Abstract
In an effort to meet ethical obligations and/or participant expectations, researchers may consider offering “raw” or uninterpreted genetic data for result return. It is therefore important to understand the motivations, behaviors, and perspectives of individuals who might choose to access raw data before such return becomes routine. In the direct-to-consumer (DTC) context, where raw data are often made available to customers, the use of third-party interpretation tools has raised concerns about genotype accuracy, data privacy, reliability of interpretation, and consumption of limited health care resources. However, relatively little is known about why individuals access raw data or what they do with the information received from third-party interpretation. Accordingly, we conducted a survey on raw data access and third-party tool usage among 1,137 DTC customers recruited through social media. Most survey respondents (89%) reported downloading their raw data. Among downloaders, 94% used at least one tool, most commonly Promethease (63%) or GEDmatch (84%). More than half (56%) used both health-related and non-health-related tools and differed significantly from those who used only one tool type in terms of demographics, participation in research, DTC tests ordered, and testing motivations. Exploratory interviews were conducted with 10 respondents and illustrated how social networking, initial lack of interesting findings, and general curiosity contributed to use of multiple tool types. These results suggest that even when initially motivated by ancestry and genealogy, consumers frequently also pursue health information in a largely unregulated and expanding suite of third-party tools, raising both challenges and opportunities for the professional genetics community.
Keywords: third-party interpretation, direct-to-consumer genomic testing, personal genomic testing, survey
Introduction
There is a widening consensus that researchers should, wherever possible, offer individual results to research participants, both to enhance autonomy, agency, and reciprocity of participants and to increase engagement with and contribution to both traditional and citizen science research initiatives.1, 2, 3 While the return of results has been the subject of scientific, ethical, and legal debate for some time,4, 5, 6 the discourse has focused mostly on interpreted results rather than “raw” or uninterpreted genetic data. Here, raw data refer to genotype calls from either array genotyping or sequencing but may also encompass upstream formats such as the sequence alignment data used to make genotype calls (e.g., BAM files).1 Empirical evidence suggests that members of the general public would want their raw genetic data if they were to participate in a genome sequencing study,7 and some investigators have begun offering such data to research participants.1 Indeed, organizers of the National Institutes of Health “All of Us” research cohort have stated plans to make raw genetic data available to participants,8, 9 and the National Academies of Sciences, Engineering, and Medicine (NASEM) have issued recommendations that are supportive of returning “individual research results” (both raw data and interpreted results) where feasible.10 Therefore, understanding how individuals make use of their raw genetic data is important for genetic researchers considering the return of such information to participants.
However, the implications of making raw genetic data available to research participants remain largely unknown. One avenue to explore potential outcomes is to examine current uses of raw genetic data from direct-to-consumer (DTC) genetic testing, which to date has been the most common route of access. Broadly, there are two types of activities DTC customers can undertake with their raw data. The first is to contribute to collective research efforts by sharing genotypic and phenotypic data on platforms such as Open Humans and openSNP.11, 12 The potential benefits of such sharing include increasing the size of datasets available to traditional researchers and creating novel opportunities for participant-driven or “citizen science” research initiatives.13, 14 The second type of activity is the pursuit of individually tailored information via third-party interpretation (TPI) tools such as Promethease or GEDmatch. As we found in a prior study,15 TPI tools are quite heterogeneous in terms of types of information returned (e.g., health, ancestry, genealogy), methods for generating that information, and cost to users. Further complicating this picture is that some TPI tools also provide opportunities to contribute to collective research efforts (e.g., openSNP and DNA.Land).
Recently, TPI tools have received growing attention in both academic and lay media discourse, where concerns have been raised about genotype accuracy, data privacy and security, reliability of health-related information, potential for false positives or false negatives, and downstream consumption of limited health care resources. For example, media reports have recounted individuals receiving distressing results from TPI, which upon clinical confirmatory testing turned out to be false positives.16, 17, 18, 19 These individuals experienced emotional and financial hardship in unnecessary follow-up, often tracing back to errors in the raw DTC genotypes. Accounts in academic literature have illustrated outcomes of bringing TPI results to genetic counselors (GCs),20 genetics specialty clinics,21 or otherwise leading to follow-up clinical sequencing.22 In addition to health-related concerns, novel uses by law enforcement of online genealogy databases such as GEDmatch are raising questions about data privacy and consent in third-party services.23, 24
However, few studies have reported on the perspectives of the tool users themselves. One survey of DTC customers found a high volume of raw data download and TPI tool use (67%, or 321/478 respondents), where the majority of tool users (81%) were satisfied with the information received.25 However, that study did not explore the relationships between use of specific tools and reactions, follow-up actions, and DTC testing motivations. Some tool developers have also surveyed their own users,12, 26 but this provides limited insight into use of raw data and TPI tools more broadly and does not measure the degree to which users access multiple tools or make distinctions among the tools that they use.
Here we contribute new information about consumers’ use of raw genetic data and TPI tools from the results of a survey of more than 1,100 DTC customers and follow-up interviews with a subset of survey respondents. Our aim was to better understand users’ motivations and behaviors from initial DTC testing through to downstream use of specific TPI tools and resulting follow-up actions, with the broader goal of understanding how raw data return in non-DTC contexts may unfold. This primarily descriptive study contributes novel insights into how individuals leverage their raw genetic data in multiple ways, often concurrently, including to learn about health risks, ancestry, and genealogy.
Subjects and Methods
Participant Recruitment
We recruited survey respondents during October and November 2017 with staggered postings to various social media venues: six genomics-related sub-Reddits, Twitter, and several Facebook groups (four genealogy groups and the DNA.Land page). Early in the 2-month period and once per venue, we posted a brief study description and link to the survey, after seeking permission from group moderators/administrators. Additionally, the survey was sent via newsletter to openSNP27 users and was posted on the first author’s academic website and the Institute for Translational Health Sciences (ITHS) Participant Portal. Recruitment messages stated eligibility criteria as being 18 years or older and having taken at least one DTC genetic test—i.e., raw data download and TPI tool use were neither mentioned nor required, though some of the groups targeted were oriented toward TPI tools (e.g., Promethease sub-Reddit or DNA.Land Facebook page). No incentives were offered for participating. The survey was closed 7 weeks after it first opened and after posting final reminders on each recruitment venue.
Survey Design and Implementation
The survey questionnaire was designed to cover three main topics: DTC testing, raw data download, and TPI tool use. Respondents were asked which DTC test(s) they had ordered, when, what motivated them to order the test(s), and whether they had downloaded their raw data. Non-downloaders were asked about reasons for not downloading while downloaders proceeded to a series of questions about TPI tools. Respondents who had used multiple tools were prompted to select one on which to base their responses. All respondents filled out a demographics section and were invited to provide contact information for voluntary follow-up interviewing.
