Abstract
Thyroid Hormone Receptor Interacting Protein 13 (TRIP13) plays a key role in regulating mitotic processes, including spindle assembly checkpoint and DNA repair pathways, which may account for Chromosome instability (CIN). As CIN is a predominant hallmark of cancer, TRIP13 may act as a tumor susceptibility locus. Amplification of TRIP13 has been observed in various human cancers and implicated in several aspects of malignant transformation, including cancer cell proliferation, drug resistance and tumor progression. Here, we discussed the functional significance of TRIP13 in cell progression, highlighted the recent findings on the aberrant expression in human cancers and emphasized its significance for the therapeutic potential.
Keywords: Cancer, CIN, TRIP13, Oncogenes
Graphical Abstract
1. Introduction
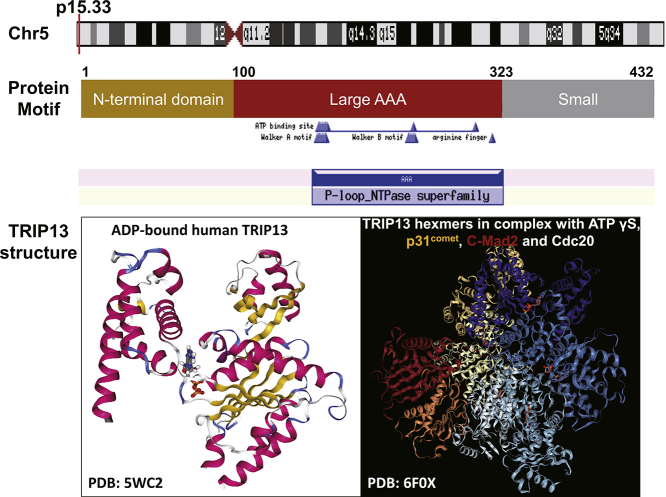
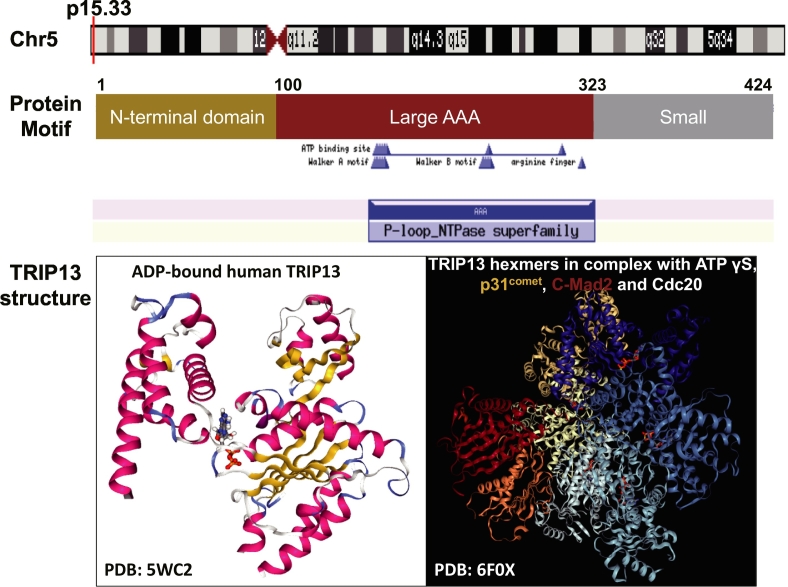
TRIP13 (Thyroid Hormone Receptor Interacting Protein 13) is one AAA (ATPase family associated with various cellular activities) protein belonging to a large AAA+ protein superfamily of ring-shaped P-loop NTPases (Pfam: PF00004) which is involved in an array of cellular processes, including the checkpoint signaling, DNA break repair and recombination, and chromosome synapsis [1,2]. The human TRIP13 gene is located on chromosome 5 and it is comprised of 14 exons coding a protein with 432 amino acid residues. TRIP13 has a small N-terminal domain putatively involved in substrate recognition and an AAA+ ATPase region containing the ATP-binding site [3]. Recently, the structures of TRIP13 protein as well as TRIP13 hexameric complex with ligands and partners are resolved, which would provide more insights into the functional study of TRIP13 [4,5] (Fig. 1). In last decades, the oncogenic roles of TRIP13 have attracted considerable attention. Accumulating researches have indicated that TRIP13 is overexpressed in multiform cancers and usually associated with poor survival [6].
Fig. 1.
Biological information for human TRIP13 gene. It is located on the short arm p15 of chromosome 5 (top pannel). The encoded protein have the N-terminal domain, putatively involved in substrate recognition, and AAA+ ATPase region which consists of large and small subdomains. The ATP binding site includes Walker A motif (GxxxxGK[T/S]) and Walker B motif (hhhh[D/E], where h is a hydrophobic residue), and numbers above the structures indicate amino acid residue positions. The 3D structures of TRIP13 protein (PDB:5WC2) bound with ADP as well as hexameric TRIP13 complex (PDB: 6F0X) with ligands and partners are illustrated (bottom pannel).
Previous studies have indicated that the spindle assembly checkpoint (SAC) is a ubiquitous safeguard that ensures the fidelity of chromosome separation in cell division [[7], [8], [9]]. A number of SAC proteins, which are highly expressed in multiple cancers, are thought to cause Chromosome instability (CIN) in tumors [[10], [11], [12]]. Moreover, telomere dysfunction [[13], [14], [15]] and defective DNA repair pathways response [16] have been demonstrated to make main contribution to CIN in cancer. Studies from different labs have corroborated that TRIP13 is one of the top genes related to CIN in human tumors [[17], [18], [19], [20]] and is associated with poor survival in various tumors. In recent years, quite a few studies focused on the roles of TRIP13 in cancer progression, and drug resistance.
In view of previous studies, we discussed the roles of TRIP13 in cell mitosis, highlighted recent findings on the aberrant expression in human cancers, and conjectured that TRIP13 may act as a promising biomarker and a potential therapeutic target for cancer diagnosis and treatment.
