Abstract
Ecological relationships between bacteria are important when considering variation in bacterial communities in humans, with such variation playing an important role in both health and disease.
Using high-throughput sequence data of the 16S rRNA marker-gene, we analysed the prevalence of taxa in the airways of a number of health- and disease-associated cohorts and determined the main drivers of community variance and bacterial co-occurrence.
A number of facultative and obligately anaerobic bacterial taxa are commonly associated with the upper airways, forming the main “core” microbiota, e.g. Streptococcus spp., Veillonella spp., Prevotella spp., Granulicatella spp. and Fusobacterium spp. Opportunistic pathogenic bacteria associated with chronic airways disease, such as Pseudomonas spp. (Pseudomonas aeruginosa), Burkholderia spp. (Burkholderia cepacia complex) and Haemophilus spp. (Haemophilus influenzae) demonstrated poor correlation with other members of their respective communities (ρ<0.5; p>0.005), indicating probable independent acquisition and colonisation. Furthermore, our findings suggest that intra-genus variation between health and disease may affect community assemblies.
Improved understanding of how bacteria assemble in time and space during health and disease will enable the future development of tailored treatment according to the patient's own signature microbiota, potentially providing benefit to patients suffering from airway diseases characterised by chronic infection.
Short abstract
Within the airways, “core” community structures are formed between microbial taxa in both health and disease, with a number of common opportunistic pathogens not being members of such core communities http://bit.ly/2Kau3ni
Introduction
In respiratory diseases characterised by chronic infection, such as cystic fibrosis (CF) and bronchiectasis (BE), persistent bacterial colonisation of the airways is a major cause of morbidity and mortality [1–3]. Studies applying classical aerobic-based microbial culture techniques have shown that people with CF and BE are frequently colonised with a range of potentially pathogenic bacteria, including Pseudomonas aeruginosa, Haemophilus influenzae, Streptococcus pneumoniae and Staphylococcus aureus [4–7]. Recent studies have also shown that these patients are frequently colonised with obligately anaerobic bacteria [6, 8–10], though their role in airways disease remains unclear.
Within the airways, the microbiota play an important role in both health and disease [11–14], with members being able to exchange or compete for nutrients, signalling molecules, or host immune evasion mechanisms [15–17]. Such microbial interactions are essential for community stability within the healthy commensal human microbiota [18–21], with dysbiosis leading to an overgrowth of competitive pathogenic species [22]. Therefore, detailed ecological characterisation of the microbiota during health is a pivotal first step in determining how changes within community structures contribute to disease progression [23–25].
In this study, we investigated the broader microbial community structures within the airways, as well as the distribution of various taxa in health and/or disease communities. Through community-wide and co-occurrence network analyses, we addressed three key questions: 1) Is there a “core” community microbiota in the airways? 2) Which taxa have the highest community-wide association (“Generalist”), strongest niche association (“Niche Specialist”), and the strongest disease association (“Disease Specialist”)? and 3) Which members are responsible for the observed variation within each community?
Methods
A detailed description of methods and statistical analysis is provided in the supplementary material. We analysed greater than 2.1 million sequences from our normalised (2000 sequences per sample) dataset, which included 1057 samples in total (table 1 and file S1).
TABLE 1.
Sample numbers studied for each cohort included in this study
| Collection site | Cohort | Samples |
| Anterior nares | Healthy | 67 |
| Attached keratinised gingiva | Healthy | 102 |
| Subgingival plaque | Healthy | 92 |
| Supragingival plaque | Healthy | 103 |
| Saliva | Healthy | 69 |
| Hard palate | Healthy | 93 |
| Tongue dorsum | Healthy | 99 |
| Buccal mucosa | Healthy | 98 |
| Throat | Healthy | 94 |
| Palatine tonsils | Healthy | 97 |
| Cystic fibrosis sputum | Disease | 94 |
| Cystic fibrosis mouthwash | Disease | 23 |
| Bronchiectasis sputum | Disease | 26 |
| Total | 1057 |
Cohorts and sample data processing
For CF and BE, samples were obtained from previous studies from our group focusing on expectorated sputum and mouthwash samples [8–10]. In addition, raw DNA sequence data from a number of CF sputum samples provided by Professor John LiPuma at the University of Michigan, USA, were analysed. 16S rRNA marker-gene datasets produced from the available resources of the Human Microbiome Project (HMP) (http://hmpdacc.org) [26] were used as a surrogate of the “normal” upper airway microbiome. All samples analysed were taken at a single time-point; the first visit for the HMP data and during a period of clinical stability for CF sputum, CF mouthwash (CFMW) and BE sputum samples. All sequence data were generated on the Roche 454-FLX Titanium platform and the analysis focused on amplicon data covering the V1V3 region of the 16S rRNA gene for all datasets, with the exception of a subset of samples belonging to the CF and CFMW that covered the V1V2 region of the 16S rRNA marker-gene.
To assess whether interstudy variation, such as differences in primer pairs (V1V2 versus V1V3) or whether niche and geographical origin explained the observed variation in our study we performed principal coordinate analysis (PCoA) using unweighted UniFrac distance metrics. Furthermore, we determined how variables associated with different cohorts (all 13 sampling sites), airway location (three groups: lungs, mouth and nose) and primer-pairing (V1V3 versus V1V2) affected the structure of the microbiota using a permutational multivariate ANOVA (PERMANOVA) test of the unweighted and weighted UniFrac distances and Bray–Curtis dissimilarities. Additionally, we performed multivariate analysis using the Adonis function (permutational multivariate ANOVA using distance matrices) as implemented within the vegan package in R with Bray–Curtis distances and 999 permutations to establish the independent effect or potential collinearity of variables under investigation. Further information on the assessment of technical variation is shown in the online supplement.
