Abstract
Radiotherapy treatment planning systems are designed for the fast calculation of dose to the tumor bed and nearby organs at risk using x-ray computed tomography (CT) images. However, CT images for a patient are typically available for only a small portion of the body, and in some cases, such as for retrospective epidemiological studies, no images may be available at all. When dose to organs that lie out-of-scan must be estimated, a convenient alternative for the unknown patient anatomy is to use a matching whole-body computational phantom as a surrogate. The purpose of the current work is to connect such computational phantoms to commercial radiotherapy treatment planning systems for retrospective organ dose estimation. A custom software with graphical user interface, called the DICOM-RT Generator, was developed in MATLAB to convert voxel computational phantoms into the Digital Imaging and Communications in Medicine radiotherapy (DICOM-RT) format, compatible with commercial treatment planning systems. DICOM CT image sets for the phantoms are created via a density-to-Hounsfield unit conversion curve. Accompanying structure sets containing the organ contours are automatically generated by tracing binary masks of user-specified organs on each phantom CT slice. The software was tested on a library of body size-dependent phantoms, the International Commission on Radiological Protection reference phantoms, and a canine voxel phantom, taking only a few minutes per conversion. The resulting DICOM-RT files were tested on several commercial treatment planning systems. As an example application, a library of converted phantoms was used to estimate organ doses for members of the National Wilms Tumor Study cohort. The converted phantom library, in DICOM format, and a standalone MATLAB-compiled executable of the DICOM-RT Generator are available for others to use for research purposes (http://ncidose.cancer.gov).
1. Introduction
Cancer survival rates have improved over the past several decades (Jemal et al 2017), with current estimates showing that around half of all cancer patients receive radiotherapy at some point during the course of their treatment (Barton et al 2014, Tyldesley et al 2011). As the cancer survivor population grows (Miller et al 2016), it becomes increasingly important to understand the late-term health effects from radiotherapy, such as second cancer. Risk projection models are needed for organs which lie in the path of the radiation treatment beams (“in-field”) as well as those which do not (“out-of-field”). Populations that have undergone radiotherapy offer unique insight into the link between radiation exposure and subsequent second cancer risk, as the radiotherapy procedure is ordinarily well-documented in medical records (Stovall et al 2006), allowing organ doses to be retrospectively calculated from treatment details. However, for many retrospective dosimetry studies, anatomical x-ray computed tomography (CT) data are either unavailable or do not cover out-of-field organs at risk. In these circumstances, computational phantoms can be a convenient surrogate for the unknown patient anatomy. Whereas CT images typically cover only part of the body and require manual segmentation of all tissues of interest, state-of-the-art computational phantoms (Xu 2014) provide whole-body anatomy for extended dosimetry with detailed organs and tissues already delineated. In collaboration with the University of Florida, the National Cancer Institute (NCI) has recently developed an extensive library of computational hybrid phantoms covering a large portion of the U.S. population in terms of age, gender, height, and weight (Geyer et al 2014, Lee et al 2010).
Such phantom resources could be used to reconstruct or extend patient anatomy for a multitude of radiotherapy applications, assuming that the phantom can first be properly defined in a file format compatible with commercial treatment planning systems (TPS). This file format, the Digital Imaging and Communications in Medicine (DICOM), is the international standard for the storage of medical image information. Though initially an implementation of image archiving, DICOM was later extended to other fields of medicine, such as radiotherapy (RT), where it is critical for diagnostic images and radiotherapy treatment details to be stored in a standardized way for seamless interaction. DICOM-RT is an extension of the DICOM standard, allowing radiotherapy objects, such as the RT-Structure, RT-Dose, and RT-Plan objects, to be added to CT image data. In DICOM, the CT files hold the image data that would be expected from each 2D slice of a CT scan and define the Hounsfield unit (HU) values at each image pixel. The RT-Structure file holds information on the contours of all organs of interest, which would typically be manually segmented using a TPS.
Previous research has established workflows using computer-aided design software to make the conversion from voxel phantom to DICOM-RT format possible, but manually rigorous and slow (Lee et al 2015). The current work aims to make this connection between phantom resources and radiotherapy software fully-automatic and more efficient. This conversion from voxel phantom to DICOM-RT format requires very little user input – only the voxel phantom, a tag dictionary file, and the phantom’s physical dimensions (matrix size and resolution) are required. All inputs are made within a newly developed software tool, named the DICOM-RT Generator, which has been compiled within the MATLAB (The Mathworks, Inc., Natick, MA) Compiler Toolkit for future distribution.
