Abstract
Neuronal intranuclear inclusion disease (NIID) is a slowly progressing neurodegenerative disease characterized by eosinophilic intranuclear inclusions in the nervous system and multiple visceral organs. The clinical manifestation of NIID varies widely, and both familial and sporadic cases have been reported. Here we have performed genetic linkage analysis and mapped the disease locus to 1p13.3-q23.1; however, whole-exome sequencing revealed no potential disease-causing mutations. We then performed long-read genome sequencing and identified a large GGC repeat expansion within human-specific NOTCH2NLC. Expanded GGC repeats as the cause of NIID was further confirmed in an additional three NIID-affected families as well as five sporadic NIID-affected case subjects. Moreover, given the clinical heterogeneity of NIID, we examined the size of the GGC repeat among 456 families with a variety of neurological conditions with the known pathogenic genes excluded. Surprisingly, GGC repeat expansion was observed in two Alzheimer disease (AD)-affected families and three parkinsonism-affected families, implicating that the GGC repeat expansions in NOTCH2NLC could also contribute to the pathogenesis of both AD and PD. Therefore, we suggest defining a term NIID-related disorders (NIIDRD), which will include NIID and other related neurodegenerative diseases caused by the expanded GGC repeat within human-specific NOTCH2NLC.
Keywords: neuronal intranuclear inclusion disease, whole-exome sequencing, linkage analysis, long-read genome sequencing, NOTCH2NLC, GGC repeat expansions
Introduction
Neuronal intranuclear inclusion disease (NIID [MIM: 603472]) is a rare multisystem neurodegenerative disease characterized by the pathology of eosinophilic intranuclear inclusions in the central, peripheral, and autonomic nervous systems, as well as in the visceral organs.1, 2 The first NIID-affected case subject was reported in 1968.3 However, until 2011, only about 40 NIID-affected case subjects had been described worldwide, which were diagnosed by post-mortem brain biopsy.1, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13 Because eosinophilic intranuclear inclusions also exist in dermal cells of NIID-affected individuals,14, 15 skin biopsy has become a useful tool to confirm NIID diagnosis, and the number of reported cases has increased to more than 100 case subjects.2
Both familial and sporadic case subjects have been described.2 The onset age of NIID varies widely and can be divided into three subgroups according to the age of onset: infant form, juvenile form, and adult form.1 The clinical manifestation of adult-onset NIID can vary widely, including dementia,9, 16 peripheral neuropathy,5, 17 autonomic dysfunction,17, 18 cerebellar ataxia,7, 8 parkinsonism,4, 19, 20 seizure,21 stroke-like episodes,22 disturbance of consciousness,21 and encephalitic episodes.4 Dementia is the most prominent symptom in sporadic NIID-affected case subjects. In familial NIID-affected case subjects, based on the initial and main symptoms, a subgrouping of adult-onset NIID-affected case subjects was suggested based on dementia-dominant and limb weakness-dominant phenotypes.2 Considering the wide range in distribution of intranuclear inclusions in the nervous system and other organs, it is possible that there are additional phenotypes of NIID. The typical symmetrical high signal seen in corticomedullary junctions using diffusion weighted imaging (DWI) could be a powerful tool for screening NIID-affected case subjects.2, 15, 23, 24 In addition, almost all dementia-dominant NIID-affected case subjects and about 40% of limb weakness-dominant NIID-affected case subjects have been seen to have remarkable leukoencephalopathy on fluid-attenuated inversion recovery (FLAIR) images and T2-weighted (T2) images, suggesting that severe white matter hyperintensity may also be an indicator for NIID.2, 15
The pathogenesis of NIID remains unknown. Immunohistochemically, the intranuclear inclusions are positive for ubiquitin and ubiquitin related proteins, including p62, SUMO1, FUS, MYO6, and OPTN-C proteins,5, 12, 13, 25, 26 indicating that the ubiquitin-proteasome system in the nucleus may play a role in NIID. In addition, some intranuclear inclusions were found to stain positive for an anti-polyglutamine antibody 1C24, 13 and also with an anti-ataxin3 antibody.11, 27 However, this could be the result of a cross-reaction. No CAG repeat expansions have been observed in NIID.27
Here we report the identification of an expanded GGC repeat in NOTCH2NLC (MIM: 618025) via long-read genome sequencing (LRS) in a five-generation Chinese Han NIID family, and the repeat expansions were validated by repeat-primed PCR (RP-PCR) and GC-rich PCR (GC-PCR) in three additional NIID-affected families as well as five sporadic NIID-affected case subjects. In addition, we found two Alzheimer disease (AD)-affected families (1.43%) and three parkinsonism-affected families (1.46%), with GGC expansions in NOTCH2NLC from a cohort that included 140 AD-affected families, 205 parkinsonism-affected families, 51 spinocerebellar ataxia (SCA)-affected families, 16 peripheral neuropathy-affected families, and 44 motor neuron disease (MND)-affected families. Further, skin biopsies confirmed the typical NIID pathology in these two AD-affected families and three parkinsonism-affected families, indicating that GGC repeat expansions could also contribute to AD and PD phenotypes. Thus, we suggest defining a term of NIID-related disorders (NIIDRD), which is a spectrum of diseases including NIID and other diseases caused by the GGC repeat expansions of NOTCH2NLC. The prevalence of NIIDRD is not rare, and screening for expanded GGC repeat within NOTCH2NLC should be done not only in dementia-affected and peripheral neuropathy-affected individuals, but also in parkinsonism-affected individuals.
Material and Methods
Study Participants
Twenty affected individuals from four NIID-affected families, five sporadic NIID-affected case subjects, and 211 healthy control subjects from mainland China were recruited for this study. All NIID-affected individuals were recruited from the National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, and subjected to a thorough neurological examination by at least two experienced neurologists. We defined familial NIID-affected case subjects as having at least one NIID-affected case subject diagnosed by skin biopsy, and the additional family member(s) had to have either high signal intensity in the corticomedullary junction using DWI or a clinical examination by a neurologist that confirmed the concordance of phenotype with the pathologically diagnosed NIID individual from the same family. All sporadic NIID-affected case subjects presented with typical high intensity in corticomedullary junction in DWI and received a skin biopsy to confirm the diagnosis. All healthy control subjects were recruited from the medical examination center of Xiangya hospital.
Besides NIID cohorts, a cohort of individuals with other neurodegenerative disorders were also recruited, including 140 AD-affected families, 205 parkinsonism-affected families, 51 SCA-affected families, 16 peripheral neuropathy-affected families, and 44 MND-affected families, of which the known pathologic genes had been excluded. All individuals were recruited from the Department of Neurology and National Clinical Research Center for Geriatric Disorders, Xiangya Hospital. All participants were subjected to a thorough neurological examination by at least two experienced neurologists.
This study was approved by the Ethics Committee of Xiangya Hospital of the Central South University in China. Written informed consent was obtained from all individuals.
Skin Biopsy, Immunohistochemistry, and Electron Microscopy
Skin biopsies were performed after local anesthesia. A 5-mm-diameter biopsy specimen was obtained at 10 cm above the lateral malleolus of the affected individual. For immunohistochemistry, all samples were fixed in 10% formalin and then were embedded in paraffin and sectioned at 6 mm thickness. Sections of all samples were stained by hematoxylin & eosin (H&E) and immunohistochemical analysis was performed. Anti-ubiquitin (3936; Cell Signaling) and anti-p62 (610833; BD Biosciences) antibodies were used for ubiquitin and p62 staining. Images were acquired by deconvolution digital microscope (Axioplan 2; Carl Zeiss).
Samples for electron microscopy were sliced into 1×1×3 mm3 size and fixed in 2.5% glutaraldehyde solution with Millonig’s phosphate buffer (pH 7.3). In the following preparations, the samples were incubated in 1% osmium tetroxide, then dehydrated with graded acetone, and embedded with resin. 50–100 nm ultrathin sections were prepared with an ultramicrotome (Leica Microsystems) and a diamond knife. After 3% uranyl acetate and lead nitrate double staining, prepared sections were examined and photographed on a Hitachi HT-7700 electron microscope.
Linkage Analysis and Whole-Exome Sequencing
Whole-genome SNP scanning on selected members in family 1 (Figure 1A) was performed using Illumina Asian Screening Array with more than 7 million polymorphic loci. Information on SNPs was extracted by the genotyping module of Genomestudio (v.2011.1). Illumina cnvPartition CNV Analysis Plug-in of Genomestudio (v.2011.1) was used for copy number variations (CNVs) analysis. Linkage analysis was performed by the parametric linkage analysis package of MERLIN (v.1.1.2). Whole-exome sequencing (WES) and analysis for the three affected individuals (F1-IV:1, F1-IV:6, and F1-IV:10) and one unaffected individual (F1-IV:3) were used to detect potential causal single nucleotide variants (SNVs) and InDels (insertions and deletions) in the mapping region. Sequencing data were subjected to quality control (QC). This was followed by Burrows-Wheeler Alignment Maximal Exact Matches (BWA-MEM) for read mapping, Samtools for SNV/InDel calling, ANNOVAR for variant annotation, and all potential disease variants were examined in Integrative Genomics Viewer.
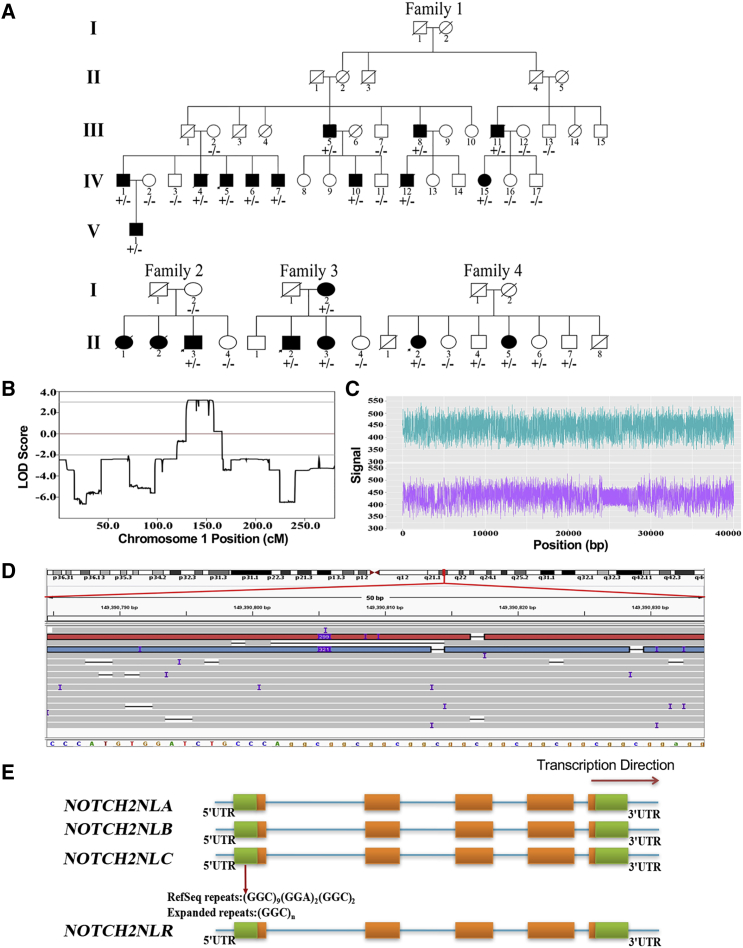
Figure 1.
