Abstract
Burkholderia cenocepacia is an opportunistic bacterial pathogen that poses a significant threat to individuals with cystic fibrosis by provoking a strong inflammatory response within the lung. It possesses a type VI secretion system (T6SS), a secretory apparatus that can perforate the cellular membrane of other bacterial species and/or eukaryotic targets, to deliver an arsenal of effector proteins. The B. cenocepacia T6SS (T6SS‐1) has been shown to be implicated in virulence in rats and contributes toward actin rearrangements and inflammasome activation in B. cenocepacia‐infected macrophages. Here, we present bioinformatics evidence to suggest that T6SS‐1 is the archetype T6SS in the Burkholderia genus. We show that B. cenocepacia T6SS‐1 is active under normal laboratory growth conditions and displays antibacterial activity against other Gram‐negative bacterial species. Moreover, B. cenocepacia T6SS‐1 is not required for virulence in three eukaryotic infection models. Bioinformatics analysis identified several candidate T6SS‐dependent effectors that may play a role in the antibacterial activity of B. cenocepacia T6SS‐1. We conclude that B. cenocepacia T6SS‐1 plays an important role in bacterial competition for this organism, and probably in all Burkholderia species that possess this system, thereby broadening the range of species that utilize the T6SS for this purpose.
Keywords: antibacterial, bacterial competition, Burkholderia, protein secretion, T6SS, type VI secretion system
1. INTRODUCTION
Bacteria utilize many systems to establish a niche, including mechanisms to exploit eukaryotic organisms and/or to compete effectively with other bacterial species colonizing the same ecosystem. Many Gram‐negative bacteria possess a protein secretion system termed the type VI secretion system (T6SS) that participates in one or both processes, depending on the species (Ho, Dong, & Mekalanos, 2014). The T6SS is found in ~25% of Gram‐negative species (Bingle, Bailey, & Pallen, 2008), including the human pathogens Pseudomonas aeruginosa (Mougous et al., 2006), Vibrio cholerae (Pukatzki et al., 2006), Serratia marcescens (Murdoch et al., 2011), and Burkholderia pseudomallei (Burtnick et al., 2011). The system is composed of multiple copies of at least thirteen different subunits (TssA‐TssM) and a single copy of the PAAR protein, which are organized into a dynamic protein injection machine containing two distinct interacting subassemblies (Basler, 2015). The first is a contractile structure that shares homology with components of the T4 bacteriophage tail and is comprised of multimers of TssD (also termed Hcp) that assemble into a tube that is sharpened at one end by a trimer of TssI (also known as VgrG) subunits capped by a monomer of the PAAR protein. The tube, in turn, is surrounded by a contractile sheath composed of polymerized TssBC subunits. The tube–sheath structure is assembled on a platform known as the baseplate that consists of the TssEFGK subunits (Brackmann, Nazarov, Wang, & Basler, 2017; Brunet, Zoued, Boyer, Douzi, & Cascales, 2015; Leiman et al., 2009; Nguyen et al., 2017). The second subassembly, composed of the TssJLM subunits, is a channel/chamber complex that spans the inner membrane, periplasm, and outer membrane, and serves to anchor the contractile machinery to the bacterial cell envelope (Brunet et al., 2015; Durand et al., 2015; Nguyen et al., 2017). The role of the TssA subunit is less certain, but it has been proposed to play roles in priming and polymerization of the tube–sheath structure or act as a baseplate component (Planamente et al., 2016; Zoued et al., 2016).
Contraction of the sheath against the baseplate drives the sharpened inner tube through the chamber complex to the exterior where it punctures the cellular membrane of a neighboring target cell. Effector proteins, which may be noncovalently associated with the TssD, TssI, or PAAR subunits (“cargo” effectors) or occur as additional domains on these proteins (“specialized” effectors), are thus delivered into the target cell where they kill or subvert the recipient (Durand, Cambillau, Cascales, & Journet, 2014). In many T6SS‐containing bacteria, these targets are other competing species of bacteria, and so the system plays a major role in bacterial competition (Diniz & Coulthurst, 2015; Hood et al., 2010; MacIntyre, Miyata, Kitaoka, & Pukatzki, 2010; Schwarz et al., 2010). Such T6SS‐dependent competition can occur in a variety of environments, including plant hosts (Ma, Hachani, Lin, Filloux, & Lai, 2014) or the mammalian gut (Chassaing & Cascales, 2018; Sana et al., 2016; Zhao, Caro, Robins, & Mekalanos, 2018). Some T6SSs also specifically target eukaryotic cells and have more of a direct role in virulence, including the T6SS‐5 of B. pseudomallei and H2‐ and H3‐T6SS of P. aeruginosa (Burtnick et al., 2011; Jiang, Waterfield, Yang, Yang, & Jin, 2014; Sana et al., 2012).
A variety of T6SS‐dependent effectors and cognate immunity proteins have now been described, including superfamilies of antibacterial effectors. These include effectors that target the peptidoglycan layer, phospholipid membrane, or host DNA/RNA, such as the amidase effector–immunity pairs termed Tae‐Tai (for type VI amidase effector/immunity; Hood et al., 2010; Russell et al., 2011; Russell et al., 2012; Fritsch et al., 2013), the type VI lipase effectors (Tle) that possess phospholipase A1, A2, or D activity (Russell et al., 2013), or the type VI DNase effectors (Tde; Ma et al., 2014), respectively. A number of anti‐eukaryotic effectors have also been described, including a P. aeruginosa effector with phospholipase D activity that can target both bacterial and eukaryotic cells (Jiang et al., 2014), the catalase effector, KatN, responsible for intramacrophage survival of enterohemorrhagic E. coli (Wan et al., 2017), and a VgrG subunit with a C‐terminal actin cross‐linking domain utilized by V. cholerae (VgrG‐1) that impairs the phagocytic activity of eukaryotic host cells (Ma, McAuley, Pukatzki, & Mekalanos, 2009; Pukatzki, Ma, Revel, Sturtevant, & Mekalanos, 2007).
The genus Burkholderia constitutes a large and diverse group of Gram‐negative bacterial species, including primary and opportunistic human pathogens, plant pathogens, and plant‐associated species with biocontrol properties (Eberl & Vandamme, 2016). Recently, the classification of the Burkholderia has undergone a proposed revision, with all members of the Burkholderia cepacia complex (Bcc) and Pseudomallei groups, together with some phytopathogenic species, remaining as Burkholderia, while all the other species (typically nonpathogenic environmental strains) have been reassigned to the new genera Paraburkholderia (Sawana, Adeolu, & Gupta, 2014) and Caballeronia (Dobritsa, Linardopoulou, & Samadpour, 2017). The Bcc is a group of at least twenty closely related species that have gained notoriety as opportunistic respiratory pathogens in cystic fibrosis (CF) patients, as some strains are highly transmissible between individuals and the resulting infections can be difficult to treat effectively and result in fatal pneumonia and septicemia (Depoorter et al., 2016; Drevinek & Mahenthiralingam, 2010). One of the most prevalent Bcc species in CF infections is B. cenocepacia. However, despite many studies investigating the virulence mechanisms of this bacterium, the molecular pathogenesis of B. cenocepacia infection is not fully understood. Numerous strategies have been proposed to account for its virulence, including its ability to invade and survive intracellularly within host cells (Burns et al., 1996; Cieri, Mayer‐Hamblett, Griffith, & Burns, 2002; Gavrilin et al., 2012; Martin & Mohr, 2000; McKeon, McClean, & Callaghan, 2010; Mesureur et al., 2017), induce pro‐inflammatory responses (Kotrange et al., 2011; Mesureur et al., 2017), scavenge iron (reviewed in Butt & Thomas, 2017), and secrete hydrolytic enzymes such as zinc metalloproteases (Corbett, Burtnick, Kooi, Woods, & Sokol, 2003; Sokol et al., 2003).
As many as eight different T6SSs have been identified across the redefined Burkholderia genus, with anywhere up to six of them being encoded in the genome of an individual species (Angus et al., 2014; Shalom, Shaw, & Thomas, 2007). The six T6SSs in B. pseudomallei have been described using two numbering systems (Schell et al., 2007; Shalom et al., 2007), with a further two T6SSs identified in other Burkholderia species referred to as T6SSa and T6SSb (Angus et al., 2014). In the present investigation, we have adopted the nomenclature of Shalom et al., 2007, and for consistency, we refer to T6SSa and T6SSb as T6SS‐7 and T6SS‐8, respectively. B. cenocepacia strains are generally considered to contain only a single T6SS that corresponds to T6SS‐1 of B. pseudomallei and B. thailandensis (Angus et al., 2014; Aubert, Flannagan, & Valvano, 2008; Aubert, Hu, & Valvano, 2015; Schwarz et al., 2010; Shalom et al., 2007).
The T6SS‐1 in the epidemic B. cenocepacia CF isolate K56‐2 was shown to contribute to bacterial survival within a rat model of chronic lung infection (Hunt, Kooi, Sokol, & Valvano, 2004). Subsequent work has suggested that T6SS‐1 is responsible for the ability of B. cenocepacia to subvert predatory eukaryotic cells, including the amoeba Dictyostelium discoideum and murine and human monocyte‐derived macrophages, and this involves actin cytoskeletal rearrangement (Aubert et al., 2008; Xu et al., 2014). The T6SS‐1 has been shown to exert its effect on cytoskeletal rearrangement through Rho GTPase inactivation (Aubert et al., 2008; Flannagan et al., 2012; Keith, Hynes, Sholdice, & Valvano, 2009; Rosales‐Reyes, Skeldon, Aubert, & Valvano, 2012). More recent studies have suggested that the T6SS‐dependent interactions between B. cenocepacia and human‐derived phagocytic cells are important for triggering an innate immune response through pyrin inflammasome activation upon GTPase inactivation, which may promote bacterial clearance and protection from potentially lethal infections in a mouse model (Aubert et al., 2016; Gavrilin et al., 2012; Xu et al., 2014). Several observations which have been attributed to T6SS‐1 activity have been obtained using a B. cenocepacia strain in which atsR, a gene encoding a hybrid sensor kinase, has been deleted. This results in upregulation of the system and allows for detection of T6SS‐1 secretion activity in a B. cenocepacia strain (Aubert et al., 2008, 2015).
Here, we present a bioinformatics analysis of the T6SS‐1 in the genus Burkholderia and related species. We demonstrate sufficient T6SS‐1 secretion activity in B. cenocepacia isolates growing under standard laboratory conditions to investigate the role of the T6SS in this Bcc species, without the need for upregulation of the system by atsR inactivation. From this, we provide evidence to support a functional role of the T6SS‐1 in B. cenocepacia in bacterial competition through a series of bacterial competition assays. The contribution of the T6SS‐1 to pathogenesis in three established eukaryotic models of B. cenocepacia infection was also investigated, but our results indicated that the system does not contribute to pathogenesis in these models.
2. MATERIALS AND METHODS
2.1. Strains, plasmids, and growth conditions
The bacterial strains and plasmids used in this study are indicated in Table A1 (Appendix 1). For cultivation of bacteria, strains were routinely grown in LB medium (E. coli, P. putida) or M9 minimal salts agar containing 0.5% glucose (B. cenocepacia) at 37°C. M9 minimal salts contained 42 mM Na2HPO4, 22 mM KH2PO4, 19 mM NH4Cl, 9 mM NaCl, 1 mM MgSO4, and 0.1 mM CaCl2. Antibiotics were used, when appropriate, at the following concentrations: ampicillin (Ap), 100 μg/ml (E. coli); chloramphenicol (Cm), 25 μg/ml (E. coli, P. putida) and 50–100 μg/ml (B. cenocepacia); kanamycin (Km), 50 μg/ml (E. coli and B. cenocepacia); rifampicin (Rf), 100 μg/ml (E. coli and B. cenocepacia); and trimethoprim (Tp), 25 μg/mL (E. coli), 25 μg/ml (B. cenocepacia H111 and Pc715j), and 100 μg/ml (B. cenocepacia K56‐2). For selection of trimethoprim resistance in E. coli, Iso‐Sensitest Agar (Oxoid) was employed, and for selection of kanamycin resistance in B. cenocepacia, Lennox agar was utilized. Dialyzed brain‐heart infusion (D‐BHI) broth was prepared according to Sokol, Ohman, and Iglewski (1979) and used as the liquid growth medium for cultures of B. cenocepacia undergoing secreted protein extraction.
2.2. DNA preparation and manipulation
Recombinant DNA techniques were performed essentially as described in Sambrook et al. (1989). DNA amplification by PCR was performed with KOD DNA polymerase enzyme (Millipore) or GoTaq G2 Flexi DNA Polymerase (Promega) according to manufacturer's instructions using boiled cell lysate as template DNA. Primers used in this study are indicated in Table A2 (Appendix 1) and were purchased from Eurogentec, Belgium. PCR products were purified from solution or by agarose gel extraction using a QIAquick PCR Purification Kit (Qiagen). DNA restriction enzymes were purchased from Promega or New England Biolabs. DNA was ligated using T4 DNA ligase (Promega). Nucleotide sequence determination was performed by the Core Genomic Facility at The University of Sheffield, UK. Genome sequencing was provided by MicrobesNG (https://www.microbesng.uk), Birmingham, UK. These sequence data have been submitted to the NCBI GenBank database under accession number MK051000. Details of data submission can be found at www.ncbi.nlm.nih.gov/genbank/.
2.3. Construction of B. cenocepacia strains and plasmids
Burkholderia cenocepacia chromosomal mutants with insertionally inactivated genes were generated by allelic replacement using the suicide vector pSHAFT2, as previously described (Shastri et al., 2017). Briefly, DNA fragments containing ~1,200 bp of the N‐terminal coding region of tssM (tssM’) and the entire tssK and tagY genes were amplified from B. cenocepacia H111 using primer pairs tssMfor and tssMrev, tssKfor and tssKrev, and tagYfor and tagYrev, respectively. Each gene/gene fragment was cloned into the vectors pBBR1MCS or pBluescriptII, where tssK was cloned between the restriction sites HindIII and BamHI, tssM’ between XbaI and XhoI, and tagY between BamHI and XhoI, generating pBBR1‐tssK, pBBR1‐tssM’, and pBluescript‐tagY. To disrupt each target gene, pBBR1‐tssK was restricted with EcoRI, pBBR1‐tssM’ with BamHI, and pBluescriptII‐tagY with ZraI, and ligated to the trimethoprim (dfrB2) resistance cassette that was excised from p34E‐Tp by EcoRI, BamHI, and SmaI, respectively. The disrupted alleles, tssK::Tp, tssM::Tp, or tagY::Tp, were then transferred to pSHAFT2 as XhoI‐NotI (tssK and tssM) or XhoI‐XbaI (tagY) fragments. pSHAFT2‐derived constructs were conjugated into B. cenocepacia strains H111, K56‐2, and Pc715j using E. coli donor strain S17‐1(λpir) according to Herrero, Lorenzo, and Timmis (1990) and de Lorenzo and Timmis (1994) and selected using M9 agar containing trimethoprim. The previously constructed pSHAFT2‐tssA::Tp plasmid was similarly introduced into K56‐2 and Pc715j. Double crossover recombinants were identified by chloramphenicol sensitivity and verified by PCR using primers pairs that annealed to genomic regions of the target gene located just outside the homologous region contained within the pSHAFT2 construct. See Appendix 2 for further details. Construction of the B. cenocepacia H111 tssM in‐frame deletion mutant has been described previously (Dix et al., 2018). The tssM complementation plasmid, pBBR1‐tssM(+), was constructed by amplifying tssM from B. cenocepacia H111 with primers tssMforAcc65I and tssMrevXbaI, and ligating the amplicon to the Acc65I and XbaI sites of pBBR1MCS, which places tssM under control of the vector lacZ promoter.
2.4. Extraction and detection of extracellular proteins
Culture supernatants were collected from 15 ml D‐BHI broth cultures of B. cenocepacia strains grown at 37°C until at OD600 of 0.6–0.8 and filter sterilized using a 0.22‐μM syringe‐driven filter unit. Sodium deoxycholate was added to supernatants to a final concentration of 0.2 mg/ml, which were then incubated on ice for 30 min. To precipitate proteins, TCA was added at 10% (w/v) final concentration and incubated overnight at −20°C. Supernatants were centrifuged to collect the protein pellets, which were then washed with acetone, collected by centrifugation, and air‐dried. Protein pellets were resolubilized with 15 μl of 1x SDS‐loading buffer (125 mM Tris‐HCl, 5% (w/v) SDS, 10% (v/v) glycerol, 5% (v/v) 2‐mercaptoethanol, 0.005% (w/v) bromophenol blue, pH 6.8). For cell‐associated protein fractions, the whole‐cell pellet was concentrated 20‐fold in PBS and combined with an equal volume of 2x SDS‐sample buffer.