2. Materials and methods
2.1. Formatting of the DICOM-RT files
DICOM files consist of data contents and a header which describes these contents through metadata elements called tag identifiers. For DICOM CT files, the header contains identifying metadata such as patient information, date and time of image acquisition, series number, image resolution (voxel size and coordinate system), etc.; the data contents are stored as an image matrix defining the HU values for each pixel. RT-Structure files, on the other hand, are all-inclusive metadata files. All of the contour data, along with other identifiers that tie this data to its CT image series, are contained within the header of the RT-Structure file. The DICOM standard (NEMA 2018) specifies the file format, including numerous metadata tags which must be defined in order to be compliant with the standard. While a voluntary standard, the DICOM file format is typically required in order for files to be compatible with commercial radiotherapy treatment planning software; however, the exact implementation of DICOM can vary by vendor and is detailed in DICOM conformance statements released with their software. The majority of these required DICOM metadata tags are not discussed within this manuscript because they relate to trivial factors (patient’s name, study date, etc.) for the conversion process. Instead, only those metadata tags deemed significant for the conversion process will be discussed. These tags have been summarized in Table 1 to provide context as to how the RT-Structure file and its constituent organ contours are linked to each CT file and are subsequently identified by the TPS.
Table 1:
Selective listing of important metadata tags for the conversion process and for DICOM compliance.
| Common Metadata Tags | |
|---|---|
| SOPClassUID | Tells the TPS what type of data is held within the DICOM file. Ex: Values of ‘1.2.840.10008.5.1.4.1.1.2’ and ‘1.2.840.10008.5.1.4.1.1.481.3’ tell software that the written data are CT images or RT-Structure files, respectively. |
| TransferSyntaxUID | The set of encoding rules used during file write-out. In the DICOM-RT Generator, the Explicit VR Little Endian syntax is used (‘1.2.840.10008.1.2.1’), as some TPS require every metadata element to have been explicitly serialized. |
| StudyInstanceUID & SeriesInstanceUID | Unique IDs for the “study” and “series” that the data belong to. Unique IDs are randomly generated in MATLAB using the built-in dicomuid function. |
| SOPInstanceUID | Unique ID for any file. This is used to link organ contours to a particular CT image slice. MATLAB generates a unique SOPInstanceUID during file write-out with the dicomwrite function. |
| FrameOfReferenceUID | Unique ID for the coordinate system for a series of images. By existing within the same Frame of Reference, the TPS knows that images must be spatially related to one another. |
| CT Specific Metadata Tags | |
| Image Position Patient (IPP) | Identifies the coordinates (mm) of the center of the voxel located in the upper-left-hand corner of the image slice. |
| Image Orientation Patient (IOP) | The directional cosines of the image’s rows and columns with respect to the IPP. |
| PixelSpacing | Physical dimensions of the phantom (mm): row spacing (y-dimension), column spacing (x-dimension). |
| SliceThickness | Physical dimension of the phantom (mm) determining the space between image slices (z-dimension). |
| RT-Structure Specific Metadata Tags | |
| StructureSetROISequence | Data structure that contains the order of the organ contours and the names for each contour. |
| RTROIObservationsSequence | Data structure that, for each organ contour, details whether it is an organ contour or external contour. |
| ROIContourSequence | Contains a large structure of organ contour data on each image slice, linked using the SOPInstanceUID. |
2.2. Creation of the phantom DICOM CT image set
As a first step, the DICOM-RT Generator converts the voxel phantom into a simulated x-ray CT image set. The voxel phantom, in binary file format, contains 8-bit unsigned integers for every voxel, with each integer representing an organ or tissue within the phantom. The organ tag integers are read into MATLAB and stored as a three-dimensional matrix. Scripts were written to convert the organ tags to their associated mass density based on a user-provided tag dictionary file. These mass densities are converted into HU values via linear interpolation of a mass density-to-HU value calibration curve which was clinically commissioned at the University of Michigan (Lee et al 2015). The default pixel data within DICOM format is stored as unsigned integers, and the minimal value for any HU is −1024 (corresponding to a density of zero). For convenience, the final data is stored in the matrix with an additional 1024 HU so that the data can be stored as unsigned 16-bit integers.
Within the DICOM CT metadata, the RescaleIntercept and RescaleSlope tags are set to −1024 and 1, respectively, to return these stored data to their original HU values when read by the TPS. Other important metadata tags are discussed within Table 1. The SOPInstanceUID is uniquely created for each CT image file and is used, in the next step, to connect the organ contours with the CT images they are created from. The image position patient (IPP) tag identifies the coordinates of the center of the voxel located in the upper-left-hand corner of the image slice; only the z-component changes from one slice to the next within an image series. The image orientation patient (IOP) tag is the direction cosines of the image’s rows and columns with respect to the IPP. For example, a value of ‘(1 0 0 0 1 0)’ demonstrates that the rows of the image follow the positive x-direction, right hand to left hand, and the columns follow the positive y-direction, anterior to posterior. Since IPP definition is in the upper left corner position, this IOP states that the patient’s right side is on the left side of the image, and the anterior of the body is at the top of the image. After metadata tags are filled, the CT data and associated CT metadata are written slice-by-slice to DICOM CT image files using MATLAB’s dicomwrite function for each slice in the z-dimension of the phantom HU matrix.
