Abstract
Arthrogryposis is a clinical finding that is present either as a feature of a neuromuscular condition or as part of a systemic disease in over 400 Mendelian conditions. The underlying molecular etiology remains largely unknown because of genetic and phenotypic heterogeneity. We applied exome sequencing (ES) in a cohort of 89 families with the clinical sign of arthrogryposis. Additional molecular techniques including array comparative genomic hybridization (aCGH) and Droplet Digital PCR (ddPCR) were performed on individuals who were found to have pathogenic copy number variants (CNVs) and mosaicism, respectively. A molecular diagnosis was established in 65.2% (58/89) of families. Eleven out of 58 families (19.0%) showed evidence for potential involvement of pathogenic variation at more than one locus, probably driven by absence of heterozygosity (AOH) burden due to identity-by-descent (IBD). RYR3, MYOM2, ERGIC1, SPTBN4, and ABCA7 represent genes, identified in two or more families, for which mutations are probably causative for arthrogryposis. We also provide evidence for the involvement of CNVs in the etiology of arthrogryposis and for the idea that both mono-allelic and bi-allelic variants in the same gene cause either similar or distinct syndromes. We were able to identify the molecular etiology in nine out of 20 families who underwent reanalysis. In summary, our data from family-based ES further delineate the molecular etiology of arthrogryposis, yielded several candidate disease-associated genes, and provide evidence for mutational burden in a biological pathway or network. Our study also highlights the importance of reanalysis of individuals with unsolved diagnoses in conjunction with sequencing extended family members.
Keywords: arthrogryposis, multilocus pathogenic variation, trio-exome, ES reanalysis, identity-by-descent, absence of heterozygosity, joint contracture, neuromuscular disease
Introduction
Arthrogryposis is a term that describes the clinical observation of joint contractures in more than one segment of the body. It does not describe a specific disease entity, but rather represents a descriptive clinical neuromuscular sign for multiple contractures associated with different medical conditions. The primary underlying cause of arthrogryposis is postulated to be decreased fetal joint mobility during intrauterine development. Lack of fetal joint mobility might result from multiple etiologies for perturbed neuromuscular function; these etiologies include intrinsic causes such as neurological disorders, muscle diseases, skeletal conditions, and abnormalities in connective and cartilage tissue and extrinsic factors such as maternal diseases, intrauterine space limitations, maternal exposures to drugs or chemicals, and decreased blood supply to the fetus.1 Thus, arthrogryposis can be thought of as a birth defect due to a malformation in some individuals and as a deformation in others or, also, as a complex trait with both genetic and environmental influences.
The prevalence of arthrogryposis ranges from ∼1/3000–5000, and it has a high (∼50%) mortality rate.2 The etiology is thought to be mostly genetic, and variants in more than 300 genes associated with over 400 Mendelian conditions have been identified.3 Copy number variants (CNVs) including microdeletions and duplications have also been implicated in individuals with arthrogryposis.4, 5, 6, 7 The molecular etiology of arthrogryposis and the potential biological pathways involved remain unknown in many individuals; molecular diagnostic rates of 47% in an Australian cohort of 38 families and 58% in a Turkish cohort of 48 families (phase I of this study) have been reported.8, 9 One of the insights into the potential genetic architecture and underlying genetic etiology of some individuals with arthrogryposis, recognized only through genome-wide screening of coding regions, is the contribution of rare variants in multiple loci of a single personal genome to the observed phenotype (i.e., multiple molecular diagnoses leading to a blended phenotype).10, 11, 12, 13, 14 Additionally, both mono- and bi-allelic variants of several genes, responsible respectively for autosomal-dominant (AD) and autosomal-recessive (AR) disease traits, are known to cause either the same or a different syndrome and are being reported more frequently with genome-wide analysis techniques such as exome sequencing (ES).8, 15, 16, 17, 18, 19
We previously reported molecular findings from 28 out of 48 arthrogryposis-affected families (a molecular diagnostic yield of 58.3%).8 In the current study, we enrolled an additional 71 families (including two families found through GeneMatcher) and performed a reanalysis of 20 undiagnosed families, including ten families for which the initial proband-exome was extended to trio exome with parental samples, from the previously reported cohort. We provide clinical and genomic evidence for 15 candidate arthrogryposis-associated genes and four candidate developmental delay and intellectual disability (DD and ID)-associated genes. Our findings suggest mutational burden might contribute to the molecular diagnosis of some individuals with arthrogryposis through the aggregation of multilocus pathogenic variation in a personal genome.
Material and Methods
Study Participants
We recruited 103 individuals and fetuses from 91 mostly Turkish families (only two families of different ethnicity were found through GeneMatcher) with the presenting clinical feature of arthrogryposis (Figure 1A). Of the 103 participants, 53 were female (51.5%), 47 were male (45.6%), and three (2.9%) had a gender that was not reported (fetus). Ages ranged from 19 gestational weeks to 42 years old. Thirty-four of the participant families were characterized as having syndromic arthrogryposis, whereas 57 had an apparent isolated neuromuscular disorder. Parental consanguinity was reported in 46 families (50.5%), and 45 families (49.5%) had no historical evidence for parental consanguinity. After all relevant family members provided written informed consent, peripheral blood was collected from affected individuals, parents, and unaffected relatives if available. Genomic DNA was extracted from blood leukocytes according to standard procedures. All genomic studies were performed on DNA samples isolated from blood. This study was approved by the institutional review board at Baylor College of Medicine (IRB protocol # H-29697).
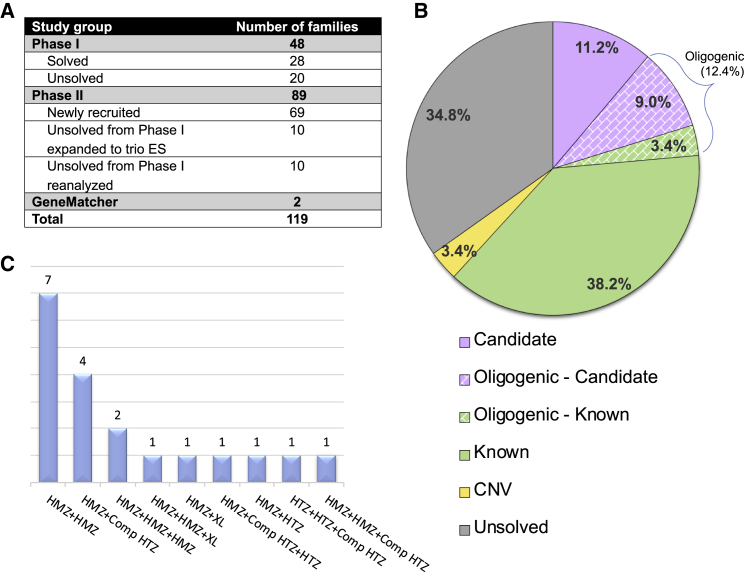
Figure 1.
Distribution and Molecular Results of Enrolled Individuals in the Arthrogryposis Cohort and Families with Multilocus Pathogenic Variation
(A) Distribution of individuals enrolled in arthrogryposis cohort.
(B) A pie chart displaying the distribution of molecular findings for each individual in this study (i.e., phase II) of the arthrogryposis cohort (89 families). Multilocus pathogenic variant model families were classified according to muscle gene status. A corresponding explanation for the colors in the pie chart is below the chart. The abbreviation CNV = copy number variation.
(C) Families affected with multilocus pathogenic variation (19 families) from phase I + II. Abbreviations are as follows: HMZ = homozygous, HTZ = heterozygous, Comp HTZ = compound heterozygous, and XL = hemizygous male.
Exome Sequencing
We applied ES to selected family members through the Baylor-Hopkins Center for Mendelian Genomics (BHCMG) research initiative.20 All experimental procedures were performed according to previously described methods.21 Briefly, genomic DNA samples underwent exome capture with Baylor College of Medicine Human Genome Sequencing Center core design (52 Mb, Roche NimbleGen, RRID: nif-0000-31466), and were then sequenced on the HiSeq platform (Illumina) with ∼100× depth of coverage. Sequence data were aligned and mapped to the human genome reference sequence (hg19) with the Mercury in-house bioinformatics pipeline.22 Variants were called with ATLAS2 (an integrative variant analysis pipeline optimized for variant discovery) and SAMTOOLS (RRID: nlx_154607; the Sequence Alignment/Map) suites, and they were annotated with an in-house-developed annotation pipeline that uses annotation of genetic variants and additional tools and databases.23, 24, 25 Variant filtering parsed nonsynonymous variants by population frequency data. The minor allele frequency of candidate variants was obtained from publicly available databases such as the 1000 Genomes Project and other large-scale exome-sequencing projects, including the Exome Variant Server; the National Heart, Lung, and Blood Institute (NHLBI) Grand Opportunity Exome Sequencing Project (ESP); the Atherosclerosis Risk in Communities Study Database (ARIC); the Genome Aggregation Database (gnomAD); the Exome Aggregation Consortium (ExAC); and our in-house-generated exome database (∼13,000 individuals) at the Baylor College of Medicine Human Genome Sequencing Center.
In order to detect disease-causing single nucleotide variants (SNVs), a stepwise analysis workflow was implemented.26 To be able to identify CNVs, we used publicly available bioinformatics tools, including XHMM (eXome-Hidden Markov Model), CoNIFER, and CoNVex programs, for larger CNVs (≥3 exon deletion and duplication), and we used an in-house-developed software, HMZDelFinder, for smaller homozygous and hemizygous intragenic CNVs.27, 28 In order to capture de novo disease-causing variants, we used another in-house-developed program called DNM (de novo mutation)-Finder.26 All candidate genes were submitted to GeneMatcher to identify additional individuals with variants likely to be damaging in the same gene.29, 30 To examine absence of heterozygosity (AOH) regions, which might represent identity-by-descent (IBD), surrounding candidate variants, we calculated B-allele frequency by using exome variant data as a ratio of variant reads to total reads.31 These data were then processed with the circular binary segmentation (CBS) algorithm32 in order to identify AOH regions. The calculated AOH intervals from BafCalculator could represent “apparent genetic transmission distortion” of individual genomic loci, resulting in runs-of-homozygosity (ROHs) for diploid alleles that can occur by: (1) IBD, (2) uniparental disomy,33 or (3) a large deletion CNV.
Sanger Sequencing
To validate exome-identified candidate variants by an orthogonal, experimental DNA-sequencing method and segregate these variants in the families, we amplified target exons from genomic DNA by using conventional PCR (HotStar TaqDNA polymerase, QIAGEN) and high-GC-content long-range PCR (TaKaRa LA Taq, Clontech), and we analyzed PCR amplification products by Sanger sequencing (DNA Sequencing Core Facility at Baylor College of Medicine). If the exome-identified variant was not confirmed by Sanger sequencing or if segregation analysis showed inconsistency with Mendelian expectations under the hypothesized genetic model, then the variant was considered to be unlikely to contribute to the arthrogryposis trait.
Chromosomal Microarray Analysis for Genome-Wide CNV Detection
For individuals in whom we detected a CNV (individual BAB7128) through genome-wide sequencing data and bioinformatic CNV-detection tools,34 or as part of the preliminary, low-resolution clinical genomics diagnostic studies such as karyotyping and low-resolution array comparative genomic hybridization (aCGH; individual BAB8145 and individual BAB9312), we performed Agilent’s custom-designed whole-genome array (Baylor Genetics Laboratory, CMA version 11, design#079906) to confirm the CNV. All array procedures, including DNA fragmentation and labeling, array hybridization, washing, and scanning, were performed according to the manufacturer’s instructions and previously described protocols35 with minor modifications. Gender-matched female control (NA15510) DNA and male control (NA10851) DNA were obtained from Coriell cell repositories (Coriell Institute for Medical Research). Data processing and analyses were done with Agilent Feature Extraction Software (version 11.5, Agilent Technologies) and Agilent Genomic Workbench (edition 7.0.4.0, Agilent Technologies).
Droplet Digital PCR Mosaicism Analysis
Variants suspected to be in a mosaic state on the basis of exome read-depth ratios (vR/tR, variant reads/total reads) were further validated via an orthogonal experimental system on a BioRad QX200 ddPCR platform. Custom-designed fluorescent TaqMan probes targeting the mutant variant (FAM fluorescence) and the reference (HEX fluorescence) were multiplexed with common forward and reverse primers and run under standard ddPCR conditions.36 The fractional abundances for the amplified mutant and reference allele were calculated to determine the percent of the mosaicism.
Human and Fruit Fly Cross-Database Mining for Candidate Gene Analyses
We evaluated orthologs and paralogs in fruit flies and humans for identified candidate disease genes. In order to determine the human paralogs for fruit fly genes, we used DRSC Integrative Ortholog Prediction Tool (DIOPT).37 We first forward checked the human gene in the fruit fly, and fly genes with the highest DIOPT scores were reverse checked to detect the number of human homologs. A DIOPT score of 3 is set as the threshold to determine the presence of paralogs or orthologs in the human or fly, respectively.
Results
Exome Sequencing and Family-Based Genomics in Arthrogryposis
We applied ES in a large cohort (Figure 1A, phase II) of 69 newly recruited families, a clinically heterogeneous group of individuals in whom the arthrogryposis phenotype is an apparent, isolated clinical feature (nonsyndromic arthrogryposis) or is part of the disease spectrum (syndromic arthrogryposis). Molecular findings from the analysis of phase I of this cohort had revealed a 58.3% (28 of 48 families studied) molecular diagnostic rate.8 In phase II of this family-based genomics study, we analyzed 91 families, including 69 newly ascertained and recruited families, 20 families who remained “molecularly undiagnosed” from phase I, and two families identified through GeneMatcher (Figure 1A).
The results were divided into the five categories presented below: (1) known disease-associated genes that have an established association with the arthrogryposis phenotype; (2) “probably causative” arthrogryposis-associated genes: rare variant, likely to be damaging, pathogenic alleles present in at least two different families; (3) “possibly causative” arthrogryposis-associated genes: pathogenic variants discovered in only one family in this study; (4) oligogenic inheritance and blended phenotypes: multilocus pathogenic variation, i.e., variants present in at least two disease-associated genes, that explains the observed set of phenotypic features including arthrogryposis; (5) CNVs causing the arthrogryposis phenotype. The molecular findings of the individuals for the entire cohort are described in Tables 1, 2, S1, and S2, and Figures S1 and S2.
Table 1.