Protein samples were separated in a 15% SDS–polyacrylamide gel, transferred onto 0.45‐μM PVDF membrane (Millipore), and incubated for 1 hr in blocking solution (5% (w/v) milk, TBS, 0.05% (v/v) Tween‐20). TssD secretion was analyzed by Western blotting as standard protocol using a custom rat antibody raised against purified recombinant TssD (The University of Sheffield Biological Services, 1:2,000) and goat anti‐rat HRP secondary antibody (SouthernBiotech, 1:5,000). RNA polymerase β‐subunit was detected as a lysis control using a monoclonal mouse anti‐RNA polymerase β‐subunit primary antibody (1:2,500, NeoClone) and rabbit anti‐mouse HRP secondary antibody (Thermo Scientific, 1:5,000).
2.5. Bacterial competition assay
Attacker (B. cenocepacia) and prey (e.g., P. putida, E. coli CC118(λpir)) strains were grown overnight in LB at 37°C. Each culture was then normalized to an OD600 of 0.5. Bacterial suspensions were combined in a 5:1 ratio of attacker:prey. Monoculture controls of target and attacker strains with LB were included using the same number of bacteria as in the attacker:prey sample, respectively. 25 μl of each coculture and control culture was spread over a 0.45‐μm nitrocellulose filter membrane on a prewarmed LB agar plate and incubated at 30°C for 4 hr. After incubation, bacteria from each filter membrane were harvested in 1 ml LB and 10−1 to 10−5 serial dilutions made. 10 μl of each dilution was spotted onto selection plates in triplicate using the surface viable count method (Miles, Misra, & Irwin, 1938). B. cenocepacia was selected by Tc resistance, P. putida by Cm resistance, E. coli CC118(λpir) by Rf resistance, and E. coli SM10(λpir) by Km resistance. Plates were incubated at either 37°C or 30°C overnight, dependent on the strain. The number of viable CFU was counted and used to calculate the CFU/mL for each coculture or control culture tested. All experiments were carried out at least three times.
2.6. Galleria mellonella larvae killing assay
Final‐instar Galleria mellonella larvae were purchased fresh from Livefood UK and maintained at 4°C before infection. For preparation of bacteria for injection, B. cenocepacia K56‐2 strains were cultured at 37°C in BHI broth until an OD600 of 0.6 was reached. The bacteria were centrifuged at 5,000 g for 2 min and resuspended in PBS to OD600 ~0.5 and serially diluted. For determination of the virulence of the strains, larvae (n = 30) were injected with 4 × 104 and 4 × 102 CFU/larvae (in 10 μl) into the hindmost left proleg semi‐automatically using a PB‐600‐1 Repeating Dispenser (Hamilton) affixed to a Gastight 500‐μL Hamilton syringe (Model 1750 RN (large hub) SYR with a 22‐gauge, large hub RN NDL, 2 inch, point style 2 needle). Three control groups (n = 20) were injected with 10 μl of sterile PBS, 10 μl heat‐killed bacteria (the lowest dilution of the bacterial culture used for infection boiled at 100°C for 10 min), or left untreated. Serial dilutions of the bacterial suspension were plated onto BHI agar and grown at 37°C overnight to estimate the bacterium inoculum. The heat‐killed bacterial suspension was also spotted onto BHI agar to check sterility. Larvae were incubated at 37°C for 26 hr in sterile plastic Petri dishes lined with filter paper discs. Larval survival was assessed from 16 to 26 hr postinfection at 2‐hr intervals. Dead larvae were classed as those that were stationary and no longer responded to touch. All experiments were carried out at least three times.
2.7. Caenorhabditis elegans killing assay
Analysis of the virulence of B. cenocepacia strains toward C. elegans N2 was performed as described in Uehlinger et al. (2009). Briefly, to form a bacterial lawn, overnight cultures of B. cenocepacia strains were adjusted to a density of approximately 1.3–1.5 × 104 CFU/ml, and 100 μl of the suspension was plated onto six‐well plates containing nematode growth medium (NGM II) and incubated at 37°C for 24 hr. Following this, approximately 20–40 hypochlorite‐synchronized L4 larvae of C. elegans Bristol N2 (obtained from the Caenorhabditis Genetics Centre, University of Minnesota, Minneapolis) were used to inoculate the plates. Plates were then incubated at 20°C and the percentage of live worms scored after 48 and 72 hr. Nematodes were considered dead when they failed to respond to touch. E. coli OP50 was used as a negative control. All experiments were carried out at least three times.
2.8. Zebrafish embryo infection assay
Infection of zebrafish (Danio rerio) embryos was performed as described in Vergunst, Meijer, Renshaw, and O’Callagha (2010), Mesureur and Vergunst (2014). Briefly, B. cenocepacia K56‐2 and the otherwise isogenic tssM::Tp and tssA:Tp mutants were grown overnight in LB containing the appropriate antibiotics. Thirty hours postfertilization, zebrafish embryos were dechorionated and anesthetized in E3 medium with 0.02% buffered tricaine methanesulfonate (MS222). Embryos (n = 20) were then microinjected with around 100 CFU of bacteria directly into the blood circulation and maintained in E3 medium at 28°C. Embryo survival was monitored at regular intervals from 40 hr postinfection (hpi). Dead embryos were scored as those without a heartbeat. The experiment was carried out twice.
From the same experiments, five infected embryos per treatment group were taken randomly at 0 and 24 hpi and subjected to bacterial enumeration as described in Mesureur & Vergunst, 2014. Statistical analysis was performed using Prism 6 (GraphPad). Survival assays are represented in Kaplan–Meier graphs and analyzed with a log‐rank (Mantel–Cox) test. In CFU count experiments, significance was determined using one‐way ANOVA, with Sidak's multiple comparison test.
2.9. Bioinformatic analysis
Relevant DNA and protein sequences were obtained from the NCBI GenBank database (Clark, Karsch‐Mizrachi, Lipman, Ostell, & Sayers, 2016). Unannotated GenBank entries were manually interrogated for coding regions and the respective protein sequences using SnapGene® software (from GSL Biotech; available at http://www.snapgene.com). All protein homology analyses were performed using NCBI blastp and the nonredundant protein sequences (nr) database. T6SS‐1 clusters were identified in a two‐step process. First, the amino acid sequences of TssH (BCAL0347) and TagX proteins (BCAL0352) from B. cenocepacia J2315 were used as search queries to identify homologous proteins. Second, the loci encoding these proteins were interrogated for the presence of other T6SS‐related genes. If homologues of the additional tag genes tagM, tagN, and tagY and the majority of core tss genes were present, the region was defined as a T6SS‐1 cluster. To identify T6SS‐7 clusters, the protein sequence of the TssH homologue in the H111 T6SS‐7 cluster (I35_RS17330) was used as the query sequence to identify homologous proteins with a percentage sequence identity ≥70% in Burkholderia and Paraburkholderia species. The surrounding loci were then interrogated. If homologues of the core tss genes (tssA‐tssM) were present in a similar genetic arrangement as that in the H111 T6SS‐7 cluster, the region was defined as a T6SS‐7 cluster.
Multiple sequence alignments were performed using Clustal W or Clustal Omega (Larkin et al., 2007; Sievers et al., 2011) and formatted for display using BoxShade (https://www.ch.embnet.org/software/BOX_form.html). The prediction of transmembrane helices within proteins was performed using TMHMM Server v.2.0 (Krogh, Larsson, Heijne, & Sonnhammer, 2001).
3. RESULTS
3.1. Comparative analysis of the T6SS‐1 in Burkholderia and non‐Burkholderia species
In a previous study, six T6SSs were identified in B. pseudomallei (Shalom et al., 2007). The only one encoded on the large chromosome (T6SS‐1) has been identified in nine other Burkholderia species and three members of the Paraburkholderia (Angus et al., 2014). We have extended this analysis to include all Burkholderia and Paraburkholderia species, and members of other related proteobacteria for which genome sequence information is available. Therefore, the amino acid sequence of protein products encoded by the T6SS‐1 gene cluster of B. cenocepacia J2315 was used in blastp searches to identify homologous proteins in other Burkholderia, Paraburkholderia, and related species, and the respective T6SS‐1 gene clusters that encoded them were identified. All members of the genus Burkholderia (i.e., the Bcc and pseudomallei groups and the phytopathogenic strains B. gladioli, B. plantarii, and B. glumae), with the exception of the recently described species B. singularis, were found to harbor the T6SS‐1 gene cluster (Table A3 in Appendix 1). In species for which a complete genome assembly was available, the T6SS‐1 was located on chromosome 1 in every case. We also found homologous loci of the Burkholderia T6SS‐1 gene cluster in many species of the closely related Paraburkholderia genus, including P. acidipaludis, P. phytofirmans, and P. fungorum (see Table A4 in Appendix 1 for additional species), several of which were located on chromosome 2 or 3 instead of chromosome 1. The T6SS‐1 cluster of P. acidipaludis is shown in Figure 1. A T6SS‐1 cluster with a similar, but not identical, genetic organization was also found in other β‐proteobacteria, including Ralstonia solanacearum, Rubrivivax gelatinosus, Achromobacter xylosoxidans, and the γ‐proteobacteria species Xanthomonas oryzae and Acinetobacter baumannii (Figure 1).
Figure 1.
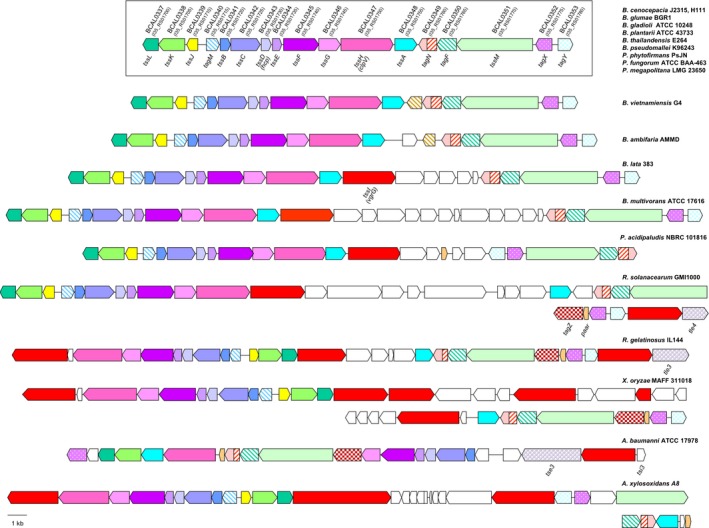
Gene arrangement and distribution of the Burkholderia T6SS‐1 gene cluster. Schematic representation of the Burkholderia T6SS‐1 gene cluster and related gene clusters in members of the Proteobacteriaceae. The box shows the genetic organization of the archetype Burkholderia T6SS‐1 gene cluster harbored by the indicated species, including B. cenocepacia (for reference, the B. cenocepacia T6SS‐1 gene cluster corresponds to BCAL0337‐BCAL0353 in strain J2315 and I35_RS01700‐I35_RS01780 in H111, as indicated). Variations on the same basic theme found in other members of the Burkholderia, related genera within the β‐proteobacteria (Achromobacter, Paraburkholderia, Ralstonia, and Rubrivivax), and some members of the γ‐proteobacteria (Acinetobacter, Xanthomonas) are shown
Most clusters were found to contain the core tss genes on three closely linked transcriptional units. However, in the majority of Bcc species, genes encoding the core T6SS subunits TssI and PAAR were not observed to be located in the T6SS‐1 gene cluster and are instead present in multiple copies at other loci distributed throughout the genome (as observed for B. cenocepacia by Aubert et al., 2015). Curiously, the T6SS‐1 gene cluster of members of the genus Acinetobacter lacked a copy of the core tssJ gene, as previously noted (Weber et al., 2013). It was also observed that several T6SS‐1 clusters contained insertions of one or more additional genes between the core genes or translocations of gene blocks, such as those in B. multivorans, P. acidipaludis, and R. solanacearum (Figure 1).
Type VI‐associated genes (tag) are conserved in some T6SSs but not others and encode proteins related to T6SS function, such as regulators or auxiliary subunits (Aschtgen, Thomas, & Cascales, 2010; Lossi et al., 2012; Shalom et al., 2007; Silverman et al., 2011). Five tag genes were recognized as being conserved in almost all T6SS‐1 clusters. These are tagF, which encodes a post‐translational regulator and is also present in some unrelated T6SSs such as the H1‐T6SS of P. aeruginosa (Lin et al., 2018; Silverman et al., 2011); tagM, encoding a putative outer membrane‐anchored lipoprotein of unknown function (Shalom et al., 2007); tagN, encoding a putative PG‐anchoring protein (Aschtgen et al., 2010); tagX, encoding a Sec‐dependent membrane‐anchored peptidoglycan hydrolase that facilitates T6SS sheath assembly through formation of holes in the peptidoglycan layer (Aubert et al., 2015; Ringel, Hu, & Basler, 2017; Weber et al., 2016); and a previously undescribed gene referred to here as tagY.
tagY corresponds to BCAL0353 in B. cenocepacia J2315 and is located upstream from tagX, but in the reverse orientation in nearly all T6SS‐1 gene clusters (Figure 1). It does not occur in unrelated T6SS gene clusters. In most Burkholderia species, tagY is likely to constitute a monocistronic operon due to the presence of a putative Rho‐independent transcription termination sequence located downstream from the tagY coding sequence, but in some non‐Burkholderia species, it constitutes the first gene of a polycistronic operon that encodes additional T6SS‐related proteins such as TssI and putative Tle effectors (Figure 1). Therefore, TagY is likely to play a role in the activity of T6SS‐1. It should be noted that tagM and tagY are not present in the Acinetobacter T6SS‐1 gene cluster. As members of this genus also appear to lack a TssJ orthologue, they are devoid of three periplasmic proteins that are present in the Burkholderia‐type T6SS‐1 in other species.
Analysis of the predicted protein product of tagY orthologues identified a putative transmembrane domain (TMD) located approximately 55 residues from the N‐terminus. The region located N‐terminal to the TMD contains two short conserved motifs separated by 10–13 amino acids (Appendix Figure A1). Based on the “positive inside rule” (Elofsson & von Heijne, 2007), the presence of amino acid residues with basic side chains immediately N‐terminal to the TMD suggests that the N‐terminal region constitutes a small cytoplasmically located domain. The TMD is followed by a long linker‐like region of low complexity, which in TagY orthologues in the Burkholderia spp. shares homology to the RnfC barrel sandwich hybrid domain (cl26195), a domain found at the N‐terminus of the RnfC electron transport complex subunit in Rhodobacter capsulatus (Biegel, Schmidt, González, & Müller, 2011; Schmehl et al.., 1993). A conserved C‐terminal region of ~40 amino acids that contain four cysteine residues was identified in most TagY orthologues (Appendix Figure A1). Due to the known role of cysteine thiols in various cellular activities, it is possible that this part of the protein, which is predicted to be located in the periplasmic space, constitutes a domain which assembles an iron–sulfur cluster. Alternatively, it may be involved in binding other transition metal ions such as zinc or copper, or act as a redox sensor.
Two additional genes are conserved in the T6SS‐1 cluster of species that are not members of the Burkholderia and Paraburkholderia genera. They correspond to RSp0764 and RSp0765 of R. solanacearum GM1000, RGE_RS12595 and RGE_RS12600 of R. gelatinosus IL144, XOO3320 and XOO3321 of X. oryzae MAFF 311018, AT699_RS16195, and an unannotated gene of A. arsenitoxydans NCTC10807, and ABAYE2409 and ABAYE2405 of A. baumannii (which were previously annotated as asaB and asaC as they were thought to be unique to the Acinetobacter T6SS; Carruthers, Nicholson, Tracy, & Munson, 2013). Bioinformatic analysis predicts that the first of each pair of genes encodes a protein possessing TMDs close to the N‐terminus (Appendix Figure A2), whereas the latter has been recognized as a putative PAAR domain‐containing protein in A. baylyi and named accordingly (Weber et al., 2016). Homologues of the asaB gene (from herein referred to as tagZ) are also present in some, but not all Burkholderia and in a single Paraburkholderia species (P. bannensis), while paar is present in all Burkholderia and Paraburkholderia species. However, both genes reside outside the T6SS‐1 cluster in these two genera and in some cases are located within a conserved gene cluster on chromosome 1 that encodes three TssI subunits and one or more effector–immunity protein pairs (Appendix Figure A3). The gene encoding the PAAR domain protein is located immediately upstream of tagZ in these T6SS‐related gene clusters, as is the case where these genes occur in the main T6SS‐1 gene cluster (Figure 1). As a number of the Burkholderia and Paraburkholderia species possess only T6SS‐1, it can be concluded that despite its location outside of the main T6SS‐1 gene cluster, the products of the paar‐tagZ gene pair play a role in the activity of T6SS‐1.