2.3. Creation of the DICOM RT-Structure file
The RT-Structure file contains metadata describing the contours for all N number of organs requested of the DICOM-RT Generator. The organs to be contoured are given to the DICOM-RT Generator through the user-supplied tag dictionary file. Perhaps the most important of all the metadata tags is the ROIContourSequence tag, which holds the physical coordinates of the organ contour boundary locations. To calculate the locations of the organ contour boundaries, the organ tag values of the original voxel phantom are read into a three-dimensional matrix. A binary mask of the organ is then found on each z-dimensional slice by searching for voxels containing that organ’s tag value(s). If an organ can be separated into left and right components (e.g. ovaries, eye lens, kidneys, etc.), the DICOM-RT Generator has the capability to define them as separate structures by considering only the left or right half of the matrix during organ mask calculation. The built-in MATLAB tracing function, bwboundaries, then traces the boundaries of the organ, including the boundaries of any hole regions, and returns a set of boundary locations that define a closed polygon for each boundary; for the same image slice, this can include one external boundary and one or more internal boundaries (e.g. ring-like structure with holes) or multiple external island boundaries (e.g. the left and right kidney as one region of interest). The boundary tracing is performed in the clockwise direction using the Moore-Neighbor tracing algorithm modified by Jacob’s stopping criteria; pixels are considered connected if their edges or corners touched (i.e. connectivity of 8 pixels). The list of boundary locations is then compressed to save file space by cutting out intermediate points on a straight contour line. Afterwards, they are translated to Cartesian positions in the IOP coordinate system using the IPP tag of the image slice and the phantom’s voxel resolution. Once these boundary locations are found, the locations are stored within the ROIContourSequence tag using a branched data structure with fields for each organ, listed Item_1 through Item_N. Within each enumerated branch, the ROIDisplayColor tag is held, which holds the RGB display color of the organ contour, as well as another structure called the ContourSequence. Inside the ContourSequence structure, further nested, enumerated structures are contained for each set of closed polygonal boundary locations found by the bwboundaries function. Here, inside these enumerated structures, the boundary locations are stored within the ContourData tag. For structures with holes or islands, the DICOM convention is to interpret multiple boundaries for an organ on the same image slice as a cumulative subtraction of overlapping regions and cumulative addition of non-overlapping regions (Cutright et al 2018).
Two other important definitions within the RT-Structure metadata are the RTROIObservationsSequence and StructureSetROISequence tags. Like the ROIContourSequence tag, both of these tags are data structures with fields, Item_1 through Item_N, for each organ requested for contouring. Within the enumerated branches of RTROIObservationsSequence, the RTROIInterpretedType tag must be filled out, categorizing the contour as either an organ or external-body contour. Within the enumerated branches of StructureSetROISequence, the ROIName tag must be filled out with the name of the contour. The ReferencedFrameOfReferenceUID tag must also be filled out with the FrameofReferenceUID of the CT image set. After other miscellaneous metadata tags are properly defined, the MATLAB structure object is finally written out as a RT-Structure file using MATLAB’s dicomwrite function.
2.4. Process of the DICOM-RT Generator
A Graphical User Interface (GUI) was created for the DICOM-RT Generator to provide a user-friendly setup for the simultaneous conversion of multiple phantoms in batch, which is helpful for users who need customized DICOM files. The GUI was created using the MATLAB GUI Development Environment program. Any voxel phantom stored in 8-bit unsigned integer raw data format can be converted to DICOM-RT format using the DICOM-RT Generator. In addition to the phantom, the DICOM-RT Generator also requires an accompanying tag dictionary file which lists the organ name and mass density for each integer tag value within the phantom. This file should also hold a list of organ contours to be created and their associated tag values. Lastly, the phantom’s voxel resolution and matrix size are requested by the code to properly scale the physical dimensions of the CT image series and its organ contours. Age and gender can also be given to the code if the phantom is part of an age- or gender-dependent series where organ mass densities vary by age. The DICOM CT image series of the phantom and an RT-Structure file of the contours for all requested organs are generated based on the methods described above. For validation purposes, multiple voxel phantoms were tested within this code: the body size-dependent phantom library (Geyer et al 2014), the UF canine phantom (Padilla et al 2008), and the International Commission on Radiological Protection (ICRP) reference adult male phantom (ICRP 2009). The resulting DICOM-RT formatted phantoms were imported into several different TPSs to verify compatibility. Organ volumes were verified by comparing the original voxel phantom organ volumes to those reported by the TPSs after conversion. The density conversion was verified by spot-checking the HU values for several organs from within the treatment planning software.