We developed survey questions primarily based on our prior study of TPI tools and developers.15 However, some items were based on existing instruments: motivations for DTC testing were adapted from the PGen (Impact of Personal Genomics) Study baseline survey,28 and questions on how the respondent learned about tools were adapted from Wang et al.25 A draft instrument was evaluated via cognitive interviewing29 with six individuals recruited from a combination of personal and professional networks. The self-administered, online questionnaire was implemented in REDCap.30 While the majority of survey questions were fixed response, some open text comment boxes were also included.
Follow-up Interviews
We purposively sampled interview participants from among those survey respondents who volunteered for follow-up contact and reported using a specific combination of tools that spanned health, ancestry, and genealogy (Promethease, DNA.Land, and GEDmatch, respectively)—or “crossover” use, described further below. Initially, 179 respondents met these criteria. Because we wanted to explore crossover in both directions (i.e., from health to non-health and vice versa), we stratified this sample by “DTC test(s) ordered” as a proxy for initial motivations for tool use among potential interviewees, and, within each bin, randomly sampled participant IDs to contact.
We conducted semi-structured interviews via phone or Skype and had audio recordings transcribed on Rev (see Web Resources). The interview guide comprised six questions focused on gaining a deeper understanding of respondents’ timing and motivations to download data and use multiple types of tools. For example, did respondents start by using GEDmatch with an interest in genealogy and go on to additionally use Promethease and if so, why? Or, conversely, did initial interest in health and use of Promethease eventually lead to use of genealogy and ancestry-focused tools as well and if so, why? We conducted a total of ten interviews, after which point we had observed multiple examples of crossover in each direction (i.e., participants initially using health tools before using non-health tools, and vice versa) and halted recruitment for this exploratory follow-up phase of the study. Interviews averaged 36 min (SD = 14 min); an interview with one deaf respondent was carried out via email.
Data Analysis
Quantitative survey data were analyzed via univariate descriptives, bivariate, and multivariate analyses (i.e., logistic regression) using all available, non-missing data. The total number of potential responses changes between survey sections due to branching logic; sporadic missing answers also affect the count of available responses for any given survey item. Therefore, throughout we report both percentages and counts. All quantitative analyses were carried out in R statistical and graphing software.31 Throughout, DTC testing motivations were dichotomized into “very important” versus “somewhat important” or “not at all important,” following precedent in analyses of the PGen survey from which those survey items were adapted.32, 33 To understand factors influencing use of specific third-party tools, we performed a series of logistic regression analyses using tool use (yes/no) as an outcome and the following covariates: each of the eight dichotomized DTC testing motivations, survey recruitment venue, and DTC test(s) ordered. The survey instrument, results dataset, and R analysis code are available on openICPSR (see Web Resources). We thematically analyzed qualitative survey data from open text boxes in Atlas.ti v8. Interview analysis was also conducted in Atlas.ti v8 and focused on understanding how and why participants came to use tools across the multiple domains of health, ancestry, and genealogy.
This study was approved by the University of Washington (UW) Institutional Review Board as minimal risk human subjects research, protocol #50238.
Results
A total of 1,137 eligible respondents took the survey (see Table 1), including 268 respondents (24%) who did not progress to the end of the survey. The most common recruitment venue was Facebook (624/1,137 or 55%), followed by Reddit (357/1,137 or 31%), Twitter (71/1,137 or 6%), and the openSNP newsletter (62/1,137 or 5%). Fewer than 20 respondents were recruited from each of the remaining venues. Below we report patterns of DTC testing and TPI tool use, including respondents’ impressions of information received and follow-up actions taken. We then analyze users by categories of TPI tools, focusing on the common phenomenon (56%, or 458/819 tool users) of using both health-related and non health-related tools.
Table 1.
Survey Respondent Characteristics
| Variable | Overall | All Tool Users | Non-health Only Tools | Crossover | Health Only Tools | Test Statistica | p Valuea |
|---|---|---|---|---|---|---|---|
| Number of respondents | 1,137 | 820b | 263 | 458 | 98 | ||
| Mean age (SD; range) | 46.4 (15; 18–>89) | 46.7 (15; 18–84) | 51.8 (14; 18–83) | 45.5 (15; 18–84) | 39.4(12; 20–73) | 27.2 | 3.87e−12 |
| Gender | |||||||
| Women (%) | 67.4 | 67.1 | 69.8 | 68.7 | 53.3 | 8.99 | 0.0111 |
| Race (%) | |||||||
| Asian | 1.8 | 1.6 | 0.9 | 1.9 | 2.2 | N/A | 0.0650 |
| Black or African American | 1.7 | 1.6 | 0.9 | 2.1 | 1.1 | ||
| Hawaiian or Pacific Islander | 0.1 | 0.1 | 0.0 | 0.2 | 0.0 | ||
| White | 81.6 | 80.6 | 76.2 | 81.7 | 86.7 | ||
| Other | 3.7 | 4.0 | 6.8 | 2.8 | 2.2 | ||
| Prefer no answer | 2.2 | 2.4 | 4.7 | 1.4 | 1.1 | ||
| Multiplec | 8.9 | 9.8 | 10.6 | 10.0 | 6.7 | ||
| Hispanic/Latino (%) | 6.7 | 6.6 | 8.1 | 6.0 | 5.6 | N/A | 0.677 |
| Lives in US (%) | 75.9 | 74.9 | 74.5 | 74.2 | 78.9 | 1.45 | 0.485 |
| Max education (%) | |||||||
| Less than high school | 1.0 | 1.1 | 2.1 | 0.5 | 1.1 | 12.3 |
0.139 |
| High school graduate or GED | 26.1 | 26.8 | 27.0 | 28.1 | 20.0 | ||
| College degree | 41.0 | 41.3 | 39.9 | 42.2 | 41.1 | ||
| Master’s degree | 23.1 | 22.8 | 21.5 | 23.2 | 24.4 | ||
| Doctorate/terminal degree |
8.9 |
8.1 |
9.4 |
6.0 |
13.3 |
||
| Occupation (%) | |||||||
| Business, financial, management, sales | 14.2 | 14.2 | 15.0 | 15.1 | 7.8 | 31.0 | 0.0556 |
| Computer, engineering, math | 16.9 | 17.1 | 16.7 | 15.5 | 25.6 | ||
| Life, physical, and social science | 9.2 | 8.2 | 6.8 | 7.2 | 15.6 | ||
| Legal | 2.5 | 2.6 | 0.9 | 3.5 | 3.3 | ||
| Education, training, library | 14.3 | 13.6 | 13.2 | 15.3 | 6.7 | ||
| Arts, design, entertainment, sports, media | 4.7 | 4.6 | 3.4 | 5.1 | 5.6 | ||
| Healthcare practitioner | 8.9 | 8.9 | 10.3 | 7.7 | 11.1 | ||
| Office, administrative support | 7.7 | 7.5 | 6.4 | 8.8 | 4.4 | ||
| Construction, maintenance, natural resources | 1.9 | 1.9 | 2.6 | 1.6 | 1.1 | ||
| Production and transportation | 1.5 | 1.7 | 2.1 | 1.6 | 1.1 | ||
| Other | 18.2 | 19.7 | 22.6 | 18.6 | 17.8 | ||
| Works in genetic research/medicine (%) | 5.1 | 3.8 | 3.0 | 2.3 | 13.3 | 24.8 | 4.11e−06 |
| Participant in genetic research (%) | 14.9 | 16.2 | 11.5 | 19.0 | 15.6 | 7.68 | 0.0215 |
Survey respondent characteristics, overall and grouped by type(s) of tools used. For categorical variables, values are given as within-group percentage, excluding NA/missing values from the denominator. Statistical tests of difference are reported for comparison between groups of tool users: users of non-health only tools, crossover users (used both health and non-health tools), and users of health-only tools. SD indicates standard deviation.
