Abstract
Chimeric antigen receptor (CAR) T cell therapies have demonstrated impressive initial response rates in hematologic malignancies. However, relapse rates are significant, and robust efficacies in other indications, such as solid tumors, will likely require novel therapeutic strategies and CAR designs. To that end, we sought to develop simple, highly selective targeting domains (D domains) that could be incorporated into complex, multifunctional therapeutics. Herein, we describe the identification and characterization of D domains specific for CD123, a therapeutic target for hematologic malignancies, including acute myelogenous leukemia (AML). CARs comprised of these D domains mediate potent T cell activation and cytolysis of CD123-expressing target cells and induce complete durable remission in two AML xenograft models. We describe a strategy of engineering less immunogenic D domains through the identification and removal of putative T cell epitopes and investigate the binding kinetics and affinity requirements of the resultant D domain CARs. Finally, we extended the utility of D domains by generating functional, bi-specific CARs comprised of a CD123-specific D domain and a CD19-specific scFv. The properties of D domains suggest that this class of targeting domain may facilitate the development of multi-functional CARs where conventional, scFv-based designs may be suboptimal.
Keywords: chimeric antigen receptor, CD123, D domain, acute myelogenous leukemia, CAR T cell
Fry and colleagues describe the development of a class of binding domain, called D domains. They identify a series of binders to CD123 (a therapeutic target for hematologic malignancies), characterize their performance as the targeting domains in chimeric antigen receptors, and suggest an alternative to conventional scFv-based CAR designs.
Introduction
In recent years, advances in chimeric antigen receptor (CAR) T cell engineering and delivery have resulted in impressive clinical efficacy against B cell malignancies. However, continued optimization of CAR function is critical, as clinical experience has revealed the safety and efficacy limitations of the current CAR design. CAR architecture has evolved from simple fusions of scFv targeting domains and T cell receptor (TCR) CD3-ζ (immunoreceptor tyrosine-based activation motif [ITAM]) signaling domains, to more complex configurations involving the addition of one or more T cell co-stimulatory domains. These newer generations are less reliant on exogenous, physiologic co-stimulation (e.g., from antigen-presenting cells [APCs]) and manifest more potent and sustained anti-tumor responses.1 Extracellular targeting and intracellular signaling domains of CARs are linked via transmembrane and hinge regions typically derived from immunoglobulins, CD8alpha, or CD28. The length, flexibility, and composition of these elements can significantly impact epitope engagement, receptor dimerization, and ultimately CAR function.2, 3
Finally, a better appreciation for what constitutes the optimal targeting domain has also emerged in recent years. Beyond the requirements of antigen selectivity and binding, considerations now include epitope specificity,4, 5 binding affinity,6 and immunogenicity.7, 8 Due to the large inventory of validated antibodies against a variety of therapeutic targets, scFv naturally emerged as the obvious and justifiable targeting domain. However, many of the characteristics that have made antibodies versatile and effective recombinant therapeutics (e.g., high affinity, bivalency, antibody-dependent cytotoxicity [ADCC], complement-dependent cytotoxicity [CDC], FcRn recycling, and low renal filtration rates) are not advantageous for membrane associated chimeric receptors. Furthermore, scFv are not native protein structures and their development, particularly as it pertains to solubility and aggregation, can be challenging.9 Indeed, Long et al.10 demonstrate that the framework regions within the scFv targeting domain of a GD2-CAR promoted antigen-independent receptor aggregation, which in turn led to constitutive CD3ζ phosphorylation, tonic signaling, and T cell exhaustion—a phenomenon that could not be remedied through CDR grafting.
To optimize the binding domain of targeted therapeutics, investigators have begun expanding the repertoire of structures beyond the scFv. Structures such as fibronectin type III repeats (adnectins), Z domains (affibodies), knottins, lipocalins (anticalins), and ankyrin repeats (DARPin) have been developed with antibody-like affinities11 and recently incorporated into CARs.12, 13 Here, we report for the first time the development and characterization of D domains as targeting agents. Derived from the de novo-designed α-helical bundle, α3D,14 D domains exhibit the requisite properties for modular targeting agents; they are relatively small (approximately 1/3 the size of a scFv), single-domain structures that lack disulfide bonds and N-linked glycosylation. These features make them amenable to manipulation and development in the full range of recombinant expression systems. In addition, the rapid folding and high thermal stability of α3D15 increases its likelihood of tolerating amino acid substitutions, such as those introduced in a combinatorial library. Finally, the predominantly α-helical structure of the D domain also presents an opportunity to explore potential paratope-epitope interfaces that may not be available to the beta sheet and loop folds of the scFv.16
Expressed on the surface of a variety of hematologic neoplasms, the interleukin-3 (IL-3) receptor alpha subunit (IL3Rα), also known as CD123, has been under investigation as a therapeutic target for hairy cell leukemia, acute myelogenous leukemia (AML), and acute lymphoblastic leukemia (ALL).17 scFvs targeting CD123 have been employed as immunotoxins,18 bi-specific antibodies,19 and CARs.20, 21 Clinical trials for AML that target CD123-expressing tumors with 32716 scFv CAR T cells are underway.22 Similar 32716-based CARs were used in pre-clinical models of ALL relapse and shown to be effective in eradicating CD12+ tumors emerging from CD19 antigen loss.23 Furthermore, simultaneously targeting both CD19 and CD123 (via bi-cistronic CAR expression vector) was superior to either the mono-specific CARs alone or pools of mono-specific CARs in their ability to kill target cells expressing both antigens. Thus, as a model therapeutic target, we sought to identify CD123-specific D domains and characterize their efficacy as drivers of CAR T cell targeting and bioactivity. In addition, the structural simplicity of the D domain led us to explore their utility in the construction of bi-specific CARs, targeting both CD19 and CD123.
Results
D Domains Direct CAR Specificity
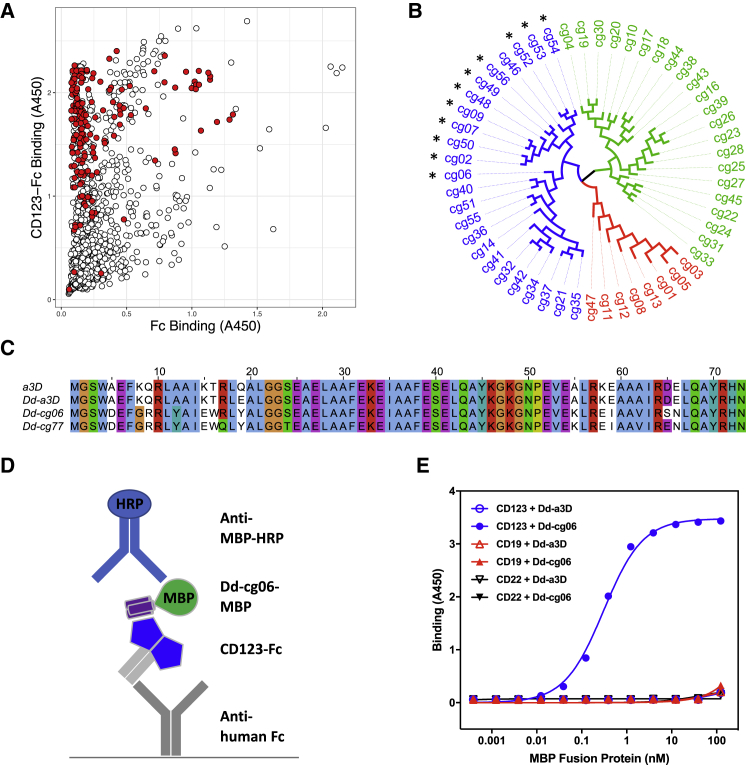
CD123-binding D domains were identified through the directed evolution of naive phage display libraries. Five different D domain library designs, containing subsets of randomized surface residues, were pooled and subjected to four rounds of selection on the extracellular domain of CD123 (Figure S1). Phage clones, representing 1,261 D domain sequences (1,052 unique), were screened by ELISA for binding to CD123 (Figure 1A). Fifty-six CD123 binding D domains, representing sequence diversity across multiple library designs, were subsequently converted to CAR constructs, using a second-generation design24 (CD8 transmembrane domain, followed by 41BB and CD3ζ intracellular domains), wherein the scFv-targeting domain was substituted with that of the D domain. (Figure 1B). D domain CARs were screened for their ability to promote nuclear factor of activated T cells (NFAT) activation in a Jurkat reporter cell line, upon exposure to CD123-expressing cells. Notably, only D domain CARs derived from the F3 library exhibited significant NFAT activation (data not shown). One such clone, Dd-cg06, and the parental D domain sequence (a variant of α3D, referred to throughout as Dd-a3D) (Figure 1C), were converted to maltose binding protein (MBP) fusions for assessment as soluble, mono-valent protein in a CD123-binding ELISA (Figure 1D). As depicted in Figure 1E, Dd-a3D exhibited no appreciable binding to CD123, CD19, or CD22. In contrast, Dd-cg06 bound CD123 with an EC50 of 0.3 nM but did not bind the unrelated receptors, CD19 or CD22.
Figure 1.
