Abstract
Motivation
Historically, gene expression has been shown to be the most informative data for drug response prediction. Recent evidence suggests that integrating additional omics can improve the prediction accuracy which raises the question of how to integrate the additional omics. Regardless of the integration strategy, clinical utility and translatability are crucial. Thus, we reasoned a multi-omics approach combined with clinical datasets would improve drug response prediction and clinical relevance.
Results
We propose MOLI, a multi-omics late integration method based on deep neural networks. MOLI takes somatic mutation, copy number aberration and gene expression data as input, and integrates them for drug response prediction. MOLI uses type-specific encoding sub-networks to learn features for each omics type, concatenates them into one representation and optimizes this representation via a combined cost function consisting of a triplet loss and a binary cross-entropy loss. The former makes the representations of responder samples more similar to each other and different from the non-responders, and the latter makes this representation predictive of the response values. We validate MOLI on in vitro and in vivo datasets for five chemotherapy agents and two targeted therapeutics. Compared to state-of-the-art single-omics and early integration multi-omics methods, MOLI achieves higher prediction accuracy in external validations. Moreover, a significant improvement in MOLI’s performance is observed for targeted drugs when training on a pan-drug input, i.e. using all the drugs with the same target compared to training only on drug-specific inputs. MOLI’s high predictive power suggests it may have utility in precision oncology.
Availability and implementation
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Precision oncology is the use of genomic data to tailor therapy for an individual cancer patient. However, response to a cancer treatment—chemotherapy or targeted drugs—is a complex phenotype and often depends on multiple factors especially the genomic profile of the patient (Lee et al., 2018). Presently, only 11% of patients treated with precision oncology can be placed in clinical trials and only 5% of patients benefit from precision oncology (Cheng et al., 2018; Marquart et al., 2018; Zehir et al., 2017). Although there are many reasons underlying this modest success rate, improved drug response prediction will significantly increase the number of patients who benefit from targeted therapy (Marquart et al., 2018) or chemotherapy, and avoid adverse side effects who will not (Gavan et al., 2018; Mishra and Verma, 2010). Various in vitro studies of cancer cell lines and patient-derived xenograft (PDX) mice models (Gao et al., 2015) have created datasets such as Genomics of Drug Sensitivity in Cancer (GDSC) (Iorio et al., 2016) and Cancer Cell Line Encyclopedia (CCLE) (Barretina et al., 2012). These datasets provide researchers with multi-omics profiles—consisting of genomic [somatic mutation and copy number aberration (CNA)], transcriptomic, proteomic and methylomic data—together with the response to a large number of targeted and chemotherapy drugs. This is different from patient datasets, which record the response only to one or a few drugs that have been administered to a patient. These in vitro datasets enable researchers to investigate the drug response mechanism at a large scale, in particular for many drugs, and all the way from various types of pre-clinical models to patients (Barretina et al., 2012; Iorio et al., 2016). Complementing in vitro studies, in silico studies have aimed at building computational methods that analyze the cumulative effects of single- or multi-omics data to accurately predict drug response (Ding et al., 2018; Geeleher et al., 2014). These studies usually measure the drug response as the drug concentration that reduces viability by 50% (IC50).
A critical challenge in drug response research is the clinical utility, i.e. whether the outcome of the study is translatable to actual patients (Geeleher et al., 2014, 2017). Ideally to achieve translatability, a computational method should be trained on in vivo data, however, available in vivo datasets such as The Cancer Genome Atlas (TCGA) datasets (Weinstein et al., 2013) do not have enough patient records with drug response information and in particular, unlike cell line datasets such as GDSC, they do not report responses to multiple drugs. For in silico drug response prediction, translatability in the simplest case means that a model with good performance (e.g. high prediction accuracy) on in vitro data—trained on more samples compared to in vivo data—should also have good performance on in vivo data.
The majority of studies suggests that gene expression data are the most effective data type for drug response prediction (Ding et al., 2016; Geeleher et al., 2014; Graim et al., 2019; Iorio et al., 2016). Geeleher et al. (2014) showed that a ridge regression model trained on GDCS gene expression data is translatable to Docetaxel, Cisplatin, Erlotinib and Bortezomib clinical trial data. They also showed that, for Docetaxel, including non-breast cancer cell lines in model training increases the predictive power of the final model compared to the model only trained on breast cancer cell lines. This ridge regression-based pipeline on gene expression also imputed the drug response for The Cancer TCGA (Geeleher et al., 2017; Weinstein et al., 2013). Despite the predictive power of gene expression, adding other omics data types can increase the predictive power especially in pan-cancer models (Iorio et al., 2016).
Multi-omics data provide a machine learning model with different views of the same sample and promise better characterization of biological processes (Argelaguet et al., 2018; Wang et al., 2014). Multi-omics data have been exploited for different problems such as driver gene identification (Dimitrakopoulos et al., 2018; Mo et al., 2013; Shrestha et al., 2017; Singh et al., 2019), patient stratification (Khakabimamaghani and Ester, 2016), survival prediction (Chaudhary et al., 2018), subgroup discovery (Liang et al., 2015) and drug response prediction (Ding et al., 2018). For the drug response prediction, Ding et al. (2018) proposed a method that concatenates mutation, CNA and gene expression data and applies autoencoders to learn features for the concatenated multi-omics cell line data. The learned features were used as the input of an elastic net classifier which predicts the binarized IC50 values. We note that the classifier was validated only on CCLE cell lines without studying its translatability to patients or PDX models.