Molecular Summary of the Known, Probably, and Possibly Causative Arthrogryposis Genes in the Cohort
| Gene | Proband | Nucleotide (Protein) | Zygosity | AOH Block Around the Gene (Mb) | Total AOH (Mb) |
|---|---|---|---|---|---|
| Known Causative Genes | |||||
| CHRNG | BAB8511 | c.753_754del (p.Val253Alafs∗44) | Hmz | 20.8 | 162.1 |
| BAB8571 | c.256C>T (p.Arg86Cys) | Hmz | 25.8 | 251.1 | |
| BAB8603 | c.256C>T (p.Arg86Cys) | Hmz | 6.7 | 154.5 | |
| BAB8685 | c.753_754del (p.Val253Alafs∗44) | Hmz | 19.0 | 248.8 | |
| BAB9851 | c.753_754del (p.Val253Alafs∗44) | Hmz | 22.3 | 123.3 | |
| BAB9942 | c.256C>T (p.Arg86Cys) | Hmz | 3.5 | 557.7 | |
| ECEL1 | BAB8223 | c.1630C>T (p.Arg544Cys) | Hmz | 3.3 | 199.2 |
| BAB9537 | c.1581+1G>A | Hmz | 14.2 | 164.4 | |
| BAB10711 | c.1916C>T (p.Ser639Phe) | Hmz | 14.7 | 238 | |
| NEB | BAB8518 | c.24988C>T (p.Arg8330∗) | Hmz | 73.4 | 151.2 |
| COL12A1 | BAB8843 | c.1488dup (p.Phe497Ilefs∗5) | Hmz | 20.8 | 297.9 |
| KLHL7 | BAB8095 | c.1051C>T (p.Arg351∗) | Hmz | 14.1 | 36.2 |
| BAB8098 | 21.7 | 96.5 | |||
| BAB8652 | c.1022del (p.Leu341Trpfs∗9) | Hmz | 15.5 | 248.5 | |
| BAB10699 | c.565C>T (p.Arg189∗) | Hmz | 26.0 | 354.3 | |
| BAB10705 | 22.9 | 238.2 | |||
| MYBPC1 | BAB8515 | c.32A>G (p.Glu11Gly) | Hmz | 25.8 | 351.9 |
| MYBPC2 | BAB9540 | c.920T>C (p.Val307Ala) | Comp Htz | – | 156.1 |
| c.3194C>T (p.Ala1065Val) | |||||
| MFN2 | BAB3941 | c.526G>A (p.Gly176Ser) | Hmz | 0.8 | 99.6 |
| SYNE1 | BAB7084 | c.2839G>A (p.Glu947Lys) | Comp Htz | – | 17.7 |
| c.21164A>G (p.Lys7055Arg) | |||||
| PLEC | BAB7079 | c.6523A>G (p.Lys2175Glu) | Comp Htz | – | 8.2 |
| c.8813C>T (p.Thr2938Met) | |||||
| POR | BAB8694 | c.859G>C (p.Ala287Pro) | Hmz | 13.5 | 102.8 |
| HSD17B4 | BAB8832 | c.1417C>T (p.Arg473Trp) | Hmz | 49.0 | 202.9 |
| BAB8835 | 60.7 | 165.3 | |||
| POMGNT1 | BAB8986 | c.461C>A (p.Pro154His) | Hmz / Hmz | 29.3 | 168.4 |
| c.550C>T (p.His184Tyr) | |||||
| FKBP10 | BAB9244 | c.890_897dup (p.Gly300∗) | Hmz | 4.3 | 154.5 |
| TGFB3 | BAB9703 | c.171del (p.Glu58Serfs∗4) | Hmz | 48.5 | 183.4 |
| BAB9704 | 30.1 | 480.7 | |||
| TTN | BAB7779 | c.2370+2T>C | Comp Htz | – | 34.2 |
| c.67279C>T (p.Arg22427∗) | |||||
| IGF1 | BAB9740 | c.156dup (p.Leu53Alafs∗5) | Hmz | 15.7 | 289.3 |
| BAB9741 | 6.6 | 208.3 | |||
| TOR1A | BAB10702 | c.506T>C (p.Phe169Ser) | Hmz | 11.8 | 196.4 |
| MYH3 | BAB3964 | c.2015G>A (p.Arg672His) | Htz | – | 30.4 |
| BAB9848 | – | 26.1 | |||
| TNNT3 | BAB3928 | c.163C>T (p.Arg55Cys) | Htz | – | 10.4 |
| SYT2 | BAB7308 | c.1081G>C (p.Asp361His) | Htz | – | 65.5 |
| PIEZO2 | BAB8789 | c.8181_8183del (p.Glu2727del) | Htz | – | 208.2 |
| TRPV4 | BAB8991 | c.806G>A (p.Arg269His) | Htz | – | 39.5 |
| PQBP1 | BAB8090 | c.463C>T (p.Arg155∗) | Hemi | – | 24.2 |
| Probably Causative Genes | |||||
| RYR3 | BAB7845 | c.2486G>A (p.Arg829His) | Hmz | 1.8 | 247.5 |
| PAED187 | c.2000A>G (p.Asp667Gly) | Comp Htz | – | 5.5 | |
| c.11164+1G>A | |||||
| BAB8988 | c.8939G>T (p.Arg2980Leu) | Hmz | 12.0 | 149.6 | |
| MYOM2 | BAB8905 | c.621C>G (p.Ser207Arg) | Hmz | 7.2 | 312.9 |
| IN076 | c.2797C>T (p.Gln933∗) | Hmz | 7.2 | 369.1 | |
| ABCA7 | BAB6807 | c.5092C>T (p.Arg1698Trp) | Hmz | 3.8 | 153.9 |
| BAB6808 | 3.0 | 180.2 | |||
| BAB10708 | c.3076C>T (p.Arg1026Cys) | Comp Htz | – | 233.8 | |
| c.4045C>T (p.Arg1349Trp) | |||||
| ERGIC1 | BAB8802 | c.782G>A (p.Gly261Asp) | Hmz | 30.8 | 274.6 |
| SPTBN4 | BAB8691 | c.6433G>A (p.Ala2145Thr) | Hmz | 3.4 | 183.9 |
| Possibly Causative Genes | |||||
| MID1IP1 | BAB8397 | c.297C>G (p.Asn99Lys) | Hemi | – | 374.7 |
| DRG1 | BAB8807 | c.118C>T (p.Arg40∗) | Hmz | 7.6 | 251.6 |
| TANC1 | c.2830C>T (p.His944Tyr) | Hmz | 10.3 | ||
| MYOM3 | BAB8532 | c.3534+56C>T | Comp Htz | – | 107.2 |
| c.1684G>A (p.Val562Ile) | |||||
| TNRC6C | BAB8688 | c.1022G>A (p.Gly341Glu) | Hmz | 7.0 | 132.0 |
| FLII | BAB7710 | c.2590C>T (p.Arg864Trp) | Hmz | 2.2 | 257.4 |
| TAF9B | BAB8400 | c.133C>A (p.Arg45Ser) | Hemi | – | 110.6 |
| MED27 | BAB8606 | c.770C>T (p.Pro257Leu) | Hmz | 11.8 | 195.2 |
| BAB8609 | 2.0 | 233.7 | |||
| CACUL1 | BAB9729 | c.910_911del (p.Leu304Ilefs∗3) | Hmz | 10.0 | 158.9 |
| FGFRL1 | BAB3944 | c.124C>T (p.Arg42Trp) | Hmz | 3.4 | 290.2 |
| TMEM214 | BAB5192 | c.764G>A (p.Arg255Gln) | Hmz | 15.9 | 220.3 |
| NR2C1 | BAB8086 | c.544+1G>C | Hmz | 4.3 | 231.7 |
| BAB8087 | 6.7 | 126.3 | |||
| PRDM2 | BAB9309 | c.4283_4295del (p.Leu1428Glnfs∗15) | Htz | – | 284.5 |
| FAT1 | BAB8356 | c.6026A>G (p.Asn2009Ser) | Hmz | 2.0 | 142.9 |
Abbreviations are as follows: Mb = megabase, AOH = absence of heterozygosity, Hmz = homozygous, Hemi = hemizygous, Htz = heterozygous, and Comp Htz = compound heterozygous.
Table 2.
Molecular Summary of the Multilocus Pathogenic Variation Cohort
| Family ID (HOU#) | Proband ID (BAB#) | Gene | Nucleotide (Protein) | Zygosity | AOH Block Around the Gene (Mb) | Total AOH (Mb) |
|---|---|---|---|---|---|---|
| HOU2523 | BAB6807 | COL6A3 | c.619C>T (p.Gln207∗) | Hmz | 7.8 | 153.9 |
| ABCA7 | c.5092C>T (p.Arg1698Trp) | Hmz | 3.8 | |||
| BAB6808 | ABCA7 | c.5092C>T (p.Arg1698Trp) | Hmz | 3.0 | 180.2 | |
| ADNP | c.775A>C (p.Asn259His) | Hmz | – | |||
| HOU2790 | BAB7710 | ECEL1 | c.505_529del (p.Gly169Serfs∗26) | Hmz | 20.1 | 257.4 |
| FLII | c.2590C>T (p.Arg864Trp) | Hmz | 2.2 | |||
| HOU2817 | BAB7845 | RYR3 | c.2486G>A (p.Arg829His) | Hmz | 1.8 | 247.5 |
| MYO18B | c.2879C>T (p.Ala960Val) | Comp Htz | – | |||
| c.3397C>T (p.Arg1133Trp) | – | |||||
| HOU3050 | BAB8397 | NEB | c.19101+5G>A | Hmz | – | 374.7 |
| MID1IP1 | c.297C>G (p.Asn99Lys) | Hemi | – | |||
| HOU3051 | BAB8400 | GBE1 | c.1864_1866del (p.Leu622del) | Hmz | 37.9 | 110.6 |
| AP4M1 | c.136C>G (p.Pro46Ala) | Hmz | 14.2 | |||
| TAF9B | c.133C>A (p.Arg45Ser) | Hemi | – | |||
| HOU3112 | BAB8532 | SPEG | c.6971T>A (p.Ile2324Asn) | Hmz | 22.6 | 107.2 |
| MYOM3 | c.1684G>A (p.Val562Ile) | Comp Htz | – | |||
| c.3534+56C>T | ||||||
| CIT | c.2651A>C:p.Gln884Pro | Htz | – | |||
| HOU3125 | BAB8600 | FBN2 | c.4094G>C (p.Cys1365Ser) | Htz | – | – |
| BAB8601 | FBN2 | – | 31.9 | |||
| COL6A3 | c.367G>A (p.Val123Met) | Hmz | 1.5 | |||
| HOU3127 | BAB8606 | COG6 | c.726del (p.Cys242Trpfs∗7) | Hmz | 16.9 | 195.2 |
| MED27 | c.770C>T (p.Pro257Leu) | Hmz | 11.8 | |||
| BAB8609 | MED27 | c.770C>T (p.Pro257Leu) | Hmz | 2.0 | 233.7 | |
| HOU3149 | BAB8688 | KLHL7 | c.1258C>T (p.Arg420Cys) | Hmz | 17.9 | 132.0 |
| HOXA11 | c.304G>A (p.Val102Met) | Hmz | 17.9 | |||
| TNRC6C | c.1022G>A (p.Gly341Glu) | Hmz | 7.0 | |||
| HOU3180 | BAB8807 | DRG1 | c.118C>T (p.Arg40∗) | Hmz | 6.7 | 251.6 |
| TANC1 | c.2830C>T (p.His944Tyr) | Hmz | 10.3 | |||
| BRWD3 | c.592-3T>C | Hemi | – | |||
| HOU3943 | BAB10708 | SPEG | c.9575C>A (p.Thr3192Asn) | Htz | – | 233.8 |
| TPM2 | c.620_631dup (p.Gln210_Ala211insValGluAlaGln) | Htz | – | |||
| ABCA7 | c.3076C>T (p.Arg1026Cys) | Comp Htz | – | |||
| c.4045C>T (p.Arg1349Trp) | – |
Abbreviations are as follows: Mb = megabase, AOH = absence of heterozygosity, Hmz = homozygous, Hemi = hemizygous, Htz = heterozygous, and Comp Htz = compound heterozygous.
The overall molecular diagnostic rate in the present phase II cohort of 89 families with arthrogryposis is 65.2% (58/89); 43 of these 89 families have variants in previously described arthrogryposis-associated genes (six out of nine oligogenic families have both known and candidate genes). 18 out of 89 families, including both single locus and multilocus families, have variants in arthrogryposis-associated candidate genes. Three families were found to have CNVs that are thought to contribute to the arthrogryposis phenotype. Considering all families studied in both phase I8 and phase II, we achieved a molecular diagnostic rate of 73.5% (86/117) when considering both known and potential disease genes, and 58.1% (68/117) of families had a molecular diagnosis involving a known disease gene.
Of note, the rate of consanguinity, as measured by AOH, was significantly higher in the molecularly diagnosed individuals compared to undiagnosed cohort individuals (60/86 = 69.8% in diagnosed cohort versus 5/31 = 16.1% in the undiagnosed cohort, p < 0.0001 Fisher’s exact two-tailed test). However, we did not observe a significant difference in evidence for consanguinity in syndromic versus non-syndromic arthrogryposis in the diagnosed compared to undiagnosed individuals (30/86 = 34.9% in the diagnosed cohort versus 8/31 = 25.8% in the undiagnosed cohort, p = 0.3824 Fisher’s exact two-tailed test). In the current study, we propose five probably causative and ten possibly causative arthrogryposis-associated genes and four candidate DD-and-ID-associated genes contributing to the disease phenotype (Figure 1B). Potential multilocus pathogenic variation, i.e., the oligogenic model, was observed in 11 families in this phase II cohort, specifically in 19.0% (11/58) of individuals with a diagnosis. The overall potential oligogenic inheritance rate including both phase I and phase II studies is 22.1% (19/86) of diagnosed individuals (Figure 1C).
Known Arthrogryposis-Associated Genes
A total of 43 families have a pathogenic variant in a gene associated with either nonsyndromic (isolated) or syndromic arthrogryposis (Tables 1, 2, and S1, and Figures S1 and S2). The most common identified molecular diagnoses and implicated genes included CHRNG (MIM: 100730), associated with Escobar syndrome and lethal multiple pterygium syndrome (EVMPS [MIM: 265000] and LMPS [MIM: 253290], respectively), in 12 of 116 Turkish families (10.3%, excluding the individual of Arabic origin in phase I), and ECEL1 (MIM: 605896), associated with distal arthrogryposis type 5D (DA5D [MIM: 615065]) in six families. The calculated total AOH after we used BafCalculator on exome-generated variant data31 was on average 218.66 Mb for these 18 families, whereas the average calculated total AOH size in the entire Turkish cohort (∼700 exomes) is 91.38 Mb. Rare variants in KLHL7 (MIM: 611119), previously associated with cold-induced sweating syndrome (CISS3 [MIM: 617055]), were identified in four unrelated families with syndromic primarily distal arthrogryposis. The syndromic features were characterized by variable involvement of the genitourinary system, DD and ID, and congenital heart disease, each in at least two families; these features represent a potential expansion of the known disease phenotypes associated with KLHL7. Additional molecular diagnoses reported in more than one family involved MYH3 (MIM: 160720), associated with several forms of distal arthrogryposis (DA2A [MIM: 193700], DA2B [MIM: 601680], and DA8 [MIM: 178110]) in two families, and SPEG (MIM: 615950), linked to centronuclear myopathy 5 (CNM5 [MIM: 615959]). In total, rare variants in 45 different known genes were found within 43 families (including oligogenic families), and these findings reveal the extensive genetic heterogeneity in the arthrogryposis cohort. Additionally, we identified a second family with variants in MYBPC2 (MIM: 160793), first proposed as a candidate arthrogryposis-associated gene in the phase I study.
Probably Causative Arthrogryposis-Associated Genes
We found evidence for five candidate genes, RYR3 (MIM: 180903), MYOM2 (MIM: 603509), ERGIC1 (MIM: 617946), SPTBN4 (MIM: 606214), and ABCA7 (MIM: 605414), in which predicted deleterious variants are likely to be contributing to and probably causing the arthrogryposis phenotype; the variants likely to be pathogenic are present in at least two families, and additional families were identified either within our cohort, through GeneMatcher, or in recent publications (molecular findings in Tables 1, 2, and S1, and Figures S1 and S2; clinical findings in the Supplemental Text). For each of these genes, additional data, such as minor allele frequencies, evolutionary conservation, and CADD-phred scores of identified variants, that support their candidacy as arthrogryposis genes are provided (Tables 1, 2, and S1). These genes were not previously linked to any human disease in OMIM, or there has been only one reported individual, family, or variant; thus, we consider the variants within these genes to be probably causative for the arthrogryposis phenotype.
