Abstract
Motile cilia and sperm flagella share an evolutionarily conserved axonemal structure. Their structural and/or functional defects are associated with primary ciliary dyskinesia (PCD), a genetic disease characterized by chronic respiratory-tract infections and in which most males are infertile due to asthenozoospermia. Among the well-characterized axonemal protein complexes, the outer dynein arms (ODAs), through ATPase activity of their heavy chains (HCs), play a major role for cilia and flagella beating. However, the contribution of the different HCs (γ−type: DNAH5 and DNAH8 and β−type: DNAH9, DNAH11, and DNAH17) in ODAs from both organelles is unknown. By analyzing five male individuals who consulted for isolated infertility and displayed a loss of ODAs in their sperm cells but not in their respiratory cells, we identified bi-allelic mutations in DNAH17. The isolated infertility phenotype prompted us to compare the protein composition of ODAs in the sperm and ciliary axonemes from control individuals. We show that DNAH17 and DNAH8, but not DNAH5, DNAH9, or DNAH11, colocalize with α-tubulin along the sperm axoneme, whereas the reverse picture is observed in respiratory cilia, thus explaining the phenotype restricted to sperm cells. We also demonstrate the loss of function associated with DNAH17 mutations in two unrelated individuals by performing immunoblot and immunofluorescence analyses on sperm cells; these analyses indicated the absence of DNAH17 and DNAH8, whereas DNAH2 and DNALI, two inner dynein arm components, were present. Overall, this study demonstrates that mutations in DNAH17 are responsible for isolated male infertility and provides information regarding ODA composition in human spermatozoa.
Keywords: DNAH17, dynein, ODA, axoneme, cilia, sperm flagellum, PCD, male infertility
Main Text
The axoneme is a highly evolutionarily conserved structure that corresponds to the microtubule-based cytoskeleton and is notably found in motile cilia and sperm flagella. This structure comprises an intricate network of protein complexes, and this network sustains the canonical conformation of nine doublets of microtubules (A and B) circularly arranged around a central pair of singlet microtubules (“9 + 2” pattern).1 Among the well-characterized axonemal protein complexes, the outer and inner dynein arms (ODAs and IDAs, respectively) play a major role in the beating of both cilia and sperm flagella.2 ODAs and IDAs are multiprotein ATPase complexes, which comprise heavy, intermediate, and light chains, and are attached to the A-microtubule of each peripheral doublet.3, 4 By anchoring to the B-microtubule of the adjacent doublet, ODAs and IDAs drive the sliding of the microtubule doublets and behave as indispensable molecular motors for the beating of those organelles.
In humans, structural and functional anomalies of motile cilia lead to primary ciliary dyskinesia (PCD [MIM: 244400]), a rare, autosomal-recessive disease characterized by chronic airway infections with bronchitis and rhinosinusitis and associated with situs inversus in half of cases (i.e., Kartagener syndrome).5, 6 At the ultrastructural level, the most frequent ciliary defects that are encountered in PCD-affected individuals concern the dynein arms (DAs) and principally the ODAs, as illustrated by the loss of ODAs or both ODAs and IDAs in about 70% of PCD-affected individuals.7, 8, 9 In human respiratory cells, ODAs are constituted of a globular head domain, which comprises β- and γ-type heavy chains (β-HCs and γ-HCs). This globular domain is associated with an intermediate domain, which comprises the DNAI1 (MIM: 604366) and DNAI2 (MIM: 605483) intermediate dynein chains. In addition, various light chains such as DNAL1 (MIM: 610062) and DNAL4 (MIM: 610565) are associated with the complex, together with a docking subunit, which anchors the ODAs to the microtubules.10, 11 Two types of ODAs have been reported in human respiratory cells, and they are classified on the basis of the composition of their dynein HCs: type 1 ODAs locate at the proximal part of the cilia and contain DNAH5 (γ-HC) (MIM: 603335) and DNAH11 (β-HC) (MIM: 603339), whereas type 2 ODAs locate at the distal part of the cilia and contain DNAH5 (γ-HC) and DNAH9 (β-HC) (MIM: 603330).12, 13
To date, most of the PCD-associated gene mutations affect the assembly, anchoring, or structure of the DAs.6, 7 As expected from the structural similarities between motile cilia and flagella, most males affected by PCD are infertile and display severe or total asthenozoospermia,14 defined by the reduction or the absence of motile sperm.15 Consequently, although the sperm phenotype of PCD-affected individuals is generally poorly investigated, many of the PCD-associated genes have been linked to male infertility.16 Hence, mutations in genes, such as DNAH5,12 DNAI1,17 DNAI218 and DNAH9,19 encoding ODA components were suggested to induce male infertility. Although motile cilia and sperm flagella definitely share a similar axonemal structure, those organelles might display some structural and/or protein content differences that could explain (1) why not all men with PCD have asthenozoospermia and (2), conversely, the existence of axonemal defects restricted to the sperm flagella. Here, we report the identification of mutations in DNAH17 (MIM: 610063), which encodes an axonemal heavy chain of the ODAs specifically expressed in the sperm cells; the DNAH17 mutations result in male infertility due to asthenozoospermia, but not in PCD.
We analyzed five male individuals who consulted for primary infertility and were enrolled in procedures of assisted reproduction technologies at the hospital of Lille, France (Centre Hospitalier Regional Universitaire de Lille). Individuals 18GM00285 (II-1 from family 1) and 18GM01926 (II-1 from family 4) originated from France, individuals 18GM01913 and 18GM01974 (II-3 and II-4, respectively, from family 2 GM03354) were two siblings from Algeria and born to a consanguineous union, and individual 18GM01932 (II-4 from family 3) originated from Morocco and was born to a consanguineous union. (Table 1). All five individuals had a normal somatic karyotype (46, XY), normal development of genital organs, and normal follicle-stimulating hormone (FSH) and testosterone levels but presented with asthenozoospermia, defined by less than 40% of sperm in the ejaculate being motile.15 In particular, the progressive motility was highly impacted because no, or only very few, progressive sperm were observed (between 0% and 5%; normal value >32%) (Table 1). All but one individual (18GM01932) showed a severe reduction of the number of spermatozoa with normal morphology compared to the normal value (between 2% to 12%; normal value >23%). Detailed sperm analysis indicated an increased number of spermatozoa with short, absent, irregularly shaped, and coiled flagella compared to the known distribution of these anomalies in fertile control individuals20 (Tables 1 and S1). The presence of such a mosaic of sperm anomalies in asthenozoospermic individuals was thoroughly described in the past and corresponds to a “short tails” phenotype,21, 22, 23 recently renamed “multiple morphological anomalies of the sperm flagellum” (MMAF).24 In addition to asthenozoospermia, which was a constant feature in all individuals, reduced sperm count or vitality was recorded for individuals 18GM00285 (II-1 from family 1) and 18GM01913 and 18GM01974 (II-3 and II-4, respectively, from family 2) (Table 1). Ultrastructure analysis of the spermatozoa was performed by transmission electron microscopy (TEM) for all five individuals. In keeping with descriptions of the so-called MMAF phenotype,21, 22, 23, 24, 25 we observed spermatozoa with an abnormal mitochondrial sheath and cytoplasmic bags containing unassembled flagellar components, together with severe axonemal disorganization, as illustrated by the absence of the central pair (“9 + 0” pattern) and outer microtubule doublets (Figure 1). In addition, on all analyzed axonemal transversal sections (including those with microtubule disorganization), the absence of the ODAs was reported (Table 1 and Figures 1 and 2). Importantly, although the absence of ODAs is a well-established ciliary defect associated with PCD, none of the five individuals displayed respiratory symptoms, nor situs inversus, according to clinical interrogations and examinations (Table 2). Furthermore, respiratory investigations (including thoracic and ear, nose, and throat (ENT) tomodensitometry; mucociliary clearance; and ciliary analysis), which were performed for both siblings from family GM03354 and the unrelated individual 18GM01932, clearly excluded cystic fibrosis and PCD (Table 2). Lastly, TEM analyses, completed on airway epithelial cells (AECs) collected by nasal brushing from individual 18GM01913 (II-3 from family 2, in Figure 3) and the unrelated individual 18GM01932 (II-4 from family 3, in Figure 3), showed a normal ultrastructure of the ciliary axoneme and, most notably, the presence of the ODAs in the cilia (Figure 2). Taken together, the above data led us to conclude that the five individuals displayed a non-PCD-associated form of asthenozoospermia due to specific loss of the ODAs in the sperm cells only. We subsequently intended to identify the genetic causes responsible for this phenotype. To this end, a signed informed consent was first obtained from all individuals to be included in the study according to the local protocols and the principles of the Declaration of Helsinki. The study is approved by the institutional review board of the French Institute of Medical Research and Health (CEEI-IRB: opinion number 15-259) and the Comité de Protection des Personnes CPP Ile de France III (record number CPP02748).
Table 1.
Phenotypic Features and Semen Characteristics of Individuals with Identified DNAH17 Mutations
| Family (origin) | Individuals | Known consanguinity | Gender (age) | Fertility | Sperm TEM |
Semen Characteristics |
Flagellar Abnormalities |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pH | Volume (ml) | Total sperm count (106) | Total motility | Progressive motility | Vitality | Absent | Short | Irregular caliber | Coiled | Multiple | Typical forms | ||||||
| GM02359 (France/Italy) | 18GM00285 | no | male (36 y) | Infertilitya | ODA loss (100%) | 8.1 | 2.5 | 20∗ | 10∗ | 1∗ | 72 | 5 | 2 | 36∗ | 27∗ | 1 | 11∗ |
| GM03354(Algeria) | 18GM01974 | yes | male (35 y) | Infertility a | ODA loss (100%) | 8 | 3.5 | 119 | 1∗ | 0∗ | 51∗ | 12∗ | 34∗ | 16∗ | 38∗ | 2∗ | 2∗ |
| 18GM01913 | yes | male (37 y) | Infertility a | ODA loss (100%) | 8 | 3.8 | 27.7∗ | 1∗ | 0∗ | 61 | 16∗ | 34∗ | 14∗ | 38∗ | 0 | 4∗ | |
| GM03364 (Morocco) | 18GM01932 | yes | male (27 y) | Infertility a | ODA loss (100%) | 7.8 | 4.5 | 88.6 | 25∗ | 5∗ | 76 | 2 | 1 | 6∗ | 18∗ | 2∗ | 22 |
| GM03360 (France) | 18GM01926 | no | male (30 y) | Infertility | ODA loss (100%) | 7.7 | 6 | 198 | 20∗ | 2∗ | 60 | 2 | 2∗ | 12∗ | 22∗ | 0 | 12∗ |
| Reference values for semen analysis | 7.2 | 1.5 | 39 | 40 | 32 | 58 | 5 | 1 | 2 | 17 | 1 | 23 | |||||
Values are percentages unless specified otherwise. Lower reference limits for semen analysis are provided according to the World Health Organization (WHO) standards15 and the distribution range of morphologically normal spermatozoa observed in 926 fertile individuals.20
Individual 18GM00285: II-1 from family 1; Individuals 18GM01913 and 18GM01974: II-3 and II-4, respectively, from family 2; Individual 18GM01932: II-4 from family 3; Individual 18GM01926: II-1 from family 4.
ICSI failure (1–3 attempts).
indicates abnormal values.
