Abstract
In order to develop a practical model of breast cancer, with in vitro and syngeneic-immune intact in vivo growth capacity, we established a primary cell line derived from a mammary carcinoma in the transgenic FVB/N-Tg(MMTV-ErbB2*)NDL2-5Mul mouse, referred to as “NDLUCD”. The line was adapted to standard cell culture and can be transplanted into syngeneic FVB/N mice. The line maintained a stable phenotype over multiple in vitro passages and rounds of in vivo transplantation. NDLUCD tumors in FVB/N mice exhibited high expression of ErbB2 and ErbB3 and signaling molecules downstream of ErbB2. The syngeneic transplant tumors elicited an immune reaction in the adjacent stroma, detected and characterized using histology, immunophenotyping, and gene expression. NDLUCD cells also expressed PD-L1 in vivo and in vitro, and in vivo transplants were reactive to anti-immune checkpoint therapy with responses conducive to immunotherapy studies. This new NDLUCD cell line model is a practical alternative to the more commonly used 4T1 cells, and our previously described FVB/N-Tg(MMTV-PyVT)634Mul derived Met-1fvb2 and FVB/NTg(MMTV-PyVTY315F/Y322F) derived DB-7fvb2 cell lines. The NDLUCD cells remained genetically and phenotypically stable over many generations, with consistent and reproducible results in immune intact preclinical cohorts.
Keywords: PD-L1, mouse model, breast cancer, immunotherapy, Erbb2, syngeneic
INTRODUCTION:
Mouse models are the primary preclinical models of human disease. Patient derived xenografts of human cancer and human cell lines are widely used in preclinical cancer research for targeted and cytotoxic therapies. However, these experimental models do not recapitulate the host’s normal and complex immune response. Thus, they are of limited value for studies of tumor-immune interaction or immunotherapy. Genetically engineered mouse models of cancer have intact immune function, but present practical limitations with respect to penetrance, latency, and heterogeneity [1]. We, and others, have pioneered the practical use of syngeneic transplants to generate nearly identical cohorts for preclinical comparisons and to investigate cancer in an immune intact host. In the face of the growing demand for experimental immunotherapy models, the available selection of syngeneic models of breast cancer is limited. The majority of mammary tumor-immunity studies utilize the “4T1” line, which develops as a rapidly growing, metastatic, mesenchymal-like phenotype, reflecting only a subset of human breast cancers [2]. In a study comparing six syngeneic cancer models, the 4T1 model did not respond to either anti-PD-1 or anti-CTLA4 therapy, two primary immunotherapeutics. Only one model (CT26 colon cancer) showed sensitivity to anti-PD-1 therapy and only two (CT26 and RENCA) to anti-CTLA4 therapy [3]. Here, we describe a novel syngeneic HER2 positive model of breast cancer, NDLUCD which expresses PD-L-1 and is reactive to anti-PD-1 therapy [4] - and we provide a detailed pathologic description of the host immune reaction and molecular phenotype of the model.
The cell line, derived from a mammary carcinoma arising in an FVB/N-Tg(MMTV-ErbB2*)NDL2-5Mul mouse, carries an in-frame deletion of the rat HER2/neu proto-oncogene in the same region as the human Δ16 splice variant, resulting in disulfide-bond stabilized, constitutively active HER2 homodimers [5]. The Δ16 splice variant in humans skips exon 16, which forms part of the extracellular domain of the protein. This variant HER2 receptor forms constitutively activated homodimers, stabilized by disulfide bonds. The Δ16 splice variant is common in human breast cancers, present in 90% of HER2 tumors at a transcript frequency of 9% on average, with some evidence that some fraction of this isoform is required for transformation [6, 7].
Immune cell infiltration in breast cancers, especially in the more aggressive HER2 positive and triple negative subtypes, has been shown to correlate with better outcome (reviewed in [8]). Constitutively active HER2 is a prototype neoantigen and known to be immuno-recognized [9]. HER2 reactive circulating T-cells are often present in HER2 positive breast cancer patients and the presence of HER2 reactive CD8 cells correlates with a better prognosis [10, 11]. HER2 positive tumors with more infiltrating lymphocytes have a greater benefit from anti-HER2 (trastuzumab) therapy [12] and the ErbB2Δ16 splice variant may specifically activate the tumor immune microenvironment [7]. The ErbB2Δ16-like/HER2 expressing NDLUCD transplant is a suitable model to investigate these tumor-host interactions, particularly in the setting of novel combined immunotherapies.
METHODS:
Animal care:
FVB/N mice were purchased from Charles River (Wilmington, MA), the FVB/N-Tg(MMTV-ErbB2*)NDL2-5Mul mice were provided by William J. Mueller (McMaster University, Hamilton, Ontario, Canada) [5]. Mice were housed in a vivarium under NIH guidelines and all animal experiments followed protocols approved by the UC Davis Institutional Animal Care and Use Committee (IACUC).
Establishing the cell line:
Primary tumors, from FVB/N-Tg(MMTV-ErbB2*)NDL2-5Mul mouse, were washed in PBS (Invitrogen Carlsbad, CA) twice, and then dissociated in serum-free DMEM:F12 (Invitrogen) buffered with HEPES (Invitrogen), supplemented with 0.5 mg/ml Penicillin/Streptomycin (Invitrogen), 2% bovine serum albumin fraction V (Invitrogen), 5 μg/ml insulin (Sigma Aldrich, Saint Louis, MO), 10 ng/ml cholera toxin (Sigma Aldrich), and 3 mg/ml collagenase (Worthington Biochemical Corp., Lakewood, NJ). The tissue was minced manually with enough serum-free digestion reagent to allow for complete coverage of the tissue. This mixture was digested in a sterile 50 mL tube with gentle agitation overnight at room temperature, then centrifuged at 80x g for 1.5 minutes, the cell pellet was washed with DMEM:F12 and centrifuged again at 80x g for 4 minutes. The remaining cell pellet was plated in DMEM supplemented with 10% FBS and 0.5 mg/ml Penicillin/Streptomycin.
Tissue culture:
Cells were maintained in DMEM supplemented with 10% FBS and 0.5 mg/ml Penicillin-Streptomycin. Differential trypsinization was used to eliminate fibroblast contamination and, after passage five, a single cell suspension was labeled with CD49f (Integrin alpha6) and sorted in a Cytomation (Fort Collins, CO) MoFlo cell sorter to exclude all non-epithelial cells. For in vitro growth evaluation, 200,000 cell/3 ml media/well (approximately 6,667 cells/ml) were plated in six well plates. Triplicate wells were harvested and three samples per well were counted with a hemocytometer each day for four days when cells reached confluency.
Cell line transplantation:
The cell line was transplanted into syngeneic FVB/N female mice by cell line injection into the mammary fat pad (orthotopically) as described previously [13, 14]. Briefly, the bilateral inguinal mammary fat pads of the mice were injected with 0.1-2 × 106 cells/20 μl PBS solution with a 25-gauge needle. Tumors were palpable 10-14 days after transplantation. Alternatively, serial transplantation of 0.5-1 mm3 frozen or fresh tumor tissue into the mammary fat pad or subcutaneous space reliably establishes new tumor grafts as described previously [13]. Growth of palpable tumors was measured with calipers.
Tail-vein injection:
To evaluate metastatic potential, tail-vein injections of the NDLUCD cell line were performed as described previously [13]. Six animals were injected with passage 15 NDLUCD cells. Cells were trypsinized (Invitrogen) and washed with PBS, then 106 cells/100 μl PBS solution per animal were injected using a 25-gauge needle.
Immunohistochemistry (IHC)
Tissue removal, preparation and immunohistochemistry was performed as previously described [14, 15]. Slides were scanned on an Aperio AT2 ScanScope (Leica Biosystems, Danvers, MA). The image analysis for counting marker-positive cells was performed with QuPath [16]. Digital images were processed using Adobe Photoshop (Adobe Systems, San Jose, CA).
