Abstract
Ischemic cardiac disease is the leading cause of morbidity and mortality in the world. Despite the great efforts and progress in cardiac research, the current treatment of cardiac ischemia reperfusion injury (I/RI) is still far from being satisfactory. This study was performed to investigate the role of long non-coding RNA (lncRNA) H19 in regulating myocardial I/RI. We found that H19 expression was downregulated in the I/R hearts of mice and cardiomyocytes treated with H2O2. Overexpression of H19 alleviated myocardial I/RI of mice and cardiomyocyte injury induced by H2O2. We found that H19 functioned as a competing endogenous RNA of miR-877-3p, which decreased the expression of miR-877-3p through the base-pairing mechanism. In parallel, miR-877-3p was upregulated in H2O2-treated cardiomyocytes and mouse ischemia reperfusion (I/R) hearts. miR-877-3p exacerbated myocardial I/RI and cardiomyocyte apoptosis. We further established Bcl-2 as a downstream target of miR-877-3p. miR-877-3p inhibited the mRNA and protein expression of Bcl-2. Furthermore, H19 decreased the Bcl-2/Bax ratio at mRNA and protein levels, cytochrome c release, and activation of caspase-9 and caspase-3 in myocardial I/RI mice, which were canceled by miR-877-3p. In summary, the H19/miR-877-3p/Bcl-2 pathway is involved in regulation of mitochondrial apoptosis during myocardial I/RI, which provided new insight into molecular mechanisms underlying regulation of myocardial I/RI.
Keywords: myocardial ischemia reperfusion injury, I/RI, lncRNA H19, miR-877-3p, Bcl-2, mitochondrial apoptosis
Graphical Abstract
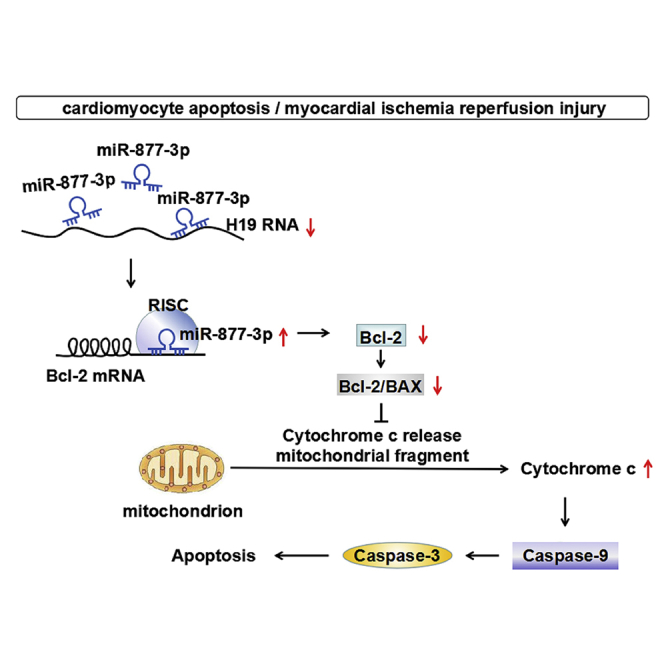
Introduction
Thrombolytic therapy or primary percutaneous coronary intervention is the most effective therapeutic intervention for patients with acute myocardial infarction and significantly reduces myocardial infarct size and mortality.1 However, the progress of myocardial reperfusion itself can induce injury to the myocardium, and such phenomenon is known as myocardial ischemia reperfusion injury (I/RI). Myocardial I/RI contributes to the final lethal myocardial infarct size and can develop into heart failure.2 Clinical treatment of myocardial I/RI is still a challenge, partly because the understanding of the mechanisms of myocardial I/RI remains to be further improved. Ischemia and reperfusion can induce cardiomyocyte necrosis and apoptosis.3 Multiple alterations occurring after reperfusion can induce cardiomyocyte apoptosis, such as a high level of intracellular ATP, calcium overload, and accumulation of free radicals in cardiac cells.4 In mammals, cell apoptosis can be initiated by the extrinsic death receptor pathway and the intrinsic mitochondrial apoptotic pathway. The mitochondrial apoptotic pathway has been recognized as playing key roles in a variety of cardiovascular disease including myocardial I/RI,5 heart failure,6 and cardiomyopathy.7, 8
Recently, long non-coding RNAs (lncRNAs), a class of non-coding RNAs that are more than 200 nt in length and possess no significant protein-coding ability, have emerged as a new layer of gene expression regulation.9 lncRNAs either negatively or positively regulate the expression of protein-coding genes through diverse modes of action. One of them is the interaction between lncRNA and microRNA, a newly defined mechanism called competing endogenous RNAs (ceRNAs).10, 11 Specifically, lncRNAs can act as ceRNAs by binding microRNAs (miRNAs) through sequence complementary to reduce the availability of functional miRNAs.12, 13 Moreover, both lncRNAs and miRNAs have been significantly implicated in a variety of biological functionalities and pathological processes, including cardiovascular diseases.14
H19 is a conserved lncRNA that is transcripted from the imprinted H19-insulin growth factor 2 (IGF2) locus15, 16 and is located in both the nucleus and the cytoplasm.17 As a ceRNA, scaffold, or guide, H19 has been established as a critical regulator in tumor initiation and progression.18, 19 H19 was also shown to play an important role in various cardiovascular diseases such as myocardial I/RI,20, 21 acute myocardial infarction,22 diabetic cardiomyopathy,23 cardiomyocyte hypertrophy,24 and calcific aortic valve disease.25 Wang et al.20 found that H19 regulates programmed necrosis and myocardial I/RI by targeting miR-103/107 and FADD. Luo et al.21 reported that the lncRNA H19/miR-675 axis regulates myocardial I/RI by targeting PPARαs. Besides, Zhou et al.22 found that H19 protects acute myocardial infarction through activating autophagy in mice. However, the mechanism of H19 on regulating apoptosis triggered by an intrinsic mitochondrial apoptotic pathway in myocardial I/RI is unknown. miR-877-3p, a mammalian miRNA with the mirtron origins,26 has been reported to be deregulated in type 2 diabetic kidney disease,27 osteosarcoma,28 Sjögren’s syndrome,29 and bilateral renal ischemia reperfusion (I/R).30 Our pilot analysis showed that both H19 and miR-877-3p were deregulated in a mouse model of myocardial I/RI, and lncRNA H19 contains a conserved binding site of miR-877-3p (http://regrna2.mbc.nctu.edu.tw/ and http://starbase.sysu.edu.cn/). Furthermore, there are two binding sites of miR-877-3p at the 3′ UTR of Bcl-2 mRNA, which plays an important role in regulating mitochondrial apoptotic pathway and apoptosis.
We therefore designed the present study to investigate the roles of H19 and miR-877-3p in a mitochondrial apoptotic pathway involved in apoptosis of myocardial I/RI. We elucidated that lncRNA H19 acted as a ceRNA of miR-877-3p to exert its anti-apoptosis effects, and Bcl-2 was the target of the miR-877-3p that mediated the effects of H19.
Results
H19 Alleviates Cardiomyocyte Injury Induced by H2O2 and Myocardial I/RI in Mice
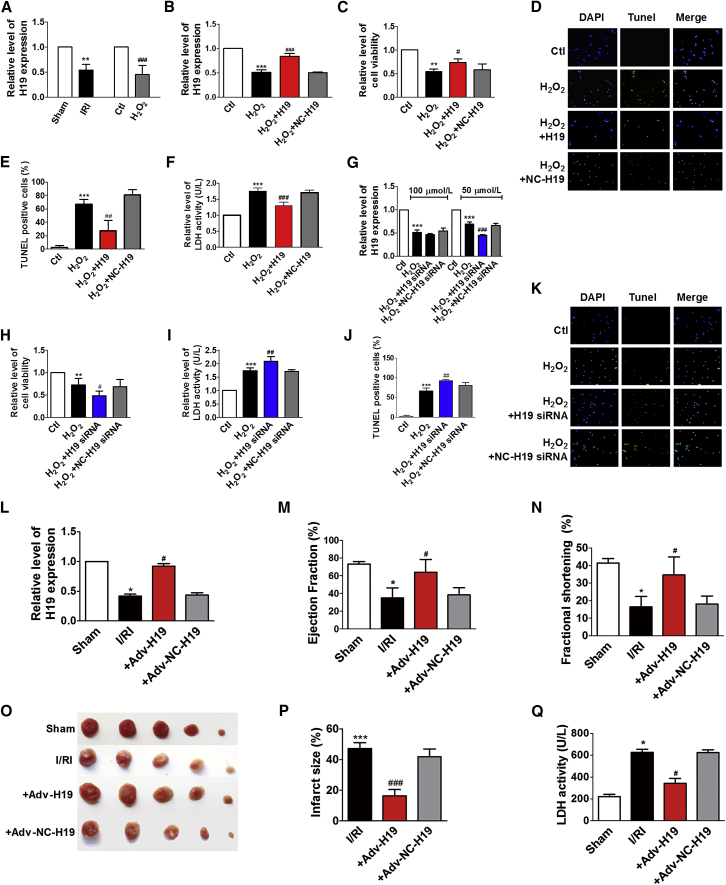
We found that the expression levels of H19 in hearts of I/RI mice and cardiomyocytes treated with 100 μM H2O2 were downregulated by 45.8% and 55.3% compared with controls (Figure 1A). Forced overexpression of H19 (Figure 1B) increased cell viability (Figure 1C) and decreased TUNEL-positive cells (Figures 1D and 1E) and lactate dehydrogenase (LDH) activity (Figure 1F) of 100 μM H2O2-treated cells. The small interfering RNA (siRNA) of H19 reduced H19 level when cardiomyocytes were treated with 50 μM, but not 100 μM H2O2 (Figure 1G). The siRNA of H19 decreased cell viability (Figure 1H) and increased LDH activity (Figure 1I) and TUNEL-positive cells (Figures 1J and 1K). The respective negative controls (NCs) of H19 or its siRNA had no effects on H19 expression and cardiomyocyte injury induced by H2O2.