A critical challenge in multi-omics data analysis is how to integrate different data types. There are two major approaches to multi-omics integration: early integration and late integration (Rappoport and Shamir, 2018; Zitnik et al., 2019). In early integration, all omics data types available for a sample are first concatenated, and then an integrated representation of the sample is created by applying some feature learning method, such as autoencoders (Goodfellow et al., 2016), to that representation. Early integration has three disadvantages: first, it disregards the unique distribution of each omics data type. Second, it requires proper normalization to avoid giving more weight to the omics data type with more dimensions. Third, it further increases the dimensionality of the input data which often is already a challenge for single-omics input data (Rappoport and Shamir, 2018). In late integration, features are learned separately for each omics data type, and these features are then integrated into one unified representation to be used as the input for a classifier or a regressor. The advantage of this approach is that it works with the unique distribution of each omics data type, it can employ single-omics normalization for each data type, and it does not increase the dimensionality of the input space.
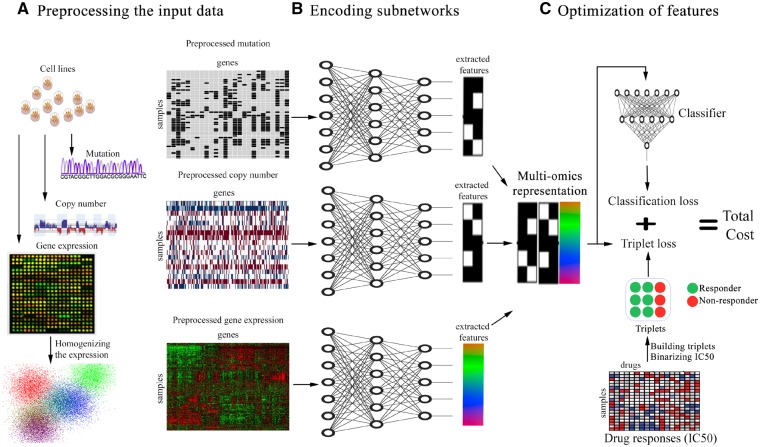
In this paper, we explore the problem of drug response prediction and propose MOLI, a multi-omics late integration method based on deep neural networks. MOLI takes somatic mutation, CNA and gene expression data as input, and predicts the response to a given drug as the output. MOLI learns features for each omics data type by type-specific encoding sub-networks and concatenates the learned features into one representation of the multi-omics profiles. To the best of our knowledge, MOLI is the first end-to-end late integration method with deep neural networks that optimizes this representation via a combined cost function consisting of a triplet loss function (Schroff et al., 2015) and a binary cross-entropy loss function. The former makes the representations of responder cell lines more similar to each and different from the representations of non-responder cell lines and the latter makes this representation predictive of the IC50 values. As another contribution, MOLI employs transfer learning to increase the size of the training dataset. It trains a drug response model on pan-drug inputs (using all the drugs with the same target) instead of drug-specific inputs. Figure 1 illustrates the workflow of MOLI.
Fig. 1.
Schematic overview of MOLI (A) pre-processing mutation, CNA and gene expression data. (B) Each encoding sub-network learns features for its omics data type and the learned features are concatenated into one representation. (C) MOLI cost function consists of a triplet loss and a classification loss, obtained from the classifier sub-network that uses the multi-omics representation to predict drug response
We validated MOLI on in vitro (PDX) and in vivo (TCGA patients) datasets for five chemotherapy agents and two targeted therapeutics. Our comparison with the state-of-the-art single-omics and early integration multi-omics methods showed that MOLI can achieve significantly better performance in terms of Area Under the receiver operating characteristic Curve (AUC) on PDX and patient data. Moreover, we observed substantial improvement in MOLI’s performance for targeted drugs when training on a pan-drug dataset compared to training on drug-specific datasets. We conclude that MOLI models trained on in vitro data translate well to in vivo data and may have utility for precision oncology. Finally, we showed that the responses predicted by MOLI—while trained on the pan-drug input for the epidermal growth factor receptor (EGFR) inhibitors—for breast, lung, kidney and prostate cancers from TCGA patients (without recorded drug response) had statistically significant associations with some of the genes in the EGFR pathway. This shows that MOLI captures biological aspects of the response.
2 Materials and methods
2.1 MOLI
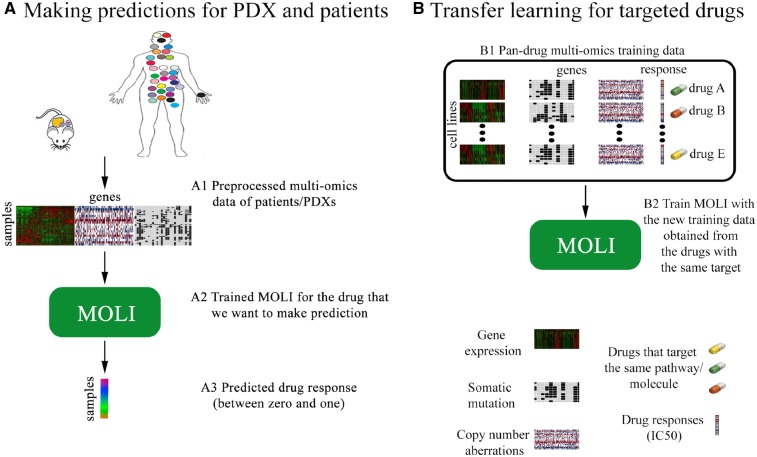
MOLI is a deep neural network that predicts the drug response for a given sample, represented by its multi-omics profile, and for a given drug. MOLI assumes that values for the same genes are provided for each omics data type. MOLI’s network consists of the following sub-networks. It has multiple feed forward encoding sub-networks, one for each input omics data type. Each encoding sub-network receives its corresponding omics data and encodes it into a learned feature space. The learned features from the encoding sub-networks are integrated into one representation by concatenation. The concatenated representation serves as input for a classification sub-network, which predicts the drug response. The entire network is trained in an end-to-end fashion using a cost function combining a classification loss and a triplet loss. Figure 1 shows MOLI’s components during training and model development, while Figure 2A shows the application of MOLI for external validation.
Fig. 2.