D Domain Selection
(A) ELISA binding data from naive phage library screen. Individual phage clones were assayed for binding to both human CD123-Fc (CD123-Fc)- and human Fc alone (Fc)-coated wells. Bound phage were detected with anti-M13-HRP. A total of 1,261 D domains were screened, yielding 1,052 unique sequences. Clones representing a diversity of sequence relatedness (shown in red) were selected for further characterization. (B) Dendrogram of D domain clone sequences converted to MBP fusions and/or CARs for further characterization. Colors indicate three distinct sequence families, referred to as F3 (blue), F1 (green), and C2 (red). CARs promoting NFAT activation in Jurkat reporter assay are indicated with an asterisk. (C) Sequence alignment of α3D (a3D), the parental D domain (Dd-a3D), the naive CD123-binding D domain (Dd-cg06), and the derived, deimmunized variant (Dd-cg77). Clustal X color scheme used for alignments in Jalview.49 (D) A schematic representation of MBP ELISA. (E) Results of MBP ELISA. Equimolar concentrations of Fc fusion of CD123, CD19, and CD22 were captured with anti-human Fc. Bound Dd-cg06-MBP (Dd-cg06) or Dd-a3D-MBP (Dd-a3D) proteins are detected with anti-MBP-HRP. Dd-cg06 binds to CD123-Fc, but not CD19-Fc or CD22-Fc. The parental D domain, Dd-a3D, is a poor binder of any target protein tested.
Dd-cg06-CART Are Activated by and Kill CD123+ Target Cells In Vitro and In Vivo
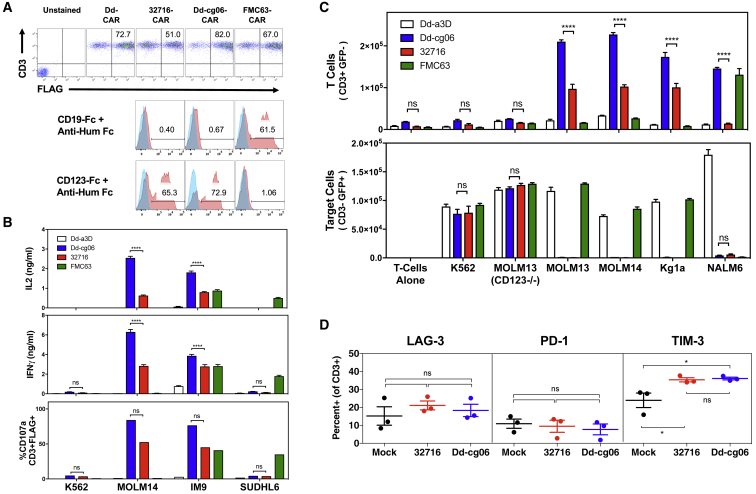
Functional characterization of D domain CARs was performed using primary human T cells transduced with CAR constructs encoded within a third-generation lentiviral vector.25 CAR expression for all constructs (Dd-a3D-CAR, Dd-cg06-CAR, 32716-CAR, and the anti-CD19 scFv, FMC63-CAR) were typically > 80% and comparable with respect to percent transduced and mean fluorescence intensity (MFI). In addition, Dd-cg06-CAR exhibited a similar selectivity for soluble target binding as that of 32716-CAR (Figure 2A). See Figure S2 for additional CAR expression results.
Figure 2.
Cytokine Secretion, Degranulation, Proliferation, and Exhaustion of Dd-cg06 CAR T Cells
(A) Primary CAR T cells assessed for expression and binding of soluble CD19-Fc and CD123-Fc 6-7 days post-transduction. CAR expression was measured for CD3+ cells using anti-FLAG antibody. Bound CD123-Fc and CD19-Fc were detected with PE labeled anti-human Fc. Blue histograms represent binding to Dd-a3D transfected T cells. (B) Primary T cells expressing Dd-cg06 CAR secrete IL-2 and IFNγ (one donor, n = 3, mean ± SEM) and degranulate (one donor, n = 1) in response to target cells. K562 are CD123−/CD19−. MOLM14 are CD123+/CD19−. IM9 are CD123+/CD19+. SUDHL6 are CD123+/CD19+. Data generated with additional donors can be found in Figure S4. (C) CAR-expressing T cells proliferate in response to target cells. Nine days after transduction, cells were cultured overnight in media without IL-2, then CFSE labeled and cultured for 4 days in the presence of untreated target cells (25,000 FLAG+ T cells and 25,000 target cells). CD3+ CAR T cells and CD3− target cells are then counted by flow cytometry. Data represents mean ± SEM, triplicates of one donor. MOLM13(CD123−/−) are CD123−/CD19−. MOLM13 and KG1a are CD123+/CD19−. NALM6 are CD123+/CD19+. Data generated with additional donors can be found in Figure S5. (D) Transduced T cells were stained with antibodies against CD3, LAG-3, PD1, and TIM3 9 days after activation with anti-CD3/anti-CD28 T cell activation beads in culture media supplemented with 40 U/mL of IL-2. Data represents mean ± SEM for three donors. One-way ANOVA analysis, using Tukey correction for multiple comparisons, was performed for LAG-3, PD1, and TIM3. ns, p > 0.05; *p < 0.01.
Engagement by CAR T cells with target cell surface antigen initiates a cascade of T cell responses, including cytokine secretion, cell proliferation, degranulation, and ultimately, target cell lysis. The specificity of Dd-cg06 CAR T cell response was assessed upon exposure to a variety of cell types with variable expression of CD123 and CD19 (see Figure S3 for target receptor characterization of cell lines used in this study). Both 32716-CAR and Dd-cg06-CAR T cells secreted interferon (IFN)γ and IL-2 in response to MOLM14 (CD123+, CD19−) and IM9 (CD123+, CD19+), with little to no response to K562 (CD123−, CD19−) and SUDHL6 (CD123−, CD19+) (Figure 2B). Conversely, FMC63-CAR T cells, specific to CD19+ target cells, secreted cytokines in response to IM9 and SUDHL6 cell lines, while Dd-CAR T cells demonstrated minimal responsiveness to any target cell. While the magnitude of response varied with T cell donor, Dd-cg06 CAR consistently exhibited robust and target-specific activation (Figure S4). Degranulation, a prerequisite to perforin-granzyme-mediated killing, was assessed by measuring CD107a/LAMP-1 on CD3+ CAR T cells. Dd-cg06 CAR T cells exhibited a degranulation pattern similar to that of cytokine secretion. Taken together, these data demonstrate that Dd-cg06 CAR can specifically recognize native, membrane-bound CD123 and initiate T cell effector and cytolytic responses.
Proliferation of T cells upon antigenic stimulus is an important component of the adaptive immune response. Co-culturing equal numbers of CAR T and tumor cells resulted in the antigen-specific proliferation of fluorescently labeled CAR T cells, coincident with the destruction of tumor cells (Figure 2C). The selectivity of this antigen-specific expansion is most notable with the MOLM13 cells; In contrast to the MOLM13 (CD123−/−) cell line, in which CD123 has been deleted (via CRISPR), the parental MOLM13 cells promote T cell proliferation and lysis by CAR T cells. While the Dd-cg06 CAR demonstrated comparable target specificity to that of 32716, the response was more consistent across donors (Figure S4) and pronounced on NALM6 cells, which express relatively lower levels of CD123.
Tonic CAR T cell activation, stemming from constitutive, antigen-independent CAR signaling, promotes premature T cell exhaustion, typically manifested by decreased proliferation, cytokine expression, cytolytic potency, and the upregulation of T cell inhibitory receptors (e.g., LAG-3, PD-1, and TIM3).10, 26 To monitor potential antigen-independent exhaustion, transduced T cells were stained with antibodies against CD3, LAG-3, PD1, and TIM3, 9 days after initial activation. Summary (Figure 2D) and representative (Figure S6) flow cytometry data indicate no significant differences in the inhibitory markers between the 32716 scFv and D domain-based CAR T cells. These in vitro findings suggest that the D domain as a targeting element does not promote inhibitory signaling.
