Abstract
Background
Genome-wide association studies (GWAS) have identified numerous loci associated with diseases and traits. However, the elucidation of disease mechanisms followed by drug development has remained a challenge owing to complex gene interactions. We performed pathway analysis with MAGENTA (Meta-Analysis Geneset Enrichment of variaNT Associations) to clarify the pathways in genetic background of AF.
Methods
The existing GWAS data were analyzed using MAGENTA. A microarray analysis was then performed for the identified pathways with human atrial tissues, followed by Gene-Set Enrichment Analysis (GSEA).
Results
MAGENTA identified two novel candidate pathways for AF pathogenesis, the CTCF (CCCTC-binding factor, p = 1.00 × 10−4, FDR q = 1.64 × 10−2) and mTOR pathways (mammalian target of rapamycin, p = 3.00 × 10−4, FDR q = 3.13 × 10−2). The microarray analysis with human atrial tissue using the GSEA indicated that the mTOR pathway was suppressed in AF cases compared with non-AF cases, validating the MAGENTA results, but not CTCF pathway.
Conclusions
MAGENTA identified a novel pathway, mTOR, followed by GSEA with human atrial tissue samples. mTOR pathway is a key interface that adapts the change of environments by pressure overload and metabolic perturbation. Our results indicate that the MTOR pathway is involved in the pathogenesis of AF, although the details of the basic mechanism remain unknown and further analysis for causal-relationship of mTOR pathway to AF is required. CTCF pathway is essential for construction of chromatin structure and transcriptional process. The gene-set components of CTCF overlap with those of mTOR in Biocarta.
Keywords: Atrial fibrillation, Pathway analysis, mTOR, MAGENTA, Transcriptome analysis
1. Introduction
Atrial fibrillation (AF) is a common arrhythmia in clinical practice [1]. The prevalence of AF was estimated 2.2 million people in the United States and 716,000 people in Japan [2]. According to the Global Burden of Disease 2010 Study, an international collaborative effort to systematically assess global data on AF, evidence from 184 different publications showed that around 33.5 million individuals have AF worldwide in 2010, with 5 million new AF cases occurring per year [3]. AF is a multifaceted condition affected by risk factors, and co-morbidities [4] such as aging [5], genetic predisposition and acquired factors including pressure overload in atrium [6], and other cardiovascular diseases. Furthermore, atrium is exposed to numerous mediators such as metabolic perturbation [7], oxidative stress [8], and inflammation [9], ectopy of pulmonary vein, electrical and structural remodeling [10]. As for electrical remodeling based on animal experiments and clinical research [11,12], the enhanced membrane repolarization through reduced ICa,L, and increased IK1 and agonist-independent constructive IK,ACh promotes action potential duration shortening and re-entry, whose alteration affects Ca2+ handling due to the perturbation of Ca2+ release from sarcoplasmic reticulum Ca2+ store. Slow and/or heterogeneous conduction through changes in the expression, function, and localization of connexins responsible for electrical cell-to-cell coupling can promote re-entry. On the other hand, structural remodeling in the atrium is related to environmental stress and growth factors such as activated renin-angiotensin-aldosterone system [13], and transforming growth factor-β [14]. In individuals with genetic risk factors, acquired and environmental factors accumulate and enhance susceptibility to atrial remodeling.
Genome-wide association studies (GWAS) have been used to identify loci associated with numerous diseases and physical properties. GWAS is a comprehensive and powerful method to detect candidate molecules underlying diseases. Recent large-scale meta-analysis of AF-GWAS identified 97 loci in trans-ethnic population and 16 loci in Japanese population [15,16]. However, it is challenging to apply these association data to the elucidation of disease mechanisms because the functions of loci are often unknown. First, functional analysis of regions and genes included in a locus associated with AF has been attempted. 4q25 region including PITX2 has been identified repetitively in GWAS and the functional analyses were performed through searching for enhancer sequence [17,18]. Second, besides these functional analysis for individual loci, pathway analysis with AF-GWAS of European population was performed and identified cardiac developmental, electrophysiological, contractile and structural pathways [15]. Bioinformatics approach including pathway analysis with GWAS data had identified new therapeutic options of existing drugs. Candidate drugs targeted for type 2 diabetes has been suggested in the approved drugs such as thiazolidinediones for rs1801282 and/or unapproved drugs such as GCK activators for rs10278336 through protein-protein interaction data and Therapeutic Targets Database using the GWAS data for type 2 diabetes [19]. The GWAS for rheumatoid arthritis also indicated existing cancer drugs as novel candidates for rheumatoid arthritis using a similar strategy [20].
Meta-Analysis Gene-set Enrichment of variaNT Associations (MAGENTA) was developed to decode the basis of complex traits [21]. MAGENTA considers all SNP data obtained by GWAS using the gene set enrichment analysis (GSEA) approach: 1) SNPs are mapped to the nearest gene, 2) SNPs with the lowest p-value are selected for each gene, 3) the p-values of the best SNPs are corrected depending on the length of the gene, 4) significant pathways are estimated by GSEA. The statistical power of MAGENTA to detect disease-associated pathways has been evaluated by computer simulation and validated using several GWAS datasets. MAGENTA was employed in the AF-GWAS in the trans-ethnic population and identified cardiac developmental pathways including PITX2 and TBX5 [15].