2. Biological Functions of TRIP13 in Cells
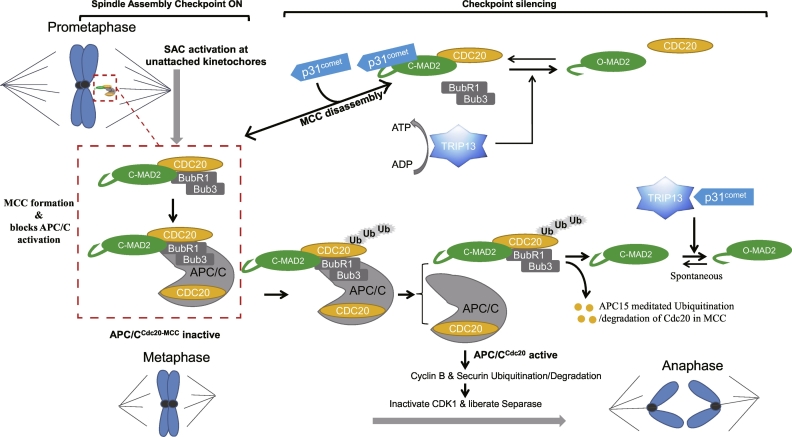
TRIP13 plays an indispensable role in cell progression, particularly with respect to the checkpoint signaling. Subcellular localization analysis shows that TRIP13 interacts with p31comet, a MAD2 (mitotic arrest deficient 2)-binding HORMA-domain protein that negatively regulates the SAC localizing to kinetochores in prometaphase, and TRIP13 co-localizes with MAD2 at kinetochores. More detailed localization studies on TRIP13 have reported that it is localized to kinetochores and co-expresses with centromere/kinetochore components [[21], [22], [23], [24]]. In addition, immunofluorescence analysis demonstrates that GFP-TRIP13 is distributed in reticulum-like structures and localizes at the nuclear envelope partially in interphase cells, while it disappears from kinetochores in metaphase and anaphase cells [22]. Several studies have shown that TRIP13 is involved in the key mechanism, SAC, an evolutionarily conserved cell-cycle checkpoint supervising the fidelity of chromosome separation in mitosis [25,26]. In further studies researchers take advantage of mitotic makers to investigate the role of TRIP13 in mitosis. Mitotic protein monocolonal 2 (MPM2) has been regarded as a mitotic marker [27]. Then flow cytometry analysis shows that TRIP13-overexpressing multiple myeloma (MM) cells have less MPM2-positive cells compared to control cells when all cells are treated with spindle toxin nocodazole [6]. In addition, phosphorylation of histone H3 at Ser10 has been considered as another mitotic marker [28]. In a similar vein, TRIP13 overexpressing cells have lower expression of phosphorylated histone H3 [6]. These results strengthen the link between the functional SAC and TRIP13. In more details, Mitotic checkpoint complex (MCC), as the SAC effector, which consists of MAD2, BubR1/Mad3 and BUB3, as well as CDC20 [29]. Meanwhile, the MCC can bind and inhibit the anaphase-promoting complex or cyclosome (APC/C) [30,31]. In vitro, TRIP13 catalyzes the conversion of closed MAD2 (C-MAD2) to open MAD2 (O-MAD2), because of its HORMA-domain [2,32,33]. During MCC assembly, O-MAD2 is recruited to unattached kinetochores, provided a catalytic platform for the conversion of C-MAD2 [[34], [35], [36], [37], [38]]. The complete MCC assembly includes two steps: firstly C-MAD2 binds to CDC20 to form MAD2-CDC20 complex, then the complex recruits BubR1 [[39], [40], [41]]. Hoi Tang Ma illuminated that TRIP13 is not only involved in MCC activation but also in MCC inactivation [42]. Although the crucial mechanism for SAC silencing is reported about the ubiquitination and degradation of CDC20 [26,43], another novel mechanism has recently been identified in which MCC disassembled through the joint action of TRIP13 and p31comet, providing a progress involving ATP hydrolysis (Fig. 2) [1,44,45]. For the two mechanisms, TRIP13 and p31comet preferentially catalyze the disassembly of free MCC that not bound to APC/CCdc20 while APC15-mediated conformational changes of the APC/C could allow ubiquitination of Cdc20 in MCC, followed by reactivation of APC/CCdc20 [46,47]. Collectively, these mechanisms reduce the MCC levels and promote the activation of APC/CCdc20 which ubiquitinates securin and cyclin B1 to inactivate CDK 1 (allowing for mitotic exit) and to liberate the protease Separase to initiate the onset of anaphase, respectively (summarized in Fig. 2).
Fig. 2.
Model for the roles of TRIP13 in disassembling the mitotic checkpoint complex (MCC) and in promoting mitotic progression. Unattached kinetochores contribute to the formation of MCC and spindle-assembly checkpoint (SAC) activation. Upon SAC activation, MCC is produced and diffuses into the cytosol to bind and inhibit APC/CCdc20. The SAC signal is negatively regulated by chromosome bi-orientation. During checkpoint silencing, the production of MCC is attenuated due to the binding of p31comet to C-Mad2 in MCC and displaces BubR1–Bub3 from MCC. TRIP13 then disassembles the C-Mad2/Cdc20 complex together with p31comet, and converts Cdc20-bound C-Mad2 to O-Mad2. Furthermore, TRIP13 and p31comet preferentially catalyze the disassembly of free MCC that not bound to APC/CCdc20. Alternatively, APC15-mediated conformational changes of the APC/C can allow ubiquitination of Cdc20 in MCC, followed by reactivation of APC/CCdc20. Collectively, the above mechanisms reduce the MCC levels and promote the activation of APC/CCdc20 which ubiquitinates securin and cyclin B1 to inactivate CDK 1 (allowing for mitotic exit) and liberate the protease separase to initiate the onset of anaphase.
Apart from its functions in the spindle assembly checkpoint of human cells, previous studies have found that TRIP13, the mouse orthologue of pachytene checkpoint 2 (Pch2), mediates the repair of Spo11-generated Double-strand breaks (DSBs) during meiotic cell divisions [[48], [49], [50], [51]]. During the meiosis, the pachytene checkpoint is the surveillant machinery which senses meiotic errors and removes cells containing unrepaired defects, and its function is similar to the spindle checkpoint in the mitosis. It monitors DSB repair and chromosome synapsis, two aspects of meiotic chromosome metabolism in S. cerevisiae and C. elegans [52,53]. As recombination is progressing, homologous chromosomes become paired prominently relied on the synaptonemal complex (SC), which comprises two axial/lateral elements and transverse filaments. In C. elegans, researchers found that various mutations of Pch2 could alleviate the checkpoint-induced meiotic arrest of certain mutants, such as Zip1, Zip2 and Dmc1 [52,54]. In addition, it has been reported that Pch2 is required for monitoring meiotic damage in budding yeast, however deletion of Pch2 suppresses the delay of meiotic progression with occurrence of aberrant SC [53]. Similarly, Pch2 was a requisite element for delaying meiosis progression when SC formation was defective in Drosophila melanogaster [55,56]. Besides the above mechanism, the DSB repair was also proposed to occur when the recombination checkpoint responsed to the defects in DSB processing. The homologous recombination (HR) and nonhomologous end-joining (NHEJ) are the two major DSB repair pathways in general [57]. Plenty of studies suggest that Pch2 could contribute to the establishment of interhomologue biased HR [[58], [59], [60]]. During the DSB progression, Pch2 acts as a modulator of Hop1 which is a specific protein in meiosis and is required for DSB formation, chromosome organization and pairing [61,62]. Meanwhile, Hop1 is a significant phosphor-substrate of Tel1A™/Mec1ATR, and Pch2 promotes its phosphorylation [60,63]. In addition, the deletion of HORMAD1/2 induced by TRIP13 is required for higher order chromosome structures in organisms [64]. Last but not the least, it was recently found the distinct role of TRIP13 in mouse spermatocytes. Besides participating in the formation of synapsis and recombination, TRIP13 was also required for proper sex body formation based on the TRIP13-dependent HORMAD1 and HORMAD2 accumulation at the unsynapsed regions of the X and Y chromosomes, subsequently the sex chromosomes silenced [51]. Furthermore TRIP13 can allow proper H2AX phosphorylation and Small ubiquitin-related modifier 1 (SUMO-1) loading [65].
TRIP13 is essential for DSB repair via NHEJ, a well-known repair pathway in mammalian cells that is active throughout the cell division [66,67]. Since Phospho-histone H2A/H2AX isoform formed following DSB signaling, γH2AX became a marker of DSBs [68]. Western blot analysis demonstrates that knockdown of TRIP13 have more expression of γH2AX in cells. Therefore, it seems that loss of TRIP13 promotes DNA damage [66]. The published evidence supports the interaction between TRIP13 and NHEJ/DNA repair group proteins included KU70, KU80 and DNA-PKcs. In addition, the NHEJ and HR florescent reporter constructs were used for quantifying the level of NHEJ and HR efficiency by flow cytometry. Cells expressing NHEJ reporter constructs with TRIP13 siRNA had less GFP+ cells than control cells [66]. All together, these findings showed that TRIP13 could take part in the NHEJ pathway, and thereby may contribute to the CIN and even human tumorigenesis.
