M., and Forbes, D.J. (2003). Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell 11, 853-864. [DOI] [PubMed]
- 27.Fichtman B., Shaulov L., Harel A. Imaging metazoan nuclear pore complexes by field emission scanning electron microscopy. Methods Cell Biol. 2014;122:41–58. doi: 10.1016/B978-0-12-417160-2.00002-3. [DOI] [PubMed] [Google Scholar]; Fichtman, B., Shaulov, L., and Harel, A. (2014). Imaging metazoan nuclear pore complexes by field emission scanning electron microscopy. Methods Cell Biol. 122, 41-58. [DOI] [PubMed]
- 28.Shaulov L., Harel A. Improved visualization of vertebrate nuclear pore complexes by field emission scanning electron microscopy. Structure. 2012;20:407–413. doi: 10.1016/j.str.2012.01.022. [DOI] [PubMed] [Google Scholar]; Shaulov, L., and Harel, A. (2012). Improved visualization of vertebrate nuclear pore complexes by field emission scanning electron microscopy. Structure 20, 407-413. [DOI] [PubMed]
- 29.Cassany A., Gerace L. Reconstitution of nuclear import in permeabilized cells. Methods Mol. Biol. 2009;464:181–205. doi: 10.1007/978-1-60327-461-6_11. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cassany, A., and Gerace, L. (2009). Reconstitution of nuclear import in permeabilized cells. Methods Mol. Biol. 464, 181-205. [DOI] [PMC free article] [PubMed]
- 30.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sobreira, N., Schiettecatte, F., Valle, D., and Hamosh, A. (2015). GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 36, 928-930. [DOI] [PMC free article] [PubMed]
- 31.Domene C., Vemparala S., Klein M.L., Vénien-Bryan C., Doyle D.A. Role of aromatic localization in the gating process of a potassium channel. Biophys. J. 2006;90:L01–L03. doi: 10.1529/biophysj.105.072116. [DOI] [PMC free article] [PubMed] [Google Scholar]; Domene, C., Vemparala, S., Klein, M.L., Venien-Bryan, C., and Doyle, D.A. (2006). Role of aromatic localization in the gating process of a potassium channel. Biophys. J. 90, L01-L03. [DOI] [PMC free article] [PubMed]
- 32.Palmieri F., Pierri C.L. Structure and function of mitochondrial carriers - role of the transmembrane helix P and G residues in the gating and transport mechanism. FEBS Lett. 2010;584:1931–1939. doi: 10.1016/j.febslet.2009.10.063. [DOI] [PubMed] [Google Scholar]; Palmieri, F., and Pierri, C.L. (2010). Structure and function of mitochondrial carriers - role of the transmembrane helix P and G residues in the gating and transport mechanism. FEBS Lett. 584, 1931-1939. [DOI] [PubMed]
- 33.Pierri C.L., Palmieri F., De Grassi A. Single-nucleotide evolution quantifies the importance of each site along the structure of mitochondrial carriers. Cell. Mol. Life Sci. 2014;71:349–364. doi: 10.1007/s00018-013-1389-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pierri, C.L., Palmieri, F., and De Grassi, A. (2014). Single-nucleotide evolution quantifies the importance of each site along the structure of mitochondrial carriers. Cell. Mol. Life Sci. 71, 349-364. [DOI] [PMC free article] [PubMed]
- 34.Schwartz T.U. Modularity within the architecture of the nuclear pore complex. Curr. Opin. Struct. Biol. 2005;15:221–226. doi: 10.1016/j.sbi.2005.03.003. [DOI] [PubMed] [Google Scholar]; Schwartz, T.U. (2005). Modularity within the architecture of the nuclear pore complex. Curr. Opin. Struct. Biol. 15, 221-226. [DOI] [PubMed]
- 35.Davis L.I., Blobel G. Identification and characterization of a nuclear pore complex protein. Cell. 1986;45:699–709. doi: 10.1016/0092-8674(86)90784-1. [DOI] [PubMed] [Google Scholar]; Davis, L.I., and Blobel, G. (1986). Identification and characterization of a nuclear pore complex protein. Cell 45, 699-709. [DOI] [PubMed]
- 36.Jarnik M., Aebi U. Toward a more complete 3-D structure of the nuclear pore complex. J. Struct. Biol. 1991;107:291–308. doi: 10.1016/1047-8477(91)90054-z. [DOI] [PubMed] [Google Scholar]; Jarnik, M., and Aebi, U. (1991). Toward a more complete 3-D structure of the nuclear pore complex. J. Struct. Biol. 107, 291-308. [DOI] [PubMed]