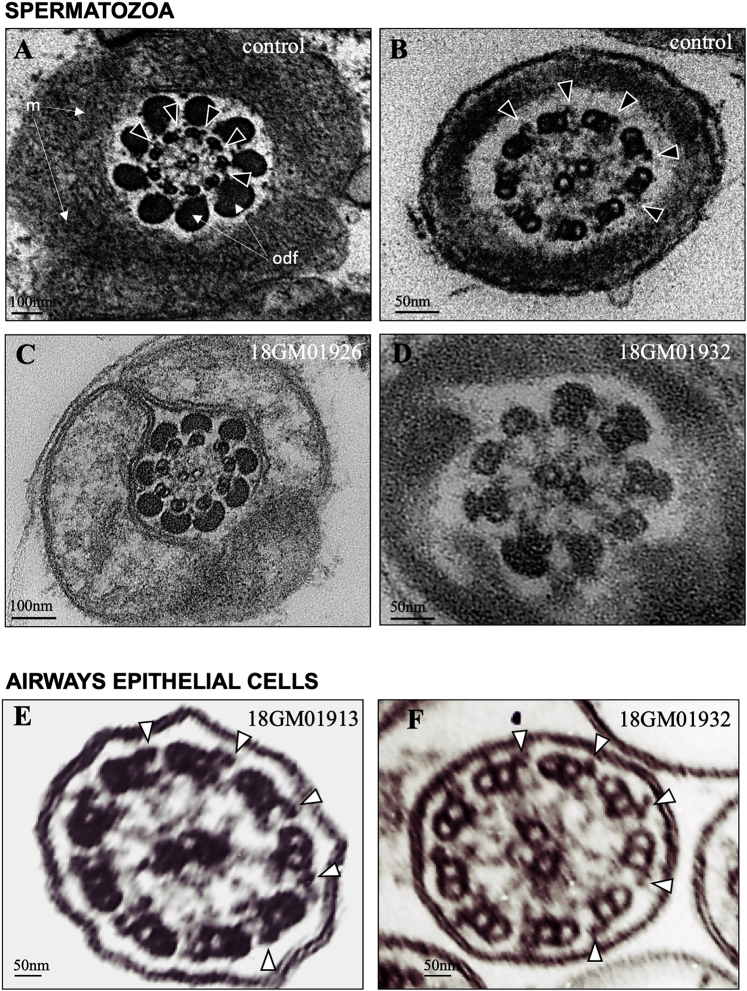
Figure 1.
TEM Analysis of Spermatozoa from Individuals 18GM01913, 18GM01932, and 18GM01926 Who Have an MMAF Phenotype
(A) Left: a longitudinal section of control spermatozoa with the head, comprising the nucleus (N) and acrosome (Ac). The flagellum extends from the head and comprises the axoneme (Ax), which is surrounded by mitochondria (M) in the midpiece, and the fibrous sheath (FS) in the principal piece. Right inset: a transversal section of control spermatozoa in the principal piece region. The axoneme comprises nine doublets of microtubules (MTD) circularly arranged around a central pair (CP) of microtubules. A few other peri-axonemal structures, such as the outer dense fibers (ODF) and the longitudinal columns (LC), are visible.
(B and C) Longitudinal sections of spermatozoa from individuals 18GM01913 (II-3 from family 2, in Figure 3) and 18GM01932 (II-4 from family 3, in Figure 3) showing cytoplasmic bags with unassembled axonemal and peri-axonemal components in due place of the flagellum.
(D and E) Transversal sections of spermatozoa from individuals 18GM01926 (II-1 from family 4, in Figure 3) and 18GM01932 (II-4 from family 3, in Figure 3) showing the lack of the central pair (9 + 0 pattern) and the lack of outer microtubule doublets. The scale bars represent 2 μm (A, B, and C) or 50 nm (D and E). Human sperm cells were processed for transmission electron microscopy (TEM) analysis as previously described.51
Figure 2.
Comparative Transmission Electron Microscopy Analysis of Spermatozoa and Airway Cilia from Individuals 18GM01913, 18GM01926, and 18GM01932
Upper panel: spermatozoa from a control individual and from individuals 18GM01926 (II-1 from family 4, in Figure 3) and 18GM01932 (II-4 from family 3, in Figure 3).
(A and C) Axonemal sections from the sperm midpiece showing mitochondria and outer dense fibers (ODF). Outer dynein arms (ODAs) are indicated by white arrow heads in the control and are absent in individual 18GM01926 (II-1 from family 4, in figure 3).
(B and D) Axonemal sections from the sperm principal piece. Outer dynein arms (ODAs) are indicated by arrow heads in the control and are absent in individual 18GM01932.
Lower panel: Cilia from airway epithelial cells (AECs) from individuals 18GM01913 (II-3 from family 2, in Figure 3) and 18GM01932.
(E and F) Axonemal sections from AEC cilia showing the presence of ODAs, indicated by arrow heads. The scale bars represent in the upper panel 100 nm (A and C) or 50 nm (B and D); and in lower panel 50 nm (E and F). Human sperm cells and a nasal biopsy were processed for TEM analysis as previously described.51, 52
Table 2.
Respiratory Phenotypic Features of Individuals with Identified DNAH17 Mutations
| Individuals | Airway disease | SI | Cilia TEM | Other Explorations |
|---|---|---|---|---|
| 18GM00285 | no | no | NA | NA |
| 18GM01974 | no | no | NA | normal thoracic/ENT TDM, normal mucociliary clearance |
| 18GM01913 | no | no | normal (ODA present) | normal thoracic/ENT TDM, normal mucociliary clearance |
| 18GM01932 | no | no | normal (ODA present) | normal ciliary beating, normal thoracic/ENT TDM, normal mucociliary clearance |
| 18GM01926 | no | no | NA | NA |
Abbreviations are as follows: SI = situs inversus; TEM = transmission electronic microscopy; NA = not available; ENT: ears, nose, and throat; and TDM: tomodensitometry.
Individuals 18GM00285: II-1 from family 1; 18GM01913: II-3 from family 2; 18GM01974: II-4 from family 2; 18GM01932: II-4 from family 3; 18GM01926: II-1 from family 4.
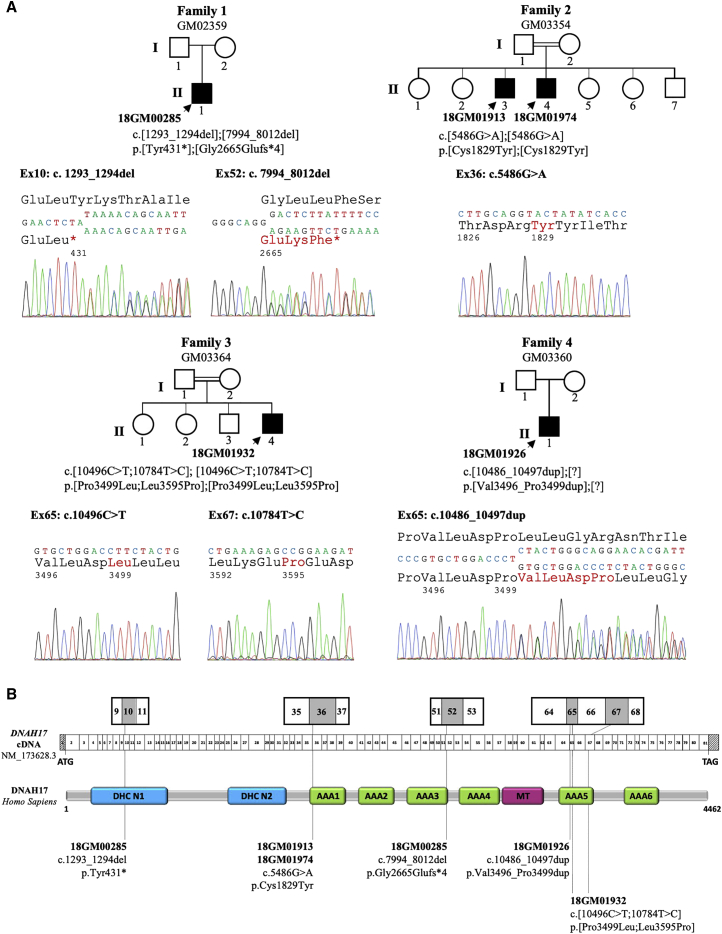
Figure 3.
Identification of DNAH17 Mutations in Five Infertile Males from Four Unrelated Families
(A) Pedigrees of the four unrelated families and Sanger sequencing data of the mutations identified in infertile individuals (solid symbols): 18GM00285 (II-1 from family 1), 18GM01913 and 18GM01974 (II-3 and II-4, respectively, from family 2), 18GM01932 (II-4 from family 3), and 18GM01926 (II-1 from family 4). The double line indicates consanguinity between parents.
(B) A schematic representation of DNAH17 exon structure (top) and predicted protein domains (bottom) according to the SMART webtool and Uniprot; the positions of the six different mutations identified in the five infertile individuals are marked. All mutations are declared and accessible in the LOVD public variation database with the following identification numbers: 0000473833 (c.1293_1294del [p.Tyr431∗]); 0000473834 (c.7994_8012del [p.Gly2665Glufs∗4]); 0000473835 (c.5486G>A [p.Cys1829Tyr]); 0000473837 (c.10496C>T [p.Pro3499Leu]); 0000473838 (c.10784T>C [p.Leu3595Pro]); and 0000473840 (c.10486_10497dup [p.Val3496_Pro3499dup]). DHC N1 and N2 (blue boxes) are conserved domains in the stem of dynein heavy chains. AAA1 to AAA6 (green boxes) indicate ATPase domains (ATPases associated with diverse cellular activities; AAAs) that are arranged in a ring-shaped molecular motor and couple ATP binding and hydrolysis to conformational changes. The microtubule-binding region (MT, red box) is made of a coiled-coil stalk with a globular tip that binds to the adjacent B microtubule.
The search for gene mutations underlying the absence of ODAs was performed in the four probands: 18GM00285, 18GM01974, 18GM01932, and 18GM01926 (II-1 from family 1, II-4 from family 2, II-4 from family 3, and II-1 from family 4, respectively, in Figure 3) by a targeted capture panel (SeqCap EZ Choice, Roche) and parallel sequencing on a MiSeq system (Illumina) with genomic DNA extracted from EDTA blood samples. The targeted panel analyzed the so-far 40 PCD-associated genes, including DNAH5, DNAH11, and DNAH9, which are associated with the absence of ODAs, and DNAH1 (MIM: 603332), a gene that was associated with male infertility due to the absence of IDAs in the sperm cells. In addition, we sequenced 250 PCD candidate genes, including the seven other axonemal dynein heavy chains: DNAH2 (MIM: 603333), DNAH3 (MIM: 603334), DNAH6 (MIM: 603336), DNAH7 (MIM: 610061), DNAH8 (MIM: 603337), DNAH10 (MIM: 605884), DNAH12 (MIM: 603340), DNAH14 (MIM: 603341), and DNAH17. The enrichment probes targeted coding and non-coding exons and their flanking intronic regions; large indels were searched by depth-ratio analysis. Raw data was computed through two independent pipelines as previously described.26 Confirmation and analysis in the relatives from the four families was performed by Sanger sequencing (Figure 3A). We identified bi-allelic variants in DNAH17, a gene not previously associated with PCD, in three probands and one brother: 18GM00285 (II-1 from family 1), 18GM01913 and 18GM01974 (II-3 and II-4, respectively, from family 2), and 18GM01932 (II-4 from family 3). In addition, a heterozygous in-frame duplication in DNAH17 was identified in the proband 18GM01926 (II-1 from family 4). Importantly, all individuals born to consanguineous unions were shown to carry the identified DNAH17 variations in the homozygous state (Figure 3A). DNAH17 (GenBank: NM_173628; Ensembl: ENSG00000187775) is located on chromosome 17 and contains 81 exons (transcript GenBank: NM_173628.3), predicting a 4,462-amino-acid protein (NCBI: NP_775899.3; UniProtKB: DYH17_Human Q9UFH2). The DNAH17 protein comprises nine principal domains: the DHC N1 and N2 conserved domains located in the stem of the dynein heavy chains and six ATPase domains (namely “ATPase associated with diverse cellular activities” domains; AAA domains) arranged in a ring-shaped molecular motor, which couples ATP binding and hydrolysis to conformational changes; in addition, the microtubule-binding region (MT) contains a coiled-coil stalk with a globular tip that binds to the B-tubule of the peripheral microtubule doublets (Figure 3B).
