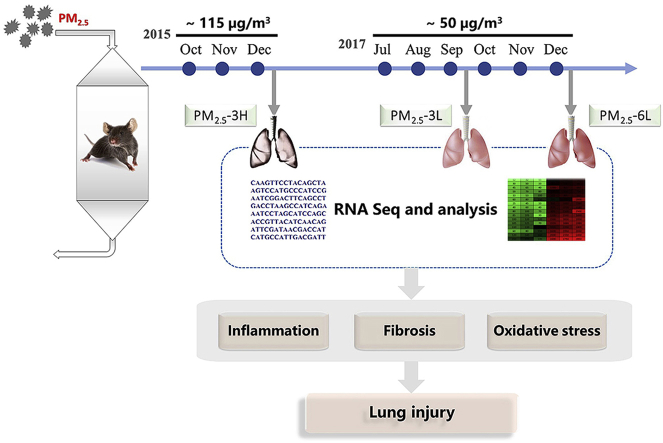
Abstract
The association between airborne fine particulate matter (PM2.5) concentration and the risk of respiratory diseases has been well documented by epidemiological studies. However, the mechanism underlying the harmful effect of PM2.5 has not been fully understood. In this study, we exposed the C57BL/6J mice to airborne PM2.5 for 3 months (mean daily concentration ~50 or ~110 μg/m3, defined as PM2.5–3L or PM2.5–3H) or 6 months (mean daily concentration ~50 μg/m3, defined as PM2.5–6L) through a whole-body exposure system. Histological and biochemical analysis revealed that PM2.5–3H exposure caused more severe lung injury than did PM2.5–3L, and the difference was greater than that of PM2.5–6L vs PM2.5–3L exposure. With RNA-sequencing technique, we found that the lungs exposed with different concentration of PM2.5 have distinct transcriptional profiles. PM2.5–3H exposure caused more differentially expressed genes (DEGs) in lungs than did PM2.5–3L or PM2.5–6L. The DEGs induced by PM2.5–3L or PM2.5–6L exposure were mainly enriched in immune pathways, including Hematopoietic cell lineage and Cytokine-cytokine receptor interaction, while the DEGs induced by PM2.5–3H exposure were mainly enriched in cardiovascular disease pathways, including Hypertrophic cardiomyopathy and Dilated cardiomyopathy. In addition, we found that upregulation of Cd5l and reduction of Hspa1 and peroxiredoxin-4 was associated with PM2.5-induced pulmonary inflammation and oxidative stress. These results may provide new insight into the cytotoxicity mechanism of PM2.5 and help to development of new strategies to attenuate air pollution associated respiratory disease.
Keywords: PM2.5, Lung injury, RNA-Sequencing, Inflammation, Oxidative stress
Graphical abstract
Highlights
-
•
High concentration of PM2.5 has a profound effect on lung injury and gene expression profile.
-
•
RNA-Seq analysis revealed that PM2.5 exposure affects immune and cardiovascular pathways.
-
•
PM2.5 increases Cd5l expression and decreases Hspa1 and Prdx4 expression in a concentration dependently manner.
Abbreviations
- 3′-NT
3-nitrotyrosine
- 4-HNE
4-hydroxynonenal
- BALF
bronchoalveolar lavage fluid
- Cd5l
CD5 antigen-like
- COPD
chronic obstructive pulmonary disease
- DCM
dilated cardiomyopathy
- DEGs
differentially expressed genes
- FA
filtered air
- Gpx
glutathione peroxidase
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- H&E
hematoxylin and eosin
- HCM
hypertrophic cardiomyopathy
- Hspa1
heat shock 70 kDa protein 1
- IL
interleukin
- ILr
interleukin receptor
- KEGG
kyoto encyclopedia of genes and genomes
- Ndufs
NADH dehydrogenase iron-sulfur protein
- PM
particulate matter
- Prdx4
peroxiredoxin-4
- qPCR
quantitative real-time polymerase chain reaction
- RNA-seq
RNA-sequencing
- ROS
reactive oxygen species
- Sod
superoxide dismutase
- TLR
Toll-like receptors
- TNFα
Tumor Necrosis Factor
- TNFrsf
TNF receptor superfamily member 14
- Trxn
thioredoxin
- Trxr2
thioredoxin reductase
1. Introduction
Currently, ambient air pollution has become a large threat to public health. Fine particulate matter (PM2.5, aerodynamic diameter ≤ 2.5 μm) is one of the most important components of outdoor air pollution. High concentration of PM2.5 increases the risk of respiratory diseases, including asthma [1], bronchitis [2], chronic obstructive pulmonary disease (COPD) [3] and lung cancer [4,5]. Using an analysis of daily time-series for the 20 largest US cities, the PM-mortality dose-response curves and threshold levels were firstly described in 2000 [6]. Then, multiple epidemiological studies have validated that there is a concentration-response relationship between airborne PM2.5 and its harmful effects on respiratory system [5,[7], [8], [9]]. In mice models, a recent study found that the severity of lung injury caused by ambient PM exposure is associated with cumulative dose [10]. Acute exposure to low doses of fine PM by intranasal instillation also induced lung inflammation and oxidative stress in a dose-dependent manner [11]. It has been suggested that PM2.5-induced inflammatory response was associated with Toll-like receptors (TLR2/TLR4) and PM2.5 can drive a Th2-biased immune response in mice [12]. Notably, during inflammation induced by PM2.5, enhanced production of reactive oxygen species (ROS) could result in DNA damage, lipid peroxidation and cell death [[13], [14], [15]]. However, the comprehensive mechanisms by which PM2.5 causes lung injury have not been full elucidated.
RNA-sequencing (RNA-seq) is a precise and sensitive tool for measuring global gene expression profiles expression. Recently, this technique has been used to investigate the mechanism of PM2.5-induce cytotoxicity in cell models, including 16HBE [16], BEAS-2B [17], A549 [18] and human non-small-cell lung cancer (H1299) cells [19]. To better understand the harmful effects of PM2.5 on respiratory system, we exposed mice to either airborne PM2.5 or filtered air (FA) for 3–6 months through a whole-body exposure system and then obtained global gene expression profiles in lungs of FA or PM2.5 exposed mice using RNA-seq.
2. Materials methods
2.1. Reagents and antibodies
BCA protein assay kit and reduced/oxidized glutathione (GSH and GSSG) kit were purchased from the Beyotime Institute of Biotechnology (#P0012, #S0053, Shanghai, China). Elisa kits for mouse tumor necrosis factor alpha (TNFα), 3′-nitrotyrosine (3′-NT) and 4-hydroxynonenal (4-HNE) were purchased from Sino Biological Inc (#SEK50349, Beijing, China), Abcam PLC (#ab116691, Cambridge, UK) and Donggeboye Biological Technology Co. LTD (#DG30947 M, Beijing, China), respectively. The Masson's trichrome stain kit and superoxide dismutase 3 (SOD3) antibody were obtained from Solarbio Science &Technology Co. LTD (#G1340, #K006598P, Beijing, China). Primary antibodies against β-tubulin, SOD1, SOD2, peroxiredoxin 3 (PRDX3), PRDX4, PRDX5 and thioredoxin reductase 2(TRXR2) were purchased from Signalway Antibody LLC (#38075, #32058, #32265, #38567, #43303, #38828, #32885, College Park, MD, USA). Anti-galectin 3 and neutrophil antibodies were purchased from Bioss Biotechnology Co. LTD (#bs-20700R, #bs-6982R, Beijing, China).
2.2. Animal experiments
As described previously [20], male C57BL/6J mice (20–22g, obtained from HFK Bioscience Co., Beijing, China) were exposed to either ambient PM2.5 or FA in a “real-world” exposure system for 12 h/day, 7 days/week, for 3–6 months (October–December 2015 or July–December 2017). The exposure system contains two chambers and ambient air was inhaled into the chambers by air pump. In the inlet valve of FA chamber, a high efficiency particulate air filter (Shanghai Liancheng Purification Equipment CO., LTD, Shanghai) was installed to remove all the microparticles. In the PM2.5 exposure chamber, PM with an aerodynamic diameter greater than 2.5 μM was removed by a swirler. The exposure system locates at Zhongguancun campus of the University of Chinese of Academy of Sciences (N39°57′39.83″E116°20′10.97″), which is ~50 m away from a traffic main artery (Sihuan Road). During the whole exposure stage, the mice were fed commercial mouse chow and distilled water ad libitum, and were housed under a controlled temperature (22 ± 2 °C) and relative humidity (40–60%) with a 12 h light/dark cycle. Animal studies were performed in accordance with the principles of laboratory animal care (NIH publication no. 85–23, revised 1985) and with approval by the University Of Chinese Academy of Sciences Animal Care and Use Committee.
2.3. Bronchoalveolar lavage
The mice were anesthetized by pentobarbital sodium after exposure. Then, the whole lungs were lavaged 3 times with 1 ml phosphate buffer solution (PBS, pH = 7.4). The bronchoalveolar lavage fluid (BALF) was collected and centrifuged at 1000 rpm for 5–10 min. Total cell number and the protein content of the BALF were measured respectively.
2.4. Histologic assessment
Mouse lungs were harvested quickly, and then washed with ice-cold PBS for three times. After fixation with 4% paraformaldehyde for 48 h, the lungs were embedded in paraffin. Tissue sections (5 μm) were stained with hematoxylin and eosin (H&E) or Masson trichrome staining kits. To identify macrophages and neutrophils, tissue sections were stained with anti-galectin 3 and anti-neutrophil monoclonal antibodies, respectively.