- 37.Bui K.H., von Appen A., DiGuilio A.L., Ori A., Sparks L., Mackmull M.T., Bock T., Hagen W., Andrés-Pons A., Glavy J.S., Beck M. Integrated structural analysis of the human nuclear pore complex scaffold. Cell. 2013;155:1233–1243. doi: 10.1016/j.cell.2013.10.055. [DOI] [PubMed] [Google Scholar]; Bui, K.H., von Appen, A., DiGuilio, A.L., Ori, A., Sparks, L., Mackmull, M.T., Bock, T., Hagen, W., Andres-Pons, A., Glavy, J.S., and Beck, M. (2013). Integrated structural analysis of the human nuclear pore complex scaffold. Cell 155, 1233-1243. [DOI] [PubMed]
- 38.Edvardson S., Kose S., Jalas C., Fattal-Valevski A., Watanabe A., Ogawa Y., Mamada H., Fedick A.M., Ben-Shachar S., Treff N.R. Leukoencephalopathy and early death associated with an Ashkenazi-Jewish founder mutation in the Hikeshi gene. J. Med. Genet. 2016;53:132–137. doi: 10.1136/jmedgenet-2015-103232. [DOI] [PubMed] [Google Scholar]; Edvardson, S., Kose, S., Jalas, C., Fattal-Valevski, A., Watanabe, A., Ogawa, Y., Mamada, H., Fedick, A.M., Ben-Shachar, S., Treff, N.R., et al. (2016). Leukoencephalopathy and early death associated with an Ashkenazi-Jewish founder mutation in the Hikeshi gene. J. Med. Genet. 53, 132-137. [DOI] [PubMed]
- 39.Imamoto N. Heat stress-induced nuclear transport mediated by Hikeshi confers nuclear function of Hsp70s. Curr. Opin. Cell Biol. 2018;52:82–87. doi: 10.1016/j.ceb.2018.02.010. [DOI] [PubMed] [Google Scholar]; Imamoto, N. (2018). Heat stress-induced nuclear transport mediated by Hikeshi confers nuclear function of Hsp70s. Curr. Opin. Cell Biol. 52, 82-87. [DOI] [PubMed]
- 40.Kose S., Furuta M., Imamoto N. Hikeshi, a nuclear import carrier for Hsp70s, protects cells from heat shock-induced nuclear damage. Cell. 2012;149:578–589. doi: 10.1016/j.cell.2012.02.058. [DOI] [PubMed] [Google Scholar]; Kose, S., Furuta, M., and Imamoto, N. (2012). Hikeshi, a nuclear import carrier for Hsp70s, protects cells from heat shock-induced nuclear damage. Cell 149, 578-589. [DOI] [PubMed]
- 41.Fahrenkrog B. Nucleoporin Gene Fusions and Hematopoietic Malignancies. New J. Sci. 2014;2014:18. [Google Scholar]; Fahrenkrog, B. (2014). Nucleoporin Gene Fusions and Hematopoietic Malignancies. New J. Sci. 2014, 18.
- 42.von Lindern M., Poustka A., Lerach H., Grosveld G. The (6;9) chromosome translocation, associated with a specific subtype of acute nonlymphocytic leukemia, leads to aberrant transcription of a target gene on 9q34. Mol. Cell. Biol. 1990;10:4016–4026. doi: 10.1128/mcb.10.8.4016. [DOI] [PMC free article] [PubMed] [Google Scholar]; von Lindern, M., Poustka, A., Lerach, H., and Grosveld, G. (1990). The (6;9) chromosome translocation, associated with a specific subtype of acute nonlymphocytic leukemia, leads to aberrant transcription of a target gene on 9q34. Mol. Cell. Biol. 10, 4016-4026. [DOI] [PMC free article] [PubMed]
- 43.van Deursen J., Boer J., Kasper L., Grosveld G. G2 arrest and impaired nucleocytoplasmic transport in mouse embryos lacking the proto-oncogene CAN/Nup214. EMBO J. 1996;15:5574–5583. [PMC free article] [PubMed] [Google Scholar]; van Deursen, J., Boer, J., Kasper, L., and Grosveld, G. (1996). G2 arrest and impaired nucleocytoplasmic transport in mouse embryos lacking the proto-oncogene CAN/Nup214. EMBO J. 15, 5574-5583. [PMC free article] [PubMed]
- 44.Egloff M., Nguyen L.S., Siquier-Pernet K., Cormier-Daire V., Baujat G., Attié-Bitach T., Bole-Feysot C., Nitschke P., Vekemans M., Colleaux L., Malan V. Whole-exome sequence analysis highlights the role of unmasked recessive mutations in copy number variants with incomplete penetrance. Eur. J. Hum. Genet. 2018;26:912–918. doi: 10.1038/s41431-018-0124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; Egloff, M., Nguyen, L.S., Siquier-Pernet, K., Cormier-Daire, V., Baujat, G., Attie-Bitach, T., Bole-Feysot, C., Nitschke, P., Vekemans, M., Colleaux, L., and Malan, V. (2018). Whole-exome sequence analysis highlights the role of unmasked recessive mutations in copy number variants with incomplete penetrance. Eur. J. Hum. Genet. 26, 912-918. [DOI] [PMC free article] [PubMed]