(A) Using MOLI to make predictions for PDX/patient inputs during external validation. (B) Combining targeted drugs that target the same pathway or molecule to make a pan-drug training dataset for MOLI
2.1.1. Learning features by encoding sub-networks
To learn features for each omics data type in the input, we design separate encoding feed forward sub-networks to map the input space to the feature space. In this paper, we focus on mutation, CNA and gene expression data. XM, XE and XC denote mutation, CNA and gene expression data, respectively, each of which are of dimensionality N × D, where N is the number of samples and D is the number of genes. We note that the proposed approach can be extended for any number of omics data types. Each encoding sub-network has a fully connected layer with Rectified linear unit (ReLU) activation functions. In addition, each sub-network employs dropout to regularize the model and batch normalization to enhance the training process. The input of each encoding sub-network is one omics data type and the output is the learned features for that omics (Fig. 1B). We denote these sub-networks as and , respectively.
2.1.2. Integrating learned features by late integration
In the integration step, we utilize a late integration approach and concatenate the learned features of the different single-omics data types to obtain one multi-omics representation. For example, if the outputs of three encoding sub-networks are three M × N feature matrices, after concatenation, the output will be one representation matrix. The integrated representation is further smoothed through an l2 normalization layer. We denote MOLI’s integration, receiving multi-omics data as input and returning the integrated representation, as follows:
| (1) |
where, ⊕ denotes the concatenation operator.
2.1.3. Optimizing the learned features by the combined cost function
The learned features will be used by a classifier that predicts the drug response. Therefore, the last sub-network of MOLI is a classification layer with the Sigmoid activation function, using dropout and weight decay for regularization (Fig. 1C). We denote this classifier as g(.). Since the MOLI network will be used for classification, i.e. drug response prediction, the cost function used for training must include a term that measures the difference between the predicted drug response and the ground truth drug response. We choose the binary cross-entropy classification loss, one of the most common classification losses, defined as follows:
where, denotes the binarized IC50 which is used as measure for the drug response.We add a triplet loss to the cost function to impose a further constraint that is necessary for accurate classification. This constraint forces responders to be more similar to each other than to non-responders. The triplet loss function was introduced in FaceNet (Schroff et al., 2015) for optimizing the mapping from a space of face images to a Euclidean space where the difference between learned features is correlated with the similarity among faces. The idea is that for the image of a given person’s face, the distance between that image’s learned features and the features of another image of the same person should be smaller than the distance between that image’s learned features and the learned features of the image of some other person. In our context, we employ the triplet loss function as follows. For T given triplets in the form of (Anchor, Positive, Negative), where the first two are (the multi-omics data of) responder cell lines to a given anti-cancer drug and the last one is (the multi-omics data of) a non-responder to that drug, we require the following condition: , where d(.) is an arbitrary distance function—we used the Euclidean distance.If we move the right hand-side to the left, we obtain:
In order to avoid the trivial zero solution, a margin is required:
We want the distance of the Anchor and the Negative to be larger than the distance of the Anchor and the Positive. Thus, the value of the triplet loss function for the i-th triplet is:
and the total triplet loss for T triplets is:
| (2) |
Generally, there are two approaches to select triplets for the triplet loss function: offline selection and online selection. The offline selection builds the triplets based on the value of the labels (in this case the drug response) before training the model. The online selection selects the triplets from samples in each mini-batch during the training. We adopted the online approach. Triplets can be built based on all possible combinations of the input samples/mini-batches (soft selection) or based only on those triplets with high triplet loss value (hard selection). Soft selection provides the model with more training triplet examples but the network might rely too much on easy cases, and as a result may be unable to perform well on hard examples (Schroff et al., 2015). Hard selection solves this problem by only relying on the hard cases in the train data to build the triplets, but this approach may suffer from having fewer training triplets especially in the case of small unbalanced datasets. We adopted the soft selection approach.
Therefore, the combined cost J is defined as follows:
| (3) |
where γ is a regularization term for the triplet loss.
2.2 Transfer learning for targeted drugs
For targeted drugs, we use transfer learning and train MOLI with a new pan-drug input. This pan-drug input consists of multi-omics profiles and drug responses for a family of targeted drugs that target the same pathway or molecule. Such drugs are expected to produce highly correlated responses in cell lines. One MOLI model is trained for a family of drugs instead of one separate model for each individual drug. This approach increases the training dataset size, since the set of the screened cell lines and the obtained responses are similar but not identical for the drugs of one family. In our experiments, we evaluate transfer learning for EGFR pathway inhibitors due to the availability of external validation data, but the approach is applicable to any family of targeted drugs. Figure 2B illustrates the idea of transfer learning for targeted drugs.
2.3 Predicting drug response for TCGA patients
To study MOLI’s performance, similar to Geeleher et al. (2017), we employ the model trained on the pan-drug input for the EGFR inhibitors to predict the drug response for patients in several TCGA datasets for which there was no drug response recorded. Since these drugs target EGFR pathway, we expect the expression status of the genes of this pathway to be strongly correlated with the predicted drug response. We obtain the list of genes in EGFR pathway from REACTOME. To study the correlation, we employ multiple linear regression between the predicted responses and the level of expression. We obtain P-values for each gene and correct them for multiple comparisons, using Bonferroni correction ().
2.4 Datasets
We use four main resources in this paper:
GDSC cell lines dataset (Iorio et al., 2016).
PDX Encyclopedia dataset (Gao et al., 2015).
TCGA patients with the drug response available in their records (Ding et al., 2016).
TCGA patients without the drug response (Weinstein et al., 2013).
The GDSC dataset (Iorio et al., 2016; Yang et al., 2012) has created a multi-omics dataset of more than a thousand cell lines from different cancer types, screened with 265 targeted and chemotherapy drugs. We use GDSC as the training dataset due to a high number of screened drugs. Multi-omics profiles and drug responses for GDSC are retrieved from ftp://ftp.sanger.ac.uk/pub/project/cancerrxgene/releases/release-7.0/.
