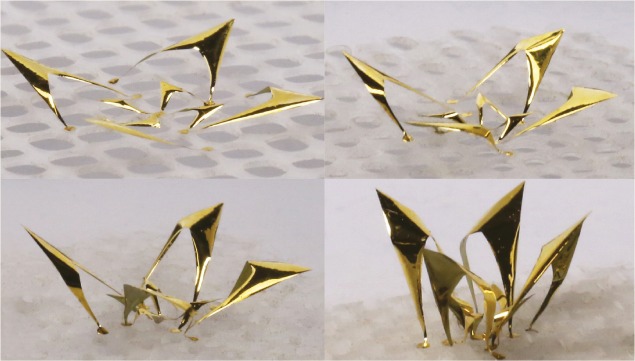
Controlled twisting of advanced materials into complex structures
3D microstructure in the form of a flower, created by buckling and twisting a 2D precursor.
Advances in 3D manufacturing have enabled the production of complex 3D architectures, but existing manufacturing techniques are limited in the types of achievable geometries. Hangbo Zhao, Kan Li, et al. (pp. 13239–13248) describe a strategy involving the controlled twisting of advanced materials to yield previously inaccessible 3D shapes, including strongly chiral and folding architectures and complex designs inspired by the paper folding and cutting techniques of origami and kirigami. The strategy uses elastomeric substrates cut to form interconnected segments that deform and rotate relative to one another when put under strain. When the strain is released, conventional buckling and controlled twisting deformations transform 2D precursors into complex 3D structures. The assembly process is controllable, enabling the use of computational techniques to guide design. The authors provide examples of more than 20 complex 3D structures built across a wide variety of scales—from micrometers to centimeters—and using a number of different materials, including metals, polymers, and inorganic semiconductors. According to the authors, the strategy could be used to create 3D structures for a variety of applications. — S.R.
Uncovering art forgeries through radiocarbon dating

Forged painting of a 19th-century village scene signed and dated “Sarah Honn May 5, 1866 AD.” Image courtesy of James Hamm (Buffalo State College, The State University of New York, Buffalo, NY).
The detection of forgeries in artworks requires increasingly sophisticated techniques. To determine whether radiocarbon (14C) dating is a reliable method to detect post-1950 art forgeries, Laura Hendriks et al. (pp. 13210–13214) dated a known forgery created in 1985 by extracting microsamples from both the pictorial layer and canvas. The authors report that 14C analysis of the canvas was consistent with an 1866 attribution, which is the signed date on the canvas. Preliminary analysis of the paint revealed the presence of inorganic pigments in a mixed binding medium coated with a layer of varnish. The paint sample cleaned from the varnish yielded as much as 20 µg carbon, in which the authors detected an excess of 14C, which is characteristic of the nuclear testing period during the 20th century. Specifically, the oil used to bind the pigments was harvested from seeds between either 1958–1961 or 1983–1989. The results contradict the dating of the support material and suggest that the forger recycled an older canvas to increase the counterfeit’s credibility. According to the authors, radiocarbon dating of micropaint samples could help detect modern forgeries if suitable sampling locations are identified. — M.S.
Targeted CRISPR knockouts in quail eggs
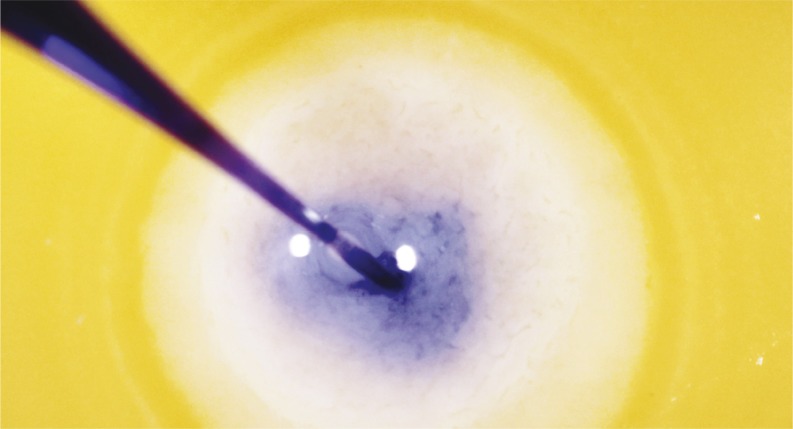
Microinjection of a blue dye into the subgerminal cavity of a quail blastoderm at stage XI.
In mammals, CRISPR/Cas9 has been used to modify zygote genomes at the single-cell stage and produce species in which specific genes have been knocked out. However, in the avian reproductive system, oocytes are fertilized within minutes of ovulation and the single-cell zygote expands rapidly to around 60,000 cells prior to egg laying. Circumventing the current strategy, which involves isolating, culturing, and modifying primordial germ cell genomes in vitro for subsequent injection into recipient embryos, Joonbum Lee et al. (pp. 13288–13292) demonstrate a technique to generate targeted knockouts by directly injecting the adenoviral CRISPR/Cas9 vector into the blastoderm of newly laid quail eggs. The authors show that the technique can produce chimeras whose offspring carry a desired mutation, namely melanophilin homozygous mutants whose gray plumage differs from that of the brown-feathered parent quails. Furthermore, the technique does not propagate foreign adenovirus DNA into the mutants’ genomes or result in off-target mutations. The study suggests that direct injection of adenovirus CRISPR/Cas9 represents a safe and convenient system for producing knockout birds to advance avian research, according to the authors. — T.J.