3. Overexpression of TRIP13 Is Associated With Human Cancer
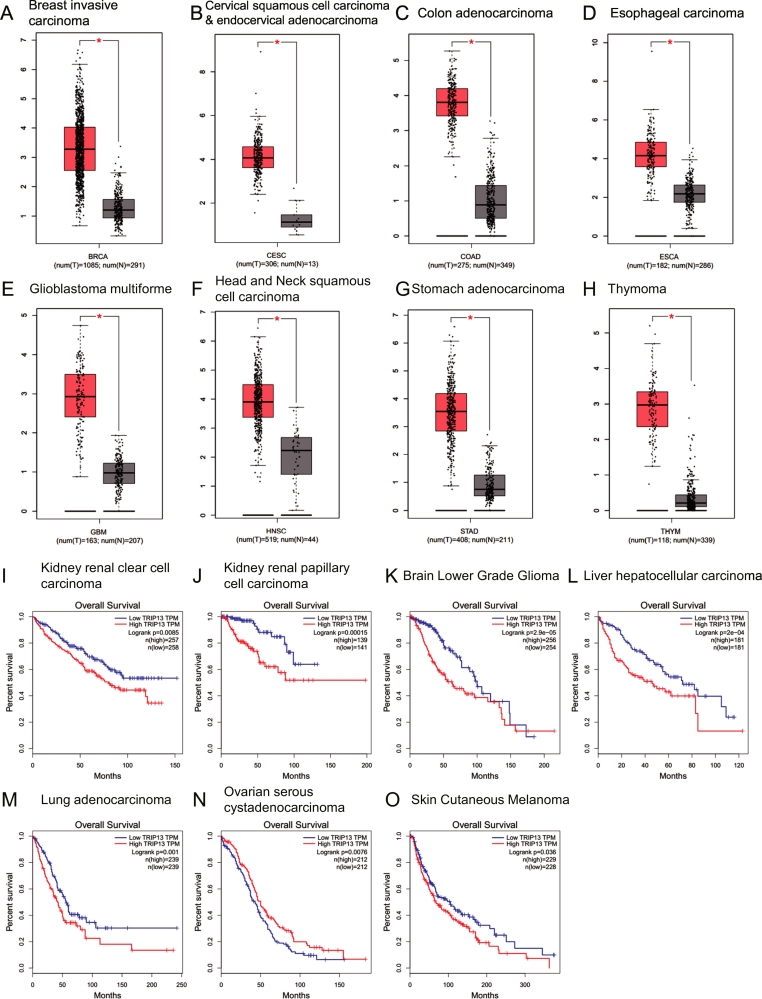
The spindle assembly checkpoint proteins are often aberrantly expressed in tumor cells. Aberrations in their expression can result in CIN and aneuploidy, potentially contributing to tumorigenesis [11,12,69]. It has been reported that TRIP13 is aberrantly expressed in various tumor cells detected by RT-PCR, Western blot and Microarray analysis (Table 1). It seems that TRIP13 overexpression may be a common phenotype in these primary tumors and cancer cell lines. To further understand the clinical outcome of TRIP13 expression, we examined and mined the data about multiple tumors from the GTEx (Genotype-Tissue Expression) and TCGA (The Cancer Genome Atlas) using GEPIA online tool (version 2017, http://gepia.cancer-pku.cn) [70] with customizable functional analysis such as tumor/normal differential gene profiling, patient survival analysis (Fig. 3A–H). We compared TRIP13 gene expression in eight kinds of tumor (breast invasive carcinoma, cervical squamous cell carcinoma and endocervical adenocarcinoma, colon adenocarcinoma, esophageal carcinoma, glioblastoma multiforme, head and neck squamous cell carcinoma, stomach adenocarcinoma, thymoma) samples with normal tissues (Fig. 3, A-H). In addition, the Overall Survival (OS) analysis revealed that high TRIP13 expression conferred inferior outcomes in other carcinomas, such as kidney renal clear cell carcinoma, kidney renal papillary cell carcinoma, brain lower grade glioma, liver hepatocellular carcinoma and so on (Fig. 3I–O). Therefore, the aberrant expression of TRIP13 is a frequent event in cancer cells, indicating a potential oncogenic role of TRIP13 in cancer development. (See Table 2.)
Table 1.
Overview of aberrant expression of TRIP13 in human primary tumors and cancer cell lines investigated.
| Tumor type | TRIP13 expression level | Detection method | Reference number |
|---|---|---|---|
| Wilms tumor | Downregulation | RT-PCR, Western blot | [19] |
| Primary cutaneous T-cell lymphoma | Overexpression | RT-PCR | [100] |
| Non-small cell lung cancer | overexpression | Microarray | [104] |
| Lung adenocarcinoma | Overexpression | Q-PCR, Western blot | [87] |
| Breast cancer | Overexpression | Microarray | [[97], [98], [99]] |
| Prostate cancer | Overexpression | Microarray, Q-PCR, Western blot | [88,101] |
| Colorectal cancer | Overexpression | RT-PCR, Microarray, Western blot | [89,102] |
| Squamous cell carcinoma of the head and neck | Overexpression | RT-PCR, Western blot | [66] |
| Chronic lymphocytic leukemia | Overexpression | Q-PCR | [86] |
| Multiple myeloma | Overexpression | Q-PCR | [6,103] |
Fig. 3.
High TRIP13 expression in tumor tissues compared with normal tissues (A-H) and its high expression linked to a poor prognosis in multiple cancers (I—O). (A-H)TRIP13-expression in cancer tissues (T) is compared with normal counterpart tissues (N), including breast invasive carcinoma (A), cervical squamous cell carcinoma, endocervical adenocarcinoma (B), colon adenocarcinoma (C), esophageal carcinoma (D), glioblastoma multiforme (E), head and neck squamous cell carcinoma (F), stomach adenocarcinoma (G) and thymoma (H). TRIP13 expression is significantly higher in all tumors examined (p < .05). (I—O) Kaplan-Meier analyses of OS revealed that high TRIP13 expression conferred inferior outcomes in kidney renal clear cell carcinoma (I), kidney renal papillary cell carcinoma (J), brain lower grade glioma (K), liver hepatocellular carcinoma (L), lung adenocarcinoma (M), ovarian serous cystadenocarcinoma (N), skin cutaneous melanoma (O). Above tumor/normal differential expression analysis and patient survival analysis are from TCGA and GTEx projects and mining using GEPIA tools (http://gepia.cancer-pku.cn) with a standard pipeline compatible with each other.
Table 2.
Summary of the correlation of TRIP13 expression and clinical outcomes.