Individual 18GM00285 (II-1 from family 1, in Figure 3) carried two heterozygous DNAH17 pathogenic mutations leading to a premature stop codon; these mutations were: a two-nucleotide deletion, c.1293_1294del (p.Tyr431∗), and a 19-nucleotide deletion, c.7994_8012del (p.Gly2665Glufs∗4). The c.1293_1294del mutation, referenced as dbSNP: rs767723684, was present at a very low prevalence in the general population: 8.741 × 10−6 (2 out of 228,814 alleles in gnomAD). The c.7994_8012del deletion was absent from the gnomAD database. Both mutations introduce a premature stop codon (positions 431 and 2,665, respectively, in the 4,462 amino acid DNAH17 protein sequence) and are therefore expected to induce nonsense-mediated mRNA decay that is likely to prevent protein production. The two siblings 18GM01913 and 18GM01974 from family 2 (II-3 and II-4, respectively, in Figure 3) were found to be homozygous for the c.5486G>A (p.Cys1829Tyr) missense variation. A DNA sample from their father was not available, but their mother was heterozygous for the variant, and their brother, who fathered two children naturally, did not carry the variant. The p.Cys1829Tyr variation is not described in gnomAD; it does not seem prone to alter splicing because it only slightly weakens the MaxEntScan score of the intron 35 acceptor splice site (10.76 versus 11.03 for the normal site). Importantly, cysteine 1,829 is highly conserved throughout evolution and conserved in 12 out the 13 axonemal dyneins (Figure S1). This residue lies 18 amino acids from the ATPase catalytic site of the AAA1 domain, which, among the six AAA domains, is the only one that retained an ATP hydrolyze activity. Altogether, these elements suggest that the p.Cys1829Tyr missense variation is likely to disrupt the stability and/or the function of the protein. Individual 18GM01932 (II-4 from family 3, in Figure 3) was homozygous for a complex allele and had two distinct missense variations, c.[10496C>T;10784T>C] (p.[Pro3499Leu;Leu3595Pro]), located in the AAA5 domain. None of these variations are described in gnomAD, and they do not create donor or acceptor splice sites (MaxEntScan tool). These two variations involve a proline residue, the most effective amino acid in disrupting regular secondary structure elements,27 and they are thus expected to strongly alter the protein structure by occasioning the anomalous presence or absence of a proline residue in the protein sequence. In addition, proline 3,499 and leucine 3,595 are conserved and highly conserved, respectively, throughout evolution; Leu3595 is also conserved among the 13 axonemal dyneins (Figure S1). Lastly, individual 18GM01926 (II-1 from family 4, in Figure 3) carried a heterozygous 12-nucleotide in-frame duplication, c.10486_10497dup (p.Val3496_Pro3499dup), and no other molecular defect was identified. This variant, which is not reported in gnomAD, does not create any donor or acceptor splice site within exon 65 (MaxEntScan); it is predicted to introduce a motif (valine-leucine-asparte-proline) within the AAA5 domain, and this may also alter the secondary structure of the region because of the introduction of a proline residue. In total, this molecular screening led to the identification of five DNAH17 bi-allelic variations, considered to be highly likely to be pathogenic, in four individuals and one heterozygous, probably pathogenic variant in the fifth individual. Given (1) the nature of the predicted protein encoded by DNAH17, (2) the above in silico data supporting the pathogenicity of all variants identified in DNAH17, (3) the familial segregation of the variants in families GM03354 and GM03364, and (4) the absence of other predicted pathogenic variants in genes encoding known components of dynein arms or in genes expressed in the testis and/or related to cilia and/or flagella, we focused the subsequent studies on DNAH17, which appeared to be the best candidate for explaining the isolated primary male infertility observed in the five individuals.
To date, very few data on ODA composition of mammalian spermatozoa have been reported in the literature. Therefore, we first intended to better define the expression profile of the established human genes, namely DNAH5, DNAH8, DNAH9, DNAH11 and DNAH17, encoding the ODA heavy chains. First, by comparing the distribution of ODA heavy chains in tissue expression databases (EMBL-EBI Expression Atlas) and performing quantitative RT-PCR analyses on human adult tissues (Figure S2), we observed that DNAH8 and DNAH17 were clearly expressed in the testis and less abundant in the lung, whereas DNAH5, DNAH9, and DNAH11 were the most abundant heavy chains detected in the lung. In order to distinguish between somatic and germline expression and to specify the stage of differentiation of the germ cells, we analyzed available quantitative single-cell RNA sequencing datasets from the human adult testis (ReproGenomics Viewer).28, 29, 30 In keeping with the expression pattern of the genes involved in spermiogenesis, DNAH8 and DNAH17 expression was plainly detected in the germ cells from the early spermatocyte to late spermatid stages but not in the somatic cells (Leydig, Sertoli cells) (Figure S3A). DNAH9 was also detected in the germ cells but with a shorter window of expression because the onset was observed in the late spermatocyte and early spermatid stages. In agreement with their tissue distribution profile, the expression levels of DNAH5 and DNAH11 were very low in both the somatic and germ cell lineages of the testis. We next analyzed quantitative proteomic data available in the literature, and these indicated that DNAH8 and DNAH17 were both categorically detectable in human sperm proteome analyses,31, 32 whereas their amounts were found to be very low in the human airway cilia proteome33 (Figure S3B). Conversely, DNAH5, DNAH9, and DNAH11 were not significantly detected in the human sperm flagellar proteome (or only with a limited number of unique peptides identified by mass spectrometry), whereas they were strongly detected in the human ciliary proteome and were even part of the most abundant ciliary proteins. Taken together, the above data indicate that DNAH8, DNAH9, and DNAH17 transcripts are expressed in the germ cells during spermiogenesis. However, only DNAH8 and DNAH17 are detected by proteomic analysis in mature spermatozoa.
In order to further confirm these data, we analyzed the presence and location of all ODA heavy chain proteins in AECs and spermatozoa from control individuals by performing immunoblot and immunofluorescence analyses. By means of an immunoblot that used antibodies directed against DNAH5, DNAH8, DNAH9, DNAH11, and DNAH17, we first show that DNAH8 and DNAH17 are detected at the expected molecular weights in sperm cells but not in AECs, whereas DNAH5, DNAH9, and DNAH11 are only detected in AECs (Figure 4A). Next, by means of immunofluorescence with co-staining of the axoneme with anti α-tubulin antibody, we confirm the above results. Hence, as previously described in AECs,12, 13 DNAH5 was detected along the full length of the cilia, whereas DNAH9 and DNAH11 distributions were restricted to the distal and proximal part of the cilia, respectively. It is noteworthy that we observed that neither DNAH8 nor DNAH17 were detected in cilia from AECs obtained from controls (Figure 4B). The opposite pictures were observed in spermatozoa from control individuals, where only DNAH8 and DNAH17 could be co-detected with the α-tubulin along the sperm flagellum, with the exception of the terminal piece, which is known to lack complete axonemal structure (Figures 4B, S4, and S5). Taken together, these data indicate that the sperm cells harbor a set of ODA heavy chains (i.e., DNAH8 and DNAH17), and this set is distinct from that (i.e., DNAH5, DNAH9, and DNAH11) found in respiratory ciliated cells. Such a distribution pattern is consistent with the absence of ODAs in individuals carrying DNAH17 mutations, which we found restricted to the sperm cells.
Figure 4.
Outer Dynein Arm Heavy Chain Protein Detection and Localization in Human Sperm Cells and Airway Epithelial Cells from Healthy Control Individuals
(A) Detection of DNAH8, DNAH9, and DNAH17 by immunoblot in spermatozoa (Spz1 and Spz2) and airway epithelial cells (AECs) (AEC1 and AEC2) from two control individuals; DNAI1 was used for loading control. AECs and spermatozoa, obtained by nasal brushing and masturbation, respectively, were suspended in lysis buffer (50 mM Tris, 150 mM NaCl, 1% Triton X-100, and complete protease inhibitor cocktail, Roche Diagnostics). Lysates were sonicated and cleared by centrifugation. Equal amounts of cells were loaded on 4%–12% gradient SDS-PAGE and transferred overnight on a PVDF membrane. The primary antibodies (SIGMA DNAH8 HPA028447, DNAH9 HPA052641, DNAH17 HPA024354, and DNAI1 HPA021649) were incubated overnight at 4°C. A secondary antibody coupled to HRP (SIGMA rabbit HRP A0545) and Amersham ECL Select Western Blotting Detection Reagent were used for detection.
(B) Immunofluorescence staining of AECs and spermatozoa from a control with ODA heavy chain antibody (in red) and α-tubulin (in green). Cells were counterstained with DAPI (blue) as a nuclei marker. The scale bars represent 5 μm. AECs obtained by airway brushing were cytocentrifugated onto glass slides and fixed for 15 min with 4% paraformaldehyde. Spermatozoa were smeared onto glass slides and fixed for 10 min with 4% paraformaldehyde and then epitope retrieval in citrate buffer (pH 6) was performed. Cells were permeabilized in Triton X-100 for 10 min (0.1% for AECs and 0.2% for spermatozoa) and labeled with rabbit polyclonal antibody directed against ODA heavy chains DNAH5 (SIGMA HPA037470), DNAH8 (SIGMA HPA028447), DNAH9 (SIGMA HPA052641), DNAH11 (SIGMA HPA045880), and DNAH17 (SIGMA HPA024354) and were co-stained with acetylated α-tubulin (mouse monoclonal Abcam ab24610) for AECs and α-tubulin (mouse monoclonal SIGMA DM1A T9026) for spermatozoa.