RYR3
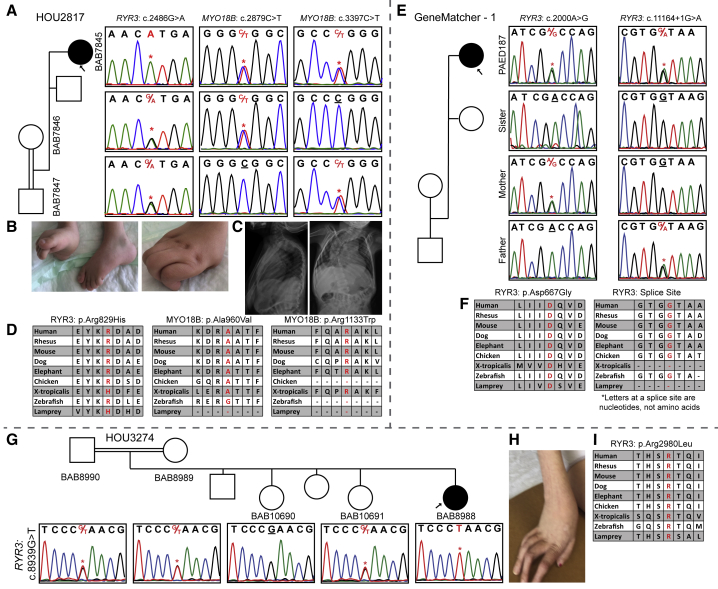
RYR3 bi-allelic variants were found in three unrelated individuals (Table 1 and Figure 2): one was found as part of a potential multilocus mutational burden disease model (RYR3 + MYO18B [MIM: 607295]) (individual BAB7845), one was identified through GeneMatcher (individual PAED187), and one was found by assuming a single-gene-single-disease model (individual BAB8988). The first individual, BAB7845, is a 6-month-old female who was born to a first degree cousin marriage with 247.5 Mb of total AOH. The proband exome revealed a compound-heterozygous variant in a known gene, MYO18B (GenBank: NM_032608; c.[2879C>T]; [3397C>T]: p.[Ala960Val]; [Arg1133Trp]), and a homozygous variant in RYR3 (GenBank: NM_001243996; c.2486G>A [p.Arg829His]) (Figures 2A–2D). The c.2879C>T MYO18B variant and the RYR3 variant were each reported as homozygous in one individual in gnomAD. On the basis of the amino acid conservation and “likely damaging prediction scores” (Table S1) of the variants identified herein, we propose that all three variants are hypomorphic alleles, and each contributes to the arthrogryposis phenotype in this individual. Variants in MYO18B were recently shown to cause a neuromuscular disease spectrum ranging from nemaline myopathy with cardiomyopathy to Klippel-Feil anomaly with myopathy (KFS4 [MIM: 616549]),38, 39 and here we report the fourth family with MYO18B variants. Individual PAED187 has compound heterozygosity for a missense variant (GenBank: NM_001036.3; c.2000A>G [p.Asp667Gly]) and a variant affecting the splice donor (c.11164+1G>A) in RYR3 (Figures 2E and 2F). Individual BAB8988 was born to first degree cousin parents with 149.6 Mb of total AOH according to BafCalculator. Trio ES revealed a homozygous nonsynonymous change in RYR3 (GenBank: NM_001243996; c.8939G>T [p.Arg2980Leu]) (Figures 2G–2I). Recently, compound-heterozygous alleles in RYR3, c.[6208A>G]; [8939G>T]: p.[Met2070Val]; [Arg2980Leu], were reported in a 22-year-old female with myopathic facies, proximal weakness in all four limbs, mild scapular winging, and type 1 muscle fiber atrophy and predominance in muscle biopsy.40 The c.8939G>T variant in the reported individual is the same variant observed in individual BAB8988.
Figure 2.
Segregation Results, Images, and Variant Distributions of the Individuals with RYR3 Mutation
(A) Sanger studies showing compound-heterozygous variants in MYO18B and a homozygous variant in RYR3 in the proband of family HOU2817.
(B) Pictures of the proband’s contractures in fingers and toes.
(C) Anterior-posterior and lateral views of spinal X-ray showing the kyphoscoliosis.
(D) Conservation of the three variants through species. Variants of interest are written with a red font.
(E) Segregation studies for both the c.2000A>G and c.11164+1G>A variants in GeneMatcher – 1.
(F) Highly conserved asparagine (D) amino acid and splice site through species.
(G) A pedigree and Sanger segregation of family HOU3274 that had a single affected female individual (BAB8988) and two unaffected siblings available for study. Both parents and one unaffected sibling are heterozygous, the second unaffected sibling is wild type, and our proband is homozygous for the c.8939G>T variant.
(H) A colored picture of the hand showing the contractures in the fingers of individual BAB8988.
(I) The arginine (R) amino acid residue is highly conserved among species and specified in red font.
The ryanodine receptors are intracellular Ca2+ release channels that play a key role in cell signaling via Ca2+. RYR1 (MIM: 180901) plays a role in the initial voltage-dependent Ca2+ release, and RYR3 functions during the prolonged Ca2+ release.41, 42 Further evidence that the pathogenicity of the variants in RYR3 probably contribute to the arthrogryposis phenotype comes from bioinformatics studies; Abath Neto et al. performed a computational data-mining study based on gene ontology (GO) terms, human phenotype ontology (HPO) terms, pathways, complexes, and protein motifs, and they found that RYR3 is highly ranked as one of the congenital myopathy genes.43 Mutations in RYR1, a paralog of RYR3, were shown to cause several neuromuscular disorders including central core disease (CCD [MIM: 117000]), myopathies (MIM: 255320 and 117000), and the complex trait of susceptibility to malignant hyperthermia (MHS1 [MIM: 145600]), a sometimes lethal disease induced by exposure to certain anesthetics during surgical interventions; deleterious variants in RYR2 (MIM: 180902) were known to cause cardiac rhythm disorders (ARVD2 [MIM: 600996] and CPVT1 [MIM: 604772]), whereas no disease has been linked to RYR3. Taken together, we propose pathogenic variants in RYR3 are causing neuromuscular disease on the basis of the presence of deleterious variants in three unrelated families and functional evidence for its involvement in neuromuscular disease.
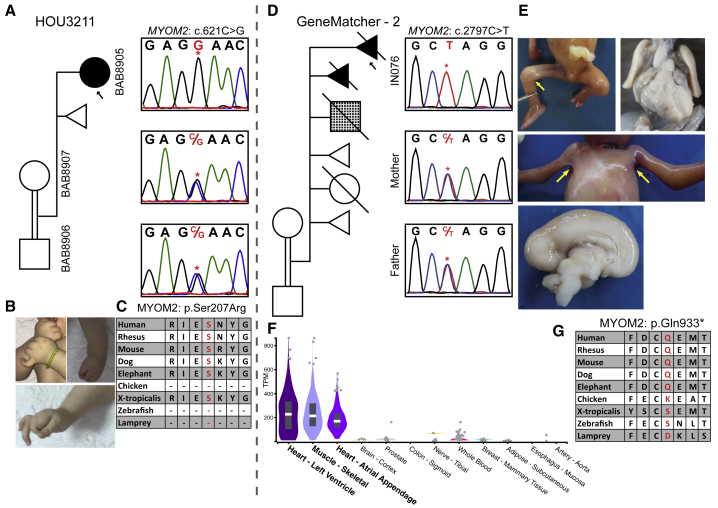
MYOM2
Two unrelated individuals carry variants in MYOM2 (Figure 3). Trio ES revealed a homozygous missense variant (GenBank: NM_003970; c.621C>G [p.Ser207Arg]) in Myomesin-2 (MYOM2) in individual BAB8905 (Figures 3A–C). The second individual, a fetus terminated at 20 weeks of gestation (found through GeneMatcher) (individual IN076), was found to have a homozygous nonsense variant and presumed null allele (GenBank: NM_003970; c.2797C>T [p.Gln933∗]) in MYOM2 (Figures 3D–G). Because MYOM2 is specifically expressed in the heart and skeletal muscle, the cardiac and arthrogryposis findings can be potentially explained by loss of function (LoF) mutation of MYOM2. Further evidence for probable pathogenicity comes from functional studies that showed that MYOM2 interacts directly with dysferlin, which plays an essential role in sarcolemma repair; abnormalities of dysferlin cause a wide variety of myopathies called dysferlinopathies.44
Figure 3.
Clinical and Molecular Details of the MYOM2 Individuals
(A) Pedigree and segregation analyses of the identified MYOM2 variant (c.621C>G) in family HOU3211. The parents are shown as consanguineous because they are from the same small village and the proband demonstrates large AOH blocks on exome data. As expected with a recessive inheritance pattern, the parents are heterozygous and the proband (individual BAB8905) is homozygous for the c.621C>G variant.
(B) Proband’s photos showing the contractures in the hands and feet.
(C) High conservation of the Ser207 amino acid in other species. The serine (S) amino acid is written with a red font color.
(D) A pedigree of the family (GeneMatcher – 2) showing a complicated medical history including affected and unaffected deceased siblings along with medically terminated or spontaneously aborted individuals in the GeneMatcher family. The fourth pregnancy is shown with a checkered box to indicate a different phenotype than neuromuscular disease (i.e., he was found to have cardiac and gastrointestinal anomalies along with intrauterine growth restriction). Sanger PCR from available individuals shows that the affected fetus is homozygous and the parents are heterozygous for the c.2797C>T variant.
(E) Pictures of the fetus showing pterygia of axillae and popliteal joints and a midsagittal cut of fetal brain showing partial agenesis of the corpus callosum and bilateral hypoplastic lungs.
(F) A genotype-tissue expression (GTEx) panel showing specific expression of MYOM2 in skeletal and heart muscles.
(G) Amino acid conservation around Gln933 among other species. The Gln (Q) amino acid is written in red font.
ABCA7
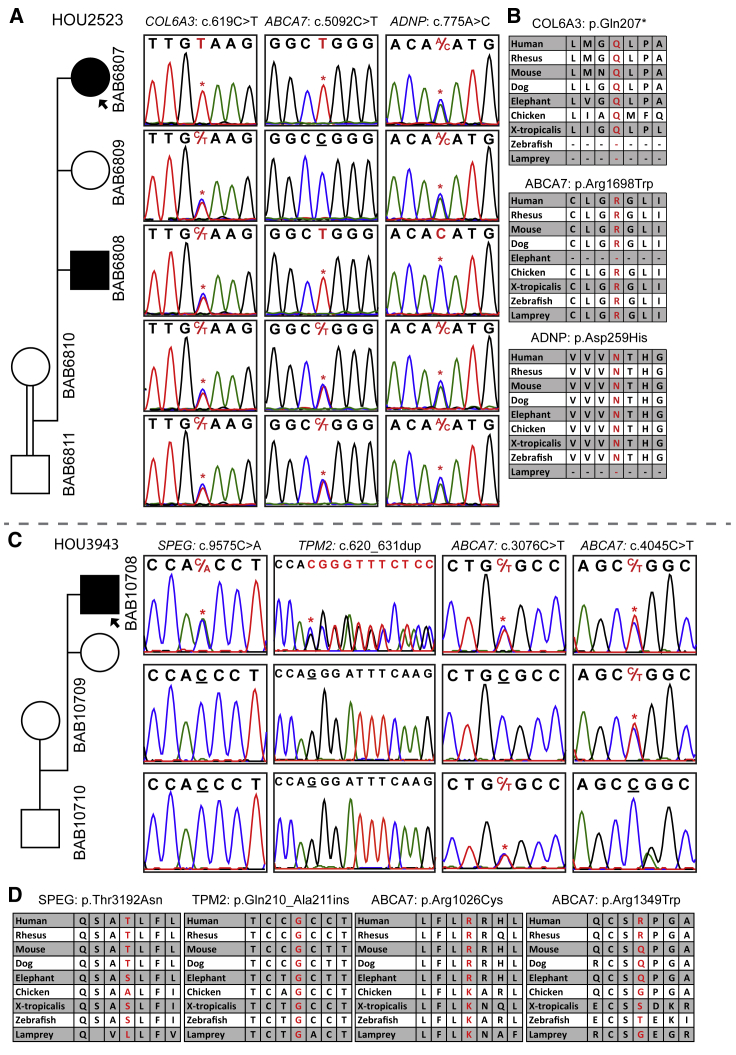
Bi-allelic ABCA7 variants were found in two unrelated families (Figure 4). In the first family, there were two affected siblings, individuals BAB6807 and BAB6808 (Figures 4A and4B). The younger female sibling was more severely affected with arthrogryposis, whereas the older male sibling, individual BAB6808, was observed to have DD and ID in addition to a neuromuscular phenotype. ES was performed on both affected siblings. Individual BAB6807 was found to have a novel homozygous stop-gain mutation (GenBank: NM_057166; c.619C>T [p.Gln207∗]) in the known COL6A3 gene (MIM: 120250), but the affected brother and healthy parents were heterozygous for this variant. Given the relative phenotypic similarity between the two affected siblings (the female is more severely affected, and the brother had additional DD and ID phenotype), we searched for variants shared by the two affected siblings and found a homozygous missense change (GenBank: NM_019112; c.5092C>T [p.Arg1698Trp]) in ATP-binding cassette, subfamily A, member 7 (ABCA7). Individual BAB6808, who has an additional DD and ID phenotype, was found to carry a deleterious homozygous variant (GenBank: NM_015339; c.775A>C [p.Asn259His]) in ADNP (MIM: 611386), in which variants are known to cause DD and ID (HVDAS [MIM: 615873]).45 The homozygous COL6A3 variant in individual BAB6807 probably explains the more severe phenotype in this individual. In the second family, individual BAB10708 is found to have variants in three different genes: (1) compound-heterozygous deleterious variants in ABCA7 (GenBank: NM_019112; c.[3076C>T]; [4045C>T]: p.[Arg1026Cys]; [Arg1349Trp]), (2) a de novo heterozygous variant in SPEG (MIM: 615950; GenBank: NM_005876; c.9575C>A [p.Thr3192Asn]), in which bi-allelic variants are known to cause centronuclear myopathy 5 (CNM5 [MIM: 615959]), and (3) a de novo, heterozygous, non-frameshift insertion in TPM2 (MIM: 190990; GenBank: NM_003289; c.620_631dup [p.Gln210_Ala211insValGluAlaGln]), in which mono-allelic variants are known to cause a broad spectrum of neuromuscular disorders including distal arthrogryposis multiplex congenita types 1 and 2B (DA1A [MIM: 108120] and DA2B [601680]) (Figures 4C and 4D). Heterozygous variants in ABCA7 have been linked to Alzheimer susceptibility (AD9 [MIM: 608907]); however, bi-allelic variants associated with recessive trait variants have not been reported in any diseases previously. ATP-binding cassette (ABC) proteins transport various molecules across extra- and intra-cellular membranes. Heterozygous variants in ABCA1 (MIM: 600046) are associated with type 2 HDL deficiency and protection against coronary artery disease in familial hypercholesterolemia (MIM: 604091 and 143890, respectively), and homozygous variants in ABCA1 are known to cause Tangier disease (TGD [MIM: 205400]), which has clinical features of distal muscle atrophy and peripheral neuropathy. As per the STRING database, there is distant interaction between ABCA1 and ABCA7.