Comparing three groups of tool users: non-health only tool users, crossover tool users, and health-only tool users. For categorical values where all cell counts >5, the test statistic and p value are from a chi-square test. For categorical values with low cell counts (i.e., race and Hispanic/Latino), the p value is from a Fisher exact test, and the test statistic is N/A. ANOVA was used to compare continuous variables; the test statistic given is the F value.
Of the 820 respondents who used at least one tool, one respondent reported using only “various R packages” and could therefore not be assigned a tool user group (non-health only; crossover; health only).
Respondents who checked more than one box for self-identified race are counted under “Multiple.” Note all American Indian/Alaska Native respondents checked more than one box and are therefore all counted under “Multiple” here.
Patterns of DTC Testing
DTC Tests Taken
Respondents reported ordering a range of DTC tests, with 36% (413/1,137) ordering multiple tests: 21% (236/1,137) ordered two tests, 12% (135/1,137) ordered three, and 4% (42/1,137) of respondents ordered four or more. While asked explicitly about 23andMe, AncestryDNA, and FamilyTreeDNA, respondents noted additional tests via an open text box, most commonly MyHeritage (n = 37), Living DNA (n = 32), National Genographic (n = 19), and Genes for Good (n = 18). The majority of DTC tests were ordered after 2015 (see Figure S1), potentially reflecting a rise in DTC test popularity and/or a recency effect in that those who recently ordered tests were more likely to be active in our online recruitment venues.
DTC Testing Motivations
The most common motivations for pursuing DTC testing were general curiosity about genetic make-up (661/972 or 68% rated as very important) and curiosity about ancestry (645/977 or 66% rated as very important; see Table 2). Less common motivations were limited information about family health history (201/972 or 21% rated as very important) and other family members pursuing testing (104/969 or 11% rated as very important). In free text responses, 20% of respondents (225/1,137) noted additional motivations, which included pharmacogenomics; being adopted; breaking through “brick walls,” or dead ends, in genealogy research by matching with new relatives;34 and professional interests (e.g., teaches genetics or is a GC). Notably, 81% of respondents (787/969) rated desire to have their raw genetic data file as either a very important or somewhat important motivation to pursue DTC testing.
Table 2.
DTC Tests Ordered and DTC Testing Motivations
| Variable | Overall | All Tool Users | Non-health Only Tools | Crossover | Health Only Tools | Test Statistica | p Valuea |
|---|---|---|---|---|---|---|---|
| Number of respondents | 1,137 | 820 | 263 | 458 | 98 | ||
| DTC Tests Ordered (%) | |||||||
| 23andMe | 62.4 | 61.7 | 40.7 | 68.1 | 87.8 | 85.3 | <2.2e−16 |
| AncestryDNA | 53.9 | 59.8 | 73.0 | 62.0 | 14.3 | 104.5 | <2.2e−16 |
| FamilyTreeDNA | 26.8 | 30.2 | 37.3 | 32.1 | 3.1 | 41.2 | 1.14e−09 |
| Rating of DTC Testing Motivationsb(% Very Important) | |||||||
| General curiosity | 68.0 | 67.6 | 55.5 | 71.4 | 82.7 | 30.7 | 2.1e−07 |
| Ancestry | 66.0 | 66.3 | 69.6 | 69.8 | 41.2 | 31.1 | 1.8e−07 |
| Find relatives | 46.9 | 49.9 | 62.4 | 50.4 | 14.3 | 66.1 | 4.4e−15 |
| Risk for specific diseases | 31.4 | 30.9 | 16.1 | 34.2 | 54.1 | 53.9 | 1.97e−12 |
| Limited family health history | 20.7 | 20.7 | 21.4 | 21.0 | 17.7 | 0.621 | 0.733 |
| Other family members are using | 10.7 | 10.7 | 10.8 | 11.0 | 8.3 | 0.656 | 0.720 |
| Participate in research | 32.3 | 31.6 | 26.3 | 33.8 | 34.7 | 4.79 | 0.0911 |
| Raw genetic data file | 50.8 | 53.1 | 44.4 | 56.4 | 60.4 | 11.9 | 0.00263 |
Comparisons of DTC tests ordered and DTC testing motivations, overall and grouped by type(s) of tools used.
Comparing non-health only tool users, crossover tool users, and health-only tool users. Test statistics and p values are from chi-square tests.
Responses to DTC testing motivations were dichotomized into “very important” versus “somewhat important” or “not at all important.”
The prevalence of raw data access as a motivation was borne out in the rate of data download: 26% (252/974) of respondents reported they had not or were unsure whether they had downloaded their raw data from at least one of the DTC tests they had taken (note respondents were asked about download separately for each DTC test taken). Of these 252, 148 had downloaded data from another one of the DTC tests they had taken, leaving 104 (11% of 974) who had not downloaded any of their available raw data files. Demographically, non-downloaders did not differ from downloaders; however, in pursing DTC testing, downloaders were more motivated to find relatives (β = 0.839, p = 0.0007) and obtain raw data (β = 1.49, p = 9.62e−08) compared to non-downloaders (see also Figure S2).
Third-Party Tool Use
Impressions and Follow-up Actions
A total of 820 respondents who downloaded raw data also reported using at least one tool and formed the basis for subsequent analyses. Most used multiple tools, with 76% (623/820) using two or more (median number of tools = 3, max number = 11). Thirteen tools were specified in fixed-response survey questions, though respondents could note additional tools via free text. The most commonly used tools were GEDmatch (n = 688 respondents), Promethease (n = 515), and DNA.Land (n = 450; see Table S1). Additional tools most frequently noted in the open text box were WeGene (n = 39), FamilyTreeDNA (n = 35), and MyHeritage (n = 34).
Respondents’ impressions of the information they received varied by tool (see Figure 1). Promethease users overwhelmingly agreed they received information about health (171/186, or 92%) and about risk of specific disease (180/186, or 97%). Notably, some respondents indicated receiving information outside a tool’s scope: 16% of GEDmatch users (83/505) agreed they received some health information while 37% of Promethease users (69/188) agreed they received results related to ancestry or genealogy. (It is possible some respondents who used multiple tools did not limit responses to the selected tool as directed in survey instructions.) Across all tools, respondents largely agreed using the tool increased their understanding of genetics in general (593/778 or 76% agreed) and of how DTC companies interpret genetic data (522/775 or 67% agreed). The majority also felt satisfied with the information received (678/770 or 88%), though some reported feeling confused (266/767 or 35%) or upset (48/758 or 6%). These reactions were relatively consistent across tools (see Figure S3).