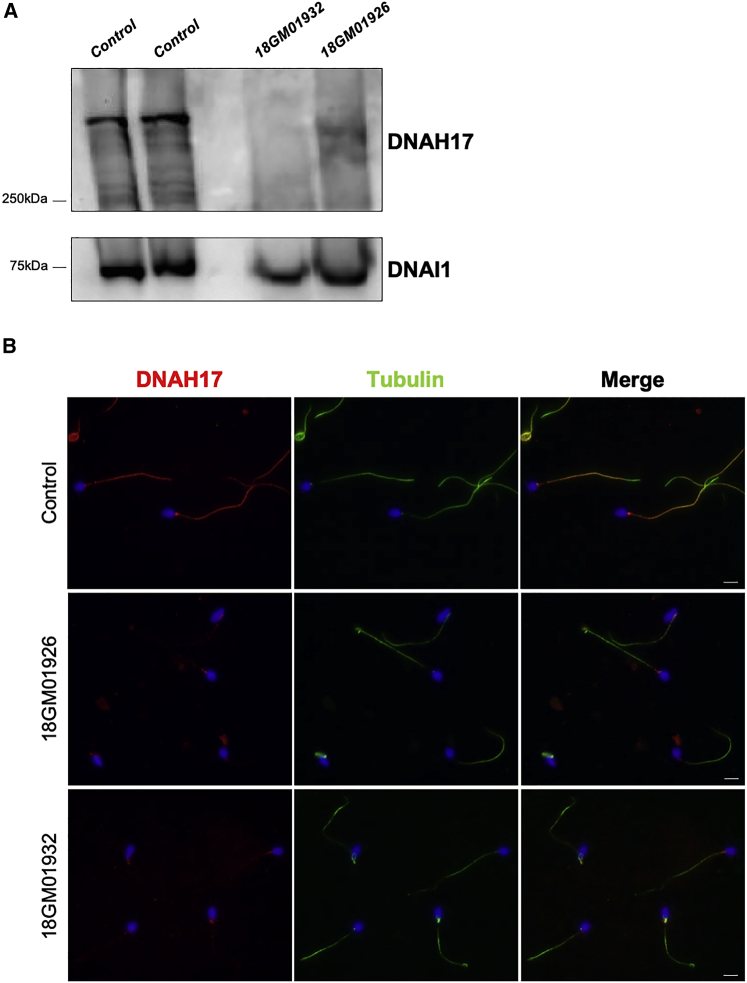
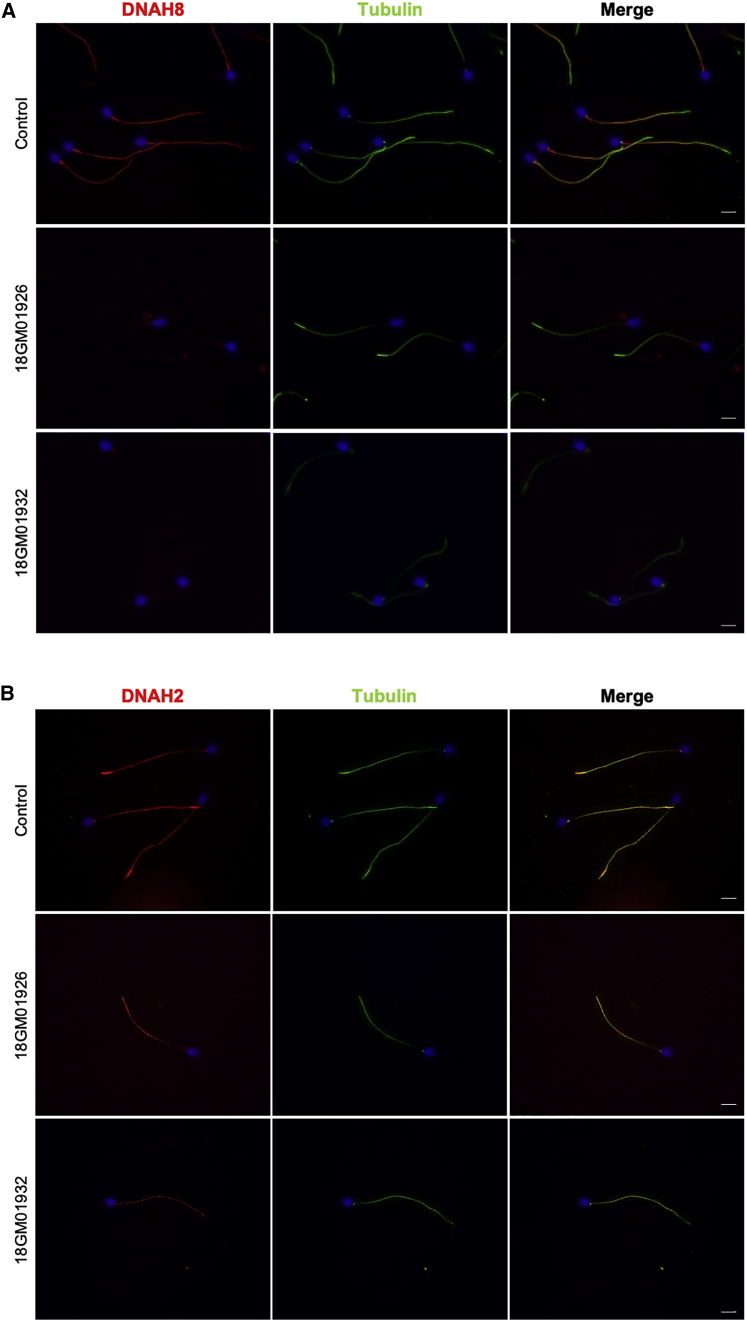
To confirm the pathogenicity of the identified DNAH17 mutations, we analyzed the sperm protein content of individuals 18GM01932 (II-4 from family 3, in Figure 3) and 18GM01926 (II-1 from family 4, in Figure 3). When performing immunoblot analysis on equal numbers of spermatozoa, DNAH17 was almost undetectable in sperm from those two individuals who carry DNAH17 mutations, whereas DNAI1 was present, although slightly diminished, in affected individuals compared to in controls (Figure 5A). The absence of DNAH17 in sperm from these two individuals was also confirmed by immunofluorescence studies; the DNAH17 signal was absent all along the flagellum and only small dots were observed in the proximal region of the flagellum, whereas α-tubulin staining was readily detectable (Figure 5B). We next analyzed other components of ODAs and IDAs to better assess the impact of DNAH17 mutations in axonemal protein complexes, structure, and assembly. Immunofluorescence studies performed on sperm cells from individuals 18GM01932 (II-4 from family 3, in Figure 3) and 18GM01926 (II-1 from family 4, in Figure 3) indicated that the heavy chain DNAH8 was totally absent from the sperm axoneme, thus further confirming the absence of ODAs shown by TEM (Figure 6A). As previously observed by immunoblot analysis (Figure 5A), DNAI1, an intermediate chain of the ODAs, was detected by immunofluorescence in sperm from individuals 18GM01932 (II-4 from family 3, in Figure 3) and 18GM01926 (II-1 from family 4, in Figure 3) but in reduced amount, suggesting that part of the ODA may subsist but not be detectable by TEM (Figure S5A). Importantly, DNAH2, and DNALI1, corresponding to a heavy and a light intermediate chain of the IDA, respectively, were both clearly detected in sperm from both individuals (Figures 6B and S5B), indicating that the IDAs are not impacted by DNAH17 mutations.
Figure 5.
DNAH17 Protein Amount and Localization in Spermatozoa from Mutation Carriers Individuals 18GM01932 and 18GM01926
(A) Detection of DNAH17 by immunoblot in spermatozoa from two controls and from individuals 18GM01932 (II-4 from family 3, in Figure 3) and 18GM01926 (II-1 from family 4, in Figure 3). DNAI1 was used as positive control. Spermatozoa were suspended in lysis buffer (50 mM Tris, 150 mM NaCl, 1% Triton X-100, and complete protease inhibitor cocktail, Roche Diagnostics). Lysates were sonicated and cleared by centrifugation. The same amount of cells was loaded on a 4%–12% gradient SDS-PAGE gel that was transferred overnight on a PVDF membrane. The membrane was incubated overnight at 4°C with primary antibodies (SIGMA DNAH17 HPA024354 and DNAI1 HPA021649). After incubation with the secondary antibody (rabbit HRP SIGMA A0545), proteins were detected with Amersham ECL Select Western Blotting Detection Reagent.
(B) Immunofluorescence staining of spermatozoa from a control individual and from individuals 18GM01926 (II-1 from family 4, in Figure 3) and18GM01932 (II-4 from family 3, in Figure 3) with DNAH17 (in red) and α-tubulin (in green). Spermatozoa were counterstained with DAPI (blue) as a nuclei marker. The scale bars represent 5 μm. Spermatozoa were smeared onto glass slides and fixed for 10 min with 4% paraformaldehyde before epitope retrieval in citrate buffer (pH 6). Spermatozoa were then permeabilized in Triton X-100, 0.2% for 10 min and blocked for 1 h with 1% bovine serum albumin (BSA). Finally, cells were labeled with rabbit polyclonal antibody directed against DNAH17 (SIGMA HPA024354) and α-tubulin (mouse monoclonal DM1A SIGMA T9026), then a secondary antibody incubation (anti-rabbit Alexa 568 and anti-mouse Alexa 488, Invitrogen) and mounting in DAPI Vectashield (Vector) were performed.
Figure 6.
Immunostaining of DNAH8 and DNAH2 in Spermatozoa from a Healthy Control and Individuals 18GM01926 and 18GM01932
(A) Immunofluorescence staining of spermatozoa from a control and from individuals 18GM01926 (II-1 from family 4, in Figure 3) and 18GM01932 (II-4 from family 3, in Figure 3) with DNAH8 (γ-HC ODA, in red) and α-tubulin (in green).
(B) Immunofluorescence of spermatozoa from a control and from individuals 18GM01926 (II-1 from family 4, in Figure 3) and 18GM01932 (II-4 from family 3, in Figure 3) with DNAH2 (a heavy chain of inner dynein arms [IDAs], in red) and α-tubulin (in green). Spermatozoa were counterstained with DAPI (blue) as a nuclei marker. The scale bars represent 5 μm. Spermatozoa were smeared onto glass slides and fixed for 10 min with 4% paraformaldehyde before epitope retrieval in citrate buffer (pH 6). Spermatozoa were then permeabilized in Triton X-100, 0.2% for 10 min and blocked for 1 h with 1% bovine serum albumin (BSA). Finally, cells were labeled with rabbit polyclonal antibody directed against DNAH8 (SIGMA HPA028447) or DNAH2 (SIGMA HPA067103) and α-tubulin (mouse monoclonal DM1A SIGMA T9026), and then secondary antibody incubation (anti-rabbit Alexa 568 and anti-mouse Alexa 488, Invitrogen) and mounting in DAPI Vectashield (Vector) were performed.
In conclusion, we identified DNAH17 mutations, which we demonstrate to be associated with a specific loss of the ODAs in the sperm cells and recessive male infertility but not PCD. Four of the mutations were identified in the homozygous state and one in the heterozygous state. Immunoblot and immunofluorescence detection in sperm from individual 18GM01932, who carried the two homozygous missense mutations p.Leu3595Pro and p.Pro3499Leu, showed the absence of DNAH17 compared to controls, strongly supporting that the mutations, which incriminate a proline residue, alter DNAH17 protein stability. To date, no crystal structure from an axonemal dynein heavy chain is available. In order to retrieve more information about the impact of some of the DNAH17 mutations that we identified on conserved residues among dynein heavy chain proteins, we used the available crystal structure of the closest human cytoplasmic heavy chain, DYNC1H1 (MIM: 600112) (PDB: 5NUG). Figure S7 presents the crystal structure of DYNC1H1 with the location of the three equivalent amino-acids targeted by the p.Cys1829Tyr, p.Pro3499Leu, and p.Leu3595Pro DNAH17 mutations. From this representation, the Cys1829 residue (1,888 in DYNC1H1) appears close to the ADP binding site, whereas the proline 3,499 (3,647 in DYNC1H1) residue is located at the end of an alpha-helix; the consequences of its change to a leucine residue are difficult to predict. The Leu3595 residue (3,745 in DYNC1H1) is located within an alpha-helix, and its mutation to a proline residue seems prone to alter this secondary structure (Figure S7). Taken together, this information retrieved from the comparison with the crystal structure of the cytosolic DYNC1H1 supports the potential pathogenic effects of the p.Cys1829Tyr and p.Leu3595Pro mutations.