Identification of Expanded GGC Repeat within NOTCH2NLC in Neuronal Intranuclear Inclusion Disease
(A) Pedigrees of neuronal intranuclear inclusion disease (NIID)-affected families and corresponding individual genotypes.
(B) Genetic linkage analysis indicated maximum logarithm of odds (LOD) scores 3.184 in chromosome 1, a 49.8-Mb region at 1p13.3-q23.1 (chr1:109260034-159016186).
(C and D) GGC expansions detected by LRS. Nanopore electric signal (C) from subject F1-IV:15 indicated GGC expansions in the lower lane compared to the normal allele in the upper lane. More than ten reads covering the causative region were seen in the Integrative Genomics Viewer for subject F1-IV:15. Two reads were determined to carry the “insertion” variation (chr1:149390803-149390842, hg38 version), corresponding to the GGC triplet expansion in NOTCH2NLC (D).
(E) Schematic representation of the causal variant in NOTCH2NLC: a certain number of GGC triplets exist in 5′ UTR of NOTCH2NLC in healthy individuals, and large expanded GGC triplets are present in affected individuals.
Long-Read Genome Sequencing
Long-read genome sequencing (LRS) on seven affected individuals (F1-IV:7, F1-IV:15, F2-II:3, F4-II:2, F5-II:1, F5-II:4, F9-II:6) and three healthy control subjects was performed on the Oxford Nanopore platform as describe previously.28 Combined analysis strategy of RepeatHMM and inScan were used for detection in the target region. Minimap2 and nanopolish calls were used for analysis of DNA methylation.
PCR Assays and GGC Repeat Size Determination
For the repeat-primed PCR (RP-PCR) assay, a fluorescein (FAM)-labeled gene-specific primer (5′-CCTCAGCCCGATACTCACCAT-3′) and repeat-containing primers (5′-TACCAATACGCATCCCGCGATTTGTCTTA(CGG)5-3′) were utilized for identifying the CGG repeat expansion. For the GC-rich PCR (GC-PCR) assay, the fluorescein (FAM)-labeled forward primer (5′-AGCGCCAGGGCCTGAGCCTTTGAAGCAG-3′) and reverse primer (5′-TCGCCCCAGGTGGCAGCCCCGGGCGCCGCGGAC-3′) were utilized for repeat size determination. Seven-deaza-2-deoxy guanosine triphosphate (deaza-dGTP) was used in place of dGTP. PCRs were performed using 50 ng of genomic DNA in a 25 μL reaction mix including 10X Expand Long Template Buffer 1, 2.5 mM MgCl2 (Thermo Scientific, Cat#F-510Mg), 2% 2,4-dimethylsulfolane (Sigma, Cat#1003-78-7), 0.2 mM each of deaza-dGTP, dATP, dCTP, and dTTP, 0.2 μM primers (BioSune, Shanghai, China) and 1 U of DNA ploymerase from Expand Long Template PCR System (Roche, Cat#11681842001), using the following thermal conditions: 98°C for 4 min, followed by 30 cycles of 98°C for 45 s, 60°C for 45 s, 72°C for 4 min, and a final extension at 72°C for 7 min. The PCR products were subjected to capillary electrophoresis using the 3500xL Genetic Analyzer for Human Identification (Applied Biosystems). Allele sizes were determined using GeneScan 1000 ROX Size Standard (Applied Biosystems). GGC repeats less than 200 were determined directly by the size of PCR product as determined by ABI 3500xL and compared with the PCR products of known FMR1 (MIM: 309550) premutation CGG repeat alleles of various sizes. For GGC repeats over 200, the size was determined using an Agilent 2100.
RNA Extraction and Quantitative RT-PCR
Human postmortem dorsolateral prefrontal cortex (DLPFC) tissue from the subjects of different ages (4, 15, 36, and 60 years old) were obtained from the NIH NeuroBioBank. Total RNA was isolated using Trizol (ThermoFisher). Before reverse transcription, total RNA was treated with ezDNase (Millipore) for 5 min at 37°C to remove contaminated genomic DNA, and then incubated at 55°C for 5 min to inactivate the ezDNase. After ezDNase treatment, total RNA was extracted with phenol: chloroform = 25:24 (pH 5.2) and precipitated with glycogen and ethanol. The reverse transcription was conducted using SuperScript (II) (ThermoFisher, Cat# Cat. No. 18064-022) to generate first strand cDNA. The first strand cDNA served as the template for real-time PCR using FAST-7500 real time PCR equipment (Thermo Fisher). GAPDH TaqMan Gene Expression Assay (Hs02786624_g1) was used as an internal control. NOTCH2NLB primers: Forward (5′-GGGAGATATGAAGGGACGCA-3′); Reverse (5′-GGCACACACCTTCCATTCTC-3′). NOTCH2NLC primers: Forward (5′-CTGACCTTTCAAGATCCTGCTTTCATCCCAGCT-3′); Reverse (5′-AAGTGCCTTACTTTGCGTAGCTGTGTGCTTGGCAGT-3′).
Statistical Analyses
For methylation analysis, the dispersion parameters were estimated through a shrinkage estimator based on a Bayesian hierarchical model, then a Wald test was performed as describe previously29 to identify differentially methylated loci (DML). For the expression data of NOTCH2NLC in the blood of both NIID-affected individuals and control subjects, statistical analysis was performed using Student’s t test. Differences with p < 0.05 were considered as being statistically significant.
Results
Identification of an Expanded GGC Repeat within Human-Specific NOTCH2NLC in a NIID-Affected Family
We recruited a five-generation Chinese NIID-affected family (family 1) with muscle weakness-dominant type (Figure 1A) and performed genetic linkage analysis. Our analysis identified a single peak with a maximum logarithm of odds (LOD) score of 3.184 at 1p13.3-q23.1 (chr1:109260034–159016186) (Figure 1B). To determine the genetic etiology of NIID in this family, we performed whole-exome sequencing (WES) in three affected individuals (F1-IV:1, F1-IV:6, and F1-IV:10) and one unaffected individual (F1-IV:3) (Figure 1A), and we were unable to identify any nonsynonymous or potentially causal variants. We went on to perform low-coverage long-read genome sequencing (LRS) on two other affected individuals (F1-IV:7, F1-IV:15) using the Oxford Nanopore platform. Intriguingly, we observed that one allele of NOTCH2NLC had an expanded GGC repeat in both affected individuals (Figures 1C and S1), which WES failed to detect.
The expanded GGC repeat is located at the 5′ end of NOTCH2NLC. NOTCH2NLC is one of the four NOTCH2 paralogs that are present only in the human genome; the other paralogs are NOTCH2NLA (MIM: 618023), NOTCH2NLB (MIM: 618024), and NOTCH2NLR (MIM: 618026). Previous studies have shown that all four NOTCH2NL paralogs contain the first four exons and introns of NOTCH2 as the result of a partial segmental duplication of the NOTCH2 ancestral gene.30 NOTCH2NLA, B, and C are highly similar to each other, with more than 98% sequence identity over NOTCH2NL exons 1–5.30 However, only NOTCH2NLC contains the GGC repeat in the reference human genome (hg38), (GGC)9(GGA)2(GGC)2, which is expanded in the NIID-affected family that we have examined here (Figures 1D and 1E).
Expanded GGC Repeat Is Associated with NIID
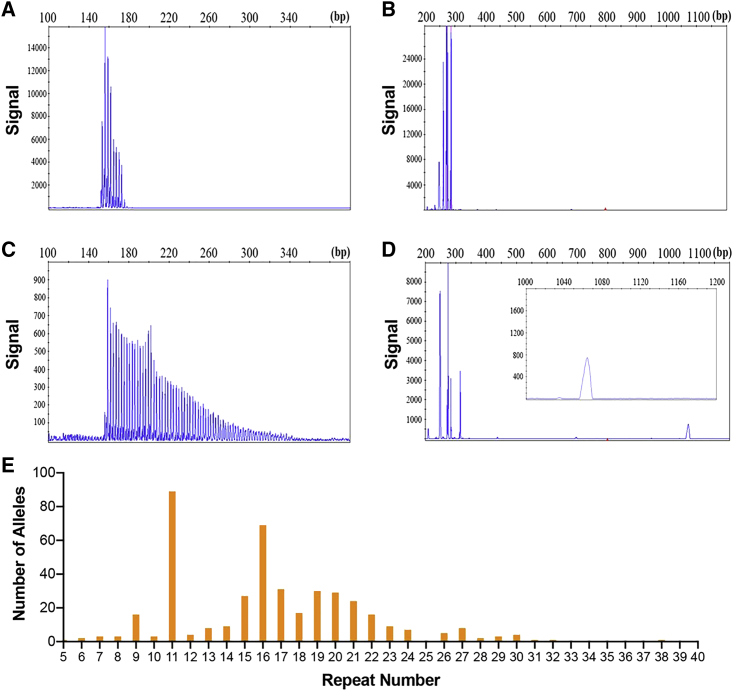
To confirm our Nanopore sequencing results, we designed primer sets for both repeat-primed PCR (RP-PCR) and GC-rich PCR (GC-PCR) assays to detect both normal and expanded GGC repeats. Among the 211 healthy control subjects that we examined, the GGC repeat sizes range from 5 to 38, with modes at 11 and 16 repeats (Figure 2E). Our RP-PCR analysis confirmed the expanded GGC repeat among the affected family members in family 1 (Figure 1A). All affected individuals in this family showed similar peak patterns using RP-PCR, indicating the presence of expanded GGC repeats (Figure 2C), while unaffected members of the pedigree carried only normal repeats (Figure 2A). We then determined the repeat size using GC-PCR and found that all the affected individuals carry alleles with greater than 100 repeats (Figure 2D). To further confirm the role of GGC repeat expansion in NIID, we examined three additional NIID-affected families (families 2–4) (Figure 1A) and five sporadic NIID-affected case subjects by RP-PCR and GC-PCR assays, some of which were further confirmed by LRS (Figure S1). All affected individuals from both familial and sporadic case subjects carried an expanded GGC repeat larger than 66. Together, these results suggest that GGC repeat expansion is indeed associated with NIID.
Figure 2.