Figure 1.
H19 Alleviates Cardiomyocyte Injury Induced by H2O2 and Myocardial I/RI in Mice
(A) The relative level of H19 expression of myocardial I/RI mice hearts and cardiomyocytes treated with 100 μM H2O2 (n = 4). The relative level of H19 expression (B, n = 4), cell viability (C, n = 3), TUNEL staining (D, n = 3), TUNEL-positive cells (E, n = 3), and LDH activity (F, n = 4) of cardiomyocytes transfected with H19 overexpression plasmids (H19) or negative control (NC) plasmids in the condition of 100 μM H2O2. (G) The relative levels of H19 expression of cardiac myocytes transfected with H19 siRNA and its NC with the treatment of 50 and 100 μM H2O2 (n = 4). Cell viability (H, n = 5), LDH activity (I, n = 4), TUNEL-positive cells (J, n = 4), TUNEL staining (K, n = 3) of cardiac myocytes transfected with H19 siRNA, and its NC in the 50 μM H2O2 condition. Data are expressed by mean ± SD. **p < 0.01, ***p < 0.001 versus control group; #p < 0.05, ##p < 0.01, ###p < 0.001 versus H2O2-treated group. The relative level of H19 expression (L, n = 8), ejection fraction (EF) (M, n = 8), fractional shortening (FS) (N, n = 8), TTC staining (O, n = 4), infarct size (%) (P, n = 4), and LDH activity (Q, n = 8) of the mice heart in different groups containing sham group (sham), myocardial I/RI group (I/RI), I/RI + Adv-H19 group: mice co-treated with myocardial I/RI operation and infection of adenovirus vectors (Adv-H19) for overexpression H19, and I/RI+Adv-NC-H19 group: mice co-treated with myocardial I/RI operation and infection of NC adenovirus vectors (Adv-NC-H19) for contrast. Data are expressed by mean ± SD. *p < 0.05, ***p < 0.001 versus sham group (sham); #p < 0.05, ###p < 0.001 versus myocardial I/RI group.
To further verify the role of H19 in myocardial I/RI in mice, we then examined the effects of H19-overexpressing adenovirus (Adv-H19) on myocardial I/RI mice. Adv-H19 effectively increased H19 expression by 2.2 ± 0.04-fold in the hearts of I/RI mice (Figure 1L). Overexpression of H19 in myocardial I/RI mice led to improved ejection fraction (EF) and fractional shortening (FS) (Figures 1M and 1N), and reduced myocardial infarct area (Figures 1O and 1P) and LDH activity (Figure 1Q).
H19 Functions as a miR-877-3p ceRNA
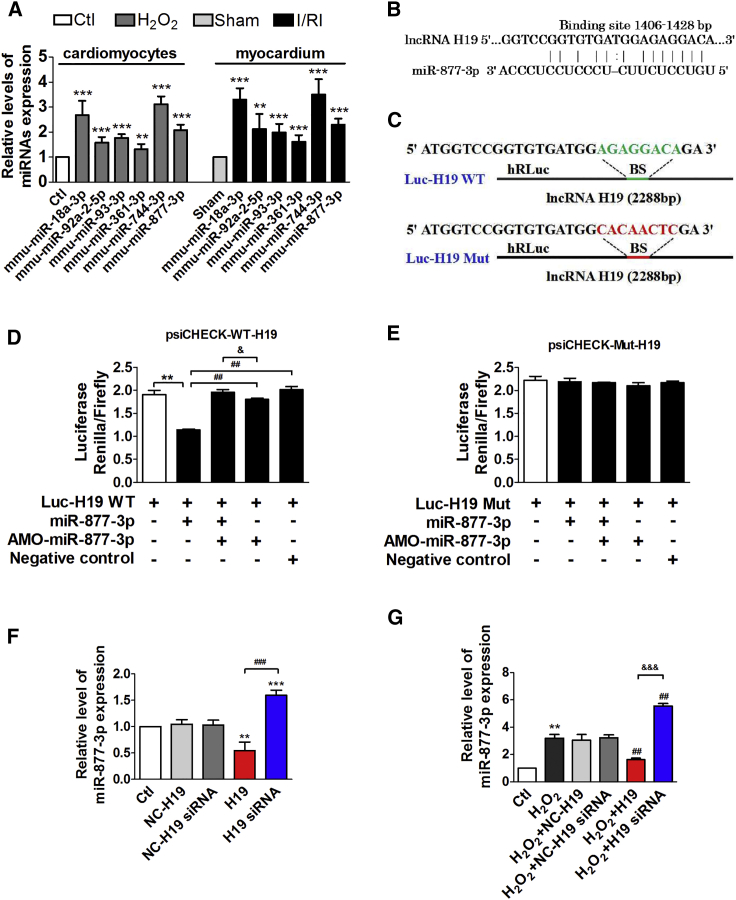
To get insight into the molecular mechanisms for regulation of H19 on myocardial I/RI, we analyzed the potential interacting miRNAs of H19 by using RegRNA2.0 (http://regrna2.mbc.nctu.edu.tw/) and starBase (http://starbase.sysu.edu.cn/), predicted the targets of miRNAs involved in cell apoptosis by using TargetScan (http://www.targetscan.org/vert_72/), and further screened the target miRNAs including miR-18a-3p, miR-92a-2-5p, miR-93-3p, miR-361-3p, miR-744-3p, and miR-877-3p according to the conservation between human and mouse species. qPCR data showed that miR-18a-3p, miR-744-3p, and miR-877-3p were the top three that are significantly upregulated in cardiomyocytes treated with H2O2 and I/RI hearts of mice (Figure 2A), and miR-877-3p is of the highest expression level (according to the cycle number of miRNAs in Figure S1). Therefore, miR-877-3p was chosen for further study. lncRNA H19 contains a target site of miR-877-3p at the position of 1,406–1,428 (Figure 2B). We constructed two luciferase plasmids carrying the full-length (2,288 bp) H19 (Luc-H19-WT [wild type]) and the mutated H19 (Luc-H19-Mut) (Figure 2C). Dual-luciferase reporter gene assay showed that the relative activity of firefly luciferase of Luc-H19-WT in HEK293 cells was significantly decreased by co-transfection of miR-877-3p, whereas it did not change the luciferase activity of Luc-H19-Mut (Figures 2D and 2E).
Figure 2.
H19 Functions as a miR-877-3p ceRNA
(A) The relative levels of miRNA expression in cardiomyocytes treated with H2O2 and I/R myocardium (n = 5). **p < 0.01, ***p < 0.001 versus control group or sham group. (B) The sequences and the position of binding site between H19 and miR-877-3p. (C) The sequences and positions of H19 containing wild type and the mutant binding site of miR-877-3p that cloned into the psiCHECK-2 luciferase reporter vector. (D) The dual-luciferase reporter assays between the wild type of H19 and miR-877-3p or AMO-miR-877-3p. (E) The dual luciferase reporter assays between the mutant luciferase plasmid of H19 and miR-877-3p or AMO-miR-877-3p (n = 3). **p < 0.01 versus control group; ##p < 0.01 versus Luc-H19 WT + miR-877-3p group; &p < 0.05 versus Luc-H19 WT + miR-877-3p + AMO-miR-877-3p group. (F) Relative level of miR-877-3p expression of cardiac myocytes transfected with the overexpression plasmids or siRNA of H19 and the respective NCs (n = 3). **p < 0.01, ***p < 0.001 versus control group; ###p < 0.001 versus H19 group. (G) Relative level of miR-877-3p expression of cardiac myocytes transfected with the overexpression plasmids or siRNA of H19 and the respective NCs in the condition of H2O2 treatment (n = 3). **p < 0.01 versus control group; ##p < 0.01 versus H2O2-treated group; &&&p < 0.001 versus H2O2 + H19 group. Data are expressed by mean ± SD.