Figure 1.
Perceived Information Received from Tools
Agreement with statements about information received from TPI tools, separately by tool, over 820 respondents who reported using at least one tool. Counts plotted to the right of x = 0 are for agreement; counts plotted to the left of x = 0 are for disagreement.
The most common follow-up actions were sharing the results with a family member (664/780 or 85% shared) or non-family member friend or loved one (552/778 or 71% shared). Other common follow-up actions included pursuing additional analysis via a different tool (430/776 or 55%), pursuing more genetic testing (253/776 or 33%), or participating in a genetic research study (275/775 or 35%). Few respondents made changes to either health insurance (7/770 or 0.9% changed) or other types of insurance (i.e., life or long-term care; 8/771 or 1% changed). Fifteen percent of respondents (116/775) indicated sharing results with a health care provider (HCP), most commonly a general practitioner (92/116, or 79% of those who shared). As might be expected based on the differing types of information offered by each tool, the reported rate of HCP sharing differed significantly by tool: 8.1% (39/484) of GEDmatch users shared while 31% (56/183) of Promethease users shared (Chi-square test statistic = 53.4, p = 2.69e−13). Notably, across all respondents who shared with a HCP, 13 (11% of 116) reported sharing results with a medical geneticist or GC. A total of 52 respondents (45% of 116) wrote in an additional type of HCP with whom they shared results, including practitioners from non-genetics specialties (e.g., cardiologist, gastroenterologist, ophthalmologist), psychiatry/psychology, and alternative medicine (e.g., naturopath, acupuncturist).
Use of Specific Third-Party Tools
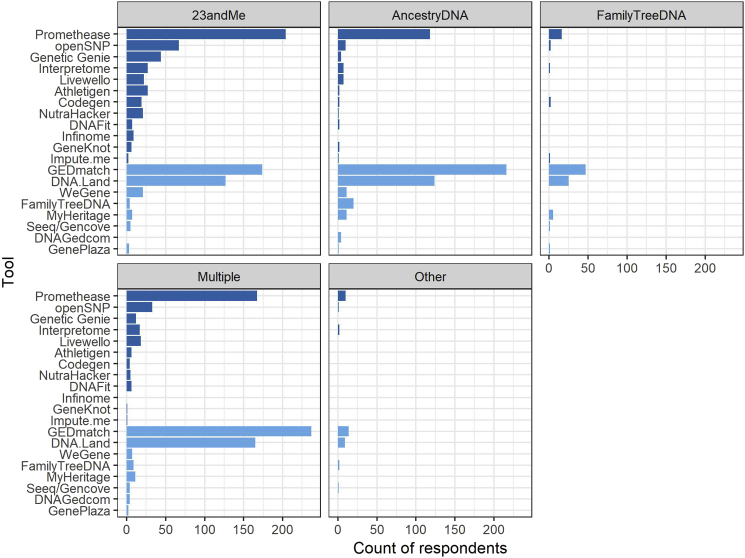
Next, we sought further understanding of what factors influenced an individual to use a given tool or combination of tools, in particular when respondents used multiple tools across the domains of health, ancestry, and genealogy. We performed a series of logistic regression analyses of tool use, separately for the 7 tools with at least 40 users: GEDmatch, Promethease, DNA.Land, openSNP, GeneticGenie, Interpretome, and Livewello (see Table S2). Desire to learn about ancestry was significantly and positively associated with GEDmatch (β = 0.957, p = 3.76e−05) and DNA.Land (β = 0.447, p = 0.00775). Desire to learn about personal risk for specific diseases was positively associated with use of Promethease (β = 0.604, p = 0.00146), Genetic Genie (β = 1.23, p = 0.000286), and Livewello (β = 1.30, p = 0.000766). Desire to obtain raw genetic data was positively associated with use of DNA.Land (β = 0.517, p = 0.00176) and Interpretome (β = 0.854, p = 0.0166). Survey recruitment venue was strongly associated with tool use, as GEDmatch and DNA.Land users were more likely to have been recruited via Facebook while Promethease users were more likely to have been recruited from Reddit or the openSNP newsletter. DTC test(s) ordered were also linked to tool use (see also Figure 2), with the most significant associations between 23andMe and Promethease (β = 0.791, p = 3.e−06) and FamilyTreeDNA and GEDmatch (β = 1.53, p = 0.000199).
Figure 2.
Tools Used Based on DTC Test(s) Taken
Results are plotted for 820 respondents who reported using at least one TPI tool. Darker blue shading on bars indicates tools that offer health-related information (see Table S1).
Categories of Third-Party Tool Use
The large proportion of respondents using multiple third-party tools, including tools spanning the disparate categories of health, ancestry, and genealogy, led us to next examine combinations of tools used. We grouped respondents into those using only health-related tools (n = 98 respondents); only non-health tools (n = 263); and those using both types (n = 458), which we refer to as the “crossover group” (see Table S1 for tool characterization). We present differences in DTC testing motivations, DTC test(s) ordered, and demographics between the three groups (health-only tool users, non-health only, and crossover group) in Tables 1 and 2. There was a linear trend in age, where non-health only tool users were oldest (mean age = 51.8), health only tool users were youngest (mean age = 39.4), and the crossover group was in between (mean age = 45.5). The health-only tool group had a significantly lower proportion of women (53% versus >68% in the other two groups) and higher proportion of respondents working in genetic research or medicine (13% versus ≤3% in the other two groups). The crossover group had a slightly higher proportion of respondents participating in (non-DTC) genetic research (19%) compared to the health only (15.6%) or non-health only (11.5%) user groups (p = 0.0215). DTC test(s) ordered were significantly different between the three groups, with the proportion of 23andMe customers increasing from the non-health group (40.7%) to crossover group (68.1%) to health-only group (87.8%) and a reverse pattern for both AncestryDNA and FamilyTreeDNA (proportion decreasing from non-health to crossover to health-only; see Table 2). Notable associations between DTC testing motivations and user group included increasing importance of general curiosity about genetics from non-health to crossover to health-only. Interest in having raw data was highest among the health-only tool group and lowest in the non-health only, suggesting that those who exclusively used health-related TPI tools were more highly motivated by raw data access when initially pursuing DTC testing.
Interviews with Crossover Tool Users
In follow-up interviews with respondents in the crossover group, we observed tool crossover in both directions: three initially used health-related tools before trying non-health tools, and four initially used non-health tools before trying health-related tools, illustrated by quotes in Box 1. In contrast to those who began with an interest in one domain then moved to another, for three interviewees who were either adopted or had an adopted parent, interest in health and genealogy were inextricably bound together in their search to learn simultaneously about their biological family and about potential implications for their own health risks.