| Tumor | The correlation of TRIP13 expression and OS | P Value |
|---|---|---|
| Adrenocortical carcinoma | Negative | 1 * 10−8 |
| Breast invasive carcinoma | NS | 0.11 |
| Cervical squamous cell carcinoma and endocervical adenocarcinoma | NS | 0.69 |
| Cholangio carcinoma | NS | 0.6 |
| Colon adenocarcinoma | NS | 0.18 |
| Lymphoid Neoplasm Diffuse Large B-cell Lymphoma | NS | 0.71 |
| Esophageal carcinoma | NS | 0.9 |
| Glioblastoma multiforme | NS | 0.24 |
| Head and Neck squamous cell carcinoma | NS | 0.94 |
| Kidney Chromophobe | Negative | 0.012 |
| Kidney renal clear cell carcinoma | Negative | 0.0085 |
| Kidney renal papillary cell carcinoma | Negative | 0.00015 |
| Acute Myeloid Leukemia | NS | 0.24 |
| Brain lower grade Glioma | Negative | 2.9 * 10−5 |
| Liver hepatocellular carcinoma | Negative | 2 * 10−4 |
| Lung adenocarcinoma | Negative | 0.001 |
| Lung squamous cell carcinoma | NS | 0.46 |
| Mesothelioma | Negative | 1.3 * 10−6 |
| Ovarian serous cystadenocarcinoma | Negative | 0.0076 |
| Pancreatic adenocarcinoma | Negative | 0.04 |
| Pheochromocytoma and Paraganglioma | NS | 0.17 |
| Prostate adenocarcinoma | NS | 0.74 |
| Rectum adenocarcinoma | NS | 0.94 |
| Sarcoma | NS | 0.063 |
| Skin Cutaneous Melanoma | Negative | 0.036 |
| Stomach adenocarcinoma | NS | 0.99 |
| Testicular Germ Cell Tumors | NS | 0.33 |
| Thyroid carcinoma | NS | 0.36 |
| Thymoma | Negative | 0.048 |
| Uterine Corpus Endometrial Carcinoma | NS | 0.75 |
| Uterine Carcinosarcoma | NS | 0.73 |
| Uveal Melanoma | NS | 0.55 |
4. TRIP13 Aberrant Expression Leads to Tumorigenesis
As mentioned above, studies have validated that TRIP13 is involved in the regulation of spindle assembly checkpoint signaling and DNA damage repair during cell division. Thus aberrant expression of TRIP13 in cancer cells can lead to chromosome segregation errors. Given the impact of mitotic errors on cell proliferation and tumorigenesis [71], the overexpression of TRIP13 may induce tumorigenesis by promoting CIN and aneuploidy. In line with this point, DNA copy number variations (CNVs) analysis illuminates that overexpression of TRIP13 in multiple myeloma cell results in CIN [6,17]. In addition, based on Oncomine (www.oncomine.org), TRIP13 mutation, DNA copy number and gene expression frequency in multiple cancers have been analyzed using meta-analysis. As a result, both TRIP13 copy number and gene expression frequency are increased in multiple cancers such as bladder cancer, breast cancer, cervical cancer, colorectal cancer and esophageal cancer [66].
Given the essential roles of TRIP13 in the regulation of SAC and DNA damage repair, several spindle proteins and signaling pathways may be involved in mediation of TRIP13 on CIN and aneuploidy. The PI3K/Akt signaling pathway is required for maintaining the appearance of supernumerary centrosomes and its uncontrolled activity has been implicated in CIN [72]. Previous study has shown that PI3K inhibitor LY294002 elevates MAD2 levels and restores sensitivity to Bortezomib in TRIP13 overexpressed cells [6]. Besides, the p53 signaling pathway plays a significant role in progression towards apoptotic cell death following injury and cell cycle status [73,74]. It was reported that TRIP13 directly interacted with a p53 co-factor called Tetratricopeptide Repeat Domain 5 (TTC5), and knockdown of TRIP13 in murine inner medullary collecting duct cells enhanced the activity of p53 at Serine 15 [75]. Additionally, it was observed that TRIP13 was higher in p53−/− NIH/3 T3 cells and over 10% of MM patients were diagnosed with the identification of p53 deletion [76]. Taken together, TRIP13 may be involved in the PI3K/Akt signaling pathway associated with CIN and tumorigenesis.
Cell aneuploidy, an important factor in tumorigenesis can be caused by aberrant expression of SAC proteins [77,78]. Abnormal expressions of MAD2 and BubR1, key components of SAC, have been reported in various cancers [12,[79], [80], [81], [82], [83]]. Previous studies have demonstrated that TRIP13 can induce proteasome degradation of MAD2. Moreover, TRIP13 regulates the activation and inactivation of SAC depended on the MAD2 [6] level maintained by p31comet. Subsequently, dysfunctional SAC induces CIN [42]. Recent studies have confirmed that the combinaction of TRIP13 and chaperonin containing TCP1 promotes the sufficient disassembly of MCC, which is essential for the inactivation of the mitotic checkpoint [84]. These studies suggest that TRIP13 is involved in the regulation of MCC induced CIN in tumor cells.
In summary, the expression and activity of TRIP13 are necessary for the accuracy of chromosome segregation. It strongly suggests that TRIP13 is an oncogene capable of monitoring the fidelity of chromosome segregation by several pathways, which is consistent with the convinced evidence of the effects of TRIP13 on cell physiology. Hence, TRIP13 may be involved in the process of tumorigenesis.
5. Elevated TRIP13 Promotes Tumor Progression
Studies in various types of cancers have demonstrated that overexpression of TRIP13 promotes cell proliferation, while its suppression with siRNA or shRNA inhibits proliferation and induces cell death [6,66,[85], [86], [87], [88], [89]]. Moreover, TRIP13-overexpressing cancer cells showed a significant increase in proliferation, invasion and migration compared with control cells [66]. In a xenograft mouse model, subcutaneous injection of TRIP13 shRNA around the tumor nodules led to reduction of tumor size compared with those of control shRNA injection [6,66]. On the contrary, in chick chorioallantoic membrane model, overexpression of TRIP13 in NIH3T3 cells resulted in significantly more cellular stratification and proliferation. In addition, high expression of TRIP13 promoted malignant transformation, enhanced repair of DNA damage as well as aggressive, treatment-resistant tumors, and TRIP13 overexpressing tumors conferred xenograft mice poorer survival than controls [66]. Meanwhile, the prognosis analysis on TCGA datasets indicate that patients with high TRIP13 expression had inferior outcomes than those patients with low TRIP13 expression (Fig. 3I–O) [6]. Furthermore, recent studies showed that TRIP13 expression was positively associated with MAD2 expression in multiple myeloma and breast cancer [6,18]. In addition, the expression of TRIP13 was positively associated with cancer grade and tumor size in breast invasive ductal carcinoma. These data all point that TRIP13 facilitates tumor progression both in vitro and in vivo, and elevated TRIP13 levels can lead to CIN and aneuploidy, which will ultimately trigger tumorigenesis and promote tumor development.
6. TRIP13 Contributes to Drug Resistance
The major reason for cancer treatment failure is the drug resistance. Recent studies have implicated that overexpression of TRIP13 exhibited less sensitivity to anticancer drugs (bortezomib and cisplatin) [6,66]. Cell viability assay showed that the number of viable cells in multiple myeloma cells transfected with TRIP13 was dramatically higher compared with control cells when treated with anticancer drugs bortezomib and etoposide [6]. Similarly, squamous cell carcinoma of the head and neck cells overexpressed TRIP13 exhibited less sensitivity to cisplatin compared with control cells [66]. Thus it is clear that TRIP13 plays a role in cancer cell drug resistance. To understand the contribution of TRIP13 to drug resistance, the researchers conducted flow cytometry to detect apoptotic cells by annexin V/Hoechst 33258 staining. The results indicated that MM cells overexpressed TRIP13 showed decreased apoptosis and protection from drug-induced cytotoxicity compared with cells transfected with empty vectors when treated with serial dosages of bortezomib. Consistently, G2/M cell cycle arrest induced by bortezomib was inhibited in MM cells overexpressed TRIP13 compared with those control cells [6]. Moreover, shRNA-mediated TRIP13 knockdown in MM cells overcame drug resistance and induced apoptosis in vitro as well as in a xenograft myeloma mouse model. Downregulation of TRIP13 in cancer cells increased the level of cleaved PARP and activation of caspase-3, indicating a possible role of TRIP13 against the apoptosis pathway [6]. Likewise, in human chronic lymphocytic leukemia the microarray data analyzed by Ingenuity Pathway Analysis “canonical pathway” module indicated that TRIP13 participated in several pathways involved in apoptosis such as “induction of apoptosis by HIV1”, “p53 signaling” and “PPAR signaling”. Furthermore, knockdown of TRIP13 induced a remarkable up-regulation of caspase 3/7 activity in Granta-519 and JVM-2 cells, both of which are B-cell Lympocytic Leukemia cell lines. The mechanism by which TRIP13 contributes to chronic lymphocytic leukemia was confirmed through the C-MYC/TRIP13/PUMA axis regulation [86]. The other group also found that TRIP13 knockdown in Squamous cell carcinoma of the head and neck cancer cells induces cell cycle arrest. There is more accumulation of DSB marker observed in cells transfected with siTRIP13. Western blot indicated that siTRIP13-mediated DSB precedes apoptosis [66]. These results strongly indicated that TRIP13 could enhance DNA repair and then induce treatment resistance. Taken together, TRIP13-induced anti-apoptosis action may contribute to the high drug resistance in cancer cells, because one of the main mechanisms of anticancer drugs used to stimulate cell death is induction of apoptosis.