2.5. RNA isolate and RNA-sequencing
Total RNA was extracted from the lungs of FA- or PM2.5-exposed mice using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA quality was measured by Agilent 2100 bioanalyzer (Thermo Fisher Scientific, MA, USA) and samples with an RNA integrity number over than 8 were used for subsequent experiments. The total RNA was further purified by digesting the double-stranded and single-stranded DNA with DNase I and remove of rRNA using Ribo-Zero method (human, mouse, plants) (Illumina, USA). The library construction and RNA sequencing were performed on a BGISEQ500 platform (BGI-Shenzhen, China).
2.6. Read mapping and differentially expressed gene analysis
The raw data were firstly counted and cleaned using SOAPnuke (BGI-Shenzhen, China) and trimmomatic [21] software to remove ligation sequence, low quality sequence and repeats. The sequencing data for clean reads generated by this study have been deposited in the NCBI Sequence Read Archive (SRA) database (accession number: PRJNA540011). Then the clean reads were mapped to the reference genome (Mus_musculus, GCF_000001635.25_GRCm38.p5) using HISAT (Hierarchical Indexing for Spliced Alignment of Transcripts) [22] or Bowtie 2 [23] software. The matched reads were calculated and then normalized to RPKM value (reads per kilo base per million mapped reads) using RESM software [24] to obtain the gene expression level. The differential expression of genes (DEGs) between two groups was screened by DEGseq [25] with the thresholds of fold change ≥ 2 and adjusted P value ≤ 0.001.
To further understand the biological functions of genes, the identified DEGs in each pair were mapped to terms in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/pathway.html). In addition, we performed enrichment analysis using the phyper function of R software. The p-value was adjusted for false discovery rate (FDR) to get q-value, and q-value ≤ 0.05 was considered as significant enrichment.
2.7. Quantitative real-time PCR analysis and western blot
The cDNA was synthesized using a PrimeScript RT Reagent Kit (#RR036B, TaKaRa, Otsu, Japan) and mRNA expression were measured by quantitative real-time polymerase chain reaction (qPCR) with the SYBR® Premix Ex Taq™ II Kit (#RR820A). The results were normalized to 18S ribosomal RNA. Primers used in this study are listed in Table S1.
Proteins were extracted from mouse lung using lysis buffer (150 mM NaCl, 100 μg/ml phenylmethylsulfonyl fluoride, 50 mM Tris-Cl and 1% Triton X-100) with protease and phosphatase inhibitor cocktail (#04693124001, #4906837001, Roche, Basel, Switzerland) on 4 °C for 20–30 min. After centrifugation at 12,000 g and 4 °C for 20 min, the supernatant was used for western blot analysis as reported previously [20].
2.8. Statistical analysis
All data were analyzed by StatView (SAS Institute Inc.) and expressed as mean ± SEM. One-way analysis of variance (ANOVA) with Tukey's correction was used to make multiple comparisons among the groups. p < 0.05 was defined statistical significance.
3. Results
3.1. Effect of exposure time and concentration on PM2.5-induced lung inflammation & fibrosis
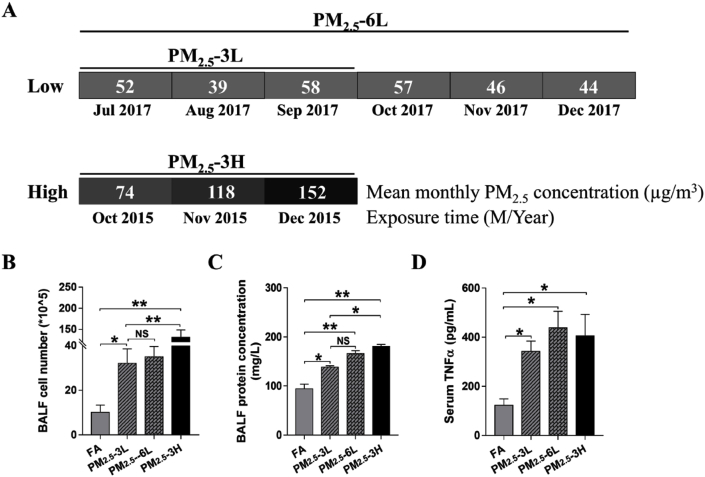
The average monthly concentration of PM2.5 during the exposed period was calculated based on the daily data from http://datacenter.mep.gov.cn/. The experiment groups were defined as PM2.5–3L group (exposed from July to September in 2017, average concentration ~50 μg/m3), PM2.5–6L group (exposed from July to December in 2017, average concentration ~50 μg/m3) and PM2.5–3H group (exposed from October to December in 2015, average concentration ~115 μg/m3) (Fig. 1A). As to FA-exposed mice, 3 months or 6 months exposure had no obvious difference in lung morphology, BALF cell flux and serum tumor necrosis factor (TNFα) levels. Therefore, the mice exposed to FA from July to September in 2017 were used as the control group.
Fig. 1.
Effect of exposure time and concentration on PM2.5-induced systematic inflammation and lung injury. PM2.5 concentration of Wanliu monitoring station was recorded during the exposure period. Mean monthly PM2.5 concentration of July, August, September, October, November and December in 2017, as well as October, November and December in 2015 were shown (A). After exposure to filtered air (FA), low concentration of PM2.5 for 3–6 months (PM2.5–3L, PM2.5–6L) or high concentration of PM2.5 (PM2.5–3H) for 3 months, total cell number (B) and protein content (C) in bronchoalveolar lavage fluid (BALF), and serum TNFα level was measured (D). Data are presented as the means ± SEM. N = 3–4. *p < 0.05; **p < 0.01; NS, not significant.
To determine whether PM2.5 concentration or exposure time affects pulmonary alveoli injury, the cell flux and protein content in BALF were measured. As shown in Fig. 1B–C, PM2.5 exposure significantly increased the cell number and protein concentration of BALF. In addition, BALF from PM2.5–3H mice had significantly more cell number and higher protein concentration than that of PM2.5–3L mice, while the differences in cell number and protein concentration of BALF between PM2.5–3L and PM2.5–6L groups were not significant (Fig. 1B–C). Although serum TNFα levels were elevated in PM2.5-exposed mice, there was no obvious difference among PM2.5–3L, PM2.5–6L and PM2.5–3H groups (Fig. 1D).
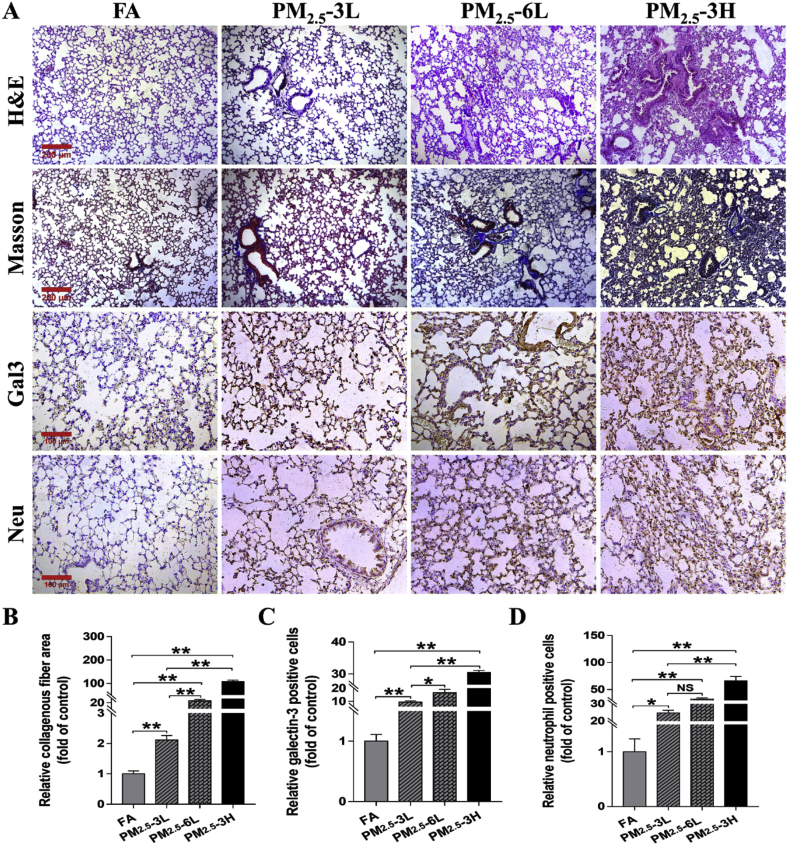
As revealed by H&E and Masson staining, PM2.5 exposure resulted in obvious lung injury & fibrosis, as indicated by the collapse of alveoli, airway epithelial thickening and collagen deposition. Histopathological analysis of lung sections further demonstrated that lungs from PM2.5–6L or PM2.5–3H group developed significantly more severe injury and fibrosis than lungs from FA or PM2.5–3L group. Immunohistochemical staining by antibodies specific for macrophage marker (galectin-3) and neutrophil also revealed that lungs from PM2.5–3H group exhibited more infiltration of macrophages and neutrophils than lungs from FA or PM2.5–3L group (Fig. 2). Together, these results indicated that the degree of lung injury was associated with the PM2.5 concentration and exposure time.
Fig. 2.