Comparison between community structures
Taxa were ranked according to their mean relative abundance into three main categories: “Generalist” defined as a taxon occurring in at least >70% of all samples, “Niche Specialists” defined as taxa showing strong niche association within a single cohort and “Chronic Airways Disease Specialists” defined as taxa representing the majority of the relative abundance and occurrence associated with chronic disease (CF or BE).
Co-occurrence analysis
To assess whether nonrandom co-occurrence patterns existed within our sample cohort, we initially performed calculations based on the C-score (checkerboard units) calculations [27] under a null model with preserved site frequencies. The C-score measures the average number of checkerboard units, or co-occurrences, between all possible OTU (operational taxonomic unit; based on sequence similarities) pairs and enabled determination of whether taxa/genera tended to aggregate together more than would be expected due to chance alone.
Network inference was generated by calculating all possible Spearmańs rank correlation coefficients (ρ) between taxon pairs. To minimise the occurrence of spurious associations we considered a valid co-occurrence between two different taxa if the Spearman's correlation coefficient (ρ) was both >0.5 and statistically significant (adjusted p<0.005; Benjamini–Hochberg–Yekutieli). In the reconstructed co-occurrence networks, all nodes represent taxa that show at least 97% identity, with the edges (i.e. connections) corresponding to a significant correlation between nodes (i.e. taxa; based on ρ and significance according to the adjusted p-value). To compare the effect each taxon had on the observed variance within its environment, we conducted principal component analysis (PCA) on the normalised OTU abundance measures.
Statistical analysis
A Spearman rank correlation coefficient (ρ) was calculated to measure the strength of association between different taxa. Community richness and diversity (Shannon–Wiener index) was compared between three or more cohorts by the Kruskal–Wallis test, followed by post hoc testing using the nonparametric Mann–Whitney test with Bonferroni adjustment to evaluate differences between two sample cohorts. A p-value <0.05 was deemed statistically significant. Further information regarding data processing and analysis is provided in the supplementary material.
Results
We analysed more than 2.1 million sequences from our normalised (2000 sequences per sample) dataset, which included 1057 samples in total (table 1 and file S1).
Airway community richness and diversity
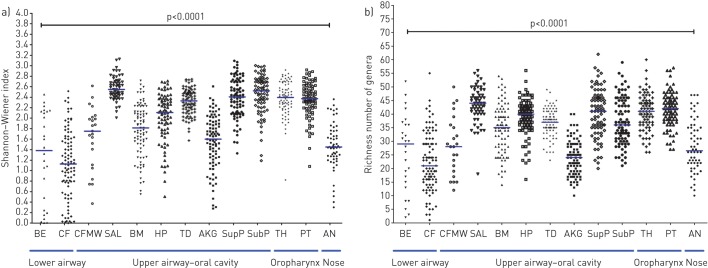
Community diversity and richness were significantly different between communities originating from different airway sites (p<0.0001, Kruskal–Wallis test) (figure 1a and b); these differences were primarily between cohorts associated with health and disease, with no differences apparent between the three disease cohorts (CF sputum, CFMW and BE sputum). Furthermore, there was no significant difference observed between the community diversity and richness of the disease-associated cohorts and several of the health-associated cohorts, which exhibited relatively low diversity and richness (p>0.05, Mann–Whitney post hoc testing with Bonferroni adjustment). Further information for pairwise comparisons between each sample cohort are shown in supplementary tables S1a and S1b, respectively.
FIGURE 1.
a) Taxa diversity (Shannon–Wiener diversity index) and b) taxa richness (counts of observed taxa per sample) for the 13 cohorts. Data are presented as intracohort spread with the horizontal line showing median values. Differences between pairs were evaluated using the Mann–Whitney test with Bonferroni adjustment (p<0.05 denoted significant differences). BE: bronchiectasis sputum; CF: cystic fibrosis sputum; CFMW: cystic fibrosis mouthwash; SAL: saliva; BM: buccal mucosa; HP: hard palate; TD: tongue dorsum; AKG: attached keratinised gingiva; SupP: supragingival plaque; SubP: subgingival plaque; TH: throat; PT: palatine tonsils; AN: anterior nares.