3. Results and discussion
3.1. The DICOM-RT Generator
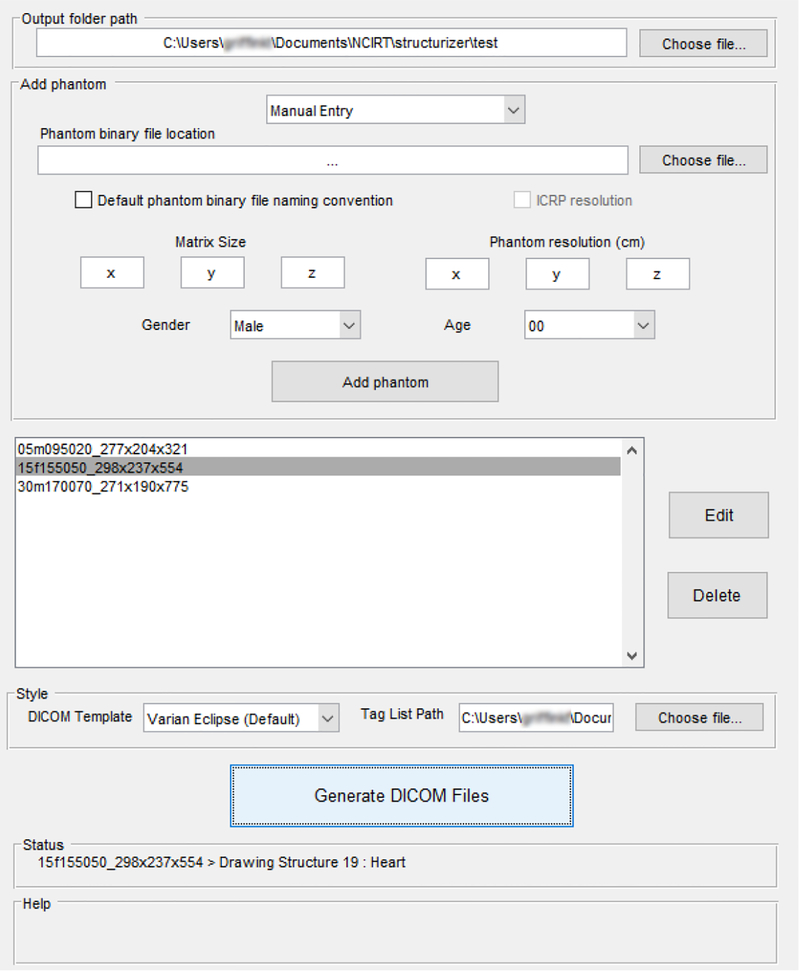
Figure 1 shows the GUI of the DICOM-RT Generator for converting voxel phantoms to DICOM-RT format. An automatic entry mode exists for submitting large numbers of phantoms within a folder to the queue in one step. While the conversion process takes place, a log file is written out to provide users helpful feedback on any errors that have been caught by MATLAB or by the DICOM-RT Generator. The software performance was tested on a variety of phantoms created in-house and gathered from external sources.
Figure 1:
Graphical User Interface of the DICOM-RT Generator, in-use converting a queue of three binary voxel phantoms to DICOM-RT format; status updates for the user are displayed. Phantom names are in “default naming convention” – 15f (age and sex) 155 (height in cm) 050 (weight in kg) _298×237×554 (matrix size).
3.2. Comprehensive phantom library in DICOM format
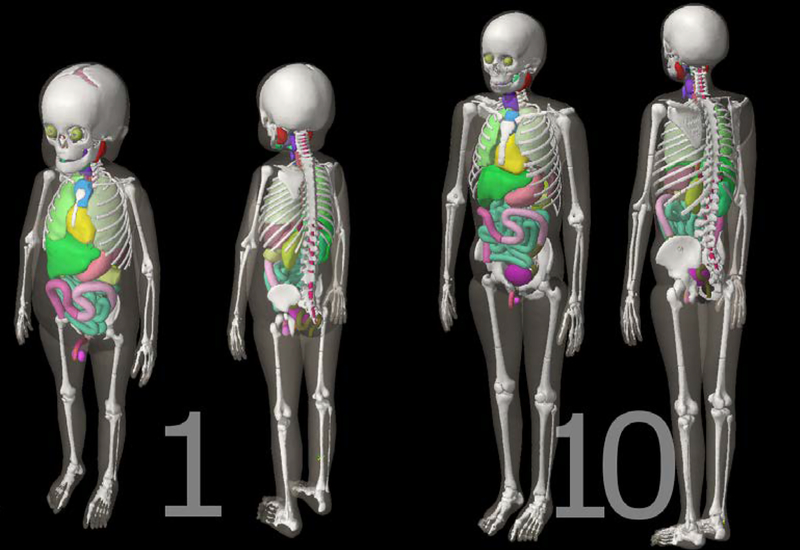
The entirety of the hybrid phantom library (Geyer et al 2014) was first converted into voxel format using an in-house MATLAB script, termed the Voxelizer (Lee et al 2007); these voxel phantoms were then converted into DICOM format using the DICOM-RT Generator, producing 351 total DICOM file sets. Fifty-five organ contours are included, such as: the brain, eyes, eye lens, spinal cord, trachea, gall bladder, colon (ascending, descending, sigmoid, and total), kidneys, lungs, and many more. Cortical and spongiosa bones, grouped into nineteen different bone sites, were also contoured for future red bone marrow dosimetry. Figures 2 and 3 show example visualizations of organ contours for two of the 351 phantoms using Eclipse TPS, in two and three dimensions respectively. Good agreement between organ boundaries and contours can be seen in Figure 2, where the contours of the esophagus, heart, humeri, lungs, ribs, scapulae, spinal cord, sternum, and thoracic vertebrae are depicted for two phantoms. Table 2 quantifies the contouring accuracy through a comparison of organ volumes between the original voxel phantom and the imported DICOM structure volumes calculated by the Varian Eclipse TPS and the CERR viewing platform for the brain, colon, heart, lungs, testes, and thyroid in the one-, five-, and fifteen-year-old male phantoms. Validation of the organ contour volume is shown to within 5% of the original volume for all structures imported into the CERR platform and for all structures (except the testes and thyroid) imported into the Varian Eclipse TPS. The differences for the testes and thyroid are attributed to how the Eclipse TPS interprets the contour data, which seemingly applies a function to the imported contour data for smoothing the contour’s surface. As seen in Figure 3, smooth transitions from one slice to the next occur within the automatically-generated contours. The result is a continuously contoured surface area for all organs, which is due in part to the phantom series’ fine voxel resolution. Run-times of the software were around seven minutes for converting each of the 351 phantoms into DICOM files, with resultant DICOM file sizes (RT-Structure file and all DICOM CT files) of around 100 MB for each phantom. Speed-up in contouring times is likely possible but was not the focus of this study.