In addition, H19 overexpression significantly inhibited miR-877-3p expression by 45.6% and 45.9% under both physiological and H2O2 treatment conditions in neonatal mouse cardiomyocytes, respectively. When H19 was silenced, miR-877-3p expression level was dramatically increased by 1.6 ± 0.1-fold compared with control cells and by 1.9 ± 0.2-fold compared with H2O2-treated cardiomyocytes (Figures 2F and 2G). Meanwhile, the respective NCs of H19 and its siRNA had no effect on miR-877-3p expression.
miR-877-3p Exacerbates H2O2-Induced Cardiomyocyte Injury and Myocardial I/RI in Mice
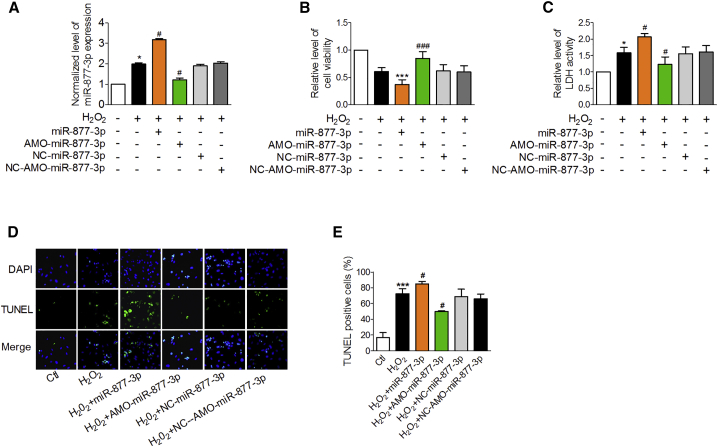
We then evaluated the effects of miR-877-3p on cardiac injury. miR-877-3p was significantly upregulated in the hearts of I/RI mice and H2O2-treated cardiomyocytes (Figure 2A). miR-877-3p overexpression (Figure 3A) markedly aggravated H2O2-induced cardiomyocyte death as revealed by decreased cell viability, and increased activity of LDH and apoptotic cell death (Figures 3B–3E). In contrast, anti-miRNA-oligos (AMOs)-miR-877-3p effectively reduced miR-877-3p expression (Figure 3A) and alleviated H2O2-induced cell injury (Figures 3B–3E).
Figure 3.
miR-877-3p Contributes to H2O2-Induced Cardiomyocyte Death
(A–E) Relative level of miR-877-3p (A, n = 3), cell viability (B, n = 6), LDH activity (C, n = 3), TUNEL staining (D, n = 3), and TUNEL-positive cells (E, n = 3) in cardiac myocytes co-treated with miR-877-3p mimic, AMO-miR-877-3p, NC-miR-877-3p, or NC-AMO-miR-877-3p transfection and H2O2. *p < 0.05, ***p < 0.001 versus control group (Ctl); #p < 0.05, ###p < 0.001 versus H2O2-treated group (H2O2). Data are expressed by mean ± SD.
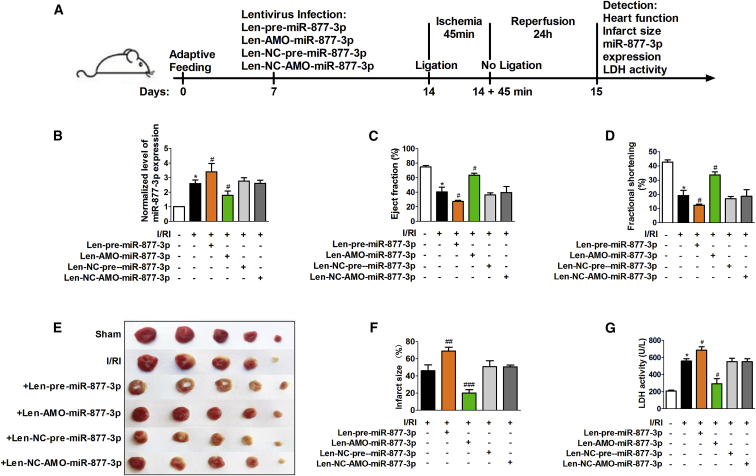
To better understand the role of miR-877-3p in cardiac injury, we performed the experiments in mice as illustrated in Figure 4A. Consistently, the expression of miR-877-3p in I/RI mouse hearts was significantly increased by 2.6 ± 0.2-fold compared with sham-operated mice (Figure 4B). Administration of lentivirus vectors expressing the precursors of miR-877-3p (Len-pre-miR-877-3p) increased the expression of mature miR-877-3p, and lentivirus vectors expressing AMO-miR-877-3p (Len-AMO-miR-877-3p) reduced miR-877-3p expression in mouse hearts, whereas the NC lentivirus vectors (Len-NC-pre-miR-877-3p and Len-NC-AMO-miR-877-3p) had no effects (Figure 4B). Echocardiographic assessment demonstrated significant deterioration of cardiac function in I/RI mice with Len-pre-miR-877-3p administration and improvement of cardiac function by Len-AMO-miR-877-3p application (Figures 4C and 4D). The infarct area of hearts was significantly increased in I/RI mice, which was further increased by 1.5 ± 0.1-fold upon Len-pre-miR-877-3p treatment, and markedly decreased by 26.0% in the Len-AMO-miR-877-3p-infected mice compared with myocardial I/RI mice (Figures 4E and 4F). Consistently, serum LDH in the myocardial I/RI mice was further reduced by Len-AMO-miR-877-3p, but elevated by Len-pre-miR-877-3p (Figure 4G). These data imply that miR-877-3p plays a detrimental role in myocardial I/RI.
Figure 4.
miR-877-3p Exacerbates Myocardial I/RI in Mice
(A) The schematic diagram of the experimental process for verifying the function of miR-877-3p in myocardial ischemia reperfusion injury (I/RI) mice. Normalized level of miR-877-3p expression (B, n = 8), EF (C, n = 8), FS (D, n = 8), hearts with TTC staining (E), infarct size (F, n = 3), and LDH activity (G, n = 8) of the mice in different groups containing sham group, myocardial I/RI group (I/RI), I/RI+Len-pre-miR-877-3p group, I/RI+Len-AMO-miR-877-3p group, I/RI+Len-NC-pre-miR-877-3p group, and I/RI+Len-NC-AMO-miR-877-3p group. *p < 0.05 versus sham group (sham); #p < 0.05, ##p < 0.01, ###p < 0.001 versus myocardial I/RI group. Data are expressed by mean ± SD.
Bcl-2 Is a Downstream Target of miR-877-3p
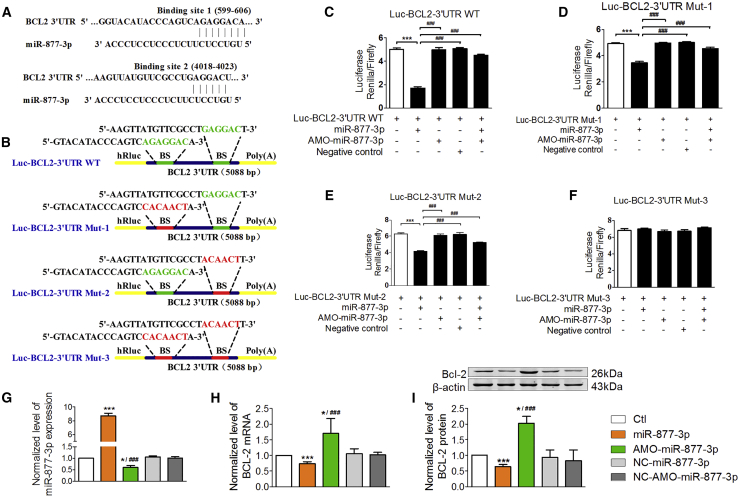
To explore the downstream signaling mechanisms for the pro-apoptotic action of miR-877-3p in cardiomyocytes, we conducted computational analysis (TargetScan, miRanda, miRDB) to search for apoptosis-related genes. The predicted result showed that there are two binding sites of miR-877-3p at the 3′ UTR of Bcl-2 mRNA (Figure 5A). We then constructed luciferase plasmids carrying the wild-type 3′ UTR (5,088 bp) of Bcl-2 mRNA (Luc-Bcl-2-3′ UTR WT), the 3′ UTR of Bcl-2 with a single-binding-site mutation at the 599–606 position (Luc-Bcl-2-3′ UTR Mut-1), the 3′ UTR of Bcl-2 with a single-binding-site mutation at the 4,018–4,023 position (Luc-Bcl-2-3′ UTR Mut-2), and the 3′ UTR of Bcl-2 with a double-binding-site mutation at both 599–606 and 4,018–4,023 positions (Luc-Bcl-2-3′ UTR Mut-3). The specific sites of mutation are shown in Figure 5B. Dual-luciferase reporter gene assay showed that the relative activity of firefly luciferase was significantly decreased with miR-877-3p co-transfected with Luc-Bcl-2-3′ UTR WT, Luc-Bcl-2-3′ UTR Mut-1, or Luc-Bcl-2-3′ UTR Mut-2 in HEK293 cells. Each of the single-binding-site mutant constructs exhibited diminished changes, whereas the double-binding-site mutant construct almost completely wiped out the changes of luciferase activities induced by miR-877-3p (Figures 5C–5F). This indicated that miR-877-3p can act on the 3′ UTR of Bcl-2 mRNA at both binding sites. In addition, knockdown of miR-877-3p by its AMO resulted in upregulation of Bcl-2 expression at both mRNA and protein levels, whereas overexpression of miR-877-3p did the opposite (Figures 5G–5I). These data demonstrated that Bcl-2 is a downstream target of miR-877-3p.
Figure 5.
Bcl-2 Is a Downstream Target of miR-877-3p
(A) The sequences and the position of two binding sites of miR-877-3p at the 3′ UTR of Bcl-2 mRNA. (B) The sequences and positions of the Bcl-2 3′ UTR containing wild-type and the mutant binding site of miR-877-3p that cloned to the psiCHECK-2 luciferase reporter vector. (C) The dual-luciferase reporter assays between the wild-type plasmid containing the whole length of the 3ʹ UTR of Bcl-2 and miR-877-3p or AMO-miR-877-3p or NC of miR-877-3p. (D) The dual-luciferase reporter assays between the mutant luciferase plasmid 1, (E)mutant luciferase plasmid 2, and (F) mutant luciferase plasmid 3 containing the whole length of the 3′ UTR of Bcl-2 and miR-877-3p or AMO-miR-877-3p or NC of miR-877-3p. ***p < 0.001 versus control group; ###p < 0.001 versus miR-877-3p group. (G–I) The normalized expression level of miR-877-3p (G, n = 3), Bcl-2 mRNA (H, n = 5), and protein (I, n = 3) of cardiac myocytes transfected with miR-877-3p mimics, AMO-miR-877-3p, NC-miR-877-3p, or NC-AMO-miR-877-3p. *p < 0.05, ***p < 0.001 versus control group; ###p < 0.001 versus miR-877-3p group.