Box 1. Interview Quotes (Participant ID in Parentheses).
Initial Use of Health-Related Tools:
|
Initial Use of Non-health Related Tools:
|
Inextricably Linked (e.g., for Adoptees):
|
The primary phenomena responsible for tool crossover were social networking, general curiosity, and initial lack of interesting findings. Facebook and Reddit were the primary social networks in which participants learned about multiple tools, including those seemingly outside the scope of the given Facebook group or sub-Reddit. Once participants learned about additional tools, they tried them often out of curiosity or general hunger to learn more, in particular to go beyond the information provided in DTC company reports. An initial lack of interesting findings in one domain also pushed some participants to seek out tools in another area.
Discussion
In this sample of DTC customers, we found high rates of raw data download and usage of TPI tools. Given some of our recruitment venues, this volume of TPI tool use may be expected; however, the scale and scope were notable. Specifically, respondents reported using on average three different TPI tools, often spanning a range of health, ancestry, and genealogy. Users of different tool categories (health, non-health, and both) differed in their demographics, motivations for DTC testing, and DTC tests taken. In follow-up interviews with a subset of crossover users (i.e., those using both health and non-health tools), we observed individuals often migrated to using tools outside their original scope of interest due to peer-to-peer sharing on social networks, general curiosity, and/or initial lack of interesting results. Below we discuss implications of these findings for the professional genetics community, including both researchers and providers.
Our findings suggest that in the research context, returning raw data to participants could present both a benefit and a liability to researchers. Raw data return may benefit the research community by increasing rates of participation in genetic research and engagement with and enthusiasm for genetics more broadly.12 In our survey, 35% of tool users reported that using third-party tools led them to participate in a genetic research study, while 76% agreed that it increased their understanding of genetics in general. The potential liability for the researcher community is that third-party tools vary widely in the quality, scope, and complexity of information returned,15, 35 in addition to having variable data security and privacy practices.36, 37 Addressing potential legal liability is outside the scope of this paper, but we suggest that researchers may bear some ethical responsibility if participants are harmed by downstream third-party tool usage in the way some DTC customers have been harmed.16, 17, 18, 19 Notably, in our results, only 6% of respondents reported feeling upset by information received from third-party tools while 35% felt confused. While not reported by a majority of our respondents, these negative outcomes merit further study and consideration on how to avoid and/or mitigate.
As with DTC testing, one concern with TPI tool usage is downstream overutilization of scarce health care resources, or “raiding of the medical commons.”38 Indeed, of the few accounts of TPI tool usage to date, several have focused on interactions with the health care system.20, 21, 22 These prior studies rightly illustrate the potentially alarming outcomes of patients’ misunderstandings of TPI reports; however, they do not indicate how often these scenarios result from “garden variety” TPI tool usage. In the current study, we observed a relatively low reported rate of sharing TPI results with HCPs (15%). This rate was lower than the 30% (of 321 surveyed) previously reported by Wang et al.,25 which may be due to our participants responding based on a specific tool rather than across all tools used. Indeed, when limiting to respondents who answered based on Promethease, our rate of HCP sharing was comparable (31%). Likely of concern to genetics professionals is that among respondents who shared with HCPs, the majority (79% of 116) did so with general practitioners or non-genetics specialists, rather than with medical geneticists or GCs. This will present challenges for primary care physicians, especially given that TPI reports are typically longer and harder to digest compared to DTC company reports.15, 20, 35
Regardless of whether they brought TPI reports to providers, respondents were frequently engaging with health information via TPI (i.e., 68% of tool users used health-related tools; see also Table S1). This was true even for respondents who were initially intent on finding relatives and receiving genetic ancestry percentages. Due to the flow of information on social media platforms such as Facebook and Reddit, respondents who started off using tools in one domain often switched or “crossed over” to using tools in another. This has implications for those whose TPI reports eventually do prompt them to interact with the health care system. For example, those initially interested in genealogy who later use health tools may be more likely to overestimate the reliability and comprehensiveness of the health information. There is relatively little uncertainty in identifying close relatives from genotyping array information; as one interviewee said about genetic genealogy, “DNA doesn’t lie.” This is in stark contrast to disease prediction based on similar data, which is far more probabilistic and uncertain. Furthermore, the raw data file is incomplete given that it is often based on array genotyping rather than genome sequencing, and may even be incorrect.22 The opposite effect is also possible: those involved in both paper trail and genetic genealogy may realize that genetics alone does not provide complete information when researching family history and so may be better equipped to understand limitations of health-related genetic information. Indeed, some individuals who primarily pursue ancestry testing over health-related testing may do so because they perceive the latter to lack accuracy and utility. Providers who better understand the course patients have taken to leverage their raw data via TPI may be better equipped to calibrate and manage patient expectations and understanding.
Another important finding is that approximately 40% of tool users agreed they had received health information, including about disease risk. This contrasts with our prior study of tool developers in which they characterized tools’ direct linking to scientific publications or variant annotation databases as merely “bridging to the literature” and hence stopping short of actual interpretation.15 However, our survey data suggest users regard TPI reports as providing personally relevant health information. At the same time, developers’ claims that “bridging” may increase understanding of genetic risk15, 35 were supported by our survey results: the majority of respondents agreed that using TPI tools increased their understanding of genetics in general (76%) and in particular how DTC companies interpret genetic data (67%).
To our knowledge, this is the largest study to date of raw data and TPI tool usage. However, our recruitment of a convenience sample via social media limits the ability to generalize findings to DTC customers more broadly, as it is difficult to know how our survey respondents compare to the larger population of DTC customers. It is reasonable to infer, however, that our respondents are likely highly motivated individuals who were active in these online forums and thus may overestimate the degree of data download and tool usage. The relatively small number of non-downloaders may not have provided enough power to detect differences with downloaders. However, we contend that limited generalizability is mitigated in part by our collection of qualitative data through open text survey responses and follow-up interviews, which generated deeper understanding of users’ motivations and experiences. Convenience sampling also allowed a rapid collection of a large number of respondents already engaged in the topics of interest, which seems appropriate for gaining preliminary insight into a relatively understudied area. Furthermore, by recruiting from social media venues where individuals were likely to be engaging with raw data and third-party interpretation, we were able to observe more of the phenomena of interest. The length of the survey may have contributed to the 24% non-completion rate and therefore potential survey item response bias, though compared to prior surveys,25 we collected more extensive and granular information about specific tool usage. Some incongruities between information reportedly received and the actual offerings of tools suggests that respondents may not have limited responses to the selected tool as directed; however, these incongruities were not widespread.