Figure 4.
Families with ABCA7 Variants
(A) A pedigree suggesting an autosomal-recessive inheritance pattern for ABCA7, COL6A3, and ADNP in family HOU2523; the affected individuals are homozygous, whereas unaffected individuals are heterozygous. Individual BAB6807 has a homozygous stop-gain mutation in COL6A3 in addition to a homozygous ABCA7 variant shared with individual BAB6808. The ADNP variant is homozygous in individual BAB6808, who has a DD and ID phenotype.
(B) Conservation profiles of all three genes.
(C) A pedigree of family HOU3943 with segregation studies showing bi-allelic variants in ABCA7 and de novo heterozygous variants in SPEG and TPM2.
(D) The amino acid alignment of the detected variants across different species.
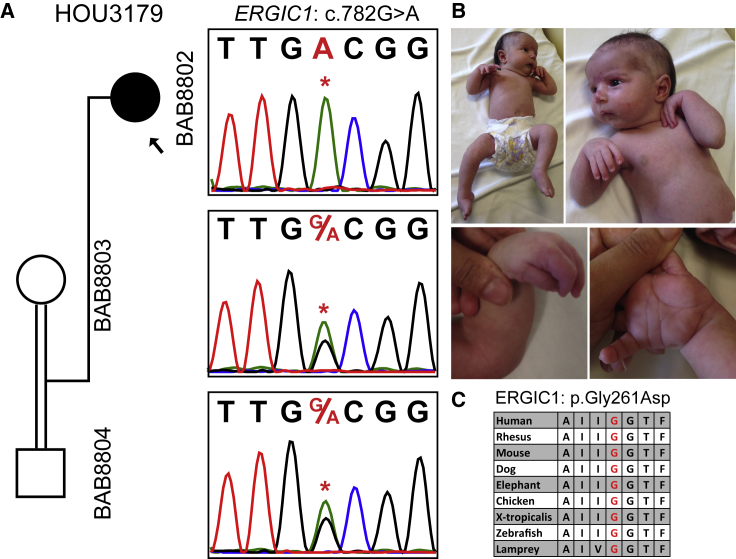
ERGIC1
Trio ES of individual BAB8802 showed a homozygous change (GenBank: NM_001031711; c.782G>A [p.Gly261Asp]) in ERGIC1 (Figure 5). This gene was reported in a large family affected with arthrogryposis multiplex congenita; in this family, 40 affected individuals were found to have c.293T>A (p.Val98Glu), cosegregating with the AR disease trait.46 However, to date, no additional families or variants in ERGIC1 have been associated with arthrogryposis. ERGIC (ER-Golgi intermediate compartment) family proteins are transmembrane proteins that are localized to the ER-Golgi intermediate compartment, but the function of ERGIC1 remains elusive.47 The observation of a second family with rare variation in ERGIC1 in association with arthrogryposis strengthens its relevance to the arthrogryposis phenotype.
Figure 5.
Segregation, Pictures, and Protein Conservation for a Homozygous ERGIC1 Variant in Individual BAB8802
(A) Segregation analyses of the exome-detected variant; the proband was homozygous and the parents were heterozygous carriers, as expected in recessive disease traits.
(B) Proband photographs showing restrictions in the wrists and fingers and the pes equinovarus deformity in the feet.
(C) A peptide alignment showing the conservation of the affected amino acid across species.
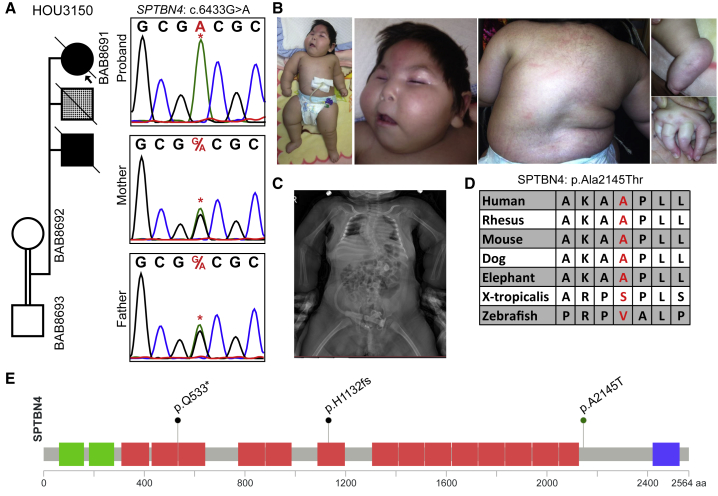
SPTBN4
Trio ES revealed a homozygous GenBank: NM_020971; c.6433G>A (p.Ala2145Thr) variant in SPTBN4 in individual BAB8691, who presented with distal contractures, hearing loss, and DD and ID (Figure 6). Recently, Knierim et al. reported a homozygous nonsense variant GenBank: NP_066022.2; p.Gln533∗ in a male with myopathy, distal arthrogryposis, scoliosis, hearing loss, severe DD and ID, increased cerebrospinal fluid (CSF) spaces on brain MRI, and peripheral neuropathy.48 An immunoblot and immunostaining showed the absence of SPTBN4, and a knock-out mouse model showed findings, including muscle weakness, decreased locomotion, quivering, ataxia, and hearing loss, that are similar to those in humans. Between these two individuals there is significant phenotypic overlap, including severe DD and ID, grossly normal brain MRI, hearing loss, distal contractures, and failure to thrive. Another homozygous variant (GenBank: NM_020971.2; c.3394del [p.His1132Thrfs∗39]) of SPTBN4 was reported by Anazi et al. in an individual who had no reported arthrogryposis but did have clinical features of global developmental delay, hypotonia, dysphasia, recurrent respiratory infections, blue sclerae, hyporeflexia, and failure to thrive.49 Although the authors did not report any electromyography or nerve conduction studies, the clinical observation of hyporeflexia and hypotonia might be features of peripheral neuropathy. SPTBN4 encodes a nonerythrocytic member of the beta-spectrin protein family that is mainly expressed in the brain, peripheral nervous system, pancreas, and skeletal muscle. Mutation of different domains might explain why each person presented with more central nervous system than neuromuscular features. Our individual has both neuromuscular and central nervous system findings and is the second individual now demonstrated to have arthrogryposis in association with a rare variant in SPTBN4; this supports that SPTBN4 is associated with both DD and ID and with the arthrogryposis phenotype.
Figure 6.
Clinical and Molecular Studies for a Homozygous SPTBN4 Variant in Individual BAB8691
(A) Segregation studies showing heterozygosity for the parents and homozygosity for the index for variant c.6433G>A. The second child, who died of an unknown lung malformation, is shown with a checkered box.
(B) Pictures of the proband showing a myopathic face, scoliosis, contractures in the fingers, and pes equinovarus deformity in the feet.
(C) An anterior-posterior view of a spine X-ray showing severe thoracic scoliosis.
(D) High conservation of the mutated amino acid residue across species (red font).
(E) Protein domains of SPTBN4, with the variant identified in the present cohort (p.Ala2145Thr) and variants reported in two individuals in the literature (p.Gln533∗ and p.His1132fs).
Possibly Causative Arthrogryposis-Associated Genes
We identified ten candidate arthrogryposis-associated genes (CACUL1, DRG1 [MIM: 603952], FAT1 [MIM: 600976], FGFRL1 [MIM: 605830], FLII [MIM: 600362], MID1IP1 [MIM: 300961], MYOM3 [MIM: 616832], NR2C1 [MIM: 601529], PRDM2 [MIM: 601196], and TMEM214 [MIM: 615301]) and four candidate DD-and-ID-associated genes (MED27 [MIM: 605044], TAF9B [MIM: 300754], TANC1 [MIM: 611397], and TNRC6C [MIM: 610741]) (Tables 1, 2, and S1, Figures S1–S4, and the Supplemental Text). Rare variants in each gene were identified in a single family, leading to their classification as candidate disease genes for which additional evidence is required before they are established as disease-associated genes. Evidence supporting plausible pathogenicity of these variant alleles includes amino acid conservation among other species, the predicted probably damaging effect of the variant made on the basis of bioinformatics tools, and interactome studies. Only two of these genes are highlighted here, and clinical and molecular results of all candidate genes are detailed in the oligogenic inheritance and blended phenotype section in Tables 1, 2, and S1, and the Supplemental Text.
Trio ES in individual BAB9729 revealed a homozygous 2 bp deletion (GenBank: NM_153810; c.910_911del [p.Leu304Ilefs∗3]) in CACUL1, encoding a cell-cycle-associated protein capable of promoting cell proliferation through the activation of CDK2 at the G1/S phase transition (Figure S5). Interacting genes, NEDD8 (MIM: 603171) and FBXW7 (MIM: 606278), have a role in muscle function. Lange et al. showed that obscurin, a binding partner of titin, contains a non-modular C terminus that interacts with muscle specific isoforms of Ankyrin-1 (small Ankyrin-1.5; sAnk1.5).50 sAnk1.5 localization and levels of sAnk1.5 expression are downregulated in obscurin knockout tissues, probably through ubiquitylation and/or neddylation of sAnk1.5.50 Another interacting gene, Fbxw7b, negatively regulates differentiation, proliferation, and migration of myoblasts and satellite cells on muscle fibers.51 The same group additionally showed that overexpression of Fbxw7b induces the expression of myogenin and major atrogene markers (atrogin-1 and MuRF-1) and eventually reduces myoblast differentiation.52
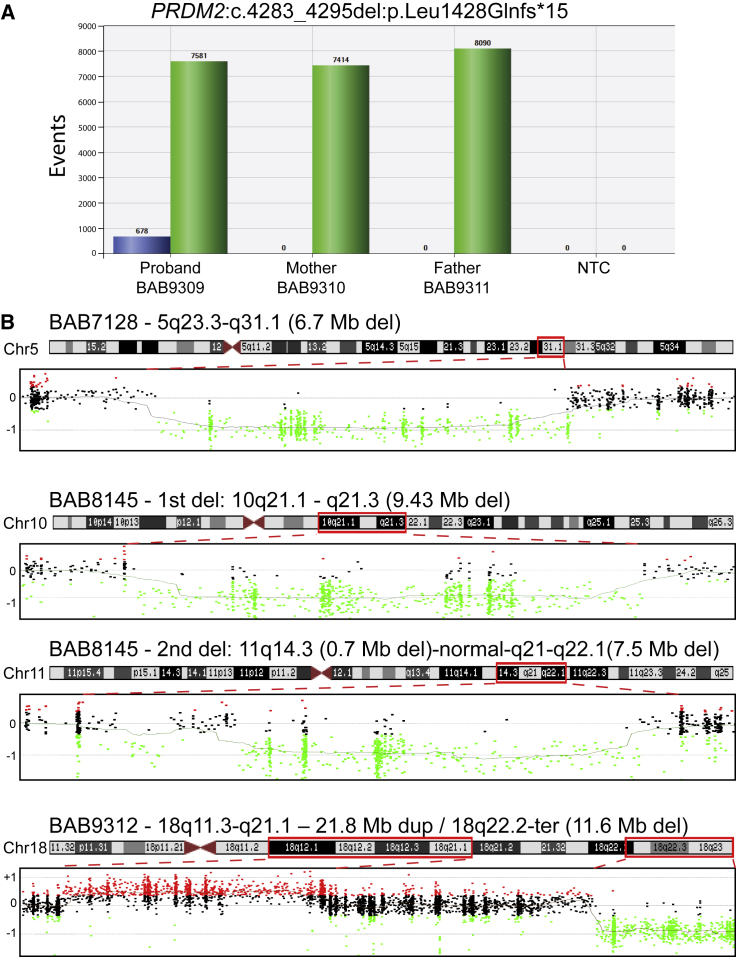
Trio ES in individual BAB9309 revealed a de novo frameshift deletion (GenBank: NM_012231; c.4283_4295del [p.Leu1428Glnfs∗15]) in PR domain-containing protein 2 (PRDM2 [MIM: 601196]). The fraction of variant reads (12/150, 8.0%) suggested somatic mosaicism, an experimental conclusion that was supported by an independent orthogonal experimental approach that used ddPCR (Figure 7A). PRDM2 is a member of a nuclear histone and protein methyltransferase superfamily and plays a major role in the regulation of myogenesis.53, 54
Figure 7.
Molecular Studies in Individual BAB9309 and Copy Number Variant Families
(A) Droplet digital PCR (ddPCR) results for the family; the results show 8% mosaicism for the PRDM2 variant in the proband, and this finding is consistent with the exome mosaicism rate (8%) and no deletion in the healthy parents.
(B) Results for aCGH analyses in three individuals are shown. Each dot indicates oligonucleotide probes: black dots represent a normal copy number, red dots represent a copy number gain, and green dots represent copy number losses as compared with a gender-matched control. The proband’s designated ID (BAB#s), the chromosomal location, and the approximate sizes of the copy number variants (CNVs) are written above each chromosome ideogram, and the chromosome number is written on the left side of ideogram. Red dashed lines determine the chromosomal region investigated by aCGH. Abbreviations are as follows: Chr = chromosome, Del = deletion, Dup = duplication, Mb = megabase, NTC = no template control, and Ter = terminal.
CNVs in Arthrogryposis
In three individuals, we found CNVs that were probably contributing to the arthrogryposis phenotype (Figure 7B and Table S2). We searched the genomic intervals defined by these CNV regions in these three individuals for possible candidate dosage-sensitive genes; however, we could not readily identify any specific potentially dosage-sensitive genes. We also examined the DECIPHER database for additional individuals with CNVs involving the identified regions and found a few individuals with overlapping features in two of these three individuals (individuals BAB7128 and BAB9312). However, the overlapping region of CNV remained substantial and had multiple annotated genes within its boundaries, precluding identification of a single, potentially etiologic gene. Individual BAB7128 was a 20-month-old male with DD and ID, hearing loss, bilateral hip dislocation, and pes equinovarus (PEV) deformity. The CNV detection tool (XHMM) showed a 6.7 Mb de novo deletion that includes 54 genes in the deletion region on chromosome 5 (hg19.g.chr5: 129673070–136365890). A DECIPHER database search revealed four individuals (DECIPHER ID#s 785, 2224, 256078, and 260667) with scoliosis, distal arthrogryposis, camptodactyly, and/or PEV. The smallest overlapping region of ∼2 Mb involving 11 genes (hg19.g.chr5: 129673070–131550089) was found in DECIPHER individual 785. This individual has camptodactyly and PEV, which partially explain our subject’s clinical features. Individual BAB8145 was a 3.5-year-old female with DD and ID, contractures in fingers and toes, and PEV. She was found to have two de novo deletions: a 9.43Mb deletion in chromosome 10 (hg19.g.chr10: 58374373–67805420) involving 29 genes and a 0.7 Mb de novo deletion in a non-coding region on chromosome 11 (hg19.g.chr11: 90798107–91499657); this was followed by an ∼1 Mb normal copy number and 7.58 Mb de novo deletion on chromosome 11 (hg19.g.chr11: 92950818–100534639) involving 39 genes in the region, and this was detected by karyotyping. Individual BAB9312 was a 7-year-old female who presented with DD and ID, hearing loss, and PEV. She was found to have a complex genomic rearrangement (CGR)55 involving chromosome 18: a 21.8 Mb duplication (hg19.g.chr18: 21901302–43716565) and an 11.6 Mb deletion (hg19.g.chr18: 66350129–78002264) (Figure 7B). Interestingly, there were three DECIPHER individuals (# 251072, 269015, and 319532) with overlapping clinical features that included DD and ID, hearing loss, PEV, and additional clinical findings, but the overlapping region was ∼10 Mb in all three individuals (hg19.g.chr18: 67223975–77120273).