We use the other publicly available multi-omics datasets for external validation as follows:
We apply PDX Encyclopedia mice models published by Gao et al. (2015). This dataset has more than 300 PDX models for different cancer types, screened with 34 targeted and chemotherapy drugs.
TCGA (Weinstein et al., 2013) data including profiles of tumor samples collected from more than 10 000 patients with different cancer types, downloaded from Firehose Broad GDAC (https://doi.org/10.7908/C11G0KM9, http://gdac.broadinstitute.org/runs/stddata__2016_01_28/). For TCGA datasets, we use clinical annotations of the drug response for some patients which were obtained from Supplementary Material of Ding et al. (2016).
We also use TCGA patients for breast (BRCA), bladder (BLCA), pancreatic (PAAD), lung (LUAD), kidney (KIRP) and prostate (PRAD) cancers. These patients are without the drug response in their records.
We note that we used only those genes which are in common for all of the omics data types in both training and external validation datasets for each drug.
The data pre-processing steps and the used resource files are summarized below and are presented in more detail in the Supplementary Material (Section S1) and Supplementary Table S1. Table 1 provides the characteristics of each dataset such as type of drug, the number of samples and the number of genes. After the pre-processing, we have the same number of genes for the training and the external validation datasets and for each of the three omics data types. We only consider samples for which all three omics data types are available.
Table 1.
List of the studied drugs from the used resources with multi-omics profiles available
| Drug | Type | Resource | Number of samplesa | Number of genesb | Usage |
|---|---|---|---|---|---|
| Afatinib | Targeted | GDSC | 828 (NR: 678, RS: 150) | 13 081 | Training |
| Cetuximab | Targeted | GDSC | 856 (NR: 735, RS: 121) | 12 346c/13 081d | Training |
| Cetuximab | Targeted | PDX | 60 (NR: 55, RS: 5) | 12 346c/13 081d | External validation |
| Cisplatin | Chemotherapy | GDSC | 829 (NR: 752, RS: 77) | 15 493 | Training |
| Cisplatin | Chemotherapy | TCGA | 66 (NR: 6, RS: 60) | 15 493 | External validation |
| Docetaxel | Chemotherapy | GDSC | 829 (NR: 764, RS: 65) | 15 016 | Training |
| Docetaxel | Chemotherapy | TCGA | 16 (NR: 8, RS: 8) | 15 016 | External validation |
| Erlotinib | Targeted | GDSC | 362 (NR: 298, RS: 64) | 12 325c/13 081d | Training |
| Erlotinib | Targeted | PDX | 21 (NR: 18, RS: 3) | 12 325c/13 081d | External validation |
| Gefitinib | Targeted | GDSC | 825 (NR: 710, RS: 115) | 13 081 | Training |
| Gemcitabine | Chemotherapy | GDSC | 844 (NR: 790, RS: 54) | 12 067/15 381 | Training |
| Gemcitabine | Chemotherapy | PDX | 25 (NR: 18, RS: 7) | 12 067 | External validation |
| Gemcitabine | Chemotherapy | TCGA | 57 (NR: 36, RS: 21) | 15 381 | External validation |
| Lapatinib | Targeted | GDSC | 387 (NR: 326, RS: 61) | 13 081 | Training |
| Paclitaxel | Chemotherapy | GDSC | 389 (NR: 363, RS: 26) | 12 482 | Training |
| Paclitaxel | Chemotherapy | PDX | 43 (NR: 38, RS: 5) | 12 482 | External validation |
| Pan-drug | Targeted | GDSC | 3258 (NR: 2747, RS: 511) | 13 081 | Training |
Note: NR, non-responder; RS, responder.
Number of screened samples with all three omics data types available.
Number of genes in common between the train data and the external validation data for each drug.
Number of genes for the drug-specific experiments.
Number of genes for the pan-drug experiments.
2.4.1. Gene expression profiles
Raw intensities are obtained from ArrayExpress (E-MTAB-3610) for GDSC dataset were RMA-normalized (Irizarry et al., 2003), log-transformed and aggregated to the level of genes. Gene expression values of PDX and all TCGA datasets are converted to TPM (Li and Dewey, 2011) and log-transformed. FPKM values for PDX samples are converted into TPM and log-transformed. To make expression profiled by different platforms comparable, we standardize gene expression and perform pairwise homogenization procedure, as described in Geeleher et al. (2014) and Johnson et al. (2007). Also, in each dataset we exclude the 5% of genes with lowest variance assuming them to be not informative.
2.4.2. Somatic copy number profiles
We remove unreliable segments from genome segmentation files for TCGA datasets and assign every gene a value corresponding to the intensity log-ratio of the segment it overlaps. If the gene overlaps more than one segment, we keep the most extreme log-ratio value. Different from TCGA, the GDSC and PDX datasets provided gene-level estimates of total copy number. In order to make these data comparable with TCGA, we compute for every gene the logarithm of its copy number divided by the ploidy of copy-neutral state in the sample. Finally, for all four datasets we binarize gene-level copy number estimates assigning zeros to copy-neutral genes and ones to all genes overlapping deletions or amplifications.
2.4.3. Somatic point mutations
Similarly with previous works (Ding et al., 2018; Iorio et al., 2016), we assign ones to genes carrying somatic point mutations and zeros to all others.
3 Results
3.1 Experimental design
In our experiments, we investigated the following questions:
Does MOLI outperform single-omics and early integration baselines in terms of prediction AUC on PDX and patient data?
Does transfer learning work for targeted drugs, i.e. does MOLI trained on pan-drug data outperform MOLI trained on drug-specific (single drug) data?
Finally, for the targeted drugs, does the predicted response by MOLI have associations with the target of that drug?