Validation of Expanded GGC Repeats and Variations of GGC Repeat Size among Normal Individuals
(A–D) Representative electropherogram of the RP-PCR assay and the GC-PCR assay showed abnormal repeat expansion in affected individuals (C and D) and negative result in control subjects (A and B).
(E) Size distribution of GGC repeat among healthy control subjects, which are usually less than 40 repeats.
However, we did note that the expanded GGC repeat was also observed in a few individuals that were initially thought to be phenotypically normal, such as F1-III:5 and F1-III:8 (Figures S2A and S2B). Upon re-examination of subject F1-III:8, we found that this individual has subclinical peripheral neuropathy and typical eosinophilic intranuclear inclusions (Figures S2C and S2D). Subject F1-III:5 did not complain of limb weakness and refused to receive further examination but presented with tremor and disturbance of consciousness, which are the classic symptoms of NIID. Nevertheless, this indicates the possibility of varying severity for expanded GGC repeat-caused NIID.
Expanded GGC Repeat Is Associated with Dementia and Parkinsonism
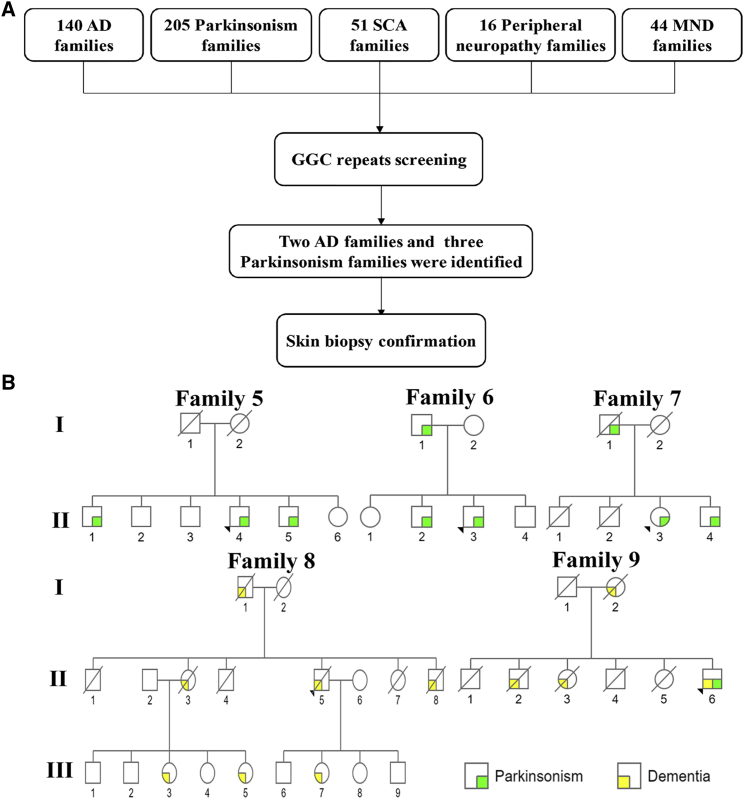
Because of the wide range of clinical manifestations associated with NIID, we further determined the size of the GGC repeat in a cohort of 140 families with AD, 205 families with parkinsonism, 51 families with SCA, 16 families with peripheral neuropathy, and 44 families with MND (Figure 3A). Surprisingly, we found affected individuals from three parkinsonism-affected families (families 5–7) and two AD-affected families (families 8 and 9) that carry the expanded GGC repeat in NOTCH2NLC (Figure 3B, Tables 1, S2, and S3). LRS was also performed on three affected individuals (F5-II:1, F5-II:4, F9-II:6), confirming the presence of expanded GGC repeat (Figure S1). Upon reexamination, we confirmed that these individuals display classic PD and AD symptoms, respectively. Intriguingly, their skin biopsies revealed typical eosinophilic, p62-positive, and ubiquitin-positive intranuclear inclusions, the pathological feature associated with classic NIID-affected individuals (Figures S3A–S3H and S3M–S3T). These findings suggest that the expanded GGC repeat could also be associated with dementia and parkinsonism.
Figure 3.
Expanded GGC Repeats Are Associated with Dementia and Parkinsonism
(A) Flow chart of GGC repeat expansion screening in a cohort of families with neurodegenerative disorders.
(B) Pedigrees of AD-affected and parkinsonism-affected families that carry expanded GGC repeats in NOTCH2NLC.
Table 1.
Summary of Clinical Features of NIID
| Sporadic Case Subjects (n = 5) |
Familial Case Subjects |
||||
|---|---|---|---|---|---|
| Total (n = 40) | Muscle Weakness (n = 15) | Parkinsonism (n = 9) | Dementia (n = 16) | ||
| Sex ratio (male/female) | 2/3 | 26/14 | 12/3 | 8/1 | 6/10 |
| Average onset age (range) | 62.0 (51–69) | 50.6 (30–78) | 35.6 (30–54) | 60.6 (37–78) | 58.1 (31–71) |
| Average disease duration (range) | 5.6 (1–14) | 12.5 (1–49) | 16.6 (3–49) | 6.0 (1–15) | 12.4 (2–30) |
| Clinical Manifestations | |||||
| Dementia | 2/5 (40.0%) | 14/40 (35.0%) | 0/15 (0%) | 0/9 (0%) | 14/16 (87.5%) |
| Abnormal behavior | 2/5 (40.0%) | 15/40 (37.5%) | 2/15 (13.3%) | 0/9 (0%) | 13/16 (81.3%) |
| Peripheral neuropathy | |||||
| Muscle weakness | 0/5 (0%) | 18/39 (46.2%) | 13/15 (86.7%) | 1/9 (11.1%) | 4/15 (26.7%) |
| Sensory disturbance | 0/5 (0%) | 13/37 (35.1%) | 4/13 (30.8%) | 4/9 (44.4%) | 5/15 (33.3%) |
| Autonomic dysfunction | |||||
| Bladder dysfunction | 3/5 (60.0%) | 22/39 (56.4%) | 5/15 (33.3%) | 5/9 (55.6%) | 12/15 (80.0%) |
| Miosis | 2/5 (40.0%) | 5/29 (17.2%) | 1/11 (9.1%) | 0/8 (0%) | 4/10 (40.0%) |
| Parkinsonism | |||||
| Tremor | 1/5 (20.0%) | 19/40 (47.5%) | 11/15 (73.3%) | 5/9 (55.6%) | 3/16 (18.8%) |
| Rigidity | 1/5 (20.0%) | 12/40 (30.0%) | 1/15 (6.7%) | 9/9 (100.0%) | 2/16 (12.5%) |
| Bradykinesia | 1/5 (20.0%) | 12/40 (30.0%) | 1/15 (6.7%) | 9/9 (100.0%) | 2/16 (12.5%) |
| Ataxia | 0/5 (0%) | 7/40 (17.5%) | 2/15 (13.3%) | 4/9 (44.4%) | 1/16 (6.3%) |
| Neurological attack | |||||
| Disturbance of consciousness | 4/5 (80.0%) | 7/40 (17.5%) | 3/15 (20.0%) | 1/9 (11.1%) | 3/16 (18.8%) |
| Stroke-like episode | 4/5 (80.0%) | 4/40 (10.0%) | 1/15 (6.7%) | 2/9 (22.2%) | 1/16 (6.3%) |
| Encephalitic episode | 3/5 (60.0%) | 2/40 (5.0%) | 1/15 (6.7%) | 0/9 (0%) | 1/16 (6.3%) |
| Brain MRI | |||||
| Severe leukoencephalopathy | 5/5 (100.0%) | 7/20 (35.0%) | 1/7 (14.3%) | 2/6 (33.3%) | 4/7 (57.1%) |
| DWI U-fiber high signal | 5/5 (100.0%) | 6/16 (37.5%) | 1/3 (33.3%) | 1/6 (16.7%) | 4/7 (57.1%) |
| Cognitive function test | |||||
| MMSE (<education matched average) | 0/2 (0%) | 4/21 (19.0%) | 0/7 (0%) | 0/7 (0%) | 4/7 (57.1%) |
| MoCA (<education matched average) | 1/2 (50.0%) | 9/16 (56.3%) | 3/7 (42.9%) | 4/5 (80.0%) | 2/4 (50.0%) |
| Nerve conduction | |||||
| MCV slowing | 3/3 (100.0%) | 19/22 (86.4%) | 8/9 (88.9%) | 6/6 (100%) | 5/7 (71.4%) |
| CMAP reduction | 1/3 (33.3%) | 14/22 (63.6%) | 7/9 (77.8%) | 2/6 (33.3%) | 5/7 (71.4%) |
| SCV slowing | 3/3 (100.0%) | 15/22 (68.2%) | 7/9 (77.8%) | 3/6 (50.0%) | 5/7 (71.4%) |
| SNAP reduction | 1/3 (33.3%) | 13/22 (59.1%) | 6/9 (66.7%) | 2/6 (33.3%) | 5/7 (71.4%) |
| Skin biopsy | 5 | 14 | 3 | 5 | 6 |
| Average no. of GGC repeats (range) | 105 (86–133) | 188 (66–517) | 272 (118–517) | 83 (66–102) | 129 (91–268) |
Expanded GGC Repeat and Phenotypic Variability
A total of 40 affected familial individuals with expanded GGC repeats, ranging from 66 to 517 repeats, were analyzed in this study. Only about 37.5% of affected familial individuals presented with the classical NIID radiological findings of symmetrical hyperintense linear lesions in the corticomedullary junction in DWI (Figure S4A) and severe white matter hyperintensity in FLAIR images/T2 weighted image (Figure S4B). However, all skin biopsies revealed eosinophilic, p62-positive, and ubiquitin-positive intranuclear inclusions in dermal cells (Figure S3). Electron microscopy imaging also revealed intranuclear inclusions lacking membranes (Figure S3H). The main clinical manifestations in these affected familial individuals included bladder dysfunction (56.4%), tremor (47.5%), muscle weakness (46.2%), abnormal behavior (37.5%), sensory disturbance (35.1%), dementia (35.0%), rigidity (30.0%), bradykinesia (30.0%), ataxia (17.5%), disturbance of consciousness (17.5%), miosis (17.2%), stroke-like episodes (10.0%), and encephalitic episodes (5.0%) (Table 1). Based on the initial and main clinical manifestations among these affected individuals with expanded repeats, we were able to divide the familial case subjects into three subgroups: muscle weakness-dominant type, parkinsonism-dominant type, and dementia-dominant type (Table 1 and Supplemental Note).
The muscle weakness-dominant type (families 1 and 2) was similar to what has been reported for classic NIID where the main and initial clinical manifestation is muscle weakness. The average onset age was around 36 (Figure 1A, Tables 1 and S1). Muscle weakness usually began in the distal part of lower limbs, then moved up to the throat muscles and face. However, the severity and position of muscle weakness could vary significantly between individuals. Tremor was frequently seen in the early stages of almost all of the affected members. The number of GGC repeats in this subgroup ranged from 118 to 517 repeats.
