Abstract
Background and Aims
Reticulate evolution, coupled with reproductive features limiting further interspecific recombinations, results in admixed mosaics of large genomic fragments from the ancestral taxa. Whole-genome sequencing (WGS) data are powerful tools to decipher such complex genomes but still too costly to be used for large populations. The aim of this work was to develop an approach to infer phylogenomic structures in diploid, triploid and tetraploid individuals from sequencing data in reduced genome complexity libraries. The approach was applied to the cultivated Citrus gene pool resulting from reticulate evolution involving four ancestral taxa, C. maxima, C. medica, C. micrantha and C. reticulata.
Methods
A genotyping by sequencing library was established with the restriction enzyme ApeKI applying one base (A) selection. Diagnostic single nucleotide polymorphisms (DSNPs) for the four ancestral taxa were mined in 29 representative varieties. A generic pipeline based on a maximum likelihood analysis of the number of read data was established to infer ancestral contributions along the genome of diploid, triploid and tetraploid individuals. The pipeline was applied to 48 diploid, four triploid and one tetraploid citrus accessions.
Key Results
Among 43 598 mined SNPs, we identified a set of 15 946 DSNPs covering the whole genome with a distribution similar to that of gene sequences. The set efficiently inferred the phylogenomic karyotype of the 53 analysed accessions, providing patterns for common accessions very close to that previously established using WGS data. The complex phylogenomic karyotypes of 21 cultivated citrus, including bergamot, triploid and tetraploid limes, were revealed for the first time.
Conclusions
The pipeline, available online, efficiently inferred the phylogenomic structures of diploid, triploid and tetraploid citrus. It will be useful for any species whose reproductive behaviour resulted in an interspecific mosaic of large genomic fragments. It can also be used for the first generations of interspecific breeding schemes.
Keywords: Citrus, reticulate evolution, mosaic genome, GBS, polyploids, SNPs, phylogenomic karyotype
INTRODUCTION
Reticulate evolution is recognized as a major evolutionary process of eukaryotes and as a source of genetic diversity (Arnold, 2006). Interspecific and introgressive hybridization, recombination between genes, horizontal gene transfer and infectious heredity are the main mechanisms involved (Posada and Crandall, 2001; Linder and Rieseberg, 2004; Makarenkov and Legendre, 2004). Hybridization of genetically distinguishable populations, groups or taxa, leading to the production of viable hybrids (Barton and Hewitt, 1985; Mallet, 2005), has long been known to be involved in the emergence of plant species (Stebbins, 1950, 1959; Rieseberg, 1997; Abbott et al., 2010, 2013). Hybridization between species or subspecies has a significant weight in evolving processes including speciation, adaptation and extinction (Dowling and Secor, 1997; Barton, 2001; Yakimowski and Rieseberg, 2014). It can lead to rapid genomic changes (Baack and Rieseberg, 2007) and is an important source of genetic variability. Stebbins (1959) suggested that a high degree of genetic variability was required for major evolutionary advances; hence interspecific hybridization appears to be a predominant evolutionary force in plants. The evolutionary history of the concerned species cannot be correctly described using phylogenetic trees, but rather appears as a network (Stebbins, 1950; Grant, 1981; Arnold, 1997; Doolittle, 1999; Otto and Whitton, 2000) or a ‘Web of life’ (Arnold and Fogarty, 2009), generating phylogenetic discordance between nuclear and cytoplasmic (mitochondrial and chloroplast) genomes, and between different regions of the same nuclear genome (Pamilo and Nei, 1988; Rieseberg and Soltis, 1991; Linder and Rieseberg, 2004; Beiko and Hamilton, 2006). Reticulations lead not only to faulty phylogenetic conclusions, but also to interspecific heterozygosity of large portions of the genome when vegetative propagation involving apomixes, bulbs, tubers, corms, suckers, etc. takes place immediately or a few generations after reticulation events as described in fern (Dyer et al., 2012), banana (Perrier et al., 2009, 2011) or citrus (Curk et al., 2014). Deciphering this type of complex genome needs appropriate analytical approaches and tools based on a whole-genome scan.