We have focused on the consumer genomics context as it is currently the most common way to access raw data. However, this work can help explore how broadening routes of access may unfold. We have discussed return of raw data from genetic research above, but another potential route is through clinical sequencing. Since 2014, the HIPAA direct access right has allowed individuals to access the contents of their designated record sets,39 which for clinical sequencing laboratories would likely include uninterpreted sequence data.40 Laboratories are not required to provide additional explanation or interpretation, which may lead recipients to seek out TPI. Indeed, many TPI tools accept the Variant Call Format (VCF) file type common to genome and exome sequencing.15 Future research should evaluate how individuals’ interactions with their raw data potentially differ across the contexts of DTC testing, clinical sequencing, and return of results from research.
In summary, moving forward individuals will have increasing routes to access their raw genetic data and leverage it in an expanding menu of largely unregulated TPI services. These activities raise a set of concerns related to but distinct from DTC genetic testing and thus merit further investigation to more fully understand potential harms and benefits. Rather than taking sides in a potential ensuing “culture war” about raw data,41 the professional genetics community has an opportunity to proactively engage with users, understand the complexity of their motivations for pursing third-party analysis, and ultimately educate them about potential limitations.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank our participants as well as the administrators of the Facebook groups and openSNP newsletter who aided in recruitment. Interview transcription was supported by funds from the UW Institute for Public Health Genetics. This work was partially supported by the National Human Genome Research Institute (NHGRI) and the National Cancer Institute (NCI) CSER Consortium, U01 HG006507 and U24 HG007307 (Jarvik, PI). This research used statistical consulting resources provided by the Center for Statistics and the Social Sciences at UW. REDCap and the Participant Portal at ITHS are supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1 TR002319.
Published: June 13, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.05.014.
Web Resources
Service used to transcribe audio recordings from participant interviews, https://www.rev.com
Survey instrument, quantitative survey dataset, and R analysis code, http://doi.org/10.3886/E105721V3
Supplemental Data
References
- 1.Thorogood A., Bobe J., Prainsack B., Middleton A., Scott E., Nelson S., Corpas M., Bonhomme N., Rodriguez L.L., Murtagh M., Kleiderman E., Participant Values Task Team of the Global Alliance for Genomics and Health APPLaUD: access for patients and participants to individual level uninterpreted genomic data. Hum. Genomics. 2018;12:7. doi: 10.1186/s40246-018-0139-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Thorogood, A., Bobe, J., Prainsack, B., Middleton, A., Scott, E., Nelson, S., Corpas, M., Bonhomme, N., Rodriguez, L.L., Murtagh, M., and Kleiderman, E.; Participant Values Task Team of the Global Alliance for Genomics and Health (2018). APPLaUD: access for patients and participants to individual level uninterpreted genomic data. Hum. Genomics 12, 7. [DOI] [PMC free article] [PubMed]
- 2.Nelson S. Geneticists should offer data to participants. Nature. 2016;539 doi: 10.1038/539007a. 7–7. [DOI] [PubMed] [Google Scholar]; Nelson, S. (2016). Geneticists should offer data to participants. Nature 539, 7-7. [DOI] [PubMed]
- 3.Lunshof J.E., Church G.M., Prainsack B. Information access. Raw personal data: providing access. Science. 2014;343:373–374. doi: 10.1126/science.1249382. [DOI] [PubMed] [Google Scholar]; Lunshof, J.E., Church, G.M., and Prainsack, B. (2014). Information access. Raw personal data: providing access. Science 343, 373-374. [DOI] [PubMed]
- 4.Beskow L.M., Burke W. Offering individual genetic research results: context matters. Sci. Transl. Med. 2010;2:38cm20. doi: 10.1126/scitranslmed.3000952. [DOI] [PMC free article] [PubMed] [Google Scholar]; Beskow, L.M., and Burke, W. (2010). Offering individual genetic research results: context matters. Sci. Transl. Med. 2, 38cm20. [DOI] [PMC free article] [PubMed]
- 5.Bredenoord A.L., Kroes H.Y., Cuppen E., Parker M., van Delden J.J.M. Disclosure of individual genetic data to research participants: the debate reconsidered. Trends Genet. 2011;27:41–47. doi: 10.1016/j.tig.2010.11.004. [DOI] [PubMed] [Google Scholar]; Bredenoord, A.L., Kroes, H.Y., Cuppen, E., Parker, M., and van Delden, J.J.M. (2011). Disclosure of individual genetic data to research participants: the debate reconsidered. Trends Genet. 27, 41-47. [DOI] [PubMed]
- 6.Wright C.F., Middleton A., Barrett J.C., Firth H.V., FitzPatrick D.R., Hurles M.E., Parker M. Returning genome sequences to research participants: Policy and practice. Wellcome Open Res. 2017;2:15. doi: 10.12688/wellcomeopenres.10942.1. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wright, C.F., Middleton, A., Barrett, J.C., Firth, H.V., FitzPatrick, D.R., Hurles, M.E., and Parker, M. (2017). Returning genome sequences to research participants: Policy and practice. Wellcome Open Res 2, 15. [DOI] [PMC free article] [PubMed]
- 7.Middleton A., Wright C.F., Morley K.I., Bragin E., Firth H.V., Hurles M.E., Parker M., DDD study Potential research participants support the return of raw sequence data. J. Med. Genet. 2015;52:571–574. doi: 10.1136/jmedgenet-2015-103119. [DOI] [PMC free article] [PubMed] [Google Scholar]; Middleton, A., Wright, C.F., Morley, K.I., Bragin, E., Firth, H.V., Hurles, M.E., and Parker, M.; DDD study (2015). Potential research participants support the return of raw sequence data. J. Med. Genet. 52, 571-574. [DOI] [PMC free article] [PubMed]
- 8.Karow J. GenomeWeb; 2018. All of Us Program Plans to Return Disease Variants, PGx Results, Primary Genomic Data. [Google Scholar]; Karow, J. (2018). All of Us Program Plans to Return Disease Variants, PGx Results, Primary Genomic Data (GenomeWeb).
- 9.National Institutes of Health (2018). Informational Webinar on the “All of Us Genetic” Counseling Resource Funding Announcement, https://allofus.nih.gov/sites/default/files/genetic_counseling_resource_webinar.pdf.