The antibodies and dilutions used were, anti-CD4 (4SM95) (eBioscience, Waltham, MA, dilution: 1:100); anti-CD8a (4SM15) (eBioscience, dilution: 1:400); anti-FOXP3 (FJK-16s) (eBioscience, dilution: 1:100); anti-PD-1 (EPR20665) (Abcam, Cambridge, UK, dilution:1:500); anti-PD-L1 (E1L3N XP) (Cell Signalling Technology, Danvers, MA, dilution: 1:200); anti-CK818 (20R-cp004) (Fitzgerald, Acton, MA, dilution: 1:2000); anti-F4/80 (BM8) (eBioscience, 1:100); anti-B220 (RA3-6B2) (BD Pharmigen, San Jose, CA, dilution: 1:800); anti-ER (MC-20) (Santa Cruz Biotechnology, Dallas, dilution: 1:1000); anti-KI67 (RB-1510-P) (Thermo Fisher Scientific, Waltham, MA, dilution: 1:800); anti-PR (A0098) (Dako, Santa Clara, CA, dilution: 1:500); anti-ERBB2 (RM-2112-5) (Thermo Fisher Scientific, dilution 1:1000); anti E-cadherin (24E10) (Cell Signalling Technology, dilution 1:1000).
CD3, PD-1 and ECAD were detected with multiplex IHC, performed with tyramide signal amplification (TSA)-based fluorescence color visualization according to the manufacturer’s protocol (PerkinElmer, Waltham, MA). Visualization of multiplexed images was performed with an LSM710 laser scanning confocal microscope (Carl Zeiss Microscopy, Oberkochen, Germany).
Western blot analysis:
Antibodies for western blotting analysis were, anti-ERBB3 (OP120) and anti-ERBB2 (OP15) (EMD Millipore, Burlington, MA); anti-SHC (PG-797) (Santa Cruz Biotechnology, Dallas, TX); anti-CDKN1A (SXM30) (BD Biosciences); anti-PD-L1 (E1L3N XP) (Cell Signalling Technology) and anti-β-Actin (AC-15) (Sigma-Aldrich).
Total protein was extracted from cells using RIPA lysis buffer (50 mM Tris (pH 7.4), 150 mM NaCl, 0.1% SDS, 1% TritonX-100, 0.5% NaDeoxycholate). After incubation on ice, cells were scraped from T25 flasks, lysates were vigorously vortexed and centrifuged at 13,000 x g for 15 minutes. Lysate supernatants were placed into new microcentrifuge tubes and protein concentrations were determined using BCA kit (Pierce, Rockford, IL, USA). A unit of 20 μg of cell lysate was denatured in 6X Laemmli sample buffer ((50 mmol 1-1 Tris-HCl (pH 6.8), 2% SDS, 10% glycerol, 0.25% β-mercaptoethanol and bromophenol blue (1 mg/ml)) at 95°C for 5 minutes. 2X Laemmli sample buffer was added to samples to bring to a final volume of 40 μl. Samples were separated on an 8% or 15% SDS-polyacrylamide gel, and then transferred to nitrocellulose membrane (Millipore, Billerica, MA). Membranes were blocked with 5% non-fat dry milk in TBST (Tris-buffered saline containing 0.05% Tween 20), and incubated with specific primary antibodies overnight at 4°C, washed with TBST, and detected with horseradish peroxidase-conjugated secondary antibodies for one hour at room temperature, and ECL (Pierce) chemiluminescence on an Alpha Innoteceh Digital Imaging Station. Blot images are a representation of three technical replicates.
RNA isolation:
Differential gene expression of the NDLUCD cell line was compared to two other mouse mammary carcinoma cell lines (Met-1fvb2[13] and SSM2UCD [17]). Cells were propagated in vitro and injected (0.1 – 2.0 × 106 cells) bilaterally into the uncleared mammary fat pads of 6- to 8-week-old female FVB/N mice. Tumors were harvested when they reached a maximum width of 5-10 mm and 1-2 mm sections were snap-frozen. RNA was isolated using the miRNeasy Mini Kit (Qiagen, Hilden, Germany). RNA concentration and purity were assessed with a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific) and quality assessments (e.g., RNA integrity) were made using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Three samples per line were then analysed with Affymetrix GeneChip Mouse Gene 1.0 ST Arrays (Thermo Fisher Scientific) (note: two Met-1fvb2 samples originated from different injection sites in the same mouse, all other samples were from individual recipient mice).
Microarray:
Microarray gene expression profiling was performed by the UC Davis Comprehensive Cancer Center Genomics Shared Resource (GSR) on Affymetrix Mouse Gene 1.0 Sense Target (ST) Arrays (Thermo Fisher Scientific). Biotinylated sense strand DNA targets were prepared from total RNA (100 ng) using the Ambion WT Expression Kit (Thermo Fisher Scientific) and Affymetrix GeneChip WT Terminal Labeling Kit. Briefly, double-stranded cDNA was prepared followed by in vitro transcription to generate antisense cRNA (aRNA), which was used as template for a second cycle cDNA synthesis in a dUTP-containing reaction mixture. The dUTP-containing cDNA was fragmented by treatment with uracil-DNA glycosylase (UDG) and apurinic/apyrimidinic endonuclease 1 (APE1). DNA was then end-labeled with biotin using the DNA Labeling Reagent and deoxynucleotidyl transferase (TdT). All microarray processing procedures, including target hybridization, washing, staining, and array scanning were performe according to Affymetrix’s standard protocols.
Microarray data analysis:
The microarray data processing and statistical analyses were performed in the R language and statistical environment [18]. For the direct comparison of the tumors that arose from three mouse mammary carcinoma cell lines (NDLUCD, Met-1fvb2, SSM2UCD), raw files were background-corrected, summarized, and normalized using the Robust Multi-array Average (RMA) method with the “oligo” Bioconductor package (version 1.40.2) [19]. Quality was checked before and after processing with the “ArrayQualityMetrics” Bioconductor package (version 3.32.0) [4] and there were no outliers identified. The Rank Product statistics were used to identify differentially expressed genes in pairwise comparison (between NDLUCD versus SSM2UCD, and NDLUCD versus Met-1fvb2; the comparison of Met-1fvb2 with SSM2UCD was not performed in this study) (significance limit: pfp<0.05) with the “rankprod” Bioconductor package (version 3.2.0). For functional annotation of the significant gene lists DAVID 6.8 were used [24, 25], searching Gene Ontology categories and relevant pathways in the KEGG pathways [20] database. Cluster 3.0 [21] was used for clustering tasks. Hierarchical clustering with average linkage was performed. The clustering was visualized with Java TreeView [22].
RNA-Sequencing
For whole-transcriptome analysis (next-generation sequencing (NGS)-based RNA-Sequencing (RNA-Seq)) total RNA-Seq sequencing libraries were prepared from snap-frozen tumor samples. Briefly, rRNA-depleted RNA was prepared from total RNA input (1-5 μg) by Ribo-Zero rRNA Removal Kit (Epicentre, Madison, WI). Directional Total RNA-Seq libraries were prepared from rRNA-depleted RNA (50 ng) using the ScriptSeq v2 RNA-Seq Library Preparation Kit (Epicentre) according to the manufacturer’s protocol. Briefly, 5’,3’-di-tagged cDNA was synthesized using tagged random hexamer-primed cDNA synthesis primers followed by annealing of 3’-terminal-tagging oligo (TTO) and extension with DNA Polymerase. cDNA was purified with Agencourt AMPure XP beads (Beckman Coulter, Pasadena, CA). Illumina adaptor sequences and indexes (barcodes) were incorporated during library amplification with the appropriate PCR primers and high-fidelity FailSafe PCR Enzyme Mix, and ScriptSeq v2 libraries were purified and quantitated with the Qubit fluorometer (Invitrogen) with insert sizes determined with the Agilent 2100 Bioanalyzer. The molar concentration of PCR-competent sequencing templates in the libraries were determined by quantitative PCR with the KAPA Library Quantification Kit (Kapa Biosystems, Wilmington, MA). The libraries were then loaded on TruSeq paired-end flow cells for cluster generation with the cBot (Illumina, San Diego, CA) followed by sequencing (40-bp single-read) with the Genome Analyzer IIx (Illumina) using Illumina TruSeq SBS Kit v5-GA kitted reagents and according to the manufacturer’s protocols [23].