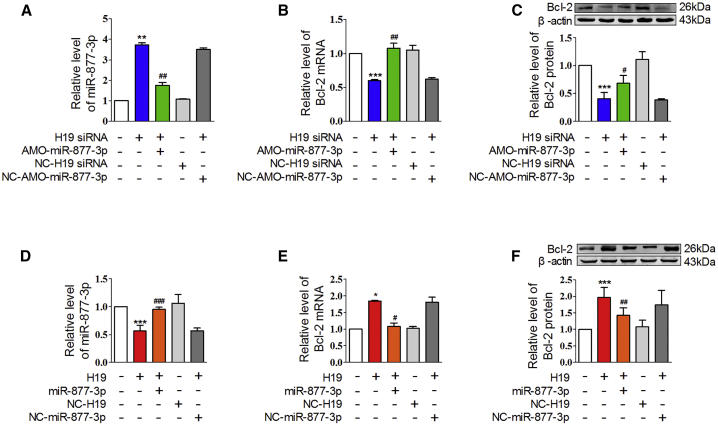
We further examined the action of H19 on Bcl-2, the downstream target of miR-877-3p. The H19 siRNA increased the expression of miR-877-3p and decreased Bcl-2 expression at both mRNA and protein levels in normal cardiomyocytes, which were canceled by co-administration of AMO-miR-877-3p (Figures 6A–6C). In contrast, H19 overexpression decreased the level of miR-877-3p and increased Bcl-2 mRNA and protein expression, which were canceled by miR-877-3p administration (Figures 6D–6F). These data indicated that H19 could regulate Bcl-2 expression as the ceRNA of miR-877-3p.
Figure 6.
H19 Regulated Bcl-2 mRNA and Protein Expression through miR-877-3p
(A–C) The relative levels of miR-877-3p (A, n = 3), Bcl-2 mRNA (B, n = 3), and protein (C, n = 4) of cardiomyocytes co-treated with H19 siRNA and AMO-miR-877-3p. **p < 0.01, ***p < 0.001 versus control group; #p < 0.05, ##p < 0.01 versus siRNA of H19 group (H19 siRNA). (D–F) The relative level of miR-877-3p (D, n = 3), Bcl-2 mRNA (E, n = 3), and Bcl-2 protein (F, n = 6) of cardiac myocytes co-treated with H19 overexpression plasmid and miR-877-3p. *p < 0.05, ***p < 0.001 versus control group; #p < 0.05, ##p < 0.01, ###p < 0.001 versus H19 group (H19).
miR-877-3p Exerts Its Function through the Bcl-2-Mediated Mitochondrial Apoptotic Pathway
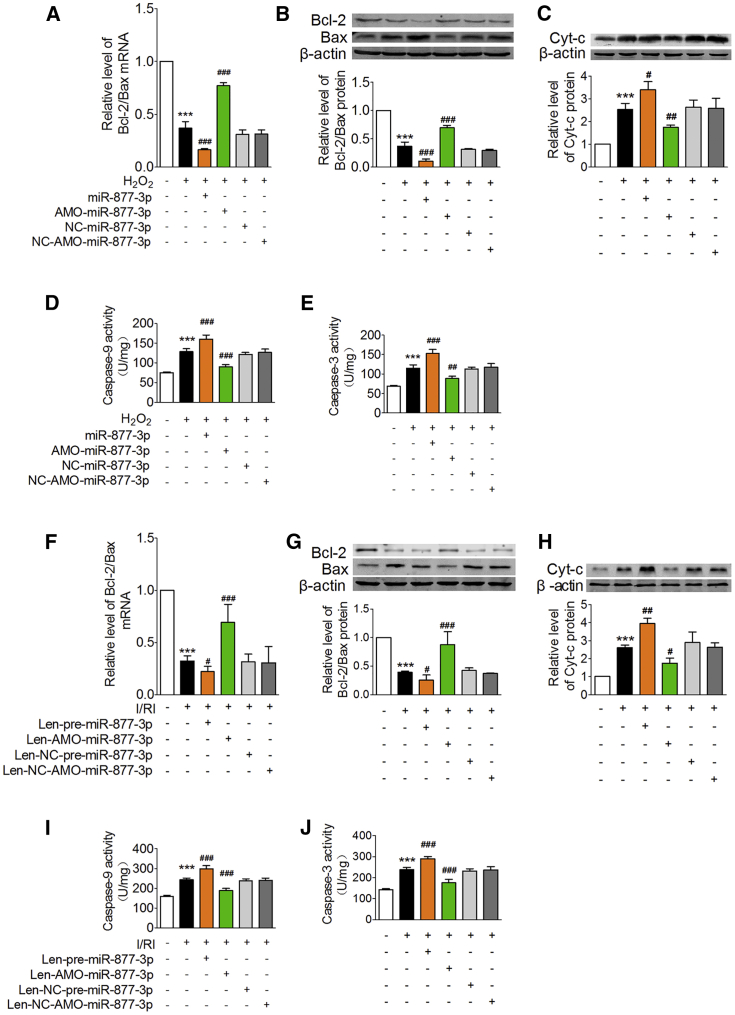
The Bcl-2 protein family determines the fate of cells to apoptosis.31 Reduction of the Bcl-2/Bax ratio can trigger cytochrome c (Cyt-c) release from mitochondria to activate caspase-9 and eventually activate caspase-3, an executioner of apoptotic cell death.32 We found that the Bcl-2/Bax ratio was significantly decreased at both mRNA and protein levels in cardiomyocytes treated with H2O2, which was aggravated by miR-877-3p overexpression and counteracted by AMO-miR-877-3p (Figures 7A and 7B). On the other hand, the protein level of Cyt-c in cytoplasm and the activity of caspase-9 and caspase-3 were increased in cardiomyocytes exposed to H2O2, and these deleterious alterations were reversed by AMO-miR-877-3p, but further exacerbated by miR-877-3p (Figures 7C–7E). NC-miR-877-3p and NC-AMO-miR-877-3p produced no effects.
Figure 7.
miR-877-3p Exerts Its Function through the Bcl-2-Mediated Mitochondrial Apoptotic Pathway
(A–E) The relative level of Bcl-2/Bax mRNA expression (A, n = 3), Bcl-2/Bax protein expression (B, n = 3), cytochrome c (Cyt-c) protein expression in cytoplasm (except Cyt-c in mitochondria) (C, n = 3), caspase-9 activity (D, n = 4), and caspase-3 activity (E, n = 4) of cardiomyocytes transfected with miR-877-3p mimic, AMO-miR-877-3p, NC-miR-877-3p, and NC-AMO-miR-877-3p in the condition of H2O2. ***p < 0.001 versus control group; #p < 0.05, ##p < 0.01, ###p < 0.001 versus H2O2-treated group. (F–J) Relative level of Bcl-2/Bax mRNA expression (F, n = 5), Bcl-2/Bax protein expression (G, n = 4), Cyt-c expression in cytoplasm (H, n = 3), caspase-9 activity (I, n = 5), and caspase-3 activity (J, n = 5) of the mice heart in different groups including sham group, myocardial I/RI group (I/RI), I/RI+Len-pre-miR-877-3p group, I/RI+Len-AMO-miR-877-3p group, I/RI+Len-NC-pre-miR-877-3p group, and Len-NC-AMO-miR-877-3p group. ***p < 0.001 versus sham group; #p < 0.05, ##p < 0.01, ###p < 0.001 versus myocardial I/RI group. Data are expressed by mean ± SD.
Then, we investigated the effect of miR-877-3p on the mitochondrial apoptotic pathway in myocardial I/RI mice. We found that the Bcl-2/Bax ratios at mRNA and protein levels were significantly decreased in the heart of I/RI mice, which were further exaggerated by Len-pre-miR-877-3p and restored by Len-AMO-miR-877-3p (Figures 7F and 7G). Concomitantly, the protein level of Cyt-c and the activity of caspase-9 and caspase-3 were remarkably increased in the heart of I/RI mice, which were further elevated by Len-pre-miR-877-3p and suppressed by Len-AMO-miR-877-3p (Figures 7H–7J). These data imply that miR-877-3p contributes to cardiomyocytes injury induced by H2O2 and myocardial I/RI via Bcl-2 and its mediated mitochondrial apoptotic pathway.