Dysfunctions in MCC surveillance system facilitate chromosome mis-segregation and failure to arrest in mitosis, ultimately leading to the development of human cancers and drug resistance in cancer [90]. Recent study has supported that overexpression of TRIP13 decreased MAD2 protein levels [6]. When the MCC surveillance system is turned on, MAD2 forms a complex with APC/C, preventing the degradation of securin and cyclin B1, and consequently arresting cells at prometaphase [91]. Interestingly, the increased expression of MAD2 protein results in subsequent CIN and drug resistance to chemotherapeutic agents that target microtubules [92]. However, the down-regulation or deletion of MAD2 also has been reported in a variety of human cancers. Moreover, down-regulation of MAD2 is shown to accelerate proliferation and enhance the drug resistance in gastric cancer cells [93]. There is evidence that the PI3K/Akt signaling pathway plays a critical role in the adjustment of proliferation, migration and drug resistance of MM cells [94]. Meanwhile, the ubiquitination, phosphorylation and degradation of other proteins can regulate tumorigenesis and chemoresistance when PI3K/Akt is activated [95,96]. It's likely that MAD2 degradation and ubiquitination are induced by TRIP13 via activating Akt signaling pathway, which further results in damaged checkpoint surveillance and consequent drug resistance [6].
7. Targeting TRIP13 May Be Perspective for the Treatment of Cancer
Given the rationale mentioned above, there is no doubt that TRIP13 contributes to tumorigenesis, tumor progression, and drug resistance in various human cancers, and it may be an ideal target for therapy in cancer. To explore the roles of TRIP13 in human breast ductal carcinoma progression, researchers correlated the expression of TRIP13 to some of the pathological characteristics in human breast ductal carcinoma. Breast cancer patients with high expression of TRIP13 showed higher mortality and recurrence rate than TRIP13 low expression patients [97,98]. In consistence, it has been corroborated that expression of TRIP13 was associated with detrimental relapse free survival (RFS) and OS in luminal tumors which are a breast cancer subtype that expresses hormone receptors [99]. In human Mycosis Fungoides Tumor, TRIP13 is highly upregulated versus control biopsies [100]. Likewise, previous study in Metastatic prostate cancer has shown that TRIP13 expression in combination with Gleason score and preoperative prostate-specific antigen (PSA) level was able to correctly predict recurrence in 85.7% of cases [101]. In line with the previous study, TRIP13 was significantly associated with OS in colorectal cancer patients [102]. Moreover, Kaplan-Meier survival analysis of OS of patients from TT2 and TT3 has validated that high expression of TRIP13 is strongly linked to poor survival in multiple myeloma [6,103]. Furthermore, the TRIP13 mRNA levels of CD19+ B cells were 4 fold higher in chronic lymphocytic leukemia patient than in the healthy person [86]. What's more, comparative genomic hybridization (CGH) study revealed that TRIP13 (13/19; 68%) was involved in genomic copy number changes (≥40% of patients) in NSCLC. Thus, the chromosomal changes induced by TRIP13 are involved in NSCLC tumorigenesis [104]. In addition, survival curves for mice with TRIP13 overexpressed tumors show inferior survival than those with control tumors in vivo [66]. Meanwhile, Kaplan-Meier analyses of various cancer samples provided by Zhang's Lab, indicated that higher expression of TRIP13 is associated with shorter OS in examined tumors (Fig. 3). Taken together, those data suggest that TRIP13 is a novel potential biomarker for diagnosis and a possible therapeutic target for cancer.
Of great potential but with a little focus is to find the cause of aberrant expression of TRIP13, such as epigenetic changes or ncRNAs, which may provide insights into promising therapeutic target on TRIP13. For example, miR192 was recently reported to target TRIP13 during colorectal cancer progression [105]. Furthermore, termed TINCR (Terminal differentiation-induced non-coding RNA) expressed in prostate cancer tissue cell lines, in a manner of negatively regulating the TRIP13 mRNA and protein, inhibiting cell proliferation, migration and invasion [106]. Considering the multifunctional roles of regulatory ncRNAs, it would pave another way to design and develop therapeutic nucleic acid drugs to treat TRIP13 aberrant expressed diseases.
8. Conclusion
TRIP13 plays a key role in several biological processes. However, high expression of TRIP13 is frequently observed in various human cancers. Silencing TRIP13 sensitizes tumor cells to chemotherapeutics. These evidences together suggest that TRIP13 may be a novel therapeutic target for human cancers. To develop specific TRIP13 inhibitor is of great importance. Clinical studies are demanded to further confirm TRIP13 as a potential therapeutic target in TRIP13High cancers.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by National Natural Science Foundation of China 81770220, 81600177, 81670200 (to CG & YY); The 2016 outstanding youth fund of Jiangsu Province BK20160048 (to YY); Natural Science Foundation of Jiangsu Province BK20161041 (to CG); National key research and development program-precision medicine sub-program 2016YFC0905900 (to YY); The Priority Academic Program Development of Jiangsu Higher Education Institutions for Chinese Medicine.