We trained MOLI on GDSC cell lines screened with Docetaxel, Cisplatin, Gemcitabine, Paclitaxel, Erlotinib and Cetuximab. We chose these drugs based on availability of PDX/patient multi-omics data for these drugs which is necessary for external validations. We trained all of the baselines for the same drugs and compared them to MOLI in terms of prediction AUC.
We compared MOLI against early integration via deep neural networks inspired by Ding et al. (2018) and early integration via non-negative matrix factorization (NMF) (Cichocki and Phan, 2009; Févotte and Idier, 2011), against the single-omics (gene expression) ridge regression method proposed by Geeleher et al. (2014), against an ordinary feed forward network with classification loss trained on the expression data, and against a version of MOLI trained only on the gene expression data. To test whether the triplet loss contributes to improve the performance, we compared MOLI to a late integration feed forward network with an architecture similar to MOLI but using only a classification loss.
Finally, to study transfer learning for the targeted drugs, we focused on drugs that target the EGFR pathway because we have Cetuximab and Erlotinib that target this pathway in the PDX dataset utilized for external validations. In addition, GDSC was screened with numerous drugs that target EGFR including: Afatinib, Cetuximab, Erlotinib, Gefitinib and Lapatinib. We used multi-omics data of all of these drugs in GDSC and created a large training set (>3000 samples). We trained MOLI on this pan-drug data and compared the results to MOLI which was trained on the drug-specific inputs.
We used 5-fold cross validation in most of the experiments to tune the hyper-parameters of the deep neural networks based on the AUC. The hyper-parameters tuned were number of nodes in the hidden layers, learning rates, mini-batch size, weight decay, the dropout rate, number of epochs and margin and regularization term (only for the triplet loss). Details on the ranges considered for each hyper-parameter and the selected settings for each drug are provided in Supplementary Tables S2 and S3. Finally, the network was re-trained on the obtained hyper-parameters on the entire dataset for that drug (train and validation). We used Adagrad for optimizing parameters in all of the deep neural networks (Duchi et al., 2011). We used the PyTorch framework to implement all deep neural networks codes. For the ridge regression pipeline, we downloaded the implemented pipeline with leave-one-out cross validation provided by the original authors (Geeleher et al., 2014) and applied it to our datasets. To make sure that both the downloaded pipeline and the way we pre-processed the gene expression data are correct, we evaluated it on the datasets from the original paper and got AUCs for Docetaxel and Bortezomib comparable to those of He et al. (2018). For early integration via NMF, we first concatenate the omics data types, and then train an NMF on the resulting matrix to learn the latent factors. Finally we train the Geeleher et al. (2014) method (using the learned factors as features) to predict the drug response. The details of this new baseline and the tuning of its hyper-parameter are presented in the Supplementary Material (Section S2).
3.2 Multi-omics integration by MOLI improves the drug response performance
Table 2 reports the performance of MOLI and the baselines in terms of AUC.
Table 2.
Performance of different versions of MOLI compared to the baselines in terms of prediction AUC across two targeted therapeutics and five chemotherapy agents
| Method | PDX | PDX | PDX | PDX | TCGA | TCGA | TCGA | Input omics |
|---|---|---|---|---|---|---|---|---|
| Drug | Paclitaxel | Gemcitabine | Cetuximab | Erlotinib | Docetaxel | Cisplatin | Gemcitabine | |
| Geeleher et al. (2014) | 0.52 | 0.59 | 0.58 | 0.67 | 0.59 | 0.62 | 0.53 | Expression |
| Early integration via NMF | 0.24 | 0.56 | 0.53 | 0.28 | 0.39 | 0.40 | 0.58 | Multi |
| Early integration via DNNs | NSC | 0.66 | NSC | NSC | 0.52 | NSC | 0.59 | Multi |
| Feed forward net | 0.68 | 0.48 | 0.43 | 0.37 | 0.69 | 0.44 | 0.65 | Expression |
| MOLI complete | 0.69 | 0.52 | 0.51 | 0.39 | 0.63 | 0.75 | 0.64 | Expression |
| MOLI with classifier | NSC | 0.55 | 0.46 | NSC | 0.58 | 0.6 | 0.69 | Multi |
| MOLI complete | 0.74 | 0.64 | 0.53 | 0.63 | 0.58 | 0.66 | 0.65 | Multi |
| MOLI complete Pan-drug | NA | NA | 0.80 | 0.72 | NA | NA | NA | Multi |
Note: NSC: no stable classifier during cross validation or final training. Boldface indicates the best method for the corresponding drug and italics indicates the second best method. NA: the corresponding drug is not for targeted therapy. Complete: when MOLI has both classification and the triplet losses; NMF, non-negative matrix factorization; DNNs, deep neural networks.
First, we compared the complete MOLI (MOLI trained on multi-omics data and using its combined cost function) to the early integration baselines. MOLI achieved better performance in six out of seven external validation datasets compared to early integration via deep neural networks. Moreover, MOLI also achieved better performance in all of the external validation datasets compared to early integration via NMF. These results indicate that MOLI outperforms multi-omics early integration. Second, we compared MOLI trained on multi-omics data with two deep neural network scenarios trained only on the gene expression data (one of them was MOLI itself). MOLI trained on multi-omics data showed better performance in four out of seven external validation datasets and tied in another dataset. These results indicate that deep neural networks trained on multi-omics data achieve better performance than those trained on single-omics data.
Third, we compared MOLI with MOLI without the triplet loss, both trained on multi-omics input. MOLI with its combined cost function obtained better performance in five out of seven external validation datasets and tied in another one. These results demonstrate the contribution of the triplet loss to improving the prediction performance.
Finally, we compared MOLI against a single-omics non-deep neural networks baseline which is the only published method for drug response prediction (Geeleher et al., 2014) that had been tested on patient data. MOLI achieved better performance in four out of seven external validation datasets and tied with this baseline in another one. These experiments show the substantial gain in predictive performance resulting from the combination of using multi-omics data, deep neural networks and the proposed cost function.