The emergence of NGS (next-generation sequencing) technologies has considerably changed ways of analysing plant evolution, moving from phylogenetics to phylogenomics. The analysis of whole-genome variability has become possible and has already provided new information on the history of domestication of some cereals (Mascher et al., 2016; Meyer et al., 2016; Ramos-Madrigal et al., 2016; Pankin et al., 2018) and fruit crops, including grapes (Zhou et al., 2017), apples (Duan et al., 2017) and citrus (Wu et al., 2014, 2018). However, whole-genome re-sequencing (WGS) remains costly for studies of large populations. Therefore, cost-effective methods combining NGS and a reduction of the complexity of genomes have been developed, such as genotyping by sequencing (GBS) (Elshire et al., 2011), restriction site-associated DNA sequencing (RADseq) (Miller et al., 2007; Baird et al., 2008; Davey and Blaxter, 2011; Peterson et al., 2012) and sequenced-based genotyping (SBG) (Truong et al., 2012). These methods allow sufficient coverage of the genomes and are robust means for sampling whole genomes. They enable the analysis of large segregating progenies and marker trait association studies based on linkage disequilibrium and even genomic selection (Baxter et al., 2011; Ma et al., 2012; Poland et al., 2012; Ward et al., 2013; Wang et al., 2016; Curtolo et al., 2017). The efficiency of these methods has been demonstrated not only by constructing genetic maps and conducting genetic associations studies, but also by carrying out diversity analyses and revealing phylogenetically informative variation (Garcia et al., 2013; Escudero et al., 2014; Penjor et al., 2014, 2016; Hamon et al., 2017; Oueslati et al., 2017; Stetter and Schmid, 2017). More specifically, GBS has been used to perform genetic studies of numerous diploid and polyploid species, including maize (Crossa et al., 2013), wheat (Poland et al., 2012; Heslot et al., 2013), barley (Poland et al., 2012; Liu et al., 2014), rice (Huang et al., 2009; Courtois et al., 2013; Spindel et al., 2013), ryegrass (Byrne et al., 2013), soybean (Sonah et al., 2013), chickpea (Verma et al., 2015), sugarcane (Almeida Balsalobre et al., 2017), banana (Martin et al., 2016) and citrus (Oueslati et al., 2017). However, for polyploid species, due to the generally low read depths at individual single nucleotide polymorphism (SNP) loci, genotyping has been limited to the identification of homozygous genotypes (nulliplex or quadriplex for a tetraploid) or heterozygous genotypes, joining the different classes of heterozygosity (simplex, duplex, triplex for a tetraploid) in the same genotyping class (Clevenger et al., 2015; Rocher et al., 2015; Almeida Balsalobre et al., 2017; Yang et al., 2017). For tetraploid potatoes, a technical solution has been proposed to improve the individual SNP read depths by combining GBS with enriched cultivar-specific DNA sequencing libraries using an in-solution hybridization method (SureSelect), reducing the genome to 807 target genes distributed across the genomes (Uitdewilligen et al., 2013). New analytical methods have also been proposed to deal with the low read depths. Rather than calling genotypes, Ashraf et al. (2014) and Sverrisdóttir et al. (2017) directly used the variant allele frequencies at each data point for association studies and genomic selection from GBS data. New pipelines have also been proposed to estimate allele doses at an individual locus (McKinney et al., 2018; Bastien et al., 2018), but it remains challenging.