In this study, we applied MAGENTA to previously obtained AF-GWAS data and identified novel disease pathways. We assessed the finding with the following microarray analysis of human atrial tissues. The combination of bioinformatics data from GWAS and microarray experiments led to the identification of an association between the mTOR pathway and AF. These findings may be a clue in understanding the complicated AF pathogenesis.
2. Materials and methods
2.1. MAGENTA
MAGENTA (Broad Institute; http://www.broadinstitute.org/mpg/magenta) software was applied to identify novel pathways associated with AF. Data from a previous Japanese GWAS (843 AF cases and 3350 controls) [22] were used for the analysis. All samples were collected by BioBank Japan. Written informed consent was obtained from each study participant. The dataset included 610K SNPs genotyped using the Affymetrix GeneChip500K, annotated with GRCh37. BIOCARTA and Ingenuity were used as informative pathway resources, including data for 214 and 81 pathways, respectively. The false discovery rate (FDR) q-value and p-value were obtained to identify significantly enriched pathways.
2.2. Human atrial tissue samples
Atrial tissue samples were collected from eleven patients who underwent cardiac surgery of the mitral valve in the Department of Cardiovascular Surgery, Tokyo Medical and Dental University Hospital. This study was approved by the Ethics Committee for Genome Research at the Medical School, Tokyo Medical and Dental University.
2.3. DNA array analysis
All participants provided written informed consent. RNA was extracted from the samples and treated with RNAlater after their excision. The RNeasy Mini Kit (Qiagen, Hilden, Germany) and 3′ IVT PLUS Array Kit (Affymetrix, Santa Clara, CA, USA) were used according to the manufacturers' protocols. Samples were applied to the GeneChip® Human Genome U133 Plus2.0 Array (Affymetrix) and expression levels were measured using GeneChip Scanner 3000 equipment after treatment. Data processing was performed using Affymetrix® Expression Console, Tiling Analysis, and Transcriptome Analysis Console according to standard protocols. Quality control procedures were performed according to the manufacturer's protocol (http://www.thermofisher.com). The quality control metrics includes hybridization controls, labeling controls, internal control genes, global array metrics, and algorithm parameters [Supplementary Fig. 1, Supplementary Fig. 2A, B, Supplementary Table 1].
2.4. Gene set enrichment analysis (GSEA)
An enrichment analysis was performed for the two pathways identified by MAGENTA, the mTOR and CTCF pathways, using GSEA software (Broad Institute, http://www.broadinstitute.org/gsea/index.jsp) [23]. In brief, after calculating the enrichment score (ES), target genes were assessed to determine whether they are randomly distributed or exhibit statistically significant differences between two biological phenotypes, AF and non-AF. The expression dataset was processed with Expression Console 1.4.1.46 software based on MAS-5 program for the background correction and the normalization of the raw data and applied for BioCarta pathway database (c2. cp. biocarta. v5.1. entrez) and the Molecular Signatures Database (MSigDB). A 100-permutation test was performed to create a histogram of ES values. Accordingly, the criteria of significant pathways and genes was based on ES at a nominal p-value of <0.05. The ES reflected the degree to which mTOR and CTCF pathway sets are overrepresented at the extremes of the entire ranked list. The scores were calculated by summing the observed frequencies of the gene sets of the two pathways in the ranked list using Kolmogorov–Smirnov statistics.
3. Results
3.1. Identification of the mTOR and CTCF pathways using MAGENTA
We identified novel biological or molecular pathways demonstrating significant enrichment using previous GWAS data for atrial fibrillation in the Japanese population (843 AF cases and 3350 control subjects) [22]. Although these data indicated associations of AF with KCNN3, PRRX1, PITX2, CAV1, C9orf3, and ZFHX3, an in silico pathway analysis with DAVID (https://david.ncifcrf.gov/) failed to identify a particular pathway including these six loci (data not shown). MAGENTA software was applied to these GWAS data using the gene sets included in BIOCARTA and Ingenuity as information resources for biological and molecular pathways; they include 214 and 81 pathways, respectively. The pathways exhibiting statistically significant associations with AF were the mTOR (p = 3.00 × 10−4, FDR q = 3.13 × 10−2) and CTCF pathways (p = 1.00 × 10−4, FDR q = 1.64 × 10−2) [Table 1] [Supplementary Table 2].
Table 1.