MOLI when trained on the pan-drug input (only applicable for targeted drugs) had significantly better performance compared to itself when it was trained on the drug-specific inputs for Erlotinib and Cetuximab. The majority of the baselines had either poor performance or no stable classifier status (NSC) for Paclitaxel and Erlotinib. NSC means that during either cross validation or final re-training with the obtained hyper-parameters the cost and/or AUC curves were fluctuating. This may be due to the small number of samples, because both of these drugs had the fewest number of cell lines (∼400). Also, we observed NSC in the early integration baseline for four drugs which may be due to the concatenation at the beginning because it increased the dimensionality substantially, which makes feature learning harder for the autoencoder and later the classifier in this method.
MOLI achieved an AUC of >0.7 for four drugs (Paclitaxel, Cetuximab, Erlotinib and Cisplatin) which may be beneficial for precision oncology particularly for the targeted drugs (Cetuximab and Erlotinib).
We also studied the area under precision–recall curve for MOLI and the main baselines including the early integration methods and the Geeleher et al. (2014) method. Compared to the early integration methods, MOLI achieved better performance in four out of seven external validation datasets and tied in two other datasets. Compared to the (Geeleher et al., 2014) single-omics baseline, MOLI had better performance in six out of seven external validation datasets. We also investigated the area under precision–recall curve for the pan-drug training data. MOLI trained on pan-drug data had better performance in one external validation dataset and had competitive performance in another one compared to MOLI trained on the drug-specific input (Supplementary Table S4). All these results again suggest that MOLI may be beneficial for precision oncology.
3.3 Transfer learning for targeted drugs improves performance significantly
We observed that for the targeted drugs (in our experiment, EGFR inhibitors), MOLI trained on the pan-drug multi-omics inputs achieved significantly better performance than MOLI trained on drug-specific inputs. Pan-drug MOLI achieved an AUC of 0.8 for Cetuximab and 0.72 for Erlotinib which were significantly higher than the drug-specific performance. This suggests that transfer learning can improve the prediction performance for the targeted drugs.
3.4 Predictions for TCGA patients by MOLI have associations with EGFR genes
We applied MOLI (trained on the pan-drug input for EGFR inhibitors) to multi-omics data without drug response downloaded from TCGA (breast, bladder, pancreatic, lung, kidney and prostate cancers) and predicted the response for these patients. According to the P-values obtained from multiple linear regression, there are a number of strong associations between EGFR genes and the responses predicted by MOLI. For breast cancer, we observed statistically significant associations between the level of expression in AP2A1 (P = 0.007), CALM2 (P = 0.01), CLTA (P = 0.0002), EGFR (), PIK3CA (P = 0.007) and UBA52 () genes and the predicted responses. For prostate cancer, we found that the predicted responses have statistically significant associations with the expression of AKT1 (P = 0.02), CDK1 (P = 0.01), RICTOR (P = 0.0002), CREB1 (P = 0.02) and CSK (P = 0.01). In kidney cancer, expression of EGFR (P = 0.04) gene had association with the predicted response. In lung cancer, we observed significant associations for CDC42 (P = 0.04), EGFR () and PRKAR2A (P = 0.01) genes. However, for bladder and pancreatic cancers, we did not observe any significant associations.
4 Discussion
In this paper, we proposed MOLI method based on deep neural networks to predict drug response. MOLI integrates somatic mutation, CNA and gene expression data and predicts the drug responses. To the best of our knowledge, MOLI is the first end-to-end method for MOLI with deep neural networks that utilize a combined cost function. Our experiments showed that MOLI with its combined cost function can achieve better performance than single-omics and early integration multi-omics methods based on deep neural networks. We also observed that transfer learning for targeted drugs improves the prediction performance compared to drug-specific inputs. To the best of our knowledge, this is the first method to use transfer learning with a pan-drug approach for targeted drugs. Finally, we analyzed MOLI’s predictions for drugs targeting the EGFR pathway on breast, kidney, lung and prostate cancer patients in TCGA. We showed that MOLI’s predictions have statistically significant associations with the level of expression for some of the genes in the EGFR pathway, including the EGFR gene itself, for breast, kidney and lung cancers.
We would like to point out the following directions for future research.
Although we used only somatic mutation, CNA and gene expression data in our experiments, MOLI can be extended for integrating other omics data types. For example, proteomics data can be a good candidate because it has been shown to be a contributing factor in pan-cancer drug response prediction (Ali et al., 2018) and is known to be in concordance with the other omics data types (Gonçalves et al., 2017; Ryan et al., 2017). We performed experiments on transfer learning only for the drugs that target EGFR, but this approach is also applicable for other families of targeted drugs if multi-omics data are available for external validation. Another advantage of the pan-drug approach is that there is no need to train separate pan-drug models for each EGFR inhibitor, and one model can be validated on different external datasets. In the drug-specific approach, we trained one model on Cetuximab data and another one on Erlotinib data, and could not validate them on each other’s external validation data. However, in the pan-drug approach, we trained one model for all of the EGFR inhibitors and validated it on both Cetuximab and Erlotinib data.
While we studied only the triplet loss for optimizing the concatenated representation, we note that this loss function can be replaced by other similar losses such as the contrastive loss function which was used in the Siamese network (Hadsell et al., 2006). We trained separate MOLI models for different drugs, but it is an interesting direction for future research to utilize multi-task learning (Yuan et al., 2016) and predict the outcome for multiple drugs at the same time. Unlike areas such as medical imaging, transfer learning is yet to be explored in genomics, especially cancer genomics (Ching et al., 2018). While in this paper we explored transferring related samples (also known as instance-transfer), other aspects of transfer learning such as relational-knowledge-transfer (Pan and Yang, 2010) should be explored in the future.