The parkinsonism-dominant type (families 5–7) presented with parkinsonism as the main clinical manifestation. The onset age was typically around 60 years old (Figure 3B, Tables 1 and S2). The severity of symptoms varied greatly, even between siblings with similar repeat sizes. No dementia was observed in either the muscle weakness or parkinsonism groups. A smaller repeat expansion, 66–102 repeats, was observed in this group.
In the dementia-dominant type (families 3, 4, 8, 9), individual characteristics were similar to the previously described dementia-dominant phenotype in NIID.2 The average onset age was 58. Dementia was the most prominent symptom (Figures 1A and 3B, Tables 1 and S3). Abnormal behavior, autonomic dysfunction, disturbance of consciousness, tremor, rigidity, limb weakness, and sensory disturbance were also seen. This subgroup repeat size ranged from 91 to 268 GGC repeats.
Methylation and Expression at NOTCH2NLC Locus
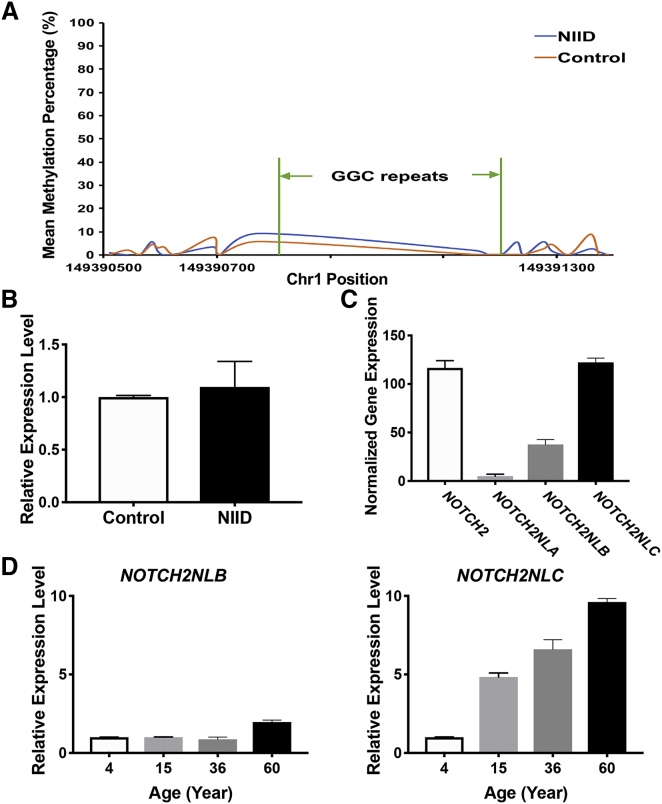
Previous works have found that more than 200 CGG repeats at the FMR1 locus can lead to hypermethylation of the CGG repeat and adjacent CpG islands, resulting in transcriptional silencing.31 Because many of the expanded repeats that we discovered at the NOTCH2NLC locus in NIID-affected case subjects were greater than 200, we examined the methylation status of GGC repeat and adjacent regions by re-analyzing the signals from Nanopore sequencing. The methylation level around the GGC repeats was very low and no significant methylation difference between NIID and control was detected (Figure 4A), which suggests that the expanded GGC repeat at the NOTCH2NLC locus is likely unmethylated. Furthermore, we determined the expression of NOTCH2NLC in the blood of both NIID-affected case subjects and control subjects, and did not detect significant changes in expression for the NIID-affected case subjects with expanded GGC repeats (Figure 4B), indicating that the expanded GGC repeat does not alter the expression of NOTCH2NLC and that the GGC repeat RNA could potentially play a role in the molecular pathogenesis of NIID.
Figure 4.
Methylation and Expression at NOTCH2NLC Locus
(A) Methylation status across expanded GGC repeats region was determined using LRS data from seven affected individuals (F1-IV:7, F1-IV:15, F2-II:3, F4-II:2, F5-II:1, F5-II:4, F9-II:6) and three healthy control subjects, and no significant methylation difference was detected between NIID-affected case subjects and control subjects. Wald test was performed for statistical analysis; ∗p < 0.05.
(B) NOTCH2NLC expression level in both NIID-affected case subjects (NIID) and normal control subjects (Control). The ezDNase-treated total RNA isolated from the blood of both NIID-affected case subjects and control subjects was reversely transcribed into cDNA followed by quantitative PCR. GAPDH was used as internal control. Error bars represent the SD; Student’s t test was performed for statistical analysis; ∗p < 0.05. p = 0.776 (ns).
(C) Expression levels of NOTCH2 and three NOTCH2NL paralogs (NOTCH2NLA, NOTCH2NLB, and NOTCH2NLC) in human adult cortex detected by RNA-seq. Significant differences were observed in the expression levels of these four genes. Shown are the normalized gene expression levels. Error bars represent the SD.
(D) Dynamic change of NOTCH2NLC expression in human brain during aging. Relative expression levels of both NOTCH2NLB (left) and NOTCH2NLC (right) in DLPFC region of human brain during aging are shown. The total RNA isolated from the DLPFC of human postmortem brains from 4-, 15-, 36-, and 60-year-old subjects were used for quantitative RT-PCR with GAPDH as internal control. Error bars represent the SD.
Previous studies have shown that NOTCH2NLA, NOTCH2NLB, and NOTCH2NLC can be transcribed in human ES cells.30, 32 We analyzed the expression of these three paralogs and NOTCH2 using the published human brain RNA-seq datasets33 and found that NOTCH2NLC displays the highest expression in the brain among these three NOTCH2 paralogs (Figure 4C). Given that NIID is a neurodegenerative disorder, we designed specific primers that could distinguish the expression of NOTCH2NLB and NOTCH2NLC by RT-PCR (NOTCH2NLA has very low expression in brain; Figure 4C) and determined the expression dynamics of both genes during human brain aging. Interestingly, while the expression of NOCTH2NLB displays only modest changes in human prefrontal cortex during aging, the expression of NOTCH2NLC increased significantly with age (Figure 4D). Thus, the increased expression of expanded GGC repeats during aging could contribute to the molecular pathogenesis of NIID.
Discussion
Neuronal intranuclear inclusion disease (NIID) is a slowly progressing neurodegenerative disease characterized by eosinophilic intranuclear inclusions in the nervous system and multiple visceral organs. The clinical manifestation of NIID varies widely, and both familial and sporadic case subjects have been reported.2 Here we report the identification of a GGC repeat expansion at the 5′ end of NOTCH2NLC as the genetic cause of NIID. NOTCH2NLC is one of the three human-specific NOTCH2-related genes (NOTCH2NLA, NOTCH2NLB, and NOTCH2NLC) in 1q21.1, highly expressed in the brain and thought to be involved in the evolutionary expansion of the human brain.30, 32 Thus, NIID represents a genetic disorder caused by an expanded repeat in a human-specific gene.
Almost 30 years ago, expansion of unstable nucleotide (microsatellite) repeats, notably trinucleotide repeats, was identified as a previously unknown mutational mechanism underlying certain human diseases.34, 35, 36, 37 In recent years the development of next-generation sequencing along with the focus on exome capture provided a massive increase in the catalog of genes with mutations in Mendelian disorders;38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50 however, in many instances the causal mutations could not be identified even when genome-wide association studies (GWASs) or genetic linkage analysis identified a specific genetic locus. Recent works, including identification of an unstable hexanucleotide repeat within C9ORF72 (MIM: 614260) as a frequent cause of frontotemporal dementia (FTD)/amyotrophic lateral sclerosis (ALS)51, 52 and CTG repeat expansion as the cause of Fuch’s endothelial corneal dystrophy (FECD),53 suggest that repeat expansions could contribute to the genetic etiology of these disorders, which could be missed by short-read sequencing. In our initial analyses, we failed to detect any causal mutations using WES despite the fact that we mapped the NIID-causing gene to 1p13.3-q23.1. Our findings presented here further highlight the importance of considering the genetic contribution of unstable repeats in disorders where traditional single nucleotide variants (SNVs) or copy number variants (CNVs) cannot be identified.
In addition to GGC repeats, we did observe different repeat units, such as GGT, through our LRS analyses (Figure S1). This is likely due to the high error rate of LRS itself and interference of sequence from paralogs. With the high error rate of Nanopore sequencing, it was challenging to acquire accurate sequence data within the expanded GGC repeat region, which would require further analyses with more accurate methodology. Despite these technical issues, most reads that we obtained were expanded GGC repeats.
The key features associated with the disorders caused by unstable nucleotide (microsatellite) repeats are genetic instability and anticipation.31 In the NIID-affected families reported in our study, a wide range (66–517) of repeat numbers were observed in affected individuals, while control subjects have fewer than 40 repeats. Anticipation was seen in a few families, while unlike fragile X-related disorders, the pioneer locus with expanded CGG/GGC repeats, increased instability of the GGC repeat size over generations was not observed in all families. Therefore, anticipation was not clearly seen in NIID-affected families, and more NIID-affected families need to be studied to explore the genetic pattern over multiple generations. Nevertheless, the number of affected individuals does seem to increase over multiple generations, suggesting a potential genetic anticipation effect. We also observed variable severity in the clinical manifestation among our affected case subjects; however, there is no apparent association between GGC repeat size and the severity or the onset age in NIID. It is possible that additional genetic or environmental factors could contribute to the pathogenesis of NIID. In addition, extensive works on the FMR1 locus have shown that there is a threshold for the methylation status of the CGG repeat.31 Typically, if the CGG repeat is below 200, it is unmethylated, but the majority of expanded CGG repeats over 200 are hypermethylated, leading to transcriptional silencing of FMR1 and fragile X syndrome.31 Thus, it has been hypothesized that loci with more than 200 CGG repeats could be recognized by DNA methyltransferase(s) and induce hypermethylation.54 Similar observations have recently been reported in Baratela-Scott syndrome (BSS), which was found due to an expanded GGC repeat in XYLT1 (MIM: 608124).55 However, as reported here, many expanded alleles associated with NIID-affected individuals exceeded 200 repeats but were not methylated. Our finding suggests that in addition to GGC/CGG repeat size, additional factor(s) or sequence(s) could also determine the methylation status of expanded GGC/CGG repeat in vivo.
Our analyses of both classic NIID and other individuals with neurological conditions revealed a broadened clinical spectrum of adult-onset NIID. Besides the suggested dementia-dominant type and muscle weakness-dominant type,2 we have identified the parkinsonism-dominant type as a clinical presentation in NIID. Among sporadic case subjects, we also identified a paroxysmal disease-dominant type (Supplemental Note, Table S4). Furthermore, GGC repeat expansion was also observed in two AD-affected families (1.43%) and three parkinsonism-affected families (1.46%), implicating that GGC repeat expansions in NOTCH2NLC could also contribute to the pathogenesis of both AD and PD. Therefore, we suggest defining a term NIID-related disorders (NIIDRD), which include NIID and other related neurodegenerative diseases caused by the expanded GGC repeat. It is likely that the prevalence of NIIDRD could be higher than initially thought. Given that only about 37% of affected individuals showed typical MRI features of NIID, genetic testing for the expanded GGC repeat could potentially be a more sensitive and accurate screening tool for NIIDRD.