- 45.Shamseldin H.E., Makhseed N., Ibrahim N., Al-Sheddi T., Alobeid E., Abdulwahab F., Alkuraya F.S. NUP214 deficiency causes severe encephalopathy and microcephaly in humans. Hum. Genet. 2018;138:221–229. doi: 10.1007/s00439-019-01979-w. [DOI] [PubMed] [Google Scholar]; Shamseldin, H.E., Makhseed, N., Ibrahim, N., Al-Sheddi, T., Alobeid, E., Abdulwahab, F., and Alkuraya, F.S. (2019). NUP214 deficiency causes severe encephalopathy and microcephaly in humans. Hum Genet 138, 221-229. [DOI] [PubMed]
- 46.Bonnin E., Cabochette P., Filosa A., Jühlen R., Komatsuzaki S., Hezwani M., Dickmanns A., Martinelli V., Vermeersch M., Supply L. Biallelic mutations in nucleoporin NUP88 cause lethal fetal akinesia deformation sequence. PLoS Genet. 2018;14:e1007845. doi: 10.1371/journal.pgen.1007845. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bonnin, E., Cabochette, P., Filosa, A., Juhlen, R., Komatsuzaki, S., Hezwani, M., Dickmanns, A., Martinelli, V., Vermeersch, M., Supply, L., et al. (2018). Biallelic mutations in nucleoporin NUP88 cause lethal fetal akinesia deformation sequence. PLoS Genet. 14, e1007845. [DOI] [PMC free article] [PubMed]
- 47.Singh R.R., Sedani S., Lim M., Wassmer E., Absoud M. RANBP2 mutation and acute necrotizing encephalopathy: 2 cases and a literature review of the expanding clinico-radiological phenotype. Eur. J. Paediatr. Neurol. 2015;19:106–113. doi: 10.1016/j.ejpn.2014.11.010. [DOI] [PubMed] [Google Scholar]; Singh, R.R., Sedani, S., Lim, M., Wassmer, E., and Absoud, M. (2015). RANBP2 mutation and acute necrotizing encephalopathy: 2 cases and a literature review of the expanding clinico-radiological phenotype. Eur. J. Paediatr. Neurol. 19, 106-113. [DOI] [PubMed]
- 48.Straussberg R., Shorer Z., Weitz R., Basel L., Kornreich L., Corie C.I., Harel L., Djaldetti R., Amir J. Familial infantile bilateral striatal necrosis: clinical features and response to biotin treatment. Neurology. 2002;59:983–989. doi: 10.1212/wnl.59.7.983. [DOI] [PubMed] [Google Scholar]; Straussberg, R., Shorer, Z., Weitz, R., Basel, L., Kornreich, L., Corie, C.I., Harel, L., Djaldetti, R., and Amir, J. (2002). Familial infantile bilateral striatal necrosis: clinical features and response to biotin treatment. Neurology 59, 983-989. [DOI] [PubMed]
- 49.Eftekharzadeh B., Daigle J.G., Kapinos L.E., Coyne A., Schiantarelli J., Carlomagno Y., Cook C., Miller S.J., Dujardin S., Amaral A.S. Tau Protein Disrupts Nucleocytoplasmic Transport in Alzheimer’s Disease. Neuron. 2018;99:925–940.e7. doi: 10.1016/j.neuron.2018.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]; Eftekharzadeh, B., Daigle, J.G., Kapinos, L.E., Coyne, A., Schiantarelli, J., Carlomagno, Y., Cook, C., Miller, S.J., Dujardin, S., Amaral, A.S., et al. (2018). Tau Protein Disrupts Nucleocytoplasmic Transport in Alzheimer’s Disease. Neuron 99, 925-940.e7. [DOI] [PMC free article] [PubMed]
- 50.Fox B.W., Tibbetts R.S. Neurodegeneration: Problems at the nuclear pore. Nature. 2015;525:36–37. doi: 10.1038/nature15208. [DOI] [PubMed] [Google Scholar]; Fox, B.W., and Tibbetts, R.S. (2015). Neurodegeneration: Problems at the nuclear pore. Nature 525, 36-37. [DOI] [PubMed]
- 51.Mikhaleva S., Lemke E.A. Beyond the Transport Function of Import Receptors: What’s All the FUS about? Cell. 2018;173:549–553. doi: 10.1016/j.cell.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mikhaleva, S., and Lemke, E.A. (2018). Beyond the Transport Function of Import Receptors: What’s All the FUS about? Cell 173, 549-553. [DOI] [PMC free article] [PubMed]
- 52.Fahrenkrog B., Harel A. Perturbations in Traffic: Aberrant Nucleocytoplasmic Transport at the Heart of Neurodegeneration. Cells. 2018;7:7. doi: 10.3390/cells7120232. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fahrenkrog, B., and Harel, A. (2018). Perturbations in Traffic: Aberrant Nucleocytoplasmic Transport at the Heart of Neurodegeneration. Cells 7, 7. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.