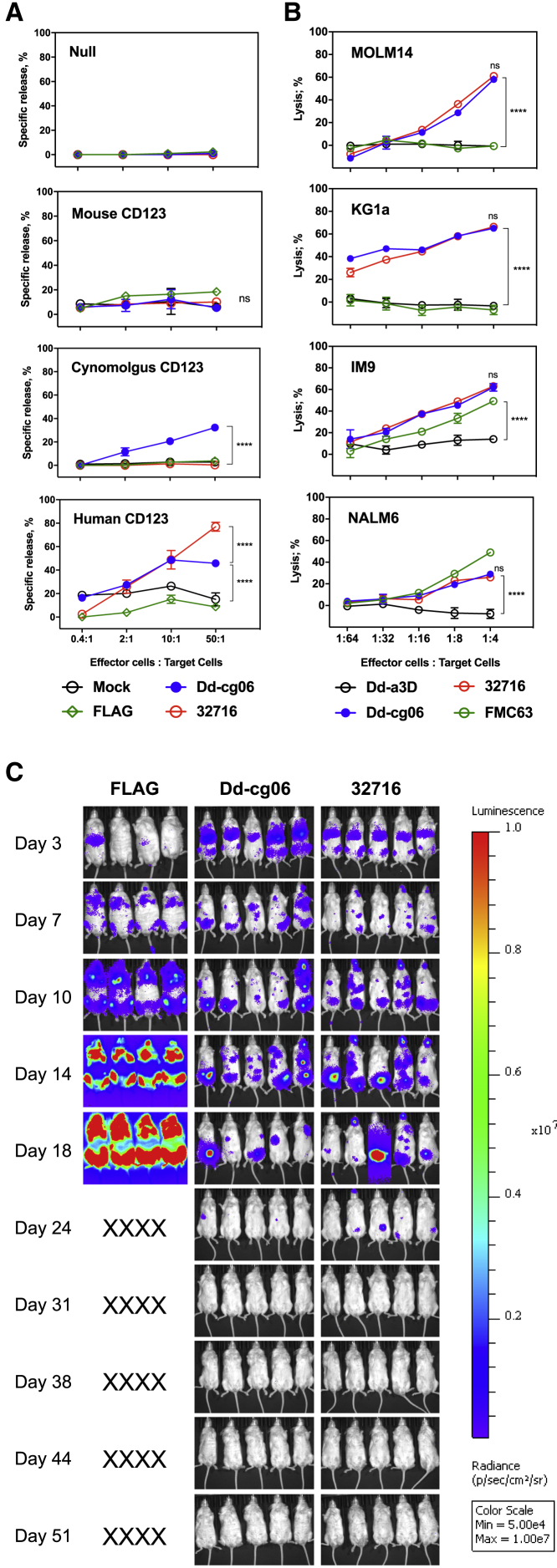
The extracellular domain of human CD123 is 87% and 31% identical to that of cynomolgus and mouse CD123, respectively. Thus, species selectivity for CD123 was characterized through the use of HEK293T target cells transfected with CD123 orthologs. Human T cells transduced with Dd-cg06-CAR are effective in killing HEK cells expressing human and cynomolgus monkey CD123, but not mouse CD123 or mock-transfected cells (Figures 3A and S7). Notably, 32716 scFv does not demonstrate cross-reactivity against cynomolgus CD123.
Figure 3.
Dd-cg06-CAR T Cells Kill Target Cells In Vitro and In Vivo
(A) Europium-release cytotoxicity assay using HEK293 target cells transfected with mouse, cynomolgus, or human CD123. Human primary T cells transfected with FLAG-CAR (FLAG), Dd-cg06-CAR (Dd-cg06), 32716-CAR (32716), or no CAR construct (Mock). CD123 ortholog expression data can be found in Figure S7. In two-way ANOVA analysis, mean value differences are not significant (ns, p > 0.05) between any treatments at E:T of 50:1 for mouse CD123. n = 2, mean ± SEM. Where indicated, ****p < 0.0001. (B) Bioluminescence cytotoxicity assay using MOLM14, KG1a (both CD123+, CD19−), NALM6, and IM9 (both CD123+, CD19+) cells expressing GFP and luciferase. Human primary T cells expressing Dd-a3D, Dd-cg06, 32716, or FMC63 CAR were co-cultured with target cells (40,000) at various E:T ratios. After 16 h, cells were washed and luciferase activity was assessed. Percent lysis was based on luciferase released in the absence of CAR T cells. In two-way ANOVA analysis, mean value differences are not significant (p > 0.05) between Dd-cg06 and 32716 at E:T of 1:4 for all target cells. n = 3, mean ± SEM. Where indicated, ****p < 0.0001. (C) Dd-cg06-CAR T cells kill target cells in vivo. NSG mice received 1 × 106 MOLM14 (GFP+, luciferase+) cells on day 0. On day 3, mice received 5 × 106 CAR T cells. FLAG-CAR (FLAG), Dd-cg06-CAR (Dd-cg06), 32716-CAR (32716). Expression data (FLAG positive) for CAR T cells and statistical analysis of bioluminescent signal flux data can be found in Figure S8.
Next, we assessed the ability of Dd-cg06-CAR T cells to kill AML (MOLM14, KG1a), ALL (NALM6), and multiple myeloma (IM9) tumor cell lines, each with varying surface expression of CD123 (Figure 3B). All tumor targets are lysed by Dd-cg06-CAR T cells at effector:target (E:T) ratios comparable to those of the conventional scFv-based CAR, 32716. No killing is observed by Dd-CAR T cells for any cell line or by FMC63 for cell lines lacking CD19 (e.g., MOLM14 and KG1a).
Finally, in vivo efficacy of the Dd-cg06 CAR was assessed using a MOLM14 tumor model. Engrafted tumors were treated with Dd-cg06-CAR, 32716-CAR, and FLAG-CAR (a negative control construct, expressing only the FLAG epitope sequence as the extracellular domain). Mice receiving Dd-cg06-CAR or 32716 scFv CAR demonstrated regression between days 18 and 24 and remained tumor-free by day 31 (Figure 3C) with comparable kinetics of tumor clearance by both CARs and no signs of graft versus host disease (GvHD). Taken together, the in vitro and in vivo results indicate that the Dd-cg06 can direct target-specific, potent T cell activation resulting in eradication of tumor xenografts.
Deimmunization of Dd-cg06 through Rational Design
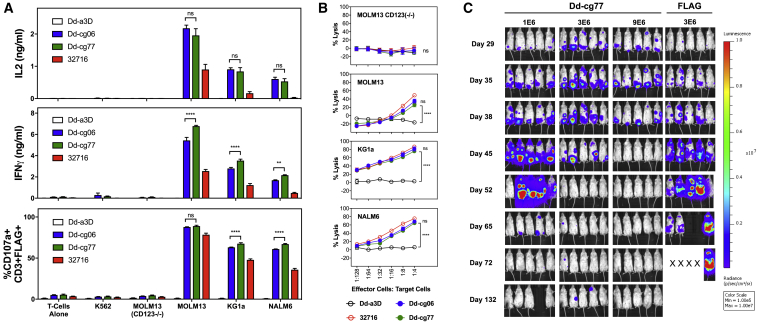
The safety and efficacy of any protein therapeutic may be compromised by potential host-mediated immunogenic responses. Therefore, we employed a virtual matrix-based T cell epitope prediction algorithm27 in the assessment of D domain sequences. While Dd-cg06 exhibited potent in vitro and in vivo activity, it nevertheless harbored three putative promiscuous T cell epitopes, which we endeavored to remove via targeted mutation. Based on the structure of α3D and our D domain library design, we identified mutagenic sites that we hypothesized would be less likely to be involved in either target binding (i.e., opposite or adjacent to the F3 face of the domain) or the formation of the domain’s hydrophobic core—the primary driver of the folding and stability.15 The introduction of three mutations (R17Q, S24T, and S65E) into Dd-cg06 resulted in a new sequence, Dd-cg77, in which the number of promiscuous epitopes had been reduced to zero (Figure S9). To assess the impact of these mutations on binding, we generated Fc fusions of Dd-cg06 and Dd-cg77 and measured the KD of their mono-valent interactions with CD123 to be 14.4 and 6.4 nM, respectively (Figure S10; Table S1). Nevertheless, the approximately 2-fold increase in mono-valent affinity of Dd-cg77 did not affect the CAR activity, as demonstrated by comparable cytokine release, degranulation (Figure 4A), and in vitro killing of CD123+ target cells (Figure 4B). Notably, the KD of the FLAG-less, Dd-cg77-Fc fusion (6.4 nM) is similar to that of the FLAG-tagged Dd-cg77 MBP fusion (7.2 nM), suggesting the presence of the tag has little impact on the binding of this D domain. In vivo, Dd-cg77-CAR T cells mediated potent and durable remission of KG1a xenografts, derived from an AML cell line, with CD123 expression levels approximately 6-fold lower than MOLM14 (Figure 4C). Thus, targeted, deimmunizing mutations introduced into a naive D domain sequence (e.g., Dd-cg06) are not detrimental to the functionality or specificity of the derived (e.g., Dd-cg77) CAR T cells.
Figure 4.
Deimmunized D-Domain CAR Exhibits Killing of CD123+ Target Cells In Vitro and In Vivo
(A) CAR T cells secrete IL-2 and IFNγ and degranulate in response to target cells. Cytokine production by CAR T cells was assessed by culturing 25,000 transduced T cells (7 days post-activation) with 25,000 CD123+ (MOLM13, KG1a, and NALM6) or CD123− (T cells alone, K562, and MOLM13(CD123−/−)) target cells in 96-well plates. Media cytokine concentrations were measured after coculturing cells for 24 h. Degranulation of CAR T cells was assessed after coculturing 1 × 105 transduced T cells (day 9 post-activation) with the 2 × 105 cells listed above for 4 h in the presence of monensin and PE-conjugated anti-CD107a/LAMP1 antibody. Data represents two donors with triplicate measurements each, mean ± SEM; ns, not significant; **p < 0.01; ****p < 0.0001. (B) Bioluminescence cytotoxicity assay utilizing CD123+ (MOLM13, KG1a, and NALM6) and CD123− (MOLM13(CD123−/−)) cells, all express GFP and luciferase. Human primary T cells expressing Dd-a3D, Dd-cg06, Dd-cg77, or 32716 CAR were cocultured at the indicated concentrations with 40,000 target cells per well. After 16 h, cells were washed and luciferase activity was assessed. Percent lysis was based on luciferase activity in target cells cultured in the absence of CAR T cells. Data represent two donors with triplicate measurements each, mean ± SEM; ns, not significant; ****p < 0.0001. (C) In vivo xenograft model. NSG mice received 6 × 106 KG1a cells on day 0 and various doses of Dd-cg77 or FLAG CAR T cells on day 29. Expression data (FLAG positive) for CAR T cells and statistical analysis of bioluminescent signal flux data can be found in Figure S8.