3.2. Identification of an additional, isolate‐specific, type VI secretion system in Burkholderia cenocepacia
During the bioinformatic analysis of the T6SS‐1 described above, an additional, complete T6SS gene cluster was identified in B. cenocepacia strain H111, a cystic fibrosis isolate (Carlier et al., 2014; Geisenberger et al., 2000). Further genome mining revealed that it was also present in B. cenocepacia strains FL‐5‐3‐30‐S1‐D7, VC12308, and DWS 37E‐2, and several additional B. cenocepacia isolates for which only contig or scaffold‐level genomic data are currently available, including D2AES, PC148, and TAtl‐371 (see Table A5 in Appendix 1 for loci). This second T6SS cluster is located on chromosome 2 in the completely sequenced strains and encodes orthologues of all the core T6SS subunits, including TssI and PAAR (Figure 2). The cluster shares a genetic arrangement that is similar to a T6SS cluster present in the plant pathogenic Burkholderia species, B. glumae, and to a T6SS gene cluster present in several Paraburkholderia species, including P. tuberum, which has been referred to as T6SSa (Angus et al., 2014), but for consistency with the established nomenclature is referred to here as the Burkholderia T6SS‐7. Our analysis also identified T6SS‐7 clusters in some but not all isolates of other Bcc species and in Cupriavidus metallidurans, a species that is closely related to the Burkholderia/Paraburkholderia clade (Table A5 in Appendix 1 and Figure 2).
Figure 2.
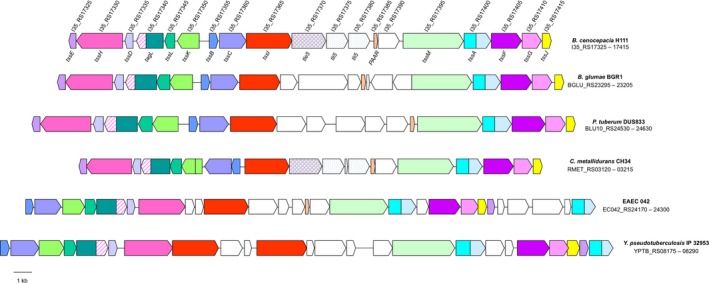
Burkholderia cenocepacia H111 possesses an additional T6SS that is present in some plant‐associated and human pathogenic bacteria. Schematic representation of a T6SS gene cluster identified in B. cenocepacia H111 (top) (I35_RS17325–I35_RS17415), which has a similar genetic organization to the T6SS‐7 cluster (also known as T6SS‐a) previously identified in B. glumae BGR1 and P. tuberum DUS833. A related T6SS cluster is also present in C. metallidurans CH34, EAEC 042 (the T6SS‐1 or sci‐1 cluster), and Y. pseudotuberculosis IP 32953 (T6SS‐2)
Burkholderia T6SS‐7 is notable in possessing a TagL orthologue which serves as an auxiliary subunit that anchors the T6SS to the peptidoglycan (Aschtgen et al., 2010). Accordingly, the genetic organization of this T6SS gene cluster is also similar to those encoding TagL‐dependent T6SSs present in human pathogens such as T6SS‐2 of Yersinia pseudotuberculosis, the T6SS of the uropathogenic E. coli strain CFT073, and the T6SS‐1 (Sci‐1 T6SS) of enteroaggregative E. coli (Figure 2).
Bioinformatic analysis of the T6SS‐7 gene cluster also suggests that it encodes a phospholipase D (PLD) effector and two corresponding Tli immunity proteins in members of the Burkholderiaceae. This particular PLD belongs to the Tle5 group of phospholipase effectors and is closely related to the PldB protein, PA5089, encoded by the H3‐T6SS of P. aeruginosa that has been shown to serve as a transkingdom effector (Russell et al., 2013; Jiang et al., 2014; Appendix Figure A4).
3.3. The Burkholderia cenocepacia T6SS‐1 is functional during growth under standard laboratory conditions
The presence of the core T6SS subunit, TssD, in bacterial culture supernatants is the hallmark of an active T6SS and can be used as a method to determine functionality of the T6SS. This assay was used to determine whether B. cenocepacia isolates possess an active T6SS‐1 during growth under standard laboratory conditions and to validate T6SS‐1 mutants prior to their use in bacterial competition and virulence assays described below. Therefore, mutants defective in the core tssA, tssK, and tssM components of T6SS‐1 were generated in strains H111, K56‐2, and Pc715j, and TssD secretion of the mutants was compared to that of the corresponding wild‐type parent strains grown in broth culture.
Western blotting showed that TssD was absent in the culture supernatant of the tssA, tssK, and tssM mutants but present in the respective wild‐type H111 and K56‐2 supernatants consistent with previous results obtained using a B. cenocepacia atsR mutant (Aubert et al., 2015; Figure 3a). The H111 and K56‐2 tssM mutants were subjected to a complementation analysis, whereby TssD secretion could be restored in both strains by introduction of a plasmid expressing tssM (Figure 3b). Together, these results indicate that B. cenocepacia isolates H111 and K56‐2 have an active T6SS‐1 under standard laboratory conditions.
Figure 3.
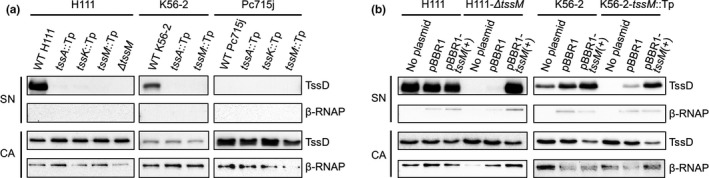
Burkholderia cenocepacia T6SS‐1 is active under standard laboratory conditions. Secretion activity of B. cenocepacia T6SS‐1 in vitro. Anti‐TssD immunoblot was performed on proteins extracted from culture supernatants (SN) and cell‐associated proteins (CA) of B. cenocepacia wild‐type (WT) strains H111, K56‐2, and Pc715j, and corresponding T6SS‐1 mutants (tssA::Tp, tssK::Tp, tssM::Tp, and/or ΔtssM) (a) and the H111 and K56‐2 WT and ΔtssM or tssM::Tp strains carrying a complementation or empty control plasmid (pBBR1‐tssM(+) and pBBR1MCS (“pBBR1”), respectively) (b). Anti‐β‐RNAP antibody was used as an indicator of bacterial cell lysis in preparations. Scales and labels as indicated. The H111 tssA::Tp mutant was included as a control
The additional B. cenocepacia isolate analyzed, Pc715j (and its T6SS‐deficient derivatives), was unable to secrete TssD into the extracellular milieu, despite detection of this protein in whole‐cell extracts (Figure 3a), indicating that TssD was being expressed but that the T6SS‐1 was incapable of firing and/or assembly in this strain. Whole‐genome sequencing of our laboratory stock of Pc715j indicated that an IS element was inserted into the tssM gene. The IS element exhibited homology to the ISUmu23 insertion sequence found in the Bcc‐specific phage KS5 (Lynch, Stothard, & Dennis, 2010), and its insertion into the tssM coding sequence was predicted to result in production of a nonfunctional truncated form of the TssM subunit that lacked the C‐terminal 447 amino acids.
The role of the candidate post‐translational regulatory protein, TagY, in T6SS‐1 activity was also explored by inactivating the tagY gene in strain H111. However, no significant difference in TssD secretion was observed between the wild‐type and the mutant strains (results not shown). These results could be explained if TagY acts to further upregulate the system in response to an unknown signal that is not present under the assay conditions.
3.4. Burkholderia cenocepacia T6SS‐1 exhibits antibacterial activity
It has been demonstrated that the T6SS can target effector proteins to other bacteria, thereby helping the organism to compete more effectively against other bacterial species in its growth environment. However, to date, the antibacterial nature of the T6SS‐1 in any member of the Bcc has not been reported. Therefore, we addressed the role of the T6SS‐1 in the ability of B. cenocepacia to compete effectively with other bacterial species.
As basal‐level TssD secretion appeared to be greater in B. cenocepacia H111 than in strain K56‐2 (Figure 3a), the former was chosen to evaluate the role of the T6SS‐1 in competition in this species. Strains H111 and H111‐ΔtssM were used as “attackers” in a bacterial competition experiment against Gram‐negative “prey” species Pseudomonas putida KT2440, Escherichia coli CC118(λpir), and E. coli SM10(λpir). Following cocultivation for four hours on solid medium, viable prey bacteria were enumerated and the number that survived attack by the wild‐type and mutant attackers were compared.
For all three prey strains tested, the number of recovered surviving prey bacteria was significantly lower (by one to two orders of magnitude) when they were cocultured with the wild‐type attacker strain in comparison with no attacker, demonstrating that B. cenocepacia can restrict the growth of E. coli and P. putida (Figure 4a). Furthermore, following coculture with the ΔtssM attacker strain, the number of surviving prey bacteria was similar to those observed when no attacker was present (Figure 4a). The number of recoverable attacking H111 bacteria was similar for both the WT and T6SS mutant strains and was unaffected by coculture with all prey strains (Appendix Figure A5). To validate these results, a complementation experiment was performed using the E. coli SM10(λpir) strain as the prey. The antibacterial activity of the tssM mutant attacker toward the E. coli strain could be restored to wild‐type levels by introduction of a plasmid expressing tssM into the mutant attacker strain (Figure 4b). These data strongly suggest that the T6SS‐1 in B. cenocepacia has antibacterial properties.
Figure 4.
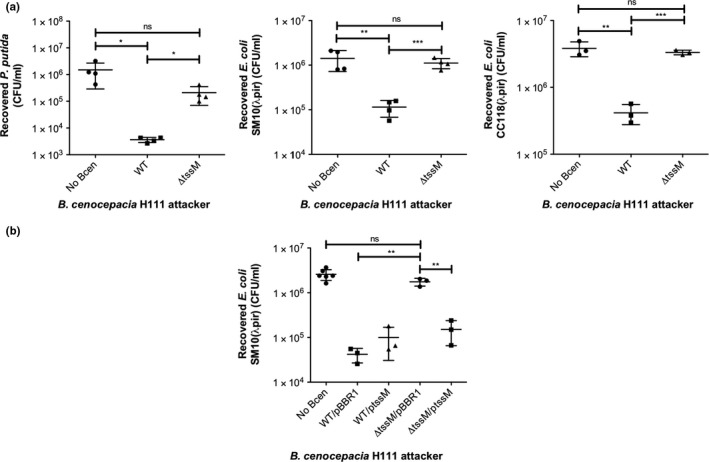
The Burkholderia cenocepacia T6SS‐1 plays a role in bacterial competition. (a) Recovery of viable P. putida, E. coli SM10(λpir) and E. coli CC118(λpir) (in CFU/ml) “prey” strains following coculture with the indicated B. cenocepacia H111 “attacker” strains for 4 hr at 30°C. (b) Comparison of recovery of E. coli SM10(λpir) prey following coculture with B. cenocepacia H111 WT or ΔtssM mutant attacker strains carrying complementation or control plasmids pBBR1‐tssM(+) (“ptssM”) and pBBR1MCS (“pBBR1”), respectively. n ≥3 and error bars indicate SD
3.5. Burkholderia cenocepacia T6SS‐1 is not required for virulence in eukaryotic models of infection
Several eukaryotic infection models have been used to identify virulence factors of B. cenocepacia, including the nematode C. elegans, larvae of the waxmoth G. mellonella, and zebrafish embryos (Seed & Dennis, 2008; Uehlinger et al., 2009; Vergunst et al., 2010). To ascertain the contribution of T6SS‐1 to the virulence of B. cenocepacia, we utilized all three of these infection models. Comparison of the survival of C. elegans infected with B. cenocepacia strain H111 and its tssA and tssK mutant derivatives for 48 and 72 hr showed that the wild‐type and mutant strains exhibited a similar killing efficiency during both time periods (Figure 5a). tssA and tssM mutants of strain K56‐2 were used to explore the role of T6SS‐1 in virulence toward G. mellonella larvae and zebrafish embryos. Comparison of the survival of G. mellonella following infection with these mutants demonstrated that they were able to kill the larvae as effectively as the wild‐type strain at 24 hr postinfection, whether high or low bacterial loads were employed (4 × 104 and 4 × 102 CFU/larvae, respectively), (Figure 5b). Wild‐type K56‐2 and its T6SS‐1 mutant derivatives were also found to be similarly virulent in the zebrafish model, both in terms of mortality and multiplication of the bacteria in the host (Figure 5c). Taken together, these results suggest that the T6SS‐1 in B. cenocepacia is primarily used to target other bacterial species. Although T6SS‐1 does not significantly contribute to virulence in the eukaryotic models tested here, it is possible that it may have an impact in other systems.
Figure 5.