Comparison between community structures
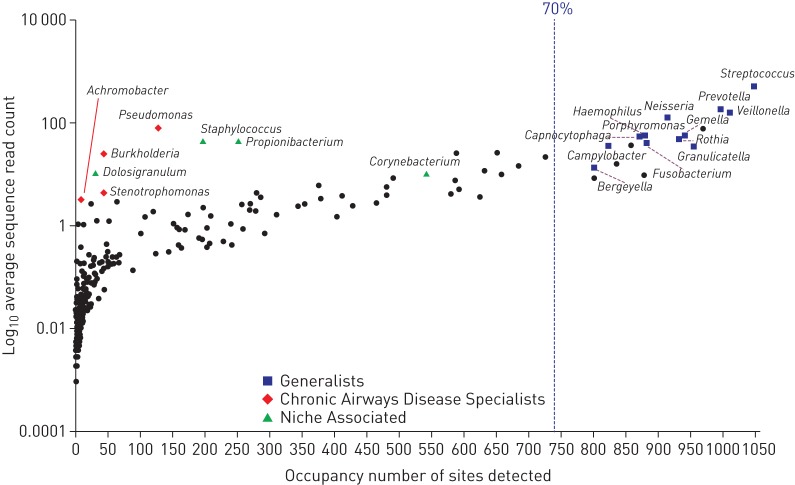
The overall community structures were dominated by relatively few taxa, with the vast majority of observed OTUs present in low occurrence and low relative abundance. Evaluation of the complete dataset revealed that bacterial taxa separated into three main groups (figure 2). The “Generalists” or the “core” community included taxa present at a relatively high frequency (occupancy) among the 13 cohorts and at a relatively high abundance. Each of the “Generalist” groups contained multiple different OTUs; however, only a few of these exceeded >70% occupancy within the overall sample cohort. The main OTUs corresponding to the “core” community were further assessed based on at least 60% prevalence in either of the health and disease-associated cohorts and in at least 70% of all samples. A number of specific OTUs formed the “core” community, including members of Streptococcus spp., Veillonella spp., Prevotella spp., Granulicatella spp. and Fusobacterium spp. CF and BE sputum samples demonstrated “skewed” community structures attributed to the presence of a small number of taxa that formed the “Chronic Airways Disease Specialists”. Taxa in this group included opportunistic bacteria such as Pseudomonas spp., Burkholderia spp., Achromobacter spp., and Stenotrophomonas spp., which demonstrated a strong shift towards predominance within their corresponding samples (figure 2). These taxa were present with low frequency but high relative abundance. In the anterior nares, “Niche Specialists” were mainly composed of Staphylococcus spp., Propionibacterium spp., Dolosigranulum spp. and Corynebacterium spp., which were present in high relative abundance (figure 2). Further information regarding taxa distribution in all 1057 samples is provided in file S2.
FIGURE 2.
Overall taxa distribution (occupancy) within the 13 cohorts included in the study. The blue dotted line represents a cut-off for taxa termed as “Generalists” determined according to specific operational taxonomic units (sequence types) observed in ≥70% of all samples.
Airway community variance
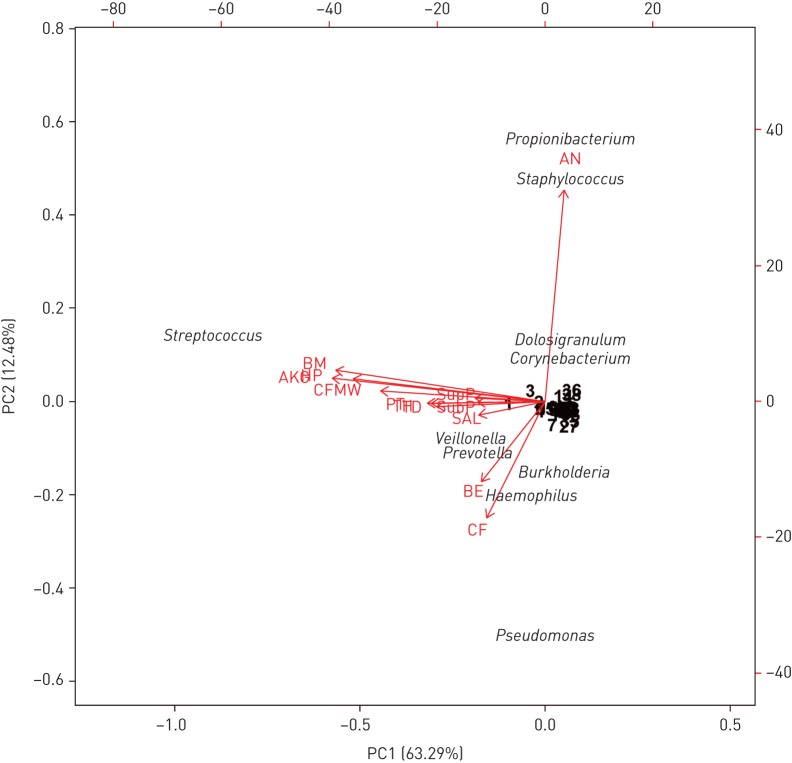
We applied PCA to determine which OTUs represented the most significant source of population variance within each community. The first two principal components in our normalised dataset accounted for 75.77% variance explained (figure 3). Variation was driven by the dominance of Pseudomonas spp. and Burkholderia spp. in CF and Pseudomonas spp. and Haemophilus spp. in BE, with these taxa having the most effect on the shape and direction of the corresponding communities (figure 3). In the upper airways (mouth and oropharynx), the principal factor contributing to the observed variance was Streptococcus spp., a taxon most frequently occupying samples from these sites. The anterior nares formed distinct community structures compared with the other sampling sites, with the variation in the samples strongly affected by predominance of either Staphylococcus spp., Propionibacterium spp., Dolosigranulum spp. or Corynebacterium spp. Furthermore, anaerobic bacteria such as Prevotella spp. and Veillonella spp. were shown to affect variation within a number of niches, driving a proportion of the observed variance in both disease-associated cohorts, as well as in saliva, throat, tongue dorsum and hard palate (figure 3).
FIGURE 3.