D Domain CAR Activity Can Be Tuned through Affinity Mutations
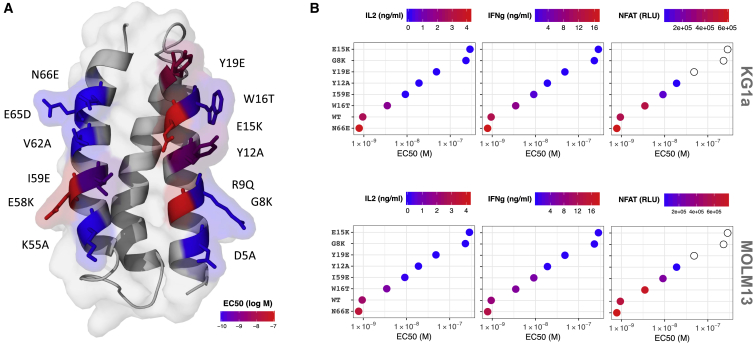
The ability of lower affinity CARs to discriminate between low (normal) and high (neoplastic) target densities has been advocated as a mechanism for reducing on-target, off-tumor toxicities.6, 28, 29 In an effort to reduce the relative high affinity of Dd-cg77, we targeted the 13 library positions (i.e., those most likely involved in binding), and in an effort to maximize proper folding, mutated them to the corresponding residue found in Dd-a3D. For 7 of the 13 mutants, surface plasmon resonance (SPR) assessment of CD123-HIS binding to MBP-captured Dd-MBP fusion proteins revealed KD values ranging from 3.2 nM (N66E) to 100 nM (W16T), with lower affinities typically associated with faster off-rates (Figure S11; Table S2). However, for the remainder of mutants (G8K, Y12A, E15K, Y19E, E58K, and I59E) we measured no appreciable binding using SPR. Nevertheless, using the CD123-Fc capture ELISA (described in Figure 1) we measured EC50 values for binding of all 13 mutants (Figure S12). Four of the six mutations with the greatest impact on binding are located in helix 1, with the other two in helix 3, (Figure 5A) and the three mutants most detrimental to binding are centrally distributed within the F3 face. A subset of these mutants, spanning the range of affinities, were converted to CARs and assessed for cytokine release and NFAT activation by target cells. Both of these cellular responses correlated with the relative binding affinity measured by ELISA and SPR (Figure 5B).
Figure 5.
Select Dd-cg77 Mutations Reduce Binding and CAR Activity
(A) Homology model50 of Dd-cg77 indicating the location and identity of 13 independent mutants. Each mutant reverts the indicated position to the corresponding residue in Dd-a3D. Residue positions are colored according to CD123 binding EC50, obtained for each mutant (Figure S12). (B) Representative mutants (spanning the range of affinities) were tested as CARs for their ability to induce the release of IL-2 and IFNγ and activate an NFAT promoter. CAR T cells were co-incubated with KG1a (top panel) and MOLM13 (bottom panel) target cells. Empty circles were not tested for NFAT activation.
Bi-specific CAR T Cells Bind and Kill Both CD19- and CD123+ Target Cells
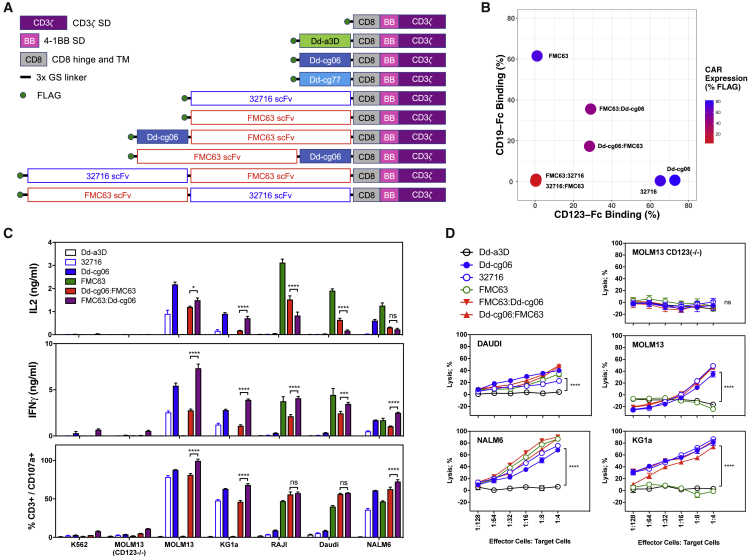
The high expression and potency of the D-domain-based CARs led us to construct Dd-cg06 CD19/CD123 bi-specific CAR constructs. Recognizing that the relative position of the two targeting domains may impact expression, target binding and T cell activation, we constructed bi-specific receptors with the FMC63 scFv and the Dd-cg06 D domain in both the N- and C-terminal positions (Figure 6A). Mono-specific CAR constructs resulted in the best expression (ranging between 70% and 95% FLAG+), whereas the bi-specific CARs composed of two scFv domains exhibited very poor expression (less than 12% of their respective T cells population) (Figures 6B and S2). Bi-specific constructs composed of both D domain and scFv improved expression over the scFv-scFv bi-specifics (approximately 45% to 72% surface FLAG expression). These CAR T cells were next assessed for their ability to bind soluble target. Mono-specific CAR T cells bound the highest levels of soluble antigen, while binding of either antigen was virtually undetectable for the bi-specifics comprised solely of scFv domains. Interestingly, despite similar expression and CD123-Fc binding, FMC63:Dd-cg06 bound significantly more CD19-Fc than the Dd-cg06:FMC63 orientation (35.5% versus 17.3%, respectively). In experiments where simultaneous binding of CD123 and CD19 to these bi-specific CARs was performed, this positional difference was even more pronounced (Figure S2).
Figure 6.
Expression and Activity of Bi-specific CAR Constructs
(A) Schematic representations of CAR designs used in this report. Receptor constructs are optionally comprised of D domain (Dd-a3D, Dd-cg06, or Dd-cg77), 32716 scFv, FMC63 scFv, the CD8 hinge, and transmembrane domains (CD8) and the 4-1BB (BB) and CD3ζ signaling domains. Extracellular domains are linked via 3xG4S peptides. For uniform detection, both scFv and D domain CAR incorporated an N-terminal FLAG epitope tag. (B) Mono- and bi-specific CAR expression (% FLAG+ of CD3+ cells) and binding to soluble CD19 and CD123. Bi-specific CARs comprised of Dd-cg06 and FMC63 scFv express and bind both CD19 and CD123, while bi-specific CARs composed exclusively of 32716 and FMC63 scFvs do not express. Shown are representative data. Histograms can be found in Figure S2. (C) Bi-specific CARs secrete cytokines and degranulate in a target-specific manner. Cytokine production by CAR T cells was assessed by culturing 25,000 transduced T cells (7 days post-activation) with 25,000 CD123−/CD19− cells (K562, MOLM13(CD123−/−)), CD123+/CD19− (MOLM13, KG1a), CD123+/CD19+ (NALM6), or CD123−/CD19− (Raji, Daudi) cells in 96-well plates. Cytokine concentrations of cultured supernatants were measured after 24 h coculturing cells. Degranulation of CAR T cells was assessed after coculturing 1 × 105 transduced T cells (day 9 post-activation) with the 2 × 105 cells listed above, for 4 h in the presence of monensin and PE-conjugated anti-CD107a/LAMP1 antibody. Data represent two donors with triplicate measurements each, mean ± SEM; ns, not significant; *p < 0.05; ***p < 0.001; ****p < 0.0001. (D) Bioluminescence cytotoxicity assay utilizing MOLM13, MOLM13(CD123−/−), KG1a, DAUDI, and NALM6 target cells, all express GFP and luciferase. Human CAR T cells were co-cultured at the indicated concentrations with 40,000 target cells per well. After 16 h, cells were washed, and luciferase activity was assessed. Percent lysis was based on luciferase activity in target cells cultured in the absence of CAR T cells. Data represents mean ± SEM for triplicate measurements using one donor. ns, not significant; ****p < 0.0001.
Bi-specific CAR T cells were also assessed for their reactivity to tumor-derived cell lines expressing varying levels of CD19 and/or CD123 (Figure 6C). As measured by IFNγ and IL-2 secretion and degranulation (CD107a+/CD3+), mono-specific Dd-cg06-CAR and 32716-CAR T cells react strongly to CD123+ cells (MOLM13, KG1a, and NALM6) and poorly to those lacking CD123 (MOLM13(CD123−/−), Daudi, and Raji). In contrast, FMC63 scFv CAR T cells react only to CD19+ cells (Daudi, Raji, and NALM6). The bi-specific CAR T cells react to target cells expressing CD19, CD123, or both. Finally, the killing of target cells expressing CD19 and/or CD123 by the bi-specific CAR T cells was assessed via bioluminescence assay (Figure 6D), demonstrating lysis by the bi-specific CAR T cells at E:T ratios that are comparable to their mono-specific counterpart but with activity against both CD19 and CD123. These data indicate that D-domain-containing bi-specific CARs can be expressed in human T cells at levels sufficient to enable CAR T cell activation and killing of target cells expressing either CD19, CD123, or both.