H19 Protects Ischemia and Reperfusion Heart through the miR-877-3p/Bcl-2/Mitochondrial Apoptotic Pathway
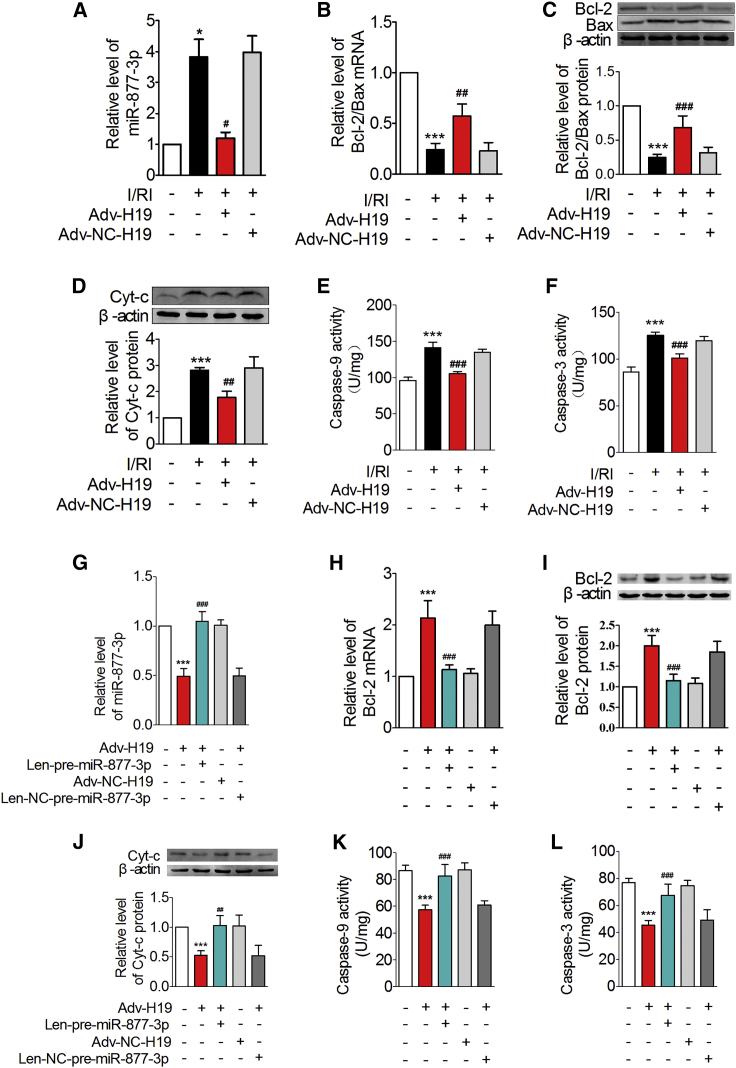
We next explored whether miR-877-3p and its target Bcl-2 mediate the effects of H19. miR-877-3p level was significantly increased in I/RI hearts relative to the normal hearts, which were inhibited by the administration of Adv-H19 (Figure 8A). Adv-H19 restored the levels of Bcl-2/Bax mRNA and protein, suppressed the protein expression level of Cyt-c, and reduced the activity of caspase-9 and caspase-3 in I/RI mice (Figures 8B–8F). The above actions of Adv-H19 on miR-877-3p/Bcl-2/mitochondrial apoptotic pathway were abolished by Len-pre-miR-877-3p (Figures 8G–8L). These data indicate that H19 inhibits the mitochondrial apoptotic pathway and alleviates myocardial I/RI through acting on miR-877-3p and its target Bcl-2 sequentially.
Figure 8.
H19 Protects I/R Heart through the miR-877-3p/Bcl-2/Mitochondrial Apoptotic Pathway
(A–F) Relative level of miR-877-3p expression (A, n = 8), Bcl-2/Bax mRNA expression (B, n = 5), Bcl-2/Bax protein expression (C, n = 5), Cyt-c protein expression in cytoplasm (D, n = 3), caspase-9 activity (E, n = 5), and caspase-3 activity (F, n = 5) of mice myocardium infected with the adenovirus that can express H19 in the pathology condition of I/RI. *p < 0.05, ***p < 0.001 versus sham group; #p < 0.05, ##p < 0.01, ###p < 0.001 versus myocardial I/R injury group (I/RI). (G–L) The relative level of miR-877-3p (G, n = 8), Bcl-2 mRNA (H, n = 8), Bcl-2 protein (I, n = 6), Cyt-c in cytoplasm (J, n = 4), caspase-9 activity (K, n = 5), and caspase-3 activity (L, n = 5) of mice heart administered with Adv-H19 and Len-pre-miR-877-3p. ***p < 0.001 versus control group (Ctl); ##p < 0.01, ###p < 0.001 versus Adv-H19 group (Adv-H19). Data are expressed by mean ± SD.
Discussion
Our present study demonstrated that lncRNA H19 participated in regulating myocardial I/RI. We identified H19 as an anti-apoptotic lncRNA by acting as a miR-877-3p ceRNA in myocardial I/RI. We established Bcl-2 as the target of miR-877-3p, which is the downstream effector of the H19/miR-877-3p axis in cardiomyocyte mitochondrion apoptosis in the setting of myocardial I/RI. Studies have demonstrated that lncRNA plays an important role in regulation of protein-coding genes involved in cancer, neurological disease, myocardial fibrosis, myocardial hypertrophy, and myocardial I/RI.33, 34, 35 Wang et al.36 reported that cardiac apoptosis-related lncRNA (CARL) inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by regulating the miR-539/PHB2 pathway. In another study, the same group found that lncRNA necrosis-related factor (NRF) regulates programmed necrosis and myocardial injury during I/R by targeting miR-873.37 H19 has also been demonstrated to be involved in the pathogenesis of other cardiovascular diseases. Liu et al.24 found that the H19-miR-675 axis is a negative regulator of cardiac hypertrophy by targeting calcium-/calmodulin-dependent protein kinase type II subunit delta (CaMKIIδ). Hadji et al.25 reported that the altered DNA methylation of H19 in calcific aortic valve disease promotes mineralization by silencing NOTCH1. Besides, H19 contributes to cardiac fibroblast proliferation and fibrosis, which act in part through repression of DUSP5/ERK1/2.38 Previous studies showed that the expression of H19 can be either increased or decreased in different myocardial diseases. For instance, H19 is upregulated in pathological cardiac hypertrophy and heart failure24, 39 and coronary artery disease,40, 41 whereas it is downregulated in diabetic cardiomyopathy.23 Although H19 was shown to promote apoptosis in the late-stage cardiac differentiation and dilated cardiomyopathy,42 it is anti-apoptosis in diabetic cardiomyopathy.23 These data indicated that the function of H19 in different cardiac diseases may be determined by the specific pathological alterations. We found that H19 was downregulated in I/RI hearts and cardiomyocytes under oxidative stress. H19 reduced apoptosis of cardiomyocytes induced by H2O2 and alleviated the myocardial I/RI in vivo. These data are consistent with other research.20, 21
Like the action mode of other lncRNAs, H19 can also work as a ceRNA by the base complementation mechanism.18 Kallen et al.43 found that the imprinted H19 lncRNA can antagonize let-7. Keniry et al.44 discovered that H19 is a developmental reservoir of miR-675 that suppresses growth and insulin-like growth factor 1 receptor (Igf1r). In this study, we found that H19 contained a binding site for miR-877-3p at the position of 1,406–1,428 bp and acted as a ceRNA to limit the functional availability of miR-877-3p through sequence complementarity. It was reported that miR-877-3p can regulate the production of IL-8 and IL-1β in mesangial cells activated by secretory immunoglobulin A (IgA) from IgA nephropathy patients.45 It is associated with myofibroblast differentiation and bleomycin-induced lung fibrosis by targeting Smad7.46 Besides, upregulation of p16 by miR-877-3p inhibits the proliferation of bladder cancer.47 Our study for the first time found that miR-877-3p participated in cardiomyocyte apoptosis and myocardial I/RI. We found that miR-877-3p was significantly upregulated and aggravated cardiomyocytes injury induced by H2O2 and myocardial I/RI. We further established Bcl-2 as the target of miR-877-3p. The upregulation of miR-877-3p repressed the expression of anti-apoptotic protein Bcl-2 in myocardial I/RI mice.
Apoptosis can be initiated by the extrinsic death receptor pathway and the intrinsic mitochondrion apoptosis pathway.48, 49 The later one has been confirmed to be involved in myocardial I/RI.2 Release of Cyt-c from mitochondria is a key step in the mitochondrial apoptosis pathway. In cardiac myocytes, the intrinsic apoptotic pathway was strictly controlled by expressing various members of the B cell lymphoma 2 (Bcl-2) family, including anti-apoptotic and pro-apoptotic Bcl-2 family proteins. Bax is a pro-apoptotic protein in the Bcl-2 family; it can promote permeabilization and release of Cyt-c and reactive oxygen species (ROS) over the membrane of mitochondrion. This pro-apoptotic protein is in turn activated by BH3-only proteins and is inhibited by Bcl-2.50 Once Cyt-c is released from mitochondria, it binds to apoptotic protease activating factor-1 (Apaf-1) and forms a complex that in turn activates caspase-9, leading to activation of caspase-3 and thereby apoptosis. On the other hand, it changes the symmetry of the plasma membrane and causes the nuclear pyknosis, DNA fragmentation, and cell death.51 Yu and Dong52 speculated that lncRNA H19 may protect cardiomyocytes against acute myocardial infarction (AMI) via anti-apoptosis. Zhang et al.53 hypothesized subsequently that the H19/miR-22-3p axis might be a potential signaling pathway of apoptosis in myocardial I/RI. However, more systematic research needed to be performed to provide ample evidence for their speculation. Furthermore, the mechanism of H19 on regulating apoptosis triggered by the intrinsic mitochondrial apoptotic pathway in myocardial I/RI is still unknown. For the first time, we demonstrated that H19, a ceRNA for miR-877-3p, alleviated cardiomyocyte apoptosis induced by H2O2 and myocardial I/RI by inhibiting the mitochondrial apoptosis pathway. H19 could reduce Cyt-c releasing from the mitochondria to the cytoplasm and inhibit caspase-9 and caspase-3 activation. Meanwhile, miR-877-3p participated in the myocardial ischemia injury (in vivo) and cardiomyocytes apoptosis induced by H2O2 (in vitro) through the function of its target Bcl-2 and the imbalance of Bcl-2/Bax in the intrinsic mitochondrial apoptosis pathway. miR-877-3p promoted Cyt-c release, which activated caspase-9 and caspase-3. Furthermore, we identified that the prominent actions of H19 could be canceled by miR-877-3p.