Families Potentially Affected by Multilocus Pathogenic Variation
Although many of the early cohort-based studies describing multiple molecular diagnoses involve known disease genes, there have been more recent reports of multiple molecular diagnoses for which one identified gene is a candidate gene proposed to explain the observed, expanded phenotype associated with a well-studied, known disease-associated gene.10, 14, 31, 56 Given the rarity of the conditions studied, it is unlikely that two or more unrelated families will share the same combination of multiple molecular diagnoses. However, apparent phenotypic expansion in the setting of a single molecular diagnosis can, in some individuals, suggest a second, or even third, molecular diagnosis explaining the observed, expanded phenotypic features.31 Considering this possibility in the present cohort, we further explored 11 families for which there was evidence of multilocus pathogenic variation potentially contributing to the phenotype, i.e., mutational burden contributing to a blended phenotype (Tables 2 and S1, Figures S1 and S2, and the Supplemental Text).10 Six out of 11 families have three genes with variants that are likely to be pathogenic and that we propose have an impact on the disease phenotype. We highlight one example from “known-known” and one example from “known-candidate” gene patterns, and the remainder of individuals with proposed multilocus pathogenic variation are detailed in Tables 2 and S1, Figure S2, and the Supplemental Text.
Trio ES in individual BAB8600 (index) and her more severely affected mother, individual BAB8601, revealed novel variants in two known genes, FBN2 (MIM: 612570) and COL6A3 (Figure S6). The variant in FBN2 was a de novo missense change (GenBank: NM_001999; c.4094G>C [p.Cys1365Ser]) in the mother that was transmitted to the less severely affected child. The COL6A3 variant (GenBank: NM_057167; c.367G>A [p.Val123Met]) was a homozygous missense change in the mother and heterozygous in the affected child and maternal grandparents. Mutations in FBN2 are known to cause autosomal-dominant contractural arachnodactyly (DA9 [MIM: 121050]), whereas deleterious variants in COL6A3 cause several neuromuscular phenotypes including dominantly and recessively inherited Bethlem myopathy 1 (BTHLM1 [MIM: 158810]). Additional features in the mother can potentially be parsimoniously explained by homozygosity for the identified variant in COL6A3.
Individual BAB8397 underwent trio ES, which showed a reported, homozygous splice-site variant (GenBank: NM_001271208; c.19101+5G>A) in NEB (MIM: 161650) and a hemizygous, i.e., X-linked in a male proband, nonsynonymous variant (GenBank: NM_001098791; c.297C>G [p.Asn99Lys]) in MID1IP1 (Figure S7). The NEB variant identified in the proband was previously reported in a compound-heterozygous state with a nonsense variant (p.Tyr1858∗) in an arthrogryposis-affected individual who died during the neonatal period.57 The more severe phenotype in that individual could be explained by loss of function of the second allele. The second gene, MID1IP1, was not linked to any human disease previously. However, there are studies showing that MID1IP1 probably plays a role in myogenesis.58, 59 Casey et al. showed that Mid1ip1 expression is increased in the skeletal muscle of rats with peroxisome proliferator-activated receptors (PPAR)-induced myopathy.58 Additionally, Bower et al. showed upregulation of Mid1ip1 both with feeding and in vitro myogenesis in salmon fish.59 The study further showed that Mid1ip1 is highly expressed in fast and slow twitch muscle fibers compared to other tissues in the body. Although the function of MID1IP1 is not fully delineated, taken together, these data provide evidence that the MID1IP1 variant is likely to be contributing to the myopathy and contracture phenotype in our study individual.
The median total AOH size (individual locus AOH regions ≥3 Mb) calculated from exome data with BafCalculator in the probands with arthrogryposis and without any evidence for multilocus pathogenic variation (n = 97) is significantly higher (89.7 Mb) than the median total AOH size in the non-Turkish probands present in the Baylor-CMG database (n = 2,340) (23.5 Mb) (Mann-Whitney U test, p value = 7.35 × 10−17). Intriguingly, the probands that were studied in this arthrogryposis cohort and had dual molecular diagnoses (n = 19) have a larger median total AOH size (159.3 Mb) than the median total AOH size of the probands without any dual molecular diagnosis (n = 97; 89.7 Mb) (Mann-Whitney U test, p value = 0.02).
Discussion
We investigated the underlying molecular etiology of a large cohort (totaling 117 families including the phase I cohort) of individuals with arthrogryposis, either as an isolated clinical entity or as part of a syndrome, by using genome-wide assays. The current phase II cohort consists of 91 families, including two families identified through GeneMatcher, and 20 families who remained undiagnosed during the phase I study: ten out of 20 families who remained undiagnosed during phase I underwent reanalysis, and we expanded the sequencing to additional family members (e.g., index ES to trio or quad ES) in the remaining ten families. Overall, the molecular diagnostic rate from the combined phase I and II families was 73.5% (86 out of 117) when including both known and candidate genes, and 58.1% (68 out of 117) when considering molecular diagnoses involving only known genes.
Some experimental analysis enhancements that were implemented in our phase II approach and that potentially contributed to the increase in the molecular diagnostic yield in the second cohort (65.1% in phase II versus 58.3% in phase I) are expanding to trio ES and reanalyzing with additional bioinformatics tools the individuals from phase I who had unsolved molecular diagnoses. Multilocus pathogenic variation, consistent with a mutational burden or a potential oligogenic inheritance model, was observed in 19.0% (11/58) of the unrelated individuals with a molecular diagnosis in the present (phase II) cohort and 22.1% (19/86) of all 86 arthrogryposis-affected individuals with a molecular diagnosis reported here and in our previous [Phase I8 + II] study. Although AOH is indeed predicted to increase the frequency of homozygous variants in this cohort, identified multiple molecular diagnoses involved at least one known disease-associated locus in each individual and included known pathogenic or premature truncating variants at six loci, and additional evidence for the pathogenicity of identified variants is provided (Table S1 and Figure S2). Parents who are carriers for multilocus variation are notably unaffected, and four families with intrafamilial phenotypic variation (families HOU2523, HOU2620, HOU2791, and HOU3125) provide an opportunity to further dissect the relationship between observed phenotypes and identified genotypes.
Our phase II study additionally contributed to the classification of five genes (MYOM2, RYR3, ERGIC1, SPTBN4, and ABCA7), in which variation is probably causative of the arthrogryposis phenotype because they have now been identified in more than one family with the same phenotype.
We previously studied human disease-associated genes by using the universality of biology and the bidirectional synergism between fly and human genetics; this showed that essential fly genes with multiple human paralogs are 8-fold more likely to be disease-causing genes.60 We applied this approach to human paralogs for our proposed candidate genes in the arthrogryposis cohort and found that 14 of 19 have a fly ortholog, and 8 of 14 genes (including DD-and-ID-associated genes) have two or more human paralogs (Table S3).
Reanalysis of Exome Data and Expansion to Include Studies of Additional Family Members
We previously showed in a pilot study (N = 74 families) that reanalysis in the research setting could potentially solve the previously unsolved clinical exomes for about half of affected individuals.26 Effective experimental approaches for improving molecular diagnostic “solved rates” included: expanding singleton ES to additional family members (trio ES or other affected family member[s]) and applying additional, in-house-developed bioinformatics tools to detect indels, CNVs, and de novo variants with more relaxed analysis parameters. In the phase II study, we also expanded singleton ES to trio ES in ten previously studied families to uncover the genetic etiology of arthrogryposis and evaluate the diagnostic value of trio ES over singleton ES. Four out of ten families (40%) were provided with a molecular diagnosis upon expansion to trio ES and the application of bioinformatics tools to identify de novo and compound heterozygous variants and CNVs.26 For the remaining ten families for whom parental samples were not available, reanalysis revealed variants that can potentially explain the molecular etiology of the observed phenotypes in five families. The overall diagnostic rate of exome reanalysis, including expansion to trio ES, in this cohort is 45% (9/20), which is consistent with our previous findings of the diagnostic utility of reanalysis.26 Crucial experimental approaches for a reanalysis cohort are detailed in Table S4.
Phenotypic Expansion
In two personal genomes from unrelated individuals, we found variants in genes previously associated with conditions that do not include arthrogryposis as a phenotype; we now propose an expansion of the known phenotypes associated with these genes. Individual BAB7308 was found to have a de novo, heterozygous SYT2 (MIM: 600104) variant (GenBank: NM_177402; c.1081G>C [p.Asp361His], Figure S8), and details of this individual are described in Tables 1 and S1 and the Supplemental Text. The second individual, BAB8356, had proband ES, which revealed a homozygous change (GenBank: NM_005245; c.6026A>G [p.Asn2009Ser]) in FAT tumor-suppressor homolog 1 - Drosophila (FAT1) (Figure S9). Clinical details of this gene are described in the Supplemental Text. Bi-allelic variants of FAT1 were shown to cause glomerulotubular nephropathy.61 Interestingly, two recent studies showed that FAT1 expression has a role in the pathogenesis of fascioscapulohumeral dystrophy (FSHD1 [MIM: 158900]).62, 63 Two additional studies showed heterozygous rare variants of FAT1 in individuals with FSHD.64, 65 FAT1 is a member of protocadherin and has a role in smooth muscle cell motility, actin accumulation, cell polarity, migrating muscle precursors, and kidney development.66, 67, 68, 69 We here propose another bi-allelic variant in this gene with a phenotype of arthrogryposis. Interestingly, there are increasing examples of Mendelian disease-gene discoveries for which bi-allelic variation associated with a recessively inherited trait is identified in a gene previously or simultaneously described in association with a dominant trait conferred by mono-allelic variation. MAB21L2 (MIM: 604357), associated with dominant and recessive forms of microphthalmia/coloboma and skeletal dysplasia syndrome (MCSKS [MIM: 615877]), and ATAD3A (MIM: 612316), associated with autosomal-dominant or recessive Harel-Yoon syndrome (HAYOS [MIM: 617183]), are examples that illustrate both de novo and homozygous variants, and they lead to dominantly and recessively inherited conditions, respectively, that are associated with a single locus,17, 18 as is now described for MFN2 (MIM: 608507), TGFB3 (MIM: 190230), CIT (MIM: 605629), SPEG, ADNP, and FAT1 in the present study.
Rare-Variant Multilocus Pathogenic Variation
In the combined phase I + phase II arthrogryposis cohort, we identified 19 families with rare variation found at more than one locus and contributing to the observed phenotype; there were 16 individuals with an AR + AR (14 individuals) or AR + AR + AR (two individuals) multilocus disease-trait pattern, representing 84.2% (16/19) of individuals with multilocus variation (Figure 1C). Of the 37 identified AR molecular loci in all 19 individuals with multiple molecular diagnoses, 31 (31/37, 83.7%) loci demonstrated homozygosity for rare variants that are private to the studied clan (the Turkish population frequency can be found in Table S1). This is in striking contrast to an analysis of multilocus variation in a cohort of North American referrals to a diagnostic laboratory; for this cohort there were 11 individuals with an AR + AR (nine individuals) or AR + AR + AR (two individuals) disease pattern, representing 10.9% (11/101) of multilocus variation individuals studied (Figure 1C).10 The relative frequency of multiple molecular diagnoses was higher (22%, 19/86) in the arthrogryposis cohort compared to a relatively outbred population (4.9%, 101/2076), and this was driven by a high rate of AR + AR diagnoses (84.2% 16/19) in the arthrogryposis cohort compared to the North American referral population (10.9%, 11/101, two-tailed p < 0.0001, Fisher’s exact test). Further evidence for IBD-driven, genome-wide accumulation of deleterious variants is shown in large cohort population studies as well.70
In summary, we provide evidence for multiple candidate genes and further evidence for multilocus pathogenic variation that contributes to the complex trait of arthrogryposis. Moreover, we provide evidence further extending the clan genomics hypothesis71 and emboldening the rare-variant multilocus-pathogenic-variation model for a complex trait. This study also highlights: (1) the value of reanalysis and exome variant analyses on additional family members and its ability to conclude a molecular diagnosis in this era of rapid gene discovery, (2) oligogenic inheritance and blended phenotypes can be responsible for up to ∼22% of the arthrogryposis diagnoses in individuals, and the majority of this can be driven by population substructure and IBD, (3) both mono-allelic and bi-allelic variants in a single gene can contribute to human disease, causing either similar or different phenotypes, (4) paralogs of known disease-associated genes can be responsible for disease traits, and (5) genetic models for complex traits can be built from underlying Mendelian principles and explored experimentally.
Declaration of Interests
Baylor College of Medicine (BCM) and Miraca Holdings have formed a joint venture with shared ownership and governance of the Baylor Genetics (BG), which performs clinical microarray analysis and clinical exome sequencing. J.R.L. serves on the scientific advisory board of the BG. J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The other authors declare no conflicts of interest.
Acknowledgments
The authors thank the families and their health care teams for participating in this project.
This work was supported in part by R35 NS105078 and MDA#512848 to J.R.L. and a jointly funded National Human Genome Research Institute (NHGRI) and National Heart, Lung, and Blood Institute (NHLBI) grant to the Baylor-Hopkins Center for Mendelian Genomics (UM1 HG006542). J.E.P. is supported by NHGRI K08 HG008986. D.P. is supported by the National Institutes of Health - Brain Disorders and Development Training Grant (T32 NS043124-17) and a Clinical Research Training Scholarship in Neuromuscular Disease partnered by the American Brain Foundation (ABF) and Muscle Study Group (MSG). This study is partly funded by Tubitak project number 217S675, Turkey to N.E. and B.T.. This study is partly funded by Indian Council of Medical Research, New Delhi, India with File no.: No.5/13/58/2015/NCD-III to A.S.
Published: June 20, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.05.015.
Accession Numbers
The dbGaP accession number for all exome sequences reported in this paper and for which informed consent for data sharing in controlled-access databases has been provided is dbGaP: phs000711.v5.p1.