Result of MAGENTA analysis. As database for comparison, Ingenuity and BIOCARTA were used.
| Database | Gene signaling | Number | MEAN | P-value | FDR | Expected | Observed |
|---|---|---|---|---|---|---|---|
| Ingenuity | Neuregulin.Signaling | 27 | 92 | 3.30E-03 | 1.27E-01 | 3 | 8 |
| Ingenuity | TR.RXR.Activation | 62 | 75 | 5.60E-03 | 9.83E-02 | 6 | 13 |
| Ingenuity | Apoptosis.Signaling | 31 | 54 | 7.30E-03 | 1.13E-01 | 3 | 8 |
| Ingenuity | Sonic.Hedgehog.Signaling | 10 | 62 | 1.11E-02 | 1.52E-01 | 1 | 4 |
| Ingenuity | Amyotrophic.Lateral.Sclerosis.Signaling | 29 | 89 | 1.73E-02 | 1.69E-01 | 3 | 7 |
| Ingenuity | PI3K.AKT.Signaling | 29 | 57 | 2.30E-02 | 1.96E-01 | 3 | 7 |
| Database | Gene signaling | Original number | MEAN | P-value | FDR | Expected | Observed |
|---|---|---|---|---|---|---|---|
| BIOCARTA | CTCF_PATHWAY | 23 | 58 | 1.00E-04 | 1.64E-02 | 2 | 9 |
| BIOCARTA | MTOR_PATHWAY | 23 | 59 | 3.00E-04 | 3.13E-02 | 2 | 9 |
| BIOCARTA | P35ALZHEIMERS_PATHWAY | 11 | 79 | 2.70E-03 | 6.02E-02 | 1 | 5 |
| BIOCARTA | IGF1MTOR_PATHWAY | 20 | 76 | 8.30E-03 | 2.18E-01 | 2 | 6 |
| BIOCARTA | WNT_PATHWAY | 26 | 72 | 1.30E-02 | 2.43E-01 | 3 | 7 |
| BIOCARTA | BCELLSURVIVAL_PATHWAY | 16 | 51 | 1.36E-02 | 2.56E-01 | 2 | 5 |
| BIOCARTA | RAS_PATHWAY | 23 | 59 | 1.52E-02 | 2.39E-01 | 2 | 6 |
| BIOCARTA | ACH_PATHWAY | 16 | 72 | 1.61E-02 | 2.69E-01 | 2 | 5 |
| BIOCARTA | PAR1_PATHWAY | 37 | 132 | 1.67E-02 | 2.57E-01 | 3 | 8 |
| BIOCARTA | NGF_PATHWAY | 18 | 50 | 2.20E-02 | 2.45E-01 | 2 | 5 |
| BIOCARTA | ALK_PATHWAY | 37 | 72 | 2.28E-02 | 2.96E-01 | 4 | 8 |
| BIOCARTA | EIF4_PATHWAY | 24 | 109 | 2.78E-02 | 3.20E-01 | 2 | 6 |
| BIOCARTA | MITOCHONDRIA_PATHWAY | 21 | 39 | 2.94E-02 | 3.08E-01 | 2 | 5 |
| BIOCARTA | BAD_PATHWAY | 26 | 80 | 3.27E-02 | 2.61E-01 | 3 | 6 |
| BIOCARTA | DC_PATHWAY | 22 | 11 | 3.32E-02 | 2.65E-01 | 2 | 5 |
| BIOCARTA | TOB1_PATHWAY | 21 | 49 | 3.36E-02 | 2.81E-01 | 2 | 5 |
| BIOCARTA | DEATH_PATHWAY | 33 | 43 | 3.58E-02 | 2.96E-01 | 3 | 7 |
| BIOCARTA | STATHMIN_PATHWAY | 19 | 82 | 3.73E-02 | 2.64E-01 | 2 | 5 |
| BIOCARTA | G1_PATHWAY | 28 | 65 | 4.41E-02 | 2.91E-01 | 3 | 6 |
Number: original numbers included in signaling.
MEAN: mean gene size.
FDR: False discovery rate.
Expected: expected number of genes.
Observed: actually observed number of genes.
The genes in Bold were significant in GSEA.