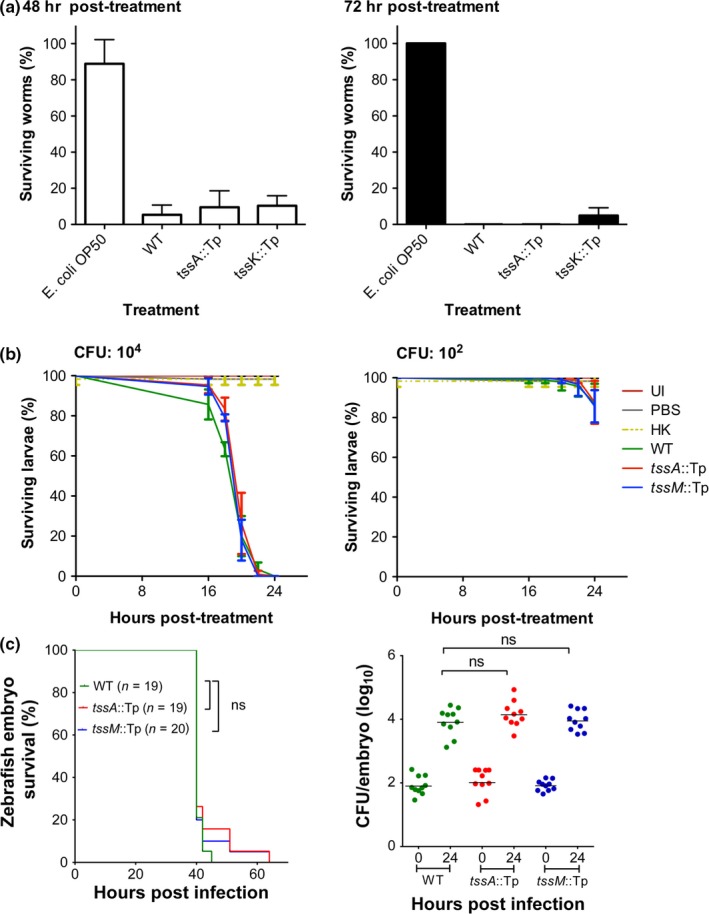
The Burkholderia cenocepacia T6SS‐1 is not required for virulence toward eukaryotes. (a) Percentage survival of C. elegans following 48‐hr (white bars, left) and 72‐hr (black bars, right) infection with the indicated B. cenocepacia H111 strains at 20°C. Twenty to 40 worms were used per condition. E. coli OP50 was used as a negative control. Each point indicates mean (n = 3), and error bars indicate SD. (b) Percentage survival of wax moth larvae following 24‐hr infection with high (1 × 104) (left) and low (1 × 102) (right) doses of B. cenocepacia K56‐2 (WT) and indicated mutant strains at 37°C. Thirty larvae were infected per condition. Uninfected (UI), heat‐killed B. cenocepacia WT (HK), and mock‐infected (PBS) controls were included. Each point indicates mean % survival (n = 3), and error bars indicate SD. (c) Zebrafish embryos were microinjected with ~100 CFU of indicated B. cenocepacia K56‐2 strains and kept at 28°C in individual wells containing E3 medium. About 20 embryos were used for determination of survival percentage over time (representative experiment shown on the left), and five embryos per indicated time point were used to determine recovery of viable B. cenocepacia K56‐2 counts (n = 5 per time point per experiment, geometric mean; right‐hand graph, showing summary of two independent experiments). ns: not significant
3.6. In silico identification of putative T6SS‐dependent effectors in B. cenocepacia
The T6SS‐1 cluster in B. cenocepacia encodes no obvious T6SS‐dependent effectors. However, an earlier bioinformatics survey of the B. cenocepacia K56‐2 genome identified ten TssI proteins encoded at other locations within the genome, of which two (BCAL1359 and BCAS0667) contain C‐terminal effector domains (Aubert et al., 2015). Here, using what is known from previously characterized T6SS‐dependent effectors, coupled with bioinformatic tools, we have identified additional putative T6SS‐dependent non‐TssI effectors and their cognate immunity proteins encoded by the B. cenocepacia genome. As T6SS‐dependent effector genes in other species are often located within close proximity to tssI genes (Lien & Lai, 2017), we used the predicted amino acid sequences of protein products encoded within close proximity to the ten intact tssI genes and one disrupted tssI gene (BCAL2503) present within the B. cenocepacia J2315 genome as queries in BLASTP searches to identify putative functional domains and homology to proteins belonging to established T6SS effector superfamilies (Appendix Figure A6). Twelve putative T6SS effectors were identified using this approach, with each of the tssI clusters in B. cenocepacia J2315 encoding at least one putative effector. Of the putative effectors identified, six were predicted to be phospholipases (encoded by BCAL1296, BCAL1358, BCAL1366, BCAL2277, BCAM0046, and BCAM0149), five of which belong to the Tle antibacterial effector superfamily (Russell et al., 2013). Of the six remaining effectors, one is a predicted peptidoglycan hydrolase (BCAL1166), two were identified as putative nuclease effectors (BCAL1298 and BCAS0663), of which the latter contains RHS repeats, an additional RHS repeat protein (BCAM2253) containing a RES‐type NAD+ glycohydrolase CTD (Skjerning, Senissar, Winther, Gerdes, & Brodersen, 2018), and two homologues of the antibacterial pore‐forming toxin Tse4 (BCAL1292 and BCAL2505; Whitney et al., 2014; LaCourse et al., 2018) were also identified. An additional putative T6SS effector was identified by using homologues of the Tae peptidoglycan hydrolase T6SS effector superfamilies as queries to search the entire translated genome of B. cenocepacia, resulting in identification of a Tae4‐Tai4 effector immunity pair (BCAM1464‐BCAM1465) located away from a tssI gene cluster. Further details of the putative T6SS effectors identified in these searches are included in Table A6 in Appendix 1, which includes the specific domains identified and putative immunity proteins. It should be noted that the previously described TecA effector (Aubert et al., 2016) is not encoded within a tssI gene cluster and was not independently identified in our analysis.
4. DISCUSSION
Although some species of bacteria, such as S. marcescens and V. cholerae V52, do exhibit high basal levels of T6SS activity during growth in laboratory media (Gerc et al., 2015; MacIntyre et al., 2010), in other cases the T6SS is observed to be inactive (Burtnick et al., 2011; Mougous et al., 2006; Zheng, Shin, Cameron, & Mekalanos, 2010), necessitating the use of bacterial strains that have a constitutively active T6SS in order to investigate the functional role of the system and aid in the identification of T6SS‐dependent substrates (Hood et al., 2010; Russell et al., 2011). Here, we demonstrate that the T6SS‐1 of B. cenocepacia is active under standard laboratory conditions with sufficient basal activity to allow detection of TssD in concentrated culture supernatant by immunoblotting. This observation is consistent with a previous proteomic study in which a protein identified as “hemolysin‐coregulated protein” (i.e., Hcp or TssD) was detected in the extracellular fraction of strain H111 through 2‐DE coupled mass spectrometry which was not recognized as a T6SS subunit at the time (Riedel, Carranza, Gehrig, Potthast, & Eberl, 2006). These results are also consistent with an investigation that demonstrated TssD secretion in strain K56‐2 could be increased upon inactivation of a global virulence regulator, atsR (Aubert et al., 2008). This study showed the presence of very small amounts of a protein corresponding in size to TssD in wild‐type K56‐2 culture supernatants by SDS‐PAGE, which was confirmed by mass spectrometry rather than immunoblotting, as in our study. Moreover, the low abundance of this protein in the secreted fraction led the authors to consider the T6SS activity to be insufficient to use the wild‐type strain in further investigations into the role of the T6SS in B. cenocepacia. It is possible that our method of sample preparation and detection in wild‐type K56‐2 was more sensitive than that used in the Aubert and co‐workers study.
The role of the T6SS in interspecies and intraspecies bacterial competition has been recognized as a prominent feature of the system in a variety of T6SS‐containing bacteria, including P. aeruginosa, V. cholerae, and S. marcescens (Hood et al., 2010; MacIntyre et al., 2010; Murdoch et al., 2011). In this study, we provide evidence to support a role for the B. cenocepacia T6SS‐1 in competition against two bacterial species, P. putida and E. coli. We have also identified an arsenal of potential antibacterial cargo effectors that could be delivered by T6SS‐1, notably including peptidoglycan hydrolases. The additional T6SS cluster we identified in B. cenocepacia H111 (T6SS‐7) is very unlikely to function in bacterial competition under the conditions tested, as the bacterial competition experiments performed in this study indicated that the level of prey survival was the same in the presence of a mutant attacker with an inactive T6SS‐1 as it was when there was no B. cenocepacia attacker strain present (Figure 4a). Our results are consistent with observations in other species that encode a Burkholderia T6SS‐1‐type secretion system. This includes the T6SS‐1 in B. thailandensis, which was found to be the sole T6SS cluster involved in bacterial competition (Schwarz et al., 2010), and the T6SS‐1 homologue in Acinetobacter spp. that was shown to contribute to interbacterial competition (Basler, Ho, & Mekalanos, 2013; Carruthers et al., 2013; Repizo et al., 2015; Weber et al., 2016). They are also consistent with recent observations in the related Paraburkholderia species P. phymatum, where two non‐T6SS‐1‐type secretion systems (T6SS‐3 and T6SS‐b (T6SS‐8)) were found to be responsible for interbacterial competition against β‐rhizobia strains in vitro and as a consequence were less efficient in root nodulation (de Campos, Lardi, Gandolfi, Eberl, & Pessi, 2017).
The H1‐T6SS in P. aeruginosa PAO1 is thought to be triggered by attacks from the T6SS (or T4SS) of neighboring cells as a defensive strategy (Basler & Mekalanos, 2012; Basler et al., 2013; Ho, Basler, & Mekalanos, 2013). As a result, P. aeruginosa does not display a fitness advantage over T6SS‐deficient competing species (Basler et al., 2013). The T6SSs in S. marcescens Db10 and V. cholerae V52, on the other hand, fire indiscriminately and do not require activation from a neighboring attacking bacterium, and thereby confer a fitness advantage on the host bacterium against various Gram‐negative competitor species, such as a E. coli, Salmonella typhimurium, and Pseudomonas fluorescens (Gerc et al., 2015; MacIntyre et al., 2010). Here, we demonstrate that the T6SS‐1 confers a fitness advantage on B. cenocepacia over both T6SS‐positive (P. putida KT2440) and T6SS‐negative (E. coli SM10(λpir)) bacterial species. This may suggest that, like S. marcescens and V. cholerae, the T6SS‐1 in B. cenocepacia is constitutively active and its activation is not stimulated by external T6SS attacks, which is also supported by the evidence of T6SS activity in wild‐type strains of B. cenocepacia H111 and K56‐2 under standard laboratory conditions. This would provide additional support for the idea that the defensive regulatory strategy used by P. aeruginosa is atypical among T6SSs (Gerc et al., 2015). In addition, one of the T6SSs in P. putida KT2440 has been shown to be highly efficient at killing phytopathogens such as X. campestris and P. syringae (Bernal, Allsopp, Filloux, & Llamas, 2017). However, our results indicate that B. cenocepacia survival is unaffected by the presence of P. putida. The constitutive activity of the T6SS‐1 we have observed in B. cenocepacia may account for this, where B. cenocepacia may be able to subvert P. putida before P. putida can attack with its own T6SS. Alternatively, B. cenocepacia may be immune to the T6SS activity of P. putida due to the presence of T6SS immunity proteins with interspecies reactivity, as seen for some Tae‐Tai and Tse‐Tsi effectors–immunity pairs in other species (Russell et al., 2012).
In comparison with the antibacterial T6SSs of other species, the fitness advantage of B. cenocepacia over the prey species tested is notably less than that observed in several other attacker species, including P. aeruginosa, V. cholerae, and S. marcescens. In these species, an active T6SS is responsible for 1,000‐ to 100,000‐fold reduction in the number of recovered prey bacteria in a bacterial competition assay (Hood et al., 2010; MacIntyre et al., 2010; Murdoch et al., 2011), whereas we only observed a modest 10‐ to 58‐fold reduction. This observation may be due to several reasons. For example, T6SS expression and activity may be lower in B. cenocepacia than these other T6SS‐positive strains, the prey strains used in our competition assay may produce their own antibacterial factors (such as bacteriocins, siderophores, or effectors secreted by other systems), or there may be inherent differences in growth rates between B. cenocepacia and the prey species. However, upon enumerating the B. cenocepacia attacker species in our bacterial competition assays, we found that B. cenocepacia survival was not affected by coculture with the prey species (Appendix Figure A5). It is also conceivable that the prey used here may have immunity toward specific T6SS effectors due to cross‐reacting T6SS‐immunity proteins between species (Russell et al., 2012). It is possible that by screening a larger panel of bacterial species, a species may be identified that is more susceptible to the T6SS‐1‐dependent antibacterial activity of B. cenocepacia. Moreover, as the T6SS‐1 cluster harbors a number of genes that potentially encode post‐translational regulators (i.e., tagF, tagM, and tagY), this system may have the capacity to be further upregulated under certain conditions.
The B. cenocepacia T6SS‐1 was first implicated in virulence toward eukaryotes in a signature‐tagged mutagenesis (STM) study carried out in a rat model of chronic lung infection in which transposon insertions within the T6SS‐1 gene cluster were associated with impaired survival of the bacterium (Hunt et al., 2004). In subsequent studies, this group demonstrated that the T6SS‐1 contributes toward cytoskeletal rearrangements and inflammasome activation in B. cenocepacia‐infected macrophages through host Rho GTPase inactivation (Aubert et al., 2008; Flannagan et al., 2012; Keith et al., 2009; Rosales‐Reyes et al., 2012; Xu et al., 2014). In contrast to the reported impaired survival of T6SS mutants during rat lung infection (Hunt et al., 2004), more recent evidence suggests that the T6SS may contribute to a pyrin inflammasome‐dependent innate immune response that promotes lung tissue inflammation and bacterial clearance in a mouse infection model (Aubert et al., 2016; Xu et al., 2014). The study by Aubert and co‐workers presented data to show that a putative T6SS effector was responsible for this mechanism.
We have tested several T6SS‐inactive strains of B. cenocepacia in three eukaryotic host–pathogen models, nematodes, larvae of the wax worm, and zebrafish larvae (Seed & Dennis, 2008; Uehlinger et al., 2009; Vergunst et al., 2010). We found no significant difference in host survival rates in comparison with infection with the WT strain in any of these infection models, suggesting that the T6SS‐1 in B. cenocepacia does not have a functional role in pathogenicity. Of note, we have performed our assays in the presence of a functional atsR, so the T6SS is not constitutively upregulated as occurs in the absence of AtsR, and instead activation above basal levels would depend on the presence of the appropriate stimulus (as yet unknown) in any of the model systems. Therefore, the T6SS is either not expressed in these models in the presence of AtsR, or does not contribute to a significant host‐induced protective immune response, as seen in mice (Aubert et al., 2016; Xu et al., 2014). We cannot exclude, however, that in the absence of atsR, a measurable effect on virulence could be detected.
To conclude, we have carried out a bioinformatic and functional analysis of the T6SS‐1 in the Bcc species B. cenocepacia. We have shown that it is encoded on the large chromosome in nearly all Burkholderia species, unlike the other T6SSs associated with members of this genus, which are not conserved in all species and are usually specified by chromosome 2. Therefore, T6SS‐1 can be considered as the ancestral Burkholderia T6SS and may serve as a marker for this genus. We also showed that T6SS‐1 was constitutively active in two representative clinical strains and could be used to compete against other bacterial species, including P. putida and E. coli. This is the first demonstration that T6SS‐1 in a Bcc member plays a role in interbacterial competition and adds to the catalogue of Gram‐negative bacteria that use the T6SS for this purpose. The natural reservoir of B. cenocepacia is within the environment, particularly in the soil around plant root systems where many other bacteria compete to establish themselves. It is therefore unsurprising that B. cenocepacia has evolved a mechanism for competitive fitness against other bacteria, in a similar manner to other ubiquitous Burkholderia and Paraburkholderia species (de Campos et al., 2017; Schwarz et al., 2010). Future work will look to identify and characterize the secreted components responsible for the T6SS‐dependent antibacterial activity of B. cenocepacia.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHORS CONTRIBUTION
H.L.S., M.S.T., A.C.V., and L.E. conceived and designed experiments, and contributed to the writing of the manuscript. H.L.S., S.S., L.Z., and S.Sch. conducted experiments.
5. ETHICS STATEMENT
Protocols and procedures employed in this investigation were reviewed and approved by the appropriate institutional review committees. Zebrafish (Danio rerio) were kept and handled in compliance with the guidelines of the European Union for handling laboratory animals (http://ec.europa.eu/environment/chemicals/lab_animals/home_en.htm). Studies performed at VBMI are approved by the Direction Départementale de la Protection des Populations (DDPP) du Gard (ID 30–189–4). Infection experiments were terminated before the larvae reached the free feeding stage and did not classify as animal experiments according to the 2010/63/EU Directive. Care and maintenance of zebrafish was as described previously (Vergunst et al., 2010).
ACKNOWLEDGEMENT
This work was supported by a BBSRC Doctoral Training Grant (BB/F016840/1) awarded to H.L.S. Genome sequencing was provided by MicrobesNG (http://www.microbesng.uk), which is supported by the BBSRC (grant number BB/L024209/1). VBMI is supported by l'Institut National de la Santé et de la Recherche Médicale INSERM and Université de Montpellier. L.Z. was a Marie‐Curie fellow in the Initial Training Network FishForPharma (PITN‐GA‐2011‐289209). L.E. is funded by the Swiss National Science Foundation (SNSF) (Project 31003A_169307).
APPENDIX 1.