Web Resources
1000 Genomes, http://www.1000genomes.org
ARIC Database, https://sites.cscc.unc.edu/aric/
BaFCalculator Browser, https://github.com/BCM-Lupskilab/BafCalculator
Cell Culture Repositories, https://ccr.coriell.org
CoNIFER, http://conifer.sourceforge.net
CoNVex, ftp://ftp.sanger.ac.uk/pub/users/pv1/CoNVex/Docs/CoNVex.pdf
DNM-Finder Browser, https://github.com/BCM-Lupskilab/DNM-Finder
ExAC Browser, http://exac.broadinstitute.org/
GeneMatcher Browser, https://genematcher.org/
gnomAD Browser, https://gnomad.broadinstitute.org/
Genotype-Tissue Expression - GTEx Portal, https://gtexportal.org/home/
HmzDelFinder Browser, https://github.com/BCM-Lupskilab/HMZDelFinder
Human-Fly Ortholog Comparison Browser, https://www.flyrnai.org/cgi-bin/DRSC_orthologs.pl
Online Mendelian Inheritance in Man, https://www.omim.org/
NHLBI GO Exome Sequencing Project, https://evs.gs.washington.edu/EVS
STRING 10.5, https://string-db.org/
UCSC Genome Browser, https://genome.ucsc.edu
UniProt, https://www.uniprot.org/
DECIPHER, https://decipher.sanger.ac.uk/
Supplemental Data
References
- 1.Hall J.G. Arthrogryposis (multiple congenital contractures): Diagnostic approach to etiology, classification, genetics, and general principles. Eur. J. Med. Genet. 2014;57:464–472. doi: 10.1016/j.ejmg.2014.03.008. [DOI] [PubMed] [Google Scholar]; Hall, J.G. (2014). Arthrogryposis (multiple congenital contractures): Diagnostic approach to etiology, classification, genetics, and general principles. Eur. J. Med. Genet. 57, 464-472. [DOI] [PubMed]
- 2.Lowry R.B., Sibbald B., Bedard T., Hall J.G. Prevalence of multiple congenital contractures including arthrogryposis multiplex congenita in Alberta, Canada, and a strategy for classification and coding. Birth Defects Res. A Clin. Mol. Teratol. 2010;88:1057–1061. doi: 10.1002/bdra.20738. [DOI] [PubMed] [Google Scholar]; Lowry, R.B., Sibbald, B., Bedard, T., and Hall, J.G. (2010). Prevalence of multiple congenital contractures including arthrogryposis multiplex congenita in Alberta, Canada, and a strategy for classification and coding. Birth Defects Res. A Clin. Mol. Teratol. 88, 1057-1061. [DOI] [PubMed]
- 3.Hall J.G., Kiefer J. Arthrogryposis as a syndrome: Gene ontology analysis. Mol. Syndromol. 2016;7:101–109. doi: 10.1159/000446617. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hall, J.G., and Kiefer, J. (2016). Arthrogryposis as a syndrome: Gene ontology analysis. Mol. Syndromol. 7, 101-109. [DOI] [PMC free article] [PubMed]
- 4.Balta B., Erdogan M., Ergul A.B., Sahin Y., Ozcan A. Interstitial deletion 5p14.1-p15.2 and 5q14.3-q23.2 in a patient with clubfoot, blepharophimosis, arthrogryposis, and multiple congenital abnormalities. Am. J. Med. Genet. A. 2017;173:2798–2802. doi: 10.1002/ajmg.a.38386. [DOI] [PubMed] [Google Scholar]; Balta, B., Erdogan, M., Ergul, A.B., Sahin, Y., and Ozcan, A. (2017). Interstitial deletion 5p14.1-p15.2 and 5q14.3-q23.2 in a patient with clubfoot, blepharophimosis, arthrogryposis, and multiple congenital abnormalities. Am. J. Med. Genet. A. 173, 2798-2802. [DOI] [PubMed]
- 5.Carrascosa-Romero M.C., Suela J., Pardal-Fernández J.M., Bermejo-Sánchez E., Vidal-Company A., MacDonald A., Tébar-Gil R., Martínez-Fernández M.L., Martínez-Frías M.L. A 2.84 Mb deletion at 21q22.11 in a patient clinically diagnosed with Marden-Walker syndrome. Am. J. Med. Genet. A. 2013;161A:2281–2290. doi: 10.1002/ajmg.a.35862. [DOI] [PubMed] [Google Scholar]; Carrascosa-Romero, M.C., Suela, J., Pardal-Fernandez, J.M., Bermejo-Sanchez, E., Vidal-Company, A., MacDonald, A., Tebar-Gil, R., Martinez-Fernandez, M.L., and Martinez-Frias, M.L. (2013). A 2.84 Mb deletion at 21q22.11 in a patient clinically diagnosed with Marden-Walker syndrome. Am. J. Med. Genet. A. 161A, 2281-2290. [DOI] [PubMed]
- 6.Lukusa T., Fryns J.P. Pure de novo 17q25.3 micro duplication characterized by micro array CGH in a dysmorphic infant with growth retardation, developmental delay and distal arthrogryposis. Genet. Couns. 2010;21:25–34. [PubMed] [Google Scholar]; Lukusa, T., and Fryns, J.P. (2010). Pure de novo 17q25.3 micro duplication characterized by micro array CGH in a dysmorphic infant with growth retardation, developmental delay and distal arthrogryposis. Genet. Couns. 21, 25-34. [PubMed]
- 7.Tabet A.C., Aboura A., Gérard M., Pilorge M., Dupont C., Gadisseux J.F., Hervy N., Pipiras E., Delahaye A., Kanafani S. Molecular characterization of a de novo 6q24.2q25.3 duplication interrupting UTRN in a patient with arthrogryposis. Am. J. Med. Genet. A. 2010;152A:1781–1788. doi: 10.1002/ajmg.a.33433. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tabet, A.C., Aboura, A., Gerard, M., Pilorge, M., Dupont, C., Gadisseux, J.F., Hervy, N., Pipiras, E., Delahaye, A., Kanafani, S., et al. (2010). Molecular characterization of a de novo 6q24.2q25.3 duplication interrupting UTRN in a patient with arthrogryposis. Am. J. Med. Genet. A. 152A, 1781-1788. [DOI] [PMC free article] [PubMed]
- 8.Bayram Y., Karaca E., Coban Akdemir Z., Yilmaz E.O., Tayfun G.A., Aydin H., Torun D., Bozdogan S.T., Gezdirici A., Isikay S. Molecular etiology of arthrogryposis in multiple families of mostly Turkish origin. J. Clin. Invest. 2016;126:762–778. doi: 10.1172/JCI84457. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bayram, Y., Karaca, E., Coban Akdemir, Z., Yilmaz, E.O., Tayfun, G.A., Aydin, H., Torun, D., Bozdogan, S.T., Gezdirici, A., Isikay, S., et al. (2016). Molecular etiology of arthrogryposis in multiple families of mostly Turkish origin. J. Clin. Invest. 126, 762-778. [DOI] [PMC free article] [PubMed]
- 9.Todd E.J., Yau K.S., Ong R., Slee J., McGillivray G., Barnett C.P., Haliloglu G., Talim B., Akcoren Z., Kariminejad A. Next generation sequencing in a large cohort of patients presenting with neuromuscular disease before or at birth. Orphanet J. Rare Dis. 2015;10:148. doi: 10.1186/s13023-015-0364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; Todd, E.J., Yau, K.S., Ong, R., Slee, J., McGillivray, G., Barnett, C.P., Haliloglu, G., Talim, B., Akcoren, Z., Kariminejad, A., et al. (2015). Next generation sequencing in a large cohort of patients presenting with neuromuscular disease before or at birth. Orphanet J. Rare Dis. 10, 148. [DOI] [PMC free article] [PubMed]
- 10.Posey J.E., Harel T., Liu P., Rosenfeld J.A., James R.A., Coban Akdemir Z.H., Walkiewicz M., Bi W., Xiao R., Ding Y. Resolution of disease phenotypes resulting from multilocus genomic variation. N. Engl. J. Med. 2017;376:21–31. doi: 10.1056/NEJMoa1516767. [DOI] [PMC free article] [PubMed] [Google Scholar]; Posey, J.E., Harel, T., Liu, P., Rosenfeld, J.A., James, R.A., Coban Akdemir, Z.H., Walkiewicz, M., Bi, W., Xiao, R., Ding, Y., et al. (2017). Resolution of disease phenotypes resulting from multilocus genomic variation. N. Engl. J. Med. 376, 21-31. [DOI] [PMC free article] [PubMed]
- 11.Tarailo-Graovac M., Shyr C., Ross C.J., Horvath G.A., Salvarinova R., Ye X.C., Zhang L.H., Bhavsar A.P., Lee J.J., Drögemöller B.I. Exome sequencing and the management of neurometabolic disorders. N. Engl. J. Med. 2016;374:2246–2255. doi: 10.1056/NEJMoa1515792. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tarailo-Graovac, M., Shyr, C., Ross, C.J., Horvath, G.A., Salvarinova, R., Ye, X.C., Zhang, L.H., Bhavsar, A.P., Lee, J.J., Drogemoller, B.I., et al. (2016). Exome sequencing and the management of neurometabolic disorders. N. Engl. J. Med. 374, 2246-2255. [DOI] [PMC free article] [PubMed]
- 12.Balci T.B., Hartley T., Xi Y., Dyment D.A., Beaulieu C.L., Bernier F.P., Dupuis L., Horvath G.A., Mendoza-Londono R., Prasad C., FORGE Canada Consortium. Care4Rare Canada Consortium Debunking Occam’s razor: Diagnosing multiple genetic diseases in families by whole-exome sequencing. Clin. Genet. 2017;92:281–289. doi: 10.1111/cge.12987. [DOI] [PubMed] [Google Scholar]; Balci, T.B., Hartley, T., Xi, Y., Dyment, D.A., Beaulieu, C.L., Bernier, F.P., Dupuis, L., Horvath, G.A., Mendoza-Londono, R., Prasad, C., et al.; FORGE Canada Consortium; Care4Rare Canada Consortium (2017). Debunking Occam’s razor: Diagnosing multiple genetic diseases in families by whole-exome sequencing. Clin. Genet. 92, 281-289. [DOI] [PubMed]
- 13.Posey J.E., Rosenfeld J.A., James R.A., Bainbridge M., Niu Z., Wang X., Dhar S., Wiszniewski W., Akdemir Z.H., Gambin T. Molecular diagnostic experience of whole-exome sequencing in adult patients. Genet. Med. 2016;18:678–685. doi: 10.1038/gim.2015.142. [DOI] [PMC free article] [PubMed] [Google Scholar]; Posey, J.E., Rosenfeld, J.A., James, R.A., Bainbridge, M., Niu, Z., Wang, X., Dhar, S., Wiszniewski, W., Akdemir, Z.H., Gambin, T., et al. (2016). Molecular diagnostic experience of whole-exome sequencing in adult patients. Genet. Med. 18, 678-685. [DOI] [PMC free article] [PubMed]
- 14.Yang Y., Muzny D.M., Xia F., Niu Z., Person R., Ding Y., Ward P., Braxton A., Wang M., Buhay C. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yang, Y., Muzny, D.M., Xia, F., Niu, Z., Person, R., Ding, Y., Ward, P., Braxton, A., Wang, M., Buhay, C., et al. (2014). Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 312, 1870-1879. [DOI] [PMC free article] [PubMed]
- 15.Monies D., Maddirevula S., Kurdi W., Alanazy M.H., Alkhalidi H., Al-Owain M., Sulaiman R.A., Faqeih E., Goljan E., Ibrahim N. Autozygosity reveals recessive mutations and novel mechanisms in dominant genes: Implications in variant interpretation. Genet. Med. 2017;19:1144–1150. doi: 10.1038/gim.2017.22. [DOI] [PubMed] [Google Scholar]; Monies, D., Maddirevula, S., Kurdi, W., Alanazy, M.H., Alkhalidi, H., Al-Owain, M., Sulaiman, R.A., Faqeih, E., Goljan, E., Ibrahim, N., et al. (2017). Autozygosity reveals recessive mutations and novel mechanisms in dominant genes: Implications in variant interpretation. Genet. Med. 19, 1144-1150. [DOI] [PubMed]
- 16.Worman H.J., Bonne G. “Laminopathies”: A wide spectrum of human diseases. Exp. Cell Res. 2007;313:2121–2133. doi: 10.1016/j.yexcr.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]; Worman, H.J., and Bonne, G. (2007). “Laminopathies”: A wide spectrum of human diseases. Exp. Cell Res. 313, 2121-2133. [DOI] [PMC free article] [PubMed]
- 17.Rainger J., Pehlivan D., Johansson S., Bengani H., Sanchez-Pulido L., Williamson K.A., Ture M., Barker H., Rosendahl K., Spranger J., UK10K. Baylor-Hopkins Center for Mendelian Genomics Monoallelic and biallelic mutations in MAB21L2 cause a spectrum of major eye malformations. Am. J. Hum. Genet. 2014;94:915–923. doi: 10.1016/j.ajhg.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rainger, J., Pehlivan, D., Johansson, S., Bengani, H., Sanchez-Pulido, L., Williamson, K.A., Ture, M., Barker, H., Rosendahl, K., Spranger, J., et al.; UK10K; Baylor-Hopkins Center for Mendelian Genomics (2014). Monoallelic and biallelic mutations in MAB21L2 cause a spectrum of major eye malformations. Am. J. Hum. Genet. 94, 915-923. [DOI] [PMC free article] [PubMed]
- 18.Harel T., Yoon W.H., Garone C., Gu S., Coban-Akdemir Z., Eldomery M.K., Posey J.E., Jhangiani S.N., Rosenfeld J.A., Cho M.T., Baylor-Hopkins Center for Mendelian Genomics. University of Washington Center for Mendelian Genomics Recurrent de novo and biallelic variation of ATAD3A, encoding a mitochondrial membrane protein, results in distinct neurological syndromes. Am. J. Hum. Genet. 2016;99:831–845. doi: 10.1016/j.ajhg.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; Harel, T., Yoon, W.H., Garone, C., Gu, S., Coban-Akdemir, Z., Eldomery, M.K., Posey, J.E., Jhangiani, S.N., Rosenfeld, J.A., Cho, M.T., et al.; Baylor-Hopkins Center for Mendelian Genomics; University of Washington Center for Mendelian Genomics (2016). Recurrent de novo and biallelic variation of ATAD3A, encoding a mitochondrial membrane protein, results in distinct neurological syndromes. Am. J. Hum. Genet. 99, 831-845. [DOI] [PMC free article] [PubMed]
- 19.Warner L.E., Mancias P., Butler I.J., McDonald C.M., Keppen L., Koob K.G., Lupski J.R. Mutations in the early growth response 2 (EGR2) gene are associated with hereditary myelinopathies. Nat. Genet. 1998;18:382–384. doi: 10.1038/ng0498-382. [DOI] [PubMed] [Google Scholar]; Warner, L.E., Mancias, P., Butler, I.J., McDonald, C.M., Keppen, L., Koob, K.G., and Lupski, J.R. (1998). Mutations in the early growth response 2 (EGR2) gene are associated with hereditary myelinopathies. Nat. Genet. 18, 382-384. [DOI] [PubMed]
- 20.Posey J.E., O’Donnell-Luria A.H., Chong J.X., Harel T., Jhangiani S.N., Coban Akdemir Z.H., Buyske S., Pehlivan D., Carvalho C.M.B., Baxter S., Centers for Mendelian Genomics Insights into genetics, human biology and disease gleaned from family based genomic studies. Genet. Med. 2019;21:798–812. doi: 10.1038/s41436-018-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; Posey, J.E., O’Donnell-Luria, A.H., Chong, J.X., Harel, T., Jhangiani, S.N., Coban Akdemir, Z.H., Buyske, S., Pehlivan, D., Carvalho, C.M.B., Baxter, S., et al.; Centers for Mendelian Genomics (2019). Insights into genetics, human biology and disease gleaned from family based genomic studies. Genet. Med. 21, 798-812. [DOI] [PMC free article] [PubMed]