Identification of the GGC repeat expansion as the genetic cause of NIID will help elucidate the molecular pathogenesis in NIID. Our results presented here suggest that the GGC repeat expansion-induced pathogenesis is likely dominant, which suggests that the expression of the expanded GGC repeat could cause the neuronal toxicity associated with NIID and its related disorders. Indeed, a dominant-negative gain-of-function model has been proposed for another neurodegenerative disorder, fragile X-associated tremor/ataxia syndrome (FXTAS [MIM: 300623]), which is also caused by the expanded GGC/CGG repeats.56 Intriguingly, it was previously discussed that NIID and FXTAS share many clinical and neuropathological features,27, 58 which could be well explained by our genetic findings presented here. Two potential mechanisms have been proposed for FXTAS: RNA toxicity and repeat-associated non-AUG (RAN) translation.59 The CGG repeat RNA could sequester key RNA binding proteins and prevent them from performing their normal physiological function.56 Also, repeat-associated non-AUG (RAN) translation of the CGG repeat into polypeptides, the predominant species being poly-Glycine (FMRpolyG peptides), could contribute to FXTAS pathogenesis.60, 61 Both FMR1 mRNA and FMRpolyG peptide have been found in the human postmortem brain inclusions that are characteristic of FXTAS pathology, suggesting that both mechanisms may contribute to the disease.60, 62 It will be important to explore both mechanisms in NIID pathogenesis in the future. In addition, it could be challenging to establish appropriate model(s) to study NIID since NOTCH2NLC is a human-specific gene.
In summary, we have identified an expanded GGC repeat within human-specific NOTCH2NLC as the genetic cause of NIIDRD, which could include dementia, parkinsonism, and peripheral neuropathy. Our finding will facilitate more accurate clinical diagnosis in the future and help us better understand the molecular pathogenesis of NIIDRD in general.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We would like to thank the affected individuals for permitting us to publish this information and NIH NeuroBioBank for providing human brain tissues. This work was funded by National Key R&D Program of China (Grant2018YFC1312003) and National Natural Science Foundation of China (Grant81430023, Grant81701263, Grant81671075, Grant81771366, and Grant81671120).
Published: June 6, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.05.013.
Contributor Information
Peng Jin, Email: peng.jin@emory.edu.
Lu Shen, Email: shenlu@csu.edu.cn.
Web Resources
Genomestudio (v.2011.1), https://support.illumina.com/array/array_software/genomestudio/downloads.html
gnomAD Browser, https://gnomad.broadinstitute.org/
MERLIN (v.1.1.2), http://csg.sph.umich.edu/abecasis/Merlin/download/
Minimap2, https://github.com/lh3/minimap2
Nanopolish call, https://github.com/jts/nanopolish
OMIM, http://www.omim.org/
RepeatHMM, https://github.com/WGLab/RepeatHMM
UCSC Genome Browser, https://genome.ucsc.edu
Supplemental Data
References
- 1.Takahashi-Fujigasaki J. Neuronal intranuclear hyaline inclusion disease. Neuropathology. 2003;23:351–359. doi: 10.1046/j.1440-1789.2003.00524.x. [DOI] [PubMed] [Google Scholar]; Takahashi-Fujigasaki, J. (2003). Neuronal intranuclear hyaline inclusion disease. Neuropathology 23, 351-359. [DOI] [PubMed]
- 2.Sone J., Mori K., Inagaki T., Katsumata R., Takagi S., Yokoi S., Araki K., Kato T., Nakamura T., Koike H. Clinicopathological features of adult-onset neuronal intranuclear inclusion disease. Brain. 2016;139:3170–3186. doi: 10.1093/brain/aww249. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sone, J., Mori, K., Inagaki, T., Katsumata, R., Takagi, S., Yokoi, S., Araki, K., Kato, T., Nakamura, T., Koike, H., et al. (2016). Clinicopathological features of adult-onset neuronal intranuclear inclusion disease. Brain 139, 3170-3186. [DOI] [PMC free article] [PubMed]
- 3.Lindenberg R., Rubinstein L.J., Herman M.M., Haydon G.B. A light and electron microscopy study of an unusual widespread nuclear inclusion body disease. A possible residuum of an old herpesvirus infection. Acta Neuropathol. 1968;10:54–73. doi: 10.1007/BF00690510. [DOI] [PubMed] [Google Scholar]; Lindenberg, R., Rubinstein, L.J., Herman, M.M., and Haydon, G.B. (1968). A light and electron microscopy study of an unusual widespread nuclear inclusion body disease. A possible residuum of an old herpesvirus infection. Acta Neuropathol. 10, 54-73. [DOI] [PubMed]
- 4.Liu Y., Mimuro M., Yoshida M., Hashizume Y., Niwa H., Miyao S., Ujihira N., Akatsu H. Inclusion-positive cell types in adult-onset intranuclear inclusion body disease: implications for clinical diagnosis. Acta Neuropathol. 2008;116:615–623. doi: 10.1007/s00401-008-0442-7. [DOI] [PubMed] [Google Scholar]; Liu, Y., Mimuro, M., Yoshida, M., Hashizume, Y., Niwa, H., Miyao, S., Ujihira, N., and Akatsu, H. (2008). Inclusion-positive cell types in adult-onset intranuclear inclusion body disease: implications for clinical diagnosis. Acta Neuropathol. 116, 615-623. [DOI] [PubMed]
- 5.Kimber T.E., Blumbergs P.C., Rice J.P., Hallpike J.F., Edis R., Thompson P.D., Suthers G. Familial neuronal intranuclear inclusion disease with ubiquitin positive inclusions. J. Neurol. Sci. 1998;160:33–40. doi: 10.1016/s0022-510x(98)00169-5. [DOI] [PubMed] [Google Scholar]; Kimber, T.E., Blumbergs, P.C., Rice, J.P., Hallpike, J.F., Edis, R., Thompson, P.D., and Suthers, G. (1998). Familial neuronal intranuclear inclusion disease with ubiquitin positive inclusions. J. Neurol. Sci. 160, 33-40. [DOI] [PubMed]
- 6.Schuffler M.D., Bird T.D., Sumi S.M., Cook A. A familial neuronal disease presenting as intestinal pseudoobstruction. Gastroenterology. 1978;75:889–898. [PubMed] [Google Scholar]; Schuffler, M.D., Bird, T.D., Sumi, S.M., and Cook, A. (1978). A familial neuronal disease presenting as intestinal pseudoobstruction. Gastroenterology 75, 889-898. [PubMed]
- 7.Michaud J., Gilbert J.J. Multiple system atrophy with neuronal intranuclear hyaline inclusions. Report of a new case with light and electron microscopic studies. Acta Neuropathol. 1981;54:113–119. doi: 10.1007/BF00689403. [DOI] [PubMed] [Google Scholar]; Michaud, J., and Gilbert, J.J. (1981). Multiple system atrophy with neuronal intranuclear hyaline inclusions. Report of a new case with light and electron microscopic studies. Acta Neuropathol. 54, 113-119. [DOI] [PubMed]
- 8.Patel H., Norman M.G., Perry T.L., Berry K.E. Multiple system atrophy with neuronal intranuclear hyaline inclusions. Report of a case and review of the literature. J. Neurol. Sci. 1985;67:57–65. doi: 10.1016/0022-510x(85)90022-x. [DOI] [PubMed] [Google Scholar]; Patel, H., Norman, M.G., Perry, T.L., and Berry, K.E. (1985). Multiple system atrophy with neuronal intranuclear hyaline inclusions. Report of a case and review of the literature. J. Neurol. Sci. 67, 57-65. [DOI] [PubMed]
- 9.Munoz-Garcia D., Ludwin S.K. Adult-onset neuronal intranuclear hyaline inclusion disease. Neurology. 1986;36:785–790. doi: 10.1212/wnl.36.6.785. [DOI] [PubMed] [Google Scholar]; Munoz-Garcia, D., and Ludwin, S.K. (1986). Adult-onset neuronal intranuclear hyaline inclusion disease. Neurology 36, 785-790. [DOI] [PubMed]
- 10.Weidenheim K.M., Dickson D.W. Intranuclear inclusion bodies in an elderly demented woman: a form of intranuclear inclusion body disease. Clin. Neuropathol. 1995;14:93–99. [PubMed] [Google Scholar]; Weidenheim, K.M., and Dickson, D.W. (1995). Intranuclear inclusion bodies in an elderly demented woman: a form of intranuclear inclusion body disease. Clin. Neuropathol. 14, 93-99. [PubMed]
- 11.Takahashi J., Tanaka J., Arai K., Funata N., Hattori T., Fukuda T., Fujigasaki H., Uchihara T. Recruitment of nonexpanded polyglutamine proteins to intranuclear aggregates in neuronal intranuclear hyaline inclusion disease. J. Neuropathol. Exp. Neurol. 2001;60:369–376. doi: 10.1093/jnen/60.4.369. [DOI] [PubMed] [Google Scholar]; Takahashi, J., Tanaka, J., Arai, K., Funata, N., Hattori, T., Fukuda, T., Fujigasaki, H., and Uchihara, T. (2001). Recruitment of nonexpanded polyglutamine proteins to intranuclear aggregates in neuronal intranuclear hyaline inclusion disease. J. Neuropathol. Exp. Neurol. 60, 369-376. [DOI] [PubMed]
- 12.Pountney D.L., Huang Y., Burns R.J., Haan E., Thompson P.D., Blumbergs P.C., Gai W.P. SUMO-1 marks the nuclear inclusions in familial neuronal intranuclear inclusion disease. Exp. Neurol. 2003;184:436–446. doi: 10.1016/j.expneurol.2003.07.004. [DOI] [PubMed] [Google Scholar]; Pountney, D.L., Huang, Y., Burns, R.J., Haan, E., Thompson, P.D., Blumbergs, P.C., and Gai, W.P. (2003). SUMO-1 marks the nuclear inclusions in familial neuronal intranuclear inclusion disease. Exp. Neurol. 184, 436-446. [DOI] [PubMed]