Table A1. Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Escherichia coli strains | ||
| JM83 | F− ara Δ(lac‐proAB) rpsL ϕ80dlacZΔM15 (SmR) | (Yanisch‐Perron et al., 1985) |
| SM10(λpir) | thi‐1 thr leu tonA lacY supE recA::RP4‐2‐Tc::Mu (KmR) (λpir) | (Simon et al., 1983) |
| S17‐1(λpir) | thi proA hsdR recA RP4‐2‐tet::Mu‐1 kan::Tn7 integrant (TpR, SmR) (λpir) | (Simon et al., 1983) |
| CC118(λpir) | araD139 Δ(ara‐leu)7697 ΔlacX74 galE galK phoA20 thi‐1 rpsE argE (am) recA1 λpir rpoB (RfR) | (Herrero et al., 1990) |
| OP50 | E. coli B uracil auxotroph; food source for C. elegans | (Brenner, 1974) |
| Pseudomonas putida strains | ||
| KT2440 | Spontaneous r‐ derivative of mt‐2 | (Bagdasarian et al., 1981) |
| Burkholderia cenocepacia strains | ||
| H111 | CF isolate | (Römling et al., 1994) |
| K56‐2 | CF isolate, ET12 lineage | (Mahenthiralingam et al., 2000) |
| Pc715j | CF sputum isolate, ET12 lineage | (McKevitt et al., 1989; Darling et al., 1998) |
| H111‐tssA::Tp | H111 with tssA disrupted by dfrB2 cassette (TpR) | (Dix et al., 2018) |
| H111‐tssK::Tp | H111 with tssK disrupted by dfrB2 cassette (TpR) | This study |
| H111‐tssM::Tp | H111 with tssM disrupted by dfrB2 cassette (TpR) | This study |
| H111‐ΔtssM | H111 with an in‐frame deletion of the internal XhoI fragment of tssM | (Dix et al., 2018) |
| H111‐tagY::Tp | H111 with tagY disrupted by dfrB2 cassette (TpR) | This study |
| K56‐2‐tssA::Tp | K56‐2 with tssA disrupted by dfrB2 cassette (TpR) | This study |
| K56‐2‐tssM::Tp | K56‐2 with tssM disrupted by dfrB2 cassette (TpR) | This study |
| Pc715j‐tssA::Tp | Pc715j with tssA disrupted by dfrB2 cassette (TpR) | This study |
| Pc715j‐tssK::Tp | Pc715j with tssK disrupted by dfrB2 cassette (TpR) | This study |
| Pc715j‐tssM::Tp | Pc715j with tssM disrupted by dfrB2 cassette (TpR) | This study |
| Plasmids | ||
| pBBR1MCS | Mobilizable BHR cloning vector, pBBR1‐replicon (CmR) | (Kovach et al., 1994) |
| pBluescriptII KS (+) | General cloning vector, ColE1‐derived phagemid, lacZα MCS (ApR) | (Alting‐Mees and Short, 1989) |
| p34E‐Tp | p34E containing dfrB2 gene (ApR, TpR) | (DeShazer and Woods, 1996) |
| pSHAFT2 | Suicide vector, R6K‐derived replicon, oriT+ (ApR, CmR) | (Shastri et al., 2017) |
| pBBR1‐tssK | pBBR1MCS containing tssK from B. cenocepacia H111 cloned between HindIII and BamHI (CmR) | This study |
| pBBR1‐tssM’ | pBBR1MCS‐1 containing 1.2 kbp N‐terminal fragment of tssM from B. cenocepacia H111 cloned between XbaI and XhoI (CmR) | This study |
| pBBR1‐tssM(+) | pBBR1MCS containing full‐length tssM from B. cenocepacia H111 cloned between Acc65I and XbaI (CmR) | This study |
| pBluescriptII‐tagY | pBluescriptII containing tagY from B. cenocepacia H111 cloned between BamHI and XhoI sites (ApR) | This study |
| pBBR1‐tssK::Tp | pBBR1MCS containing tssK disrupted by dfrB2 cassette at EcoRI site in same orientation as tssK (CmR, TpR) | This study |
| pBBR1‐tssM’::Tp | pBBR1MCS containing tssM’ disrupted by dfrB2 cassette at BamHI site in reverse orientation as tssM’ (CmR, TpR) | This study |
| pBluescriptII‐tagY::Tp | pBluescriptII containing tagY disrupted by the dfrB2 cassette at ZraI site in the same orientation as tagY (ApR) | This study |
| pSHAFT2‐tssA::Tp | pSHAFT2 containing tssA::Tp allele from pBBR1‐tssA::Tp cloned between XhoI and NotI (ApR, CmR, TpR) | (Dix et al., 2018) |
| pSHAFT2‐tssK::Tp | pSHAFT2 containing tssK::Tp allele from pBBR1‐tssK::Tp cloned between XhoI and NotI (ApR, CmR, TpR) | This study |
| pSHAFT2‐tssM’::Tp | pSHAFT2 containing tssM’::Tp allele from pBBR1‐tssM’::Tp cloned between XhoI and NotI (ApR, CmR, TpR) | This study |
| pSHAFT2‐tagY::Tp | pSHAFT2 containing tagY::Tp allele from pBluescriptII‐tagY::Tp cloned between SalI and XbaI (ApR, CmR, TpR) | This study |
Abbreviations: ApR, ampicillin‐resistant; CmR, chloramphenicol‐resistant; KmR, kanamycin‐resistant; RfR, rifampicin‐resistant; SmR, streptomycin‐resistant; TcR, tetracycline‐resistant; TpR, trimethoprim‐resistant; BHR, broad host range.
Table A2. Primers used in this study
| Primer ID | Primer sequencea |
|---|---|
| iotAfor | 5’‐GCGCAAGCTTCACGCGACATCTCATGCATC |
| iotArev2 | 5’‐ATCACGAAGAGCATTCCGCC |
| tssKfor | 5’‐GCGCAAGCTTCCACATTAACCGGATTTGAC |
| tssKrev | 5’‐GCGCGGATCCTCATGATGTGACCGCGATCA |
| tssK‐OPfor | 5’‐GCGATTTAATTCGGGCACGA |
| tssK‐OPrev | 5’‐ACAGCAAATCGAGCAGCGAA |
| tssMfor | 5’‐GCGCTCTAGAGGAACCTGAACGTCCTATGC |
| tssMrev | 5’‐GCGCCTCGAGCTGTTGGTTTCGCCTTCCTG |
| tssMforAcc65I | 5’ ‐GCGCGGTACCTTAAAATCGCACCGGAACCTGAAC |
| tssMrevXbaI | 5’‐GCGCTCTAGAATTTGCGCCTGTACGGTTTG |
| tssM‐OPfor | 5’‐TCATCCCGTTTGACAGCATG |
| tssM‐OPrev | 5’‐AGAAGCCGTTCTTCGAGAAC |
| tagYfor | 5’‐GCGCCTCGAGTAAAGTGCGCCGGAAAATTCAAA |
| tagYrev | 5’‐GCGCGGATCCCAGTGTCACGCGACATCATA |
| tagY‐OPfor | 5’‐GCGCTCTAGAATCCCCGGAAATTGGAATTG |
| tagY‐OPrev | 5’‐GCGCAAGCTTGGTAAGGAAAGGAGACGTAT |
aSequences specifying restriction endonuclease cleavage sites are underlined.
Table A3. T6SS‐1 gene loci in the Burkholderia genus
| Species | Strain | Chrb | Locusa | Old locus/aliasa |
|---|---|---|---|---|
| Burkholderia cepacia complex | ||||
| B. ambifaria | AMMD | 1 | BAMB_RS01920‐BAMB_RS02010 | Bamb_0377‐Bamb_0395 |
| B. anthina | AZ‐4‐2‐10‐S1‐D7 | 1 | WS64_RS00645‐WS64_RS00755 | WS64_00645‐WS64_00755 |
| B. catarinensis | 89 | n/a | BFF94_26275‐BFF94_35355 | n/a |
| B. cenocepacia | J2315 | 1 | QU43_RS38220‐QU43_RS38300 | BCAL0337‐BCAL0353 |
| B. cepacia | UCB 717 | 1 | APZ15_RS05925‐APZ15_RS06005 | APZ15_05925‐APZ15_06005 |
| B. contaminans | 170816 | 1 | C3743_RS13835‐ C3743_RS13945 | C3743_28295‐C3743_28405 |
| B. diffusa | RF2‐non‐BP9 | 1 | WI26_RS01890‐WI26_RS01990 | WI26_01890‐WI26_01990 |
| B. dolosa | AU0158 | 1 | AK34_RS26290‐AK34_RS26210 | AK34_2669‐AK34_2653 |
| B. lata | 383 | 1 | BCEP18194_RS07870‐BCEP18194_RS07970 | Bcep18194_A3555‐Bcep18194_A3577 |
| B. latens | AU17928 | 1 | WK25_RS00385‐WK25_RS00305 | WK25_00385‐WK25_00305 |
| B. metallica | FL‐6‐5‐30‐S1‐D7 | 1 | WJ16_RS01935‐WJ16_RS02050 | WJ16_01935‐WJ16_02050 |
| B. multivorans | ATCC 17616 | 1 | BMULJ_RS01495‐BMULJ_RS01645 | BMULJ_00300‐BMULJ_00329 |
| B. paludis | MSh1 | n/a | GQ56_0123510‐GQ56_0123430 | n/a |
| B. pseudomultivorans | SUB‐INT23‐BP2 | 1 | WS57_RS19835‐WS57_RS19915 | WS57_19795‐WS57_19875 |
| B. puraquae | CAMPA 1040 | n/a | B7G54_RS33210‐B7G54_RS33130 | B7G54_33195‐B7G54_33115 |
| B. pyrrocinia | DSM 10685 | 1 | ABD05_RS07950‐ABD05_RS08030 | ABD05_07950‐ABD05_08030 |
| B. seminalis | FL‐5‐4‐10‐S1‐D7 | 1 | WJ12_RS01985‐WJ12_RS02085 | WJ12_01985‐WJ12_02085 |
| B. stabilis | ATCC BAA‐67 | 1 | BBJ41_RS12095‐BBJ41_RS12210 | BBJ41_12095‐BBJ41_12210 |
| B. stagnalis | MSMB735 | 1 | WT74_RS02265‐WT74_RS02350 | WT74_02260‐WT74_02345 |
| B. territorii | RF8‐non‐BP5 | 1 | WS51_RS12720‐WS51_RS12820 | WS51_12715‐WS51_12815 |
| B. ubonensis | MSMB22 | 1 | BW23_RS21305‐BW23_RS21205 | BW23_1274‐BW23_1254 |
| B. vietnamiensis | G4 | 1 | BCEP1808_RS02265‐BCEP1808_RS02350 | Bcep1808_0456‐Bcep1808_0473 |
| Burkholderia pseudomallei group | ||||
| B. humptydooensis | MSMB122 | 1 | WS76_02215‐WS76_02295 | n/a |
| B. mallei c | NCTC 10229 | 1 | BMA10229_RS17595‐BMA10229_RS17645 | BMA10229_A1710‐BMA10229_A1720 |
| B. oklahomensis | EO147 | 1 | DM82_RS14115‐DM82_RS14035 | DM82_2790‐DM82_2774 |
| B. pseudomallei | K96243 | 1 | BPSL3111‐BPSL3095 | AQ15_RS22375‐AQ15_RS22455 |
| B. thailandensis | E254 | 1 | BTH_RS27330‐BTH_RS27250 | BTH_I2968‐BTH_I2951 |
| B. singularis | LMG 28154 | ‐ | ‐ | |
| Phytopathogens | ||||
| B. gladioli | ATCC 10248 | 1 | BM43_RS25670‐BM43_RS25750 | BM43_1793‐BM43_1809 |
| B. glumae | BGR1 | 1 | BGLU_RS01925‐BGLU_RS02005 | bglu_1g03850‐bglu_1g04010 |
| B. plantarii | ATCC 43733 | 1 | bpln_RS01775‐bpln_RS01855 | bpln_1g03440‐bpln_1g03600 |
Abbreviations: Chr, chromosome; n/a, not applicable; ‐, not present.
aGene loci refer to the first (tssL) and last (tagY) genes in the T6SS‐1 gene cluster in these species as shown in Figure 1.
bIf n/a is stated, chromosome location was not available as the loci coordinates were obtained from draft assemblies consisting of contigs.
cTruncated cluster that lacks tssL‐tssD.
Table A4. Paraburkholderia species containing homologous loci of the Burkholderia T6SS‐1 cluster identified through bioinformatics analysis
| Species | Strain | Chrb | Locusa | Old locus/aliasa |
|---|---|---|---|---|
| P. acidipaludis | NBRC 101816 | n/a | BAC01S_RS24625‐BAC01S_RS24720 | n/a |
| P. aspalathi | LMG 27731 | n/a | BM438_RS23205‐BM438_RS23285 | SAMN05192563_101653‐SAMN05192563_101669 |
| P. bannensis | NBRC 103871 | n/a | BBA01S_RS03935‐BBA01S_RS03850 | n/a |
| P. bryophila | 376MFSha3.1 | n/a | H281_RS0127575‐H281_RS0127655 | n/a |
| P. caledonica | NBRC 102488 | n/a | BCA01S_RS25625‐BCA01S_RS25545 | n/a |
| P. caribensis | DSM 13236 | 3 | C2L66_RS31465‐C2L66_RS31545 | C2L66_31465‐C2L66_31545 |
| P. caryophylli | Ballard 720 | n/a | C0Z17_RS09175‐C0Z17_RS09265 | C0Z17_09180‐C0Z17_09270 |
| P. dilworthii | WSM3556 | n/a | F759_RS0111885‐F759_RS0111970 | n/a |
| P. eburnea | LMG 29537 | n/a | BX588_RS11555‐BX588_RS30870 | BX588_10514‐BX588_1421 |
| P. fungorum | GAS106B | n/a | BLS41_RS32405‐BLS41_RS32325 | SAMN05443245_6595‐SAMN05443245_6579 |
| P. ginsengiterrae | DCY85 | n/a | A6V36_RS34655‐A6V36_RS34735 | A6V36_13555‐A6V36_13635 |
| P. graminis | C4D1M | n/a | BGRAMDRAFT_RS31300‐BGRAMDRAFT_RS31220 | BgramDRAFT_6363‐BgramDRAFT_6347 |
| P. insulsa | LMG 28183 | n/a | BX589_RS19295‐BX589_RS19215 | BX589_111106‐BX589_11190 |
| P. kururiensis | M130 | n/a | G118_RS0127045‐G118_RS0127125 | n/a |
| P. megapolitana | LMG 23650 | n/a | BM166_RS27120‐BM166_RS27200 | SAMN05192543_109158‐SAMN05192543_109174 |
| P. nodosa | CNPSo 1341 | n/a | BFD71_RS23080‐BFD71_RS22965 | n/a |
| P. oxyphila | NBRC 105797 | n/a | BO1_RS31110‐BO1_RS23945 | n/a |
| P. phenazinium | GAS95 | n/a | BUS12_RS10920‐BUS12_RS10830 | SAMN05444165_2229‐SAMN05444165_2211 |
| P. phenoliruptrix | BR3459a | 2 | BUPH_RS28500‐BUPH_RS28580 | BUPH_06127‐BUPH_06111 |
| P. phytofirmans | PsJN | 2 | BPHYT_RS24375‐BPHYT_RS24455 | Bphyt_4909‐Bphyt_4925 |
| P. rhynchosiae | WSM 3937 | n/a | C0Z16_RS18310‐C0Z16_RS18230 | C0Z16_18290‐C0Z16_18210 |
| P. sediminicola | LMG 24238 | n/a | BLT79_RS22400‐BLT79_RS22480 | SAMN05192547_102957‐SAMN05192547_102973 |
| P. soli | GP25‐8 | n/a | C0Z19_RS08700‐C0Z19_RS08620 | C0Z19_08720‐C0Z19_08640 |
| Paraburkholderia sp. | BL18I3N2 | n/a | B0G75_RS24645‐B0G75_RS24565 | B0G75_110100‐B0G75_11084 |
| Paraburkholderia sp. | BL21I4N1 | n/a | B0G83_RS22810‐B0G83_RS22905 | B0G83_109119‐B0G83_109138 |
| Paraburkholderia sp. | BL25I1N1 | n/a | B0G73_RS11290‐B0G73_RS11370 | B0G73_106134‐B0G73_106151 |
| Paraburkholderia sp. | C35 | n/a | DK391_RS17735‐DK391_RS17655 | n/a |
| Paraburkholderia sp. | GV068 | n/a | C8K18_RS26440‐C8K18_RS26520 | C8K18_115102‐C8K18_115118 |
| Paraburkholderia sp. | GV072 | n/a | C8K19_RS26210‐C8K19_RS26130 | C8K19_11551‐C8K19_11535 |
| P. symbiotica | JPY 581 | n/a | C0Z20_RS18355‐C0Z20_RS18280 | C0Z20_18350‐C0Z20_18275 |
| P. terricola | LMG 20594 | n/a | BUE39_RS22490‐BUE39_RS22570 | SAMN05192548_102957‐SAMN05192548_102973 |
| P. tropica | LMG 22274 | n/a | BMY06_RS23990‐BMY06_RS23890 | SAMN05216550_114137‐SAMN05216550_114117 |
Abbreviations: Chr, chromosome; n/a, not applicable.
aGene loci refer to the first (tssL) and last (tagY) genes in the T6SS‐1 gene cluster in these species as shown in Figure 1. P. acidipaludis is an exception to this, where the gene loci refer to tssL to tagN.
bIf n/a is stated, chromosome location was not available as the loci coordinates were obtained from draft assemblies consisting of contigs.