Importantly, although a second molecular defect was not identified in individual 18GM01926 (II-1 from family 4, in Figure 3), DNAH17 was barely detectable in sperm from this individual by immunoblot or immunofluorescence. The absence of DNAH17 in sperm from individual 18GM01926 could be explained by (1) a mutation in the non-coding regions by a second DNAH17 allele, (2) a mutation in DNAH8 because herein, conversely, we show DNAH17 mutations are associated with the loss of both DNAH17 and DNAH8 proteins, or (3) a dominant-negative effect of the identified heterozygous DNAH17 mutation. Unfortunately, DNA samples from parents of individual 18GM01926 were not available, preventing us from directly testing some of the above hypotheses. In addition, the limited quantity of biological samples from individual 18GM01926 did not allow us to perform RT-PCR analyses. Nevertheless, from our genetic analyses, we can formally exclude the hypothesis of mutations in DNAH8 because no pathogenic variants were identified for individual 18GM01926, nor for other individuals reported in this study (Table S2). Considering that the mutation carried by individual 18GM01926 is an in-frame duplication of four amino acids and contains a proline residue, it is possible that the mutated protein acts through a dominant-negative effect; a modification of the secondary structure could induce instability and degradation of the mutated protein together with that of associated proteins (i.e., the wild-type DNAH17 encoded by the second allele and DNAH8). This would imply dimerization and/or physical interaction of DNAH8 and DNAH17, but importantly, also a transmission of the mutation by the mother or a neo-mutation in the proband; unfortunately, as mentioned above, we could not test this hypothesis. Alternatively, the phenotype of individual 18GM01926 and the absence of DNAH17 could be attributed to a deep intronic mutation or a deletion in the regulatory sequences of DNAH17, but such molecular defects were not evidenced by analyses of the coding and flanking splice-site sequences. Because, to date, PCD-associated mutations in dynein genes were associated with an autosomal-recessive mode of transmission, we much more favor this later hypothesis.
Taken together, these genetic findings indicate the importance of DNAH17 in the composition of sperm ODAs, an observation that is supported by its expression profile. Hence, by characterizing the expression profile of ODA heavy chains, we propose a model in which the sperm flagellum is composed of only one type of ODA, containing DNAH8 (γ-HC) and DNAH17 (β-HC), which both localize all along the axoneme (Figure 7). In this model, we exclude any contribution of the ciliary DNAH5 (γ-HC) and DNAH11 (β-HC) proteins. Furthermore, no restricted localizations of DNAH8 nor DNAH17 were observed along the flagellum, in contrast to respiratory cilia12, 13 (Figure 7). Surprisingly, DNAH9 transcripts were highly detected in germ cells by single-cell RNA sequencing,30 but the protein was not reported in mass spectrometry analyses, and we did not detect DNAH9 in human spermatozoa by immunofluorescence nor by immunoblot assays, in contrast to a previous report based on immunofluorescence.12 Considering the high amounts of transcripts detected during the last stage of spermiogenesis, DNAH9 (β-HC) might fulfill a transient function in the spermatid cells, and the protein amount, if present in mature spermatozoa, might be very low.
Figure 7.
Schematic Model of Outer Dynein Arm Heavy Chain Distribution in Human Airway Epithelial Cell and Sperm Flagellum Axonemes
Longitudinal view of the axoneme from airway epithelial cell (AEC) cilia and a sperm flagellum illustrating outer dynein arm (ODA) heavy chain composition. In respiratory cilia, γ-heavy chains are composed of DNAH5 (in orange), which locates along the full length of the axoneme. The β-heavy chains (in blue) are composed of DNAH11 (light blue) in the proximal part and DNAH9 (blue) in the distal part of the axoneme. In spermatozoa, we propose a model in which ODAs are exclusively composed by the γ-HC DNAH8 and the β-HC DNAH17, both of which localize all along the axoneme.
Although the loss of the ODAs in sperm cells from all individuals carrying mutations in DNAH17 was highlighted by TEM analyses, we observed by immunoblot and immunofluorescence analyses that DNAH17 and DNAH8 were both absent, but DNAI1 was present, although its amount was found to be reduced. This suggests that the mutant DNAH17 proteins might not be assembled into the axoneme and could be eliminated in the residual bodies during spermiogenesis or subjected to degradation if not properly folded. In either of these two situations, DNAH8, the expected (γ-HC)-associated chain within the globular domain of the ODA, faces the same fate. The presence of DNAI1 in the sperm flagellum of individuals carrying mutations in DNAH17 suggests that the entirety of the ODAs might not be lost and that the intermediate domain of the ODAs might succeed in being assembled or stabilized in the absence of the heavy chains, as was recently demonstrated in the case of DNAH9 mutations.19
To date, about 40 genes associated with PCD have been identified,7 and their mutations affect most often the dyneins themselves or the pre-assembly, addressing, or anchoring of the arms. Hence, mutations in DNAH5,34, 35 DNAI1,36 DNAI2,18 DNAL1,37 ARMC4 (MIM: 615408),38 CCDC114 (MIM: 615038),39, 40 and CCDC151 (MIM: 615956)41 were identified in several PCD-affected individuals with an absence of ODAs. Mutations in DNAH9 were also identified but do not affect all ODAs due to the restricted distribution of DNAH9 to the distal part of the cilia, and these mutations result in a PCD phenotype with moderate symptoms.19, 42 Finally, DNAH11 mutations were associated with PCD without ultrastructural defects.43, 44, 45 Although the sperm phenotypes were poorly investigated in these studies, mutations in some of the above dynein genes (DNAH5,12 DNAI1,17 DNAI218 and DNAH919) were also suggested to be responsible for male infertility. Herein, we demonstrate that mutations in DNAH17 are not associated with PCD but are exclusively responsible for male infertility. So far, this situation was only described once in the literature, in the case of mutations in DNAH1, which encodes an IDA dynein HC.24, 46, 47, 48 Similarly to what we observed in the case of DNAH17 mutations, previous work identified DNAH1 mutations that are associated with severe asthenozoospermia due to an MMAF phenotype and the loss of IDAs.24 In both cases, the sperm-restricted phenotype is explainable by nearly specific or preferential gene expression in the testis and sperm cell precursors. Despite this highly preferential tissue distribution, it can be difficult to state that mutations in those genes result in isolated infertility and to exclude a mild PCD phenotype when no other functional and structural analyses are performed. Herein, our data are strongly supported by the TEM analyses of both the sperm cells and respiratory cells of the individuals, together with clinical examinations, therefore allowing us to exclude a PCD phenotype. On the basis of the study of several cohorts from various ethnical origins, DNAH1 accounts for one of the most frequent genetic causes of the MMAF phenotype.25, 49 The outcome (i.e., fertilization, pregnancy, and delivery rates) of assisted reproduction technology procedures, which were carried out for individuals with DNAH1 mutations, was not reported to be affected compared to non-DNAH1 mutated MMAF-affected individuals.50 Herein, we observed that among the five individuals carrying mutations in DNAH17, four individuals did not succeed in achieving pregnancy by using intracytoplasmic sperm injection (ICSI) (Table 1), thus suggesting that DNAH17 mutations could impede ICSI outcomes. Further studies of MMAF-affected individuals, together with genotype/phenotype correlations, should provide an estimation of the prevalence of DNAH17 mutations and a prognosis for ICSI outcomes in cases of DNAH17 mutations.
Lastly and intriguingly, although the loss of dynein arms does not induce structural disorganization of the axoneme in ciliated cells, in sperm cells, the loss of IDAs and ODAs (induced by DNAH1 and DNAH17, respectively) was associated with a global and severe disorganization of the axonemal structure, as illustrated by the absence of microtubule doublets. The pathophysiological mechanisms underlying this sperm-specific phenotype are not known but probably result from axonemal assembly and/or stability defects, and they are in line with expected differences in the mode of assembly and organization of cilia and sperm flagella. This study therefore emphasizes that despite evolutionary conservation, the axonemes of mammalian cilia and sperm flagella display specific sets of homologous proteins, which might, in addition, harbor properties specific to each organelle.
Overall, our study provides a comprehensive analysis of ODA heavy chain distribution in human spermatozoa and demonstrates the importance of DNAH17 (β-HC) in sperm flagellum structure and motility. We demonstrate that DNAH17 (β-HC) mutations are associated with a loss of ODAs and male infertility but not with PCD. Our results reinforce the importance of better characterizing PCD-affected individuals regarding their fertility status. As shown in this study, the axonemes of respiratory cilia and sperm flagella are not strictly identical; this therefore precludes the generalization of infertility in male individuals who carry mutations associated with PCD. Particular attention should be given to the fertility status of PCD-affected male individuals; the fertility status must be ascertained on the basis of spermocytograms and the exclusion of any female infertility component. Conversely, male individuals consulting for infertility ought to be specifically questioned in order to highlight possible respiratory phenotypes. In the future, such refined clinical procedures should help in improving our understanding of the genetic causes of male infertility as an isolated disease or as part of the PCD phenotypic continuum.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank all the individuals and their families for their cooperation, as well as all the referring physicians and Denise Escalier for initial TEM analyses. This work was supported by the Institut National de la Santé et de la Recherche Médicale (INSERM), the Centre National de la Recherche Scientifique (CNRS), the Université Paris Descartes, the Université Pierre et Marie Curie, the French National Research Agency (grant ANR MASFLAGELLA 14-CE15-0002-03), the Fondation pour la Recherche Médicale (grant DEQ20120323689), RaDiCo (funded by the French National Research Agency under the specific program “Investments for the Future” [cohort grant agreement ANR-10-COHO-0003]), the Legs Poix grant from the Chancellerie des Universités of Sorbonne Université, and the BEAT-PCD (Better Experimental Approaches to Treat Primary Ciliary Dyskinesia) COST (European Collaboration in Science and Technology) action.
Published: June 6, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.04.015.