RNA-Sequencing data analysis
Image processing, base calling, and quality scoring (Phred) were executed by SCS 2.6/RTA 1.6 software (Illumina). The TopHat-Cufflinks workflow was utilized for analysis of gene and transcript isoform expression [24]. For this, read alignment to the reference mouse genomic sequence (July 2007 NCBI37/mm9) and splice junction mapping were performed with TopHat [4] and allowing for a maximum of two mismatches. Cufflinks was used for transcript assembly, quantitation, and differential expression [5]. Transcript abundance values were expressed as fragments per kilobase of transcript per million fragments mapped (FPKM). Mapped sequence data was visualized using the Integrative Genomics Viewer [6].
Statistics:
Statistical analysis was performed with GraphPad Prism or R. Statistics of the gene expression analysis are discussed above under microarray data analysis. One-way ANOVA with two-stage step-up method of Benjamini, Krieger and Yekutieli [25] for controlling false discovery ratio was used to compare immune cell infiltration. Spearman’s rank correlation was used to assess correlation between the density of different immune cells in the tumor and the stroma. Significance limit was set as p<0.05, no multiple testing correction was performed.
RESULTS
Establishing the cell line
Primary tumor cell dissociation and adaptation to adherent cell culture was implemented, with purification utilized both traditional and flow sorting methods as described [13, 26] to eliminate non-epithelial cells. The cells grew in vitro, adherent to the dish surface, with a uniform epithelial phenotype that did not change or drift over many passages (more than 30).
Growth rate
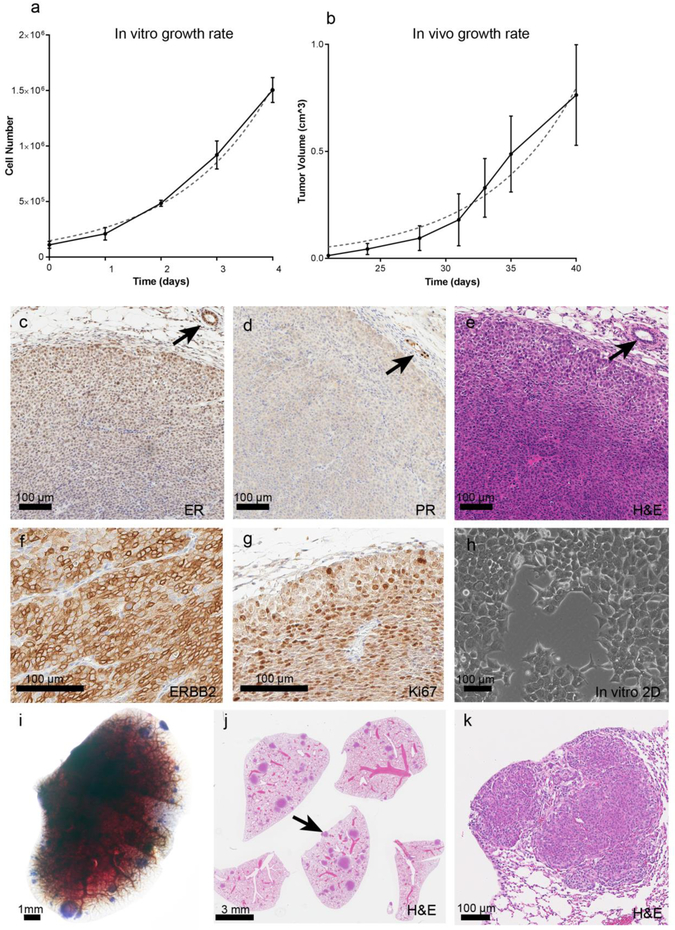
In vitro and in vivo serial transplant growth rates were evaluated. The NDLUCD cell in 2D culture showed a slightly slower growth compared to many other frequently used mouse cell lines with an in vitro population doubling time of 1.18 days (in vitro growth of B16F10 melanoma line = 0.49 days, 4T1 breast line = 0.95 days [27].) The in vivo doubling rate of NDLUCD tumors was 4.871 days (data used for calculating in vivo population doubling has been previously published [4]). In vitro growth (Figure1. a) h)) and in vivo growth (Figure1 b)) are shown.
Figure 1:
In vitro and in vivo growth and phenotype of NDLUCD. a-b) Growth attributes of the NDLUCD cell line, a) In vitro growth rate in 2D culture. b) In vivo growth rate of transplanted NDLUCD tumor (this data has been previously published [4]). c-g) Immunohistochemistry of the NDLUCD transplant tumors; c) ER, d) PR, e) H&E, f) ERBB2, g) KI67. Arrows indicate normal mammary glands. h) In vitro 2D culture showing cobblestone epithelial phenotype. i-k) Lung metastasis, example of a lung showing high multiplicity of NDLUCD tumor outgrowth after tail-vein injection. i) Whole-mount photograph of a lung lobe with multiple NDLUCD outgrowths. j) high tumor multiplicity, arrow indicates tumor shown at a higher magnification in k). k) The lung outgrowths show characteristic NDLUCD phenotype.
Pathology
Primary mammary carcinomas in the FVB/N-Tg(MMTV-ErbB2*)NDL2-5Mul mouse have a consistent “signature” phenotype (as described in [28] and [29]) characterized by a nodular growth pattern, with high vascularity, and minimal central necrosis. Three distinct structural zones have been described [30]. A zone of differentiation is located at the periphery of the nodules, approximately 3-5 cell layers in diameter. Immediately subjacent to this peripheral zone is a proliferative area which has the most intense ERBB2 membrane expression. In the innermost central zone, the cells remain epithelioid without metaplastic (spindled or squamous) features, but here the cells have smaller nuclei and less cytoplasm with lower levels of ERBB2 expression. The centers of larger tumors frequently have areas of necrosis.
The orthotopic mammary fat pad transplanted NDLUCD cells recapitulated the “signature” tumor phenotype described for the native transgenic tumors (Figure 1. e) H&E). These were solid, nodular carcinomas, well vascularized with the same zonation areas and the same proliferation rates, (Figure 1. g)), and ERBB2 expression levels (Figure 1. f)). The ER and PR immunoreactivity were weak (Figure 1. c) d)).
Metastatic potential
After tail vein injection of 106 NDLUCD cells into the syngeneic FVB/N female mice metastasis lesion were histologically confirmed in 3 out of 6 mice. The detected metastases invaded into the alveolar parenchyma, characteristic of “true” metastases as compared to intravascular embolic deposits [31]. (Figure 1. i) j) k)).
Erbb2 pathway activation
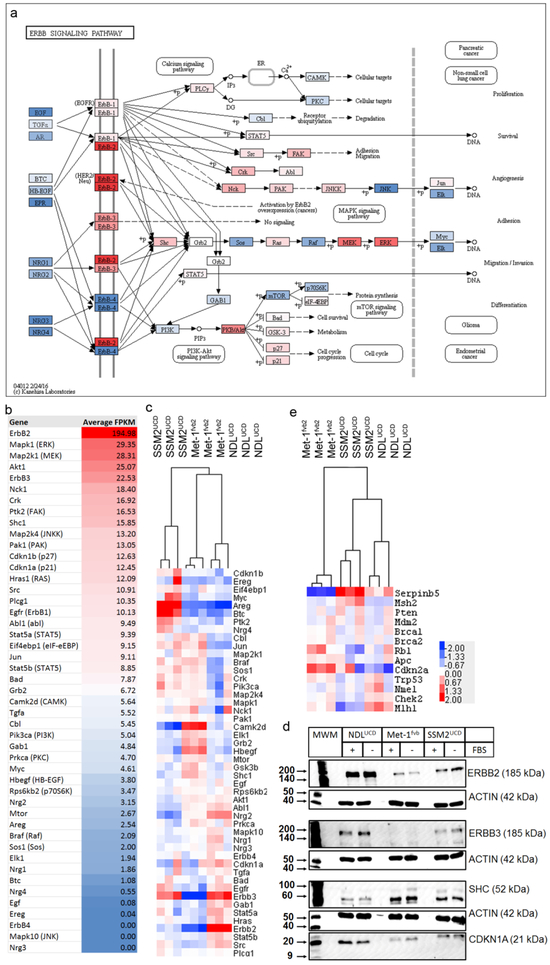
Erbb2 was highly expressed as detected by IHC, western blot, and mRNA expression analyses (microarray and RNA-Seq (average of 195 FPKM)) (Figure 2). Components of the Erbb2 activation pathway were highly expressed including Mapk1, Map2k1, Akt1, Erbb3, Nck1, Ptk2, Shc1, Map2k4, p27, p21, Ras and Src. Protein expression of ERBB2, ERBB3, CDKN1A and SHC were validated in western blot from the 2D cultured cells. The protein expression pattern of these genes in Met-1fvb2, NDLUCD and SSM2UCD cultured cells reflected the RNA expression levels observed in tumor samples.