Table A5. Strain‐specific T6SS‐7 gene loci in Burkholderia and related species
| Species | Strain | Chrb | Locusa | Old locus/aliasa |
|---|---|---|---|---|
| Burkholderia | ||||
| B. cenocepacia | H111 | 2 | I25_RS17325‐I35_RS17415 | I35_0565‐I35_0547 |
| DWS 37E‐2 | 2 | DM40_RS13140‐DM40_RS13060 | DM40_4776‐DM40_4759 | |
| FL‐5‐3‐30‐S1‐D7 | 2 | WJ11_00625‐WJ11_00725 | n/a | |
| VC12308 | 2 | A8E75_RS00805‐A8E75_RS00910 | A8E75_00815‐A8E75_00910 | |
| D2AES | n/a | W5I_RS0113450‐W5I_RS0113540 | n/a | |
| PC184 Mulks | 1c | B9Z07_RS01130‐B9Z07_RS01225 | B9Z07_01130‐B9Z07_01230 | |
| TAtl‐371 | n/a | BLS50_RS24480‐BLS50_RS24390 | SAMN05443026_4796‐SAMN05443026_4778 | |
| B. ambifaria | RZ2MS16 | n/a | AS146_RS14130‐AS146_RS14210 | n/a |
| B. cepacia | LK29 14 | n/a | VL15_RS08940‐VL15_RS08850 | VL15_08935‐VL15_08845 |
| B. diffusa | MSMB0010 | n/a | WJ30_RS22370‐WJ30_RS22295 | WJ30_23825‐WJ30_23750 |
| B. dolosa | AU0158 | 2 | AK34_RS03975‐AK34_RS04060 | AK34_3963‐AK34_3981 |
| B. latens | AU17928 | 2 | WK25_RS26375‐WK25_RS26295 | WK25_26365‐WK25_26285 |
| B. metallica | FL‐6‐5‐30‐S1‐D7 | 2 | WJ16_RS22925‐WJ16_RS23015 | WJ16_22900‐WJ16_22990 |
| B. pseudomultivorans | MSMB574 | n/a | WT57_RS16705‐WT57_RS16620 | WT57_21995‐WT57_21910 |
| B. puraquae | CAMPA 1040 | n/a | B7G54_RS20355‐B7G54_RS20270 | B7G54_20345‐B7G54_20260 |
| B. pyrrocinia | MSMB1755 | n/a | WJ63_RS05815‐WJ63_RS05755 | WJ63_27610‐WJ63_27550 |
| B. stabilis | LA20W | n/a | BSLA_02f3182‐BSLA_02r3154 | n/a |
| B. stagnalis | MSMB1147 | n/a | WT05_RS32155‐WT05_RS32230 | WT05_32120‐WT05_32195 |
| B. territorii | MSMB1917 | n/a | WT40_RS07200‐WT40_RS07125 | WT40_07845‐WT40_07770 |
| B. ubonesis | MSMB2006 | n/a | WK05_RS31815‐WK05_RS31895 | WK05_23690‐WK05_23770 |
| B. vietnamiensis | FL‐2‐2‐30‐S1‐D0 | n/a | WJ01_RS25680‐WJ01_RS25595 | WJ01_26115‐WJ01_26030 |
| B. glumae | BGR1 | 2 | BGLU_RS23295‐BGLU_RS23205 | bglu_2g11110‐bglu_2g10910 |
| Species related to Burkholderia | ||||
| P. tuberum | DUS833 | n/a | BLU10_RS24530‐BLU10_RS24630 | SAMN05445850_5072‐SAMN05445850_5093 |
| C. metallidurans | CH34 | n/a | RMET_RS03120‐RMET_RS03215 | Rmet_0617‐Rmet_0637 |
Abbreviations: Chr, chromosome; n/a, not applicable.
aGene loci refer to the first (tssE) and last (tssJ) genes in the T6SS‐7 gene cluster in these species as shown in Figure 2.
bIf n/a is stated, chromosome location was not available as the loci coordinates were obtained from draft assemblies consisting of contigs.
cChromosome designation may be incorrect for this genome assembly as the largest chromosome has been designated chromosome 3 and second largest chromosome 1. Furthermore, the nucleotide sequence of chromosome 2 of PC184 Mulks shares significant sequence homology with chromosome 1 of B. cenocepacia J2315 and H111, indicating that chromosome 3 should be designated as chromosome 2, which would mean the T6SS‐7 cluster is located on chromosome 2 in this strain, as observed in other B. cenocepacia strains.
Table A6. Putative T6SS‐dependent effectors localised to tssI gene clusters in B. cenocepacia J2315 identified in silico
| Effector locusa | Chr | tssI locusa | Predicted activity | Functional domain/homologyb | Immunity locusa |
|---|---|---|---|---|---|
| BCAL1166 (QU43_RS42350) | 1 | BCAL1165 (QU43_RS42345) | Peptidoglycan hydrolase | M23 peptidase (PF01551), SH3_3 (PF08239), Lysozyme‐like CTD (cl00222) | BCAL1167 (QU43_RS73515) |
| BCAL1292 (QU43_RS42955) | 1 | BCAL1294 (QU43_RS42965) | Pore‐forming | Tse4 | BCAL1291 (QU43_RS42950) |
| BCAL1296 (QU43_RS42975) | 1 | BCAL1294 (QU43_RS42965) | Phospholipase | PAAR_like/DUF4150 (PF13665), Lipase_3 (cd00519) | BCAL1297 (QU43_RS42980) |
| BCAL1298 (QU43_RS75480) | 1 | BCAL1294 (QU43_RS42965) | Nuclease | GH‐E HNH/ENDO VII nuclease (PF14410) | BCAL1299 (QU43_RS42990) |
| BCAL1358 (QU43_RS43285) | 1 | BCAL1355 (QU43_RS43270) | Phospholipase | Tle1/DUF2235 (PF09994) | BCAL1357 (QU43_RS43280) |
| BCAL1366 (QU43_RS43320) | 1 | BCAL1362 (QU43_RS43300) | Phospholipase | Tle1/DUF2235 (PF09994) | BCAL1365 (QU43_RS43315) |
| BCAL2277 (QU43_RS48015) | 1 | BCAL2279 (QU43_RS48025) | Phospholipase | Tle3, DUF3274 (PF11678) | BCAL2276 (QU43_RS48010), BCAL2274 (QU43_RS48000), BCAL2272 (QU43_RS47990) |
| BCAL2504 (QU43_RS49175) | 1 | BCAL2503c (QU43_RS49165) | Pore‐forming | Tse4 | BCAL2505 (QU43_RS49180) |
| BCAM1464 (QU43_RS74705) | 2 | N/Ad | Peptidoglycan hydrolase | Tae4 | BCAM1465 (QU43_RS61680) |
| BCAM0046 (QU43_RS54630) | 2 | BCAM0043 (QU43_RS54615) | Phospholipase | Tle1/DUF2235 (PF09994) | BCAM0045 (QU43_RS54625) |
| BCAM0149 (QU43_RS55145) | 2 | BCAM0148 (QU43_RS55140) | Phospholipase | Tle5/PLDc_SF (cl15239) | BCAM0150 (QU43_RS55150), BCAM0152 (QU43_RS55160) |
| BCAM2253 (QU43_RS65635) | 2 | BCAM2254 (QU43_RS65645) | NAD+ glycohydrolase | PAAR_RHS (cd14742), RhsA (COG3209), RES CTD (smart00953,214933) | QU43_RS75580 |
| BCAM2252 (QU43_RS65630) | 2 | BCAM2254 (QU43_RS65645) | Unknown | RhsA (COG3209) | BCAM2251a (QU43_RS65625) |
| BCAS0663 (QU43_RS71975) | 3 | BCAS0667 (QU43_RS71995) | Nuclease | PAAR_RHS (cd14742), RhsA (COG3209), HNH/ENDO VII superfamily with WHH CTD (PF14414) | BCAS0662 (QU43_RS71970), BCAS0661c (QU43_RS75295) |
aNew/alias locus names given in parentheses.
bPfam or Conserved Domain Database accession numbers of the domains are given in parentheses.
cDisrupted tssI gene that is very likely to be non‐functional as the encoded product is truncated at the N‐terminus and fused to a tssF orthologue.
dEffector not located near a tssI cluster.
APPENDIX 2.
Further details regarding construction of B. cenocepacia mutants
Primer pairs tssA‐OPfor and tssA‐OPrev, tssK‐OPfor and tssK‐OPrev, tssM‐OPfor and tssM‐OPrev and tagY‐OPfor and tagY‐OPrev were used for PCR screening of candidate tssA::Tp, tssK::Tp, tssM::Tp and tagY::Tp mutants, respectively.
Figure A1.
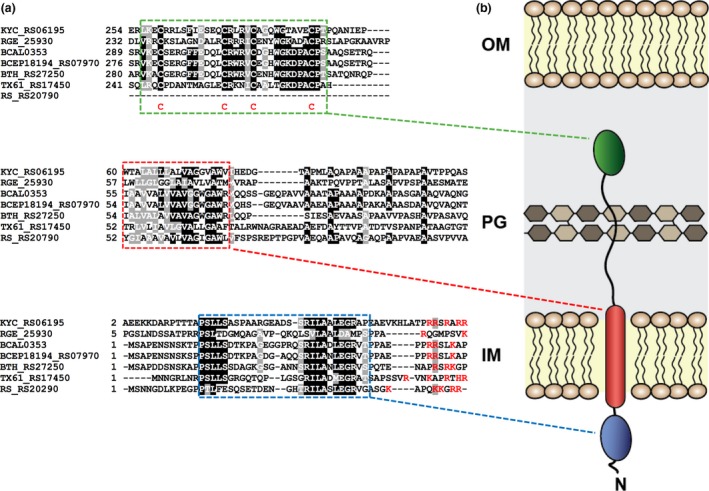
tagY encodes a predicted transmembrane protein with a periplasmically located C‐terminal domain containing four cysteine residues. (A) Amino acid sequence alignment of TagY homologues. The amino acid sequences of TagY homologues identified in B. cenocepacia J2315 (BCAL0353), A. arsenitoxydans SY8 (KYC_RS0619), R. gelatinosus IL144 (RGE_25930), B. lata 383 (BCEP18194_RS07970), B. thailandensis E264 (BTH_RS27250), X. oryzae MAFF 311018 (TX61_RS17450) and R. solanacearum GMI1000 (RS_RS20790) were aligned using Clustal W. Note that the homologue in R. solanacearum does not contain the cysteine rich C ‐terminal domain. Sequences constituting the predicted N‐terminal domain (NTD), transmembrane domain (TMD), and cysteine‐rich periplasmic C‐terminal domain (CTD) are indicated by blue, red and green boxes, respectively. Positively charged amino acids located N‐terminal to the TMD are shown in red font. (B) Schematic representation of the predicted domain arrangement of TagY. The small cytoplasmic NTD (blue), TMD (red), long linker region (black) and cysteine‐rich periplasmic CTD (green) are shown.
Figure A2.
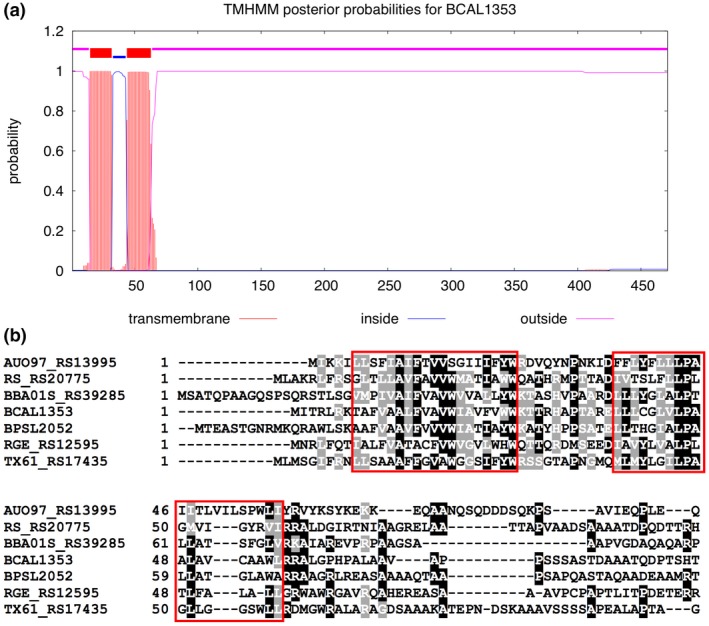
tagZ encodes a putative cytoplasmic membrane protein. (A) Schematic representation of the predicted topology and transmembrane helices within TagZ of B. cenocepacia J2315 (encoded by BCAL1353), as predicted by TMHMM Server (v. 2.0). (B) N‐terminal amino acid sequence alignment of TagZ homologues in B. cenocepacia J2315 (BCAL1353), B. pseudomallei K96243 (BPSL2052), R. gelatinosus IL144 (RGE_RS12595), X. oryzae MAFF 311018 (TX61_RS17434), P. bannensis NBRC 103871 (BBA01S_RS39285), R. solanacearum GMI1000 (RS_RS20775) and A. baumannii ATCC 17978 (AUO97_RS13995). The predicted transmembrane helices are enclosed in red boxes.
Figure A3.
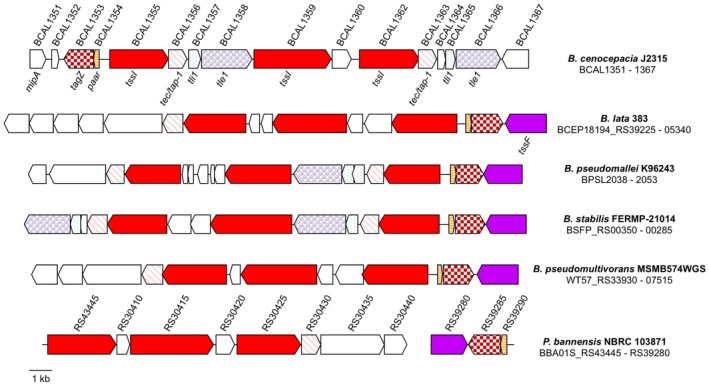
tagZ and paar genes associated with the Burkholderia T6SS‐1 are often located within a triple tssI cluster.Schematic representation of triple tssI gene clusters identified in the indicated Burkholderia spp. and Paraburkholderia bannensis NBRC 103871, which contain the T6SS‐1‐associated tagZ and paar genes. Genes encoding putative T6SS‐lipase effectors (tle1), associated lipase immunity protein(s) (tli1) and T6SS adaptor proteins (tec/tap‐1) are often present within the cluster.
Figure A4.

The T6SS‐7 cluster of B. cenocepacia H111 encodes a putative T6SS type‐5 lipase effector (Tle5) with characteristic dual HxKhhhhD sequence motifs.Amino acid sequence alignment of the two regions containing HxKhhhhD catalytic motifs present in putative T6SS‐dependent type‐5 lipase effector (Tle5) homologues from B. cenocepacia H111 (encoded within the T6SS‐7 cluster at I35_RS17370), B. cenocepacia J2315 (BCAM0149), R. solanacearum GMI1000 and CFBP2957 (RS_RS20135 and RCFBP_mp20221), C. violaceum ATCC 12472 (CV_RS06020), C. metallidurans CH42 (RMET_RS03180), P. aeruginosa PAO1 (PA5089 and PA3487), A. xylosoxidans A8 (AXYL_RS28255) and S. maltophilia SKK35 (SMSKK35_0837). The dual HxKhhhhD motif (where h is an amino acid with a hydrophobic side chain) is characteristic of phospholipase D (PLD) enzymes.
Figure A5.
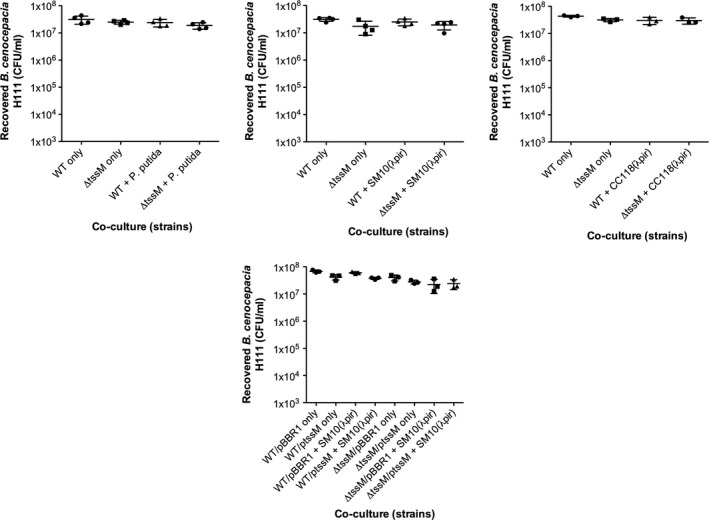
Enumeration of viable B. cenocepacia attacker strains in bacterial competition assay.(A) Recovery of viable B. cenocepacia H111 ‘attacker’ strains following co‐culture with the indicated prey strains or medium control for 4 hours at 30°C. (B) Comparison of recovery of B. cenocepacia H111 WT or ΔtssM mutant attacker strains carrying a complementation or control plasmid (pBBR1‐tssM (ptssM) and pBBR1) following co‐culture with E. coli SM10(λpir) prey or medium control. n ≥ 3 and error bars indicate SD.