- 21.Lupski J.R., Reid J.G., Gonzaga-Jauregui C., Rio Deiros D., Chen D.C., Nazareth L., Bainbridge M., Dinh H., Jing C., Wheeler D.A. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N. Engl. J. Med. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lupski, J.R., Reid, J.G., Gonzaga-Jauregui, C., Rio Deiros, D., Chen, D.C., Nazareth, L., Bainbridge, M., Dinh, H., Jing, C., Wheeler, D.A., et al. (2010). Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N. Engl. J. Med. 362, 1181-1191. [DOI] [PMC free article] [PubMed]
- 22.Reid J.G., Carroll A., Veeraraghavan N., Dahdouli M., Sundquist A., English A., Bainbridge M., White S., Salerno W., Buhay C. Launching genomics into the cloud: Deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics. 2014;15:30. doi: 10.1186/1471-2105-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]; Reid, J.G., Carroll, A., Veeraraghavan, N., Dahdouli, M., Sundquist, A., English, A., Bainbridge, M., White, S., Salerno, W., Buhay, C., et al. (2014). Launching genomics into the cloud: Deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics 15, 30. [DOI] [PMC free article] [PubMed]
- 23.Challis D., Yu J., Evani U.S., Jackson A.R., Paithankar S., Coarfa C., Milosavljevic A., Gibbs R.A., Yu F. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics. 2012;13:8. doi: 10.1186/1471-2105-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Challis, D., Yu, J., Evani, U.S., Jackson, A.R., Paithankar, S., Coarfa, C., Milosavljevic, A., Gibbs, R.A., and Yu, F. (2012). An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics 13, 8. [DOI] [PMC free article] [PubMed]
- 24.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., Marth, G., Abecasis, G., and Durbin, R.; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078-2079. [DOI] [PMC free article] [PubMed]
- 25.Wang K., Li M., Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang, K., Li, M., and Hakonarson, H. (2010). ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164. [DOI] [PMC free article] [PubMed]
- 26.Eldomery M.K., Coban-Akdemir Z., Harel T., Rosenfeld J.A., Gambin T., Stray-Pedersen A., Küry S., Mercier S., Lessel D., Denecke J. Lessons learned from additional research analyses of unsolved clinical exome cases. Genome Med. 2017;9:26. doi: 10.1186/s13073-017-0412-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Eldomery, M.K., Coban-Akdemir, Z., Harel, T., Rosenfeld, J.A., Gambin, T., Stray-Pedersen, A., Kury, S., Mercier, S., Lessel, D., Denecke, J., et al. (2017). Lessons learned from additional research analyses of unsolved clinical exome cases. Genome Med. 9, 26. [DOI] [PMC free article] [PubMed]
- 27.Karaca E., Harel T., Pehlivan D., Jhangiani S.N., Gambin T., Coban Akdemir Z., Gonzaga-Jauregui C., Erdin S., Bayram Y., Campbell I.M. Genes that affect brain structure and function identified by rare variant analyses of Mendelian neurologic disease. Neuron. 2015;88:499–513. doi: 10.1016/j.neuron.2015.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]; Karaca, E., Harel, T., Pehlivan, D., Jhangiani, S.N., Gambin, T., Coban Akdemir, Z., Gonzaga-Jauregui, C., Erdin, S., Bayram, Y., Campbell, I.M., et al. (2015). Genes that affect brain structure and function identified by rare variant analyses of Mendelian neurologic disease. Neuron 88, 499-513. [DOI] [PMC free article] [PubMed]
- 28.Gambin T., Akdemir Z.C., Yuan B., Gu S., Chiang T., Carvalho C.M.B., Shaw C., Jhangiani S., Boone P.M., Eldomery M.K. Homozygous and hemizygous CNV detection from exome sequencing data in a Mendelian disease cohort. Nucleic Acids Res. 2017;45:1633–1648. doi: 10.1093/nar/gkw1237. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gambin, T., Akdemir, Z.C., Yuan, B., Gu, S., Chiang, T., Carvalho, C.M.B., Shaw, C., Jhangiani, S., Boone, P.M., Eldomery, M.K., et al. (2017). Homozygous and hemizygous CNV detection from exome sequencing data in a Mendelian disease cohort. Nucleic Acids Res. 45, 1633-1648. [DOI] [PMC free article] [PubMed]
- 29.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sobreira, N., Schiettecatte, F., Valle, D., and Hamosh, A. (2015). GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 36, 928-930. [DOI] [PMC free article] [PubMed]
- 30.Sobreira N., Schiettecatte F., Boehm C., Valle D., Hamosh A. New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum. Mutat. 2015;36:425–431. doi: 10.1002/humu.22769. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sobreira, N., Schiettecatte, F., Boehm, C., Valle, D., and Hamosh, A. (2015). New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum. Mutat. 36, 425-431. [DOI] [PMC free article] [PubMed]
- 31.Karaca E., Posey J.E., Coban Akdemir Z., Pehlivan D., Harel T., Jhangiani S.N., Bayram Y., Song X., Bahrambeigi V., Yuregir O.O. Phenotypic expansion illuminates multilocus pathogenic variation. Genet. Med. 2018;20:1528–1537. doi: 10.1038/gim.2018.33. [DOI] [PMC free article] [PubMed] [Google Scholar]; Karaca, E., Posey, J.E., Coban Akdemir, Z., Pehlivan, D., Harel, T., Jhangiani, S.N., Bayram, Y., Song, X., Bahrambeigi, V., Yuregir, O.O., et al. (2018). Phenotypic expansion illuminates multilocus pathogenic variation. Genet. Med. 20, 1528-1537. [DOI] [PMC free article] [PubMed]
- 32.Olshen A.B., Venkatraman E.S., Lucito R., Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]; Olshen, A.B., Venkatraman, E.S., Lucito, R., and Wigler, M. (2004). Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 5, 557-572. [DOI] [PubMed]
- 33.Spence J.E., Perciaccante R.G., Greig G.M., Willard H.F., Ledbetter D.H., Hejtmancik J.F., Pollack M.S., O’Brien W.E., Beaudet A.L. Uniparental disomy as a mechanism for human genetic disease. Am. J. Hum. Genet. 1988;42:217–226. [PMC free article] [PubMed] [Google Scholar]; Spence, J.E., Perciaccante, R.G., Greig, G.M., Willard, H.F., Ledbetter, D.H., Hejtmancik, J.F., Pollack, M.S., O’Brien, W.E., and Beaudet, A.L. (1988). Uniparental disomy as a mechanism for human genetic disease. Am. J. Hum. Genet. 42, 217-226. [PMC free article] [PubMed]
- 34.Gambin T., Yuan B., Bi W., Liu P., Rosenfeld J.A., Coban-Akdemir Z., Pursley A.N., Nagamani S.C.S., Marom R., Golla S. Identification of novel candidate disease genes from de novo exonic copy number variants. Genome Med. 2017;9:83. doi: 10.1186/s13073-017-0472-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gambin, T., Yuan, B., Bi, W., Liu, P., Rosenfeld, J.A., Coban-Akdemir, Z., Pursley, A.N., Nagamani, S.C.S., Marom, R., Golla, S., et al. (2017). Identification of novel candidate disease genes from de novo exonic copy number variants. Genome Med. 9, 83. [DOI] [PMC free article] [PubMed]
- 35.Pehlivan D., Hullings M., Carvalho C.M., Gonzaga-Jauregui C.G., Loy E., Jackson L.G., Krantz I.D., Deardorff M.A., Lupski J.R. NIPBL rearrangements in Cornelia de Lange syndrome: Evidence for replicative mechanism and genotype-phenotype correlation. Genet. Med. 2012;14:313–322. doi: 10.1038/gim.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pehlivan, D., Hullings, M., Carvalho, C.M., Gonzaga-Jauregui, C.G., Loy, E., Jackson, L.G., Krantz, I.D., Deardorff, M.A., and Lupski, J.R. (2012). NIPBL rearrangements in Cornelia de Lange syndrome: Evidence for replicative mechanism and genotype-phenotype correlation. Genet. Med. 14, 313-322. [DOI] [PMC free article] [PubMed]
- 36.Tsai E.A., Grochowski C.M., Falsey A.M., Rajagopalan R., Wendel D., Devoto M., Krantz I.D., Loomes K.M., Spinner N.B. Heterozygous deletion of FOXA2 segregates with disease in a family with heterotaxy, panhypopituitarism, and biliary atresia. Hum. Mutat. 2015;36:631–637. doi: 10.1002/humu.22786. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tsai, E.A., Grochowski, C.M., Falsey, A.M., Rajagopalan, R., Wendel, D., Devoto, M., Krantz, I.D., Loomes, K.M., and Spinner, N.B. (2015). Heterozygous deletion of FOXA2 segregates with disease in a family with heterotaxy, panhypopituitarism, and biliary atresia. Hum. Mutat. 36, 631-637. [DOI] [PMC free article] [PubMed]
- 37.Hu Y., Flockhart I., Vinayagam A., Bergwitz C., Berger B., Perrimon N., Mohr S.E. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 2011;12:357. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hu, Y., Flockhart, I., Vinayagam, A., Bergwitz, C., Berger, B., Perrimon, N., and Mohr, S.E. (2011). An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics 12, 357. [DOI] [PMC free article] [PubMed]
- 38.Malfatti E., Böhm J., Lacène E., Beuvin M., Romero N.B., Laporte J. A premature stop codon in MYO18B is associated with severe nemaline myopathy with cardiomyopathy. J. Neuromuscul. Dis. 2015;2:219–227. doi: 10.3233/JND-150085. [DOI] [PMC free article] [PubMed] [Google Scholar]; Malfatti, E., Bohm, J., Lacene, E., Beuvin, M., Romero, N.B., and Laporte, J. (2015). A premature stop codon in MYO18B is associated with severe nemaline myopathy with cardiomyopathy. J. Neuromuscul. Dis. 2, 219-227. [DOI] [PMC free article] [PubMed]
- 39.Alazami A.M., Kentab A.Y., Faqeih E., Mohamed J.Y., Alkhalidi H., Hijazi H., Alkuraya F.S. A novel syndrome of Klippel-Feil anomaly, myopathy, and characteristic facies is linked to a null mutation in MYO18B. J. Med. Genet. 2015;52:400–404. doi: 10.1136/jmedgenet-2014-102964. [DOI] [PubMed] [Google Scholar]; Alazami, A.M., Kentab, A.Y., Faqeih, E., Mohamed, J.Y., Alkhalidi, H., Hijazi, H., and Alkuraya, F.S. (2015). A novel syndrome of Klippel-Feil anomaly, myopathy, and characteristic facies is linked to a null mutation in MYO18B. J. Med. Genet. 52, 400-404. [DOI] [PubMed]
- 40.Nilipour Y., Nafissi S., Tjust A.E., Ravenscroft G., Hossein Nejad Nedai H., Taylor R.L., Varasteh V., Pedrosa Domellöf F., Zangi M., Tonekaboni S.H. Ryanodine receptor type 3 (RYR3) as a novel gene associated with a myopathy with nemaline bodies. Eur. J. Neurol. 2018;25:841–847. doi: 10.1111/ene.13607. [DOI] [PubMed] [Google Scholar]; Nilipour, Y., Nafissi, S., Tjust, A.E., Ravenscroft, G., Hossein Nejad Nedai, H., Taylor, R.L., Varasteh, V., Pedrosa Domellof, F., Zangi, M., Tonekaboni, S.H., et al. (2018). Ryanodine receptor type 3 (RYR3) as a novel gene associated with a myopathy with nemaline bodies. Eur. J. Neurol. 25, 841-847. [DOI] [PubMed]
- 41.Bertocchini F., Ovitt C.E., Conti A., Barone V., Schöler H.R., Bottinelli R., Reggiani C., Sorrentino V. Requirement for the ryanodine receptor type 3 for efficient contraction in neonatal skeletal muscles. EMBO J. 1997;16:6956–6963. doi: 10.1093/emboj/16.23.6956. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bertocchini, F., Ovitt, C.E., Conti, A., Barone, V., Scholer, H.R., Bottinelli, R., Reggiani, C., and Sorrentino, V. (1997). Requirement for the ryanodine receptor type 3 for efficient contraction in neonatal skeletal muscles. EMBO J. 16, 6956-6963. [DOI] [PMC free article] [PubMed]
- 42.Jeyakumar L.H., Copello J.A., O’Malley A.M., Wu G.M., Grassucci R., Wagenknecht T., Fleischer S. Purification and characterization of ryanodine receptor 3 from mammalian tissue. J. Biol. Chem. 1998;273:16011–16020. doi: 10.1074/jbc.273.26.16011. [DOI] [PubMed] [Google Scholar]; Jeyakumar, L.H., Copello, J.A., O’Malley, A.M., Wu, G.M., Grassucci, R., Wagenknecht, T., and Fleischer, S. (1998). Purification and characterization of ryanodine receptor 3 from mammalian tissue. J. Biol. Chem. 273, 16011-16020. [DOI] [PubMed]
- 43.Abath Neto O., Tassy O., Biancalana V., Zanoteli E., Pourquié O., Laporte J. Integrative data mining highlights candidate genes for monogenic myopathies. PLoS ONE. 2014;9:e110888. doi: 10.1371/journal.pone.0110888. [DOI] [PMC free article] [PubMed] [Google Scholar]; Abath Neto, O., Tassy, O., Biancalana, V., Zanoteli, E., Pourquie, O., and Laporte, J. (2014). Integrative data mining highlights candidate genes for monogenic myopathies. PLoS ONE 9, e110888. [DOI] [PMC free article] [PubMed]
- 44.Flix B., de la Torre C., Castillo J., Casal C., Illa I., Gallardo E. Dysferlin interacts with calsequestrin-1, myomesin-2 and dynein in human skeletal muscle. Int. J. Biochem. Cell Biol. 2013;45:1927–1938. doi: 10.1016/j.biocel.2013.06.007. [DOI] [PubMed] [Google Scholar]; Flix, B., de la Torre, C., Castillo, J., Casal, C., Illa, I., and Gallardo, E. (2013). Dysferlin interacts with calsequestrin-1, myomesin-2 and dynein in human skeletal muscle. Int. J. Biochem. Cell Biol. 45, 1927-1938. [DOI] [PubMed]
- 45.Helsmoortel C., Vulto-van Silfhout A.T., Coe B.P., Vandeweyer G., Rooms L., van den Ende J., Schuurs-Hoeijmakers J.H., Marcelis C.L., Willemsen M.H., Vissers L.E. A SWI/SNF-related autism syndrome caused by de novo mutations in ADNP. Nat. Genet. 2014;46:380–384. doi: 10.1038/ng.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]; Helsmoortel, C., Vulto-van Silfhout, A.T., Coe, B.P., Vandeweyer, G., Rooms, L., van den Ende, J., Schuurs-Hoeijmakers, J.H., Marcelis, C.L., Willemsen, M.H., Vissers, L.E., et al. (2014). A SWI/SNF-related autism syndrome caused by de novo mutations in ADNP. Nat. Genet. 46, 380-384. [DOI] [PMC free article] [PubMed]
- 46.Reinstein E., Drasinover V., Lotan R., Gal-Tanamy M., Bolocan Nachman I., Eyal E., Jaber L., Magal N., Shohat M. Mutations in ERGIC1 cause arthrogryposis multiplex congenita, neuropathic type. Clin. Genet. 2018;93:160–163. doi: 10.1111/cge.13018. [DOI] [PubMed] [Google Scholar]; Reinstein, E., Drasinover, V., Lotan, R., Gal-Tanamy, M., Bolocan Nachman, I., Eyal, E., Jaber, L., Magal, N., and Shohat, M. (2018). Mutations in ERGIC1 cause arthrogryposis multiplex congenita, neuropathic type. Clin. Genet. 93, 160-163. [DOI] [PubMed]