The Citrus genus is a good example of a gene pool resulting from reticulate evolution, where apomixes and vegetative propagation have fixed ancient reticulation events and limited further interspecific recombination, resulting in mosaics of large genome fragments from different species (Nicolosi et al., 2000; Wu et al., 2014, 2018; Curk et al., 2016). Molecular marker analyses enabled the main lines of the phylogeny of the different cultivated species of Citrus to be drawn and the identification of the various domestication events (Federici et al., 1998; Nicolosi et al., 2000; Barkley et al., 2006; Li et al., 2010; Garcia-Lor et al., 2012, 2013; Ollitrault et al., 2012a, b; Ramadugu et al., 2013; Curk et al., 2016). Four taxa [C. medica L. (citron), C. reticulata Blanco (mandarin), C. maxima (Burm.) Merr. (pummelo) and C. micrantha Wester (papeda)] have been identified as being the ancestors of most of the cultivated citrus (Nicolosi et al., 2000; Garcia-Lor et al., 2012; Ollitrault et al., 2012b; Ramadugu et al., 2013; Curk et al., 2014, 2015; Wu et al., 2018). These four ancestral taxa, which are still sexually compatible, were differentiated by foundation effects and allopatric evolution in four South-east Asian geographic regions ranging from the southern Himalayas to Indonesia. Pummelos originated in the Malay Archipelago and Indonesia. Citrons evolved in north-eastern India and in the nearby areas of Myanmar and China. Mandarins were diversified over a wide region which includes Vietnam, southern China and Japan, while C. micrantha is endemic to the Philippine islands (Wester, 1915; Tanaka, 1954; Webber et al., 1967; Scora, 1975). Secondary species [C. sinensis (L.) Osb. (sweet orange), C. aurantium L. (sour orange), C. paradisi Macf. (grapefruit), C. limon (L.) Burm. (lemon) and C. aurantiifolia (Christm.) Swing. (lime)] and modern cultivars are the result of hybridizations between the four basic taxa (Nicolosi et al., 2000; Garcia-Lor et al., 2013; Curk et al., 2016) engendering the wide genetic and phenotypic diversity observed among them. In terms of morphological characteristics (Ollitrault et al., 2003), carotenoid compositions (Fanciullino et al., 2006) and the distribution of coumarins and furanocoumarins (Dugrand-Judek et al., 2015), the structure of phenotypic variability is closely linked with the reticulate evolution of the gene pool. Therefore, in parallel with the search for the origin of cultivated forms and the optimization of genetic resources management, deciphering the phylogenomic structures of modern cultivars will open the way for association studies based on ancestral haplotypes and phylogenomic-based reconstruction breeding strategies (Rouiss et al., 2018). The accurate study of citrus interspecific mosaic genomes started with the release of the first high-quality citrus reference haploid genome by the International Citrus Genomics Consortium (ICGC; Wu et al., 2014). WGS data revealed Citrus maxima introgressions in traditional mandarin genomes (Wu et al., 2014) and the interspecific mosaic structure of sweet orange (Xu et al., 2013; Wu et al., 2014), sour orange and clementine (Wu et al., 2014). More recently, WGS data (Wu et al., 2018), including the four Citrus ancestral species and modern varieties, revealed the mosaic genome structures of the other most important horticultural groups, such as grapefruit, lemon and lime, and confirmed C. maxima introgressions in all domesticated mandarins.
A GBS approach was recently applied to analyse the interspecific admixture of diploid secondary species and modern varieties resulting from two Citrus gene pools, C. reticulata and C. maxima (Oueslati et al., 2017). To date, the phylogenomic structures of the citrus polyploid germplasm remain unpublished.
The objectives of the present work were to (1) develop a GBS approach in Citrus with a dense genotyping and a good depth, to decipher – at limited cost – the phylogenomic structures of large diploid and polyploid populations originating from a limited number of interspecific recombinations between C. reticulata, C. maxima, C. medica and C. micrantha gene pools; (2) provide a reference matrix of diagnostic SNP (DSNP) markers for the four Citrus ancestral taxa; (3) implement a generic workflow for mosaic genome analysis from GBS data of diploid and polyploid populations resulting from reticulate evolution; and (4) analyse the phylogenomic structure of modern varieties of the main citrus diploid and polyploid horticultural groups. As proof of concept, 53 citrus accessions, including several varieties already analysed using WGS (Wu et al., 2014, 2018), were sequenced in a single Illumina Hiseq 2000 line, using the restriction enzyme ApeKI and a selective PCR for GBS library preparation. Close to 16 000 DSNPs were identified and successfully used to decipher the complex genomes of the 53 accessions, using a workflow based on maximum likelihood analysis of multilocus ancestral read numbers. The GBS approach we developed combined with the reference DSNP matrix will be useful for any study of germplasm and hybrids resulting from breeding within the Citrus genus. The implemented workflow for the analysis of mosaic genomes is available online and will be useful for species with any number of identified ancestral taxa, for diploid, triploid and tetraploid accessions.