3.2. Microarray analysis using human atrial tissue samples
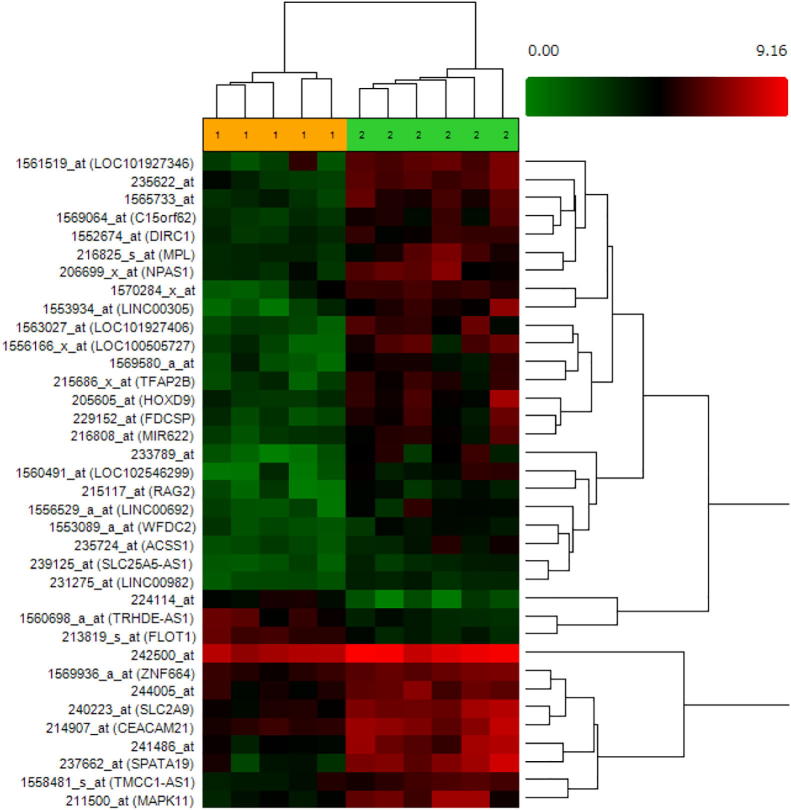
We investigated the enrichment of genes in the mTOR and CTCF pathways in left atrial tissues of AF and non-AF patients using GSEA software. The clinical background of the patients is summarized in Table 2. All patients were diagnosed with mitral valve disease: a mean age of 68.7 ± 2 years, a mean left atrial diameter of 52.6 ± 2.3 mm, and different atrial fibrillation risk factors, including hypertension and heart failure. A microarray analysis was performed to estimate differential gene expression between the non-AF group and AF group Among 54,613 genes on the microarray, 1575 genes were differentially expressed (p < 0.05, fold change <−2 or >2), including 214 up-regulated genes and 1361 down-regulated genes in the comparison between the AF and non-AF groups (Supplementary Fig. 2A and B, Supplementary Table 1). A hierarchical clustering analysis after filtering data with p < 0.001 showed the significant difference in the top core gene sets between AF and non-AF samples (Fig. 1).
Table 2.
Clinical characteristics of patients.
| Phenotypes | Non-AF |
AF |
Total |
p-Value |
|---|---|---|---|---|
| n = 5 | n = 6 | n = 11 | ||
| Age (years) | 72.0 ± 4.1 | 66.0 ± 7.2 | 68.7 ± 2.0 | NS |
| Female, n (%) | 1(20.0) | 3(50.0) | 4(36.4) | NS |
| Drinking history, n (%) | 1(20.0) | 4(66.7) | 5(45.5) | NS |
| Smoking history, n (%) | 3(60.1) | 2(33.4) | 5(45.5) | NS |
| LA diameter (mm) | 50.6 ± 3.0 | 54.3 ± 10.2 | 52.6 ± 2.3 | NS |
| Dyslipidemia, n (%) | 2(40.0) | 1(16.7) | 3(27.3) | NS |
| History of CAD, n (%) | 2(40.0) | 2(33.4) | 4(36.4) | NS |
| Diabetes mellitus, n (%) | 0(0.0) | 0(0.0) | 0(0.0) | NS |
| Heart failure, n (%) | 3(60.1) | 5(83.3) | 8(72.7) | NS |
| Hypertension, n (%) | 3(60.2) | 2(33.4) | 5(45.5) | NS |
| HCM, n (%) | 0(0.0) | 1(16.7) | 1(9.1) | NS |
| CKD, n (%) | 1(20.0) | 0(0.0) | 1(9.1) | NS |
| COPD, n (%) | 1(20.0) | 0(0.0) | 1(9.1) | NS |
LA diameter, left atrial diameter; BNP, B-type natriuretic peptide; MVD, mitral valve disease; CAD, coronary atherosclerotic heart disease; HCM, hypertrophic cardiac myopathy; CKD, chronic kidney disease; COPD, chronic obstructive pulmonary disease.
Fig. 1.
Hierarchical clustering.
All the genes under condition (p-value<0.001, fold change<−2 or >2) were clustered according to the distance between the data of two genes. The number 1 and 2 indicate non-AF group and AF group, respectively.
3.3. Gene set enrichment analysis
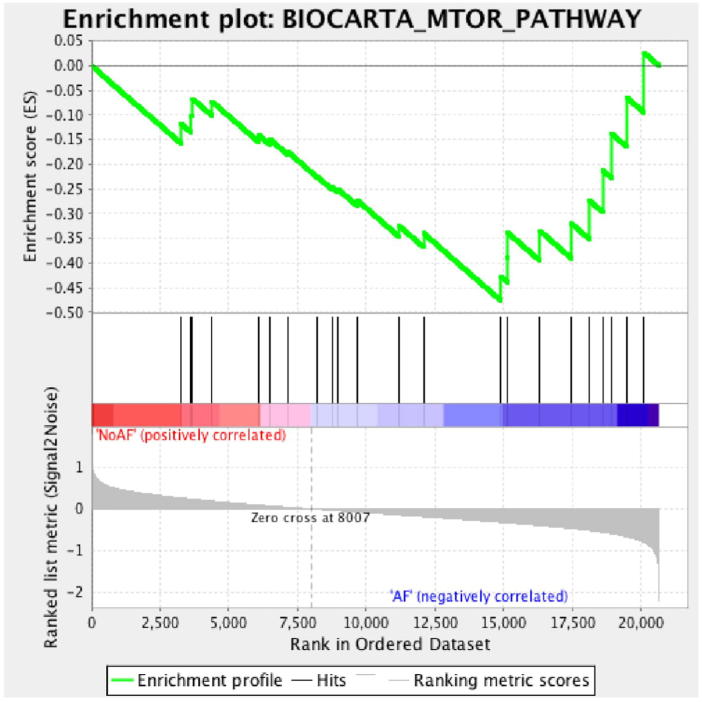
We investigated the up- or down-regulated gene sets in the overall dataset including mTOR and CTCF pathways in left atrial tissues of AF and non-AF patients using GSEA software according to the recommended parameters. All genes in the data set were ranked according to the correlation between their expression and functional category (Supplementary Table 3, Supplementary Table 4). We used BioCarta as a biological pathway database and determined whether members of the gene sets were randomly distributed in the observed gene set. Based on ES estimates, we demonstrated that the mTOR pathway was significantly enriched in AF tissues with a nominal p-value <0.05, and was down-regulated in the AF phenotype. Most of the gene sets in the mTOR pathway were down-regulated in the AF group compared with the non-AF group (Table 3, Fig. 2). On the other hand, the CTCF pathway, which is related to signaling in immature B cells by B-cell receptor and transforming growth factor β (TGF-β), was randomly distributed with low ES.