Figure 2:
Expression of ErbB pathway molecules in the NDLUCD model. a) The ErbB pathway as presented in KEGG pathways [20] colored, based on the level of expression of the indicated molecule in the RNA-Seq analysis of NDLUCD transplant tumors (n=3). b) Expression values in the RNASeq analysis used for color coding in the “a” panel. c) Comparison of expression of ErbB pathway genes in three transplantable mouse models: NDLUCD, SSM2UCD, Met-1fvb2 (Gene expression profiling was performed with Affymetrix Mouse Gene 1.0 ST Arrays (n=6/each model)). Log2 transformed values were utilized for hierarchical clustering and the results are visualized with a heat map. d) Comparison of expression of main tumor suppressor genes in the NDLUCD, SSM2UCD, Met-1fvb2 models. e) Western blot of ErbB2 pathway proteins from cell cultures of three transplantable mouse models (NDLUCD, SSM2UCD,Met-1fvb2) (MWM: Molecular Weight Marker).
Immune cell infiltration
The host reaction in primary tumors from the parental Tg(MMTV-ErbB2*)NDL2-5Mul mouse and transplants from the NDLUCD cell line were analyzed and compared.
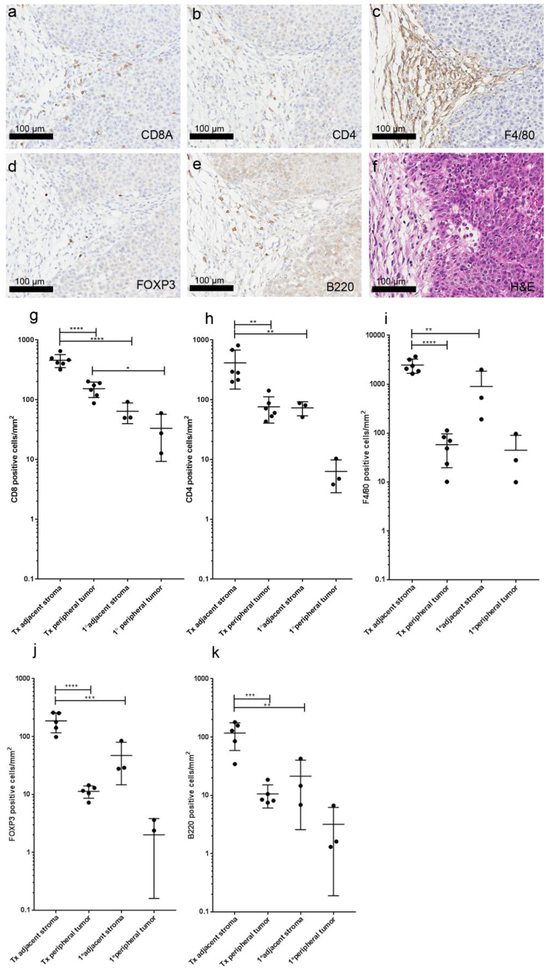
A significant host reaction was present in primary Tg(MMTV ErbB2*)NDL2-5Mul samples characterized as predominantly fibroblastic with collagen stroma replacing adipose adjacent to the in situ early tumors, increasing around larger masses. Syngeneic transplant NDLUCD tumors elicited a similar fibroblast component and collagen stroma (Figure 3). A scattered mixed inflammatory component was observed in the surrounding stroma, with predominantly macrophages and T-lymphocytes, and few B-lymphocytes. CD8 and CD4 positive T-lymphocytes and occasional macrophages were found infiltrating the tumors. Scattered FOXP3 positive cells (regulatory T-cells) and B-lymphocytes were also present.
Figure 3:
Immune cell distribution in syngeneic transplant of NDLUCD. a-f) Representative images of immunohistochemistry on an NDLUCD transplant tumor showing the same region: a) CD8A, b) CD4, c) F4/80, d) FOXP3, e) B220, f) H&E. g-k): Immune cell marker positive cells in the tumor adjacent stroma and in the peripheral tumor of transplanted NDLUCD tumors and primary tumors from the GEMM. n=6 transplant tumors from 3 animals (Tx) n=3 primary tumors from 3 animals (1°). * q<0.05, **q<0.01, ***q<0.001, ****q<0.0001
The differences between the host response to primary and transplant tumors and their respective tumor-immune phenotypes [32] were compared. The immune cell accumulation in the i) tumor adjacent stroma, defined as a band (250 μm wide) immediately adjacent to the periphery of the tumor; and in the ii) peripheral tumor, defined as the most peripheral area of tumor (250 μm wide) were evaluated. In each tumor, the surrounding tumor adjacent stroma had higher density of immune cells than the peripheral tumor infiltrating cells. This difference, however, was statistically significant only in the NDLUCD tumors, suggesting tumor cell/area immune suppression or the so-called “immune-excluded” phenotype [32]. The overall host reaction was more robust in the NDLUCD tumors compared to the primary tumors. Transplants had significantly higher CD4, CD8, FOXP3, B-cell and macrophage density in the tumor adjacent stroma than the primary tumors (Figure 3).
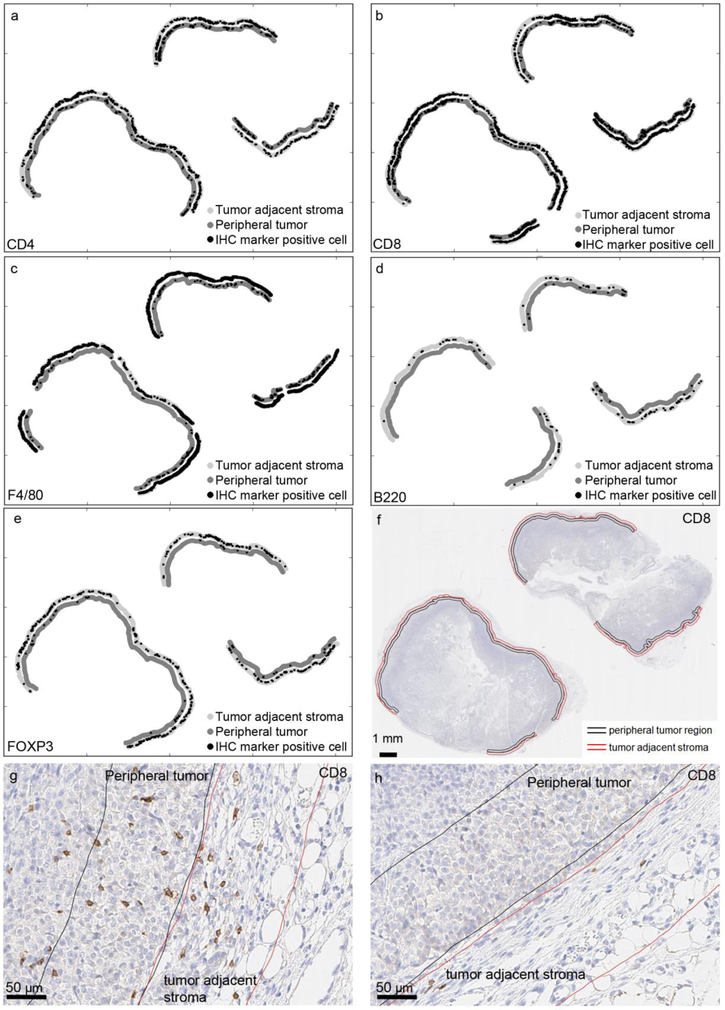
The spatial distribution of immune cell infiltrates around the tumors was not uniform but concentrated in discontinuous areas or “pockets”. The immunocytic stromal pockets corresponded with the highest density of immune cell infiltration into the tumor (Figure 4).