- 47.Breuza L., Halbeisen R., Jenö P., Otte S., Barlowe C., Hong W., Hauri H.P. Proteomics of endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membranes from brefeldin A-treated HepG2 cells identifies ERGIC-32, a new cycling protein that interacts with human Erv46. J. Biol. Chem. 2004;279:47242–47253. doi: 10.1074/jbc.M406644200. [DOI] [PubMed] [Google Scholar]; Breuza, L., Halbeisen, R., Jeno, P., Otte, S., Barlowe, C., Hong, W., and Hauri, H.P. (2004). Proteomics of endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membranes from brefeldin A-treated HepG2 cells identifies ERGIC-32, a new cycling protein that interacts with human Erv46. J. Biol. Chem. 279, 47242-47253. [DOI] [PubMed]
- 48.Knierim E., Gill E., Seifert F., Morales-Gonzalez S., Unudurthi S.D., Hund T.J., Stenzel W., Schuelke M. A recessive mutation in beta-IV-spectrin (SPTBN4) associates with congenital myopathy, neuropathy, and central deafness. Hum. Genet. 2017;136:903–910. doi: 10.1007/s00439-017-1814-7. [DOI] [PubMed] [Google Scholar]; Knierim, E., Gill, E., Seifert, F., Morales-Gonzalez, S., Unudurthi, S.D., Hund, T.J., Stenzel, W., and Schuelke, M. (2017). A recessive mutation in beta-IV-spectrin (SPTBN4) associates with congenital myopathy, neuropathy, and central deafness. Hum. Genet. 136, 903-910. [DOI] [PubMed]
- 49.Anazi S., Maddirevula S., Salpietro V., Asi Y.T., Alsahli S., Alhashem A., Shamseldin H.E., AlZahrani F., Patel N., Ibrahim N. Expanding the genetic heterogeneity of intellectual disability. Hum. Genet. 2017;136:1419–1429. doi: 10.1007/s00439-017-1843-2. [DOI] [PubMed] [Google Scholar]; Anazi, S., Maddirevula, S., Salpietro, V., Asi, Y.T., Alsahli, S., Alhashem, A., Shamseldin, H.E., AlZahrani, F., Patel, N., Ibrahim, N., et al. (2017). Expanding the genetic heterogeneity of intellectual disability. Hum. Genet. 136, 1419-1429. [DOI] [PubMed]
- 50.Lange S., Ouyang K., Meyer G., Cui L., Cheng H., Lieber R.L., Chen J. Obscurin determines the architecture of the longitudinal sarcoplasmic reticulum. J. Cell Sci. 2009;122:2640–2650. doi: 10.1242/jcs.046193. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lange, S., Ouyang, K., Meyer, G., Cui, L., Cheng, H., Lieber, R.L., and Chen, J. (2009). Obscurin determines the architecture of the longitudinal sarcoplasmic reticulum. J. Cell Sci. 122, 2640-2650. [DOI] [PMC free article] [PubMed]
- 51.Shin K., Hwang S.G., Choi I.J., Ko Y.G., Jeong J., Kwon H. Fbxw7β, E3 ubiquitin ligase, negative regulation of primary myoblast differentiation, proliferation and migration. Anim. Sci. J. 2017;88:712–719. doi: 10.1111/asj.12687. [DOI] [PubMed] [Google Scholar]; Shin, K., Hwang, S.G., Choi, I.J., Ko, Y.G., Jeong, J., and Kwon, H. (2017). Fbxw7β, E3 ubiquitin ligase, negative regulation of primary myoblast differentiation, proliferation and migration. Anim. Sci. J. 88, 712-719. [DOI] [PubMed]
- 52.Shin K., Ko Y.G., Jeong J., Kwon H. Skeletal muscle atrophy is induced by Fbxw7β via atrogene upregulation. Cell Biol. Int. 2017;41:213–220. doi: 10.1002/cbin.10713. [DOI] [PubMed] [Google Scholar]; Shin, K., Ko, Y.G., Jeong, J., and Kwon, H. (2017). Skeletal muscle atrophy is induced by Fbxw7β via atrogene upregulation. Cell Biol. Int. 41, 213-220. [DOI] [PubMed]
- 53.Cheedipudi S., Puri D., Saleh A., Gala H.P., Rumman M., Pillai M.S., Sreenivas P., Arora R., Sellathurai J., Schrøder H.D. A fine balance: Epigenetic control of cellular quiescence by the tumor suppressor PRDM2/RIZ at a bivalent domain in the cyclin a gene. Nucleic Acids Res. 2015;43:6236–6256. doi: 10.1093/nar/gkv567. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cheedipudi, S., Puri, D., Saleh, A., Gala, H.P., Rumman, M., Pillai, M.S., Sreenivas, P., Arora, R., Sellathurai, J., Schroder, H.D., et al. (2015). A fine balance: Epigenetic control of cellular quiescence by the tumor suppressor PRDM2/RIZ at a bivalent domain in the cyclin a gene. Nucleic Acids Res. 43, 6236-6256. [DOI] [PMC free article] [PubMed]
- 54.Hodgson B., Mafi R., Mafi P., Khan W. The regulation of differentiation of mesenchymal stem-cells into skeletal muscle: A look at signalling molecules involved in myogenesis. Curr. Stem Cell Res. Ther. 2018;13:384–407. doi: 10.2174/1574888X11666170907113151. [DOI] [PubMed] [Google Scholar]; Hodgson, B., Mafi, R., Mafi, P., and Khan, W. (2018). The regulation of differentiation of mesenchymal stem-cells into skeletal muscle: A look at signalling molecules involved in myogenesis. Curr. Stem Cell Res. Ther. 13, 384-407. [DOI] [PubMed]
- 55.Carvalho C.M., Lupski J.R. Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 2016;17:224–238. doi: 10.1038/nrg.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]; Carvalho, C.M., and Lupski, J.R. (2016). Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 17, 224-238. [DOI] [PMC free article] [PubMed]
- 56.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A., Braxton A., Beuten J., Xia F., Niu Z. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yang, Y., Muzny, D.M., Reid, J.G., Bainbridge, M.N., Willis, A., Ward, P.A., Braxton, A., Beuten, J., Xia, F., Niu, Z., et al. (2013). Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 369, 1502-1511. [DOI] [PMC free article] [PubMed]
- 57.Böhm J., Vasli N., Malfatti E., Le Gras S., Feger C., Jost B., Monnier N., Brocard J., Karasoy H., Gérard M. An integrated diagnosis strategy for congenital myopathies. PLoS ONE. 2013;8:e67527. doi: 10.1371/journal.pone.0067527. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bohm, J., Vasli, N., Malfatti, E., Le Gras, S., Feger, C., Jost, B., Monnier, N., Brocard, J., Karasoy, H., Gerard, M., et al. (2013). An integrated diagnosis strategy for congenital myopathies. PLoS ONE 8, e67527. [DOI] [PMC free article] [PubMed]
- 58.Casey W.M., Brodie T., Yoon L., Ni H., Jordan H.L., Cariello N.F. Correlation analysis of gene expression and clinical chemistry to identify biomarkers of skeletal myopathy in mice treated with PPAR agonist GW610742X. Biomarkers. 2008;13:364–376. doi: 10.1080/13547500801903545. [DOI] [PubMed] [Google Scholar]; Casey, W.M., Brodie, T., Yoon, L., Ni, H., Jordan, H.L., and Cariello, N.F. (2008). Correlation analysis of gene expression and clinical chemistry to identify biomarkers of skeletal myopathy in mice treated with PPAR agonist GW610742X. Biomarkers 13, 364-376. [DOI] [PubMed]
- 59.Bower N.I., Johnston I.A. Discovery and characterization of nutritionally regulated genes associated with muscle growth in Atlantic salmon. Physiol. Genomics. 2010;42A:114–130. doi: 10.1152/physiolgenomics.00065.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bower, N.I., and Johnston, I.A. (2010). Discovery and characterization of nutritionally regulated genes associated with muscle growth in Atlantic salmon. Physiol. Genomics 42A, 114-130. [DOI] [PMC free article] [PubMed]
- 60.Yamamoto S., Jaiswal M., Charng W.L., Gambin T., Karaca E., Mirzaa G., Wiszniewski W., Sandoval H., Haelterman N.A., Xiong B. A drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell. 2014;159:200–214. doi: 10.1016/j.cell.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yamamoto, S., Jaiswal, M., Charng, W.L., Gambin, T., Karaca, E., Mirzaa, G., Wiszniewski, W., Sandoval, H., Haelterman, N.A., Xiong, B., et al. (2014). A drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell 159, 200-214. [DOI] [PMC free article] [PubMed]
- 61.Gee H.Y., Sadowski C.E., Aggarwal P.K., Porath J.D., Yakulov T.A., Schueler M., Lovric S., Ashraf S., Braun D.A., Halbritter J. FAT1 mutations cause a glomerulotubular nephropathy. Nat. Commun. 2016;7:10822. doi: 10.1038/ncomms10822. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gee, H.Y., Sadowski, C.E., Aggarwal, P.K., Porath, J.D., Yakulov, T.A., Schueler, M., Lovric, S., Ashraf, S., Braun, D.A., Halbritter, J., et al. (2016). FAT1 mutations cause a glomerulotubular nephropathy. Nat. Commun. 7, 10822. [DOI] [PMC free article] [PubMed]
- 62.Caruso N., Herberth B., Bartoli M., Puppo F., Dumonceaux J., Zimmermann A., Denadai S., Lebossé M., Roche S., Geng L. Deregulation of the protocadherin gene FAT1 alters muscle shapes: Implications for the pathogenesis of facioscapulohumeral dystrophy. PLoS Genet. 2013;9:e1003550. doi: 10.1371/journal.pgen.1003550. [DOI] [PMC free article] [PubMed] [Google Scholar]; Caruso, N., Herberth, B., Bartoli, M., Puppo, F., Dumonceaux, J., Zimmermann, A., Denadai, S., Lebosse, M., Roche, S., Geng, L., et al. (2013). Deregulation of the protocadherin gene FAT1 alters muscle shapes: Implications for the pathogenesis of facioscapulohumeral dystrophy. PLoS Genet. 9, e1003550. [DOI] [PMC free article] [PubMed]
- 63.Mariot V., Roche S., Hourdé C., Portilho D., Sacconi S., Puppo F., Duguez S., Rameau P., Caruso N., Delezoide A.L. Correlation between low FAT1 expression and early affected muscle in facioscapulohumeral muscular dystrophy. Ann. Neurol. 2015;78:387–400. doi: 10.1002/ana.24446. [DOI] [PubMed] [Google Scholar]; Mariot, V., Roche, S., Hourde, C., Portilho, D., Sacconi, S., Puppo, F., Duguez, S., Rameau, P., Caruso, N., Delezoide, A.L., et al. (2015). Correlation between low FAT1 expression and early affected muscle in facioscapulohumeral muscular dystrophy. Ann. Neurol. 78, 387-400. [DOI] [PubMed]
- 64.Puppo F., Dionnet E., Gaillard M.C., Gaildrat P., Castro C., Vovan C., Bertaux K., Bernard R., Attarian S., Goto K. Identification of variants in the 4q35 gene FAT1 in patients with a facioscapulohumeral dystrophy-like phenotype. Hum. Mutat. 2015;36:443–453. doi: 10.1002/humu.22760. [DOI] [PubMed] [Google Scholar]; Puppo, F., Dionnet, E., Gaillard, M.C., Gaildrat, P., Castro, C., Vovan, C., Bertaux, K., Bernard, R., Attarian, S., Goto, K., et al. (2015). Identification of variants in the 4q35 gene FAT1 in patients with a facioscapulohumeral dystrophy-like phenotype. Hum. Mutat. 36, 443-453. [DOI] [PubMed]
- 65.Park H.J., Lee W., Kim S.H., Lee J.H., Shin H.Y., Kim S.M., Park K.D., Lee J.H., Choi Y.C. FAT1 gene alteration in facioscapulohumeral muscular dystrophy type 1. Yonsei Med. J. 2018;59:337–340. doi: 10.3349/ymj.2018.59.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]; Park, H.J., Lee, W., Kim, S.H., Lee, J.H., Shin, H.Y., Kim, S.M., Park, K.D., Lee, J.H., and Choi, Y.C. (2018). FAT1 gene alteration in facioscapulohumeral muscular dystrophy type 1. Yonsei Med. J. 59, 337-340. [DOI] [PMC free article] [PubMed]
- 66.Moeller M.J., Soofi A., Braun G.S., Li X., Watzl C., Kriz W., Holzman L.B. Protocadherin FAT1 binds Ena/VASP proteins and is necessary for actin dynamics and cell polarization. EMBO J. 2004;23:3769–3779. doi: 10.1038/sj.emboj.7600380. [DOI] [PMC free article] [PubMed] [Google Scholar]; Moeller, M.J., Soofi, A., Braun, G.S., Li, X., Watzl, C., Kriz, W., and Holzman, L.B. (2004). Protocadherin FAT1 binds Ena/VASP proteins and is necessary for actin dynamics and cell polarization. EMBO J. 23, 3769-3779. [DOI] [PMC free article] [PubMed]
- 67.Cho E., Feng Y., Rauskolb C., Maitra S., Fehon R., Irvine K.D. Delineation of a Fat tumor suppressor pathway. Nat. Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]; Cho, E., Feng, Y., Rauskolb, C., Maitra, S., Fehon, R., and Irvine, K.D. (2006). Delineation of a Fat tumor suppressor pathway. Nat. Genet. 38, 1142-1150. [DOI] [PubMed]
- 68.Hou R., Sibinga N.E. Atrophin proteins interact with the Fat1 cadherin and regulate migration and orientation in vascular smooth muscle cells. J. Biol. Chem. 2009;284:6955–6965. doi: 10.1074/jbc.M809333200. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hou, R., and Sibinga, N.E. (2009). Atrophin proteins interact with the Fat1 cadherin and regulate migration and orientation in vascular smooth muscle cells. J. Biol. Chem. 284, 6955-6965. [DOI] [PMC free article] [PubMed]
- 69.Skouloudaki K., Puetz M., Simons M., Courbard J.R., Boehlke C., Hartleben B., Engel C., Moeller M.J., Englert C., Bollig F. Scribble participates in Hippo signaling and is required for normal zebrafish pronephros development. Proc. Natl. Acad. Sci. USA. 2009;106:8579–8584. doi: 10.1073/pnas.0811691106. [DOI] [PMC free article] [PubMed] [Google Scholar]; Skouloudaki, K., Puetz, M., Simons, M., Courbard, J.R., Boehlke, C., Hartleben, B., Engel, C., Moeller, M.J., Englert, C., Bollig, F., et al. (2009). Scribble participates in Hippo signaling and is required for normal zebrafish pronephros development. Proc. Natl. Acad. Sci. USA 106, 8579-8584. [DOI] [PMC free article] [PubMed]
- 70.Szpiech Z.A., Mak A.C., White M.J., Hu D., Eng C., Burchard E.G., Hernandez R.D. Ancestry-dependent enrichment of deleterious homozygotes in runs of homozygosity. bioRxiv. 2018;382721 doi: 10.1016/j.ajhg.2019.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; Szpiech, Z.A., Mak, A.C., White, M.J., Hu, D., Eng, C., Burchard, E.G., and Hernandez, R.D. (2018). Ancestry-dependent enrichment of deleterious homozygotes in runs of homozygosity. bioRxiv 382721. 10.1101/382721. [DOI] [PMC free article] [PubMed]
- 71.Lupski J.R., Belmont J.W., Boerwinkle E., Gibbs R.A. Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lupski, J.R., Belmont, J.W., Boerwinkle, E., and Gibbs, R.A. (2011). Clan genomics and the complex architecture of human disease. Cell 147, 32-43. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.