Biplot of principal components analysis conducted for the total community data set (sites and taxa). Per cent variation covered by each principal component is indicated in parentheses in the axis titles, with the first two components accounting for 75.77% total variance explained. Arrows indicate direction and magnitude of loadings (sites) on the first principal component. Factors and taxa names that had limited effect on any of the sites (i.e. clustered near the centre axis) are displayed as their assigned rank number for clarity. BE: bronchiectasis sputum; CF: cystic fibrosis sputum; CFMW: cystic fibrosis mouthwash; SAL: saliva; BM: buccal mucosa; HP: hard palate; TD: tongue dorsum; AKG: attached keratinised gingiva; SupP: supragingival plaque; SubP: subgingival plaque; TH: throat; PT: palatine tonsils; AN: anterior nares.
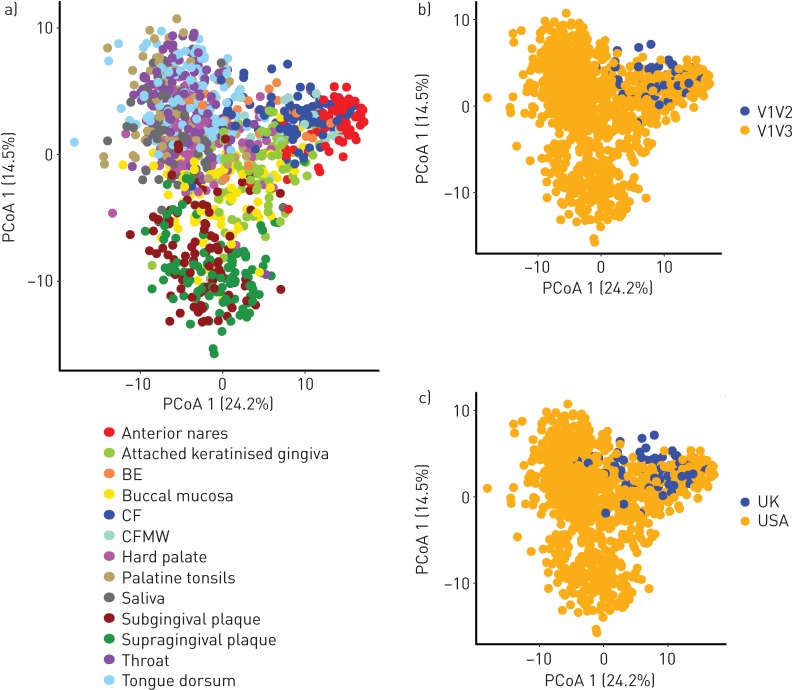
The unweighted UniFrac PCoA plots demonstrated that clustering by primer choice or geographical origin of the samples did not have a significant overall effect on the sample clustering, while there was a strong clustering by body site (figure 4 a–c). Factors potentially affecting the community structuring (i.e. cohort (sampling site), airway location (three groups: lungs, mouth and nose) and primer-pairing (V1V3 versus V1V2)) demonstrated that the different primer-pairing accounted for 3–6% (p<0.0001) of the explained community variance dependent on the metric used (tables S3 and S4). Furthermore, multivariate analysis showed a significant effect of sampling site (R2=0.37, p<0.001), as well as significant effect of primer choice and airway location (R2=0.01, p<0.001 and R2=0.16, p<0.001, respectively). However, when exclusively looking at the CF group, as this group included the overlapping primer regions for a subset of samples, we observed effect due to the primer group (PERMANOVA: R2 0.07, p<0.001), confirming that ∼7% of the variance in the CF group could be explained by primer choice.
FIGURE 4.
Unweighted UniFrac principal coordinate analysis (PCoA) plots illustrating the relationship between the bacterial diversity in samples from the current study. a) All 13 cohorts, b) V1V3 versus V1V2 overlapping primer pairs and c) geographical sampling location. BE: bronchiectasis sputum; CF: cystic fibrosis sputum; CFMW: cystic fibrosis mouthwash.
Co-occurrence network analysis of airway associated microbiota
Based on ecological measurements of checkerboard units (C-Score) [27, 28], we observed that the whole community was based on nonrandom co-occurrence patterns (C-score 185.40, p<0.01).
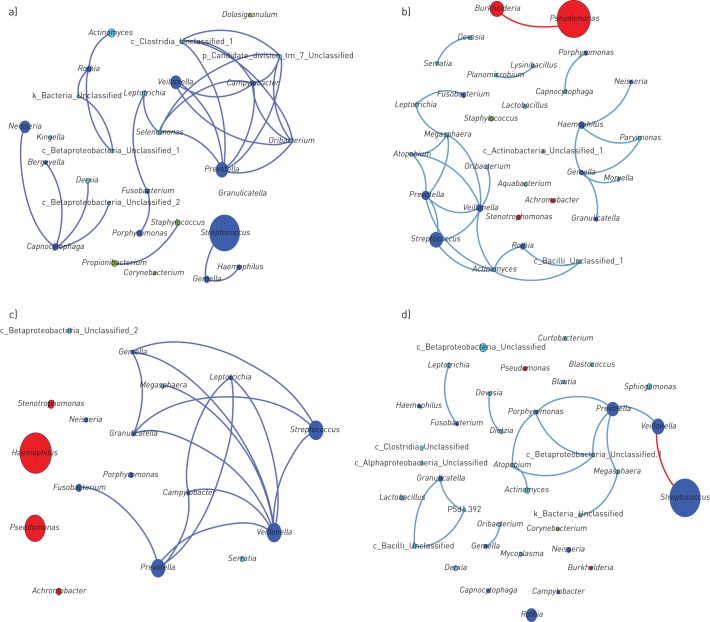
Co-occurrence network analysis for the different airway sites revealed a “core” community structure that consisted of a number of bacterial taxa commonly associated with the upper airway microbiota (figure 5a). In the overall sample cohort, we detected five main subnetworks of co-occurring taxa. The main subnetwork demonstrated a strong correlation (ρ>0.5; adjusted p<0.005) between several taxa, with many belonging to the “Generalist” group. Members of this subnetwork included anaerobic and microaerophilic genera such as Prevotella spp., Veillonella spp., Campylobacter spp., Leptotrichia spp., Fusobacterium spp. and Selenomonas spp. (figure 5a).