Figure 4:
Illustration of heterogeneity in immune cell infiltration. a-e) Distribution of immune cells in the peripheral tumor and the tumor adjacent stroma; a) CD4, b) CD8A, c) F4/80, d) B220, e) FOXP3. f) The analyzed area shown in the tissue overview, stromal area borders shown in red, peripheral tumor area borders shown in black. g) Example of an area of high CD8 positive cell density in the peripheral tumor and in the tumor adjacent stroma. h) Example of an area with low CD8 positive cell density in the peripheral tumor and in the tumor adjacent stroma from the same tumor as g).
Significant deviations were observed within the same biological groups (transplants or primary tumors) in the measured immune cell densities as shown in Figure 3. We hypothesized that this deviation originated from individual variance between immunogenicity of the tumors (inter-tumoral heterogeneity of immune cell attraction). To investigate tumor to tumor immunogenicity, the correlation of the density of different lymphocytes throughout the six NDLUCD transplant tumors (from three animals) and three primary tumors (from three animals) was analyzed by Spearman’s rank correlation (correlation defined as p<0.05, multiple testing correction was not performed). The density of CD4 positive cells in the tumor adjacent stroma correlated with the T-reg and B-cell density in the tumor adjacent stroma as well as peripheral tumor infiltrating CD4 cell density. Meanwhile, tumor adjacent stroma CD8 density correlated with the tumor adjacent stroma T-reg and B-cell density as well as CD8 and T-reg peripheral tumor infiltration. Tumor adjacent stroma T-reg density correlated with stromal B-cell, CD4 and CD8 density and peripheral tumor infiltrating CD4 and T-reg density. The tumor adjacent stroma macrophage density correlated with the peripheral tumor infiltrating CD4 and macrophage density. Between individual tumors there was a range of total inflammatory infiltrate, but the composition and relative proportions of different cell subsets were similar across all tumors.
PD-1 and PD-L-1 expression
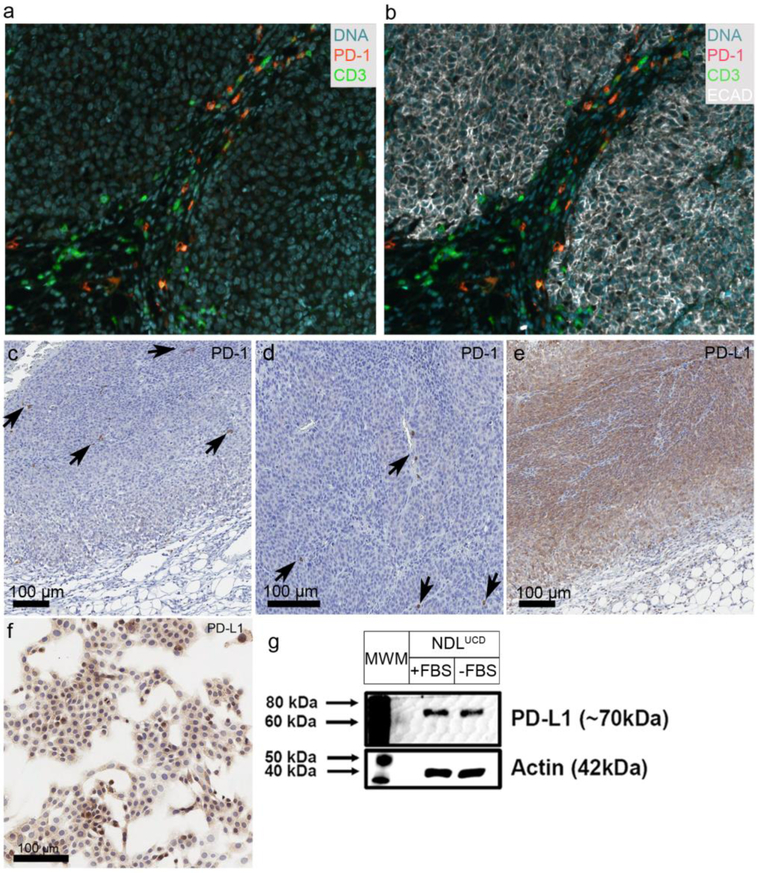
NDLUCD cells were previously found to be reactive to anti-PD-1 [5]. Therefore, PD-1 and PD-L1 were evaluated using IHC. PD-L1 IHC documented membrane accentuated cytoplasmic immunoreactivity in NDLUCD tumor cells (Figure 5). Expression intensity varied by tumor zone, with enlarged cells with lower intensity of staining in the tumor edge, and stronger PD-L1 staining in the proliferative zone. The NDLUCD cells retained PD-L1 positivity in vitro, demonstrated by both IHC (both nuclear and weaker cytoplasmic/membrane immunoreactivity) and western blot of total cell lysates. PD-1 IHC showed scattered T-cells with strong positive staining, and no expression by tumor cells (Figure 5).
Figure 5:
PD-1 and PD-L1 expression in the NDLUCD model. a) b) PD-1, CD3, ECAD expression stained with multiplex IHC. c) d) PD-1 expressing immune cells. The arrows indicate PD-1 positive cells. e) Positive PD-L1 staining, with typical zonal distribution of staining intensity. f) PD-L1 IHC on cell culture. g) PD-L1 western blot for cultured NDLUCD cells.
Gene expression
NDLUCD tumors were directly compared to two previously described mouse mammary carcinoma cell lines: SSM2UCD [17], derived from Stat1−/− estrogen receptor positive, luminal B tumors, and Met-1fvb2[13] derived from PyVT (Polyomavirus middle T-antigen) tumors, respectively. Pairwise comparisons with rank product statistics were performed comparing NDLUCD tumors to the two other cell lines. 483 genes were significantly overexpressed in NDLUCD over Met-1fvb2, 149 genes were significantly overexpressed in Met-1fvb2 over NDL(UCD). 426 genes were significantly overexpressed in NDLUCD over SSM2UCD and 111 genes overexpressed in the SSM2UCD over NDLUCD (at least two-fold expression change.) The full list of the significant differentially expressed genes is available in Supplementary Material 1. The identified gene lists were substantially overlapping with 42.5% of the NDLUCD-Met-1fvb2 and 50.1% of the NDLUCD-SSM2UCD differentially expressed genes shared. Functional annotation of the differentially expressed genes were performed using DAVID Bioinformatics Resources (version 6.8) [33, 34], the identified GO (Gene Ontology) categories and KEGG pathways [20] (Benjamini<0.1) are listed in Supplementary Material 2.
The data suggest that drug metabolism through glutathione or cytochrome P450 was activated in the NDLUCD tumors compared to SSM2UCD or Met-1fvb2. Genes such as Cyp3a11, Gsta3, Gstt1, Gstt3, Gstm4, Gstm5, etc. were overexpressed in NDLUCD. The activation of these xenobiotic metabolism pathways can cause resistance to certain cytotoxic chemotherapeutics. Resistance against almost 20 anti-cancer drugs has been associated with elevated levels of glutathione S-transferases (Gst-s), including drugs widely used in breast cancer chemotherapy such as topoisomerase 1 and 2 inhibitors or carboplatin [35]. NDLUCD might be a model for studying chemoresistance through GST activation and overexpression. This is the subject of ongoing work in the lab.
Met-1fvb2 is a highly metastatic cell line, it’s in vitro phenotype is described by long thin pseudopodia and philophodia suggesting a high propensity for migration, which was also shown previously with a pore migration assay [36] [13]. NDLUCD cells showed less migratory and metastatic potential, and these observations are supported by the gene expression results. The NDLUCD tumors were overexpressing many extracellular matrix-cell interaction molecules such as collagens (Col1a1, Col1a2, Col4a6, Col5a2), Integrin beta 8, Laminin gamma 2 and others. NDLUCD tumors were overexpressing Claudin 1, Claudin 3, Tight junction protein 2 and Occludin and other molecules involved in tight junctions. In contrast, genes which were identified in epithelial to mesenchymal transformation, cell migration, angiogenesis and metastatic phenotype were overexpressed in the Met-1fvb2 tumors, such as Zeb1, Vegfa, Cdh2, Bmp2 and many others. A panel of tumor suppressor genes important in breast cancer [37] were also compared and visualized in Figure 2. e).