- 10.NASEM . National Academies Press; Washington, D.C.: 2018. Returning Individual Research Results to Participants: Guidance for a New Research Paradigm. [PubMed] [Google Scholar]; NASEM (2018). Returning Individual Research Results to Participants: Guidance for a New Research Paradigm (Washington, D.C.: National Academies Press). [PubMed]
- 11.Haeusermann T., Fadda M., Blasimme A., Tzovaras B.G., Vayena E. Genes wide open: Data sharing and the social gradient of genomic privacy. AJOB Empir. Bioeth. 2018;9:207–221. doi: 10.1080/23294515.2018.1550123. [DOI] [PubMed] [Google Scholar]; Haeusermann, T., Fadda, M., Blasimme, A., Tzovaras, B.G., and Vayena, E. (2018). Genes wide open: Data sharing and the social gradient of genomic privacy. AJOB Empir. Bioeth. 9, 207-221. [DOI] [PubMed]
- 12.Haeusermann T., Greshake B., Blasimme A., Irdam D., Richards M., Vayena E. Open sharing of genomic data: Who does it and why? PLoS ONE. 2017;12:e0177158. doi: 10.1371/journal.pone.0177158. [DOI] [PMC free article] [PubMed] [Google Scholar]; Haeusermann, T., Greshake, B., Blasimme, A., Irdam, D., Richards, M., and Vayena, E. (2017). Open sharing of genomic data: Who does it and why? PLoS ONE 12, e0177158. [DOI] [PMC free article] [PubMed]
- 13.Ball M.P., Bobe J.R., Chou M.F., Clegg T., Estep P.W., Lunshof J.E., Vandewege W., Zaranek A., Church G.M. Harvard Personal Genome Project: lessons from participatory public research. Genome Med. 2014;6:10. doi: 10.1186/gm527. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ball, M.P., Bobe, J.R., Chou, M.F., Clegg, T., Estep, P.W., Lunshof, J.E., Vandewege, W., Zaranek, A., and Church, G.M. (2014). Harvard Personal Genome Project: lessons from participatory public research. Genome Med. 6, 10. [DOI] [PMC free article] [PubMed]
- 14.Swan M. Crowdsourced health research studies: an important emerging complement to clinical trials in the public health research ecosystem. J. Med. Internet Res. 2012;14:e46. doi: 10.2196/jmir.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]; Swan, M. (2012). Crowdsourced health research studies: an important emerging complement to clinical trials in the public health research ecosystem. J. Med. Internet Res. 14, e46. [DOI] [PMC free article] [PubMed]
- 15.Nelson S.C., Fullerton S.M. “Bridge to the Literature”? Third-Party Genetic Interpretation Tools and the Views of Tool Developers. J. Genet. Couns. 2018;27:770–781. doi: 10.1007/s10897-018-0217-9. [DOI] [PubMed] [Google Scholar]; Nelson, S.C., and Fullerton, S.M. (2018). “Bridge to the Literature”? Third-Party Genetic Interpretation Tools and the Views of Tool Developers. J. Genet. Couns. 27, 770-781. [DOI] [PubMed]
- 16.Kolata, G. (2018). The Online Gene Test Finds a Dangerous Mutation. It May Well Be Wrong. New York Times, July 3, 2018. D1. https://www.nytimes.com/2018/07/02/health/gene-testing-disease-nyt.html.
- 17.Almendrala, A. (2018). Home Genetic Tests May Be Riddled With Errors, And Companies Aren’t Keeping Track. Huffingt. Post, April 3, 2018. https://www.huffpost.com/entry/home-genetic-test-false-positives_n_5ac27188e4b04646b6451c42.
- 18.Hercher, L. (2018). 23andMe Said He Would Lose His Mind. Ancestry Said the Opposite. Which Was Right? New York Times, September 16, 2018. SR7. https://www.nytimes.com/2018/09/15/opinion/sunday/23andme-ancestry-alzheimers-genetic-testing.html.
- 19.Matloff, E. (2019). I Had Lynch Syndrome For 30 Hours. Forbes, February 12, 2019. https://www.forbes.com/sites/ellenmatloff/2019/02/12/i-had-lynch-syndrome-for-30-hours-2/#66ed644a2567.
- 20.Allen C.G., Gabriel J., Flynn M., Cunningham T.N., Wang C. The impact of raw DNA availability and corresponding online interpretation services: A mixed-methods study. Transl. Behav. Med. 2018;8:105–112. doi: 10.1093/tbm/ibx009. [DOI] [PubMed] [Google Scholar]; Allen, C.G., Gabriel, J., Flynn, M., Cunningham, T.N., and Wang, C. (2018). The impact of raw DNA availability and corresponding online interpretation services: A mixed-methods study. Transl. Behav. Med. 8, 105-112. [DOI] [PubMed]
- 21.Moscarello T., Murray B., Reuter C.M., Demo E. Direct-to-consumer raw genetic data and third-party interpretation services: more burden than bargain? Genet. Med. 2019;21:539–541. doi: 10.1038/s41436-018-0097-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; Moscarello, T., Murray, B., Reuter, C.M., and Demo, E. (2019). Direct-to-consumer raw genetic data and third-party interpretation services: more burden than bargain? Genet. Med. 21, 539-541. [DOI] [PMC free article] [PubMed]
- 22.Tandy-Connor S., Guiltinan J., Krempely K., LaDuca H., Reineke P., Gutierrez S., Gray P., Tippin Davis B. False-positive results released by direct-to-consumer genetic tests highlight the importance of clinical confirmation testing for appropriate patient care. Genet. Med. 2018;20:1515–1521. doi: 10.1038/gim.2018.38. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tandy-Connor, S., Guiltinan, J., Krempely, K., LaDuca, H., Reineke, P., Gutierrez, S., Gray, P., and Tippin Davis, B. (2018). False-positive results released by direct-to-consumer genetic tests highlight the importance of clinical confirmation testing for appropriate patient care. Genet. Med. 20, 1515-1521. [DOI] [PMC free article] [PubMed]
- 23.Erlich Y., Shor T., Pe’er I., Carmi S. Identity inference of genomic data using long-range familial searches. Science. 2018;362:690–694. doi: 10.1126/science.aau4832. [DOI] [PMC free article] [PubMed] [Google Scholar]; Erlich, Y., Shor, T., Pe’er, I., and Carmi, S. (2018). Identity inference of genomic data using long-range familial searches. Science 362, 690-694. [DOI] [PMC free article] [PubMed]
- 24.Ram N., Guerrini C.J., McGuire A.L. Genealogy databases and the future of criminal investigation. Science. 2018;360:1078–1079. doi: 10.1126/science.aau1083. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ram, N., Guerrini, C.J., and McGuire, A.L. (2018). Genealogy databases and the future of criminal investigation. Science 360, 1078-1079. [DOI] [PMC free article] [PubMed]
- 25.Wang C., Cahill T.J., Parlato A., Wertz B., Zhong Q., Cunningham T.N., Cummings J.J. Consumer use and response to online third-party raw DNA interpretation services. Mol. Genet. Genomic Med. 2018;6:35–43. doi: 10.1002/mgg3.340. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang, C., Cahill, T.J., Parlato, A., Wertz, B., Zhong, Q., Cunningham, T.N., and Cummings, J.J. (2018). Consumer use and response to online third-party raw DNA interpretation services. Mol. Genet. Genomic Med. 6, 35-43. [DOI] [PMC free article] [PubMed]
- 26.Yuan J., Gordon A., Speyer D., Aufrichtig R., Zielinski D., Pickrell J., Erlich Y. DNA.Land is a framework to collect genomes and phenomes in the era of abundant genetic information. Nat. Genet. 2018;50:160–165. doi: 10.1038/s41588-017-0021-8. [DOI] [PubMed] [Google Scholar]; Yuan, J., Gordon, A., Speyer, D., Aufrichtig, R., Zielinski, D., Pickrell, J., and Erlich, Y. (2018). DNA.Land is a framework to collect genomes and phenomes in the era of abundant genetic information. Nat. Genet. 50, 160-165. [DOI] [PubMed]
- 27.Greshake B., Bayer P.E., Rausch H., Reda J. openSNP--a crowdsourced web resource for personal genomics. PLoS ONE. 2014;9:e89204. doi: 10.1371/journal.pone.0089204. [DOI] [PMC free article] [PubMed] [Google Scholar]; Greshake, B., Bayer, P.E., Rausch, H., and Reda, J. (2014). openSNP--a crowdsourced web resource for personal genomics. PLoS ONE 9, e89204. [DOI] [PMC free article] [PubMed]
- 28.Carere D.A., Couper M.P., Crawford S.D., Kalia S.S., Duggan J.R., Moreno T.A., Mountain J.L., Roberts J.S., Green R.C., PGen Study Group Design, methods, and participant characteristics of the Impact of Personal Genomics (PGen) Study, a prospective cohort study of direct-to-consumer personal genomic testing customers. Genome Med. 2014;6:96. doi: 10.1186/s13073-014-0096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; Carere, D.A., Couper, M.P., Crawford, S.D., Kalia, S.S., Duggan, J.R., Moreno, T.A., Mountain, J.L., Roberts, J.S., and Green, R.C.; PGen Study Group (2014). Design, methods, and participant characteristics of the Impact of Personal Genomics (PGen) Study, a prospective cohort study of direct-to-consumer personal genomic testing customers. Genome Med. 6, 96. [DOI] [PMC free article] [PubMed]
- 29.Krosnick J.A., Presser S. Question and Questionnaire Design. In: Marsden P.V., Wright J.D., editors. Handbook of Survey Research. Emerald; 2010. pp. 263–313. [Google Scholar]; Krosnick, J.A., and Presser, S. (2010). Question and Questionnaire Design. In Handbook of Survey Research, P.V. Marsden and J.D. Wright, eds. (Emerald), pp. 263-313.