FIGURE 5.
Microbial co-occurrence network for the main taxa from a) 13 sites of the greater airways (taxa accounting for >0.5% of the total sequences in the 778 samples of the combined upper airways), b) co-occurring microbial taxa from the cystic fibrosis (CF) subset (taxa >0.1% of total sequences), c) co-occurring microbial taxa from the bronchiectasis subset (taxa >0.1% of total sequences), and d) co-occurring microbial taxa from the CF mouthwash subset (taxa >0.1% of total sequences). Each node denotes a particular taxon within the network and each line (edge) a significant co-occurrence relationship (Spearman's rank correlation coefficient >0.5 for positive correlations (blue lines) and <−0.5 for negative correlations (red lines); adjusted p-value <0.005 (false discovery rate correction for multiple testing according to the Benjamini–Hochberg–Yekutieli procedure). Nodes are coloured according to “Generalists” (blue), “Chronic Airways Disease Specialists” (red), “Niche Associated” (green) and not belonging to any of the former three groups (aqua).
Within the CF sputum (figure 5b), there was a strong correlation between members of Veillonella spp. and Prevotella spp. (ρ=0.730; adjusted p<1.00×10−10) which formed the main subnetwork with members of Streptococcus spp. CF sputum communities were dominated by Pseudomonas spp. and Burkholderia spp., with neither demonstrating a significant correlation with any other members of their communities.
For BE sputum (figure 5c), members of the obligately anaerobic Prevotella spp., and Veillonella spp. and facultative anaerobic Streptococcus spp. and Campylobacter spp., formed the main subnetwork, showing significant correlations with a large number of other members of this community. BE sputum communities were dominated by Pseudomonas spp. and Haemophilus spp.
In CFMW, the community and taxa distributions were considerably different from any other cohorts within both the upper and lower airways. The main community subnetwork consisted mainly of taxa associated with the upper airways or oral cavity (figure 5d) with Streptococcus spp. the predominant taxon present; however, Streptococci did not show a positive correlation with any other members of the community. Further information regarding correlation scores between community members within the all cohorts is shown in table S2a–m and additional co-occurrence networks are shown in figure S1a–j.
Community differences within sites: species assignment and distribution
Despite inherent limitations associated with the common methodologies used to assign taxonomic ranking to the species level in studies using short amplicon reads, we attempted species level approximation for most common genera within the 13 cohorts to provide a snapshot of potential differences that may occur between health and disease-associated communities. The results show that the main taxa (i.e. Streptococcus spp., Prevotella spp., and Veillonella spp.), as well as taxa important in chronic airways diseases (i.e. Pseudomonas spp., and Haemophilus spp.) were notably different in their distribution between groups (supplementary table S5 a–e). Further information regarding putative differences in speciation between cohorts is provided in the online supplement.
Discussion
Previous studies have shown that different sampling sites within the airways of healthy individuals, as well as those suffering from chronic airway diseases, harbour a diverse community of bacteria [9, 10, 12, 19, 21, 29, 30]. Within airway microbial communities, there is inherent variance between individuals with respect to the dominant taxa, often making it difficult to decipher which signature or “core” taxa are associated with a particular community and/or condition [31–33]. However, our results clearly demonstrate that human respiratory tract communities tend to aggregate and that microbial communities are composed of interacting bacterial taxa and not of randomly assembled bacterial consortia. Furthermore, within the airways, “core” community structures are formed between taxa in both health and disease. Bacteria in these communities exhibit the capability to inhabit similar ecological niches such as obligate anaerobes and bacteria that are able to adjust to reduction in oxygen levels within the surrounding environment, such as facultative anaerobic streptococci.
Though frequently reported as part of the normal human microbiota, and thus often not regarded as “true pathogens”, a number of obligate and facultatively anaerobic bacteria have been reported to play a role in disease progression. Opportunistic behaviour may arise from the selective advantage some members of the population have, resulting in an acute overgrowth under certain conditions [34] that influences the balance of the community as a whole. A number of other members of the “core” community have previously been shown to act as opportunistic pathogens and have been associated with a variety of pathological processes [35–37]. In CF, Streptococcus species have been associated with both clinical stability [38] and shown to contribute to pathological processes within the airways [39]. Therefore, as intra-genus changes in bacterial populations may cause a shift in phenotypic properties and community balance leading to disease progression, better understanding of the role members of the “core” community play, at the species or strain level, in progression of chronic airways disease is needed.
As reported previously, [12, 29, 30, 40] significant similarities were observed between the “core” bacteria in communities from the lungs and the oropharynx and/or oral cavity. This was not unexpected as these sites are anatomically contiguous, and it is likely that various members of the upper airway communities may be dispersed to the lower part of the airways through micro-aspiration and/or mucosal secretions. However, analysis of community composition in the anterior nares revealed a distinctive microbiome signature similar to that previously reported in subjects with chronic rhinitis [34] and healthy individuals [41].