Several genes related to inflammation and immune processes were overexpressed in Met-1fvb2 and SSM2UCD compared to NDLUCD including several pro-inflammatory cytokines and cytokine receptors such as Il33, Il1rl1 (in SSM2UCD), Ccl12, Ccr2, Cxcl9,Il18 (in Met-1fvb2). Key genes encoding enzymes of prostaglandin synthesis (Ptgs2, Hpgd) were also underexpressed in NDLUCD. These molecules are part of the chronic inflammatory environment which is a hallmark attribute of most cancers and is also characteristic of the SSM2UCD Stat1−/− model [38]. Inflammation can be tumor promoting in many contexts, and may correlated with poor prognosis [39]. NDLUCD compared to Met-1fvb2 and SSM2UCD showed lower expression of genes typically associated with a pro-tumor inflammatory environment.
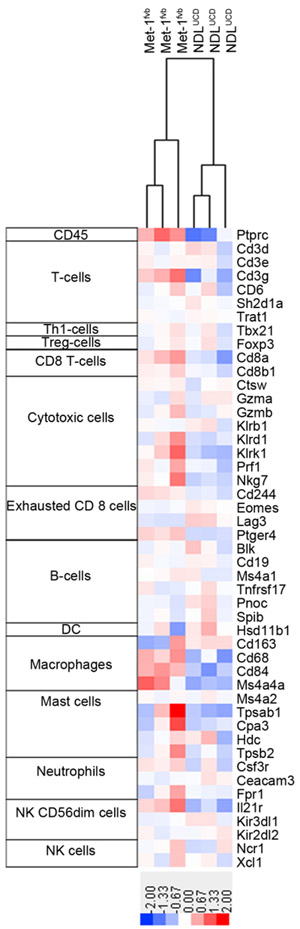
When the samples were clustered according to a list of genes proposed to identify immune infiltration from gene expression data in humans [40] 47 out of the 60 listed human genes could be evaluated (13 genes do not have mouse ortholog). The NDLUCD and Met-1fvb2 tumors clustered close together in terms of immune infiltration, while the SSM2UCD cells were more distant. Notably, this may reflect host strain (129SvEv) differences. For the purpose of the comparison, we show here only the NDLUCDand Met-1fvb2 lines, both grown in FVB/N immune intact strain background recipient mice (Figure 6.). The pan-leukocyte marker CD45 had lower expression in NDLUCD transplant samples. NDLUCDhad higher expression of genes characteristic of B-cells, dendritic cells, a cluster of cytotoxic cell markers, NK cells and T-cell markers.
Figure 6:
Immune reaction-associated gene expression in two mouse models. Gene expression profiling of NDLUCD and Met-1fvb2 transplanted tumors was performed using Affymetrix Gene 1.0 ST Arrays. Samples were clustered using a homologous gene list based on identification of immune infiltration from gene expression data in humans [40]. Log2 transformed and mean-centered values are illustrated.
DISCUSSION
The NDLUCD cell line has been used by colleagues working on mouse mammary carcinoma [4, 14, 26, 41-43] and the origin of the cell line has been briefly discussed in Miller et al. [26], However, a cohesive and comprehensive documentation of the in vitro and in vivo characteristics has not been previously published. Silvestrini et al. [4] established that the NDLUCD model is responsive to immunotherapy (combination of CpG and anti-PD-1 effectively treats tumor bearing animals). To our knowledge there are no other syngeneic mouse models of breast cancer with sensitivity to anti-PD-1 therapy. This enhances the utility of the model and prompted this more complete characterization.
With rapid advances in tumor immunology and immunotherapy there is a growing demand for transplantable cancer cell lines which can be investigated in the immunocompetent syngeneic mouse [1]. These mouse models are essential for experimental immunotherapy studies. Many common preclinical models, such as human tissue culture cell line xenografts and “patient derived xenografts” (PDX) mice must be grown in vivo in immunodeficient mouse strains. Meanwhile, genetically engineered mouse models (GEMM) develop tumors in a comparatively appropriate anatomic and histologic surrounding, with an intact tumor immune microenvironment. However, practical disadvantages of these models include variable penetrance and latency, as well as the multiplicity of tumor onset resulting from diffuse oncogene expression in the target tissue [44]. The effect of high tumor multiplicity on immunosurveillance/immunoediting is not well understood. Transplantable (subcutaneous or orthotopic) syngeneic mouse models present another option for studying immuno-oncology in an experimental setting. The number of mouse cancer cell lines is far smaller than the number of available human cell lines, with many, like 4T1, derived from carcinogen induced tumors and fewer from GEMMs (a legacy of the infamous Oncomouse patent [45]). Transplantable tumor models are usually fast growing, and have a lower cost than GEMMs, contributing to greater practical utility. Utilizing cell lines opens up numerous experimental possibilities such as transfection of expression constructs, editing of the genome, exposing the cells to carcinogens or radiation to study mutational heterogeneity, labeling the cells, etc. Despite the high demand for appropriate experimental models, the literature has only a few examples of detailed description of the immune landscape or reactivity to immune therapies in syngeneic mouse models. Among those studies the breast cancer model is almost exclusively the 4T1 model [46, 47], which is characterized by aggressive, metastatic tumors with a mesenchymal phenotype in vitro and spindle cell morphology in vivo. These are not typical attributes of most human breast cancers.
Even more importantly the 4T1 model proved to be resistant to both anti-CTL-4 and anti-PD-1 therapies unlike our NDLUCD model [3, 46] which proved to be sensitive to combined intratumoral adjuvant (CpG) and anti-PD-1 checkpoint inhibition immunotherapy. This therapy induced an intense antitumor immune reaction in the NDLUCD tumors, but it was unknown at that time if the NDLUCD cells or any of the surrounding immune cells express PD-L1 [4]. PD-1 receptor is expressed primarily in T-cells and the ligand PD-L1 is primarily expressed in antigen presenting cells but may also be expressed in stromal cells. Some cancers aberrantly express PD- L1, a mechanism selected for immune evasion. Anti-PD-1/PD-L1 checkpoint inhibitor therapies have proven to be highly effective in a variety of human tumors, particularly when PD-L1 is expressed by tumor cells in lung cancers and melanoma. PD-L1 epithelial expression in breast cancers was reported with frequencies ranging from 1.7% to 80% in IHC based studies depending on antibodies and different thresholds used [48], with cytoplasmic and membranous localization on tumor cells [49-59].
The NDLUCD cell line expresses cytoplasmic and membranous PD-L1 and therefore provides a suitable preclinical model for PD-L1 positive breast cancer. Anti-PD1 and PD-L1 therapies showed around 20% response rate in various phase I clinical trials as single agents in triple negative breast cancer and the response rates are better when PD-L1 is expressed in the tumor [60]. In the first phase III trial conducted in triple negative breast cancer the anti-PD-1 therapy in combination with chemotherapy significantly improved progression-free and overall survivals compared to chemotherapy alone, especially among patients with PD-L1 positive tumors [61]. Combinational therapies for HER2 and luminal breast cancers are currently under investigation. We suspect based on current and previous [4] results that HER2 positive patients would benefit from anti-PD-1 combination therapies.
Debate continues over the relative validity of primary tumors in GEMMs compared to syngeneic transplant for immunology studies. Although syngeneic transplants are fast and easy to use, some suggested that they might be more immunogenic than spontaneous tumors and their fast growth alters the immunologic burden on the host body [62]. Other studies failed to identify differences in immunogenicity when comparing transplanted and primary tumors [47]. Here, we documented through histologic/IHC quantitation that tumor surrounding and infiltrating lymphocytes are more abundant in the NDLUCD cell line transplants compared to the primary tumors in the genetically engineered mouse, but are similar in composition.
In summary, the NDLUCD cell line, an ErbB2 driven cell line, maintains a stable phenotype in vitro and in vivo as orthotopic transplants in the immune intact FVB/N mouse. The model expresses PD-L1 in cell culture and in vivo orthografts, with cytoplasmic and membrane expression similar to human breast cancers. We propose that NDLUCD is a useful model to study preclinical cancer-immunology, in particular for combination strategies to augment PD-1/PD-L1 checkpoint inhibition.
DATA AVAILABILITY
The microarray and RNASeq datasets generated during the current study are available in the NCBI Gene Expression Omnibus repository, under accession number GSE113380. The scanned slides of immunohistochemical stainings are available from the corresponding author on reasonable request. All further data are published in the manuscript.