Discussion
In this report, we describe, for the first time, the use of the de novo-designed α-helical bundle, α3D as a scaffold for the generation of targeting domains (D domains). Panning of phage libraries on CD123 yielded binders derived from each scaffold design tested. However, the F3 library yielded the greatest number of hits and often resulted in the highest signal-to-noise ratio (data not shown). Furthermore, upon conversion to CARs, the F3-derived domains proved most potent in activating NFAT. Although the sequencing of naive libraries suggests that library quality is not a factor (data not shown), it remains unclear whether the prolific nature of F3 is a function of the specific target (i.e., CD123), our panning conditions, limited screening, or some other intrinsic characteristic of the F3 design (e.g., unique topology or thermal stability). Empirical structure analysis and experience with other targets will help answer this.
Our targeted mutational analysis of Dd-cg77 identifies critical residue positions in both helix 1 and helix 3, suggesting a binding interface that extends across the F3 face of the α-helical bundle. The engagement of discontiguous helices of this D domain is in contrast with the structurally related affibody (Z domain), in which residues from helix 1 and 2 dominate the binding interface.30 As with affibodies, we anticipate that target contact residues will not be limited to those defined by the library design or even within the alpha helical regions of the domain. As a result of its unique topology, the D domain may offer an opportunity to target epitopes not available to scFv CDR loops.
As the targeting element of a CAR, we found D domains to perform as well as the 32716 scFv in virtually all respects. D domain CARs, either transiently transfected into HEK cells or virally transduced in primary human T cells, express at comparable levels to those of the scFv CAR. Cytokine secretion, degranulation, proliferation, and cytotoxicity are all comparable in magnitude and specificity with that of 32716. Relative to the 32716 scFv, we observed no increase in expression of the inhibitory receptors LAG-3, PD-1, and TIM3. These observations support the expectation that D domains promote the expression of well-formed (i.e., non-aggregating) chimeric receptors that do not contribute to antigen-independent exhaustion. In vivo we found D domain CARs were able to eradicate tumors with potency and kinetics similar to that of the 32716. Highlighting the functional persistence of the D domain CAR T cells, we observed prolonged mouse survival and tumor clearance through days 51 and 132 in the MOLM14 and KG1a xenograft models, respectively. While the contribution of allogeneic reactivity is difficult to quantify in these models (we observed no signs of GvHD in the D domain CAR cohorts), it is not ultimately sufficient to control tumor growth—as observed in the negative-control (FLAG) T cell cohorts. Dose escalation is a fundamental tenet of clinical safety and essential for the management of toxicities associated with high-dose CAR T administration.31, 32 The dose response observed in the KG1a model suggests that D domain CAR T therapeutics could similarly benefit from an escalated approach.
Our naive phage libraries yielded D domains that bound to CD123 with high affinity. The 6.4 nM KD of Dd-cg77 is similar in magnitude to what others have measured for CD123 monoclonal antibodies derived through conventional hybridoma methodology.18, 33 The ability to kill cells expressing low levels of target (e.g., KG1a) demonstrates the sensitivity of these Dd CARs and likely reflects the relative high-affinity and high-expression levels of these receptors. While affinity maturation is often necessary for the development of potent soluble targeting agents such as antibodies, high affinity in the context of high-avidity CARs can lead to on-target, off-tumor toxicities. Indeed, Arcangeli et al.33 demonstrated that reducing the affinity of the CD123 scFv, improves the selectivity for CD123 overexpressing tumor cells over normal tissue with lower target antigen expression.
To identify lower affinity variants as well as better understand D domain topology, we generated a series of Dd-cg77 mutants. Five of the thirteen mutants (G8K, Y12A, E15K, Y19E, and E58K) completely abolished activity in any of the assays we tested and binding itself could only be assessed via high-density ELISA. We observed a T cell activation affinity threshold for Dd-cg77; a KD less than 100 nM (e.g., W16T) appears to be required for consistent response to both MOLM13 and KG1a cells. The lower affinity D domain mutants (for which we obtained SPR data) were typically associated with faster off-rates, similar to those described for a Her2 scFv CAR.34 This is in contrast to analogous work performed on the CD123 scFv clone 7G333 (unrelated in sequence to 32716), in which lower affinity mutants exhibited slower on-rates.
The de novo origin of α3D demands a strategy to address the potential for D-domain-derived immunogenicity. The safety and efficacy of clinical immunogenicity associated with antibody and recombinant protein therapeutics is well documented. Factors such as species of origin, protein aggregation, route of administration, dosing schedule, and immune tolerance are critical considerations in the development of these therapies. However, much less is understood regarding the immunogenicity of CAR therapies; insight into the anti-CAR immunogenic response has been complicated by the often immune-compromised patient population—including lymphodepletion therapy, and in the case of CD19-CART, the complete ablation of the B cell population. While a humanized scFv domain is the rational first choice in designing CAR therapeutics for clinical use, this does not guarantee against an anti-idiotypic immune response.35 Second, as the name implies, these chimeric receptors are fusions that potentially introduce neo-epitopes into otherwise human sequences.
A variety of in silico and in vitro methods have been developed to help identify potential T-cell-reactive epitopes in therapeutic proteins, that may lead to clinical immunogenicity36—the utility of which has been demonstrated with CARs.8 We combined high-throughput screening, sequence analysis, in silico T cell epitope prediction, and structural knowledge of the D domain scaffold to select for clones that were amenable to deimmunizing mutations. As a model for this approach, we selected Dd-cg06, a domain with potent CAR activity and three promiscuous MHC class II epitopes. These potential epitopes were removed by introducing mutations in positions predicted to be less involved in the formation of either the binding interface or the hydrophobic core of the domain. Using this rational approach, we have demonstrated that it is possible to remove putative T cell epitopes from D domains, while simultaneously preserving bioactivity. Whether these putative epitopes are in fact T cell reactive will require confirmation via in vitro assessment.37 While in this study we chose to utilize a virtual matrix method for identification of class II epitopes, the same principles can be substituted or combined with alternative predictive algorithms, for example those focused on class I epitopes.
Despite the impressive response rates for CD19-CAR T therapy in ALL (complete remission rates of 70%∼94%), about half of patients receiving such therapy will eventually relapse.38 One mechanism driving relapse is epitope loss and subsequent “escape” by the tumor cells.39 By simultaneous targeting of both CD19 and CD123 with bi-cistronic CARs, Ruella et al.23 were able to demonstrate effective relapse prevention in pre-clinical models of ALL. This led us to explore the generation of CD19/CD123 bi-specific CARs, utilizing Dd-cg06. Our inability to express FMC63/32716-derived bi-specifics, demonstrates the utility of alternative targeting domains. While other bispecific CARs have been reported,40, 41, 42 to our knowledge, this is the first demonstration of a CD19/CD123 bi-specific CAR. Interestingly, while both bi-specific orientations manifest similar expression and soluble CD123 binding levels, one orientation (Dd-cg06:FMC63) binds considerably less CD19, suggesting that the scFv may be more susceptible to positional orientation than the D domain. While we can demonstrate that individual T cells can simultaneously bind to soluble CD19 and CD123, it is not clear if an individual receptor can do the same. Nor do we have insight regarding simultaneous engagement of a membrane-bound target. Nevertheless, both orientations of bi-specific CAR T release cytokines, degranulate, and kill target cells at rates and specificities comparable to that of the monospecific receptors. These bi-specific receptors require either CD19 or CD123 for activation. Other bi-specific CAR indications may demand the presence of two or more target antigens prior to T cell activation. The development of such and-gated CAR T cells would likely require fine tuning of both targeting affinities.
Relatively few cancers are effectively treated by monospecific therapies, either because unique targets are rare or because the cancer evolves mechanisms to escape or neutralize such therapies. As compared to hematologic malignancies, solid tumors present a significantly greater number of challenges43, 44, 45 that will likely require a new generation of CAR T strategies to overcome. That D domains are highly competent targeting agents, capable of driving potent anti-tumor activates of CAR T cells, suggests an alternative means of creating complex, multifunctional therapeutics, where scFv-based designs may be less viable.
Materials and Methods
Recombinant Proteins and Cell Lines
Recombinant proteins were obtained from Sino Biological (hCD123-Fc) and R&D Systems (hCD19-Fc, hCD22-Fc, CD123-His). Lenti-X 293T cells were obtained from Clontech. K562, Molm14, Raji, KG1a, BDCM, NALM6, IM9, SUDHL1, SUDHL6, Molm13, and Daudi cell lines were obtained from the American Type Culture Collection (ATCC) and cultured using recommended media formulations and methods. Bioluminescent target cell lines, expressing GFP and luciferase, were generated by transducing cell lines with the MSCV-luciferase-EF1α-copGFP-T2A-Puro BLIV 2.0 Lentivector (System Biosciences). Lentivirus was generated as described below. Cell lines were spinoculated in the presence of lentivirus and protamine sulfate for 2 h at 1,000 × g and cultured for 48 h. Transduced cells were expanded in the presence of puromycin, and integration was assessed via GFP expression. The CD123− MOLM13 cell line was generated via CRISPR by transducing the cells with LentiCRISPRv2 transfer vector46 containing the guide RNA (gRNA) sequence “5′-AGTTCCCACATCCTGGTGCG-3′,” via spinoculation, as described above. Remaining CD123+ cells were then depleted by binding cells with biotinylated anti-CD123 antibody and anti-biotin microbeads (Miltenyi) and running the cells over an LS column.