Furthermore, Pseudomonas spp. (i.e. Pseudomonas aeruginosa) the main taxa associated with both CF and BE community cohorts, did not correlate with other members of the “core” community, suggesting independent acquisition and adaptation within the airways of patients with chronic airways disease and is in an agreement with previous observations by our group for chronic obstructive pulmonary disease [42]. The prevalence of “typical” pathogens associated with CF and BE, such as P. aeruginosa and Haemophilus spp. (i.e. H. influenzae), in mouthwash samples and healthy upper airways was low, indicating that the oropharynx was an unlikely source of these pathogens. This is in agreement with the findings of previous studies that suggested a link between P. aeruginosa [43, 44] and H. influenzae [45] acquisition in the lower airways and primary colonisation of the paranasal sinuses and the nasopharynx.
Using network inference to investigate co-occurrence of microbes within communities generated from high-throughput short amplicon sequencing is a relatively unique way to assess how microbial communities are constructed in space and time. Interpreting why the observed co-occurrence patterns occur is difficult as they may be as a result of a number of factors including competition and/or sharing of resources, co-colonisation, co-aggregation, or due to an overlapping niche that both members inhabit. Questions of how host genetics, prolonged antibiotic exposure, and inclusion/exclusion capabilities that the resident microbiota have on other members and how these shape the assembly of the community still need to be addressed.
Our methodological approach has a number of important limitations that need highlighted. Firstly, given the large cohort of samples compared, it is inevitable that a number of different 454-FLX sequencing machines have been employed. This, along with potential differences in sample processing, such as in DNA extraction methods, may lead to a certain degree of bias in amplicon output and subsequent that is difficult to counteract. We attempted to limit the effect of spurious and low-quality sequences on the subsequent taxonomic assignments through careful quality control prior to downstream analysis. Sequences were filtered to exclude low-quality reads, amplicons of fewer than 200 nucleotides and potential chimeric sequences. Moreover, all sequences were inferred against a number of nonbacterial databases to eliminate any amplicons of a nonbacterial origin. Secondly, a subset of samples from the CF and CFMW cohort employed oligonucleotide primers covering the V1V2 region of the 16S rRNA marker-gene which may introduce a certain bias. However, despite the fact that our analysis demonstrated that different primer-pairings significantly affected the community delineation, the effect on the overall variability was relatively low. Any potential differences may have been reduced by the fact that both primer-pairings spanning the same variable region of the 16S rRNA marker-gene rather than covering two nonoverlapping regions, which would have required the use of the closed reference OTU picking method. Moreover, all samples employed the same forward primer and further analysis revealed no obvious differences in the ability to assign amplicons to their appropriate rank. Thirdly, in the current study we did not take into account the differences in disease stage (e.g. differences in lung function) for those samples that originated from CF and BE. This may potentially be important as significant changes in the airway microbiome have been associated with more severe lung disease [46–48]. However, whether lung disease severity affected the community composition and clustering of those cohorts was beyond the scope of the current study, but we did observe significantly higher degree of similarities within each of the sampling groups. Furthermore, samples belonging to the CFMW cohort aligned closer to the communities of the upper airways and the main “core” community structure within all the cohorts remained similar, suggesting minimal bias due to sample preparation or amplicon length. Fourthly, the availability of matched sequence data from the upper airways of patients with chronic respiratory disease and the lower airways of healthy subjects is limited in the published literature. In the current dataset, the only available comparators between the sample types from the same cohort were matched CFMW and CF sputum samples. This lack of comprehensive matched niche-based coverage limited the ability to elucidate the origin of the colonising microbiota of the lower airways in subjects with chronic respiratory disease. However, including the CFMW cohort allowed us to investigate differences relating to the main community protagonists involved in chronic bacterial infections of the lower airways in CF and BE. Next, different sampling techniques (i.e. upper airway swabs, saliva, mouthwash and sputum) may be confounded by differences in biomass, and furthermore it may be difficult to determine the exact point of origin for every sputum sample as this omits direct sampling of the lower airways, as would be the case with samples collected during bronchoscopic procedures. However, in the current study our results indicated that site-specific microbial similarities play an important role in validating our observations, in addition to lower richness and taxonomic diversity being associated with individuals that provided sputum samples but had poorer clinical status. Finally, our effort to speciate the main genera within our communities potentially provides putative indication as to how main OTUs are distributed within different cohorts. However, speciation based on high-throughput short amplicon sequencing studies is problematic and highlights need to utilise methodologies that accurately assess the distribution of species or strains in complex microbial communities. Moreover, without prior knowledge of the functional properties of individual community members, their causal role in their role in the progression of chronic diseases will be difficult to elucidate.