Table 3.
The components of mTOR pathway.
| Entrez_ID | Gene symbol | Rank in gene list | Rank metric score | Running ES | Core enrichment |
|---|---|---|---|---|---|
| 1982 | EIF4G2 | 20,129 | −0.797821045 | 0.026012314 | Yes |
| 5515 | PPP2CA | 19,489 | −0.661122859 | −0.06417525 | Yes |
| 7248 | TSC1 | 18,951 | −0.589762688 | −0.13854751 | Yes |
| 5295 | PIK3R1 | 18,619 | −0.557331443 | −0.21206315 | Yes |
| 8569 | MKNK1 | 18,131 | −0.51423192 | −0.27309778 | Yes |
| 5170 | PDPK1 | 17,484 | −0.466402262 | −0.31988618 | Yes |
| 1978 | EIF4EBP1 | 16,332 | −0.396058679 | −0.33494985 | Yes |
| 207 | AKT1 | 15,162 | −0.332502306 | −0.33846062 | Yes |
| 8672 | EIF4G3 | 15,146 | −0.331991702 | −0.38822106 | Yes |
| 2475 | FRAP1 | 14,916 | −0.321033925 | −0.42753774 | Yes |
| 1981 | EIF4G1 | 3683 | 0.23271291 | −0.06741249 | No |
| 5164 | PDK2 | 4377 | 0.189100042 | −0.07226868 | No |
| 7249 | TSC2 | 3634 | 0.235626712 | −0.1004224 | No |
| 1975 | EIF4B | 3258 | 0.26200974 | −0.11803479 | No |
| 1977 | EIF4E | 6078 | 0.093864225 | −0.14036475 | No |
| 2280 | FKBP1A | 6491 | 0.073707983 | −0.14913033 | No |
| 1973 | EIF4A1 | 7156 | 0.040029906 | −0.17521651 | No |
| 8661 | EIF3S10 | 8235 | −0.010132063 | −0.22589663 | No |
| 5728 | PTEN | 8753 | −0.032501176 | −0.24600525 | No |
| 6194 | RPS6 | 8968 | −0.042265035 | −0.24995396 | No |
| 5290 | PIK3CA | 9683 | −0.07499864 | −0.2731525 | No |
| 6198 | RPS6KB1 | 11,216 | −0.145169243 | −0.32532048 | No |
| 1974 | EIF4A2 | 12,089 | −0.18551822 | −0.33939135 | No |
The genes in Bold were significant in GSEA.
Fig. 2.
Enrichment plot of mTOR pathway.
Enrichment score of genes included in mTOR pathway correlated positively in non-AF tissue while correlated negatively in AF tissue.
4. Discussion
A pathway analysis of previously generated AF-GWAS data with MAGENTA identified the association between the mTOR pathway and AF, and this association was confirmed by microarray data of human atrial tissues obtained from AF and non-AF patients. In our study, we used two different datasets: (1) GWAS data for 850 AF patients and 3350 controls in Japan and (2) microarray transcriptome data for eleven mitral valve disease patients. As for the present results, mTOR pathway was identified through GWAS screening with lone AF as the possible mechanism. Microarray analysis using human atrial tissues which were exposed by pressure overload of mitral stenosis revealed that mTOR pathway was suppressed in AF. Although samples in the two studies were different, it indicated that mTOR pathway is associated with AF pathogenesis.