Contributor Information
C. Gu, Email: guchunyan@njucm.edu.cn.
Y. Yang, Email: yangye876@sina.com.
References
- 1.Miniowitz-Shemtov S., Eytan E., Kaisari S., Sitry-Shevah D., Hershko A. Mode of interaction of TRIP13 AAA-ATPase with the Mad2-binding protein p31comet and with mitotic checkpoint complexes. Proc Natl Acad Sci U S A. 2015;112(37):11536–11540. doi: 10.1073/pnas.1515358112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vader G. Pch2(TRIP13): controlling cell division through regulation of HORMA domains. Chromosoma. 2015;124(3):333–339. doi: 10.1007/s00412-015-0516-y. [DOI] [PubMed] [Google Scholar]
- 3.Ye Q., Kim D.H., Dereli I., Rosenberg S.C., Hagemann G., Herzog F. The AAA+ ATPase TRIP13 remodels HORMA domains through N-terminal engagement and unfolding. EMBO J. 2017;36(16):2419–2434. doi: 10.15252/embj.201797291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brulotte M.L., Jeong B.C., Li F.X., Li B., Yu E.B., Wu Q. Mechanistic insight into TRIP13-catalyzed Mad2 structural transition and spindle checkpoint silencing. Nat Commun. 2017;8 doi: 10.1038/s41467-017-02012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alfieri C., Chang L.F., Barford D. Mechanism for remodelling of the cell cycle checkpoint protein MAD2 by the ATPase TRIP13. Nature. 2018;559(7713) doi: 10.1038/s41586-018-0281-1. [274−+] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tao Y., Yang G., Yang H., Song D., Hu L., Xie B. TRIP13 impairs mitotic checkpoint surveillance and is associated with poor prognosis in multiple myeloma. Oncotarget. 2017;8(16):26718–26731. doi: 10.18632/oncotarget.14957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Musacchio A., Salmon E.D. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8(5):379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 8.Lara-Gonzalez P., Westhorpe F.G., Taylor S.S. The spindle assembly checkpoint. Curr Biol. 2012;22(22):R966–R980. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Chao W.C., Kulkarni K., Zhang Z., Kong E.H., Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484(7393):208–213. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- 10.de Carcer G., Malumbres M. A centrosomal route for cancer genome instability. Nat Cell Biol. 2014;16(6):504–506. doi: 10.1038/ncb2978. [DOI] [PubMed] [Google Scholar]
- 11.Sotillo R., Schvartzman J.M., Socci N.D., Benezra R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature. 2010;464(7287):436–440. doi: 10.1038/nature08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bargiela-Iparraguirre J., Prado-Marchal L., Pajuelo-Lozano N., Jimenez B., Perona R., Sanchez-Perez I. Mad2 and BubR1 modulates tumourigenesis and paclitaxel response in MKN45 gastric cancer cells. Cell Cycle. 2014;13(22):3590–3601. doi: 10.4161/15384101.2014.962952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tusell L., Pampalona J., Soler D., Frias C., Genesca A. Different outcomes of telomere-dependent anaphase bridges. Biochem Soc Trans. 2010;38(6):1698–1703. doi: 10.1042/BST0381698. [DOI] [PubMed] [Google Scholar]
- 14.Stewenius Y., Gorunova L., Jonson T., Larsson N., Hoglund M., Mandahl N. Structural and numerical chromosome changes in colon cancer develop through telomere-mediated anaphase bridges, not through mitotic multipolarity. Proc Natl Acad Sci U S A. 2005;102(15):5541–5546. doi: 10.1073/pnas.0408454102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bailey S.M., Murnane J.P. Telomeres, chromosome instability and cancer. Nucleic Acids Res. 2006;34(8):2408–2417. doi: 10.1093/nar/gkl303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mills K.D., Ferguson D.O., Alt F.W. The role of DNA breaks in genomic instability and tumorigenesis. Immunol Rev. 2003;194:77–95. doi: 10.1034/j.1600-065x.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhou W., Yang Y., Xia J., Wang H., Salama M.E., Xiong W. NEK2 induces drug resistance mainly through activation of efflux drug pumps and is associated with poor prognosis in myeloma and other cancers. Cancer Cell. 2013;23(1):48–62. doi: 10.1016/j.ccr.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K., Sturt-Gillespie B., Hittle J.C., Macdonald D., Chan G.K., Yen T.J. Thyroid hormone receptor interacting protein 13 (TRIP13) AAA-ATPase is a novel mitotic checkpoint-silencing protein. J Biol Chem. 2014;289(34):23928–23937. doi: 10.1074/jbc.M114.585315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yost S., de Wolf B., Hanks S., Zachariou A., Marcozzi C., Clarke M. Biallelic TRIP13 mutations predispose to Wilms tumor and chromosome missegregation. Nat Genet. 2017;49(7):1148–1151. doi: 10.1038/ng.3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter S.L., Eklund A.C., Kohane I.S., Harris L.N., Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38(9):1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 21.Yang M., Li B., Tomchick D.R., Machius M., Rizo J., Yu H. p31comet blocks Mad2 activation through structural mimicry. Cell. 2007;131(4):744–755. doi: 10.1016/j.cell.2007.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tipton A.R., Wang K., Oladimeji P., Sufi S., Gu Z., Liu S.T. Identification of novel mitosis regulators through data mining with human centromere/kinetochore proteins as group queries. BMC Cell Biol. 2012;13:15. doi: 10.1186/1471-2121-13-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Habu T., Kim S.H., Weinstein J., Matsumoto T. Identification of a MAD2-binding protein, CMT2, and its role in mitosis. EMBO J. 2002;21(23):6419–6428. doi: 10.1093/emboj/cdf659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mapelli M., Filipp F.V., Rancati G., Massimiliano L., Nezi L., Stier G. Determinants of conformational dimerization of Mad2 and its inhibition by p31comet. EMBO J. 2006;25(6):1273–1284. doi: 10.1038/sj.emboj.7601033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S., Yu H. Mutual regulation between the spindle checkpoint and APC/C. Semin Cell Dev Biol. 2011;22(6):551–558. doi: 10.1016/j.semcdb.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Musacchio A. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr Biol. 2015;25(20):R1002–R1018. doi: 10.1016/j.cub.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 27.Shin S.B., Woo S.U., Yim H. Differential cellular effects of Plk1 inhibitors targeting the ATP-binding domain or polo-box domain. J Cell Physiol. 2015;230(12):3057–3067. doi: 10.1002/jcp.25042. [DOI] [PubMed] [Google Scholar]
- 28.Wilkins B.J., Rall N.A., Ostwal Y., Kruitwagen T., Hiragami-Hamada K., Winkler M. A cascade of histone modifications induces chromatin condensation in mitosis. Science. 2014;343(6166):77–80. doi: 10.1126/science.1244508. [DOI] [PubMed] [Google Scholar]
- 29.Sudakin V., Chan G.K., Yen T.J. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154(5):925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pines J. Cubism and the cell cycle: the many faces of the APC/C. Nat Rev Mol Cell Biol. 2011;12(7):427–438. doi: 10.1038/nrm3132. [DOI] [PubMed] [Google Scholar]
- 31.Jia L., Kim S., Yu H. Tracking spindle checkpoint signals from kinetochores to APC/C. Trends Biochem Sci. 2013;38(6):302–311. doi: 10.1016/j.tibs.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 32.Ye Q., Rosenberg S.C., Moeller A., Speir J.A., Su T.Y., Corbett K.D. TRIP13 is a protein-remodeling AAA+ ATPase that catalyzes MAD2 conformation switching. eLife. 2015;4 doi: 10.7554/eLife.07367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eytan E., Wang K., Miniowitz-Shemtov S., Sitry-Shevah D., Kaisari S., Yen T.J. Disassembly of mitotic checkpoint complexes by the joint action of the AAA-ATPase TRIP13 and p31(comet) Proc Natl Acad Sci U S A. 2014;111(33):12019–12024. doi: 10.1073/pnas.1412901111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Antoni A., Pearson C.G., Cimini D., Canman J.C., Sala V., Nezi L. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol. 2005;15(3):214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 35.Mapelli M., Musacchio A. MAD contortions: conformational dimerization boosts spindle checkpoint signaling. Curr Opin Struct Biol. 2007;17(6):716–725. doi: 10.1016/j.sbi.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 36.Howell B.J., Moree B., Farrar E.M., Stewart S., Fang G., Salmon E.D. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr Biol. 2004;14(11):953–964. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- 37.Shah J.V., Botvinick E., Bonday Z., Furnari F., Berns M., Cleveland D.W. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol. 2004;14(11):942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- 38.Ciliberto A., Shah J.V. A quantitative systems view of the spindle assembly checkpoint. EMBO J. 2009;28(15):2162–2173. doi: 10.1038/emboj.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faesen A.C., Thanasoula M., Maffini S., Breit C., Muller F., van Gerwen S. Basis of catalytic assembly of the mitotic checkpoint complex. Nature. 2017;542(7642):498–502. doi: 10.1038/nature21384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simonetta M., Manzoni R., Mosca R., Mapelli M., Massimiliano L., Vink M. The influence of catalysis on mad2 activation dynamics. PLoS Biol. 2009;7(1) doi: 10.1371/journal.pbio.1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kulukian A., Han J.S., Cleveland D.W. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev Cell. 2009;16(1):105–117. doi: 10.1016/j.devcel.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma H.T., Poon R.Y. TRIP13 regulates both the activation and inactivation of the spindle-assembly checkpoint. Cell Rep. 2016;14(5):1086–1099. doi: 10.1016/j.celrep.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 43.Primorac I., Musacchio A. Panta rhei: the APC/C at steady state. J Cell Biol. 2013;201(2):177–189. doi: 10.1083/jcb.201301130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miniowitz-Shemtov S., Teichner A., Sitry-Shevah D., Hershko A. ATP is required for the release of the anaphase-promoting complex/cyclosome from inhibition by the mitotic checkpoint. Proc Natl Acad Sci U S A. 2010;107(12):5351–5356. doi: 10.1073/pnas.1001875107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eytan E., Sitry-Shevah D., Teichner A., Hershko A. Roles of different pools of the mitotic checkpoint complex and the mechanisms of their disassembly. Proc Natl Acad Sci U S A. 2013;110(26):10568–10573. doi: 10.1073/pnas.1308928110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma H.T., Poon R.Y.C. TRIP13 functions in the establishment of the spindle assembly checkpoint by replenishing O-MAD2. Cell Rep. 2018;22(6):1439–1450. doi: 10.1016/j.celrep.2018.01.027. [DOI] [PubMed] [Google Scholar]
- 47.Kim D.H., Han J.S., Ly P., Ye Q., McMahon M.A., Myung K. TRIP13 and APC15 drive mitotic exit by turnover of interphase- and unattached kinetochore-produced MCC. Nat Commun. 2018;9(1):4354. doi: 10.1038/s41467-018-06774-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X.C., Schimenti J.C. Mouse pachytene checkpoint 2 (trip13) is required for completing meiotic recombination but not synapsis. PLoS Genet. 2007;3(8) doi: 10.1371/journal.pgen.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li X.C., Bolcun-Filas E., Schimenti J.C. Genetic evidence that synaptonemal complex axial elements govern recombination pathway choice in mice. Genetics. 2011;189(1):71–82. doi: 10.1534/genetics.111.130674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keeney S. Spo11 and the formation of DNA double-Strand breaks in meiosis. Genome Dyn Stab. 2008;2:81–123. doi: 10.1007/7050_2007_026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pacheco S., Marcet-Ortega M., Lange J., Jasin M., Keeney S., Roig I. The ATM signaling cascade promotes recombination-dependent pachytene arrest in mouse spermatocytes. PLoS Genet. 2015;11(3) doi: 10.1371/journal.pgen.1005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhalla N., Dernburg A.F. A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science. 2005;310(5754):1683–1686. doi: 10.1126/science.1117468. [DOI] [PubMed] [Google Scholar]
- 53.Wu H.Y., Burgess S.M. Two distinct surveillance mechanisms monitor meiotic chromosome metabolism in budding yeast. Curr Biol. 2006;16(24):2473–2479. doi: 10.1016/j.cub.2006.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.San-Segundo P.A., Roeder G.S. Pch2 links chromatin silencing to meiotic checkpoint control. Cell. 1999;97(3):313–324. doi: 10.1016/s0092-8674(00)80741-2. [DOI] [PubMed] [Google Scholar]
- 55.Joyce E.F., McKim K.S. Drosophila PCH2 is required for a pachytene checkpoint that monitors double-strand-break-independent events leading to meiotic crossover formation. Genetics. 2009;181(1):39–51. doi: 10.1534/genetics.108.093112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Joyce E.F., KS McKim. Chromosome axis defects induce a checkpoint-mediated delay and interchromosomal effect on crossing over during Drosophila meiosis. PLoS Genet. 2010;6(8) doi: 10.1371/journal.pgen.1001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chapman J.R., Taylor M.R., Boulton S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47(4):497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 58.Joshi N., Brown M.S., Bishop D.K., Borner G.V. Gradual implementation of the meiotic recombination program via checkpoint pathways controlled by global DSB levels. Mol Cell. 2015;57(5):797–811. doi: 10.1016/j.molcel.2014.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zanders S., Sonntag Brown M., Chen C., Alani E. Pch2 modulates chromatid partner choice during meiotic double-strand break repair in Saccharomyces cerevisiae. Genetics. 2011;188(3):511–521. doi: 10.1534/genetics.111.129031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ho H.C., Burgess S.M. Pch2 acts through Xrs2 and Tel1/ATM to modulate interhomolog bias and checkpoint function during meiosis. PLoS Genet. 2011;7(11) doi: 10.1371/journal.pgen.1002351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim Y., Rosenberg S.C., Kugel C.L., Kostow N., Rog O., Davydov V. The chromosome axis controls meiotic events through a hierarchical assembly of HORMA domain proteins. Dev Cell. 2014;31(4):487–502. doi: 10.1016/j.devcel.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Panizza S., Mendoza M.A., Berlinger M., Huang L., Nicolas A., Shirahige K. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell. 2011;146(3):372–383. doi: 10.1016/j.cell.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 63.Lo Y.H., Chuang C.N., Wang T.F. Pch2 prevents Mec1/Tel1-mediated Hop1 phosphorylation occurring independently of Red1 in budding yeast meiosis. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0085687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roig I., Dowdle J.A., Toth A., de Rooij D.G., Jasin M., Keeney S. Mouse TRIP13/PCH2 is required for recombination and normal higher-order chromosome structure during meiosis. PLoS Genet. 2010;6(8) doi: 10.1371/journal.pgen.1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marcet-Ortega M., Pacheco S., Martinez-Marchal A., Castillo H., Flores E., Jasin M. p53 and TAp63 participate in the recombination-dependent pachytene arrest in mouse spermatocytes. PLoS Genet. 2017;13(6) doi: 10.1371/journal.pgen.1006845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Banerjee R., Russo N., Liu M., Basrur V., Bellile E., Palanisamy N. TRIP13 promotes error-prone nonhomologous end joining and induces chemoresistance in head and neck cancer. Nat Commun. 2014;5:4527. doi: 10.1038/ncomms5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deriano L., Roth D.B. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu Rev Genet. 2013;47:433–455. doi: 10.1146/annurev-genet-110711-155540. [DOI] [PubMed] [Google Scholar]
- 68.Li A.Y., Boo L.M., Wang S.Y., Lin H.H., Wang C.C., Yen Y. Suppression of nonhomologous end joining repair by overexpression of HMGA2. Cancer Res. 2009;69(14):5699–5706. doi: 10.1158/0008-5472.CAN-08-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bakhoum S.F., Compton D.A. Chromosomal instability and cancer: a complex relationship with therapeutic potential. J Clin Invest. 2012;122(4):1138–1143. doi: 10.1172/JCI59954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Levine M.S., Holland A.J. The impact of mitotic errors on cell proliferation and tumorigenesis. Genes Dev. 2018;32(9–10):620–638. doi: 10.1101/gad.314351.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nam H.J., Chae S., Jang S.H., Cho H., Lee J.H. The PI3K-Akt mediates oncogenic met-induced centrosome amplification and chromosome instability. Carcinogenesis. 2010;31(9):1531–1540. doi: 10.1093/carcin/bgq133. [DOI] [PubMed] [Google Scholar]