Figure 2:
Axial CT images (not to scale) of the one- (left) and ten-year-old (right) male phantoms, visualized within the contouring module of the Varian Eclipse TPS. Contours for organs have been labeled (L/R: left/right; T: thoracic). The external contour can be seen in green around the phantoms.
Figure 3:
Three-dimensional renderings (not to scale) of the one- (left) and ten-year-old (right) male phantoms, visualized within the external beam planning module of the Varian Eclipse TPS. Major organ and skeletal contours are shown. The voxel resolutions (x,y,z) are (0.0663 cm, 0.0663 cm, 0.14 cm) and (0.099 cm, 0.099 cm, 0.2425 cm) for the one- and ten-year-old, respectively.
Table 2:
Organ volume comparisons between the original voxel volume and the volume of the imported DICOM structures as reported by the Varian Eclipse TPS and the CERR viewing platform for six organs in three different-aged male voxel phantoms
| Organ | Age (m) | Original Voxel Volume | Eclipse-Reported Volume | % difference | CERR-Reported Volume | % difference |
|---|---|---|---|---|---|---|
| Brain | 1 | 920.96 | 925.07 | 0.4% | 927.79 | 0.7% |
| 5 | 1195.69 | 1185.87 | −0.8% | 1198.34 | 0.2% | |
| 15 | 1359.89 | 1346.09 | −1.0% | 1362.44 | 0.2% | |
| Colon | 1 | 98.95 | 94.52 | −4.5% | 99.09 | 0.1% |
| 5 | 204.99 | 195.57 | −4.6% | 205.25 | 0.1% | |
| 15 | 525.57 | 502.98 | −4.3% | 525.87 | 0.1% | |
| Heart | 1 | 93.42 | 93.22 | −0.2% | 94.99 | 1.7% |
| 5 | 208.79 | 205.37 | −1.6% | 209.23 | 0.2% | |
| 15 | 624.55 | 615.29 | −1.5% | 625.37 | 0.1% | |
| Lungs | 1 | 374.86 | 376.44 | 0.4% | 380.18 | 1.4% |
| 5 | 711.50 | 698.79 | −1.8% | 713.48 | 0.3% | |
| 15 | 2622.27 | 2585.90 | −1.4% | 2622.71 | 0.0% | |
| Testes | 1 | 1.44 | 1.37 | −4.9% | 1.48 | 2.8% |
| 5 | 1.63 | 1.36 | −16.6% | 1.64 | 0.6% | |
| 15 | 15.41 | 14.01 | −9.1% | 15.42 | 0.1% | |
| Thyroid | 1 | 1.71 | 1.49 | −12.9% | 1.78 | 4.1% |
| 5 | 3.23 | 2.76 | −14.6% | 3.24 | 0.3% | |
| 15 | 11.46 | 10.06 | −12.2% | 11.47 | 0.1% |
3.3. Example Applications
3.3.1. UF canine and ICRP reference phantoms
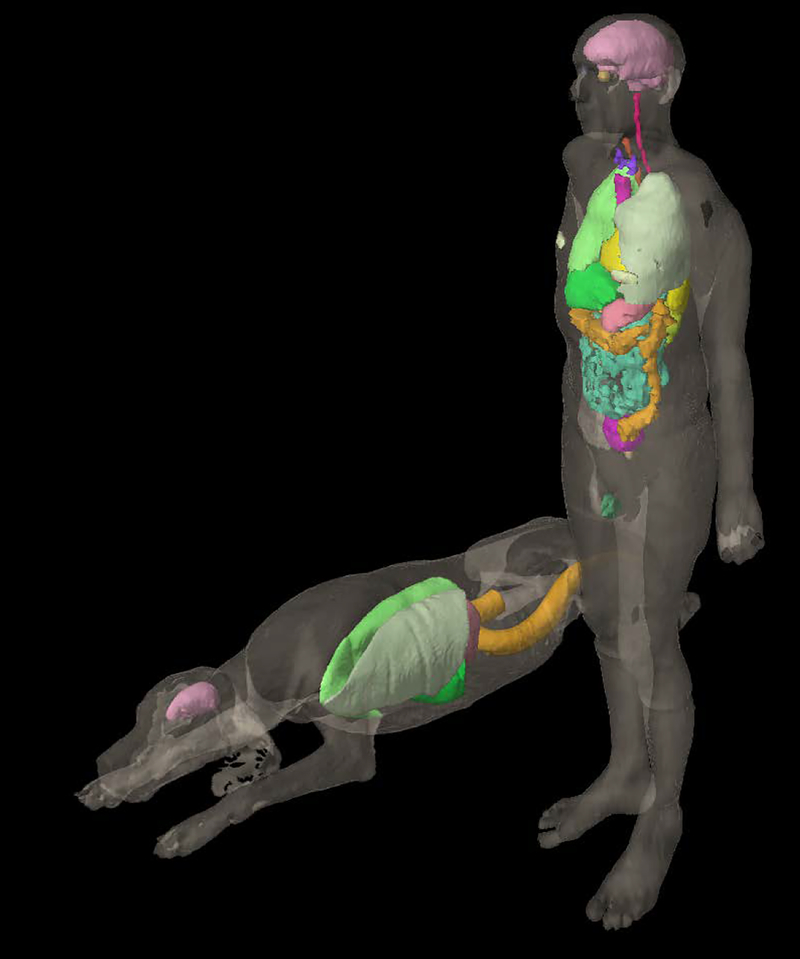
Though initially designed for use with an age-dependent human phantom series, the DICOM-RT Generator has been generalized to work with any type of voxel phantom -- even non-human phantoms. For demonstration purposes, the UF canine phantom (Padilla et al 2008) and the ICRP reference adult male phantom (ICRP 2009) were converted to DICOM-RT format and imported into Eclipse, as seen in Figure 4. A total of 6 and 24 organ contours were included for the canine and adult male cases, respectively. The effects of coarser voxel resolution in the