MAGENTA and GSEA were applied to GWAS data and microarray data, respectively. Both MAGENTA and GSEA are used to statistically compare the observed number of significant genes that differ between two groups in a certain gene-set with the predicted number. The previous AF-GWAS in Japanese have detected only the 4q25 locus in the case of the strict criterion of p < 5 × 10−8 [22]. Using the MAGENTA, we identified two new disease pathways, mTOR (mammalian target of rapamycin) and CTCF (CCCTC-bindngi factor). Second, we used microarray transcriptome data to confirm the MAGENTA results with a different dataset. Atrial tissues were excised from patients undergoing cardiac surgery for mitral valve disease, whose atrial tissues must receive high intracardiac pressure, transiently or persistently. We obtained both patients with AF and without AF, comparing their gene expression levels. Significant number of genes involved in the mTOR pathway were suppressed in the atrial tissues of AF patients and an enrichment analysis with GSEA showed that the pathway was also significantly suppressed. The GWAS missed the component molecules in mTOR pathway as candidates, because no variant in mTOR locus was identified. In our study, pathway analysis enabled to identify the associated pathway. The association of mTOR pathway with AF was confirmed in the transcriptome analysis of atrial tissue, while we failed to replicate the results for the CTCF pathway obtained by MAGENTA with microarray data. CTCF expression was not associated in atrial tissue with AF under the influence of pressure overload because the difference between lone AF and AF with mitral stenosis might affect the result. CTCF pathway is essential for construction of chromatin structure and transcriptional process of gene expression change. The gene-set components of CTCF overlap those of mTOR in Biocarta. MAGENTA found CTCF pathway due to the similar composition.
mTOR is a protein kinase that is conserved in a wide variety of species as an interface of various metabolic pathways. mTOR plays crucial roles in cell metabolism, a fundamental biological reaction involved in cell development, growth, maintenance and survival [24]. The kinase is activated by extracellular stress and growth factor signaling and enhances the cytoplasmic translational process and the protein synthesis. mTOR pathway is involved in cardiac homeostasis against stress such as pressure overload in myocardium [25]. The effect of mTOR inhibition and deficiency on myocardium differs in the physiological and pathological status [26]. The cardiac-specific mTOR knockout mice are lethal in the embryonic and neonatal stage mainly due to left ventricular dilatation and dysfunction, suppressing cell growth, protein synthesis, mitochondrial function and sarcomere structure maintenance [17,18]. mTOR pathway contributes to cell proliferation, survival, and embryogenesis of cardiomyocytes from embryonic to neonatal stage. On the other hand, the knockout and inhibition of mTOR in aged mice have resulted in the prolonged survival interval and the suppressive effect of cardiac hypertrophy. The gradual maladaptation of mTOR in the process of aging causes ROS formation, accumulation of unfolding proteins, cellular senescence, energy expenditure, and mitochondrial insufficiency. Furthermore, mTOR inhibition reactivates cardiac autophagy and acts on myocardium in a protective manner [27].
In the current study, we found that mTOR pathway identified from GWAS with lone AF was suppressed in human left atrial tissue obtained from elderly patients of their 60s–70s. Although it is generally thought that in the myocardial senescence, the quality control of proteins that contributes to the cell maintenance through metabolic integration in the young myocardium is impaired to maladaptive state, the result was contrary to expectations. The left atrial tissue in our microarray analysis was exposed to high pressure overload, which means that the observed changes in gene expression may not always reflect AF-related modeling. At least, this result showed that expression of several genes in mTOR pathway changed. Our findings indicated the association of mTOR dysfunction with AF. As a risk factor for atrial fibrillation, aging, cardiac hypertrophy, and metabolic perturbation are thought to be under quality control of mTOR. The microarray analysis indicated the suppression of mTOR pathway in myocardium as a result of the protective process against atrial remodeling.
One of the important findings in our study is that mTOR pathway had not been identified even in the several larger GWASs, which means that we need the careful interpretation for our results, thinking of the specificity of the population. First, we used lone AF cases. The characteristics of this phenotype of lone AF could affect the difference from GWAS including AF cases with many complications. Second, we used Japanese population. According to stroke GWAS, GWAS of cardiogenic and atherosclerotic stroke identified the different loci of overall stroke [28]. Our study using GWAS with these specific population with three characteristics could provide different results. The next one is that the pathway of cardiac development including PITX2 and TBX5 failed to be identified in our study. The cardiac developmental pathway is important and was identified in larger GWAS [15,16]. Our existing GWAS was thought to be purely underpowered. The components of the developmental pathways were under the level of significance.
In conclusion, MAGENTA identified a novel pathway, mTOR, followed by GSEA with human atrial tissue samples, although the interpretation of the study result might be quite limited. mTOR pathway is a key interface that adapts the change of environments by pressure overload and metabolic perturbation. mTOR pathway could be associated with AF pathogenesis.
4.1. Limitation
This study has several limitations. First, to minimize bias, we recruited participants with AF who had no complication except hypertension. On the other hand, the smaller sample size indicated the possibility of false positive result. Second, the study population consisted predominantly of Japanese patients. As for these two points, multi-ethnic pathway analysis with larger sample size should be required. Third, recent study with larger population did not detect this pathway. The inconsistency is the difference of the AF property of participants in each study. Fourth, the atrial tissue samples of surgery for validation study might be different from the participants in the existing GWAS. Fifth, mTOR pathway was suppressed in the atrial tissue with MVD contrary to our expectation. The background mechanism remains unclear and is required for further basic experiments.
The following are the supplementary data related to this article.