Effect of exposure time and concentration on PM2.5-induced pulmonary fibrosis and inflammation. (A) Representative lung sections from FA, PM2.5–3L, PM2.5–6L and PM2.5–3H exposed mice were stained with hematoxylin and eosin (H&E, Scale bar = 200 μm), Masson trichrome (Scale bar = 200 μm), and antibodies specific for macrophages (galetin-3, Gal-3) and neutrophils (brown staining). Scale bar = 100 μm. The relative collagenous fiber area (B), Gal-3 (C) or neutrophil (D) positive cell number were quantified by Image J software. Data were shown as means ± SEM. N = 4, *p < 0.05; ** indicates p < 0.01; NS, not significant. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. PM2.5 exposure changed gene expression profile in mice lungs
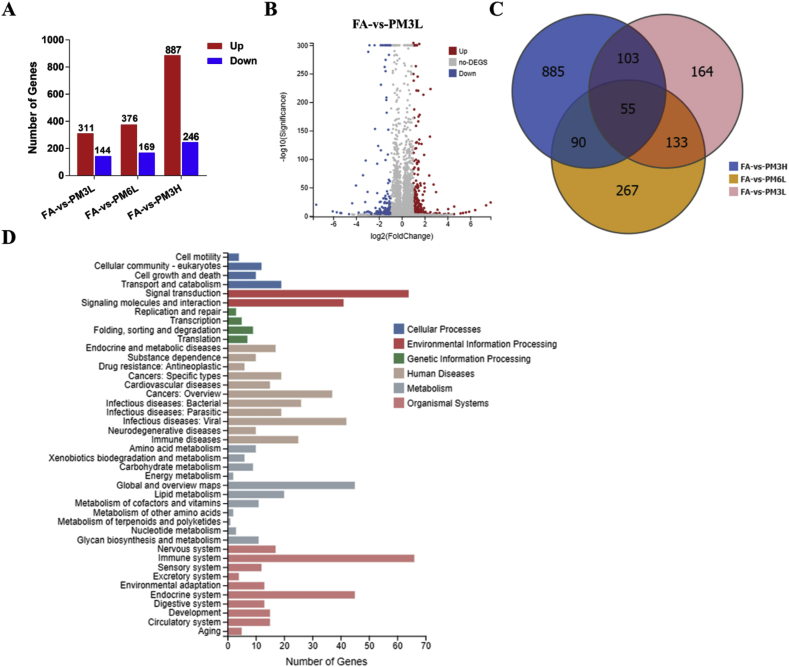
To investigate the molecular mechanism by which PM2.5 exposure causes lung injury in mice, RNA-sequencing was performed to analysis the whole-genome expression profiling changes in PM2.5-exposed lungs. Boxplot showed the general distribution of genes expression of mice lungs from FA, PM2.5–3L, PM2.5–6L and PM2.5–3H groups (Fig. S1). A total of 455 DEGs (311 up-regulated and 144 down-regulated) were identified in comparison of FA and PM2.5–3L exposed lungs, and the fold change of these DEGs were visualized by Volcano plot (Fig. 3A–B). The top 10 up- and down-regulated genes were listed in Table S2. Among these genes, Prok2, Ccl19, Ucp1, Gm13277, Pet117, Tat, Nkx6-2 and Bnc1 are involved in inflammation and oxidative stress related biological process. We also identified 545 DEGs (376 up-regulated and 169 down-regulated) from FA vs PM2.5–6L group and 1133 DEGs (887 up-regulated and 246 down-regulated) from FA vs PM2.5–3H group (Fig. 3A). The fold change of these DEGs from FA vs PM2.5–6L and FA vs PM2.5–3H groups were presented in Supplemental Figs. 2–3 and the top 10 up- and down-regulated genes of each pair were listed in Supplemental Tables 3–4. Among these genes, Gm1987, Ccl19, LOC100043921, DXBay18, Pet117, 2310045n01rik-mef2b, Mpo, Hspa1b, Gm13305 (from FA vs PM2.5–6L group), Sim2, Ifag, Spag11b, Lep, Lhx1 (from FA vs PM2.5–3H group) and 2310045n01rik-mef2b (from both) may participate in regulation of oxidative stress and inflammation related pathway. Interestingly, the top 8 up-regulated genes, including Mybpc1, Myh8, Myh4, Tnnc2, Myot, Myh1, Acta1 and Jsrp1, are involved in of muscle contraction regulation.
Fig. 3.
Analysis of differentially expressed genes between FA and PM2.5-exposed mice lungs. (A) Summery of the number of up- and down-regulated genes in the PM2.5-3L-, PM2.5-6L-, and PM2.5-3H-exposed lungs versus FA-exposed lung. Fold change ≥ 2 and adjusted p value ≤ 0.001 were used as the threshold to judge the significance of gene expression difference. (B) The fold change of differentially expressed genes (DEGs) of FA vs PM2.5–3L were visualized by Volcano plot. (C) The number of different and overlapped DEGs from FA vs PM2.5–3L, FA vs PM2.5–6L and FA vs PM2.5–3H group were illustrated in Venn diagram. (D) KEGG classification of DEGs from FA vs PM2.5–3L group. The functions of genes identified cover six main categories: cellular processes, environmental information processing, genetic information processing, human disease, metabolism and organismal system.
Next, we created a Venn diagram to visual depiction of the similarities and differences between the DEGs in each pair. As shown in Fig. 3C, there were 188 overlapped DEGs between FA vs PM2.5–3L and FA vs PM2.5–6L, 158 overlapped DEGs between FA vs PM2.5–3L and FA vs PM2.5–3H, and only 55 overlapped DEGs among the three groups. According to the KEGG annotation and official classification, we mapped the DEGs of each pair to KEGG pathways, and the results were presented in Fig. 3D and Figs. S2–S3. Among these perturbed pathways, signal transduction and immune system have the highest numbers of DEGs. We then performed KEGG pathway enrichment analysis and found that many DEGs from FA vs PM2.5–3L or FA vs PM2.5–6L were significantly enriched in immune pathways, including hematopoietic cell lineage, cytokine-cytokine receptor interaction and B cell receptor signaling pathway (Table 1, Table 2). We also found that the DEGs of FA vs PM2.5–3H group were significantly enriched in inflammation and immune pathways, including Malaria, and Amoebiasis. However, the top 3 most significantly enriched KEGG pathways (Hypertrophic cardiomyopathy (HCM), Dilated cardiomyopathy (DCM) and Cardiac muscle contraction) are associated with cardiovascular disease (Table 3). The DEGs were also significantly enriched in some metabolic pathways, including PPAR signaling pathway, Insulin secretion, Pancreatic secretion, Glycolysis/Gluconeogenesis and Adipocytokine signaling pathway.
Table 1.
Significantly enriched KEGG pathway of DEGs in FA vs PM2.5–3L group.
| Pathway ID | Pathway Name | Gene Number | Rich Ratioa | Q value |
|---|---|---|---|---|
| ko04640 | Hematopoietic cell lineage | 14 | 0.11864407 | 4.91E-05 |
| ko04060 | Cytokine-cytokine receptor interaction | 20 | 0.05464481 | 0.01323701 |
| ko04662 | B cell receptor signaling pathway | 9 | 0.09 | 0.01786184 |
| ko05150 | Staphylococcus aureus infection | 8 | 0.10126582 | 0.01786184 |
| ko04612 | Antigen processing and presentation | 11 | 0.07142857 | 0.02054528 |
| ko04672 | Intestinal immune network for IgA production | 6 | 0.12244898 | 0.02054528 |
| ko04710 | Circadian rhythm | 5 | 0.15151515 | 0.02054528 |
| ko05323 | Rheumatoid arthritis | 9 | 0.08181818 | 0.02054528 |
| ko04711 | Circadian rhythm - fly | 3 | 0.33333333 | 0.02206418 |
| ko04514 | Cell adhesion molecules (CAMs) | 14 | 0.05555556 | 0.02559195 |
Rich ratio is defined as amount of differentially expressed genes enriched in the pathway/amount of all genes in background gene set.
Table 2.
Significantly enriched KEGG pathway of DEGs in FA vs PM2.5–6L group.
| Pathway ID | Pathway Name | Gene Number | Rich Ratio | Q value |
|---|---|---|---|---|
| ko04640 | Hematopoietic cell lineage | 16 | 0.13559322 | 1.05E-05 |
| ko04060 | Cytokine-cytokine receptor interaction | 27 | 0.073770492 | 7.06E-05 |
| ko05310 | Asthma | 9 | 0.2 | 0.000127211 |
| ko04662 | B cell receptor signaling pathway | 11 | 0.11 | 0.00239461 |
| ko04711 | Circadian rhythm - fly | 4 | 0.444444444 | 0.00239461 |
| ko04062 | Chemokine signaling pathway | 18 | 0.071428571 | 0.003007857 |
| ko03320 | PPAR signaling pathway | 11 | 0.094017094 | 0.006579429 |
| ko05134 | Legionellosis | 8 | 0.103896104 | 0.02277741 |
| ko04672 | Intestinal immune network for IgA production | 6 | 0.12244898 | 0.03628709 |
| ko04710 | Circadian rhythm | 5 | 0.151515152 | 0.03628709 |
| ko04612 | Antigen processing and presentation | 11 | 0.071428571 | 0.04275478 |
Table 3.