In all of the experiments and utilized datasets, we used pan-cancer inputs. The advantage of using pan-cancer multi-omics input is that it can address, to some degree, the challenge of inter-tumor heterogeneity (Almendro et al., 2013). However, these datasets are not suitable for addressing intra-tumor heterogeneity, which would require other resources such as single cell data. Geeleher et al. (2014) showed that training on non-breast cancer cell lines in addition to breast cancer cell lines leads to improved prediction accuracy. However, predictions were tested only on breast cancer clinical trial cohort data and only for Docetaxel, a primary treatment for breast cancer. Because some drug-cancer event associations are specific to the tissue of origin and are less detectable in pan-cancer settings (Iorio et al., 2016), we believe that further research in this area is required to study the performance of pan-cancer versus cancer-specific training data for a more diverse range of cancer types and for more drugs.
We would like to point out the following limitations of this study:
The datasets used were from different resources were not in the same format and required substantial pre-processing and standardization (see the Supplementary Material). For example, different studies used different pipelines to detect CNA and reported different estimates of copy number which could not be compared directly. A similar issue was also observed for the drug response. While the GDSC cell lines used IC50 as the response measure, the majority of datasets used other metrics to measure the response. For example, the PDX dataset used tumor volume based on RECIST criteria to define responders and non-responders. Therefore, lack of standardization on both the input and the output side adds extra challenges to the drug response prediction task.
In this study we focused on monotherapy and did not explore the effect of the combination of drugs.
We did not discriminate between driver and passenger events in the somatic mutation and CNA data and treated all of them similarly. However, in reality, the majority of these genomic alterations seems to have no impact on cancer development (Vogelstein et al., 2013) and might appear just by chance. Therefore, in future work, we plan to use another format for these data types to distinguish between potential driver and passenger events.
All of the datasets used suffered from severely unbalanced class distributions, since the number of responders was much smaller than the number of non-responders. We addressed this problem by oversampling the minority class. However, this approach often causes overfitting particularly for deep neural networks with many parameters. We reduced overfitting with strong regularization such as high dropout rate and weight decay. Moreover, using triplets as input of the network increased the number of samples and led to a more stable network, due to the large number of different combinations for triplets.
As a first investigation of late integration, we did not consider interactions between genes in different omics data types in MOLI or the compared baselines. In reality, genes do not function in isolation and work in biological networks and interact with each other. Recently, Ma et al. (2018) have shown that incorporating biological domain knowledge from the Gene Ontology leads to more interpretable neural networks with performance comparable to those of purely data-driven neural networks. Therefore, incorporating domain expert knowledge to MOLI via deep neural networks is a promising future direction.
5 Conclusion
In this paper, we proposed MOLI, a method for drug response prediction based on deep neural networks and MOLI. We trained MOLI on a pan-cancer cell line dataset and successfully validated it on PDX and patient data for five chemotherapy agents and two targeted therapeutics.
Our results suggest four major findings:
MOLI outperforms single-omics (gene expression) prediction performance in terms of AUC and area under precision–recall curve.
MOLI outperforms deep neural networks using early integration in terms of AUC and area under precision–recall curve.
MOLI with its combined cost function outperforms single- and multi-omics baselines with only the classification loss.
MOLI trained on the pan-drug inputs, employing transfer learning, outperforms MOLI trained on drug-specific inputs for targeted therapeutics that target EGFR.
Finally, we analyzed the biological significance of MOLI and found substantial evidence that the responses predicted by MOLI have statistically significant associations with the expression level of numerous genes in the EGFR pathway for TCGA patients with breast, kidney, lung and prostate cancers.
In conclusion, our experimental results suggest MOLI may have a role in precision oncology where currently only ∼5% of all patients benefit from precision oncology.
Supplementary Material
Acknowledgements
We would like to thank Hossein Asghari, Baraa Orabi, Raunak Shrestha and Yen-Yi Lin (Vancouver Prostate Centre), Anne-Marie Therien-Daniel, Nazanin Mehrasa and Mehrdad Mansouri (Simon Fraser University), Paul Geeleher (St Jude Children’s Research Hospital) and Lukas Folkman (CeMM) for their kind supports. We also would like to thank Compute Canada, WestGrid and Vancouver Prostate Centre for providing us with the computational resources for this research.
Authors’ contributions
Study concept and design: H.S.-N., C.C.C., M.E.
Deep learning design, implementations and analysis: H.S.-N.
Data pre-processing, analysis and interpretation: O.Z.
Analysis and interpretation of results: H.S.-N., O.Z.
Drafting of the manuscript: All authors read and approved the final manuscript.
Supervision: C.C.C., M.E.
Funding
This work was supported by Canada Foundation for Innovation (33440 to C.C.C.), The Canadian Institutes of Health Research (PJT-153073 to C.C.C.), Terry Fox Foundation (201012TFF to C.C.C.), International DFG Research Training Group GRK 1906 (to support O.Z.) and a Discovery Grant from the National Science and Engineering Research Council of Canada R611347 (to H.S.-N. and M.E.).
Conflict of Interest: none declared.