Figure A6.
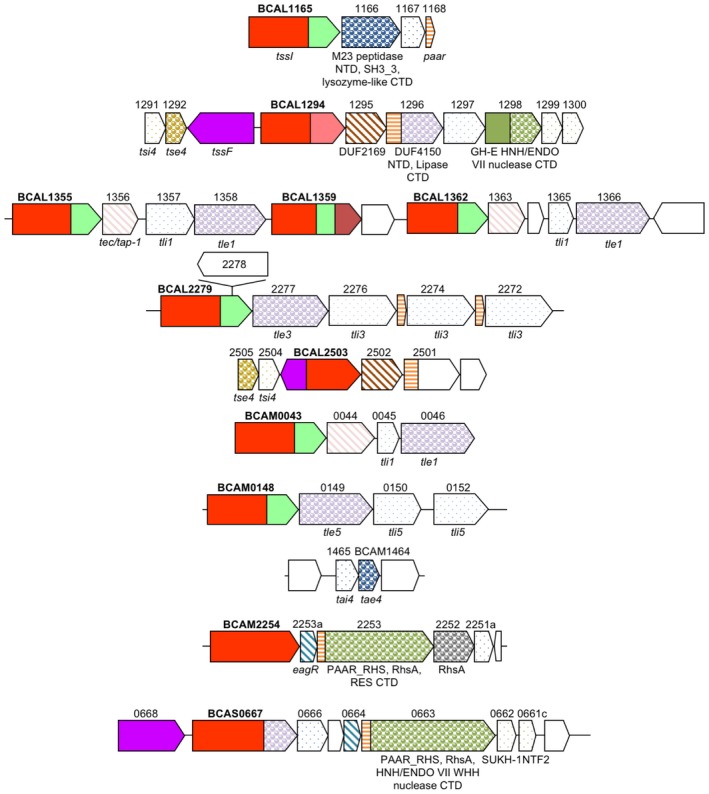
T6SS‐1 effector loci in the B. cenocepacia J2315 genome.Schematic representation of tssI gene clusters identified in B. cenocepacia J2315 and putative T6SS‐dependent effector‐immunity pairs encoded within the clusters (tssI genes are shown in red with C‐terminal coding sequences coloured green (DUF2345 domain), brown (metalloprotease domain) or with spheres (lipase_3 domain) as observed previously (Aubert et al., 2015). The BCAL2503 tssI gene contains a deletion that results in its fusion to a divergently orientated tssF gene and is presumably non‐functional. Genes encoding the putative effectors and associated immunity‐protein are labelled and filled‐in with coloured spheres and dots, respectively. Genes encoding recognized T6SS‐adaptor proteins, Tec/Tap‐1, DUF2169‐encoding proteins and EagR, and PAAR‐containing proteins are also labelled and filled with diagonal coloured stripes and orange horizontal stripes, respectively. tssF homologues are also shown (purple). The BCAM1464‐BCAM1465 tae4‐tai4 effector‐immunity gene pair is not associated with a tssI gene cluster.
Spiewak HL, Shastri S, Zhang L, et al. Burkholderia cenocepacia utilizes a type VI secretion system for bacterial competition. MicrobiologyOpen. 2019;8:e774 10.1002/mbo3.774
DATA ACCESSIBILITY
All data are provided in full in the results section of this paper apart from the DNA sequence contig encompassing B. cenocepacia Pc715j T6SS‐1 gene cluster which is available at www.ncbi.nlm.nih.gov/genbank/ under accession number MK051000.
REFERENCES
- Angus, A. A. , Agapakis, C. M. , Fong, S. , Yerrapragada, S. , Estrada‐de los Santos, P. , Yang, P. , … Hirsch, A. M. (2014). Plant‐associated symbiotic Burkholderia species lack hallmark strategies required in mammalian pathogenesis. PLoS ONE, 9, e83779 10.1371/journal.pone.0083779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschtgen, M.‐S. , Thomas, M. S. , & Cascales, E. (2010). Anchoring the type VI secretion system to the peptidoglycan: TssL, TagL, TagP…what else? Virulence, 1, 535–540. 10.4161/viru.1.6.13732 [DOI] [PubMed] [Google Scholar]
- Aubert, D. F. , Flannagan, R. S. , & Valvano, M. A. (2008). A novel sensor kinase‐response regulator hybrid controls biofilm formation and type VI secretion system activity in Burkholderia cenocepacia . Infection and Immunity, 76, 1979–1991. 10.1128/IAI.01338-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert, D. F. , Hu, S. , & Valvano, M. A. (2015). Quantification of Type VI secretion system activity in macrophages infected with Burkholderia cenocepacia . Microbiology, 161, 2161–2173. 10.1099/mic.0.000174 [DOI] [PubMed] [Google Scholar]
- Aubert, D. F. , Xu, H. , Yang, J. , Chen, S. , Valvano, M. A. , Shao, F. , … Chen, S. (2016). A Burkholderia type VI effector deamidates Rho GTPases to activate the Pyrin inflammasome and trigger inflammation. Cell Host & Microbe, 19, 664–674. 10.1016/j.chom.2016.04.004 [DOI] [PubMed] [Google Scholar]
- Basler, M. (2015). Type VI secretion system: Secretion by a contractile nanomachine. Philosophical Transactions of the Royal Society B: Biological Sciences, 370, 20150021 10.1098/rstb.2015.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler, M. , Ho, B. T. , & Mekalanos, J. J. (2013). Tit‐for‐tat: Type VI secretion system counterattack during bacterial cell‐cell interactions. Cell, 152, 884–894. 10.1016/j.cell.2013.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler, M. , & Mekalanos, J. J. (2012). Type 6 secretion dynamics within and between bacterial cells. Science, 337, 815 10.1126/science.1222901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal, P. , Allsopp, L. P. , Filloux, A. , & Llamas, M. A. (2017). The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME Journal, 11, 972–987. 10.1038/ismej.2016.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biegel, E. , Schmidt, S. , González, J. M. , & Müller, V. (2011). Biochemistry, evolution and physiological function of the Rnf complex, a novel ion‐motive electron transport complex in prokaryotes. Cellular and Molecular Life Sciences, 68, 613–634. 10.1007/s00018-010-0555-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingle, L. E. , Bailey, C. M. , & Pallen, M. J. (2008). Type VI secretion: A beginner’s guide. Current Opinion in Microbiology, 11, 3–8. 10.1016/j.mib.2008.01.006 [DOI] [PubMed] [Google Scholar]
- Brackmann, M. , Nazarov, S. , Wang, J. , & Basler, M. (2017). Using force to punch holes: Mechanics of contractile nanomachines. Trends in Cell Biology, 27, 623–632. 10.1016/j.tcb.2017.05.003 [DOI] [PubMed] [Google Scholar]
- Brunet, Y. R. , Zoued, A. , Boyer, F. , Douzi, B. , & Cascales, E. (2015). The type VI secretion TssEFGK‐VgrG phage‐like baseplate is recruited to the TssJLM membrane complex via multiple contacts and serves as assembly platform for tail tube/sheath polymerization. PLOS Genetics, 11, e1005545 10.1371/journal.pgen.1005545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns, J. L. , Jonas, M. , Chi, E. Y. , Clark, D. K. , Berger, A. , & Griffith, A. (1996). Invasion of respiratory epithelial cells by Burkholderia (Pseudomonas) cepacia . Infection and Immunity, 64, 4054–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtnick, M. N. , Brett, P. J. , Harding, S. V. , Ngugi, S. A. , Ribot, W. J. , Chantratita, N. , … Deshazer, D. (2011). The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei . Infection and Immunity, 79, 1512–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt, A. T. , & Thomas, M. S. (2017). Iron acquisition mechanisms and their role in the virulence of Burkholderia species. Frontiers in Cellular and Infection Microbiology, 7, 460 10.3389/fcimb.2017.00460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier, A. , Agnoli, K. , Pessi, G. , Suppiger, A. , Jenul, C. , Schmid, N. , … Eberl, L. (2014). Genome sequence of Burkholderia cenocepacia H111, a cystic fibrosis airway isolate. Genome Announcements, 2, e00298‐14 10.1128/genomeA.00298-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carruthers, M. D. , Nicholson, P. A. , Tracy, E. N. , & Munson, R. S. (2013). Acinetobacter baumannii utilizes a type VI secretion system for bacterial competition. PLoS ONE, 8, 59388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassaing, B. , & Cascales, E. (2018). Antibacterial weapons: Targeted destruction in the microbiota. Trends in Microbiology, 26, 329–338. 10.1016/j.tim.2018.01.006 [DOI] [PubMed] [Google Scholar]
- Cieri, M. V. , Mayer‐Hamblett, N. , Griffith, A. , & Burns, J. L. (2002). Correlation between an in vitro invasion assay and a murine model of Burkholderia cepacia lung infection. Infection and Immunity, 70, 1081–1086. 10.1128/IAI.70.3.1081-1086.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, K. , Karsch‐Mizrachi, I. , Lipman, D. J. , Ostell, J. , & Sayers, E. W. (2016). GenBank. Nucleic Acids Research, 44(D1), D67–D72. 10.1093/nar/gkv1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett, C. R. , Burtnick, M. N. , Kooi, C. , Woods, D. E. , & Sokol, P. A. (2003). An extracellular zinc metalloprotease gene of Burkholderia cepacia . Microbiology, 149, 2263–2271. 10.1099/mic.0.26243-0 [DOI] [PubMed] [Google Scholar]
- de Campos, S. B. , Lardi, M. , Gandolfi, A. , Eberl, L. , & Pessi, G. (2017). Mutations in two Paraburkholderia phymatum type VI secretion systems cause reduced fitness in interbacterial competition. Frontiers in Microbiology, 8, 2473 10.3389/fmicb.2017.02473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo, V. , & Timmis, K. N. (1994). Analysis and construction of stable phenotypes in gram‐negative bacteria with Tn5‐ and Tn10‐derived minitransposons. Methods in Enzymology, 235, 386–405. [DOI] [PubMed] [Google Scholar]
- Depoorter, E. , Bull, M. J. , Peeters, C. , Coenye, T. , Vandamme, P. , & Mahenthiralingam, E. (2016). Burkholderia: An update on taxonomy and biotechnological potential as antibiotic producers. Applied Microbiology and Biotechnology, 100, 5215–5229. 10.1007/s00253-016-7520-x [DOI] [PubMed] [Google Scholar]
- Diniz, J. A. , & Coulthurst, S. J. (2015). Intraspecies competition in Serratia marcescens is mediated by type VI‐secreted Rhs effectors and a conserved effector‐associated accessory protein. Journal of Bacteriology, 197, 2350–2360. 10.1128/JB.00199-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix, S. R. , Owen, H. J. , Sun, R. , Ahmad, A. , Shastri, S. , Spiewak, H. L. , … Thomas, M. S. (2018). Structural insights into the function of type VI secretion system TssA subunits. Nature Communications, 9, 4765 10.1038/s41467-018-07247-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa, A. P. , Linardopoulou, E. V. , & Samadpour, M. (2017). Transfer of 13 species of the genus Burkholderia to the genus Caballeronia and reclassification of Burkholderia jirisanensis as Paraburkholderia jirisanensis comb. nov. International Journal of Systematic and Evolutionary Microbiology, 67, 3846–3853. 10.1099/ijsem.0.002202 [DOI] [PubMed] [Google Scholar]
- Drevinek, P. , & Mahenthiralingam, E. (2010). Burkholderia cenocepacia in cystic fibrosis: Epidemiology and molecular mechanisms of virulence. Clinical Microbiology and Infection, 16, 821–830. 10.1111/j.1469-0691.2010.03237.x [DOI] [PubMed] [Google Scholar]
- Durand, E. , Cambillau, C. , Cascales, E. , & Journet, L. (2014). VgrG, Tae, Tle, and beyond: The versatile arsenal of Type VI secretion effectors. Trends in Microbiology, 22(9), 498–507. [DOI] [PubMed] [Google Scholar]
- Durand, E. , Nguyen, V. S. , Zoued, A. , Logger, L. , Péhau‐Arnaudet, G. , Aschtgen, M.‐S. , … Fronzes, R. (2015). Biogenesis and structure of a type VI secretion membrane core complex. Nature, 523, 555–560. 10.1038/nature14667 [DOI] [PubMed] [Google Scholar]
- Eberl, L. , & Vandamme, P. (2016). Members of the genus Burkholderia: Good and bad guys. F1000Research, 5, 1007 10.12688/f1000research.8221.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elofsson, A. , & von Heijne, G. (2007). Membrane protein structure: Prediction versus reality. Annual Reviews in Biochemistry, 76, 125–140. 10.1146/annurev.biochem.76.052705.163539 [DOI] [PubMed] [Google Scholar]
- Flannagan, R. S. , Jaumouillé, V. , Huynh, K. K. , Plumb, J. D. , Downey, G. P. , Valvano, M. A. , & Grinstein, S. (2012). Burkholderia cenocepacia disrupts host cell actin cytoskeleton by inactivating Rac and Cdc42. Cellular Microbiology, 14, 239–254. 10.1111/j.1462-5822.2011.01715.x [DOI] [PubMed] [Google Scholar]
- Fritsch, M. J. , Trunk, K. , Alcoforado Diniz, J. , Guo, M. , Trost, M. , & Coulthurst, S. J. (2013). Proteomic identification of novel secreted anti‐bacterial toxins of the Serratia marcescens type VI secretion system. Molecular and Cellular Proteomics, 12, 2735–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilin, M. A. , Abdelaziz, D. H. A. , Mostafa, M. , Abdulrahman, B. A. , Grandhi, J. , Akhter, A. , … Amer, A. O. (2012). Activation of the pyrin inflammasome by intracellular Burkholderia cenocepacia . Journal of Immunology, 188, 3469–3477. 10.4049/jimmunol.1102272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisenberger, O. , Givskov, M. , Riedel, K. , Høiby, N. , Tümmler, B. , & Eberl, L. (2000). Production of N‐acyl‐L‐homoserine lactones by P. aeruginosa isolates from chronic lung infections associated with cystic fibrosis. FEMS Microbiology Letters, 184, 273–278. [DOI] [PubMed] [Google Scholar]
- Gerc, A. J. , Diepold, A. , Trunk, K. , Porter, M. , Rickman, C. , Armitage, J. P. , … Coulthurst, S. J. (2015). Visualization of the Serratia type VI secretion system reveals unprovoked attacks and dynamic assembly. Cell Reports, 12, 2131–2142. 10.1016/j.celrep.2015.08.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero, M. , de Lorenzo, V. , & Timmis, K. N. (1990). Transposon vectors containing non‐antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram‐negative bacteria. Journal of Bacteriology, 172, 6557–6567. 10.1128/jb.172.11.6557-6567.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, B. T. , Basler, M. , & Mekalanos, J. J. (2013). Type 6 secretion system‐mediated immunity to type 4 secretion system‐mediated gene transfer. Science, 342, 250–253. 10.1126/science.1243745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, B. T. , Dong, T. G. , & Mekalanos, J. J. (2014). A view to a kill: The bacterial type VI secretion system. Cell Host & Microbe, 15, 9–21. 10.1016/j.chom.2013.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, R. D. , Singh, P. , Hsu, F. , Güvener, T. , Carl, M. A. , Trinidad, R. R. , … Mougous, J. D. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host & Microbe, 7, 25–37. 10.1016/j.chom.2009.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, T. A. , Kooi, C. , Sokol, P. A. , & Valvano, M. A. (2004). Identification of Burkholderia cenocepacia genes required for bacterial survival in vivo. Infection and Immunity, 72, 4010–4022. 10.1128/IAI.72.7.4010-4022.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, F. , Waterfield, N. R. , Yang, J. , Yang, G. , & Jin, Q. (2014). A Pseudomonas aeruginosa type VI secretion phospholipase D effector targets both prokaryotic and eukaryotic cells. Cell Host & Microbe, 15, 600–610. 10.1016/j.chom.2014.04.010 [DOI] [PubMed] [Google Scholar]
- Keith, K. E. , Hynes, D. W. , Sholdice, J. E. , & Valvano, M. A. (2009). Delayed association of the NADPH oxidase complex with macrophage vacuoles containing the opportunistic pathogen Burkholderia cenocepacia . Microbiology, 155, 1004–1015. 10.1099/mic.0.026781-0 [DOI] [PubMed] [Google Scholar]
- Kotrange, S. , Kopp, B. , Akhter, A. , Abdelaziz, D. , Abu Khweek, A. , Caution, K. , … Amer, A. O. (2011). Burkholderia cenocepacia O polysaccharide chain contributes to caspase‐1‐dependent IL‐1beta production in macrophages. Journal of Leukocyte Biology, 89, 481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh, A. , Larsson, B. , Von Heijne, G. , & Sonnhammer, E. L. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. Journal of Molecular Biology, 305, 567–580. [DOI] [PubMed] [Google Scholar]