Significantly enriched KEGG pathway of DEGs in FA-vs-PM2.5–3H group.
| Pathway ID | Pathway Name | Gene Number | Rich Ratio | Q value |
|---|---|---|---|---|
| ko05410 | Hypertrophic cardiomyopathy (HCM) | 21 | 0.165354331 | 0.000145656 |
| ko05414 | Dilated cardiomyopathy (DCM) | 21 | 0.1640625 | 0.000145656 |
| ko04260 | Cardiac muscle contraction | 17 | 0.184782609 | 0.000195424 |
| ko03320 | PPAR signaling pathway | 18 | 0.153846154 | 0.001089685 |
| ko05144 | Malaria | 12 | 0.176470588 | 0.006066107 |
| ko04261 | Adrenergic signaling in cardiomyocytes | 22 | 0.114583333 | 0.008971435 |
| ko04510 | Focal adhesion | 32 | 0.09495549 | 0.01093564 |
| ko04964 | Proximal tubule bicarbonate reclamation | 7 | 0.25 | 0.011916956 |
| ko04974 | Protein digestion and absorption | 18 | 0.119205298 | 0.014059568 |
| ko05146 | Amoebiasis | 19 | 0.113095238 | 0.017217277 |
| ko04512 | ECM-receptor interaction | 17 | 0.114864865 | 0.025215623 |
| ko04010 | MAPK signaling pathway | 33 | 0.084398977 | 0.033563343 |
| ko04911 | Insulin secretion | 13 | 0.12745098 | 0.033563343 |
| ko04972 | Pancreatic secretion | 15 | 0.11627907 | 0.033563343 |
| ko05412 | Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 13 | 0.125 | 0.033563343 |
| ko00010 | Glycolysis/Gluconeogenesis | 12 | 0.129032258 | 0.035681392 |
| ko04920 | Adipocytokine signaling pathway | 12 | 0.127659574 | 0.03683053 |
3.3. Effect of exposure time and concentration on gene expression profile in PM2.5-exposed lungs
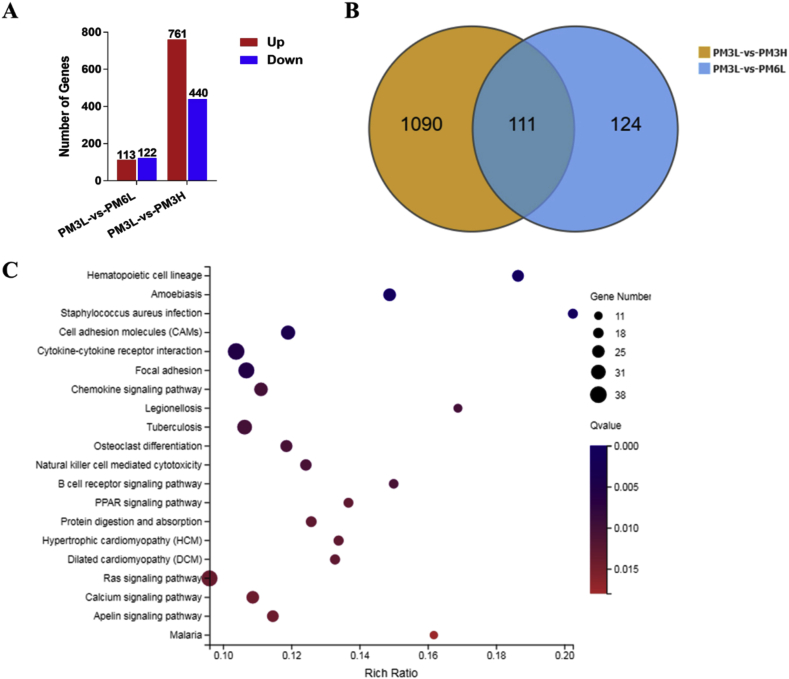
To investigate the effect of exposure time and concentration on gene expression profile in PM2.5-exposed lungs, we also identified 235 DEGs (113 up-regulated and 122 down-regulated) from PM2.5–3L vs PM2.5–6L group and 1201 DEGs (761 up-regulated and 440 down-regulated) from PM2.5–3L vs PM2.5–3H group, respectively (Fig. 4A). The fold change of these DEGs from PM2.5–3L vs PM2.5–6L and PM2.5–3L vs PM2.5–3H group were visualized by Volcano plot (Figs. S4–S5). Among all the DEGs in the volcano plots, top 10 up- and down-regulated genes of PM2.5–3L vs PM2.5–6L and PM2.5–3L vs PM2.5–3H were listed in Tables S5 and S6, respectively. Although Venn diagram showed that there were 111 overlapped DEGs (Fig. 4B), the top 10 up-regulated of genes in PM2.5 3L-vs PM2.5–3H are totally different from those of PM2.5–3L vs PM2.5–6L. There are three overlapped top down-regulated genes, including Rps27rt, GM40369 and Pcdha9. The functions of Rps27rt and GM40369 remain unclear, while Pcdha9 mutation is associated with Hirschsprung's disease [26].
Fig. 4.
Effect of exposure time and concentration on gene expression profile in PM2.5-exposed lungs. (A) Summery of the number of up- and down-regulated genes in the PM2.5-6L-, and PM2.5-3H-exposed lungs vs PM2.5–3L -exposed lung. Fold change ≥ 2 and adjusted p value ≤ 0.001 were used as the threshold to judge the significance of gene expression difference. (B) Venn diagram shows the number of different and overlapped DEGs from PM2.5–3L vs PM2.5–6L and PM2.5–3L vs PM2.5–3H group. (C) Advanced bubble chart shows the top 20 significantly enriched KEGG pathways of DEGs in PM2.5–3L vs PM2.5–3H group. Y-axis label represents pathway, and X-axis label represents rich ratio. Size and color of the bubble represent amount of DEGs enriched in pathway and enrichment significance, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We also mapped the DEGs of each pair to KEGG pathways, and most of DEGs belonged to signal transduction and immune pathways (Figs. S4–S5). KEGG pathway enrichment analysis demonstrated that the DEGs of PM2.5–3L vs PM2.5–6L were only significantly enriched in Cytokine-cytokine receptor interaction pathway, while the DEGs of PM2.5–3L vs PM2.5–3H were significantly enriched in inflammation and immune pathways (Hematopoietic cell lineage, Amoebiasis, Staphylococcus aureus infection, Cytokine-cytokine receptor interaction, and etc.) and cardiovascular disease pathways (Hypertrophic cardiomyopathy, Dilated cardiomyopathy and Cardiac muscle contraction) (Fig. 4C, Table S6).
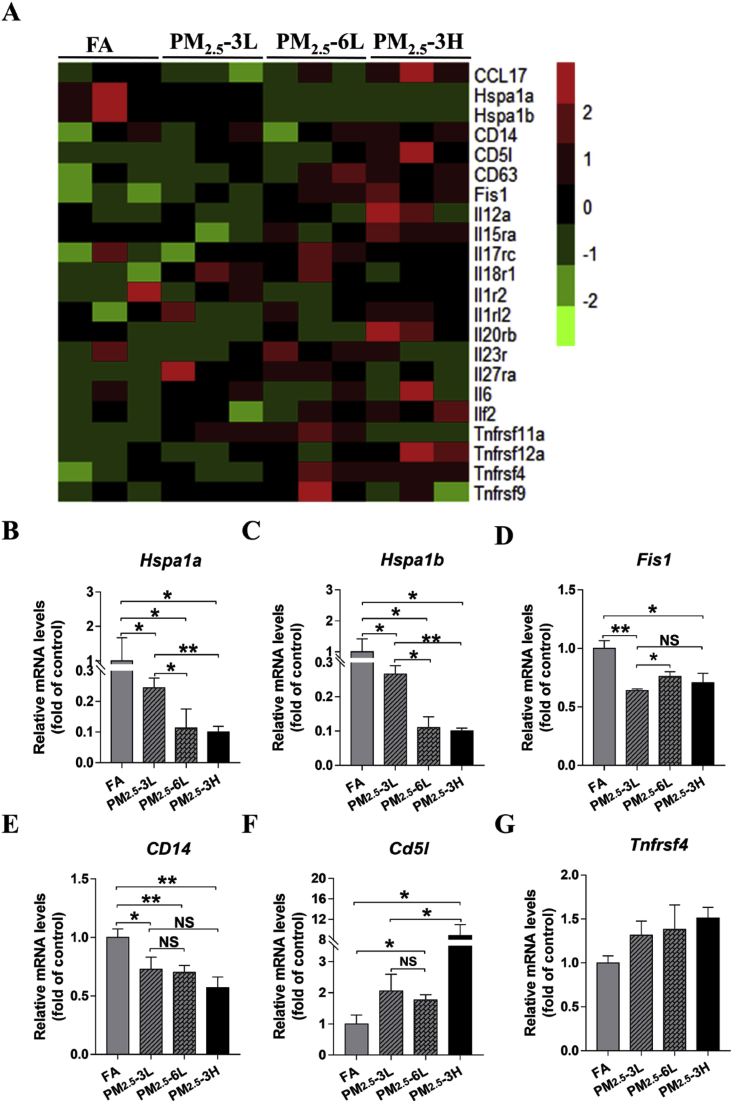
3.4. Down-regulation of Hspa1 and up-regulation of Cd5l were associated with PM2.5-induced pulmonary inflamation
To explore the mechanism for the activated immune pathway in PM2.5-exposed lungs, the expression profile of some inflammatory response related genes, including chemokine (C–C motif) ligand (CCL), interleukin (IL), interleukin receptor (ILr), tumor necrosis factor receptor superfamily (Tnfrsf), heat shock 70 kDa protein (Hspa) and cluster of differentiation (CD), were shown in the heat map (Fig. 5A). We also performed real-time qPCR to validate the changes of some genes, including heat shock protein family A member 1A (Hspa1a) and Hspa1b, Fis1 (mitochondrial fission), CD14 (endotoxin receptor), Tnfrsf4 and CD5 antigen-like (Cd5l). We found that Hspa1a, Hspa1b, Fis1 and CD14 were significantly down-regulated in PM2.5-exposed lungs (Fig. 5B–E). In addition, the mRNA levels of Hspa1a and Hsap1b were further markedly decreased in PM2.5–6L and PM2.5–3H lungs (Fig. 5B–C). We also found that PM2.5 exposure significantly increased the mRNA levels of Cd5l via a concentration-dependent manner (Fig. 5F). Previous reports demonstrated that Hspa1 protects against TNFα-induced lethal inflammatory shock and cell death [27,28], while overexpression of Cd5l in alveolar type II epithelial cells induces spontaneous lung adenocarcinoma [29]. Thus, it is likely that the reduction of Hspa1a and Hsap1b expression, as well as upregulation of Cd5l, might be important factors for the lung injury and pulmonary inflammation induced by PM2.5–3H exposure. Although heat map indicated that the expression of Tnfrsf4 was increased in lungs of PM2.5-exposed mice, qPCR results demonstrated that the up-regulation of Tnfrsf4 was not statistic significant (Fig. 5G).