Web Resources
Clustal Omega Software, https://www.ebi.ac.uk/Tools/msa/clustalo/
EMBL EBI Expression Atlas, https://www.ebi.ac.uk/gxa/home
GnomAD Browser, https://gnomad.broadinstitute.org/
Leiden Open Variation Database (LOVD), https://databases.lovd.nl/shared/variants
MaxEntScan, https://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html
Online Mendelian Inheritance in Man (OMIM), https://omim.org/
Provean, http://provean.jcvi.org/index.php
Pymol Software, https://pymol.org/2/
ReproGenomics Viewer, https://rgv.genouest.org/app/#/
Uniprot, https://www.uniprot.org/
Supplemental Data
References
- 1.Ishikawa T. Axoneme structure from motile cilia. Cold Spring Harb. Perspect. Biol. 2017;9:a028076. doi: 10.1101/cshperspect.a028076. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ishikawa, T. (2017). Axoneme structure from motile cilia. Cold Spring Harb. Perspect. Biol. 9, a028076. [DOI] [PMC free article] [PubMed]
- 2.King S.M. Axonemal dynein arms. Cold Spring Harb. Perspect. Biol. 2016;8:a028100. doi: 10.1101/cshperspect.a028100. [DOI] [PMC free article] [PubMed] [Google Scholar]; King, S.M. (2016). Axonemal dynein arms. Cold Spring Harb. Perspect. Biol. 8, a028100. [DOI] [PMC free article] [PubMed]
- 3.Viswanadha R., Sale W.S., Porter M.E. Ciliary motility: Regulation of axonemal dynein motors. Cold Spring Harb. Perspect. Biol. 2017;9:a018325. doi: 10.1101/cshperspect.a018325. [DOI] [PMC free article] [PubMed] [Google Scholar]; Viswanadha, R., Sale, W.S., and Porter, M.E. (2017). Ciliary motility: Regulation of axonemal dynein motors. Cold Spring Harb. Perspect. Biol. 9, a018325. [DOI] [PMC free article] [PubMed]
- 4.Pazour G.J., Agrin N., Walker B.L., Witman G.B. Identification of predicted human outer dynein arm genes: Candidates for primary ciliary dyskinesia genes. J. Med. Genet. 2006;43:62–73. doi: 10.1136/jmg.2005.033001. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pazour, G.J., Agrin, N., Walker, B.L., and Witman, G.B. (2006). Identification of predicted human outer dynein arm genes: Candidates for primary ciliary dyskinesia genes. J. Med. Genet. 43, 62-73. [DOI] [PMC free article] [PubMed]
- 5.Bisgrove B.W., Yost H.J. The roles of cilia in developmental disorders and disease. Development. 2006;133:4131–4143. doi: 10.1242/dev.02595. [DOI] [PubMed] [Google Scholar]; Bisgrove, B.W., and Yost, H.J. (2006). The roles of cilia in developmental disorders and disease. Development 133, 4131-4143. [DOI] [PubMed]
- 6.Mitchison H.M., Valente E.M. Motile and non-motile cilia in human pathology: From function to phenotypes. J. Pathol. 2017;241:294–309. doi: 10.1002/path.4843. [DOI] [PubMed] [Google Scholar]; Mitchison, H.M., and Valente, E.M. (2017). Motile and non-motile cilia in human pathology: From function to phenotypes. J. Pathol. 241, 294-309. [DOI] [PubMed]
- 7.Lucas J.S., Barbato A., Collins S.A., Goutaki M., Behan L., Caudri D., Dell S., Eber E., Escudier E., Hirst R.A. European Respiratory Society guidelines for the diagnosis of primary ciliary dyskinesia. Eur. Respir. J. 2017;49:1601090. doi: 10.1183/13993003.01090-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lucas, J.S., Barbato, A., Collins, S.A., Goutaki, M., Behan, L., Caudri, D., Dell, S., Eber, E., Escudier, E., Hirst, R.A., et al. (2017). European Respiratory Society guidelines for the diagnosis of primary ciliary dyskinesia. Eur. Respir. J. 49, 1601090. [DOI] [PMC free article] [PubMed]
- 8.Papon J.F., Coste A., Roudot-Thoraval F., Boucherat M., Roger G., Tamalet A., Vojtek A.M., Amselem S., Escudier E. A 20-year experience of electron microscopy in the diagnosis of primary ciliary dyskinesia. Eur. Respir. J. 2010;35:1057–1063. doi: 10.1183/09031936.00046209. [DOI] [PubMed] [Google Scholar]; Papon, J.F., Coste, A., Roudot-Thoraval, F., Boucherat, M., Roger, G., Tamalet, A., Vojtek, A.M., Amselem, S., and Escudier, E. (2010). A 20-year experience of electron microscopy in the diagnosis of primary ciliary dyskinesia. Eur. Respir. J. 35, 1057-1063. [DOI] [PubMed]
- 9.Noone P.G., Leigh M.W., Sannuti A., Minnix S.L., Carson J.L., Hazucha M., Zariwala M.A., Knowles M.R. Primary ciliary dyskinesia: Diagnostic and phenotypic features. Am. J. Respir. Crit. Care Med. 2004;169:459–467. doi: 10.1164/rccm.200303-365OC. [DOI] [PubMed] [Google Scholar]; Noone, P.G., Leigh, M.W., Sannuti, A., Minnix, S.L., Carson, J.L., Hazucha, M., Zariwala, M.A., and Knowles, M.R. (2004). Primary ciliary dyskinesia: Diagnostic and phenotypic features. Am. J. Respir. Crit. Care Med. 169, 459-467. [DOI] [PubMed]
- 10.Nicastro D., Schwartz C., Pierson J., Gaudette R., Porter M.E., McIntosh J.R. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]; Nicastro, D., Schwartz, C., Pierson, J., Gaudette, R., Porter, M.E., and McIntosh, J.R. (2006). The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313, 944-948. [DOI] [PubMed]
- 11.Hom E.F., Witman G.B., Harris E.H., Dutcher S.K., Kamiya R., Mitchell D.R., Pazour G.J., Porter M.E., Sale W.S., Wirschell M. A unified taxonomy for ciliary dyneins. Cytoskeleton (Hoboken) 2011;68:555–565. doi: 10.1002/cm.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hom, E.F., Witman, G.B., Harris, E.H., Dutcher, S.K., Kamiya, R., Mitchell, D.R., Pazour, G.J., Porter, M.E., Sale, W.S., Wirschell, M., et al. (2011). A unified taxonomy for ciliary dyneins. Cytoskeleton (Hoboken) 68, 555-565. [DOI] [PMC free article] [PubMed]
- 12.Fliegauf M., Olbrich H., Horvath J., Wildhaber J.H., Zariwala M.A., Kennedy M., Knowles M.R., Omran H. Mislocalization of DNAH5 and DNAH9 in respiratory cells from patients with primary ciliary dyskinesia. Am. J. Respir. Crit. Care Med. 2005;171:1343–1349. doi: 10.1164/rccm.200411-1583OC. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fliegauf, M., Olbrich, H., Horvath, J., Wildhaber, J.H., Zariwala, M.A., Kennedy, M., Knowles, M.R., and Omran, H. (2005). Mislocalization of DNAH5 and DNAH9 in respiratory cells from patients with primary ciliary dyskinesia. Am. J. Respir. Crit. Care Med. 171, 1343-1349. [DOI] [PMC free article] [PubMed]
- 13.Dougherty G.W., Loges N.T., Klinkenbusch J.A., Olbrich H., Pennekamp P., Menchen T., Raidt J., Wallmeier J., Werner C., Westermann C. DNAH11 localization in the proximal region of respiratory cilia defines distinct outer dynein arm complexes. Am. J. Respir. Cell Mol. Biol. 2016;55:213–224. doi: 10.1165/rcmb.2015-0353OC. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dougherty, G.W., Loges, N.T., Klinkenbusch, J.A., Olbrich, H., Pennekamp, P., Menchen, T., Raidt, J., Wallmeier, J., Werner, C., Westermann, C., et al. (2016). DNAH11 localization in the proximal region of respiratory cilia defines distinct outer dynein arm complexes. Am. J. Respir. Cell Mol. Biol. 55, 213-224. [DOI] [PMC free article] [PubMed]
- 14.Shapiro A.J., Zariwala M.A., Ferkol T., Davis S.D., Sagel S.D., Dell S.D., Rosenfeld M., Olivier K.N., Milla C., Daniel S.J., Genetic Disorders of Mucociliary Clearance Consortium Diagnosis, monitoring, and treatment of primary ciliary dyskinesia: PCD foundation consensus recommendations based on state of the art review. Pediatr. Pulmonol. 2016;51:115–132. doi: 10.1002/ppul.23304. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shapiro, A.J., Zariwala, M.A., Ferkol, T., Davis, S.D., Sagel, S.D., Dell, S.D., Rosenfeld, M., Olivier, K.N., Milla, C., Daniel, S.J., et al.; Genetic Disorders of Mucociliary Clearance Consortium (2016). Diagnosis, monitoring, and treatment of primary ciliary dyskinesia: PCD foundation consensus recommendations based on state of the art review. Pediatr. Pulmonol. 51, 115-132. [DOI] [PMC free article] [PubMed]
- 15.Cooper T.G., Noonan E., von Eckardstein S., Auger J., Baker H.W., Behre H.M., Haugen T.B., Kruger T., Wang C., Mbizvo M.T., Vogelsong K.M. World Health Organization reference values for human semen characteristics. Hum. Reprod. Update. 2010;16:231–245. doi: 10.1093/humupd/dmp048. [DOI] [PubMed] [Google Scholar]; Cooper, T.G., Noonan, E., von Eckardstein, S., Auger, J., Baker, H.W., Behre, H.M., Haugen, T.B., Kruger, T., Wang, C., Mbizvo, M.T., and Vogelsong, K.M. (2010). World Health Organization reference values for human semen characteristics. Hum. Reprod. Update 16, 231-245. [DOI] [PubMed]
- 16.Vanaken G.J., Bassinet L., Boon M., Mani R., Honoré I., Papon J.F., Cuppens H., Jaspers M., Lorent N., Coste A. Infertility in an adult cohort with primary ciliary dyskinesia: Phenotype-gene association. Eur. Respir. J. 2017;50:1700314. doi: 10.1183/13993003.00314-2017. [DOI] [PubMed] [Google Scholar]; Vanaken, G.J., Bassinet, L., Boon, M., Mani, R., Honore, I., Papon, J.F., Cuppens, H., Jaspers, M., Lorent, N., Coste, A., et al. (2017). Infertility in an adult cohort with primary ciliary dyskinesia: Phenotype-gene association. Eur. Respir. J. 50, 1700314. [DOI] [PubMed]
- 17.Guichard C., Harricane M.C., Lafitte J.J., Godard P., Zaegel M., Tack V., Lalau G., Bouvagnet P. Axonemal dynein intermediate-chain gene (DNAI1) mutations result in situs inversus and primary ciliary dyskinesia (Kartagener syndrome) Am. J. Hum. Genet. 2001;68:1030–1035. doi: 10.1086/319511. [DOI] [PMC free article] [PubMed] [Google Scholar]; Guichard, C., Harricane, M.C., Lafitte, J.J., Godard, P., Zaegel, M., Tack, V., Lalau, G., and Bouvagnet, P. (2001). Axonemal dynein intermediate-chain gene (DNAI1) mutations result in situs inversus and primary ciliary dyskinesia (Kartagener syndrome). Am. J. Hum. Genet. 68, 1030-1035. [DOI] [PMC free article] [PubMed]
- 18.Loges N.T., Olbrich H., Fenske L., Mussaffi H., Horvath J., Fliegauf M., Kuhl H., Baktai G., Peterffy E., Chodhari R. DNAI2 mutations cause primary ciliary dyskinesia with defects in the outer dynein arm. Am. J. Hum. Genet. 2008;83:547–558. doi: 10.1016/j.ajhg.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; Loges, N.T., Olbrich, H., Fenske, L., Mussaffi, H., Horvath, J., Fliegauf, M., Kuhl, H., Baktai, G., Peterffy, E., Chodhari, R., et al. (2008). DNAI2 mutations cause primary ciliary dyskinesia with defects in the outer dynein arm. Am. J. Hum. Genet. 83, 547-558. [DOI] [PMC free article] [PubMed]