Quality control of DNA array analysis. We evaluated the samples with Affymetrix probe with Expression Console™1.4.1.46 to observe Probe Log2 Intensity Histogram, Signal Histogram of MAS5, Log Probe Cell Intensity, Relative Log Probe Cell Intensity, Log Expression Signal of MAS5, and Relative Log Expression Signal of MAS5. According to this result, all the samples were included in our study.
The result of array analysis. A. Scatter plot of array analysis was shown filtered by P < 0.001 in all the genes. Horizontal axis shows the intensity of genes in AF tissue and vertical axis shows the intensity of genes in non-AF tissue. B. Volcano plot of array analysis was shown filtered by P < 0.001 and fold change <−2, 2<. Horizontal axis shows Fold Change, and Vertical axis shows Significance (P value).
Transcriptome data of GeneChip® Human Genome U133Plus2.0 Array (Affymetrix).
The elements of the identified pathways through MAGENTA.
All the Enrichment Score of genes through GSEA.
All the Enrichment Score of the pathways in GSEA.
Fund support
This work was supported by a Grant-in-Aid for Scientific Research (C) (17K07251) from Japan Society for the Promotion of Science (JSPS).
Declaration of Competing Interest
No conflicts of interest to disclose.
Acknowledgement
We would also like to express our gratitude to the study participants and the doctors and research staff at the three study sites.
References
- 1.Kannel W.B., Abbott R.D., Savage D.D., McNamara P.M. Epidemiologic features of chronic atrial fibrillation: the Framingham study. N. Engl. J. Med. 1982;306:1018–1022. doi: 10.1056/NEJM198204293061703. [DOI] [PubMed] [Google Scholar]
- 2.Feinberg W.M., Blackshear J.L., Laupacis A., Kronmal R., Hart R.G. Prevalence, age distribution, and gender of patients with atrial fibrillation. Arch. Intern. Med. 1995;155:469–473. [PubMed] [Google Scholar]
- 3.Chugh S.S., Havmoeller R., Narayanan K., Singh D., Rienstra M., Benjamin E.J., Gillum R.F., Kim Y.H., McAnulty J.H., Jr., Zheng Z.J., Forouzanfar M.H., Naghavi M., Mensah G.A., Ezzati M., Murray C.J. Worldwide epidemiology of atrial fibrillation: a Global Burden of Disease 2010 Study. Circulation. 2014;129(8):837–847. doi: 10.1161/CIRCULATIONAHA.113.005119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrade Jason, Khairy Paul, Dobrev Dobromir, Nattel Stanley. The clinical profile and pathophysiology of atrial fibrillation-relationships among clinical features, epidemiology, and mechanisms. Circ. Res. 2014;114:1453?1468. doi: 10.1161/CIRCRESAHA.114.303211. [DOI] [PubMed] [Google Scholar]
- 5.Feinberg W.M., Blackshear J.L., Laupacis A., Kronmal R., Hart R.G. Prevalence, age distribution, and gender of patients with atrial fibrillation. Analysis and implications. Arch. Intern. Med. 1995;155:469–473. [PubMed] [Google Scholar]
- 6.Verdecchia P., Reboldi G., Gattobigio R., Bentivoglio M., Borgioni C., Angeli F. Atrial fibrillation in hypertension: predictors and outcome. Hypertension. 2003;41:218–223. doi: 10.1161/01.hyp.0000052830.02773.e4. [DOI] [PubMed] [Google Scholar]
- 7.Huxley R.R1., Filion K.B., Konety S., Alonso A. Meta-analysis of cohort and case-control studies of type 2 diabetes mellitus and risk of atrial fibrillation. Am. J. Cardiol. 2011;108:56–62. doi: 10.1016/j.amjcard.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mihm M.J., Yu F., Carnes C.A., Reiser P.J., McCarthy P.M., Van Wagoner D.R. Impaired myofibrillar energetics and oxidative injury during human atrial fibrillation. Circulation. 2001;104:174–180. doi: 10.1161/01.cir.104.2.174. [DOI] [PubMed] [Google Scholar]
- 9.Aviles R.J., Martin D.O., Apperson-Hansen C., Houghtaling P.L., Rautaharju P., Kronmal R.A. Inflammation as a risk factor for atrial fibrillation. Circulation. 2003;108:3006–3010. doi: 10.1161/01.CIR.0000103131.70301.4F. [DOI] [PubMed] [Google Scholar]
- 10.Heijman J., Algalarrondo V., Voigt N., Melka J., Wehrens X.H.T., Dobrev D., Nattel S. The value of basic research insights into atrial fibrillation mechanisms as a guide to therapeutic innovation: a critical analysis. Cardiovasc. Res. 2016;109(4):467?479. doi: 10.1093/cvr/cvv275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Armouche A., Boknik P., Eschenhagen T., Carrier L., Knaut M., Ravens U. Molecular determinants of altered Ca2+ handling in human chronic atrial fibrillation. Circulation. 2006;114:670–680. doi: 10.1161/CIRCULATIONAHA.106.636845. [DOI] [PubMed] [Google Scholar]
- 12.Heijman J., Voigt N., Nattel S., Dobrev D. Cellular and molecular electrophysiology of atrial fibrillation initiation, maintenance, and progression. Circ. Res. 2014;114(9):1483–1499. doi: 10.1161/CIRCRESAHA.114.302226. [DOI] [PubMed] [Google Scholar]