References
- Ali M. et al. (2018) Global proteomics profiling improves drug sensitivity prediction: results from a multi-omics, pan-cancer modeling approach. Bioinformatics, 34, 1353–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almendro V. et al. (2013) Cellular heterogeneity and molecular evolution in cancer. Ann. Rev. Pathol., 8, 277–302. [DOI] [PubMed] [Google Scholar]
- Argelaguet R. et al. (2018) Multi-omics factor analysis a framework for unsupervised integration of multi-omics data sets. Mol. Syst. Biol., 14, e8124.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J. et al. (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature, 483, 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary K. et al. (2018) Deep learning-based multi-omics integration robustly predicts survival in liver cancer. Clin. Cancer Res., 24, 1248–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M.L. et al. (2018) Clinical tumour sequencing for precision oncology: time for a universal strategy. Nat. Rev. Cancer, 18, 527–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching T. et al. (2018) Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface, 15, 20170387.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichocki A., Phan A.-H. (2009) Fast local algorithms for large scale nonnegative matrix and tensor factorizations In: IEICE Transactions on Fundamentals of Electronics, Communications and Computer Sciences. Vol. 92, The Institute of Electronics, Information and Communication Engineers, Tokyo, Japan, pp. 708–721. [Google Scholar]
- Dimitrakopoulos C. et al. (2018) Network-based integration of multi-omics data for prioritizing cancer genes. Bioinformatics, 34, 2441–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M.Q. et al. (2018) Precision oncology beyond targeted therapy: combining omics data with machine learning matches the majority of cancer cells to effective therapeutics. Mol. Cancer Res., 16, 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z. et al. (2016) Evaluating the molecule-based prediction of clinical drug responses in cancer. Bioinformatics, 32, 2891–2895. [DOI] [PubMed] [Google Scholar]
- Duchi J. et al. (2011) Adaptive subgradient methods for online learning and stochastic optimization. J. Mach. Learn. Res., 12, 2121–2159. [Google Scholar]
- Févotte C., Idier J. (2011) Algorithms for nonnegative matrix factorization with the β-divergence. Neural Comput., 23, 2421–2456. [Google Scholar]
- Gao H. et al. (2015) High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med., 21, 1318–1325. [DOI] [PubMed] [Google Scholar]
- Gavan S.P. et al. (2018) The economic case for precision medicine. Expert Rev. Precis. Med. Drug Dev., 3, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeleher P. et al. (2014) Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol., 15, R47.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeleher P. et al. (2017) Discovering novel pharmacogenomic biomarkers by imputing drug response in cancer patients from large genomics studies. Genome Res., 27, 1743–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves E. et al. (2017) Widespread post-transcriptional attenuation of genomic copy-number variation in cancer. Cell Syst., 5, 386–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I. et al. (2016) Deep Learning. Vol. 1 MIT Press, Cambridge. [Google Scholar]
- Graim K. et al. (2019) PLATYPUS: a multiple-view learning predictive framework for cancer drug sensitivity prediction. Pac. Symp. Biocomput., 24, 136–147. [PMC free article] [PubMed] [Google Scholar]
- Hadsell R. et al. (2006) Dimensionality reduction by learning an invariant mapping. In: IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR'06), New York, NY, pp. 1735–1742.
- He X. et al. (2018) Kernelized rank learning for personalized drug recommendation. Bioinformatics, 34, 2808–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio F. et al. (2016) A landscape of pharmacogenomic interactions in cancer. Cell, 166, 740–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R.A. et al. (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res., 31, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W.E. et al. (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics, 8, 118–127. [DOI] [PubMed] [Google Scholar]
- Khakabimamaghani S., Ester M. (2016) Bayesian biclustering for patient stratification In: CC BY: PSB 2016 proceedings are published as Open Access chapters by World Scientific Publishing Company and distributed under the terms of the Creative Commons Attribution (CC BY) 4.0 License. World Scientific, pp. 345–356. [Google Scholar]
- Lee J.-K. et al. (2018) Pharmacogenomic landscape of patient-derived tumor cells informs precision oncology therapy. Nat. Genet., 50, 1399–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. (2011) RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics, 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M. et al. (2015) Integrative data analysis of multi-platform cancer data with a multimodal deep learning approach. IEEE/ACM Trans. Comput. Biol. Bioinform., 12, 928–937. [DOI] [PubMed] [Google Scholar]
- Ma J. et al. (2018) Using deep learning to model the hierarchical structure and function of a cell. Nat. Methods, 15, 290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquart J. et al. (2018) Estimation of the percentage of US patients with cancer who benefit from genome-driven oncology. JAMA Oncol., 4, 1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A., Verma M. (2010) Cancer biomarkers: are we ready for the prime time? Cancers, 2, 190–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Q. et al. (2013) Pattern discovery and cancer gene identification in integrated cancer genomic data. Proc. Natl. Acad. Sci. USA, 110, 4245–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S.J., Yang Q. (2010) A survey on transfer learning. IEEE Trans. Knowl. Data Eng., 22, 1345–1359. [Google Scholar]
- Rappoport N., Shamir R. (2018) Multi-omic and multi-view clustering algorithms: review and cancer benchmark. Nucleic Acids Res., 46, 10546–10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan C.J. et al. (2017) A compendium of co-regulated protein complexes in breast cancer reveals collateral loss events. Cell Syst., 5, 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroff F. et al. (2015) FaceNet: a unified embedding for face recognition and clustering In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, pp. 815–823. [Google Scholar]
- Shrestha R. et al. (2017) HIT’nDRIVE: patient-specific multidriver gene prioritization for precision oncology. Genome Res., 27, 1573–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. et al. (2019) DIABLO: an integrative approach for identifying key molecular drivers from multi-omic assays. Bioinformatics, [Epub ahead of print, doi:10.1093/bioinformatics/bty1054, January 18, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B. et al. (2013) Cancer genome landscapes. Science, 339, 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B. et al. (2014) Similarity network fusion for aggregating data types on a genomic scale. Nat. Methods, 11, 333–337. [DOI] [PubMed] [Google Scholar]
- Weinstein J.N. et al. (2013) The Cancer Genome Atlas pan-cancer analysis project. Nature Genet., 45, 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. et al. (2012) Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res., 41, D955–D961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H. et al. (2016) Multitask learning improves prediction of cancer drug sensitivity. Sci. Rep., 6, 31619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehir A. et al. (2017) Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10, 000 patients. Nature Medicine, 23, 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitnik M. et al. (2019) Machine learning for integrating data in biology and medicine: principles, practice, and opportunities. Inf. Fusion, 50, 71–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.