- 19.Fassad M.R., Shoemark A., Legendre M., Hirst R.A., Koll F., le Borgne P., Louis B., Daudvohra F., Patel M.P., Thomas L. Mutations in outer dynein arm heavy chain DNAH9 cause motile cilia defects and situs inversus. Am. J. Hum. Genet. 2018;103:984–994. doi: 10.1016/j.ajhg.2018.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fassad, M.R., Shoemark, A., Legendre, M., Hirst, R.A., Koll, F., le Borgne, P., Louis, B., Daudvohra, F., Patel, M.P., Thomas, L., et al. (2018). Mutations in outer dynein arm heavy chain DNAH9 cause motile cilia defects and situs inversus. Am. J. Hum. Genet. 103, 984-994. [DOI] [PMC free article] [PubMed]
- 20.Auger J., Jouannet P., Eustache F. Another look at human sperm morphology. Hum. Reprod. 2016;31:10–23. doi: 10.1093/humrep/dev251. [DOI] [PubMed] [Google Scholar]; Auger, J., Jouannet, P., and Eustache, F. (2016). Another look at human sperm morphology. Hum. Reprod. 31, 10-23. [DOI] [PubMed]
- 21.Chemes H.E., Brugo S., Zanchetti F., Carrere C., Lavieri J.C. Dysplasia of the fibrous sheath: An ultrastructural defect of human spermatozoa associated with sperm immotility and primary sterility. Fertil. Steril. 1987;48:664–669. doi: 10.1016/s0015-0282(16)59482-5. [DOI] [PubMed] [Google Scholar]; Chemes, H.E., Brugo, S., Zanchetti, F., Carrere, C., and Lavieri, J.C. (1987). Dysplasia of the fibrous sheath: An ultrastructural defect of human spermatozoa associated with sperm immotility and primary sterility. Fertil. Steril. 48, 664-669. [DOI] [PubMed]
- 22.Escalier D., Albert M. New fibrous sheath anomaly in spermatozoa of men with consanguinity. Fertil Steril. 2006;86:219.e1–219.e9. doi: 10.1016/j.fertnstert.2005.12.042. [DOI] [PubMed] [Google Scholar]; Escalier, D., and Albert, M. (2006). New fibrous sheath anomaly in spermatozoa of men with consanguinity. Fertil Steril 86, 219e1-219.e9. https://www.ncbi.nlm.nih.gov/pubmed/16750828 [DOI] [PubMed]
- 23.Escalier D. Arrest of flagellum morphogenesis with fibrous sheath immaturity of human spermatozoa. Andrologia. 2006;38:54–60. doi: 10.1111/j.1439-0272.2006.00711.x. [DOI] [PubMed] [Google Scholar]; Escalier, D. (2006). Arrest of flagellum morphogenesis with fibrous sheath immaturity of human spermatozoa. Andrologia 38, 54-60 [DOI] [PubMed]
- 24.Ben Khelifa M., Coutton C., Zouari R., Karaouzène T., Rendu J., Bidart M., Yassine S., Pierre V., Delaroche J., Hennebicq S. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am. J. Hum. Genet. 2014;94:95–104. doi: 10.1016/j.ajhg.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ben Khelifa, M., Coutton, C., Zouari, R., Karaouzene, T., Rendu, J., Bidart, M., Yassine, S., Pierre, V., Delaroche, J., Hennebicq, S., et al. (2014). Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am. J. Hum. Genet. 94, 95-104. [DOI] [PMC free article] [PubMed]
- 25.Nsota Mbango J.-F., Coutton C., Arnoult C., Ray P., Toure A. Genetic causes of male infertility: Snapshot on morphological abnormalities of the sperm flagellum. Basic Clin Androl. 2019;29 doi: 10.1186/s12610-019-0083-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nsota Mbango, J.-F., Coutton, C., Arnoult, C., Ray, P., and Toure, A. (2019). Genetic causes of male infertility: Snapshot on morphological abnormalities of the sperm flagellum. Basic Clin Androl 29. [DOI] [PMC free article] [PubMed]
- 26.Jeanson L., Thomas L., Copin B., Coste A., Sermet-Gaudelus I., Dastot-Le Moal F., Duquesnoy P., Montantin G., Collot N., Tissier S. Mutations in GAS8, a gene encoding a nexin-dynein regulatory complex subunit, cause primary ciliary dyskinesia with axonemal disorganization. Hum. Mutat. 2016;37:776–785. doi: 10.1002/humu.23005. [DOI] [PubMed] [Google Scholar]; Jeanson, L., Thomas, L., Copin, B., Coste, A., Sermet-Gaudelus, I., Dastot-Le Moal, F., Duquesnoy, P., Montantin, G., Collot, N., Tissier, S., et al. (2016). Mutations in GAS8, a gene encoding a nexin-dynein regulatory complex subunit, cause primary ciliary dyskinesia with axonemal disorganization. Hum. Mutat. 37, 776-785. [DOI] [PubMed]
- 27.Krieger F., Möglich A., Kiefhaber T. Effect of proline and glycine residues on dynamics and barriers of loop formation in polypeptide chains. J. Am. Chem. Soc. 2005;127:3346–3352. doi: 10.1021/ja042798i. [DOI] [PubMed] [Google Scholar]; Krieger, F., Moglich, A., and Kiefhaber, T. (2005). Effect of proline and glycine residues on dynamics and barriers of loop formation in polypeptide chains. J. Am. Chem. Soc. 127, 3346-3352. [DOI] [PubMed]
- 28.Darde T.A., Lecluze E., Lardenois A., Stévant I., Alary N., Tüttelmann F., Collin O., Nef S., Jégou B., Rolland A.D., Chalmel F. The ReproGenomics Viewer: A multi-omics and cross-species resource compatible with single-cell studies for the reproductive science community. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz047. [DOI] [PubMed] [Google Scholar]; Darde, T.A., Lecluze, E., Lardenois, A., Stevant, I., Alary, N., Tuttelmann, F., Collin, O., Nef, S., Jegou, B., Rolland, A.D., and Chalmel, F. (2019). The ReproGenomics Viewer: A multi-omics and cross-species resource compatible with single-cell studies for the reproductive science community. Bioinformatics. [DOI] [PubMed]
- 29.Darde T.A., Sallou O., Becker E., Evrard B., Monjeaud C., Le Bras Y., Jégou B., Collin O., Rolland A.D., Chalmel F. The ReproGenomics Viewer: An integrative cross-species toolbox for the reproductive science community. Nucleic Acids Res. 2015;43(W1) doi: 10.1093/nar/gkv345. W109-16. [DOI] [PMC free article] [PubMed] [Google Scholar]; Darde, T.A., Sallou, O., Becker, E., Evrard, B., Monjeaud, C., Le Bras, Y., Jegou, B., Collin, O., Rolland, A.D., and Chalmel, F. (2015). The ReproGenomics Viewer: An integrative cross-species toolbox for the reproductive science community. Nucleic Acids Res. 43 (W1), W109-16. [DOI] [PMC free article] [PubMed]
- 30.Wang M., Liu X., Chang G., Chen Y., An G., Yan L., Gao S., Xu Y., Cui Y., Dong J. Single-cell RNA sequencing analysis reveals sequential cell fate transition during human spermatogenesis. Cell Stem Cell. 2018;23:599–614.e4. doi: 10.1016/j.stem.2018.08.007. [DOI] [PubMed] [Google Scholar]; Wang, M., Liu, X., Chang, G., Chen, Y., An, G., Yan, L., Gao, S., Xu, Y., Cui, Y., Dong, J., et al. (2018). Single-cell RNA sequencing analysis reveals sequential cell fate transition during human spermatogenesis. Cell Stem Cell 23, 599-614.e4. [DOI] [PubMed]
- 31.Wang G., Guo Y., Zhou T., Shi X., Yu J., Yang Y., Wu Y., Wang J., Liu M., Chen X. In-depth proteomic analysis of the human sperm reveals complex protein compositions. J. Proteomics. 2013;79:114–122. doi: 10.1016/j.jprot.2012.12.008. [DOI] [PubMed] [Google Scholar]; Wang, G., Guo, Y., Zhou, T., Shi, X., Yu, J., Yang, Y., Wu, Y., Wang, J., Liu, M., Chen, X., et al. (2013). In-depth proteomic analysis of the human sperm reveals complex protein compositions. J. Proteomics 79, 114-122. [DOI] [PubMed]
- 32.Amaral A., Castillo J., Estanyol J.M., Ballescà J.L., Ramalho-Santos J., Oliva R. Human sperm tail proteome suggests new endogenous metabolic pathways. Mol. Cell. Proteomics. 2013;12:330–342.. doi: 10.1074/mcp.M112.020552. [DOI] [PMC free article] [PubMed] [Google Scholar]; Amaral, A., Castillo, J., Estanyol, J.M., Ballesca, J.L., Ramalho-Santos, J., and Oliva, R. (2013). Human sperm tail proteome suggests new endogenous metabolic pathways. Mol. Cell. Proteomics 12, 330-342. [DOI] [PMC free article] [PubMed]
- 33.Blackburn K., Bustamante-Marin X., Yin W., Goshe M.B., Ostrowski L.E. Quantitative proteomic analysis of human airway cilia identifies previously uncharacterized proteins of high abundance. J. Proteome Res. 2017;16:1579–1592.. doi: 10.1021/acs.jproteome.6b00972. [DOI] [PMC free article] [PubMed] [Google Scholar]; Blackburn, K., Bustamante-Marin, X., Yin, W., Goshe, M.B., and Ostrowski, L.E. (2017). Quantitative proteomic analysis of human airway cilia identifies previously uncharacterized proteins of high abundance. J. Proteome Res. 16, 1579-1592. [DOI] [PMC free article] [PubMed]
- 34.Hornef N., Olbrich H., Horvath J., Zariwala M.A., Fliegauf M., Loges N.T., Wildhaber J., Noone P.G., Kennedy M., Antonarakis S.E. DNAH5 mutations are a common cause of primary ciliary dyskinesia with outer dynein arm defects. Am. J. Respir. Crit. Care Med. 2006;174:120–126. doi: 10.1164/rccm.200601-084OC. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hornef, N., Olbrich, H., Horvath, J., Zariwala, M.A., Fliegauf, M., Loges, N.T., Wildhaber, J., Noone, P.G., Kennedy, M., Antonarakis, S.E., et al. (2006). DNAH5 mutations are a common cause of primary ciliary dyskinesia with outer dynein arm defects. Am. J. Respir. Crit. Care Med. 174, 120-126. [DOI] [PMC free article] [PubMed]
- 35.Olbrich H., Häffner K., Kispert A., Völkel A., Volz A., Sasmaz G., Reinhardt R., Hennig S., Lehrach H., Konietzko N. Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left-right asymmetry. Nat. Genet. 2002;30:143–144. doi: 10.1038/ng817. [DOI] [PubMed] [Google Scholar]; Olbrich, H., Haffner, K., Kispert, A., Volkel, A., Volz, A., Sasmaz, G., Reinhardt, R., Hennig, S., Lehrach, H., Konietzko, N., et al. (2002). Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left-right asymmetry. Nat. Genet. 30, 143-144. [DOI] [PubMed]
- 36.Pennarun G., Escudier E., Chapelin C., Bridoux A.M., Cacheux V., Roger G., Clément A., Goossens M., Amselem S., Duriez B. Loss-of-function mutations in a human gene related to Chlamydomonas reinhardtii dynein IC78 result in primary ciliary dyskinesia. Am. J. Hum. Genet. 1999;65:1508–1519. doi: 10.1086/302683. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pennarun, G., Escudier, E., Chapelin, C., Bridoux, A.M., Cacheux, V., Roger, G., Clement, A., Goossens, M., Amselem, S., and Duriez, B. (1999). Loss-of-function mutations in a human gene related to Chlamydomonas reinhardtii dynein IC78 result in primary ciliary dyskinesia. Am. J. Hum. Genet. 65, 1508-1519. [DOI] [PMC free article] [PubMed]
- 37.Mazor M., Alkrinawi S., Chalifa-Caspi V., Manor E., Sheffield V.C., Aviram M., Parvari R. Primary ciliary dyskinesia caused by homozygous mutation in DNAL1, encoding dynein light chain 1. Am. J. Hum. Genet. 2011;88:599–607. doi: 10.1016/j.ajhg.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mazor, M., Alkrinawi, S., Chalifa-Caspi, V., Manor, E., Sheffield, V.C., Aviram, M., and Parvari, R. (2011). Primary ciliary dyskinesia caused by homozygous mutation in DNAL1, encoding dynein light chain 1. Am. J. Hum. Genet. 88, 599-607. [DOI] [PMC free article] [PubMed]