- 30.Harris P.A., Taylor R., Thielke R., Payne J., Gonzalez N., Conde J.G. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; Harris, P.A., Taylor, R., Thielke, R., Payne, J., Gonzalez, N., and Conde, J.G. (2009). Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 42, 377-381. [DOI] [PMC free article] [PubMed]
- 31.R Core Team . R Foundation for Statistical Computing; 2013. R: A language and environment for statistical computing. [Google Scholar]; R Core Team (2013). R: A language and environment for statistical computing (R Foundation for Statistical Computing).
- 32.Baptista N.M., Christensen K.D., Carere D.A., Broadley S.A., Roberts J.S., Green R.C. Adopting genetics: motivations and outcomes of personal genomic testing in adult adoptees. Genet. Med. 2016;18:924–932. doi: 10.1038/gim.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]; Baptista, N.M., Christensen, K.D., Carere, D.A., Broadley, S.A., Roberts, J.S., and Green, R.C. (2016). Adopting genetics: motivations and outcomes of personal genomic testing in adult adoptees. Genet. Med. 18, 924-932. [DOI] [PMC free article] [PubMed]
- 33.Koeller D.R., Uhlmann W.R., Carere D.A., Green R.C., Roberts J.S., PGen Study Group Utilization of Genetic Counseling after Direct-to-Consumer Genetic Testing: Findings from the Impact of Personal Genomics (PGen) Study. J. Genet. Couns. 2017;26:1270–1279. doi: 10.1007/s10897-017-0106-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; Koeller, D.R., Uhlmann, W.R., Carere, D.A., Green, R.C., and Roberts, J.S.; PGen Study Group (2017). Utilization of Genetic Counseling after Direct-to-Consumer Genetic Testing: Findings from the Impact of Personal Genomics (PGen) Study. J. Genet. Couns. 26, 1270-1279. [DOI] [PMC free article] [PubMed]
- 34.Kirkpatrick B.E., Rashkin M.D. Ancestry Testing and the Practice of Genetic Counseling. J. Genet. Couns. 2017;26:6–20. doi: 10.1007/s10897-016-0014-2. [DOI] [PubMed] [Google Scholar]; Kirkpatrick, B.E., and Rashkin, M.D. (2017). Ancestry Testing and the Practice of Genetic Counseling. J. Genet. Couns. 26, 6-20. [DOI] [PubMed]
- 35.Badalato L., Kalokairinou L., Borry P. Third party interpretation of raw genetic data: an ethical exploration. Eur. J. Hum. Genet. 2017;25:1189–1194. doi: 10.1038/ejhg.2017.126. [DOI] [PMC free article] [PubMed] [Google Scholar]; Badalato, L., Kalokairinou, L., and Borry, P. (2017). Third party interpretation of raw genetic data: an ethical exploration. Eur. J. Hum. Genet. 25, 1189-1194. [DOI] [PMC free article] [PubMed]
- 36.Ney P.M., Ceze L., Kohno T. Computer Security Risks of Distant Relative Matching in Consumer Genetic Databases. ArXiv. 2018 1810.02895. [Google Scholar]; Ney, P.M., Ceze, L., and Kohno, T. (2018). Computer Security Risks of Distant Relative Matching in Consumer Genetic Databases. ArXiv, 1810.02895.
- 37.Hazel J.W., Slobogin C. Who Knows What, and When?: A Survey of the Privacy Policies Proffered by U.S. Direct-to-Consumer Genetic Testing Companies. Cornell J. Law Public Policy. 2018;28:35–66. [PubMed] [Google Scholar]; Hazel, J.W., and Slobogin, C. (2018). Who Knows What, and When?: A Survey of the Privacy Policies Proffered by U.S. Direct-to-Consumer Genetic Testing Companies. Cornell J. Law Public Policy 28, 35-66. [PubMed]
- 38.McGuire A.L., Burke W. An unwelcome side effect of direct-to-consumer personal genome testing: raiding the medical commons. JAMA. 2008;300:2669–2671. doi: 10.1001/jama.2008.803. [DOI] [PMC free article] [PubMed] [Google Scholar]; McGuire, A.L., and Burke, W. (2008). An unwelcome side effect of direct-to-consumer personal genome testing: raiding the medical commons. JAMA 300, 2669-2671. [DOI] [PMC free article] [PubMed]
- 39.U.S. DHHS (2014). CLIA Program and HIPAA Privacy Rule; Patients’ Access to Test Reports. 42 CFR § 493, 45 CFR § 164. [PubMed]
- 40.U.S. DHHS (2016). Individuals’ right under HIPAA to access their health information 45 CFR § 164.524.
- 41.Evans J.P., Green R.C. Direct to consumer genetic testing: Avoiding a culture war. Genet. Med. 2009;11:568–569. doi: 10.1097/GIM.0b013e3181afbaed. [DOI] [PMC free article] [PubMed] [Google Scholar]; Evans, J.P., and Green, R.C. (2009). Direct to consumer genetic testing: Avoiding a culture war. Genet. Med. 11, 568-569. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.