In summary, we for the first time clarified the effect of lncRNA H19 on mitochondrial apoptosis of myocardial I/RI. We demonstrated that lncRNA H19 is an anti-apoptotic molecule in myocardial I/RI by acting as a ceRNA of miR-877-3p. We found that miR-877-3p participated in cardiomyocytes apoptosis during myocardial I/RI. H19 alleviated cardiomyocyte apoptosis and myocardial I/RI via inhibiting the miR-877-3p-Bcl-2-mediated mitochondrial apoptotic pathway. Our findings provide new insights in understanding the mechanism of myocardial I/RI and offer a new strategy for intervening myocardial I/RI.
Conclusions
lncRNA H19 alleviated cardiomyocyte apoptosis and myocardial I/RI through suppressing the miR-877-3p-Bcl-2-mediated mitochondrial apoptotic pathway.
Materials and Methods
Mouse Model of Myocardial I/RI and Virus Vectors Delivery
Healthy male C57BL/6 mice (weighing 21–25 g, 12 weeks old) were purchased from Vital River (Beijing, China). Food and water were freely accessible by the mice. All experimental procedures were performed in accordance with and approved by the Institutional Animal Care and Use Committee of the Harbin Medical University.
Mice were anesthetized by Avertin (200 mg/kg intraperitoneally [i.p.]; Sigma-Aldrich, USA) and ventilated with an animal ventilator. The chest was opened at the fourth intercostal space, and the heart was exposed. The left anterior descending coronary artery of the mice was ligated for 45 min with 7/0 nylon suture and then released for 24 h for reperfusion. Sham-operated mice received the same operation without ligation. For intracoronary delivery of adenovirus or lentivirus vectors, adenovirus (1 × 109 titers) or lentivirus (1 × 108 titers) was injected into the left ventricular cavity while the aorta artery was cross-clamped. After 3 days of adenovirus infection or 7 days of lentivirus infection, mice were subjected to myocardial I/R surgery.
Neonatal Mouse Ventricular Cells (NMVCs) Culture and Treatments
First, the isolated hearts from 1- to 3-day-old neonatal mice were cut into l.0-mm pieces and digested in 0.25% trypsin at 37°C. The cell suspension was stored at 4°C until the heart tissue was digested, followed by filtering with a 200-mesh sterile strainer. Subsequently, cells were collected by centrifugation (1,500 rpm/min for 5 min) and then cultured in DMEM (Hyclone, UT, USA) supplemented with 10% fetal bovine serum (Biological Industries, Kibbutz Beit Haemek, Israel), 100 U/mL penicillin (Beyotime, Shanghai, China), and 100 μg/mL streptomycin (Beyotime, Shanghai, China) for 1.5 h. After fibroblasts adherence, the non-adherent cells in the supernatant were replanted into cell culture dishes at a cell density of 1 × 106 cells/mL. Forty-eight hours later, cells were treated with H2O2 (Tianli, Tianjin, China) to induce cardiomyocyte apoptosis or transfected with the corresponding plasmids, siRNAs, miRNA mimics, AMOs, and NCs. As shown in Figure S2, H2O2 reduced cardiomyocyte viability in a concentration-dependent manner, and 24-h treatment with 100 μM H2O2 induced a moderate injury and was chosen for the subsequent experiments except observation of the effects of H19 in cardiomyocytes treated with H19 siRNA transfection and 50 μM H2O2 treatment.
Construction and Transfection of the Overexpression Plasmids or siRNAs
For the experiments involving H19, the full length of H19 was amplified by PCR and was cloned into a pcDNA3.1 vector (Invitrogen, Shanghai, China). The siRNAs of H19 were synthesized by Guangzhou RiboBio (Guangzhou, China). The base sequences of siRNAs and NC were shown in Table S1. When NMVCs were ready for transfection, H19 overexpression and NC plasmids were transfected into cardiomyocytes with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) at a final concentration of 200 ng/mL following the manufacturer’s protocols. Meanwhile, H19 siRNAs were transfected at a concentration of 100 nmol/L by using X-treme GENE siRNA Transfection Reagent (Roche, Mannheim, Germany). RNAi experiments were performed following the manufacturer’s protocols.
Synthesis and Transfection of miRNA Mimics and AMOs
miRNA mimics and AMO of miR-877-3p (AMO-miR-877-3p) were synthesized by Guangzhou RiboBio (Guangzhou, China). Scrambled RNAs were used as NCs of miR-877-3p (NC-miR-877-3p) and AMO-miR-877-3p (NC-AMO-miR-877-3p), respectively. Transfection was accomplished by using the X-treme GENE siRNA Transfection Reagent (Roche, Mannheim, Germany). The experiments were performed following the manufacturer’s protocols at a final concentration of 100 nmol/L miR-877-3p, AMO-miR-877-3p, NC-miR-877-3p, or NC-AMO-miR-877-3p. Cardiomyocytes were collected for qRT-PCR and western blot analysis 48 h after transfection.
Cell Viability Assay
Cell viability was assessed by measuring the mitochondrial-dependent reduction of 3-(4,5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2-H-tetrazolium bromide (MTT). NMVCs were cultured in 96-well plates with 1 × 104 cells/well. After corresponding treatments, NMVCs were subjected to 20 μL MTT (Sigma-Aldrich, St. Louis, MO, USA) solution (5 mg/mL) in each well and incubated at 37°C for 4 h in the dark. Then 200 μL DMSO (Fuyu, Tianjin, China) was added into each well to dissolve the formazan. The absorbance value of each well was measured by a microplate reader (BioTek, Richmond, VA, USA) at 490 nm.
TUNEL Assay
Terminal-deoxynucleotidyl transferase-mediated nick end labeling (TUNEL) assay was performed by using the In Situ Cell Death Detection Kit (Roche, Mannheim, Germany) according to the manufacturer’s instructions. After TUNEL staining, the nucleus was stained with DAPI (Biosharp, Hefei, China). The samples were observed under a fluorescence microscope (Olympus, Tokyo, Japan). The percentage of TUNEL-positive cells meant the ratio of TUNEL-positive cells/DAPI-positive nucleus.
Detection of LDH Activity
LDH activity in the supernatant of NMVCs or serum of mice was measured using a LDH detection kit (Nanjing Jiancheng, Jiangsu, China) according to the manufacturer’s instructions. The absorbance value of each well was measured by a microplate reader (BioTek, Richmond, VA, USA) at 450 nm.
Quantitative Real-Time RT-PCR
Total RNA samples were extracted from cardiomyocytes or myocardium using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The expression levels of H19, miRNAs including miR-18a-3p, miR-92a-2-5p, miR-93-3p, miR-361-3p, miR-744-3p, and miR-877-3p, Bcl-2, and Bax mRNA were tested by using SYBR Green I (TOYOBO, Osaka, Japan) through the ABI 7500 fast Real Time PCR system (Applied Biosystems, Foster City, CA, USA). β-Actin was used as an internal control of H19, Bcl-2, and Bax. U6 served as an internal control of miRNAs. The relative quantitative expression was determined using the 2−ΔΔCT method. The primers were synthesized by Invitrogen and were listed in Tables S2 and S3.
Western Blot Analysis
Total proteins were extracted from cardiomyocytes or myocardium with radio immunoprecipitation assay (RIPA) lysis buffer (Beyotime, Shanghai, China). Then, the lysates were centrifuged at 13,500 rpm/min at 4°C for 15 min. The supernatant was collected, and protein concentrations were determined by using the BCA Protein Assay Kit (Beyotime, Shanghai, China). Proteins were separated by SDS-PAGE, subsequently transferred onto the nitrocellulose membrane (Millipore, Bedford, MA, USA), and blocked by using the 5% non-fat milk at room temperature for 2 h. The membranes were incubated with the primary antibody for Bcl-2 (Cell Signaling, MA, USA), Bax (Cell Signaling, MA, USA), Cyt-c (Abcam, Cambridge, USA), and β-actin (Zhongshanjinqiao, Beijing, China) overnight at 4°C. The membranes were then conjugated with a secondary anti-rabbit or anti-mouse (LI-COR, Lincoln, NE, USA) polyclonal antibody at room temperature for 1 h in the dark. Protein expression levels were detected by the Odyssey infrared scanning system (LI-COR, Lincoln, NE, USA), and the protein bands were quantified using Odyssey 3.0 software.