- 38.Hjeij R., Lindstrand A., Francis R., Zariwala M.A., Liu X., Li Y., Damerla R., Dougherty G.W., Abouhamed M., Olbrich H. ARMC4 mutations cause primary ciliary dyskinesia with randomization of left/right body asymmetry. Am. J. Hum. Genet. 2013;93:357–367. doi: 10.1016/j.ajhg.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hjeij, R., Lindstrand, A., Francis, R., Zariwala, M.A., Liu, X., Li, Y., Damerla, R., Dougherty, G.W., Abouhamed, M., Olbrich, H., et al. (2013). ARMC4 mutations cause primary ciliary dyskinesia with randomization of left/right body asymmetry. Am. J. Hum. Genet. 93, 357-367. [DOI] [PMC free article] [PubMed]
- 39.Knowles M.R., Leigh M.W., Ostrowski L.E., Huang L., Carson J.L., Hazucha M.J., Yin W., Berg J.S., Davis S.D., Dell S.D., Genetic Disorders of Mucociliary Clearance Consortium Exome sequencing identifies mutations in CCDC114 as a cause of primary ciliary dyskinesia. Am. J. Hum. Genet. 2013;92:99–106. doi: 10.1016/j.ajhg.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; Knowles, M.R., Leigh, M.W., Ostrowski, L.E., Huang, L., Carson, J.L., Hazucha, M.J., Yin, W., Berg, J.S., Davis, S.D., Dell, S.D., et al.; Genetic Disorders of Mucociliary Clearance Consortium (2013). Exome sequencing identifies mutations in CCDC114 as a cause of primary ciliary dyskinesia. Am. J. Hum. Genet. 92, 99-106. [DOI] [PMC free article] [PubMed]
- 40.Onoufriadis A., Paff T., Antony D., Shoemark A., Micha D., Kuyt B., Schmidts M., Petridi S., Dankert-Roelse J.E., Haarman E.G., UK10K Splice-site mutations in the axonemal outer dynein arm docking complex gene CCDC114 cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2013;92:88–98. doi: 10.1016/j.ajhg.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Onoufriadis, A., Paff, T., Antony, D., Shoemark, A., Micha, D., Kuyt, B., Schmidts, M., Petridi, S., Dankert-Roelse, J.E., Haarman, E.G., et al.; UK10K (2013). Splice-site mutations in the axonemal outer dynein arm docking complex gene CCDC114 cause primary ciliary dyskinesia. Am. J. Hum. Genet. 92, 88-98. [DOI] [PMC free article] [PubMed]
- 41.Hjeij R., Onoufriadis A., Watson C.M., Slagle C.E., Klena N.T., Dougherty G.W., Kurkowiak M., Loges N.T., Diggle C.P., Morante N.F., UK10K Consortium CCDC151 mutations cause primary ciliary dyskinesia by disruption of the outer dynein arm docking complex formation. Am. J. Hum. Genet. 2014;95:257–274. doi: 10.1016/j.ajhg.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hjeij, R., Onoufriadis, A., Watson, C.M., Slagle, C.E., Klena, N.T., Dougherty, G.W., Kurkowiak, M., Loges, N.T., Diggle, C.P., Morante, N.F., et al.; UK10K Consortium (2014). CCDC151 mutations cause primary ciliary dyskinesia by disruption of the outer dynein arm docking complex formation. Am. J. Hum. Genet. 95, 257-274. [DOI] [PMC free article] [PubMed]
- 42.Loges N.T., Antony D., Maver A., Deardorff M.A., Güleç E.Y., Gezdirici A., Nöthe-Menchen T., Höben I.M., Jelten L., Frank D. Recessive DNAH9 loss-of-function mutations cause laterality defects and subtle respiratory ciliary-beating defects. Am. J. Hum. Genet. 2018;103:995–1008.. doi: 10.1016/j.ajhg.2018.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Loges, N.T., Antony, D., Maver, A., Deardorff, M.A., Guleç, E.Y., Gezdirici, A., Nothe-Menchen, T., Hoben, I.M., Jelten, L., Frank, D., et al. (2018). Recessive DNAH9 loss-of-function mutations cause laterality defects and subtle respiratory ciliary-beating defects. Am. J. Hum. Genet. 103, 995-1008. [DOI] [PMC free article] [PubMed]
- 43.Bartoloni L., Blouin J.L., Pan Y., Gehrig C., Maiti A.K., Scamuffa N., Rossier C., Jorissen M., Armengot M., Meeks M. Mutations in the DNAH11 (axonemal heavy chain dynein type 11) gene cause one form of situs inversus totalis and most likely primary ciliary dyskinesia. Proc. Natl. Acad. Sci. USA. 2002;99:10282–10286. doi: 10.1073/pnas.152337699. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bartoloni, L., Blouin, J.L., Pan, Y., Gehrig, C., Maiti, A.K., Scamuffa, N., Rossier, C., Jorissen, M., Armengot, M., Meeks, M., et al. (2002). Mutations in the DNAH11 (axonemal heavy chain dynein type 11) gene cause one form of situs inversus totalis and most likely primary ciliary dyskinesia. Proc. Natl. Acad. Sci. USA 99, 10282-10286. [DOI] [PMC free article] [PubMed]
- 44.Schwabe G.C., Hoffmann K., Loges N.T., Birker D., Rossier C., de Santi M.M., Olbrich H., Fliegauf M., Failly M., Liebers U. Primary ciliary dyskinesia associated with normal axoneme ultrastructure is caused by DNAH11 mutations. Hum. Mutat. 2008;29:289–298.. doi: 10.1002/humu.20656. [DOI] [PubMed] [Google Scholar]; Schwabe, G.C., Hoffmann, K., Loges, N.T., Birker, D., Rossier, C., de Santi, M.M., Olbrich, H., Fliegauf, M., Failly, M., Liebers, U., et al. (2008). Primary ciliary dyskinesia associated with normal axoneme ultrastructure is caused by DNAH11 mutations. Hum. Mutat. 29, 289-298. [DOI] [PubMed]
- 45.Knowles M.R., Leigh M.W., Carson J.L., Davis S.D., Dell S.D., Ferkol T.W., Olivier K.N., Sagel S.D., Rosenfeld M., Burns K.A., Genetic Disorders of Mucociliary Clearance Consortium Mutations of DNAH11 in patients with primary ciliary dyskinesia with normal ciliary ultrastructure. Thorax. 2012;67:433–441.. doi: 10.1136/thoraxjnl-2011-200301. [DOI] [PMC free article] [PubMed] [Google Scholar]; Knowles, M.R., Leigh, M.W., Carson, J.L., Davis, S.D., Dell, S.D., Ferkol, T.W., Olivier, K.N., Sagel, S.D., Rosenfeld, M., Burns, K.A., et al.; Genetic Disorders of Mucociliary Clearance Consortium (2012). Mutations of DNAH11 in patients with primary ciliary dyskinesia with normal ciliary ultrastructure. Thorax 67, 433-441. [DOI] [PMC free article] [PubMed]
- 46.Amiri-Yekta A., Coutton C., Kherraf Z.E., Karaouzène T., Le Tanno P., Sanati M.H., Sabbaghian M., Almadani N., Sadighi Gilani M.A., Hosseini S.H. Whole-exome sequencing of familial cases of multiple morphological abnormalities of the sperm flagella (MMAF) reveals new DNAH1 mutations. Hum. Reprod. 2016;31:2872–2880. doi: 10.1093/humrep/dew262. [DOI] [PubMed] [Google Scholar]; Amiri-Yekta, A., Coutton, C., Kherraf, Z.E., Karaouzene, T., Le Tanno, P., Sanati, M.H., Sabbaghian, M., Almadani, N., Sadighi Gilani, M.A., Hosseini, S.H., et al. (2016). Whole-exome sequencing of familial cases of multiple morphological abnormalities of the sperm flagella (MMAF) reveals new DNAH1 mutations. Hum. Reprod. 31, 2872-2880. [DOI] [PubMed]
- 47.Sha Y., Yang X., Mei L., Ji Z., Wang X., Ding L., Li P., Yang S. DNAH1 gene mutations and their potential association with dysplasia of the sperm fibrous sheath and infertility in the Han Chinese population. Fertil Steril. 2017;107:1312–1318.e2. doi: 10.1016/j.fertnstert.2017.04.007. [DOI] [PubMed] [Google Scholar]; Sha, Y., Yang, X., Mei, L., Ji, Z., Wang, X., Ding, L., Li, P., and Yang, S. (2017). DNAH1 gene mutations and their potential association with dysplasia of the sperm fibrous sheath and infertility in the Han Chinese population. Fertil Steril 107, 1312-1318.e2. [DOI] [PubMed]
- 48.Wang X., Jin H., Han F., Cui Y., Chen J., Yang C., Zhu P., Wang W., Jiao G., Wang W. Homozygous DNAH1 frameshift mutation causes multiple morphological anomalies of the sperm flagella in Chinese. Clin. Genet. 2017;91:313–321. doi: 10.1111/cge.12857. [DOI] [PubMed] [Google Scholar]; Wang, X., Jin, H., Han, F., Cui, Y., Chen, J., Yang, C., Zhu, P., Wang, W., Jiao, G., Wang, W., et al. (2017). Homozygous DNAH1 frameshift mutation causes multiple morphological anomalies of the sperm flagella in Chinese. Clin. Genet. 91, 313-321. [DOI] [PubMed]
- 49.Coutton C., Martinez G., Kherraf Z.E., Amiri-Yekta A., Boguenet M., Saut A., He X., Zhang F., Cristou-Kent M., Escoffier J. Bi-allelic mutations in ARMC2 lead to severe astheno-teratozoospermia due to sperm flagellum malformations in humans and mice. Am. J. Hum. Genet. 2019;104:331–340. doi: 10.1016/j.ajhg.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; Coutton, C., Martinez, G., Kherraf, Z.E., Amiri-Yekta, A., Boguenet, M., Saut, A., He, X., Zhang, F., Cristou-Kent, M., Escoffier, J., et al. (2019). Bi-allelic mutations in ARMC2 lead to severe astheno-teratozoospermia due to sperm flagellum malformations in humans and mice. Am. J. Hum. Genet. 104, 331-340. [DOI] [PMC free article] [PubMed]
- 50.Wambergue C., Zouari R., Fourati Ben Mustapha S., Martinez G., Devillard F., Hennebicq S., Satre V., Brouillet S., Halouani L., Marrakchi O. Patients with multiple morphological abnormalities of the sperm flagella due to DNAH1 mutations have a good prognosis following intracytoplasmic sperm injection. Hum. Reprod. 2016;31:1164–1172. doi: 10.1093/humrep/dew083. [DOI] [PubMed] [Google Scholar]; Wambergue, C., Zouari, R., Fourati Ben Mustapha, S., Martinez, G., Devillard, F., Hennebicq, S., Satre, V., Brouillet, S., Halouani, L., Marrakchi, O., et al. (2016). Patients with multiple morphological abnormalities of the sperm flagella due to DNAH1 mutations have a good prognosis following intracytoplasmic sperm injection. Hum. Reprod. 31, 1164-1172. [DOI] [PubMed]
- 51.Lhuillier P., Rode B., Escalier D., Lorès P., Dirami T., Bienvenu T., Gacon G., Dulioust E., Touré A. Absence of annulus in human asthenozoospermia: Case report. Hum. Reprod. 2009;24:1296–1303. doi: 10.1093/humrep/dep020. [DOI] [PubMed] [Google Scholar]; Lhuillier, P., Rode, B., Escalier, D., Lores, P., Dirami, T., Bienvenu, T., Gacon, G., Dulioust, E., and Toure, A. (2009). Absence of annulus in human asthenozoospermia: Case report. Hum. Reprod. 24, 1296-1303. [DOI] [PubMed]
- 52.Tamalet A., Clement A., Roudot-Thoraval F., Desmarquest P., Roger G., Boulé M., Millepied M.C., Baculard T.A., Escudier E. Abnormal central complex is a marker of severity in the presence of partial ciliary defect. Pediatrics. 2001;108:E86.. doi: 10.1542/peds.108.5.e86. [DOI] [PubMed] [Google Scholar]; Tamalet, A., Clement, A., Roudot-Thoraval, F., Desmarquest, P., Roger, G., Boule, M., Millepied, M.C., Baculard, T.A., and Escudier, E. (2001). Abnormal central complex is a marker of severity in the presence of partial ciliary defect. Pediatrics 108, E86. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.