Phage Library Construction and Screenings
D domain libraries were designed as described in Figure S1 and generated by Kunkel mutagenesis of pCOMB phagemid “stop templates,” encoding an N-terminal fusion of Dd-a3D (containing termination codons at library-specific positions) to truncated pIII. Kunkel mutagenesis was performed using degenerate trimer oligonucleotides derived from phosphoramidite mixtures (excluding termination, proline, and cysteine codons) (Ella Biotech). Phage library pools of all five designs were subjected to four rounds of binding selection with biotinylated recombinant human CD123-His (R&D Systems) captured by M-280 Streptavidin Dynabeads (Invitrogen) with increasing levels of stringency. Successive rounds of panning utilized 200 nM, 100 nM, 75 nM, and 50 nM of biotinylated target antigen with Tween 20 concentrations of 0.1%, 0.1%, 0.2%, and 0.5%, respectively. Bound phages were eluted with 0.2 M glycine (pH 2.2), neutralized with 1 M Tris (pH 9.0), and propagated in TG1 cells (Lucigen) with VCSM13 helper phage (Agilent Technologies). Amplified output phage from each round was then precipitated with polyethylene glycol (PEG)/NaCl and resuspended in PBS to serve as the input library for the next round. After four rounds of biopanning, individual phage clones were isolated and screened for binding to recombinant human CD123-Fc-His (Sino Biological), captured by goat polyclonal anti-human Fc antibody coated on Immulon 4HBX microtiter plates. Bound phage was detected using anti-M13-HRP (GE Healthcare Biosciences), followed by 3,3',5,5'-tetramethylbenzidine (TMB; SeraCare Life Sciences).
Expression Constructs
CAR constructs utilized either pcDNA3 (for transient expression) or pELNS25 (for lentiviral expression) and were assembled by PCR with DNA fragments encoding the FMC63 scFv,47 32716 scFv,20 CD8 hinge and transmembrane domain (NCBI RefSeq: NP_001759.3, 138 to 206) and the cytoplasmic domains of 4-1BB (NCBI RefSeq: NP_001552.2, 214 to 255) and CD3 zeta (NCBI RefSeq: NP_932170.1, 52 to 164). For uniform, pre-clinical assessment of expression, all CAR constructs encoded an N-terminal FLAG epitope sequence followed by a 2xG4S linker. Chymotrypsinogen signal peptide (NCBI RefSeq: NP_001020371.3, residues 1 to 18) was chosen based on superior signal peptide processing prediction48 when used in the context of an N-terminal FLAG epitope tag. Fusion of Dd-cg06, Dd-cg77, and 32716 to human Fc were meditated via a 2xG4S linker and did not contain the N-terminal FLAG sequence.
MBP Expression and Direct Binding ELISA
MBP fusions were cloned into the pET24 (New England Biolabs) using an OmpC signal peptide (UniProtKB-P06996, 1 to 21), followed by FLAG epitope sequence, a 2xG4S linker, a D domain, 3xG4S linker, and MBP (accession WP_040064394.1, residues 27 to 392). BL21(DE3) cells containing pET24 MBP fusion constructs were induced using isopropyl β-D-1-thiogalactopyranoside (IPTG) for 4 h at 30°C. Cell pellets were harvested, washed with PBS, and then resuspended in 200 mM Tris (pH 7.4), 20 mM EDTA, and incubated overnight at 30°C to allow diffusion from periplasm to buffer. MBP protein in the supernatant was quantified by ELISA using an MBP standard (Abcam), anti-MBP polyclonal (Abcam) capture, and anti-MBP-HRP (NEB) detection. Target binding ELISA was performed using 96-well plates coated with anti-human-immunoglobulin G (IgG) Fc (Thermo Pierce) at 3.2 μg/mL overnight, followed by blocking (ELISA Blocking Buffer, Thermo Fisher) and the addition of equimolar concentrations of Fc fusion. Detection of bound protein was done using anti-MBP-horseradish peroxidase (HRP) (NEB), followed by TMB (SeraCare Life Sciences).
Jurkat NFAT Activation Reporter Assay
The Jurkat NFAT reporter cell line was generated using SureENTRY transduction reagent (QIAGEN) to transduce the Jurkat E6.1 cell line (Sigma) with the Cignal Lenti NFAT reporter, containing the NFAT enhancer coupled to the luciferase reporter gene (QIAGEN). Single-cell cloning was performed through selection with puromycin (5 μg/mL). Highly responsive clones were identified and selected by activation with anti-CD3 (OKT3). Reporter assays were performed as follows. CAR constructs were electroporated into the Jurkat NFAT reporter cell line. Twenty-four hours post-electroporation, CAR expression was assessed via anti-FLAG monoclonal antibody. CAR-expressing Jurkat cells were then co-cultured with target cells for 6 h, after which NFAT-mediated signaling was measured through the addition of luciferase assay reagent BriteLite (PerkinElmer), followed by quantitation of relative luminescence units (RLUs).
Lentiviral Production and Transduction
HEK293T cells were transiently transfected with third-generation lentiviral packaging vectors (pRSV-REV, pMDLg/pRRE, and pMD2.G) with pELNS vectors encoding CARs using Lipofectamine 3000. Six hours post-transfection, the media was changed, then lentivirus-containing media was collected at 30 and 54 h post-transfection, pooled, then centrifuged to remove cell debris. Lentivirus was then aliquoted and stored at −80°C until used for viral transduction. Transduction of human T cells with CAR lentivirus was performed using total human peripheral blood mononuclear cells (PBMCs), activated with Dynabeads Human T-Activator CD3/CD28 beads (Life Technologies) in culture media supplemented with 40 U/mL of IL2. After 24 h, 2 × 106 PBMCs were plated per well in a 6-well tissue culture plate with 1 mL of culture media and 3 mL of lentivirus containing media supplemented with 40 U/mL of IL-2 and protamine sulfate. Plates were then centrifuged for 2 h at 1,000 × g at 32°C and then incubated overnight 37°C. The following day, the lentivirus transduction procedure was repeated with fresh culture media and lentivirus-containing media. 72 hours after the initial cell activation, T cell activation beads were removed, then T cells were cultured for expansion at ∼0.25-0.5 × 106 T cells/mL in fresh media supplemented with 100 U/mL of IL-2. Every 2–3 days, T cells were supplemented with additional T cell media and IL-2 until they were used for various assays 7–10 days after the initial activation.
Cytokine Production
Cytokine production in response to antigen expression was assessed by culturing 25,000 FLAG+ T cells (7 days post-activation) with 25,000 target tumor cells per well, in 96-well plates. After 24 h, culture supernatants were collected and diluted 1:5 in ELISA buffer, and cytokine production was assessed by ELISA (Ready-Set-Go human IL-2 and IFNγ uncoated ELISA kits, eBioscience/Thermo Fisher). Values are reported as ng/mL in undiluted medium.
Degranulation
Degranulation of CAR T cells were assessed 8–9 days post-activation. In brief, 1 × 105 CAR T cells were cultured alone or in the presence of 2 × 105 target cells in T cell media for 4 h in the presence of monensin and PE-conjugated anti-CD107a/LAMP1 antibody. T cells were then washed and stained for CD3 expression.
Flow Cytometry and Binding Experiments
Flow cytometry experiments were performed on a Bio-Rad S3E or a BD Celesta equipped with a high-throughput sampler (HTS). Antibodies used in these studies were as follows: CD3 (UCHT1, Biolegend), FLAG (L5, Biolegend), LAG-3 (3D52234, eBioscience), PD1 (ebioJ105, eBioscience), TIM3 (F38.2E2, eBioscience), anti-human IgG Fc (HP6017, Biolegend), CD107a (H4A3, Biolegend), CD19 (SJ25C1, Biolegend), and CD123 (6H6, Biolegend). In most experiments, 105 T cells were stained for 20 min at 4°C, then washed and run on the flow cytometer. For the assessment of CD123 or CD19 expression, 105 target cells were Fc-blocked with human TruStain FcX (Biolegend) for 5 min at room temperature, then stained with anti-CD19, anti-CD123, or the appropriate isotype control. In some experiments, human, mouse, and cynomolgus CD123 was transiently expressed on HEK293T cells. Under these circumstances, after overnight transfection cells were harvested from cell culture plates non-enzymatically using CellStripper dissociation reagent (Gibco), washed, then stained with anti-CD123 (clone 7G3) for human and cynomolgus or anti-CD123 (clone 5B11) for mouse. Mock-transfected cells were used as the expression controls. For binding flow cytometry experiments, 105 T cells were washed and incubated with 0.5 μg of hCD19-Fc or hCD123-Fc chimeric proteins (Sino and R&D Systems, respectively) for 20 min at 4°C in flow cytometry buffer, washed twice, then binding detected with PE anti-human Fc. In some experiments, T cells were co-incubated with hCD19-Fc and biotinylated-hCD123-His (biotinylated in-house) simultaneously, washed twice, and binding detected with streptavidin-PE and A488 anti-human Fc.
T Cell Proliferation
CAR T cells were washed and cultured overnight in IL-2 free media. The following day, T cells were labeled with the proliferation dye, VPD450 (BD Biosciences), and co-cultured with equal numbers of cells (25,000 CAR+ T cells and 25,000 target cells) in round-bottom 96-well plates in 0.2 mL of T cell media (without IL-2). After 96 h, cells were spun and stained with anti-CD3 antibody. Total cells were enumerated per well by flow cytometry using an HTS, collecting 150 μL of a 200 μL sample. T cells were gated as live CD3+ GFP− cells. Target cells were gated as live CD3− GFP+ cells.