Dual-Luciferase Gene Reporter Assay
For the dual-luciferase gene reporter assay between H19 and miR-877-3p, the full length of wild-type H19 was amplified by PCR, and then the PCR products were subcloned into psiCHECK-2 luciferase reporter vector (Luc-H19-WT; Promega, WI, USA). Luc-H19 mutant (Luc-H19-Mut) was also constructed, which contained a mutated H19 at the binding site of miR-877-3p (5′-AGAGGACA-3′ mutated to 5′-CACAACTC-3′). For the dual-luciferase gene reporter assay between miR-877-3p and Bcl-2, the 3′ UTR of Bcl-2 was obtained by PCR. Two critical binding sites between Bcl-2 3′ UTR and miR-877-3p were mutated from 5′-AGAGGAC-3′ to 5′-CACAACT-3′ and from 5′-TGAGGAC-3′ to 5′-AACAACT-3′. The wild-type and mutant 3′ UTR of Bcl-2 mRNAs were subcloned into the psiCHECK-2 luciferase reporter vectors. HEK293 cells were cultured in 24-well plates and transfected with the corresponding plasmids, miR-877-3p mimics, AMO-miR-877-3p, or their respective NC. Forty-eight hours later, Renilla and firefly luciferase activities were measured with the Dual-Luciferase Reporter Assay System (Roche, Mannheim, Germany) and GloMax Luminometry System (Promega, WI, USA).
Construction of the Overexpression or Downregulation Virus Vectors
The full length of H19 and its NCs were synthesized and subcloned into adenoviral vectors (Invitrogen, Shanghai, China). The vectors were packaged into viral particles in 293T cells by co-transfection with packaging-defective helper plasmids. Virus-containing supernatants were collected 48 h later, and the adenoviral titer was determined by high-content screening. The lentivirus-expressing miR-877-3p, AMO-877-3p, or their respective NCs controls were generated using PHY-310 plasmid (Hanyinbt, Shanghai, China). Lentiviruses were packed in HEK293 cells, and the lentiviral titer was determined by high-content screening. The above adenovirus and lentivirus were constructed by Invitrogen Corporation (Shanghai, China).
Detection of the Cyt-c Protein Releasing
To study the release of Cyt-c from mitochondria into the cytoplasm, we isolated cytoplasm proteins by removing mitochondria. Mitochondria were separated from cardiomyocytes or myocardium by using the Cell Mitochondria Isolation Kit (Beyotime, Shanghai, China) or the Tissue Mitochondria Isolation Kit (Beyotime, Shanghai, China). The experimental operations were in accordance with the manual provided by the company. Then the protein expression of Cyt-c in cytoplasm was detected by western blot analysis that was described in detail in Western Blot Analysis.
Detection of Caspase-9 and Caspase-3 Activity
The activity of caspase-9 and caspase-3 in cardiomyocytes or myocardium was measured by the Caspase 9 Activity Assay Kit (C1158; Beyotime, Shanghai, China) and the Caspase 3 Activity Assay Kit (C1116; Beyotime, Shanghai, China), respectively. The experimental operations were in accordance with the manual provided by the company.
2, 3, 5-Triphenyl Tetrazolium Chloride (TTC) Staining
In order to determine the infarct area of mice, we stained the hearts with 2.0% TTC (Solarbio, Beijing, China). In detail, the heart was rapidly excised and sliced into five slices. The slices were washed by 0.9% saline and placed into TTC solution at 37°C in the dark for 30 min. Then, photos were taken by Canon CanoScan 4400F (Canon, Tokyo Japan). Image Pro-Plus software was used to measure the percentage of infarct myocardium in cardiac slices.
Echocardiographic Measurement
Transthoracic echocardiography was performed to monitor changes of the left ventricular function by using the Vevo2100 High-Resolution Imaging System (VisualSonics, Toronto, ON, Canada) equipped with a 10-MHz phased-array transducer with the M-mode recording. Ventricular parameters including EF and FS were measured and analyzed. The mice were then sacrificed, and the hearts were quickly isolated and stored in −80°C for other detection.
Statistical Analysis
Data are presented as the mean ± SD. One-way ANOVA was performed for multiple-group analysis, and two-sided Student’s t test was used to compare differences between two groups by GraphPad Prism 5.0. p < 0.05 was considered to indicate a statistical significance.
Author Contributions
Y.L., Z.P., and X.L. contributed conception and design of the study; X.L. and S.L. organized the database; X.L. and S.L. performed the statistical analysis; Y.L., Z.P., X.L., and S.L. wrote the first draft of the manuscript; S.L., J.Z., and H.W. performed the animal experiments; S.L. and X.D. performed caspase-9 and caspase-3 activity detection, LDH activity assay, dual-luciferase gene reporter assay, TTC staining, and echocardiographic measurement; W.J., Y.Y., and H.Z. performed the western blot analysis and the detection of Cyt-c protein expression in cytoplasm; R.S. and Y.X. performed the NMVCs culture, H2O2 treatment, transfection, qRT-PCR experiments, and TUNEL assay; and L.Z. performed cell viability assay. All authors contributed to manuscript revision, and read and approved the submitted version.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grants 81503070 and 81872871); Key Project of the National Natural Science Foundation of China (grant 81530010); Innovation Fund Foundation Research Project of Harbin Medical University (grant 2017JCZX56); Innovative Talent Training Program of the Undergraduate Higher Institutions of Heilongjiang Province (grant UNPYSCT-2016051); and Postdoctoral Funding of Heilongjiang Province (grant LBH-Z15137).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.05.031.
Contributor Information
Zhenwei Pan, Email: panzw@ems.hrbmu.edu.cn.
Yanjie Lu, Email: yjlu@hrbmu.edu.cn.
Supplemental Information
References
- 1.Hausenloy D.J., Yellon D.M. Myocardial ischemia-reperfusion injury: a neglected therapeutic target. J. Clin. Invest. 2013;123:92–100. doi: 10.1172/JCI62874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yellon D.M., Hausenloy D.J. Myocardial reperfusion injury. N. Engl. J. Med. 2007;357:1121–1135. doi: 10.1056/NEJMra071667. [DOI] [PubMed] [Google Scholar]
- 3.Li X., Liu M., Sun R., Zeng Y., Chen S., Zhang P. Protective approaches against myocardial ischemia reperfusion injury. Exp. Ther. Med. 2016;12:3823–3829. doi: 10.3892/etm.2016.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giorgi C., Baldassari F., Bononi A., Bonora M., De Marchi E., Marchi S., Missiroli S., Patergnani S., Rimessi A., Suski J.M. Mitochondrial Ca(2+) and apoptosis. Cell Calcium. 2012;52:36–43. doi: 10.1016/j.ceca.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morciano G., Giorgi C., Bonora M., Punzetti S., Pavasini R., Wieckowski M.R., Campo G., Pinton P. Molecular identity of the mitochondrial permeability transition pore and its role in ischemia-reperfusion injury. J. Mol. Cell. Cardiol. 2015;78:142–153. doi: 10.1016/j.yjmcc.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 6.Knowlton A.A., Liu T.T. Mitochondrial dynamics and heart failure. Compr. Physiol. 2015;6:507–526. doi: 10.1002/cphy.c150022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao J., Yin M., Deng H., Jin F.Q., Xu S., Lu Y., Mastrangelo M.A., Luo H., Jin Z.G. Cardiac Gab1 deletion leads to dilated cardiomyopathy associated with mitochondrial damage and cardiomyocyte apoptosis. Cell Death Differ. 2016;23:695–706. doi: 10.1038/cdd.2015.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steiner J.L., Lang C.H. Etiology of alcoholic cardiomyopathy: Mitochondria, oxidative stress and apoptosis. Int. J. Biochem. Cell Biol. 2017;89:125–135. doi: 10.1016/j.biocel.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Ulitsky I., Bartel D.P. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kopp F., Mendell J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dey B.K., Mueller A.C., Dutta A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription. 2014;5:e944014. doi: 10.4161/21541272.2014.944014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomson D.W., Dinger M.E. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 14.Jiang X., Ning Q. The emerging roles of long noncoding RNAs in common cardiovascular diseases. Hypertens. Res. 2015;38:375–379. doi: 10.1038/hr.2015.26. [DOI] [PubMed] [Google Scholar]
- 15.Ratajczak M.Z. Igf2-H19, an imprinted tandem gene, is an important regulator of embryonic development, a guardian of proliferation of adult pluripotent stem cells, a regulator of longevity, and a ‘passkey’ to cancerogenesis. Folia Histochem. Cytobiol. 2012;50:171–179. doi: 10.5603/fhc.2012.0026. [DOI] [PubMed] [Google Scholar]