Fig. 5.
Effect of PM2.5expose time and concentration on gene expression of inflammatory factors. (A) Heat map of gene expression of main inflammatory factors in FA-, PM2.5-3L- and PM2.5-3H-exposed lungs. To verify the RNA-seq results, mRNA levels of Hspa1a (B), Hspa1b (C), Fis1 (D), CD14 (E), Cd5l(F) and Tnfrsf4 (G) were measured by real-time qPCR. N = 3, data are shown as mean ± SEM. *p < 0.05; **p < 0.01; NS, not significant.
3.5. Down-regulation of Prdx4 was associated with PM2.5-induced pulmonary oxidative stress
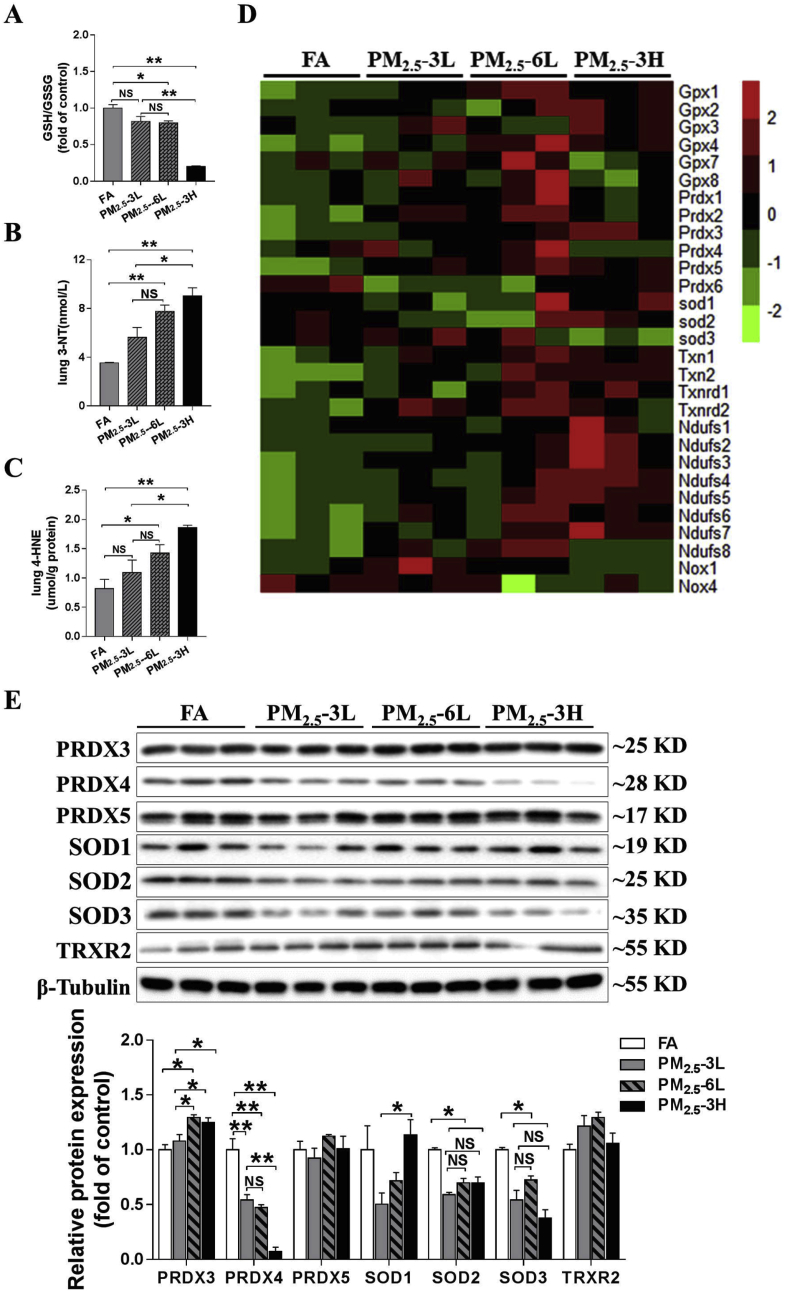
PM2.5–3L exposure resulted in slightly decrease in the GSH/GSSG ratio and moderately increase of 3′-NT and 4-HNE levels, and the changes in GSH/GSSG ratio and 4-HNE levels were not significant. However, compared with FA-exposed lungs, PM2.5-6L- and PM2.5-3H-exposed lungs exhibited significantly lower GSH/GSSG ratio and higher 3′-NT and 4-HNE levels (Fig. 6A–C). We also compared the oxidative stress degree in lung of PM2.5-exposed mice. There was no significantly difference between PM2.5–6L and PM2.5–3L groups. However, PM2.5–3H lungs exhibited significantly lower GSH/GSSG ratio and higher levels of 3′-NT and 4-HNE than those of PM2.5–3L lungs (Fig. 6A–C), indicating that PM2.5 exposure causes pulmonary oxidative stress via a concentration-dependent manner.
Fig. 6.
PM2.5induced pulmonary oxidative stress in a concentration-dependent manner. After exposed to low or high concentration of PM2.5 for 3–6 months, mice lungs were collected, and the GSH/GSSG ratio (A), 3′-NT (B) and 4-HNE (C) levels were measured. The expression of oxidative stress-related genes expression in FA-, PM2.5-3L- and PM2.5-3H-exposed lungs were listed as heat map (D). Lysates of lungs were examined by western blotting for the expression of peroxiredoxin 3 (PRDX3), PRDX4, PRDX5, superoxide dismutase 1 (SOD1), SOD2, SOD3 and thioredoxin reductase 2 (TRXR2). β-tubulin was used as a loading control (E). N = 3–5, data are shown as mean ± SEM; * indicates p < 0.05, ** indicates p < 0.01, NS, not significant.
To investigate the underlying mechanism for the increased oxidative stress in PM2.5–3H lungs, some oxidative-related genes (Gpx, Prdx, Sod, Txn, Txnrd, and Ndufs) expression profile were demonstrated as the heat map (Fig. 6D). From the heat map, we found that only Prdx4 and Sod3 were down-regulated, while other anti-oxidative genes were up regulated in PM2.5–3H lungs. To confirm the changes in these anti-oxidant genes, we also examined the protein expression of PRDX (as PRDX3, PRDX4, and PRDX5), SOD (SOD1, SOD2, and SOD3) and TRXR2 in lung lysates by western blot. As shown in Fig. 6E, PM2.5 exposure significantly decreased SOD2, SOD3 and PRDX4 expression in all groups, whereas had no obvious effect on PRDX5 and TRXR2 expression. PRDX3 expression was slightly increased in PM2.5–3L lungs, and was significantly elevated in PM2.5–6L and PM2.5–3H lungs. SOD1 expression was reduced in PM2.5–3L lungs, while the reduction was diminished in PM2.5–6L and PM2.5–3H lungs. Compared to PM2.5–3L lungs, PM2.5–6L lungs exhibited higher levels of PRDX3 and similar levels of other antioxidant enzymes, while PM2.5–3H lungs exhibited significantly higher levels of PRDX1 and SOD1, and lower levels of PRDX4. Considering that PRDX4 was the only identified antioxidant enzyme that was reduced by PM2.5 exposure in a concentration-dependent manner, we postulated that PRDX4 reduction may play an important role in PM2.5-induced pulmonary oxidative stress.