In summary, this study has shown that within the airways, “core” community structures are formed between taxa in both health and disease with opportunistic pathogens such as P. aeruginosa and H. influenzae, not being members of such “core” communities. Better understanding of how different members of local microbiota interact with each other will allow us to characterise how members of the “core” microbiota affect the overall composition of their niche during the development of chronic airway diseases. Furthermore, advancement in sequencing technologies, such as whole genome sequencing may allow determination of specific strain distribution within the community, as well as allowing culture-independent in silico antimicrobial susceptibility testing to determine how treatment may affect the community as a whole. This should enable the future development of tailored treatment according to the signature of the patients own microbial community, providing greater benefits to patients suffering from airway diseases characterised by chronic infection.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
FIGURE S1 00128-2017.figureS1 (884.3KB, pdf)
FIGURE S2 00128-2017.figureS2 (1.7MB, pdf)
FILE S1 00128-2017.fileS1 (92KB, txt)
FILE S2 00128-2017.fileS2 (948.8KB, csv)
Detailed methods 00128-2017.supp (253.8KB, pdf)
TABLE S1 00128-2017.tableS1 (21.1KB, pdf)
TABLE S2 00128-2017.tableS2 (357.7KB, xlsx)
TABLE S3 00128-2017.tableS3 (11.2KB, pdf)
TABLE S4 00128-2017.tableS4 (8.6KB, pdf)
TABLE S5 00128-2017.tableS5 (595KB, pdf)
Acknowledgements
We would like to thank Frank Emmert-Streib and Ricardo de Matos Simoes at the School of Medicine, Dentistry and Biomedical Sciences, Queen's University Belfast (Belfast, UK) for their help and time during the setup and running of cluster computing for bioinformatics analysis during this study.
Footnotes
This article has supplementary material available from openres.ersjournals.com
Author contributions: G.G. Einarsson, M.M. Tunney and J.S. Elborn conceived and designed research; G.G. Einarsson and J. Zhao performed the research; G.G. Einarsson analysed the data; G.G. Einarssson, M.M. Tunney, J.S. Elborn, J.J. LiPuma and J. Zhao interpreted the data; D.G. Downey made intellectual contributions; and G.G. Einarsson, M.M. Tunney, J.S. Elborn, J.J. LiPuma and J. Zhao wrote the paper. All authors read and approved the final manuscript.
Conflict of interest: G.G. Einarsson reports a grant from EU FP7 (CFMATTERS) during the conduct of the study.
Conflict of interest: J. Zhao has nothing to disclose.
Conflict of interest: J.J. LiPuma has nothing to disclose.
Conflict of interest: D.G. Downey has nothing to disclose.
Conflict of interest: M.M. Tunney reports grants from EU FP7 (CFMATTERS) during the conduct of the study.
Conflict of interest: J.S. Elborn has nothing to disclose.
Support statement: This work was supported by a UK Cystic Fibrosis Trust grant (PJ533), Northern Ireland Chest Heart Stroke grant (20081080) and grants from the US National Institutes of Health (HL084934 and HL100809). G.G. Einarsson, M.M. Tunney and J.S. Elborn were supported through a US–Ireland Project Partnership Grant by the HSC Research and Development, Public Health Agency, Northern Ireland and the Medical Research Council. M.M. Tunney was supported by a Health and Social Care Research and Development, Public Health Agency, Northern Ireland-funded UK National Institute for Health Research Career Scientist Award. J.J. LiPuma was supported by the US CF Foundation. Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Dodge JA, Lewis PA, Stanton M, et al. Cystic fibrosis mortality and survival in the UK: 1947–2003. Eur Respir J 2007; 29: 522–526. [DOI] [PubMed] [Google Scholar]
- 2.Bilton D. Update on non-cystic fibrosis bronchiectasis. Curr Opin Pulm Med 2008; 14: 595–599. [DOI] [PubMed] [Google Scholar]
- 3.VandenBranden SL, McMullen A, Schechter MS, et al. Lung function decline from adolescence to young adulthood in cystic fibrosis. Pediatr Pulmonol 2011; 47: 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costerton JW. Cystic fibrosis pathogenesis and the role of biofilms in persistent infection. Trends Microbiol 2001; 9: 50–52. [DOI] [PubMed] [Google Scholar]
- 5.Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev 2002; 15: 194–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris JK, De Groote MA, Sagel SD, et al. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc Natl Acad Sci USA 2007; 104: 20529–20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King PT, Holdsworth SR, Freezer NJ, et al. Microbiologic follow-up study in adult bronchiectasis. Respir Med 2007; 101: 1633–1638. [DOI] [PubMed] [Google Scholar]
- 8.Tunney MM, Klem ER, Fodor AA, et al. Use of culture and molecular analysis to determine the effect of antibiotic treatment on microbial community diversity and abundance during exacerbation in patients with cystic fibrosis. Thorax 2011; 66: 579–584. [DOI] [PubMed] [Google Scholar]
- 9.Fodor AA, Klem ER, Gilpin DF, et al. The adult cystic fibrosis airway microbiota is stable over time and infection type, and highly resilient to antibiotic treatment of exacerbations. PLoS One 2012; 7: e45001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tunney MM, Einarsson GG, Wei L, et al. Lung microbiota and bacterial abundance in patients with bronchiectasis when clinically stable and during exacerbation. Am J Respir Crit Care Med 2013; 187: 1118–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erb-Downward JR, Thompson DL, Han MK, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One 2011; 6: e16384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Gast CJ, Walker AW, Stressmann FA, et al. Partitioning core and satellite taxa from within cystic fibrosis lung bacterial communities. ISME J 2011; 5: 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abreu NA, Nagalingam NA, Song Y, et al. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci Transl Med 2012; 4: 151ra124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sze MA, Dimitriu PA, Suzuki M, et al. The host response to the lung microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woyke T, Teeling H, Ivanova NN, et al. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature 2006; 443: 950–55. [DOI] [PubMed] [Google Scholar]