- 13.Mori F., Miki Y., Tanji K., Ogura E., Yagihashi N., Jensen P.H., Wakabayashi K. Incipient intranuclear inclusion body disease in a 78-year-old woman. Neuropathology. 2011;31:188–193. doi: 10.1111/j.1440-1789.2010.01150.x. [DOI] [PubMed] [Google Scholar]; Mori, F., Miki, Y., Tanji, K., Ogura, E., Yagihashi, N., Jensen, P.H., and Wakabayashi, K. (2011). Incipient intranuclear inclusion body disease in a 78-year-old woman. Neuropathology 31, 188-193. [DOI] [PubMed]
- 14.Sone J., Tanaka F., Koike H., Inukai A., Katsuno M., Yoshida M., Watanabe H., Sobue G. Skin biopsy is useful for the antemortem diagnosis of neuronal intranuclear inclusion disease. Neurology. 2011;76:1372–1376. doi: 10.1212/WNL.0b013e3182166e13. [DOI] [PubMed] [Google Scholar]; Sone, J., Tanaka, F., Koike, H., Inukai, A., Katsuno, M., Yoshida, M., Watanabe, H., and Sobue, G. (2011). Skin biopsy is useful for the antemortem diagnosis of neuronal intranuclear inclusion disease. Neurology 76, 1372-1376. [DOI] [PubMed]
- 15.Sone J., Kitagawa N., Sugawara E., Iguchi M., Nakamura R., Koike H., Iwasaki Y., Yoshida M., Takahashi T., Chiba S. Neuronal intranuclear inclusion disease cases with leukoencephalopathy diagnosed via skin biopsy. J. Neurol. Neurosurg. Psychiatry. 2014;85:354–356. doi: 10.1136/jnnp-2013-306084. [DOI] [PubMed] [Google Scholar]; Sone, J., Kitagawa, N., Sugawara, E., Iguchi, M., Nakamura, R., Koike, H., Iwasaki, Y., Yoshida, M., Takahashi, T., Chiba, S., et al. (2014). Neuronal intranuclear inclusion disease cases with leukoencephalopathy diagnosed via skin biopsy. J. Neurol. Neurosurg. Psychiatry 85, 354-356. [DOI] [PubMed]
- 16.Araki K., Sone J., Fujioka Y., Masuda M., Ohdake R., Tanaka Y., Nakamura T., Watanabe H., Sobue G. Memory Loss and Frontal Cognitive Dysfunction in a Patient with Adult-onset Neuronal Intranuclear Inclusion Disease. Intern. Med. 2016;55:2281–2284. doi: 10.2169/internalmedicine.55.5544. [DOI] [PubMed] [Google Scholar]; Araki, K., Sone, J., Fujioka, Y., Masuda, M., Ohdake, R., Tanaka, Y., Nakamura, T., Watanabe, H., and Sobue, G. (2016). Memory Loss and Frontal Cognitive Dysfunction in a Patient with Adult-onset Neuronal Intranuclear Inclusion Disease. Intern. Med. 55, 2281-2284. [DOI] [PubMed]
- 17.Sone J., Hishikawa N., Koike H., Hattori N., Hirayama M., Nagamatsu M., Yamamoto M., Tanaka F., Yoshida M., Hashizume Y. Neuronal intranuclear hyaline inclusion disease showing motor-sensory and autonomic neuropathy. Neurology. 2005;65:1538–1543. doi: 10.1212/01.wnl.0000184490.22527.90. [DOI] [PubMed] [Google Scholar]; Sone, J., Hishikawa, N., Koike, H., Hattori, N., Hirayama, M., Nagamatsu, M., Yamamoto, M., Tanaka, F., Yoshida, M., Hashizume, Y., et al. (2005). Neuronal intranuclear hyaline inclusion disease showing motor-sensory and autonomic neuropathy. Neurology 65, 1538-1543. [DOI] [PubMed]
- 18.Nakamura M., Ueki S., Kubo M., Yagi H., Sasaki R., Okada Y., Akiguchi I., Kusaka H., Kondo T. Two cases of sporadic adult-onset neuronal intranuclear inclusion disease preceded by urinary disturbance for many years. J. Neurol. Sci. 2018;392:89–93. doi: 10.1016/j.jns.2018.07.012. [DOI] [PubMed] [Google Scholar]; Nakamura, M., Ueki, S., Kubo, M., Yagi, H., Sasaki, R., Okada, Y., Akiguchi, I., Kusaka, H., and Kondo, T. (2018). Two cases of sporadic adult-onset neuronal intranuclear inclusion disease preceded by urinary disturbance for many years. J. Neurol. Sci. 392, 89-93. [DOI] [PubMed]
- 19.O’Sullivan J.D., Hanagasi H.A., Daniel S.E., Tidswell P., Davies S.W., Lees A.J. Neuronal intranuclear inclusion disease and juvenile parkinsonism. Mov. Disord. 2000;15:990–995. doi: 10.1002/1531-8257(200009)15:5<990::aid-mds1035>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]; O’Sullivan, J.D., Hanagasi, H.A., Daniel, S.E., Tidswell, P., Davies, S.W., and Lees, A.J. (2000). Neuronal intranuclear inclusion disease and juvenile parkinsonism. Mov. Disord. 15, 990-995. [DOI] [PubMed]
- 20.Motoki M., Nakajima H., Sato T., Tada M., Kakita A., Arawaka S. Neuronal intranuclear inclusion disease showing intranuclear inclusions in renal biopsy 12 years earlier. Neurology. 2018;91:884–886. doi: 10.1212/WNL.0000000000006480. [DOI] [PubMed] [Google Scholar]; Motoki, M., Nakajima, H., Sato, T., Tada, M., Kakita, A., and Arawaka, S. (2018). Neuronal intranuclear inclusion disease showing intranuclear inclusions in renal biopsy 12 years earlier. Neurology 91, 884-886. [DOI] [PubMed]
- 21.Toyota T., Huang Z., Nohara S., Okada K., Kakeda S., Korogi Y., Nakayama T., Sone J., Sobue G., Adachi H. Neuronal intranuclear inclusion disease manifesting with new-onset epilepsy in the elderly. Neurol. Clin. Neurosci. 2015;3:238–240. [Google Scholar]; Toyota, T., Huang, Z., Nohara, S., Okada, K., Kakeda, S., Korogi, Y., Nakayama, T., Sone, J., Sobue, G., and Adachi, H. (2015). Neuronal intranuclear inclusion disease manifesting with new-onset epilepsy in the elderly. Neurol. Clin. Neurosci. 3, 238-240.
- 22.Fujita K., Osaki Y., Miyamoto R., Shimatani Y., Abe T., Sumikura H., Murayama S., Izumi Y., Kaji R. Neurologic attack and dynamic perfusion abnormality in neuronal intranuclear inclusion disease. Neurol. Clin. Pract. 2017;7:e39–e42. doi: 10.1212/CPJ.0000000000000389. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fujita, K., Osaki, Y., Miyamoto, R., Shimatani, Y., Abe, T., Sumikura, H., Murayama, S., Izumi, Y., and Kaji, R. (2017). Neurologic attack and dynamic perfusion abnormality in neuronal intranuclear inclusion disease. Neurol. Clin. Pract. 7, e39-e42. [DOI] [PMC free article] [PubMed]
- 23.Abe K., Fujita M. Over 10 years MRI observation of a patient with neuronal intranuclear inclusion disease. BMJ Case Rep. 2017;2017 doi: 10.1136/bcr-2016-218790. bcr2016218790. [DOI] [PMC free article] [PubMed] [Google Scholar]; Abe, K., and Fujita, M. (2017). Over 10 years MRI observation of a patient with neuronal intranuclear inclusion disease. BMJ Case Rep. 2017, bcr2016218790. [DOI] [PMC free article] [PubMed]
- 24.Sugiyama A., Sato N., Kimura Y., Maekawa T., Enokizono M., Saito Y., Takahashi Y., Matsuda H., Kuwabara S. MR Imaging Features of the Cerebellum in Adult-Onset Neuronal Intranuclear Inclusion Disease: 8 Cases. AJNR Am. J. Neuroradiol. 2017;38:2100–2104. doi: 10.3174/ajnr.A5336. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sugiyama, A., Sato, N., Kimura, Y., Maekawa, T., Enokizono, M., Saito, Y., Takahashi, Y., Matsuda, H., and Kuwabara, S. (2017). MR Imaging Features of the Cerebellum in Adult-Onset Neuronal Intranuclear Inclusion Disease: 8 Cases. AJNR Am. J. Neuroradiol. 38, 2100-2104. [DOI] [PMC free article] [PubMed]
- 25.Nakamura M., Murray M.E., Lin W.-L., Kusaka H., Dickson D.W. Optineurin immunoreactivity in neuronal and glial intranuclear inclusions in adult-onset neuronal intranuclear inclusion disease. Am. J. Neurodegener. Dis. 2014;3:93–102. [PMC free article] [PubMed] [Google Scholar]; Nakamura, M., Murray, M.E., Lin, W.-L., Kusaka, H., and Dickson, D.W. (2014). Optineurin immunoreactivity in neuronal and glial intranuclear inclusions in adult-onset neuronal intranuclear inclusion disease. Am. J. Neurodegener. Dis. 3, 93-102. [PMC free article] [PubMed]
- 26.Mori F., Tanji K., Kon T., Odagiri S., Hattori M., Hoshikawa Y., Kono C., Yasui K., Yokoi S., Hasegawa Y. FUS immunoreactivity of neuronal and glial intranuclear inclusions in intranuclear inclusion body disease. Neuropathol. Appl. Neurobiol. 2012;38:322–328. doi: 10.1111/j.1365-2990.2011.01217.x. [DOI] [PubMed] [Google Scholar]; Mori, F., Tanji, K., Kon, T., Odagiri, S., Hattori, M., Hoshikawa, Y., Kono, C., Yasui, K., Yokoi, S., Hasegawa, Y., et al. (2012). FUS immunoreactivity of neuronal and glial intranuclear inclusions in intranuclear inclusion body disease. Neuropathol. Appl. Neurobiol. 38, 322-328. [DOI] [PubMed]
- 27.Gelpi E., Botta-Orfila T., Bodi L., Marti S., Kovacs G., Grau-Rivera O., Lozano M., Sánchez-Valle R., Muñoz E., Valldeoriola F. Neuronal intranuclear (hyaline) inclusion disease and fragile X-associated tremor/ataxia syndrome: a morphological and molecular dilemma. Brain. 2017;140:e51. doi: 10.1093/brain/awx156. [DOI] [PubMed] [Google Scholar]; Gelpi, E., Botta-Orfila, T., Bodi, L., Marti, S., Kovacs, G., Grau-Rivera, O., Lozano, M., Sanchez-Valle, R., Muñoz, E., Valldeoriola, F., et al. (2017). Neuronal intranuclear (hyaline) inclusion disease and fragile X-associated tremor/ataxia syndrome: a morphological and molecular dilemma. Brain 140, e51. [DOI] [PubMed]
- 28.Zeng S., Zhang M.-y., Wang X., Hu Z., Li J., Li N., Wang J., Liang F., Yang Q., Liu Q. Long-read sequencing identified intronic repeat expansions in SAMD12 from Chinese pedigrees affected with familial cortical myoclonic tremor with epilepsy. J. Med. Genet. 2019;56:265–270. doi: 10.1136/jmedgenet-2018-105484. [DOI] [PubMed] [Google Scholar]; Zeng, S., Zhang, M.-y., Wang, X., Hu, Z., Li, J., Li, N., Wang, J., Liang, F., Yang, Q., Liu, Q., et al. (2019). Long-read sequencing identified intronic repeat expansions in SAMD12 from Chinese pedigrees affected with familial cortical myoclonic tremor with epilepsy. J. Med. Genet. 56, 265-270. [DOI] [PubMed]