4. Discussion
Nowadays, air pollution or PM2.5 has become a big threat to the respiratory and cardiovascular system [[30], [31], [32]]. Studies by others and ourselves have suggested that immune and inflammatory response, oxidative stress and DNA damage are potential mechanisms responsible for the adverse health effect of PM2.5 [20,[33], [34], [35]]. In addition, transcriptomic analyses of PM2.5-exposed 16HBE cells demonstrated that PM2.5-induced DEGs were involved in inflammatory and immune response pathways, response to xenobiotic stimuli and metabolic response [16]. Another study revealed that PM2.5 triggers infectious disease, cancers, cardiovascular diseases, and immune pathways in A549 cells [18]. It is therefore no strange that most of DEGs in PM2.5–3L vs FA group were enriched in immune system and infectious disease pathways, such as Cytokine-cytokine receptor interaction, B cell receptor signaling pathway, Staphylococcus aureus infection, Antigen processing and presentation and Intestinal immune network for IgA production in the present study. Surprisingly, we found that the most significantly enriched KEGG pathway of DEGs in FA vs PM2.5–3L or PM2.5–6L groups is hematopoietic cell lineage. A revolutionary research study recently identified the lungs as a site of platelet biogenesis and a reservoir for hematopoietic progenitors [36]. Furthermore, previous studies have demonstrated that oxidative stress and inflammation are key regulators of hematopoietic stem cell fate in health and disease [[37], [38], [39]]. Thus, our findings indicate that the detrimental effect of PM2.5 exposure may include hematopoiesis dysregulation.
It is well documented that the cytotoxicity of PM2.5 is dependent on its concentration. Epidemiological studies provide evidence that each 10 μg/m3 elevation in PM2.5 concentration was associated with significantly increased risk of all-cause mortality, COPD, asthma and cardiovascular disease [40]. In vitro experiments showed that PM2.5 dose-dependently decreases cell viability in BEAS-2B [41], A549 [42], 16HBE [16] and macrophages [43]. In the present study, we also demonstrated that PM2.5–3H exposure caused more severe lung injury and higher number of DEGs than did PM2.5–3L. Interestingly, the top 2 most significantly enriched KEGG pathways of DEGs in FA vs PM2.5–3H group were HCM and DCM, which are two common clinical subtypes of cardiomyopathy. Such pathway enrichment is in agreement with previous epidemiological studies, which confirmed that high PM2.5 concentration is significantly associated with increased cardiovascular risk, including ischemic heart disease, heart failure, arrhythmias, and cardiac death [44,45]. Using mouse model, we recently found that short-term PM2.5 exposure is sufficient to induce a robust lung inflammation, vascular remodeling, and promote transition from left ventricular failure to right ventricular hypertrophy [34]. In addition, as the DEGs (Mybpc1, Myh8/4/1, Myot, and etc; Table S4) enriched in HCM and DCM pathways were mainly related to myopathy, we speculated that the activation of HCM and DCM pathways in PM2.5-3H-exposed lungs might be associated with pulmonary vessel remodeling and muscularization. Hspa1 encodes a 70 kDa heat shock protein, which exhibits a broad protective role in multiple diseases, including sepsis, insulin resistance and liver injury [46]. The anti-inflammatory mechanisms of Hspa1 involves inhibition of NF-κB activation [47] and high-mobility group box 1 (HMGB1) release [48]. Furthermore, Hspa1 facilitates DNA repair in Benzo[a]pyrene exposed 16HBE cells [49]. Cd5l, also called apoptosis inhibitor of macrophage (AIM), plays an important role in the control of immune homeostasis and inflammatory disease [50]. The finding shows that Cd5l is concentration-dependently upregulated in PM2.5-exposed lung which is in agreement with previous study which demonstrated Cd5l was one of the significantly upregulated genes in welding fumes exposed rat lungs [51]. It has been reported that deletion or blockade of Cd5l attenuates the inflammatory response in acute myocardial infarction [52] and experimental sepsis [53]. By contrast, overexpression of Cd5l in myeloid or alveolar type II epithelial cells induces systemic inflammation and adenocarcinoma in the lung [29,54]. Therefore, the significantly repressed Hspa1 expression and dramatically up-regulated Cd5l may contribute to the enhanced pulmonary inflammation and fibrosis in PM2.5-3H-exposed lungs.
Previous studies have demonstrated that PM2.5 exposure could promote ROS production, which causes inflammatory cytokines release and DNA damage, and then leads to cell death and lung injury [1,11,55]. In the present study, we found that GSH/GSSG ratio and expression of some antioxidants enzymes (SOD2, SOD3 and PRDX4) were decreased, whereas 3′-NT and 4-HNE levels were increased in PM2.5-exposed lungs. Consistent with our previous finding that PM2.5 dose-dependently increases intracellular ROS in A549 cells [56], here we found that PM2.5–3H caused greater pulmonary oxidative stress than did PM2.5–3L. PRDX4 is an antioxidant enzyme located in the endoplasmic reticulum (ER) and plays an important role in H2O2 scavenging and protein folding in the ER [57]. PRDX4 deficiency has been found to exacerbate diethylnitrosamine-induced hepatic oxidative stress [58] and dextran sulfate sodium-induced intestinal inflammation [59]. Thus, the reduction of PRDX4 expression might be an important contributor for the elevated oxidative stress in PM2.5-3H-exposed lungs.
In vitro experiments consistently demonstrated that PM2.5 can affect cell viability, inflammation response and intracellular ROS in a time-dependent manner [55,60]. However, we found that there were no significant difference in system inflammation, lung injury, pulmonary GSH/GSSH ratio, 3′-NT and 4-HNE levels and antioxidant enzymes (PRDX4, PRDX5, SOD1, SOD2, SOD3 and TRXR2) expression between PM2.5–3L and PM2.5–6L groups, indicating that exposure time had no obvious effect on PM2.5-induced inflammation and oxidative stress. This may due to the adaptive response triggered by PM2.5 exposure. First, cytokines induced by early PM2.5 exposure may contribute to mount an adaptive immune response to affect cytokines expression at late stage. For example, Ccl17 expression was lower in PM2.5–6L lungs than that of PM2.5–3L lungs (Tables S2–S3). The finding that the DEGs were only significantly enriched in cytokine-cytokine receptor interaction pathways between PM2.5–3L vs PM2.5–6L, suggesting that the interaction among different cytokines may contribute to the similarity of lung injury. Second, PM2.5 exposure could promote nuclear factor erythroid-2-related factor 2 (NRF2) nuclear translocation to orchestrate the antioxidant and detoxification genes [42,61]. The activation of NRF2 then protects cell against PM2.5-induced cytotoxicity [42]. This kind of adaptive response may also explain why some antioxidant enzymes were upregulated in PM2.5-exposed lungs, especially in lungs of PM2.5–6L.
The present study has several limitations. It is well acknowledged that there are discrepancies between the mRNA and protein levels for some genes [62]. In this study, the profile of RNA-seq only provides a comprehensive overview of the entire transcriptome. However, the expression and activity levels of protein obtained from the study should be further examined. Moreover, additional analysis should be carried out to identify all the genes which have a trend for dose- or time-dependent changes in expression profile. These genes/pathways may be important for elucidating the harmful effect of PM2.5 in respiratory system. Further studies are necessary in order to clarify the underlying mechanism whether the changes in gene expression profile are due to the direct or indirect/systematic effects of PM2.5 on lung tissue.
In summary, our study indicated a strong concentration-response relationship between airborne PM2.5 and lung injury. The effect of exposure time and concentration of airborne PM2.5 on whole transcriptome profiling of mice lungs were illuminated by RNA-seq. As revealed by KEGG enrichment analysis, PM2.5 mainly induced immune pathways, especially hematopoietic cell lineage pathway. Furthermore, our results suggest that Hspa1, Cd5l and Prdx4 may play important roles in PM2.5-induced pulmonary inflammation and oxidative stress. These data may provide deeper insight into the molecular mechanism for the harmful effect of PM2.5.
Author disclosure statement
No competing financial interests exist.
Acknowledgments
This study was supported by grants from National Natural Science Foundation of China (91743104and 91643206) and University of Chinese Academy of Sciences. We would like to thank Fang Li, Shasha Zuo and Dandan Sun from University of Chinese Academy of Sciences for their kindly help in instrument operation.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101264.
Contributor Information
Wenjun Ding, Email: dingwj@ucas.ac.cn.