- 13.Goette A., Staack T., Röcken C., Arndt M., Geller J.C., Huth C. Increased expression of extracellular signal-regulated kinase and angiotensin-converting enzyme in human atria during atrial fibrillation. J. Am. Coll. Cardiol. 2000;35:1669–1677. doi: 10.1016/s0735-1097(00)00611-2. [DOI] [PubMed] [Google Scholar]
- 14.Verheule S., Sato T., Everett T., IV, Engle S.K., Otten D., Rubart-von der Lohe M. Increased vulnerability to atrial fibrillation in transgenic mice with selective atrial fibrosis caused by overexpression of TGF-beta1. Circ. Res. 2004;94:1458–1465. doi: 10.1161/01.RES.0000129579.59664.9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roselli C., Chaffin M.D., Weng L.C. Multi-ethnic genome-wide association study for atrial fibrillation. Nat. Genet. 2018;50(9):1225–1233. doi: 10.1038/s41588-018-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Low S.K., Takahashi A., Ebana Y., Ozaki K., Christophersen I.E., Ellinor P.T., AFGen Consortium Ogishima S., Yamamoto M., Satoh M., Sasaki M., Yamaji T., Iwasaki M., Tsugane S., Tanaka K., Naito M., Wakai K., Tanaka H., Furukawa T., Kubo M., Ito K., Kamatani Y., Tanaka T. Identification of six new genetic loci associated with atrial fibrillation in the Japanese population. Nat. Genet. 2017;49(6):953–958. doi: 10.1038/ng.3842. [DOI] [PubMed] [Google Scholar]
- 17.Ye J., Tucker N.R., Weng L.C., Clauss S., Lubitz S.A., Ellinor P.T. A functional variant associated with atrial fibrillation regulates PITX2c expression through TFAP2a. Am. J. Hum. Genet. 2016;99(6):1281–1291. doi: 10.1016/j.ajhg.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ebana Y., Ozaki K., Liu L., Hachiya H., Hirao K., Isobe M., Kubo M., Tanaka T., Furukawa T. Clinical utility and functional analysis of variants in atrial fibrillation-associated locus 4q25. J. Cardiol. 2017;70(4):366–373. doi: 10.1016/j.jjcc.2016.11.016. [DOI] [PubMed] [Google Scholar]
- 19.Imamura M., Takahashi A., Yamauchi T., Hara K., Yasuda K., Grarup N. Genome-wide association studies in the Japanese population identify seven novel loci for type 2 diabetes. Nat. Commun. 2016;7 doi: 10.1038/ncomms10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okada Yukinori, Di Wu Gosia Trynka, Raj Towfique. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506(7488):376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Segrè A.V., DIAGRAM Consortium, MAGIC Investigators, Groop L., Mootha V.K., Daly M.J. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellinor P.T., Lunetta K.L., Albert C.M., Glazer N.L., Ritchie M.D., Smith A.V. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat. Genet. 2012;44:670–675. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang D., Contu R., Latronico M.V., Zhang J., Rizzi R., Catalucci D. MTORC1 regulates cardiac function and myocyte survival through 4E-BP1 inhibition in mice. J. Clin. Invest. 2010;120:2805–2816. doi: 10.1172/JCI43008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sciarretta S1., Volpe M., Sadoshima J. Mammalian target of rapamycin signaling in cardiac physiology and disease. Circ. Res. 2014;114:549–564. doi: 10.1161/CIRCRESAHA.114.302022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sciarretta S., Forte M., Frati G., Sadoshima J. New insights into the role of mTOR signaling in the cardiovascular system. Circ. Res. 2018;122:489–505. doi: 10.1161/CIRCRESAHA.117.311147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malik R., Chauhan G., Traylor M., Sargurupremraj M. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet. 2018 Apr;50(4):524–537. doi: 10.1038/s41588-018-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quality control of DNA array analysis. We evaluated the samples with Affymetrix probe with Expression Console™1.4.1.46 to observe Probe Log2 Intensity Histogram, Signal Histogram of MAS5, Log Probe Cell Intensity, Relative Log Probe Cell Intensity, Log Expression Signal of MAS5, and Relative Log Expression Signal of MAS5. According to this result, all the samples were included in our study.
The result of array analysis. A. Scatter plot of array analysis was shown filtered by P < 0.001 in all the genes. Horizontal axis shows the intensity of genes in AF tissue and vertical axis shows the intensity of genes in non-AF tissue. B. Volcano plot of array analysis was shown filtered by P < 0.001 and fold change <−2, 2<. Horizontal axis shows Fold Change, and Vertical axis shows Significance (P value).
Transcriptome data of GeneChip® Human Genome U133Plus2.0 Array (Affymetrix).
The elements of the identified pathways through MAGENTA.
All the Enrichment Score of genes through GSEA.
All the Enrichment Score of the pathways in GSEA.