ICRP reference adult male can be seen in the rougher contours of the geometrically complex organs, such as the bowels (turquoise) and colon (orange). Conversion times for the ICRP and UF phantoms were around two minutes each, with resulting DICOM file sizes under 30 MB. The phantom resolution and amount of requested organ contouring are the primary factors affecting the resulting DICOM file size and the execution time of the code, respectively.
Figure 4:
The UF canine anatomic phantom (prone) and ICRP 110 reference male phantom, with select organ contours visualized within the external beam planning module of the Varian Eclipse TPS. The voxel resolutions (x,y,z) were (0.20 cm, 0.20 cm, 0.20 cm ) and (0.2137 cm, 0.2137 cm, 0.80 cm) for the UF canine and ICRP 110 phantom, respectively.
3.3.2. National Wilms Tumor Study (NWTS) application
Using the DICOM-RT Generator, sample cases of retrospective organ dose estimations were performed for two patients from the National Wilms Tumor Study (NWTS) cohort. Patient A, a five-year-old female (105 cm height, 15 kg weight), and patient B, a one-year-old female (85 cm height, 15 kg weight), received radiotherapy for Wilms Tumor shortly after diagnosis. As CT data are unavailable for these patients, patient-matched phantoms were selected from the phantom library (Geyer et al 2014) as surrogate anatomies based on age, gender, height, and weight. Several near and in-field organs were specified within the tag list file for contouring.
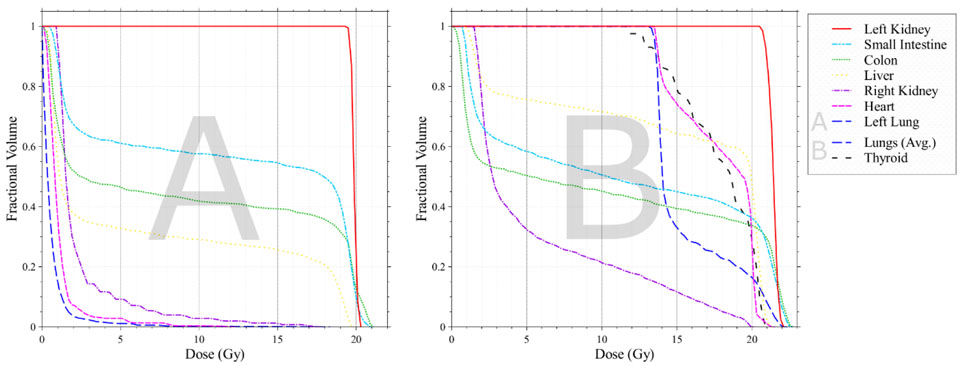
After processing the phantoms in the DICOM-RT Generator, the resulting DICOM-RT files were imported into the Philips Pinnacle TPS at Northwestern University, where patient-specific RT fields were then reconstructed (including photon energy, field-type, and prescribed dose) based on scanned paper medical records available from the NWTS database (Kalapurakal et al 2018). These radiotherapy plans were created by a practicing radiation oncologist and senior medical physicist who carefully reviewed the available medical records and portal films. Both patients were treated for Wilms tumor (nephroblastoma) of the left kidney with nephrectomy followed by adjuvant radiation and chemotherapy. Patient A received a 19.8 Gy left flank field to the left kidney cavity. Patient B, who had lung metastasis, received 12 Gy whole lung field, a 12 Gy left flank field, and an 8 Gy boost to the left flank. The calculated dose maps for these radiotherapy plans were written out from the TPS to a DICOM RT-Dose file for analysis. Dose-volume histograms for selected organs were calculated within the system and are shown in Figure 5.