Zhongbing Lu, Email: luzhongbing@ucas.ac.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Li N., Hao M., Phalen R.F., Hinds W.C., Nel A.E. Particulate air pollutants and asthma. A paradigm for the role of oxidative stress in PM-induced adverse health effects. Clin. Immunol. 2003;109(3):250–265. doi: 10.1016/j.clim.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Hertz-Picciotto I., Baker R.J., Yap P.S., Dostal M., Joad J.P., Lipsett M., Greenfield T., Herr C.E., Benes I., Shumway R.H., Pinkerton K.E., Sram R. Early childhood lower respiratory illness and air pollution. Environ. Health Perspect. 2007;115(10):1510–1518. doi: 10.1289/ehp.9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sint T., Donohue J.F., Ghio A.J. Ambient air pollution particles and the acute exacerbation of chronic obstructive pulmonary disease. Inhal. Toxicol. 2008;20(1):25–29. doi: 10.1080/08958370701758759. [DOI] [PubMed] [Google Scholar]
- 4.Hamra G.B., Guha N., Cohen A., Laden F., Raaschou-Nielsen O., Samet J.M., Vineis P., Forastiere F., Saldiva P., Yorifuji T., Loomis D. Outdoor particulate matter exposure and lung cancer: a systematic review and meta-analysis. Environ. Health Perspect. 2014;122(9):906–911. doi: 10.1289/ehp/1408092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pun V.C., Kazemiparkouhi F., Manjourides J., Suh H.H. Long-term PM2.5 exposure and respiratory, cancer, and cardiovascular mortality in older US adults. Am. J. Epidemiol. 2017;186(8):961–969. doi: 10.1093/aje/kwx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniels M.J., Dominici F., Samet J.M., Zeger S.L. Estimating particulate matter-mortality dose-response curves and threshold levels: an analysis of daily time-series for the 20 largest US cities. Am. J. Epidemiol. 2000;152(5):397–406. doi: 10.1093/aje/152.5.397. [DOI] [PubMed] [Google Scholar]
- 7.Chen C., Li C., Li Y., Liu J., Meng C., Han J., Zhang Y., Xu D. Short-term effects of ambient air pollution exposure on lung function: a longitudinal study among healthy primary school children in China. Sci. Total Environ. 2018;645:1014–1020. doi: 10.1016/j.scitotenv.2018.07.154. [DOI] [PubMed] [Google Scholar]
- 8.Dominici F., Peng R.D., Bell M.L., Pham L., McDermott A., Zeger S.L., Samet J.M. Fine particulate air pollution and hospital admission for cardiovascular and respiratory diseases. JAMA. 2006;295(10):1127–1134. doi: 10.1001/jama.295.10.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zanobetti A., Franklin M., Koutrakis P., Schwartz J. Fine particulate air pollution and its components in association with cause-specific emergency admissions. Environ. Health : A Glob. Access Sci. Source. 2009;8:58. doi: 10.1186/1476-069X-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang S., Zhou J., Zhang J., Du X., Zeng X., Pan K., Xie Y., Kan H., Sun Q., Cai J., Zhao J. The severity of lung injury and metabolic disorders induced by ambient PM2.5 exposure is associated with cumulative dose. Inhal. Toxicol. 2018:1–8. doi: 10.1080/08958378.2018.1508258. [DOI] [PubMed] [Google Scholar]
- 11.Riva D.R., Magalhaes C.B., Lopes A.A., Lancas T., Mauad T., Malm O., Valenca S.S., Saldiva P.H., Faffe D.S., Zin W.A. Low dose of fine particulate matter (PM2.5) can induce acute oxidative stress, inflammation and pulmonary impairment in healthy mice. Inhal. Toxicol. 2011;23(5):257–267. doi: 10.3109/08958378.2011.566290. [DOI] [PubMed] [Google Scholar]
- 12.Zhao C., Liao J.P., Chu W.L., Wang S.X., Yang T.S., Tao Y.H., Wang G.F. Involvement of TLR2 and TLR4 and Th1/Th2 shift in inflammatory responses induced by fine ambient particulate matter in mice. Inhal. Toxicol. 2012;24(13):918–927. doi: 10.3109/08958378.2012.731093. [DOI] [PubMed] [Google Scholar]
- 13.Vattanasit U., Navasumrit P., Khadka M.B., Kanitwithayanun J., Promvijit J., Autrup H., Ruchirawat M. Oxidative DNA damage and inflammatory responses in cultured human cells and in humans exposed to traffic-related particles. Int. J. Hyg Environ. Health. 2014;217(1):23–33. doi: 10.1016/j.ijheh.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Oh S.M., Kim H.R., Park Y.J., Lee S.Y., Chung K.H. Organic extracts of urban air pollution particulate matter (PM2.5)-induced genotoxicity and oxidative stress in human lung bronchial epithelial cells (BEAS-2B cells) Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2011;723(2):142–151. doi: 10.1016/j.mrgentox.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Cachon B.F., Firmin S., Verdin A., Ayi-Fanou L., Billet S., Cazier F., Martin P.J., Aissi F., Courcot D., Sanni A., Shirali P. Proinflammatory effects and oxidative stress within human bronchial epithelial cells exposed to atmospheric particulate matter (PM2.5 and PM > 2.5) collected from Cotonou, Benin. Environ. Pollut. 2014;185:340–351. doi: 10.1016/j.envpol.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Z., Liu Y., Duan F., Qin M., Wu F., Sheng W., Yang L., Liu J., He K. Transcriptomic analyses of the biological effects of airborne PM2.5 exposure on human bronchial epithelial cells. PLoS One. 2015;10(9) doi: 10.1371/journal.pone.0138267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng L., Liu S., Zhuang G., Xu J., Liu Q., Zhang X., Deng C., Guo Z., Zhao W., Liu T., Wang Y., Zhang Y., Lin J., Wang Q., Sui G. Signal transductions of BEAS-2B cells in response to carcinogenic PM2.5 exposure based on a microfluidic system. Anal. Chem. 2017;89(10):5413–5421. doi: 10.1021/acs.analchem.7b00218. [DOI] [PubMed] [Google Scholar]
- 18.Lei X., Muscat J.E., Huang Z., Chen C., Xiu G., Chen J. Differential transcriptional changes in human alveolar epithelial A549 cells exposed to airborne PM2.5 collected from Shanghai, China. Environ. Sci. Pollut. Res. Int. 2018;25(33):33656–33666. doi: 10.1007/s11356-018-3090-z. [DOI] [PubMed] [Google Scholar]
- 19.Yang B., Li X.M., Chen D.M., Xiao C.L. Effects of fine air particulates on gene expression in non-small-cell lung cancer. Adv. Med. Sci. 2017;62(2):295–301. doi: 10.1016/j.advms.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Wang H., Shen X., Tian G., Shi X., Huang W., Wu Y., Sun L., Peng C., Liu S., Huang Y., Chen X., Zhang F., Chen Y., Ding W., Lu Z. AMPKalpha2 deficiency exacerbates long-term PM2.5 exposure-induced lung injury and cardiac dysfunction. Free Radical Biol. Med. 2018;121:202–214. doi: 10.1016/j.freeradbiomed.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 21.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L., Feng Z., Wang X., Wang X., Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 26.Shen Q., Zhang H., Su Y., Wen Z., Zhu Z., Chen G., Peng L., Du C., Xie H., Li H., Lv X., Lu C., Xia Y., Tang W. Identification of two novel PCDHA9 mutations associated with Hirschsprung's disease. Gene. 2018;658:96–104. doi: 10.1016/j.gene.2018.02.054. [DOI] [PubMed] [Google Scholar]
- 27.Van Molle W., Wielockx B., Mahieu T., Takada M., Taniguchi T., Sekikawa K., Libert C. HSP70 protects against TNF-induced lethal inflammatory shock. Immunity. 2002;16(5):685–695. doi: 10.1016/s1074-7613(02)00310-2. [DOI] [PubMed] [Google Scholar]
- 28.Nishimura H., Emoto M., Kimura K., Yoshikai Y. Hsp70 protects macrophages infected with Salmonella choleraesuis against TNF-alpha-induced cell death. Cell Stress Chaperones. 1997;2(1):50–59. doi: 10.1379/1466-1268(1997)002<0050:hpmiws>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y., Qu P., Wu L., Li B., Du H., Yan C. Api6/AIM/Spalpha/CD5L overexpression in alveolar type II epithelial cells induces spontaneous lung adenocarcinoma. Cancer Res. 2011;71(16):5488–5499. doi: 10.1158/0008-5472.CAN-10-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xing Y.F., Xu Y.H., Shi M.H., Lian Y.X. The impact of PM2.5 on the human respiratory system. J. Thorac. Dis. 2016;8(1):E69–E74. doi: 10.3978/j.issn.2072-1439.2016.01.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao J., Bo L., Gong C., Cheng P., Kan H., Xie Y., Song W. Preliminary study to explore gene-PM2.5 interactive effects on respiratory system in traffic policemen. Int. J. Occup. Med. Environ. Health. 2015;28(6):971–983. doi: 10.13075/ijomeh.1896.00370. [DOI] [PubMed] [Google Scholar]
- 32.Xu T., Hou J., Cheng J., Zhang R., Yin W., Huang C., Zhu X., Chen W., Yuan J. Estimated individual inhaled dose of fine particles and indicators of lung function: a pilot study among Chinese young adults. Environ. Pollut. 2018;235:505–513. doi: 10.1016/j.envpol.2017.12.074. [DOI] [PubMed] [Google Scholar]