How natural selection leads to speciation
Ecological speciation is a process in which local adaptation to different environments leads to reproductive isolation as a byproduct of natural selection. However, direct demonstrations of the underlying mechanisms have been elusive, especially in natural populations. Scott Villa et al. (pp. 13440–13445) report that the experimental evolution of parasite body size over approximately 60 generations leads to reproductive isolation in natural populations of feather lice placed on captive birds. The authors transferred lice from wild-caught feral pigeons to giant runts, which are domesticated pigeons that are 3 times larger, or to feral pigeon controls. Over the course of 4 years, lice on giant runts increased in size relative to lice on feral pigeon controls. The size difference may reduce the ability of lice from different hosts to mate and reproduce. In support of this idea, pairs of lice with typical sexual dimorphism in body size spent more time copulating, were more likely to produce eggs, and had more offspring than mismatched pairs with more or less dimorphism. The findings show that local adaptation of louse body size to hosts of different sizes triggers reproductive isolation. According to the authors, the study demonstrates that ecological speciation may evolve rapidly under natural conditions in real time when divergent selection acts on a single trait that governs both survival and mating success. — J.W.
Morphine tolerance and gut microbiome
The gut microbiome may play a role in morphine tolerance. Image courtesy of Flickr/John Campbell.
Opioid use in the United States has risen significantly over the past decade, leading to the development of opioid analgesic tolerance in a subset of the population and reducing the effectiveness of opioids for chronic pain treatment. Using mouse models, Li Zhang et al. (pp. 13523–13532) explored the role of gut microbiota in the development of morphine tolerance. Mice treated with repeated escalating doses of morphine for 8 days developed analgesic tolerance to morphine. However, compared with control mice, tolerance was significantly lower in germfree (GF) mice, which never develop microbiota, or mice treated with antibiotics to deplete their microbiota. When the microbiota of GF and antibiotic-treated mice were restored via fecal microbial transplantation from control mice prior to morphine treatment, both types of mice developed morphine tolerance. Bacterial communities in the microbiota differed significantly between morphine-tolerant and control mice, with tolerant mice being depleted in Bifidobacteria and Lactobacillaeae. Treatment with probiotics restored the depleted microbial communities, and mice pretreated with probiotics developed significantly reduced morphine tolerance compared with controls. The results indicate a role for the gut microbiota in morphine tolerance and suggest that probiotic pretreatment could prevent the development of morphine tolerance and prolong the efficacy of morphine for pain management, according to the authors. — B.D.
Controlling receptors in the intact brain
Protein function is often studied with genetic or pharmacological methods, but such approaches have limited spatial and temporal resolution and can produce off-target or compensatory effects that confound experimental results. An alternative technique is photopharmacology, in which light-regulated molecules allow direct, fast, and reversible control of protein activity, with spatiotemporal resolution set by the illumination method. Silvia Pittolo et al. (pp. 13680–13689) demonstrate the feasibility of applying photopharmacology to receptors in 3D tissues. A microscope equipped with a pulsed near-infrared laser enabled 2-photon excitation of alloswitch, a potent, diffusible, light-sensitive molecule that selectively binds to and can rapidly modulate metabotropic glutamate 5 (mGlu5) receptors. The receptors play an important role in neuronal communication and learning and memory and are involved in several neurological conditions. In the absence of illumination, alloswitch hampered mGlu5 activity. Two-photon excitation released the brake exerted by alloswitch on the receptors, resulting in mGlu5-dependent calcium activity within cells in rat brain slices. Receptor activity was controlled at the 10-µm scale, which is approximately the size of a single cell body in the brain. According to the authors, 2-photon pharmacology using alloswitch is a useful addition to the molecular toolbox to study how mGlu5 receptors function in the intact brain. — J.W.
Polar bear evolution and gene copy number variation

Polar bear. Image courtesy of the US Fish and Wildlife Service.
Polar bears and brown bears occupy distinct ecologies, despite having diverged comparatively recently—less than 500,000 years ago. Differences between individuals in the number of times a DNA segment repeats in a genome, known as copy number variation (CNV), can drive ecological adaptation over short time periods. To examine the extent to which polar bear/brown bear divergence was driven by CNV, David Rinker, Natalya Specian, et al. (pp. 13446–13451) analyzed gene copy numbers in 17 individual polar bears and 9 brown bears. The authors identified nearly 200 genes whose copy numbers differed significantly between polar bears and brown bears. Nearly half of these genes encoded olfactory receptors. Other genes whose copy numbers differed between species include the gene for salivary amylase, an enzyme involved in digesting starch, and genes involved in fatty acid metabolism. The results suggest that copy number differences between the species reflect differences in dietary requirements between polar bears, which have a primarily carnivorous diet, and brown bears, which have an omnivorous diet. According to the authors, natural selection may have driven rapid gene copy loss in polar bears, playing an integral role in polar bear adaptation to the arctic environment. — B.D.