Figure 5:
Dose volume histograms for patients A and B for Wilms tumor of the left kidney, calculated by the TPS from the radiotherapy fields reconstructed on surrogate phantom anatomies. Patient A received a 19.8 Gy left flank field. Patient B had a lung metastasis and recevied a 12 Gy whole lung field, a 12 Gy left flank field, and an 8 Gy boost to the left flank.
3.4. Future applications and limitations
Given its relatively quick run-time and ease-of-use, the DICOM-RT Generator can make a significant impact to future investigations. Our group intends to use the phantom conversion function of this software to connect our computational phantom resources to cohort studies where patient-specific DICOM data may not exist or are too expensive to retrieve. In clinical practice, however, this methodology may not truly capture all the anatomical variability between different patients. The use of a large phantom library, such as the NCI phantom library (Geyer et al 2014), does encapsulate differences in body mass and height; however, it does not account for the small differences in organ mass and positioning within the body between patients of similar body mass and height. For example, in the study of secondary coronary abnormalities after breast or esophageal cancer RT treatment, a minor change in heart size estimation within the model could cause drastic differences in calculated heart dose, since only a small portion of the heart might lie within the radiotherapy beam field (Taylor et al 2007). These patient-specific issues can cause inaccurate estimation of dose in sensitive cases; nevertheless, the current methodology represents the best available solution to patient representation when no images exist.
Beyond just epidemiological studies of late-effect health risks, the DICOM-RT Generator can also be helpful for improving dose calculations to organs near the start or end of the CT image scan, where insufficient anatomy may cause dosimetric inaccuracies due to an incomplete modeling of the scattered dose. The use of computational phantoms can help model more of this anatomy. Recent work has shown methods to extend a partial-body CT by incorporating the CT image set into a matched computational phantom (Kuzmin et al 2018). Regardless of how the anatomy is extended, it should be noted that a well-performed extension of the patient’s anatomy does not mean that out-of-field organ dosimetry is accurate. Depending on the algorithm, as well as the distance beyond the treatment field edge, TPSs are known to provide inaccurate dosimetry far outside of the treatment field (Mille et al 2018, Howell et al 2010). However, an extended-anatomy phantom could still be used within a secondary dose computation that accurately models dosimetry outside the treatment field edge (e.g. within Monte Carlo code) to properly optimize the primary TPS plan for out-of-field patient dose reduction.
The results of this work showcase extensive testing within the Varian Eclipse TPS, but one additional limitation lies in the unknown capabilities of the DICOM-RT Generator to create DICOM files that are compatible with other TPSs. To confront this issue, multiple other popular systems and DICOM viewers were used to test general compatibility of the resultant DICOM files. Thanks to collaborative efforts with other TPS users, output DICOM CT and RT-Structure files from the DICOM-RT Generator have been confirmed to properly import into the Philips Pinnacle, Elekta Monaco, and RaySearch RayStation TPS. In addition, these output files were correctly imported within the open-source DICOMpyler and CERR viewing platforms. As seen in Table 2, various DICOM platforms can interpret the RT-Structure data differently and apply volume calculations in a non-standardized manner. Future work will therefore continue to test converted DICOM files in these systems to clarify differences in contour data interpretation between platforms.
5. Conclusions
This work constitutes a novel method to automatically convert binary voxel phantom files into DICOM-RT format, complete with contours for any specified organ within the phantom. Extensive testing has shown its flexibility to convert voxel phantoms into the standard DICOM-RT files compatible with several different commercial TPSs. Auto-generation of contours can be significantly helpful for those organs which are difficult and time-consuming for dosimetrists to segment, such as the skeleton and colon; DICOM RT-Structure files can now be generated for a given phantom within minutes, which makes this tool ideal for use in large-scale dose reconstruction tasks. Multiple examples of phantom conversion have been shown and discussed, demonstrating the strength and robustness behind this tool. The converted phantom library, in DICOM format, and a standalone MATLAB-compiled executable of the DICOM-RT Generator are available for others to use for research purposes (http://ncidose.cancer.gov).
Acknowledgments
The authors would like to thank Niek Schreuder, M.Sc. DABR, of the ProvisionCares Proton Therapy Center for helping to test the converted phantoms within the Raystation TPS. The authors would also like to thank Pascal Hauri, PhD, of the University of Switzerland for testing the converted phantoms within the Eclipse TPS. The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health. Certain commercially available software is identified in this manuscript to foster understanding and should not be construed as a recommendation.