- 73.Flynn R.L., Zou L. ATR: a master conductor of cellular responses to DNA replication stress. Trends Biochem Sci. 2011;36(3):133–140. doi: 10.1016/j.tibs.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim Y.J., Kim T.W., Park S.R., Kim H.T., Jung D.Y., Ryu S.Y. Deletion of NAD(P)H:quinone oxidoreductase 1 represses Mre11-Rad50-Nbs1 complex protein expression in cisplatin-induced nephrotoxicity. Toxicol Lett. 2016;243:22–30. doi: 10.1016/j.toxlet.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 75.Pressly J.D., Hama T., Brien S.O., Regner K.R., Park F. TRIP13-deficient tubular epithelial cells are susceptible to apoptosis following acute kidney injury. Sci Rep. 2017;7:43196. doi: 10.1038/srep43196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Teoh P.J., Chung T.H., Sebastian S., Choo S.N., Yan J., Ng S.B. p53 haploinsufficiency and functional abnormalities in multiple myeloma. Leukemia. 2014;28(10):2066–2074. doi: 10.1038/leu.2014.102. [DOI] [PubMed] [Google Scholar]
- 77.Kops G.J., Weaver B.A., Cleveland D.W. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5(10):773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 78.Fang X., Zhang P. Aneuploidy and tumorigenesis. Semin Cell Dev Biol. 2011;22(6):595–601. doi: 10.1016/j.semcdb.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hanks S., Coleman K., Reid S., Plaja A., Firth H., Fitzpatrick D. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet. 2004;36(11):1159–1161. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 80.Wan X., Yeung C., Kim S.Y., Dolan J.G., Ngo V.N., Burkett S. Identification of FoxM1/Bub1b signaling pathway as a required component for growth and survival of rhabdomyosarcoma. Cancer Res. 2012;72(22):5889–5899. doi: 10.1158/0008-5472.CAN-12-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fu X., Chen G., Cai Z.D., Wang C., Liu Z.Z., Lin Z.Y. Overexpression of BUB1B contributes to progression of prostate cancer and predicts poor outcome in patients with prostate cancer. Onco Targets Ther. 2016;9:2211–2220. doi: 10.2147/OTT.S101994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yamamoto Y., Matsuyama H., Chochi Y., Okuda M., Kawauchi S., Inoue R. Overexpression of BUBR1 is associated with chromosomal instability in bladder cancer. Cancer Genet Cytogenet. 2007;174(1):42–47. doi: 10.1016/j.cancergencyto.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 83.Scully R. The spindle-assembly checkpoint, aneuploidy, and gastrointestinal cancer. N Engl J Med. 2010;363(27):2665–2666. doi: 10.1056/NEJMe1008017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kaisari S., Sitry-Shevah D., Miniowitz-Shemtov S., Teichner A., Hershko A. Role of CCT chaperonin in the disassembly of mitotic checkpoint complexes. Proc Natl Acad Sci U S A. 2017;114(5):956–961. doi: 10.1073/pnas.1620451114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kurita K., Maeda M., Mansour M., Kokuryo T., Uehara K., Yokoyama Y. TRIP13 is expressed in colorectal cancer and promotes cancer cell invasion. Oncol Lett. Dec 2016;12(6):5240–5246. doi: 10.3892/ol.2016.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou K., Zhang W., Zhang Q., Gui R., Zhao H., Chai X. Loss of thyroid hormone receptor interactor 13 inhibits cell proliferation and survival in human chronic lymphocytic leukemia. Oncotarget. 2017;8(15):25469–25481. doi: 10.18632/oncotarget.16038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li W., Zhang G., Li X., Wang X., Li Q., Hong L. Thyroid hormone receptor interactor 13 (TRIP13) overexpression associated with tumor progression and poor prognosis in lung adenocarcinoma. Biochem Biophys Res Commun. 2018;499(3):416–424. doi: 10.1016/j.bbrc.2018.03.129. [DOI] [PubMed] [Google Scholar]
- 88.Dong L., Ding H., Li Y., Xue D., Li Z., Liu Y. TRIP13 is a predictor for poor prognosis and regulates cell proliferation, migration and invasion in prostate cancer. Int J Biol Macromol. 2018;121:200–206. doi: 10.1016/j.ijbiomac.2018.09.168. [DOI] [PubMed] [Google Scholar]
- 89.Sheng N., Yan L., Wu K., You W., Gong J., Hu L. TRIP13 promotes tumor growth and is associated with poor prognosis in colorectal cancer. Cell Death Dis. 2018;9(3):402. doi: 10.1038/s41419-018-0434-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McGranahan N., Burrell R.A., Endesfelder D., Novelli M.R., Swanton C. Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep. 2012;13(6):528–538. doi: 10.1038/embor.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lischetti T., Nilsson J. Regulation of mitotic progression by the spindle assembly checkpoint. Mol Cell Oncol. 2015;2(1) doi: 10.4161/23723548.2014.970484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schvartzman J.M., Sotillo R., Benezra R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat Rev Cancer. 2010;10(2):102–115. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Furlong F., Fitzpatrick P., O'Toole S., Phelan S., McGrogan B., Maguire A. Low MAD2 expression levels associate with reduced progression-free survival in patients with high-grade serous epithelial ovarian cancer. J Pathol. 2012;226(5):746–755. doi: 10.1002/path.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu J., Wang M., Cao B., Hou T., Mao X. Targeting the phosphatidylinositol 3-kinase/AKT pathway for the treatment of multiple myeloma. Curr Med Chem. 2014;21(27):3173–3187. doi: 10.2174/0929867321666140601204513. [DOI] [PubMed] [Google Scholar]
- 95.Lee M.S., Jeong M.H., Lee H.W., Han H.J., Ko A., Hewitt S.M. PI3K/AKT activation induces PTEN ubiquitination and destabilization accelerating tumourigenesis. Nat Commun. 2015;6:7769. doi: 10.1038/ncomms8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Abedini M.R., Muller E.J., Bergeron R., Gray D.A., Tsang B.K. Akt promotes chemoresistance in human ovarian cancer cells by modulating cisplatin-induced, p53-dependent ubiquitination of FLICE-like inhibitory protein. Oncogene. 2010;29(1):11–25. doi: 10.1038/onc.2009.300. [DOI] [PubMed] [Google Scholar]
- 97.Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci U S A. 2004;101(25):9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Martin K.J., Patrick D.R., Bissell M.J., Fournier M.V. Prognostic breast cancer signature identified from 3D culture model accurately predicts clinical outcome across independent datasets. PLoS One. 2008;3(8) doi: 10.1371/journal.pone.0002994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nieto-Jimenez C., Alcaraz-Sanabria A., Paez R., Perez-Pena J., Corrales-Sanchez V., Pandiella A. DNA-damage related genes and clinical outcome in hormone receptor positive breast cancer. Oncotarget. 2017;8(38):62834–62841. doi: 10.18632/oncotarget.10886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van Kester M.S., Borg M.K., Zoutman W.H., Out-Luiting J.J., Jansen P.M., Dreef E.J. A meta-analysis of gene expression data identifies a molecular signature characteristic for tumor-stage mycosis fungoides. J Invest Dermatol. 2012;132(8):2050–2059. doi: 10.1038/jid.2012.117. [DOI] [PubMed] [Google Scholar]
- 101.Larkin S.E., Holmes S., Cree I.A., Walker T., Basketter V., Bickers B. Identification of markers of prostate cancer progression using candidate gene expression. Br J Cancer. 2012;106(1):157–165. doi: 10.1038/bjc.2011.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Abdul Aziz N.A., Mokhtar N.M., Harun R., Mollah M.M., Mohamed Rose I., Sagap I. A 19-gene expression signature as a predictor of survival in colorectal cancer. BMC Med Genomics. 2016;9(1):58. doi: 10.1186/s12920-016-0218-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Heuck C.J., Qu P., van Rhee F., Waheed S., Usmani S.Z., Epstein J. Five gene probes carry most of the discriminatory power of the 70-gene risk model in multiple myeloma. Leukemia. 2014;28(12):2410–2413. doi: 10.1038/leu.2014.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kang J.U., Koo S.H., Kwon K.C., Park J.W., Kim J.M. Gain at chromosomal region 5p15.33, containing TERT, is the most frequent genetic event in early stages of non-small cell lung cancer. Cancer Genet Cytogenet. 2008;182(1):1–11. doi: 10.1016/j.cancergencyto.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 105.Agarwal S.V.B., Chakravarthi D.S., Chandrashekar, Guo Rong Jun, Pran Datta S.V., Manne Upender. Proceedings of the American association for cancer research annual meeting 2018. 2018 Apr 14–18. Trip13, a target of miR192, facilitates colorectal cancer progression through WNT/β-catenin signaling [abstract] (Cancer Res). [Chicago, IL; 2018] [Google Scholar]
- 106.Dong L.M., Ding H.L., Li Y.P., Xue D.W., Liu Y.L. LncRNA TINCR is associated with clinical progression and serves as tumor suppressive role in prostate cancer. Cancer Manag Res. 2018;10:2799–2807. doi: 10.2147/CMAR.S170526. [DOI] [PMC free article] [PubMed] [Google Scholar]