Cytolytic Assays
Bioluminescence cytotoxicity assays were performed using target cells expressing GFP and luciferase cells (their generation is described above). Briefly, 40,000 target cells were combined with 2-fold, serially diluted CAR T cells; starting at an E:T of 1:4. Control wells had target cells only and represent maximal luminescence signal. After approximately 16–18 h incubation, cells were pelleted and resuspended in Dulbecco’s PBS and BriteLite reagent (PerkinElmer). Upon complete lysis (3–5 minutes), cell lysates were transferred to 96-well white OptiPlates (96-well white opaque Microplate, PerkinElmer) and luminescence was read on a Synergy 2 instrument (Biotek) using autoscaling relative luminescence units. Percent lysis is calculated according to the following formula: %lysis = ([maximal RLU − experimental RLU]/max RLU)*100%. Europium-based cytolytic assays were performed as follows. Target cells were pre-loaded with BATDA (bis(acetoxymethyl)2,2′:6′,2″′-terpyridine-6,6″′-dicarboxylate) (PerkinElmer) and co-cultured with CAR T cells for 2 h at various E:T ratios. Upon cytolysis of target cells, Europium solution (Eu3+) was added, and fluorescent EuTDA is measured using a Synergy 2-time-resolved fluorometer (Biotek). Percent of specific lysis is calculated as % specific release = ([experimental release − spontaneous release]/maximal release − spontaneous release) × 100%.
Tumor Models
Animal experiments were carried out under protocols approved by the NCI Bethesda Animal Care and Use Committee. The luciferase-expressing AML lines MOLM14 and KG1a were intravenously injected via tail vein into NSG mice (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ, Jackson Laboratories). Tumor burden was detected by florescence imaging using the Xenogen IVIS Lumina (Caliper Life Sciences). Mice were injected intraperitoneally with 3 mg D-luciferin (Caliper Life Sciences) and were imaged 4 min later with an exposure time 1 min. Living Image version 4.3.1 SP2 software (Caliper Life Sciences) was used to analyze the bioluminescent signal flux (photons/s/cm2/sr, scaled at 105–107) for each mouse. The animals were treated with CAR-T cells via tail-vein injection once the tumor burden reached the desirable level and imaged weekly.
Statistical Analysis
Data were analyzed using GraphPad Prism 7 software (GraphPad). Unless otherwise indicated, two-way ANOVA statistical tests were performed for grouped statistics, using Tukey correction for multiple comparisons. Values are reported as mean ± SEM.
Author Contributions
Conceptualization, T.J.F., D.M.H., D.W.L.; Methodology, H.Q., J.P.E., L.Z., A.G., C.J.M., T.J.F., D.M.H., D.W.L.; Investigation, H.Q., J.P.E., L.Z., A.G., C.J.M.; Writing – Original Draft, D.W.L.; Writing – Review and Editing, H.Q., J.P.E., L.Z., A.G., C.J.M., T.J.F., D.M.H., D.W.L.; Funding Acquisition, T.J.F., D.M.H.; Supervision, T.J.F., D.M.H., D.W.L.
Conflicts of Interest
J.P.E., L.Z., A.G., C.J.M., D.M.H., and D.W.L are employees of Arcellx, Inc., Germantown, MD, USA.
Acknowledgments
We thank Janine Buonato, Sinnie Ng, Laura Richman, Marianna Sabatino, Angela Shen, and Jeff Swers for their careful review of this manuscript. This work was supported by funding from Arcellx, Inc., Germantown, MD, USA. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.04.010.
Supplemental Information
References
- 1.Kawalekar O.U., O’Connor R.S., Fraietta J.A., Guo L., McGettigan S.E., Posey A.D., Jr., Patel P.R., Guedan S., Scholler J., Keith B. Distinct Signaling of Coreceptors Regulates Specific Metabolism Pathways and Impacts Memory Development in CAR T Cells. Immunity. 2016;44:380–390. doi: 10.1016/j.immuni.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 2.Alabanza L., Pegues M., Geldres C., Shi V., Wiltzius J.J.W., Sievers S.A., Yang S., Kochenderfer J.N. Function of Novel Anti-CD19 Chimeric Antigen Receptors with Human Variable Regions Is Affected by Hinge and Transmembrane Domains. Mol. Ther. 2017;25:2452–2465. doi: 10.1016/j.ymthe.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guest R.D., Hawkins R.E., Kirillova N., Cheadle E.J., Arnold J., O’Neill A., Irlam J., Chester K.A., Kemshead J.T., Shaw D.M. The role of extracellular spacer regions in the optimal design of chimeric immune receptors: evaluation of four different scFvs and antigens. J. Immunother. 2005;28:203–211. doi: 10.1097/01.cji.0000161397.96582.59. [DOI] [PubMed] [Google Scholar]
- 4.Hombach A.A., Schildgen V., Heuser C., Finnern R., Gilham D.E., Abken H. T cell activation by antibody-like immunoreceptors: the position of the binding epitope within the target molecule determines the efficiency of activation of redirected T cells. J. Immunol. 2007;178:4650–4657. doi: 10.4049/jimmunol.178.7.4650. [DOI] [PubMed] [Google Scholar]
- 5.Long A.H., Haso W.M., Orentas R.J. Lessons learned from a highly-active CD22-specific chimeric antigen receptor. OncoImmunology. 2013;2:e23621. doi: 10.4161/onci.23621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caruso H.G., Hurton L.V., Najjar A., Rushworth D., Ang S., Olivares S., Mi T., Switzer K., Singh H., Huls H. Tuning sensitivity of CAR to EGFR density limits recognition of normal tissue while maintaining potent antitumor activity. Cancer Res. 2015;75:3505–3518. doi: 10.1158/0008-5472.CAN-15-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maus M.V., Haas A.R., Beatty G.L., Albelda S.M., Levine B.L., Liu X., Zhao Y., Kalos M., June C.H. T Cells Expressing Chimeric Antigen Receptors Can Cause Anaphylaxis in Humans. Cancer Immunol. Res. 2013;1:26–31. doi: 10.1158/2326-6066.CIR-13-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamers C.H.J., Willemsen R., van Elzakker P., van Steenbergen-Langeveld S., Broertjes M., Oosterwijk-Wakka J., Oosterwijk E., Sleijfer S., Debets R., Gratama J.W. Immune responses to transgene and retroviral vector in patients treated with ex vivo-engineered T cells. Blood. 2011;117:72–82. doi: 10.1182/blood-2010-07-294520. [DOI] [PubMed] [Google Scholar]
- 9.Gil D., Schrum A.G. Strategies to stabilize compact folding and minimize aggregation of antibody-based fragments. Adv. Biosci. Biotechnol. 2013;4(4a):73–84. doi: 10.4236/abb.2013.44A011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long A.H., Haso W.M., Shern J.F., Wanhainen K.M., Murgai M., Ingaramo M., Smith J.P., Walker A.J., Kohler M.E., Venkateshwara V.R. 4-1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat. Med. 2015;21:581–590. doi: 10.1038/nm.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hey T., Fiedler E., Rudolph R., Fiedler M. Artificial, non-antibody binding proteins for pharmaceutical and industrial applications. Trends Biotechnol. 2005;23:514–522. doi: 10.1016/j.tibtech.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 12.Han X., Cinay G.E., Zhao Y., Guo Y., Zhang X., Wang P. Adnectin-Based Design of Chimeric Antigen Receptor for T Cell Engineering. Mol. Ther. 2017;25:2466–2476. doi: 10.1016/j.ymthe.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammill J.A., VanSeggelen H., Helsen C.W., Denisova G.F., Evelegh C., Tantalo D.G.M., Bassett J.D., Bramson J.L. Designed ankyrin repeat proteins are effective targeting elements for chimeric antigen receptors. J. Immunother. Cancer. 2015;3:55. doi: 10.1186/s40425-015-0099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walsh S.T., Cheng H., Bryson J.W., Roder H., DeGrado W.F. Solution structure and dynamics of a de novo designed three-helix bundle protein. Proc. Natl. Acad. Sci. USA. 1999;96:5486–5491. doi: 10.1073/pnas.96.10.5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu Y., Alonso D.O.V., Maki K., Huang C.-Y., Lahr S.J., Daggett V., Roder H., DeGrado W.F., Gai F. Ultrafast folding of alpha3D: a de novo designed three-helix bundle protein. Proc. Natl. Acad. Sci. USA. 2003;100:15486–15491. doi: 10.1073/pnas.2136623100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sha F., Salzman G., Gupta A., Koide S. Monobodies and other synthetic binding proteins for expanding protein science. Protein Sci. 2017;26:910–924. doi: 10.1002/pro.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Testa U., Pelosi E., Frankel A. CD 123 is a membrane biomarker and a therapeutic target in hematologic malignancies. Biomark. Res. 2014;2:4. doi: 10.1186/2050-7771-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du X., Ho M., Pastan I. New immunotoxins targeting CD123, a stem cell antigen on acute myeloid leukemia cells. J. Immunother. 2007;30:607–613. doi: 10.1097/CJI.0b013e318053ed8e. [DOI] [PubMed] [Google Scholar]