- 16.Konopka A. What is microbial community ecology? ISME J 2009; 3: 1223–1230. [DOI] [PubMed] [Google Scholar]
- 17.Marx CJ. Getting in touch with your friends. Science 2009; 324: 1150–1151. [DOI] [PubMed] [Google Scholar]
- 18.Aas JA, Paster BJ, Stokes LN, et al. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol 2005; 43: 5721–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaura E, Keijser B, Huse S, et al. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol 2009; 9: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature 2011: 473: 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faust K, Sathirapongsasuti JF, Izard J, et al. Microbial co-occurrence relationships in the human microbiome. PLoS Comp Biol 2012; 8: e1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silverman MS, Davis I, Pillai DR. Success of self-administered home fecal transplantation for chronic Clostridium difficile infection. Clin Gastroenterol Hepatol 2010; 8: 471–473. [DOI] [PubMed] [Google Scholar]
- 23.Ley RE, Bäckhed F, Turnbaugh P, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 2005; 102: 11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 2007; 104: 13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young VB. The intestinal microbiota in health and disease. Curr Opin Gastroenterol 2012; 28: 63–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Methé BA, Nelson KE, Pop M, et al. A framework for human microbiome research. Nature 2012; 486: 215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stone L, Roberts A. The checkerboard score and species distributions. Oecologia 1990; 85: 74–79. [DOI] [PubMed] [Google Scholar]
- 28.Barberan A, Bates ST, Casamayor EO, et al. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J 2012; 6: 343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogers GB, Carroll MP, Serisier DJ, et al. Use of 16S rRNA gene profiling by terminal restriction fragment length polymorphism analysis to compare bacterial communities in sputum and mouthwash samples from patients with cystic fibrosis. J Clin Microbiol 2006; 44: 2601–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris A, Beck JM, Schloss PD, et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med 2013; 187: 1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huse SM, Ye Y, Zhou Y, et al. A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS One 2012; 7: e34242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature 2012; 486: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ursell LK, Clemente JC, Rideout JR, et al. The interpersonal and intrapersonal diversity of human-associated microbiota in key body sites. J Allergy Clin Immunol 2012; 129: 1204–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramakrishnan VR, Feazel LM, Gitomer SA, et al. The microbiome of the middle meatus in healthy adults. PLoS One 2013; 8: e85507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Downes J, Olsvik B, Hiom SJ, et al. Bulleidia extructa gen. nov., sp. nov., isolated from the oral cavity. Int J Syst Evol Microbiol 2000; 50: 979–983. [DOI] [PubMed] [Google Scholar]
- 36.Eribe ERK, Olsen I. Leptotrichia species in human infections. Anaerobe 2008; 14: 131–137. [DOI] [PubMed] [Google Scholar]
- 37.Cheung MK, Lam WY, Fung WYW, et al. Sputum microbiota in tuberculosis as revealed by 16S rRNA pyrosequencing. PLoS One 2013; 8: e54574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Filkins LM, Hampton TH, Gifford AH, et al. Prevalence of Streptococci and increased polymicrobial diversity associated with cystic fibrosis patient stability. J Bacteriol 2012; 194: 4709–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parkins MD, Sibley CD, Surette MG, et al. The Streptococcus milleri group—An unrecognized cause of disease in cystic fibrosis: A case series and literature review. Pediatr Pulmonol 2008; 43: 490–497. [DOI] [PubMed] [Google Scholar]
- 40.Segal LN, Alekseyenko AV, Clemente JC, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome 2013; 1: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frank DN, Feazel LM, Bessesen MT, et al. The human nasal microbiota and Staphylococcus aureus carriage. PLoS One 2010; 5: e10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Einarsson G, Comer D, McIlreavey L, et al. Community dynamics and the lower airway microbiota in stable chronic obstructive pulmonary disease, smokers and healthy non-smokers. Thorax 2016; 71: 795–803. [DOI] [PubMed] [Google Scholar]
- 43.Mainz JG, Naehrlich L, Schien M, et al. Concordant genotype of upper and lower airways P. aeruginosa and S. aureus isolates in cystic fibrosis. Thorax 2009; 64: 535–540. [DOI] [PubMed] [Google Scholar]
- 44.Hansen SK, Rau MH, Johansen HK, et al. Evolution and diversification of Pseudomonas aeruginosa in the paranasal sinuses of cystic fibrosis children have implications for chronic lung infection. ISME J 2012; 6: 31–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.King P. Haemophilus influenzae and the lung (Haemophilus and the lung). Clin Transl Med 2012; 1: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cox MJ, Allgaier M, Taylor B, et al. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS One 2010; 5: e11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao J, Schloss PD, Kalikin LM, et al. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci USA 2012; 109: 5809–5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zemanick ET, Harris JK, Wagner BD, et al. Inflammation and airway microbiota during cystic fibrosis pulmonary exacerbations. PLoS One 2013; 8: e62917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
FIGURE S1 00128-2017.figureS1 (884.3KB, pdf)
FIGURE S2 00128-2017.figureS2 (1.7MB, pdf)
FILE S1 00128-2017.fileS1 (92KB, txt)
FILE S2 00128-2017.fileS2 (948.8KB, csv)
Detailed methods 00128-2017.supp (253.8KB, pdf)
TABLE S1 00128-2017.tableS1 (21.1KB, pdf)
TABLE S2 00128-2017.tableS2 (357.7KB, xlsx)
TABLE S3 00128-2017.tableS3 (11.2KB, pdf)
TABLE S4 00128-2017.tableS4 (8.6KB, pdf)
TABLE S5 00128-2017.tableS5 (595KB, pdf)