- 29.Feng H., Conneely K.N., Wu H. A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acids Res. 2014;42:e69. doi: 10.1093/nar/gku154. [DOI] [PMC free article] [PubMed] [Google Scholar]; Feng, H., Conneely, K.N., and Wu, H. (2014). A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acids Res. 42, e69. [DOI] [PMC free article] [PubMed]
- 30.Fiddes I.T., Lodewijk G.A., Mooring M., Bosworth C.M., Ewing A.D., Mantalas G.L., Novak A.M., van den Bout A., Bishara A., Rosenkrantz J.L. Human-Specific NOTCH2NL Genes Affect Notch Signaling and Cortical Neurogenesis. Cell. 2018;173:1356–1369.e22. doi: 10.1016/j.cell.2018.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fiddes, I.T., Lodewijk, G.A., Mooring, M., Bosworth, C.M., Ewing, A.D., Mantalas, G.L., Novak, A.M., van den Bout, A., Bishara, A., Rosenkrantz, J.L., et al. (2018). Human-Specific NOTCH2NL Genes Affect Notch Signaling and Cortical Neurogenesis. Cell 173, 1356-1369.e22. [DOI] [PMC free article] [PubMed]
- 31.Nelson D.L., Orr H.T., Warren S.T. The unstable repeats--three evolving faces of neurological disease. Neuron. 2013;77:825–843. doi: 10.1016/j.neuron.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nelson, D.L., Orr, H.T., and Warren, S.T. (2013). The unstable repeats--three evolving faces of neurological disease. Neuron 77, 825-843. [DOI] [PMC free article] [PubMed]
- 32.Suzuki I.K., Gacquer D., Van Heurck R., Kumar D., Wojno M., Bilheu A., Herpoel A., Lambert N., Cheron J., Polleux F. Human-Specific NOTCH2NL Genes Expand Cortical Neurogenesis through Delta/Notch Regulation. Cell. 2018;173:1370–1384.e16. doi: 10.1016/j.cell.2018.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]; Suzuki, I.K., Gacquer, D., Van Heurck, R., Kumar, D., Wojno, M., Bilheu, A., Herpoel, A., Lambert, N., Cheron, J., Polleux, F., et al. (2018). Human-Specific NOTCH2NL Genes Expand Cortical Neurogenesis through Delta/Notch Regulation. Cell 173, 1370-1384.e16. [DOI] [PMC free article] [PubMed]
- 33.Wang T., Pan Q., Lin L., Szulwach K.E., Song C.X., He C., Wu H., Warren S.T., Jin P., Duan R., Li X. Genome-wide DNA hydroxymethylation changes are associated with neurodevelopmental genes in the developing human cerebellum. Hum. Mol. Genet. 2012;21:5500–5510. doi: 10.1093/hmg/dds394. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang, T., Pan, Q., Lin, L., Szulwach, K.E., Song, C.X., He, C., Wu, H., Warren, S.T., Jin, P., Duan, R., and Li, X. (2012). Genome-wide DNA hydroxymethylation changes are associated with neurodevelopmental genes in the developing human cerebellum. Hum. Mol. Genet. 21, 5500-5510. [DOI] [PMC free article] [PubMed]
- 34.Verkerk A.J.M.H., Pieretti M., Sutcliffe J.S., Fu Y.-H., Kuhl D.P.A., Pizzuti A., Reiner O., Richards S., Victoria M.F., Zhang F.P. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]; Verkerk, A.J.M.H., Pieretti, M., Sutcliffe, J.S., Fu, Y.-H., Kuhl, D.P.A., Pizzuti, A., Reiner, O., Richards, S., Victoria, M.F., Zhang, F.P., et al. (1991). Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65, 905-914. [DOI] [PubMed]
- 35.Vincent A., Heitz D., Petit C., Kretz C., Oberlé I., Mandel J.-L. Abnormal pattern detected in fragile-X patients by pulsed-field gel electrophoresis. Nature. 1991;349:624–626. doi: 10.1038/349624a0. [DOI] [PubMed] [Google Scholar]; Vincent, A., Heitz, D., Petit, C., Kretz, C., Oberle, I., and Mandel, J.-L. (1991). Abnormal pattern detected in fragile-X patients by pulsed-field gel electrophoresis. Nature 349, 624-626. [DOI] [PubMed]
- 36.La Spada A.R., Wilson E.M., Lubahn D.B., Harding A.E., Fischbeck K.H. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]; La Spada, A.R., Wilson, E.M., Lubahn, D.B., Harding, A.E., and Fischbeck, K.H. (1991). Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature 352, 77-79. [DOI] [PubMed]
- 37.Orr H.T., Chung M.Y., Banfi S., Kwiatkowski T.J., Jr., Servadio A., Beaudet A.L., McCall A.E., Duvick L.A., Ranum L.P.W., Zoghbi H.Y. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat. Genet. 1993;4:221–226. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]; Orr, H.T., Chung, M.Y., Banfi, S., Kwiatkowski, T.J., Jr., Servadio, A., Beaudet, A.L., McCall, A.E., Duvick, L.A., Ranum, L.P.W., and Zoghbi, H.Y. (1993). Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat. Genet. 4, 221-226. [DOI] [PubMed]
- 38.Brook J.D., McCurrach M.E., Harley H.G., Buckler A.J., Church D., Aburatani H., Hunter K., Stanton V.P., Thirion J.-P., Hudson T. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]; Brook, J.D., McCurrach, M.E., Harley, H.G., Buckler, A.J., Church, D., Aburatani, H., Hunter, K., Stanton, V.P., Thirion, J.-P., Hudson, T., et al. (1992). Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell 68, 799-808. [DOI] [PubMed]
- 39.MacDonald M.E., Ambrose C.M., Duyao M.P., Myers R.H., Lin C., Srinidhi L., Barnes G., Taylor S.A., James M., Groot N., The Huntington’s Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]; MacDonald, M.E., Ambrose, C.M., Duyao, M.P., Myers, R.H., Lin, C., Srinidhi, L., Barnes, G., Taylor, S.A., James, M., Groot, N., et al.; The Huntington’s Disease Collaborative Research Group (1993). A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 72, 971-983. [DOI] [PubMed]
- 40.Knight S.J.L., Flannery A.V., Hirst M.C., Campbell L., Christodoulou Z., Phelps S.R., Pointon J., Middleton-Price H.R., Barnicoat A., Pembrey M.E. Trinucleotide repeat amplification and hypermethylation of a CpG island in FRAXE mental retardation. Cell. 1993;74:127–134. doi: 10.1016/0092-8674(93)90300-f. [DOI] [PubMed] [Google Scholar]; Knight, S.J.L., Flannery, A.V., Hirst, M.C., Campbell, L., Christodoulou, Z., Phelps, S.R., Pointon, J., Middleton-Price, H.R., Barnicoat, A., Pembrey, M.E., et al. (1993). Trinucleotide repeat amplification and hypermethylation of a CpG island in FRAXE mental retardation. Cell 74, 127-134. [DOI] [PubMed]
- 41.Nagafuchi S., Yanagisawa H., Sato K., Shirayama T., Ohsaki E., Bundo M., Takeda T., Tadokoro K., Kondo I., Murayama N. Dentatorubral and pallidoluysian atrophy expansion of an unstable CAG trinucleotide on chromosome 12p. Nat. Genet. 1994;6:14–18. doi: 10.1038/ng0194-14. [DOI] [PubMed] [Google Scholar]; Nagafuchi, S., Yanagisawa, H., Sato, K., Shirayama, T., Ohsaki, E., Bundo, M., Takeda, T., Tadokoro, K., Kondo, I., Murayama, N., et al. (1994). Dentatorubral and pallidoluysian atrophy expansion of an unstable CAG trinucleotide on chromosome 12p. Nat. Genet. 6, 14-18. [DOI] [PubMed]
- 42.Kawaguchi Y., Okamoto T., Taniwaki M., Aizawa M., Inoue M., Katayama S., Kawakami H., Nakamura S., Nishimura M., Akiguchi I. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat. Genet. 1994;8:221–228. doi: 10.1038/ng1194-221. [DOI] [PubMed] [Google Scholar]; Kawaguchi, Y., Okamoto, T., Taniwaki, M., Aizawa, M., Inoue, M., Katayama, S., Kawakami, H., Nakamura, S., Nishimura, M., Akiguchi, I., et al. (1994). CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat. Genet. 8, 221-228. [DOI] [PubMed]
- 43.Lindblad K., Savontaus M.L., Stevanin G., Holmberg M., Digre K., Zander C., Ehrsson H., David G., Benomar A., Nikoskelainen E. An expanded CAG repeat sequence in spinocerebellar ataxia type 7. Genome Res. 1996;6:965–971. doi: 10.1101/gr.6.10.965. [DOI] [PubMed] [Google Scholar]; Lindblad, K., Savontaus, M.L., Stevanin, G., Holmberg, M., Digre, K., Zander, C., Ehrsson, H., David, G., Benomar, A., Nikoskelainen, E., et al. (1996). An expanded CAG repeat sequence in spinocerebellar ataxia type 7. Genome Res. 6, 965-971. [DOI] [PubMed]
- 44.Pulst S.-M., Nechiporuk A., Nechiporuk T., Gispert S., Chen X.-N., Lopes-Cendes I., Pearlman S., Starkman S., Orozco-Diaz G., Lunkes A. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat. Genet. 1996;14:269–276. doi: 10.1038/ng1196-269. [DOI] [PubMed] [Google Scholar]; Pulst, S.-M., Nechiporuk, A., Nechiporuk, T., Gispert, S., Chen, X.-N., Lopes-Cendes, I., Pearlman, S., Starkman, S., Orozco-Diaz, G., Lunkes, A., et al. (1996). Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat. Genet. 14, 269-276. [DOI] [PubMed]
- 45.Ishikawa K., Tanaka H., Saito M., Ohkoshi N., Fujita T., Yoshizawa K., Ikeuchi T., Watanabe M., Hayashi A., Takiyama Y. Japanese families with autosomal dominant pure cerebellar ataxia map to chromosome 19p13.1-p13.2 and are strongly associated with mild CAG expansions in the spinocerebellar ataxia type 6 gene in chromosome 19p13.1. Am. J. Hum. Genet. 1997;61:336–346. doi: 10.1086/514867. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ishikawa, K., Tanaka, H., Saito, M., Ohkoshi, N., Fujita, T., Yoshizawa, K., Ikeuchi, T., Watanabe, M., Hayashi, A., Takiyama, Y., et al. (1997). Japanese families with autosomal dominant pure cerebellar ataxia map to chromosome 19p13.1-p13.2 and are strongly associated with mild CAG expansions in the spinocerebellar ataxia type 6 gene in chromosome 19p13.1. Am. J. Hum. Genet. 61, 336-346. [DOI] [PMC free article] [PubMed]