- 19.Al-Hussaini M., Rettig M.P., Ritchey J.K., Karpova D., Uy G.L., Eissenberg L.G., Gao F., Eades W.C., Bonvini E., Chichili G.R. Targeting CD123 in acute myeloid leukemia using a T-cell-directed dual-affinity retargeting platform. Blood. 2016;127:122–131. doi: 10.1182/blood-2014-05-575704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mardiros A., Dos Santos C., McDonald T., Brown C.E., Wang X., Budde L.E., Hoffman L., Aguilar B., Chang W.C., Bretzlaff W. T cells expressing CD123-specific chimeric antigen receptors exhibit specific cytolytic effector functions and antitumor effects against human acute myeloid leukemia. Blood. 2013;122:3138–3148. doi: 10.1182/blood-2012-12-474056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan M., Li M., Gao L., Geng S., Wang J., Wang Y., Yan Z., Yu L. Chimeric antigen receptors for adoptive T cell therapy in acute myeloid leukemia. J. Hematol. Oncol. 2017;10:151. doi: 10.1186/s13045-017-0519-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Budde L., Song J.Y., Kim Y., Blanchard S., Wagner J., Stein A.S., Weng L., Del Real M., Hernandez R., Marcucci E. Remissions of Acute Myeloid Leukemia and Blastic Plasmacytoid Dendritic Cell Neoplasm Following Treatment with CD123-Specific CAR T Cells: A First-in-Human Clinical Trial. Blood. 2017;130(Suppl 1):811. [Google Scholar]
- 23.Ruella M., Barrett D.M., Kenderian S.S., Shestova O., Hofmann T.J., Perazzelli J., Klichinsky M., Aikawa V., Nazimuddin F., Kozlowski M. Dual CD19 and CD123 targeting prevents antigen-loss relapses after CD19-directed immunotherapies. J. Clin. Invest. 2016;126:3814–3826. doi: 10.1172/JCI87366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sadelain M., Brentjens R., Rivière I. The basic principles of chimeric antigen receptor design. Cancer Discov. 2013;3:388–398. doi: 10.1158/2159-8290.CD-12-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carpenito C., Milone M.C., Hassan R., Simonet J.C., Lakhal M., Suhoski M.M., Varela-Rohena A., Haines K.M., Heitjan D.F., Albelda S.M. Control of large, established tumor xenografts with genetically retargeted human T cells containing CD28 and CD137 domains. Proc. Natl. Acad. Sci. USA. 2009;106:3360–3365. doi: 10.1073/pnas.0813101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasakovski D., Xu L., Li Y. T cell senescence and CAR-T cell exhaustion in hematological malignancies. J. Hematol. Oncol. 2018;11:91. doi: 10.1186/s13045-018-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh H., Raghava G.P. ProPred: prediction of HLA-DR binding sites. Bioinformatics. 2001;17:1236–1237. doi: 10.1093/bioinformatics/17.12.1236. [DOI] [PubMed] [Google Scholar]
- 28.Park S., Shevlin E., Vedvyas Y., Zaman M., Park S., Hsu Y.S., Min I.M., Jin M.M. Micromolar affinity CAR T cells to ICAM-1 achieves rapid tumor elimination while avoiding systemic toxicity. Sci. Rep. 2017;7:14366. doi: 10.1038/s41598-017-14749-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X., Jiang S., Fang C., Yang S., Olalere D., Pequignot E.C., Cogdill A.P., Li N., Ramones M., Granda B. Affinity-tuned ErbB2 or EGFR chimeric antigen receptor T cells exhibit an increased therapeutic index against tumors in mice. Cancer Res. 2015;75:3596–3607. doi: 10.1158/0008-5472.CAN-15-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eigenbrot C., Ultsch M., Dubnovitsky A., Abrahmsén L., Härd T. Structural basis for high-affinity HER2 receptor binding by an engineered protein. Proc. Natl. Acad. Sci. USA. 2010;107:15039–15044. doi: 10.1073/pnas.1005025107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Titov A., Petukhov A., Staliarova A., Motorin D., Bulatov E., Shuvalov O., Soond S.M., Piacentini M., Melino G., Zaritskey A., Barlev N.A. The biological basis and clinical symptoms of CAR-T therapy-associated toxicites. Cell Death Dis. 2018;9:897. doi: 10.1038/s41419-018-0918-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonifant C.L., Jackson H.J., Brentjens R.J., Curran K.J. Toxicity and management in CAR T-cell therapy. Mol. Ther. Oncolytics. 2016;3:16011. doi: 10.1038/mto.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arcangeli S., Rotiroti M.C., Bardelli M., Simonelli L., Magnani C.F., Biondi A., Biagi E., Tettamanti S., Varani L. Balance of Anti-CD123 Chimeric Antigen Receptor Binding Affinity and Density for the Targeting of Acute Myeloid Leukemia. Mol. Ther. 2017;25:1933–1945. doi: 10.1016/j.ymthe.2017.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chmielewski M., Hombach A., Heuser C., Adams G.P., Abken H. T cell activation by antibody-like immunoreceptors: increase in affinity of the single-chain fragment domain above threshold does not increase T cell activation against antigen-positive target cells but decreases selectivity. J. Immunol. 2004;173:7647–7653. doi: 10.4049/jimmunol.173.12.7647. [DOI] [PubMed] [Google Scholar]
- 35.Hege K.M., Bergsland E.K., Fisher G.A., Nemunaitis J.J., Warren R.S., McArthur J.G., Lin A.A., Schlom J., June C.H., Sherwin S.A. Safety, tumor trafficking and immunogenicity of chimeric antigen receptor (CAR)-T cells specific for TAG-72 in colorectal cancer. J. Immunother. Cancer. 2017;5:22. doi: 10.1186/s40425-017-0222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baker M.P., Reynolds H.M., Lumicisi B., Bryson C.J. Immunogenicity of protein therapeutics: The key causes, consequences and challenges. Self Nonself. 2010;1:314–322. doi: 10.4161/self.1.4.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joubert M.K., Deshpande M., Yang J., Reynolds H., Bryson C., Fogg M., Baker M.P., Herskovitz J., Goletz T.J., Zhou L. Use of in vitro assays to assess immunogenicity risk of antibody-based biotherapeutics. PLoS ONE. 2016;11:e0159328. doi: 10.1371/journal.pone.0159328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park J.H., Rivière I., Gonen M., Wang X., Sénéchal B., Curran K.J., Sauter C., Wang Y., Santomasso B., Mead E. Long-Term Follow-up of CD19 CAR Therapy in Acute Lymphoblastic Leukemia. N. Engl. J. Med. 2018;378:449–459. doi: 10.1056/NEJMoa1709919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z., Wu Z., Liu Y., Han W. New development in CAR-T cell therapy. J. Hematol. Oncol. 2017;10:53. doi: 10.1186/s13045-017-0423-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zah E., Lin M.Y., Silva-Benedict A., Jensen M.C., Chen Y.Y. T Cells Expressing CD19/CD20 Bispecific Chimeric Antigen Receptors Prevent Antigen Escape by Malignant B Cells. Cancer Immunol. Res. 2016;4:498–508. doi: 10.1158/2326-6066.CIR-15-0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Munter S., Ingels J., Goetgeluk G., Bonte S., Pille M., Weening K., Kerre T., Abken H., Vandekerckhove B. Nanobody based dual specific CARs. Int. J. Mol. Sci. 2018;19:1–11. doi: 10.3390/ijms19020403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young P.A., Yamada R.E., Trinh K.R., Vasuthasawat A., De Oliveira S., Yamada D.H., Morrison S.L., Timmerman J.M. Activity of Anti-CD19 Chimeric Antigen Receptor T Cells Against B Cell Lymphoma Is Enhanced by Antibody-Targeted Interferon-Alpha. J. Interferon Cytokine Res. 2018;38:239–254. doi: 10.1089/jir.2018.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D’Aloia M.M., Zizzari I.G., Sacchetti B., Pierelli L., Alimandi M. CAR-T cells: The long and winding road to solid tumors. Cell Death Dis. 2018;15:282. doi: 10.1038/s41419-018-0278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y., Luo F., Yang J., Zhao C., Chu Y. New chimeric antigen receptor design for solid tumors. Front. Immunol. 2017;8:1934. doi: 10.3389/fimmu.2017.01934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knochelmann H.M., Smith A.S., Dwyer C.J., Wyatt M.M., Mehrotra S., Paulos C.M. CAR T Cells in Solid Tumors: Blueprints for Building Effective Therapies. Front. Immunol. 2018;9:1740. doi: 10.3389/fimmu.2018.01740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zola H., MacArdle P.J., Bradford T., Weedon H., Yasui H., Kurosawa Y. Preparation and characterization of a chimeric CD19 monoclonal antibody. Immunol. Cell Biol. 1991;69:411–422. doi: 10.1038/icb.1991.58. [DOI] [PubMed] [Google Scholar]
- 48.Nielsen H. Predicting Secretory Proteins with SignalP. Methods Mol. Biol. 2017;1611:59–73. doi: 10.1007/978-1-4939-7015-5_6. [DOI] [PubMed] [Google Scholar]
- 49.Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guex N., Peitsch M.C., Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis. 2009;30(Suppl 1):S162–S173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.