- 16.Brannan C.I., Dees E.C., Ingram R.S., Tilghman S.M. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabory A., Jammes H., Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. BioEssays. 2010;32:473–480. doi: 10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- 18.Necsulea A., Soumillon M., Warnefors M., Liechti A., Daish T., Zeller U., Baker J.C., Grützner F., Kaessmann H. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 19.van Heesch S., van Iterson M., Jacobi J., Boymans S., Essers P.B., de Bruijn E., Hao W., MacInnes A.W., Cuppen E., Simonis M. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 2014;15:R6. doi: 10.1186/gb-2014-15-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J.X., Zhang X.J., Li Q., Wang K., Wang Y., Jiao J.Q., Feng C., Teng S., Zhou L.Y., Gong Y. MicroRNA-103/107 Regulate Programmed Necrosis and Myocardial Ischemia/Reperfusion Injury Through Targeting FADD. Circ. Res. 2015;117:352–363. doi: 10.1161/CIRCRESAHA.117.305781. [DOI] [PubMed] [Google Scholar]
- 21.Luo H., Wang J., Liu D., Zang S., Ma N., Zhao L., Zhang L., Zhang X., Qiao C. The lncRNA H19/miR-675 axis regulates myocardial ischemic and reperfusion injury by targeting PPARα. Mol. Immunol. 2019;105:46–54. doi: 10.1016/j.molimm.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 22.Zhou M., Zou Y.G., Xue Y.Z., Wang X.H., Gao H., Dong H.W., Zhang Q. Long non-coding RNA H19 protects acute myocardial infarction through activating autophagy in mice. Eur. Rev. Med. Pharmacol. Sci. 2018;22:5647–5651. doi: 10.26355/eurrev_201809_15831. [DOI] [PubMed] [Google Scholar]
- 23.Li X., Wang H., Yao B., Xu W., Chen J., Zhou X. lncRNA H19/miR-675 axis regulates cardiomyocyte apoptosis by targeting VDAC1 in diabetic cardiomyopathy. Sci. Rep. 2016;6:36340. doi: 10.1038/srep36340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu L., An X., Li Z., Song Y., Li L., Zuo S., Liu N., Yang G., Wang H., Cheng X. The H19 long noncoding RNA is a novel negative regulator of cardiomyocyte hypertrophy. Cardiovasc. Res. 2016;111:56–65. doi: 10.1093/cvr/cvw078. [DOI] [PubMed] [Google Scholar]
- 25.Hadji F., Boulanger M.C., Guay S.P., Gaudreault N., Amellah S., Mkannez G., Bouchareb R., Marchand J.T., Nsaibia M.J., Guauque-Olarte S. Altered DNA methylation of long noncoding RNA H19 in calcific aortic valve disease promotes mineralization by silencing notch1. Circulation. 2016;134:1848–1862. doi: 10.1161/CIRCULATIONAHA.116.023116. [DOI] [PubMed] [Google Scholar]
- 26.Sibley C.R., Seow Y., Saayman S., Dijkstra K.K., El Andaloussi S., Weinberg M.S., Wood M.J. The biogenesis and characterization of mammalian microRNAs of mirtron origin. Nucleic Acids Res. 2012;40:438–448. doi: 10.1093/nar/gkr722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie Y., Jia Y., Cuihua X., Hu F., Xue M., Xue Y. Urinary exosomal microRNA profiling in incipient type 2 diabetic kidney disease. J. Diabetes Res. 2017;2017:6978984. doi: 10.1155/2017/6978984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu W., Zhang J., Zou C., Xie X., Wang Y., Wang B., Zhao Z., Tu J., Wang X., Li H. Microarray expression profile and functional analysis of circular RNAs in osteosarcoma. Cell. Physiol. Biochem. 2017;43:969–985. doi: 10.1159/000481650. [DOI] [PubMed] [Google Scholar]
- 29.Williams A.E., Choi K., Chan A.L., Lee Y.J., Reeves W.H., Bubb M.R., Stewart C.M., Cha S. Sjögren’s syndrome-associated microRNAs in CD14(+) monocytes unveils targeted TGFβ signaling. Arthritis Res. Ther. 2016;18:95. doi: 10.1186/s13075-016-0987-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bellinger M.A., Bean J.S., Rader M.A., Heinz-Taheny K.M., Nunes J.S., Haas J.V., Michael L.F., Rekhter M.D. Concordant changes of plasma and kidney microRNA in the early stages of acute kidney injury: time course in a mouse model of bilateral renal ischemia-reperfusion. PLoS ONE. 2014;9:e93297. doi: 10.1371/journal.pone.0093297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Czabotar P.E., Lessene G., Strasser A., Adams J.M. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 32.Bagci E.Z., Vodovotz Y., Billiar T.R., Ermentrout G.B., Bahar I. Bistability in apoptosis: roles of bax, bcl-2, and mitochondrial permeability transition pores. Biophys. J. 2006;90:1546–1559. doi: 10.1529/biophysj.105.068122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L.L. Linking long noncoding RNA localization and function. Trends Biochem. Sci. 2016;41:761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 34.Klattenhoff C.A., Scheuermann J.C., Surface L.E., Bradley R.K., Fields P.A., Steinhauser M.L., Ding H., Butty V.L., Torrey L., Haas S. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y., Li G., Lu H., Li W., Li X., Liu H., Li X., Li T., Yu B. Expression profiling and ontology analysis of long noncoding RNAs in post-ischemic heart and their implied roles in ischemia/reperfusion injury. Gene. 2014;543:15–21. doi: 10.1016/j.gene.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 36.Wang K., Long B., Zhou L.Y., Liu F., Zhou Q.Y., Liu C.Y., Fan Y.Y., Li P.F. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat. Commun. 2014;5:3596. doi: 10.1038/ncomms4596. [DOI] [PubMed] [Google Scholar]
- 37.Wang K., Liu F., Liu C.Y., An T., Zhang J., Zhou L.Y., Wang M., Dong Y.H., Li N., Gao J.N. The long noncoding RNA NRF regulates programmed necrosis and myocardial injury during ischemia and reperfusion by targeting miR-873. Cell Death Differ. 2016;23:1394–1405. doi: 10.1038/cdd.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tao H., Cao W., Yang J.J., Shi K.H., Zhou X., Liu L.P., Li J. Long noncoding RNA H19 controls DUSP5/ERK1/2 axis in cardiac fibroblast proliferation and fibrosis. Cardiovasc. Pathol. 2016;25:381–389. doi: 10.1016/j.carpath.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 39.Greco S., Zaccagnini G., Perfetti A., Fuschi P., Valaperta R., Voellenkle C., Castelvecchio S., Gaetano C., Finato N., Beltrami A.P. Long noncoding RNA dysregulation in ischemic heart failure. J. Transl. Med. 2016;14:183. doi: 10.1186/s12967-016-0926-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Z., Gao W., Long Q.Q., Zhang J., Li Y.F., Liu D.C., Yan J.J., Yang Z.J., Wang L.S. Increased plasma levels of lncRNA H19 and LIPCAR are associated with increased risk of coronary artery disease in a Chinese population. Sci. Rep. 2017;7:7491. doi: 10.1038/s41598-017-07611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bitarafan S., Yari M., Broumand M.A., Ghaderian S.M.H., Rahimi M., Mirfakhraie R., Azizi F., Omrani M.D. Association of Increased Levels of lncRNA H19 in PBMCs with Risk of Coronary Artery Disease. Cell J. 2019;20:564–568. doi: 10.22074/cellj.2019.5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y., Zhang M., Xu W., Chen J., Zhou X. The long non-coding RNA H19 promotes cardiomyocyte apoptosis in dilated cardiomyopathy. Oncotarget. 2017;8:28588–28594. doi: 10.18632/oncotarget.15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kallen A.N., Zhou X.B., Xu J., Qiao C., Ma J., Yan L., Lu L., Liu C., Yi J.S., Zhang H. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keniry A., Oxley D., Monnier P., Kyba M., Dandolo L., Smits G., Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang Y., Zhao G., Tang L., Zhang J., Li T., Liu Z. MiR-100-3p and miR-877-3p regulate overproduction of IL-8 and IL-1β in mesangial cells activated by secretory IgA from IgA nephropathy patients. Exp. Cell Res. 2016;347:312–321. doi: 10.1016/j.yexcr.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 46.Wang C., Gu S., Cao H., Li Z., Xiang Z., Hu K., Han X. miR-877-3p targets Smad7 and is associated with myofibroblast differentiation and bleomycin-induced lung fibrosis. Sci. Rep. 2016;6:30122. doi: 10.1038/srep30122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li S., Zhu Y., Liang Z., Wang X., Meng S., Xu X., Xu X., Wu J., Ji A., Hu Z. Up-regulation of p16 by miR-877-3p inhibits proliferation of bladder cancer. Oncotarget. 2016;7:51773–51783. doi: 10.18632/oncotarget.10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baines C.P., Molkentin J.D. STRESS signaling pathways that modulate cardiac myocyte apoptosis. J. Mol. Cell. Cardiol. 2005;38:47–62. doi: 10.1016/j.yjmcc.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 49.Xia P., Liu Y., Cheng Z. Signaling pathways in cardiac myocyte apoptosis. BioMed Res. Int. 2016;2016:9583268. doi: 10.1155/2016/9583268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lalier L., Cartron P.F., Juin P., Nedelkina S., Manon S., Bechinger B., Vallette F.M. Bax activation and mitochondrial insertion during apoptosis. Apoptosis. 2007;12:887–896. doi: 10.1007/s10495-007-0749-1. [DOI] [PubMed] [Google Scholar]
- 51.Chawla-Sarkar M., Leaman D.W., Borden E.C. Preferential induction of apoptosis by interferon (IFN)-beta compared with IFN-alpha2: correlation with TRAIL/Apo2L induction in melanoma cell lines. Clin. Cancer Res. 2001;7:1821–1831. [PubMed] [Google Scholar]
- 52.Yu B.Y., Dong B. LncRNA H19 regulates cardiomyocyte apoptosis and acute myocardial infarction by targeting miR-29b. Int. J. Cardiol. 2018;271:25. doi: 10.1016/j.ijcard.2018.04.108. [DOI] [PubMed] [Google Scholar]
- 53.Zhang B.F., Chen J., Jiang H. LncRNA H19 ameliorates myocardial ischemia-reperfusion injury by targeting miR-22-3P. Int. J. Cardiol. 2019;278:224. doi: 10.1016/j.ijcard.2018.11.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.