Supplementary Material
Acknowledgments
FUNDING:
The UC Davis Comprehensive Cancer Center Genomics Shared Resource is supported by Cancer Center Support Grant (P30CA093373) from the National Cancer Institute. The study was supported by U01 CA196406 and U01 CA141582 from the National Cancer Institute’s Mouse Models of Human Cancers Consortium.
Footnotes
CONFLICT OF INTEREST:
The authors declare that they have no conflict of interest.
REFERENCES
- 1.Zitvogel L, Pitt JM, Daillere R, Smyth MJ, Kroemer G: Mouse models in oncoimmunology. Nature reviews Cancer 2016, 16(12):759–773. [DOI] [PubMed] [Google Scholar]
- 2.Aslakson CJ, Miller FR: Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer research 1992, 52(6):1399–1405. [PubMed] [Google Scholar]
- 3.Mosely SI, Prime JE, Sainson RC, Koopmann JO, Wang DY, Greenawalt DM, Ahdesmaki MJ, Leyland R, Mullins S, Pacelli L et al. : Rational Selection of Syngeneic Preclinical Tumor Models for Immunotherapeutic Drug Discovery. Cancer immunology research 2017, 5(1):29–41. [DOI] [PubMed] [Google Scholar]
- 4.Silvestrini MT, Ingham ES, Mahakian LM, Kheirolomoom A, Liu Y, Fite BZ, Tam SM, Tucci ST, Watson KD, Wong AW et al. : Priming is key to effective incorporation of image-guided thermal ablation into immunotherapy protocols. JCI insight 2017, 2(6):e90521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel PM, Ryan ED, Cardiff RD, Muller WJ: Elevated expression of activated forms of Neu/ErbB-2 and ErbB-3 are involved in the induction of mammary tumors in transgenic mice: implications for human breast cancer. The EMBO journal 1999, 18(8):2149–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castiglioni F, Tagliabue E, Campiglio M, Pupa SM, Balsari A, Menard S: Role of exon-16-deleted HER2 in breast carcinomas. Endocr Relat Cancer 2006, 13(1):221–232. [DOI] [PubMed] [Google Scholar]
- 7.Turpin J, Ling C, Crosby EJ, Hartman ZC, Simond AM, Chodosh LA, Rennhack JP, Andrechek ER, Ozcelik J, Hallett M et al. : The ErbB2DeltaEx16 splice variant is a major oncogenic driver in breast cancer that promotes a pro-metastatic tumor microenvironment. Oncogene 2016, 35(47):6053–6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savas P, Salgado R, Denkert C, Sotiriou C, Darcy PK, Smyth MJ, Loi S: Clinical relevance of host immunity in breast cancer: from TILs to the clinic. Nat Rev Clin Oncol 2016, 13(4):228–241. [DOI] [PubMed] [Google Scholar]
- 9.Peoples GE, Goedegebuure PS, Smith R, Linehan DC, Yoshino I, Eberlein TJ: Breast and ovarian cancer-specific cytotoxic T lymphocytes recognize the same HER2/neu-derived peptide. Proc Natl Acad Sci U S A 1995, 92(2):432–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bailur JK, Derhovanessian E, Gueckel B, Pawelec G: Prognostic impact of circulating Her-2-reactive T-cells producing pro- and/or anti-inflammatory cytokines in elderly breast cancer patients. J Immunother Cancer 2015, 3:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailur JK, Gueckel B, Derhovanessian E, Pawelec G: Presence of circulating Her2-reactive CD8 + T-cells is associated with lower frequencies of myeloid-derived suppressor cells and regulatory T cells, and better survival in older breast cancer patients. Breast cancer research : BCR 2015, 17:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loi S, Michiels S, Salgado R, Sirtaine N, Jose V, Fumagalli D, Kellokumpu-Lehtinen PL, Bono P, Kataja V, Desmedt C et al. : Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial. Ann Oncol 2014, 25(8):1544–1550. [DOI] [PubMed] [Google Scholar]
- 13.Borowsky AD, Namba R, Young LJ, Hunter KW, Hodgson JG, Tepper CG, McGoldrick ET, Muller WJ, Cardiff RD, Gregg JP: Syngeneic mouse mammary carcinoma cell lines: two closely related cell lines with divergent metastatic behavior. Clinical & experimental metastasis 2005, 22(1):47–59. [DOI] [PubMed] [Google Scholar]
- 14.Cardiff RD, Hubbard NE, Engelberg JA, Munn RJ, Miller CH, Walls JE, Chen JQ, Velasquez-Garcia HA, Galvez JJ, Bell KJ et al. : Quantitation of fixative-induced morphologic and antigenic variation in mouse and human breast cancers. Lab Invest 2013, 93(4):480–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen JQ, Mori H, Cardiff RD, Trott JF, Hovey RC, Hubbard NE, Engelberg JA, Tepper CG, Willis BJ, Khan IH et al. : Abnormal Mammary Development in 129:STAT1-Null Mice is Stroma-Dependent. PloS one 2015, 10(6):e0129895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG et al. : QuPath: Open source software for digital pathology image analysis. Scientific reports 2017, 7(1):16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan SR, Vermi W, Luo J, Lucini L, Rickert C, Fowler AM, Lonardi S, Arthur C, Young LJ, Levy DE et al. : STAT1-deficient mice spontaneously develop estrogen receptor alpha-positive luminal mammary carcinomas. Breast Cancer Res 2012, 14(1):R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Team RDC: R: A language and environment for statistical computing . R Foundation for Statistical Computing, Vienna, Austria: 2008. [Google Scholar]
- 19.Carvalho BS, Irizarry RA: A framework for oligonucleotide microarray preprocessing. Bioinformatics 2010, 26(19):2363–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K: KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic acids research 2017, 45(D1):D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Hoon MJ, Imoto S, Nolan J, Miyano S: Open source clustering software. Bioinformatics 2004, 20(9):1453–1454. [DOI] [PubMed] [Google Scholar]
- 22.Saldanha AJ: Java Treeview--extensible visualization of microarray data. Bioinformatics 2004, 20(17):3246–3248. [DOI] [PubMed] [Google Scholar]
- 23.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR et al. : Accurate whole human genome sequencing using reversible terminator chemistry. Nature 2008, 456(7218):53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L: Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols 2012, 7(3):562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjamini Y, Krieger AM, Yekutieli D: Adaptive linear step-up procedures that control the false discovery rate. Biometrika 2006, 93(3):491–507. [Google Scholar]
- 26.Miller JK, Shattuck DL, Ingalla EQ, Yen L, Borowsky AD, Young LJ, Cardiff RD, Carraway KL 3rd, Sweeney C: Suppression of the negative regulator LRIG1 contributes to ErbB2 overexpression in breast cancer. Cancer research 2008, 68(20):8286–8294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yerlikaya A, Erin N: Differential sensitivity of breast cancer and melanoma cells to proteasome inhibitor Velcade. International journal of molecular medicine 2008, 22(6):817–823. [PubMed] [Google Scholar]
- 28.Rosner A, Miyoshi K, Landesman-Bollag E, Xu X, Seldin DC, Moser AR, MacLeod CL, Shyamala G, Gillgrass AE, Cardiff RD: Pathway pathology: histological differences between ErbB/Ras and Wnt pathway transgenic mammary tumors. The American journal of pathology 2002, 161(3):1087–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borowsky AD: Choosing a mouse model: experimental biology in context--the utility and limitations of mouse models of breast cancer. Cold Spring Harb Perspect Biol 2011, 3(9):a009670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DiGiovanna MP, Lerman MA, Coffey RJ, Muller WJ, Cardiff RD, Stern DF: Active signaling by Neu in transgenic mice. Oncogene 1998, 17(14):1877–1884. [DOI] [PubMed] [Google Scholar]
- 31.Cardiff RD, Kenney N: A compendium of the mouse mammary tumor biologist: from the initial observations in the house mouse to the development of genetically engineered mice. Cold Spring Harb Perspect Biol 2011, 3(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen DS, Mellman I: Elements of cancer immunity and the cancer-immune set point. Nature 2017, 541(7637):321–330. [DOI] [PubMed] [Google Scholar]