- 46.Campuzano V., Montermini L., Moltò M.D., Pianese L., Cossée M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]; Campuzano, V., Montermini, L., Molto, M.D., Pianese, L., Cossee, M., Cavalcanti, F., Monros, E., Rodius, F., Duclos, F., Monticelli, A., et al. (1996). Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271, 1423-1427. [DOI] [PubMed]
- 47.Holmes S.E., O’Hearn E.E., McInnis M.G., Gorelick-Feldman D.A., Kleiderlein J.J., Callahan C., Kwak N.G., Ingersoll-Ashworth R.G., Sherr M., Sumner A.J. Expansion of a novel CAG trinucleotide repeat in the 5′ region of PPP2R2B is associated with SCA12. Nat. Genet. 1999;23:391–392. doi: 10.1038/70493. [DOI] [PubMed] [Google Scholar]; Holmes, S.E., O’Hearn, E.E., McInnis, M.G., Gorelick-Feldman, D.A., Kleiderlein, J.J., Callahan, C., Kwak, N.G., Ingersoll-Ashworth, R.G., Sherr, M., Sumner, A.J., et al. (1999). Expansion of a novel CAG trinucleotide repeat in the 5′ region of PPP2R2B is associated with SCA12. Nat. Genet. 23, 391-392. [DOI] [PubMed]
- 48.Koob M.D., Moseley M.L., Schut L.J., Benzow K.A., Bird T.D., Day J.W., Ranum L.P.W. An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8) Nat. Genet. 1999;21:379–384. doi: 10.1038/7710. [DOI] [PubMed] [Google Scholar]; Koob, M.D., Moseley, M.L., Schut, L.J., Benzow, K.A., Bird, T.D., Day, J.W., and Ranum, L.P.W. (1999). An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8). Nat. Genet. 21, 379-384. [DOI] [PubMed]
- 49.Nakamura K., Jeong S.Y., Uchihara T., Anno M., Nagashima K., Nagashima T., Ikeda S., Tsuji S., Kanazawa I. SCA17, a novel autosomal dominant cerebellar ataxia caused by an expanded polyglutamine in TATA-binding protein. Hum. Mol. Genet. 2001;10:1441–1448. doi: 10.1093/hmg/10.14.1441. [DOI] [PubMed] [Google Scholar]; Nakamura, K., Jeong, S.Y., Uchihara, T., Anno, M., Nagashima, K., Nagashima, T., Ikeda, S., Tsuji, S., and Kanazawa, I. (2001). SCA17, a novel autosomal dominant cerebellar ataxia caused by an expanded polyglutamine in TATA-binding protein. Hum. Mol. Genet. 10, 1441-1448. [DOI] [PubMed]
- 50.Jacquemont S., Hagerman R.J., Leehey M.A., Hall D.A., Levine R.A., Brunberg J.A., Zhang L., Jardini T., Gane L.W., Harris S.W. Penetrance of the fragile X-associated tremor/ataxia syndrome in a premutation carrier population. JAMA. 2004;291:460–469. doi: 10.1001/jama.291.4.460. [DOI] [PubMed] [Google Scholar]; Jacquemont, S., Hagerman, R.J., Leehey, M.A., Hall, D.A., Levine, R.A., Brunberg, J.A., Zhang, L., Jardini, T., Gane, L.W., Harris, S.W., et al. (2004). Penetrance of the fragile X-associated tremor/ataxia syndrome in a premutation carrier population. JAMA 291, 460-469. [DOI] [PubMed]
- 51.DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; DeJesus-Hernandez, M., Mackenzie, I.R., Boeve, B.F., Boxer, A.L., Baker, M., Rutherford, N.J., Nicholson, A.M., Finch, N.A., Flynn, H., Adamson, J., et al. (2011). Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245-256. [DOI] [PMC free article] [PubMed]
- 52.Renton A.E., Majounie E., Waite A., Simón-Sánchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., van Swieten J.C., Myllykangas L., ITALSGEN Consortium A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; Renton, A.E., Majounie, E., Waite, A., Simon-Sanchez, J., Rollinson, S., Gibbs, J.R., Schymick, J.C., Laaksovirta, H., van Swieten, J.C., Myllykangas, L., et al.; ITALSGEN Consortium (2011). A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257-268. [DOI] [PMC free article] [PubMed]
- 53.Mootha V.V., Gong X., Ku H.-C., Xing C. Association and familial segregation of CTG18.1 trinucleotide repeat expansion of TCF4 gene in Fuchs’ endothelial corneal dystrophy. Invest. Ophthalmol. Vis. Sci. 2014;55:33–42. doi: 10.1167/iovs.13-12611. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mootha, V.V., Gong, X., Ku, H.-C., and Xing, C. (2014). Association and familial segregation of CTG18.1 trinucleotide repeat expansion of TCF4 gene in Fuchs’ endothelial corneal dystrophy. Invest. Ophthalmol. Vis. Sci. 55, 33-42. [DOI] [PMC free article] [PubMed]
- 54.Jin P., Alisch R.S., Warren S.T. RNA and microRNAs in fragile X mental retardation. Nat. Cell Biol. 2004;6:1048–1053. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]; Jin, P., Alisch, R.S., and Warren, S.T. (2004). RNA and microRNAs in fragile X mental retardation. Nat. Cell Biol. 6, 1048-1053. [DOI] [PubMed]
- 55.LaCroix A.J., Stabley D., Sahraoui R., Adam M.P., Mehaffey M., Kernan K., Myers C.T., Fagerstrom C., Anadiotis G., Akkari Y.M., University of Washington Center for Mendelian Genomics GGC Repeat Expansion and Exon 1 Methylation of XYLT1 Is a Common Pathogenic Variant in Baratela-Scott Syndrome. Am. J. Hum. Genet. 2019;104:35–44. doi: 10.1016/j.ajhg.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; LaCroix, A.J., Stabley, D., Sahraoui, R., Adam, M.P., Mehaffey, M., Kernan, K., Myers, C.T., Fagerstrom, C., Anadiotis, G., Akkari, Y.M., et al.; University of Washington Center for Mendelian Genomics (2019). GGC Repeat Expansion and Exon 1 Methylation of XYLT1 Is a Common Pathogenic Variant in Baratela-Scott Syndrome. Am. J. Hum. Genet. 104, 35-44. [DOI] [PMC free article] [PubMed]
- 56.Kong H.E., Zhao J., Xu S., Jin P., Jin Y. Fragile X-Associated Tremor/Ataxia Syndrome: From Molecular Pathogenesis to Development of Therapeutics. Front. Cell. Neurosci. 2017;11:128. doi: 10.3389/fncel.2017.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kong, H.E., Zhao, J., Xu, S., Jin, P., and Jin, Y. (2017). Fragile X-Associated Tremor/Ataxia Syndrome: From Molecular Pathogenesis to Development of Therapeutics. Front. Cell. Neurosci. 11, 128. [DOI] [PMC free article] [PubMed]
- 58.Sone J., Nakamura T., Koike H., Katsuno M., Tanaka F., Iwasaki Y., Yoshida M., Sobue G. Reply: Neuronal intranuclear (hyaline) inclusion disease and fragile X-associated tremor/ataxia syndrome: a morphological and molecular dilemma. Brain. 2017;140:e52. doi: 10.1093/brain/awx158. [DOI] [PubMed] [Google Scholar]; Sone, J., Nakamura, T., Koike, H., Katsuno, M., Tanaka, F., Iwasaki, Y., Yoshida, M., and Sobue, G. (2017). Reply: Neuronal intranuclear (hyaline) inclusion disease and fragile X-associated tremor/ataxia syndrome: a morphological and molecular dilemma. Brain 140, e52. [DOI] [PubMed]
- 59.Glineburg M.R., Todd P.K., Charlet-Berguerand N., Sellier C. Repeat-associated non-AUG (RAN) translation and other molecular mechanisms in Fragile X Tremor Ataxia Syndrome. Brain Res. 2018;1693(Pt A):43–54. doi: 10.1016/j.brainres.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; Glineburg, M.R., Todd, P.K., Charlet-Berguerand, N., and Sellier, C. (2018). Repeat-associated non-AUG (RAN) translation and other molecular mechanisms in Fragile X Tremor Ataxia Syndrome. Brain Res. 1693 (Pt A), 43-54. [DOI] [PMC free article] [PubMed]
- 60.Todd P.K., Oh S.Y., Krans A., He F., Sellier C., Frazer M., Renoux A.J., Chen K.C., Scaglione K.M., Basrur V. CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron. 2013;78:440–455. doi: 10.1016/j.neuron.2013.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]; Todd, P.K., Oh, S.Y., Krans, A., He, F., Sellier, C., Frazer, M., Renoux, A.J., Chen, K.C., Scaglione, K.M., Basrur, V., et al. (2013). CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron 78, 440-455. [DOI] [PMC free article] [PubMed]
- 61.Sellier C., Buijsen R.A.M., He F., Natla S., Jung L., Tropel P., Gaucherot A., Jacobs H., Meziane H., Vincent A. Translation of Expanded CGG Repeats into FMRpolyG Is Pathogenic and May Contribute to Fragile X Tremor Ataxia Syndrome. Neuron. 2017;93:331–347. doi: 10.1016/j.neuron.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sellier, C., Buijsen, R.A.M., He, F., Natla, S., Jung, L., Tropel, P., Gaucherot, A., Jacobs, H., Meziane, H., Vincent, A., et al. (2017). Translation of Expanded CGG Repeats into FMRpolyG Is Pathogenic and May Contribute to Fragile X Tremor Ataxia Syndrome. Neuron 93, 331-347. [DOI] [PMC free article] [PubMed]
- 62.Tassone F., Iwahashi C., Hagerman P.J. FMR1 RNA within the intranuclear inclusions of fragile X-associated tremor/ataxia syndrome (FXTAS) RNA Biol. 2004;1:103–105. doi: 10.4161/rna.1.2.1035. [DOI] [PubMed] [Google Scholar]; Tassone, F., Iwahashi, C., and Hagerman, P.J. (2004). FMR1 RNA within the intranuclear inclusions of fragile X-associated tremor/ataxia syndrome (FXTAS). RNA Biol. 1, 103-105. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.