- 33.Liu S., Zhang W., Zhang F., Roepstorff P., Yang F., Lu Z., Ding W. TMT-based quantitative proteomics analysis reveals airborne PM2.5-induced pulmonary fibrosis. Int. J. Environ. Res. Public Health. 2018;16(1) doi: 10.3390/ijerph16010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yue W., Tong L., Liu X., Weng X., Chen X., Wang D., Dudley S.C., Weir E.K., Ding W., Lu Z., Xu Y., Chen Y. Short term Pm2.5 exposure caused a robust lung inflammation, vascular remodeling, and exacerbated transition from left ventricular failure to right ventricular hypertrophy. Redox Biol. 2019;22:101161. doi: 10.1016/j.redox.2019.101161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Oliveira A.A.F., de Oliveira T.F., Dias M.F., Medeiros M.H.G., Di Mascio P., Veras M., Lemos M., Marcourakis T., Saldiva P.H.N., Loureiro A.P.M. Genotoxic and epigenotoxic effects in mice exposed to concentrated ambient fine particulate matter (PM2.5) from Sao Paulo city, Brazil. Part. Fibre Toxicol. 2018;15(1):40. doi: 10.1186/s12989-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lefrancais E., Ortiz-Munoz G., Caudrillier A., Mallavia B., Liu F.C., Sayah D.M., Thornton E.E., Headley M.B., David T., Coughlin S.R., Krummel M.F., Leavitt A.D., Passegue E., Looney M.R. The lung is a site of platelet biogenesis and a reservoir for haematopoietic progenitors. Nature. 2017;544(7648):105–+. doi: 10.1038/nature21706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pietras E.M. Inflammation: a key regulator of hematopoietic stem cell fate in health and disease. Blood. 2017;130(15):1693–1698. doi: 10.1182/blood-2017-06-780882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan D.Q., Suda T. Reactive oxygen species and mitochondrial homeostasis as regulators of stem cell fate and function. Antioxidants Redox Signal. 2018;29(2):149–168. doi: 10.1089/ars.2017.7273. [DOI] [PubMed] [Google Scholar]
- 39.Jung H., Choi I. Thioredoxin-interacting protein, hematopoietic stem cells, and hematopoiesis. Curr. Opin. Hematol. 2014;21(4):265–270. doi: 10.1097/MOH.0000000000000037. [DOI] [PubMed] [Google Scholar]
- 40.Pope C.A., 3rd, Burnett R.T., Thun M.J., Calle E.E., Krewski D., Ito K., Thurston G.D. Lung cancer, cardiopulmonary mortality, and long-term exposure to fine particulate air pollution. JAMA. 2002;287(9):1132–1141. doi: 10.1001/jama.287.9.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y., Duan J., Yang M., Li Y., Jing L., Yu Y., Wang J., Sun Z. Transcriptomic analyses of human bronchial epithelial cells BEAS-2B exposed to atmospheric fine particulate matter PM2.5. Toxicol. Vitro : Int. J. Publ. Assoc. BIBRA. 2017;42:171–181. doi: 10.1016/j.tiv.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 42.Wang H., Guo Y., Liu L., Guan L., Wang T., Zhang L., Wang Y., Cao J., Ding W., Zhang F., Lu Z. DDAH1 plays dual roles in PM2.5 induced cell death in A549 cells. Biochim. Biophys. Acta. 2016;1860(12):2793–2801. doi: 10.1016/j.bbagen.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y., Wang S., Zhu J., Li C., Zhang T., Liu H., Xu Q., Ye X., Zhou L., Ye L. Effect of atmospheric PM2.5 on expression levels of NF-kappaB genes and inflammatory cytokines regulated by NF-kappaB in human macrophage. Inflammation. 2018;41(3):784–794. doi: 10.1007/s10753-018-0732-8. [DOI] [PubMed] [Google Scholar]
- 44.Xie W., Li G., Zhao D., Xie X., Wei Z., Wang W., Wang M., Li G., Liu W., Sun J., Jia Z., Zhang Q., Liu J. Relationship between fine particulate air pollution and ischaemic heart disease morbidity and mortality. Heart. 2015;101(4):257–263. doi: 10.1136/heartjnl-2014-306165. [DOI] [PubMed] [Google Scholar]
- 45.Pope C.A., 3rd, Turner M.C., Burnett R.T., Jerrett M., Gapstur S.M., Diver W.R., Krewski D., Brook R.D. Relationships between fine particulate air pollution, cardiometabolic disorders, and cardiovascular mortality. Circ. Res. 2015;116(1):108–115. doi: 10.1161/CIRCRESAHA.116.305060. [DOI] [PubMed] [Google Scholar]
- 46.Levada K., Guldiken N., Zhang X., Vella G., Mo F.R., James L.P., Haybaeck J., Kessler S.M., Kiemer A.K., Ott T., Hartmann D., Huser N., Ziol M., Trautwein C., Strnad P. Hsp72 protects against liver injury via attenuation of hepatocellular death, oxidative stress, and JNK signaling. J. Hepatol. 2018;68(5):996–1005. doi: 10.1016/j.jhep.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheppard P.W., Sun X., Khammash M., Giffard R.G. Overexpression of heat shock protein 72 attenuates NF-kappaB activation using a combination of regulatory mechanisms in microglia. PLoS Comput. Biol. 2014;10(2) doi: 10.1371/journal.pcbi.1003471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang D., Kang R., Xiao W., Wang H., Calderwood S.K., Xiao X. The anti-inflammatory effects of heat shock protein 72 involve inhibition of high-mobility-group box 1 release and proinflammatory function in macrophages. J. Immunol. 2007;179(2):1236–1244. doi: 10.4049/jimmunol.179.2.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duan Y.Y., Huang S.L., Yang J., Niu P.Y., Gong Z.Y., Liu X.Y., Xin L.L., Currie R.W., Wu T.C. HspA1A facilitates DNA repair in human bronchial epithelial cells exposed to Benzo[a]pyrene and interacts with casein kinase 2. Cell Stress Chaperones. 2014;19(2):271–279. doi: 10.1007/s12192-013-0454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanjurjo L., Aran G., Roher N., Valledor A.F., Sarrias M.R. AIM/CD5L: a key protein in the control of immune homeostasis and inflammatory disease. J. Leukoc. Biol. 2015;98(2):173–184. doi: 10.1189/jlb.3RU0215-074R. [DOI] [PubMed] [Google Scholar]
- 51.Oh J.H., Yang M.J., Heo J.D., Yang Y.S., Park H.J., Park S.M., Kwon M.S., Song C.W., Yoon S., Yu I.J. Inflammatory response in rat lungs with recurrent exposure to welding fumes: a transcriptomic approach. Toxicol. Ind. Health. 2012;28(3):203–215. doi: 10.1177/0748233711410906. [DOI] [PubMed] [Google Scholar]
- 52.Nishikido T., Oyama J., Shiraki A., Komoda H., Node K. Deletion of apoptosis inhibitor of macrophage (AIM)/CD5L attenuates the inflammatory response and infarct size in acute myocardial infarction. J. Am. Heart Assoc. 2016;5(4) doi: 10.1161/JAHA.115.002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao X., Yan X., Yin Y., Lin X., Zhang Q., Xia Y., Cao J. Therapeutic targeting of apoptosis inhibitor of macrophage/CD5L in sepsis. Am. J. Respir. Cell Mol. Biol. 2019;60(3):323–334. doi: 10.1165/rcmb.2018-0272OC. [DOI] [PubMed] [Google Scholar]
- 54.Qu P., Du H., Li Y., Yan C. Myeloid-specific expression of Api6/AIM/Sp alpha induces systemic inflammation and adenocarcinoma in the lung. J. Immunol. 2009;182(3):1648–1659. doi: 10.4049/jimmunol.182.3.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deng X., Zhang F., Rui W., Long F., Wang L., Feng Z., Chen D., Ding W. PM2.5-induced oxidative stress triggers autophagy in human lung epithelial A549 cells. Toxicol. Vitro : Int. J. Publ. Assoc. BIBRA. 2013;27(6):1762–1770. doi: 10.1016/j.tiv.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 56.Deng X., Zhang F., Wang L., Rui W., Long F., Zhao Y., Chen D., Ding W. Airborne fine particulate matter induces multiple cell death pathways in human lung epithelial cells. Apoptosis : Int. J. Program. Cell Death. 2014;19(7):1099–1112. doi: 10.1007/s10495-014-0980-5. [DOI] [PubMed] [Google Scholar]
- 57.Zito E. PRDX4, an endoplasmic reticulum-localized peroxiredoxin at the crossroads between enzymatic oxidative protein folding and nonenzymatic protein oxidation. Antioxidants Redox Signal. 2013;18(13):1666–1674. doi: 10.1089/ars.2012.4966. [DOI] [PubMed] [Google Scholar]
- 58.Guo X., Noguchi H., Ishii N., Homma T., Hamada T., Hiraki T., Zhang J., Matsuo K., Yokoyama S., Ishibashi H., Fukushige T., Kanekura T., Fujii J., Uramoto H., Tanimoto A., Yamada S. The association of peroxiredoxin 4 with the initiation and progression of hepatocellular carcinoma. Antioxidants Redox Signal. 2019;30(10):1271–1284. doi: 10.1089/ars.2017.7426. [DOI] [PubMed] [Google Scholar]
- 59.Takagi T., Homma T., Fujii J., Shirasawa N., Yoriki H., Hotta Y., Higashimura Y., Mizushima K., Hirai Y., Katada K., Uchiyama K., Naito Y., Itoh Y. Elevated ER stress exacerbates dextran sulfate sodium-induced colitis in PRDX4-knockout mice. Free Radical Biol. Med. 2018;134:153–164. doi: 10.1016/j.freeradbiomed.2018.12.024. [DOI] [PubMed] [Google Scholar]
- 60.Abbas I., Badran G., Verdin A., Ledoux F., Roumie M., Lo Guidice J.M., Courcot D., Garcon G. In vitro evaluation of organic extractable matter from ambient PM2.5 using human bronchial epithelial BEAS-2B cells: cytotoxicity, oxidative stress, pro-inflammatory response, genotoxicity, and cell cycle deregulation. Environ. Res. 2019;171:510–522. doi: 10.1016/j.envres.2019.01.052. [DOI] [PubMed] [Google Scholar]
- 61.Deng X., Rui W., Zhang F., Ding W. PM2.5 induces Nrf2-mediated defense mechanisms against oxidative stress by activating PIK3/AKT signaling pathway in human lung alveolar epithelial A549 cells. Cell Biol. Toxicol. 2013;29(3):143–157. doi: 10.1007/s10565-013-9242-5. [DOI] [PubMed] [Google Scholar]
- 62.Wang D. Discrepancy between mRNA and protein abundance: insight from information retrieval process in computers. Comput. Biol. Chem. 2008;32(6):462–468. doi: 10.1016/j.compbiolchem.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.