References
- Barton MB, Jacob S, Shafiq J, Wong K, Thompson SR, Hanna TP and Delaney GP 2014. Estimating the demand for radiotherapy from the evidence: A review of changes from 2003 to 2012 Radiotherapy and Oncology 112 140–4 [DOI] [PubMed] [Google Scholar]
- Cutright D, Gopalakrishnan M, Roy A, Panchal A and Mittal BB 2018. DVH Analytics: A DVH database for clinicians and researchers J Appl Clin Med Phys 19 413–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer AM, O’Reilly S, Lee C, Long DJ and Bolch WE 2014. The UF/NCI family of hybrid computational phantoms representing the current US population of male and female children, adolescents, and adults—application to CT dosimetry Phys. Med. Biol. 59 5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell RM, Scarboro SB, Kry SF and Yaldo DZ 2010. Accuracy of out-of-field dose calculations by a commercial treatment planning system Physics in Medicine and Biology 55 6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICRP 2009. Adult Reference Computational Phantoms ICRP Publication 110, Ann. ICRP 39 1–166 [DOI] [PubMed] [Google Scholar]
- Jemal A, Ward EM, Johnson CJ, Cronin KA, Ma J, Ryerson AB, Mariotto A, Lake AJ, Wilson R, Sherman RL, Anderson RN, Henley SJ, Kohler BA, Penberthy L, Feuer EJ and Weir HK 2017. Annual Report to the Nation on the Status of Cancer, 1975–2014, Featuring Survival J Natl Cancer Inst 109 Online: https://academic.oup.com/jnci/article/109/9/djx030/3092246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalapurakal JA, Gopalakrishnan M, Mille M, Helenowski I, Peterson S, Rigsby C, Laurie F, Jung JW, Fitzgerald TJ and Lee C 2018. Feasibility and accuracy of UF/NCI phantoms and Monte Carlo retrospective dosimetry in children treated on National Wilms Tumor Study protocols Pediatr Blood Cancer e27395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin GA, Mille MM, Jung JW, Lee C, Pelletier C, Akabani G and Lee C 2018. A Novel Method to Extend a Partial-Body CT for the Reconstruction of Dose to Organs beyond the Scan Range Radiation Research 189 618–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Jung JW, Pelletier C, Pyakuryal A, Lamart S, Kim JO and Lee C 2015. Reconstruction of organ dose for external radiotherapy patients in retrospective epidemiologic studies Physics in Medicine and Biology 60 2309–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Lodwick D, Hasenauer D, Williams JL, Lee C and Bolch WE 2007. Hybrid computational phantoms of the male and female newborn patient: NURBS-based whole-body models Physics in Medicine and Biology 52 3309–33 [DOI] [PubMed] [Google Scholar]
- Lee C, Lodwick D, Hurtado J, Pafundi D, Williams JL and Bolch WE 2010. The UF family of reference hybrid phantoms for computational radiation dosimetry. Physics in Medicine and Biology 55 339–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mille MM, Jung JW, Lee C, Kuzmin GA and Lee C 2018. Comparison of normal tissue dose calculation methods for epidemiological studies of radiotherapy patients Journal of Radiological Protection 38 775–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A 2016. Cancer treatment and survivorship statistics, 2016 CA: A Cancer Journal for Clinicians 66 271–89 [DOI] [PubMed] [Google Scholar]
- NEMA. NEMA PS3 / ISO 12052, Digital Imaging and Communications in Medicine (DICOM) Standard. 2018.
- Padilla L, Lee C, Milner R, Shahlaee A and Bolch WE 2008. A canine anatomical phantom for preclinical dosimetry in molecular radiotherapy Journal of Nuclear Medicine 49 446–52 [DOI] [PubMed] [Google Scholar]
- Stovall M, Weathers R, Kasper C, Smith SA, Travis L, Ron E and Kleinerman R 2006. Dose Reconstruction for Therapeutic and Diagnostic Radiation Exposures: Use in Epidemiological Studies Radiation Research 166 141–57 [DOI] [PubMed] [Google Scholar]
- Taylor CW, Nisbet A, McGale P and Darby SC 2007. Cardiac Exposures in Breast Cancer Radiotherapy: 1950s–1990s International Journal of Radiation Oncology*Biology*Physics 69 1484–95 [DOI] [PubMed] [Google Scholar]
- Tyldesley S, Delaney G, Foroudi F, Barbera L, Kerba M and Mackillop W 2011. Estimating the Need for Radiotherapy for Patients With Prostate, Breast, and Lung Cancers: Verification of Model Estimates of Need With Radiotherapy Utilization Data From British Columbia International Journal of Radiation Oncology*Biology*Physics 79 1507–15 [DOI] [PubMed] [Google Scholar]
- Xu XG 2014. An exponential growth of computational phantom research in radiation protection, imaging, and radiotherapy: a review of the fifty-year history Physics in Medicine and Biology 59 R233–302 [DOI] [PMC free article] [PubMed] [Google Scholar]