- 33.Huang da W, Sherman BT, Lempicki RA: Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols 2009, 4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 34.Huang da W, Sherman BT, Lempicki RA: Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research 2009, 37(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Townsend DM, Tew KD: The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene 2003, 22(47):7369–7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jessen KA, Liu SY, Tepper CG, Karrim J, McGoldrick ET, Rosner A, Munn RJ, Young LJ, Borowsky AD, Cardiff RD et al. : Molecular analysis of metastasis in a polyomavirus middle T mouse model: the role of osteopontin. Breast cancer research : BCR 2004, 6(3):R157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliveira AM, Ross JS, Fletcher JA: Tumor suppressor genes in breast cancer: the gatekeepers and the caretakers. Am J Clin Pathol 2005, 124 Suppl:S16–28. [DOI] [PubMed] [Google Scholar]
- 38.Mori H, Chen JQ, Cardiff RD, Penzvalto Z, Hubbard NE, Schuetter L, Hovey RC, Trott JF, Borowsky AD: Pathobiology of the 129:Stat1 (−/−) mouse model of human age-related ER-positive breast cancer with an immune infiltrate-excluded phenotype. Breast cancer research : BCR 2017, 19(1):102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mantovani A, Allavena P, Sica A, Balkwill F: Cancer-related inflammation. Nature 2008, 454(7203):436–444. [DOI] [PubMed] [Google Scholar]
- 40.Danaher P, Warren S, Dennis L, D'Amico L, White A, Disis ML, Geller MA, Odunsi K, Beechem J, Fling SP: Gene expression markers of Tumor Infiltrating Leukocytes. J Immunother Cancer 2017, 5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu X, Kheirolomoom A, Mahakian LM, Beegle JR, Kruse DE, Lam KS, Ferrara KW: Insonation of targeted microbubbles produces regions of reduced blood flow within tumor vasculature. Invest Radiol 2012, 47(7):398–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Engelberg JA, Giberson RT, Young LJ, Hubbard NE, Cardiff RD: The use of mouse models of breast cancer and quantitative image analysis to evaluate hormone receptor antigenicity after microwave-assisted formalin fixation. J Histochem Cytochem 2014, 62(5):319–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rowson-Hodel AR, Berg AL, Wald JH, Hatakeyama J, VanderVorst K, Curiel DA, Leon LJ, Sweeney C, Carraway KL 3rd: Hexamethylene amiloride engages a novel reactive oxygen species- and lysosome-dependent programmed necrotic mechanism to selectively target breast cancer cells. Cancer Lett 2016, 375(1):62–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pardoll DM: The blockade of immune checkpoints in cancer immunotherapy. Nature reviews Cancer 2012, 12(4):252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smaglik P: NIH cancer researchers to get free access to 'OncoMouse'. Nature 2000, 403(6768):350. [DOI] [PubMed] [Google Scholar]
- 46.Grosso JF, Jure-Kunkel MN: CTLA-4 blockade in tumor models: an overview of preclinical and translational research. Cancer Immun 2013, 13:5. [PMC free article] [PubMed] [Google Scholar]
- 47.Lechner MG, Karimi SS, Barry-Holson K, Angell TE, Murphy KA, Church CH, Ohlfest JR, Hu P, Epstein AL: Immunogenicity of murine solid tumor models as a defining feature of in vivo behavior and response to immunotherapy. J Immunother 2013, 36(9):477–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ilie M, Hofman V, Dietel M, Soria JC, Hofman P: Assessment of the PD-L1 status by immunohistochemistry: challenges and perspectives for therapeutic strategies in lung cancer patients. Virchows Arch 2016, 468(5):511–525. [DOI] [PubMed] [Google Scholar]
- 49.Ali HR, Glont SE, Blows FM, Provenzano E, Dawson SJ, Liu B, Hiller L, Dunn J, Poole CJ, Bowden S et al. : PD-L1 protein expression in breast cancer is rare, enriched in basal-like tumours and associated with infiltrating lymphocytes. Ann Oncol 2015, 26(7):1488–1493. [DOI] [PubMed] [Google Scholar]
- 50.Bae SB, Cho HD, Oh MH, Lee JH, Jang SH, Hong SA, Cho J, Kim SY, Han SW, Lee JE et al. : Expression of Programmed Death Receptor Ligand 1 with High Tumor-Infiltrating Lymphocytes Is Associated with Better Prognosis in Breast Cancer. J Breast Cancer 2016, 19(3):242–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baptista MZ, Sarian LO, Derchain SF, Pinto GA, Vassallo J: Prognostic significance of PD-L1 and PD-L2 in breast cancer. Hum Pathol 2016, 47(1):78–84. [DOI] [PubMed] [Google Scholar]
- 52.Beckers RK, Selinger CI, Vilain R, Madore J, Wilmott JS, Harvey K, Holliday A, Cooper CL, Robbins E, Gillett D et al. : Programmed death ligand 1 expression in triple-negative breast cancer is associated with tumour-infiltrating lymphocytes and improved outcome. Histopathology 2016, 69(1):25–34. [DOI] [PubMed] [Google Scholar]
- 53.Dill EA, Gru AA, Atkins KA, Friedman LA, Moore ME, Bullock TN, Cross JV, Dillon PM, Mills AM: PD-L1 Expression and Intratumoral Heterogeneity Across Breast Cancer Subtypes and Stages: An Assessment of 245 Primary and 40 Metastatic Tumors. Am J Surg Pathol 2017, 41(3):334–342. [DOI] [PubMed] [Google Scholar]
- 54.Gatalica Z, Snyder C, Maney T, Ghazalpour A, Holterman DA, Xiao N, Overberg P, Rose I, Basu GD, Vranic S et al. : Programmed cell death 1 (PD-1) and its ligand (PD-L1) in common cancers and their correlation with molecular cancer type. Cancer Epidemiol Biomarkers Prev 2014, 23(12):2965–2970. [DOI] [PubMed] [Google Scholar]
- 55.Mittendorf EA, Philips AV, Meric-Bernstam F, Qiao N, Wu Y, Harrington S, Su X, Wang Y, Gonzalez-Angulo AM, Akcakanat A et al. : PD-L1 expression in triple-negative breast cancer. Cancer immunology research 2014, 2(4):361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mori H, Kubo M, Yamaguchi R, Nishimura R, Osako T, Arima N, Okumura Y, Okido M, Yamada M, Kai M et al. : The combination of PD-L1 expression and decreased tumor-infiltrating lymphocytes is associated with a poor prognosis in triple-negative breast cancer. Oncotarget 2017, 8(9):15584–15592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park IH, Kong SY, Ro JY, Kwon Y, Kang JH, Mo HJ, Jung SY, Lee S, Lee KS, Kang HS et al. : Prognostic Implications of Tumor-Infiltrating Lymphocytes in Association With Programmed Death Ligand 1 Expression in Early-Stage Breast Cancer. Clin Breast Cancer 2016, 16(1):51–58. [DOI] [PubMed] [Google Scholar]
- 58.Qin T, Zeng YD, Qin G, Xu F, Lu JB, Fang WF, Xue C, Zhan JH, Zhang XK, Zheng QF et al. : High PD-L1 expression was associated with poor prognosis in 870 Chinese patients with breast cancer. Oncotarget 2015, 6(32):33972–33981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soliman H, Khalil F, Antonia S: PD-L1 expression is increased in a subset of basal type breast cancer cells. PloS one 2014, 9(2):e88557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bertucci F, Goncalves A: Immunotherapy in Breast Cancer: the Emerging Role of PD-1 and PD-L1. Curr Oncol Rep 2017, 19(10):64. [DOI] [PubMed] [Google Scholar]
- 61.Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, Dieras V, Hegg R, Im SA, Wright GS et al. : LBA1_PR IMpassion130: Results from a global, randomised, double-blind, phase III study of atezolizumab (atezo) + nab -paclitaxel (nab -P) vs placebo + nab -P in treatment-naive, locally advanced or metastatic triple-negative breast cancer (mTNBC). Annals of Oncology, Volume 29 (Supplement 8). [Google Scholar]
- 62.Ostrand-Rosenberg S: Animal models of tumor immunity, immunotherapy and cancer vaccines. Curr Opin Immunol 2004, 16(2):143–150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray and RNASeq datasets generated during the current study are available in the NCBI Gene Expression Omnibus repository, under accession number GSE113380. The scanned slides of immunohistochemical stainings are available from the corresponding author on reasonable request. All further data are published in the manuscript.