- LaCourse, K. D. , Peterson, S. B. , Kulasekara, H. D. , Radey, M. C. , Kim, J. , & Mougous, J. D. (2018). Conditional toxicity and synergy drive diversity among antibacterial effectors. Nature Microbiology, 3, 440–446. 10.1038/s41564-018-0113-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, M. A. , Blackshields, G. , Brown, N. P. , Chenna, R. , McGettigan, P. A. , McWilliam, H. , … Higgins, D. G. (2007). Clustal W and Clustal X version 2.0. Bioinformatics, 23, 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Leiman, P. G. , Basler, M. , Ramagopal, U. A. , Bonanno, J. B. , Sauder, J. M. , Pukatzki, S. , … Mekalanos, J. J. (2009). Type VI secretion apparatus and phage tail‐associated protein complexes share a common evolutionary origin. Proceedings of the National Academy of Sciences of the United States of America, 106, 4154–4159. 10.1073/pnas.0813360106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien, Y.‐W. , & Lai, E.‐M. (2017). Type VI secretion effectors: Methodologies and biology. Frontiers in Cellular and Infection Microbiology, 7, 254 10.3389/fcimb.2017.00254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J.‐S. , Pissaridou, P. , Wu, H.‐H. , Tsai, M.‐D. , Filloux, A. , & Lai, E.‐M. (2018). TagF‐mediated repression of bacterial type VI secretion systems involves a direct interaction with the cytoplasmic protein Fha. Journal of Biological Chemistry, 293, 8829–8842. 10.1074/jbc.RA117.001618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossi, N. S. , Manoli, E. , Simpson, P. , Jones, C. , Hui, K. , Dajani, R. , … Filloux, A. (2012). The archetype Pseudomonas aeruginosa proteins TssB and TagJ form a novel subcomplex in the bacterial type VI secretion system. Molecular Microbiology, 86, 437–456. [DOI] [PubMed] [Google Scholar]
- Lynch, K. H. , Stothard, P. , & Dennis, J. J. (2010). Genomic analysis and relatedness of P2‐like phages of the Burkholderia cepacia complex. BMC Genomics, 11, 599 10.1186/1471-2164-11-599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, A. T. , McAuley, S. , Pukatzki, S. , & Mekalanos, J. J. (2009). Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host & Microbe, 5, 234–243. 10.1016/j.chom.2009.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L.‐S. , Hachani, A. , Lin, J.‐S. , Filloux, A. , & Lai, E.‐M. (2014). Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta . Cell Host & Microbe, 16, 94–104. 10.1016/j.chom.2014.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntyre, D. L. , Miyata, S. T. , Kitaoka, M. , & Pukatzki, S. (2010). The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proceedings of the National Academy of Sciences of the United States of America, 107, 19520–19524. 10.1073/pnas.1012931107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, D. W. , & Mohr, C. D. (2000). Invasion and intracellular survival of Burkholderia cepacia . Infection and Immunity, 68, 24–29. 10.1128/IAI.68.1.24-29.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon, S. , McClean, S. , & Callaghan, M. (2010). Macrophage responses to CF pathogens: JNK MAP kinase signaling by Burkholderia cepacia complex lipopolysaccharide. FEMS Immunology and Medical Microbiology, 60, 36–43. 10.1111/j.1574-695X.2010.00712.x [DOI] [PubMed] [Google Scholar]
- Mesureur, J. , & Vergunst, A. C. (2014). Zebrafish embryos as a model to study bacterial virulence. Methods in Molecular Biology, 1197, 41–66. [DOI] [PubMed] [Google Scholar]
- Mesureur, J. , Feliciano, J. R. , Wagner, N. , Gomes, M. C. , Zhang, L. , Blanco‐Gonzalez, M. , … Vergunst, A. C. (2017). Macrophages, but not neutrophils, are critical for proliferation of Burkholderia cenocepacia and ensuing host‐damaging inflammation. PLoS Path, 13, e1006437 10.1371/journal.ppat.1006437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles, A. A. , Misra, S. S. , & Irwin, J. O. (1938). The estimation of the bactericidal power of the blood. The Journal of Hygiene, 38(6), 732–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougous, J. D. , Cuff, M. E. , Raunser, S. , Shen, A. , Zhou, M. , Gifford, C. A. , … Mekalanos, J. J. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science, 312, 1526–1530. 10.1126/science.1128393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch, S. L. , Trunk, K. , English, G. , Fritsch, M. J. , Pourkarimi, E. , & Coulthurst, S. J. (2011). The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. Journal of Bacteriology, 193, 6057–6069. 10.1128/JB.05671-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, V. S. , Logger, L. , Spinelli, S. , Legrand, P. , Huyen Pham, T. T. , Nhung Trinh, T. T. , … Cambillau, C. (2017). Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor‐binding proteins and evolved to bind the membrane complex. Nature Microbiology, 2, 17103 10.1038/nmicrobiol.2017.103 [DOI] [PubMed] [Google Scholar]
- Planamente, S. , Salih, O. , Manoli, E. , Albesa‐Jové, D. , Freemont, P. S. , & Filloux, A. (2016). TssA forms a gp6‐like ring attached to the type VI secretion sheath. EMBO Journal, 28, 821–829. 10.15252/embj.201694024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki, S. , Ma, A. T. , Revel, A. T. , Sturtevant, D. , & Mekalanos, J. J. (2007). Type VI secretion system translocates a phage tail spike‐like protein into target cells where it cross‐links actin. Proceedings of the National Academy of Sciences of the United States of America, 104, 15508–15513. 10.1073/pnas.0706532104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki, S. , Ma, A. T. , Sturtevant, D. , Krastins, B. , Sarracino, D. , Nelson, W. C. , … Mekalanos, J. J. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proceedings of the National Academy of Sciences of the United States of America, 103, 1528–1533. 10.1073/pnas.0510322103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repizo, G. D. , Gagné, S. , Foucault‐Grunenwald, M.‐L. , Borges, V. , Charpentier, X. , Limansky, A. S. , … Salcedo, S. P. (2015). Differential role of the T6SS in Acinetobacter baumannii virulence. PLoS ONE, 10, e0138265 10.1371/journal.pone.0138265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel, K. , Carranza, P. , Gehrig, P. , Potthast, F. , & Eberl, L. (2006). Towards the proteome of Burkholderia cenocepacia H111: Setting up a 2‐DE reference map. Proteomics, 6, 207–216. [DOI] [PubMed] [Google Scholar]
- Ringel, P. D. , Hu, D. , & Basler, M. (2017). The role of type VI secretion system effectors in target cell lysis and subsequent horizontal gene transfer. Cell Reports, 21, 3927–3940. 10.1016/j.celrep.2017.12.020 [DOI] [PubMed] [Google Scholar]
- Rosales‐Reyes, R. , Skeldon, A. M. , Aubert, D. F. , & Valvano, M. A. (2012). The type VI secretion system of Burkholderia cenocepacia affects multiple Rho family GTPases disrupting the actin cytoskeleton and the assembly of NADPH oxidase complex in macrophages. Cellular Microbiology, 14, 255–273. 10.1111/j.1462-5822.2011.01716.x [DOI] [PubMed] [Google Scholar]
- Russell, A. B. , Hood, R. D. , Bui, N. K. , LeRoux, M. , Vollmer, W. , & Mougous, J. D. (2011). Type VI secretion delivers bacteriolytic effectors to target cells. Nature, 475, 343–347. 10.1038/nature10244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, A. B. , LeRoux, M. , Hathazi, K. , Agnello, D. M. , Ishikawa, T. , Wiggins, P. A. , … Mougous, J. D. (2013). Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature, 496, 508–512. 10.1038/nature12074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, A. B. , Singh, P. , Brittnacher, M. , Bui, N. K. , Hood, R. D. , Carl, M. A. , … Mougous, J. D. (2012). A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host & Microbe, 11, 538–549. 10.1016/j.chom.2012.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sana, T. G. , Flaugnatti, N. , Lugo, K. A. , Lam, L. H. , Jacobson, A. , Baylot, V. , … Monack, D. M. (2016). Salmonella typhimurium utilizes a T6SS‐mediated antibacterial weapon to establish in the host gut. Proceedings of the National Academy of Sciences of the United States of America, 113, E5044–E5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sana, T. G. , Hachani, A. , Bucior, I. , Soscia, C. , Garvis, S. , Termine, E. , … Bleves, S. (2012). The second type VI secretion system of Pseudomonas aeruginosa strain PAO1 is regulated by quorum sensing and Fur and modulates internalization in epithelial cells. Journal of Biological Chemistry, 287, 27095–27105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawana, A. , Adeolu, M. , & Gupta, R. S. (2014). Molecular signatures and phylogenomic analysis of the genus Burkholderia: Proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Frontiers in Genetics, 5, 429 10.3389/fgene.2014.00429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell, M. A. , Ulrich, R. L. , Ribot, W. J. , Brueggemann, E. E. , Hines, H. B. , Chen, D. , … Deshazer, D. (2007). Type VI secretion is a major virulence determinant in Burkholderia mallei. Molecular Microbiology, 64, 1466–1485. [DOI] [PubMed] [Google Scholar]
- Schmehl, M. , Jahn, A. , Meyer zu Vilsendorf, A. , Hennecke, S. , Masepohl, B. , Schuppler, M. , … Klipp, W. (1993). Identification of a new class of nitrogen fixation genes in Rhodobacter capsalatus: A putative membrane complex involved in electron transport to nitrogenase. Molecular and General Genetics, 241, 602–615. 10.1007/BF00279903 [DOI] [PubMed] [Google Scholar]
- Schwarz, S. , West, T. E. , Boyer, F. , Chiang, W.‐C. , Carl, M. A. , Hood, R. D. , … Mougous, J. D. (2010). Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Path, 6, e1001068 10.1371/journal.ppat.1001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed, K. D. , & Dennis, J. J. (2008). Development of Galleria mellonella as an alternative infection model for the Burkholderia cepacia complex. Infection and Immunity, 76, 1267–1275. 10.1128/IAI.01249-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalom, G. , Shaw, J. G. , & Thomas, M. S. (2007). In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology, 153, 2689–2699. 10.1099/mic.0.2007/006585-0 [DOI] [PubMed] [Google Scholar]
- Shastri, S. , Spiewak, H. L. , Sofoluwe, A. , Eidsvaag, V. A. , Asghar, A. H. , Pereira, T. , … Thomas, M. S. (2017). An efficient system for the generation of marked genetic mutants in members of the genus Burkholderia . Plasmid, 89, 49–56. 10.1016/j.plasmid.2016.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers, F. , Wilm, A. , Dineen, D. , Gibson, T. J. , Karplus, K. , Li, W. , … Higgins, D. G. (2011). Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology, 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman, J. M. , Austin, L. S. , Hsu, F. , Hicks, K. G. , Hood, R. D. , & Mougous, J. D. (2011). Separate inputs modulate phosphorylation‐dependent and ‐independent type VI secretion activation. Molecular Microbiology, 82, 1277–1290. 10.1111/j.1365-2958.2011.07889.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skjerning, R. B. , Senissar, M. , Winther, K. , Gerdes, K. , & Brodersen, D. E. (2018). The RES domain toxins of RES‐Xre toxin‐antitoxin modules induce cell stasis by degrading NAD. Molecular Microbiology. 10.1111/mmi.14150 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Sokol, P. A. , Ohman, D. E. , & Iglewski, B. H. (1979). A more sensitive plate assay for detection of protease production by Pseudomanas aeruginosa . Journal of Clinical Microbiology, 9, 538–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol, P. A. , Sajjan, U. , Visser, M. B. , Gingues, S. , Forstner, J. , & Kooi, C. (2003). The CepIR quorum‐sensing system contributes to the virulence of Burkholderia cenocepacia respiratory infections. Microbiology, 149, 3649–3658. 10.1099/mic.0.26540-0 [DOI] [PubMed] [Google Scholar]
- Uehlinger, S. , Schwager, S. , Bernier, S. P. , Riedel, K. , Nguyen, D. T. , Sokol, P. A. , & Eberl, L. (2009). Identification of specific and universal virulence factors in Burkholderia cenocepacia strains by using multiple infection hosts. Infection and Immunity, 77, 4102–4110. 10.1128/IAI.00398-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergunst, A. C. , Meijer, A. H. , Renshaw, S. A. , & O’Callaghan, D. (2010). Burkholderia cenocepacia creates an intramacrophage replication niche in zebrafish embryos, followed by bacterial dissemination and establishment of systemic infection. Infection and Immunity, 78, 1495–1508. 10.1128/IAI.00743-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, B. , Zhang, Q. , Ni, J. , Li, S. , Wen, D. , Li, J. , … Yao, Y.‐F. (2017). Type VI secretion system contributes to Enterohemorrhagic Escherichia coli virulence by secreting catalase against host reactive oxygen species (ROS). PLoS Path, 13, e1006246 10.1371/journal.ppat.1006246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, B. S. , Miyata, S. T. , Iwashkiw, J. A. , Mortensen, B. L. , Skaar, E. P. , Pukatzki, S. , & Feldman, M. F. (2013). Genomic and functional analysis of the type VI secretion system in Acinetobacter . PLoS ONE, 8, e55142 10.1371/journal.pone.0055142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, B. S. , Hennon, S. W. , Wright, M. S. , Scott, N. E. , de Berardinis, V. , Foster, L. J. , … Feldman, M. F. (2016). Genetic dissection of the type VI secretion system in Acinetobacter and identification of a novel peptidoglycan hydrolase, TagX, required for its biogenesis. Mbio, 7, e01253‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney, J. C. , Beck, C. M. , Goo, Y. A. , Russell, A. B. , Harding, B. N. , De Leon, J. A. , … Mougous, J. D. (2014). Genetically distinct pathways guide effector export through the type VI secretion system. Molecular Microbiology, 92, 529–542. 10.1111/mmi.12571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H. , Yang, J. , Gao, W. , Li, L. , Li, P. , Zhang, L. , … Shao, F. (2014). Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature, 513, 237–241. 10.1038/nature13449 [DOI] [PubMed] [Google Scholar]
- Zhao, W. , Caro, F. , Robins, W. , & Mekalanos, J. J. (2018). Antagonism toward the intestinal microbiota and its effect on Vibrio cholerae virulence. Science, 359, 210–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, J. , Shin, O. S. , Cameron, D. E. , & Mekalanos, J. J. (2010). Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proceedings of the National Academy of Sciences of the United States of America, 107, 21128–21133. 10.1073/pnas.1014998107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoued, A. , Durand, E. , Brunet, Y. R. , Spinelli, S. , Douzi, B. , Guzzo, M. , … Cascales, E. (2016). Priming and polymerization of a bacterial contractile tail structure. Nature, 531(7592), 59–63. 10.1038/nature17182 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are provided in full in the results section of this paper apart from the DNA sequence contig encompassing B. cenocepacia Pc715j T6SS‐1 gene cluster which is available at www.ncbi.nlm.nih.gov/genbank/ under accession number MK051000.