MATERIALS AND METHODS
Plant material
The study covered 53 accessions from the collection of the Inra-Cirad Citrus Biological Resource Center in San-Giuliano, Corsica, France (Luro et al., 2018). The varieties belong to the Citrus genus, and 29 of them are representative of the four ancestral taxa: 15 mandarins, six pummelos, six citrons and two papedas. They were used to identify diagnostic markers of the basic taxa. The other varieties, which are diploid, triploid and tetraploid, came from admixtures of the four ancestral taxa: two sour oranges (C. aurantium), two sweet oranges (C. sinensis), five lemons (C. limon, C. limonia Osb., C. meyeri Y. Tan. and C. jambhiri Lush.), eight limes (C. aurantiifolia, C. latifolia Tan., C. excelsa Wester, C. limettioïdes Tan.), one ‘Alemow’ (C. macrophylla Wester), three grapefruits (C. paradisi), one bergamot (C. bergamia Risso & Poit.), one clementine (C. clementina Hort. ex Tan.) and one limonette (C. limetta Risso). In order to validate our method of deciphering the citrus interspecific mosaic structure, we included some accessions already described from WGS data by Wu et al. (2014, 2018). A summary list of the varieties analysed with their classification in two widely used taxonomic systems [the Tanaka (1954) and Swingle and Reece (1967) systems] is available in Supplementary Data Table S1. Recent genetic and genomic studies demonstrated the limits of both systems resulting from reticulate evolution of the citrus gene pool and vegetative propagation of interspecific combination by apomictic seeds (Curk et al., 2016; Wu et al., 2018). Herein we refer to the Tanaka system for the secondary species (the types issued from interspecific combinations); indeed, although they cannot be considered as true species, the Tanaka classification has the advantage of distinguishing secondary taxa that have arisen from different reticulation events. Supplementary Data Table S1 also specifies whether the phylogenomic structure of each accession has already been analysed from WGS (Wu et al., 2014, 2018) or GBS (Oueslati et al., 2017) or was analysed for the first time in the present study.
GBS analysis
Library preparation and sequencing
Following the protocol of Oueslati et al. (2017), genomic DNA was isolated using the Plant DNAeasy® kit (Qiagen), according to the manufacturer’s instructions. Several in silico tests were carried out using numerous types of restriction enzymes and selective primers. The method selected consists of using the restriction enzyme ApeKI and adding a selective base (A) during the PCR step of GBS library preparation as it was found to provide a good combination of tag density and read numbers per tag. ApeKI also has the advantage of cutting DNA preferentially in gene sequences enabling better quality genotype calling (Oueslati et al., 2017). The genomic DNA concentration was adjusted to 20 ng μL–1, and ApeKI GBS libraries were prepared following the protocol described by Eslhire et al. (2011). DNA of each sample (200 ng) was digested with ApeKI (New England Biolabs, Hitchin, UK). Digestion took place at 75 °C for 2 h and then at 65 °C for 20 min to inactivate the enzyme. The ligation reaction was completed in the same plate as the digestion, again using T4 DNA ligase (New England Biolabs) at 22 °C for 1 h, and the ligase was inactivated prior to pooling the samples by holding it at 65 °C for 20 min. Ligated samples were pooled and PCR-amplified in a single tube. Complexity was further reduced using PCR primers with one selective base (A) as performed by Sonah et al. (2013). Single-end sequencing was performed on a single lane of an Illumina HiSeq2000. The Illumina Hiseq 2000 sequencing raw data are available in the NCBI SRA (Sequence Read Archive), under the accession numbers SRP109295 for the 21 mandarin, pummelo, orange, grapefruit and clementine sequences already published in Oueslati et al. (2017; Supplementary Data Table S1) and PRJNA388540 for the 32 new citrus accessions. Keygene N.V. owns patents and patent applications protecting its Sequence Based Genotyping technologies.
SNP genotype calling for diploid germplasm
The Tassel 4.0 pipeline (Glaubitz et al., 2014) was used to call SNPs from the DNA sequence reads from the Illumina raw data (unfiltered fastq file). The Tassel 4.0 GBS pipeline identified good quality, unique, sequence reads with barcodes. These sequence tags were aligned to the C. clementina 1.0 reference genome (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Cclementina) using Bowtie2 v2.2.6 (Langmead and Salzberg, 2012). For genotype calling, five reads were considered as a minimum below which they were considered as missing data (Danecek et al., 2011). We finally only considered diallelic polymorphic positions with <30 % of missing data for the 29 representatives of C. reticulata, C. maxima, C. medica and C. micrantha, and a minor allele frequency (MAF) >0.05.
Genetic parameters
The following parameters were used to describe the genetic diversity within and between the ancestral taxa: Ho, the observed heterozygosity; He, the expected proportion of heterozygous loci per individual under Hardy–Weinberg equilibrium defined as He = 1 − Σpi2, with pi the frequency of a given allele in the sub-population concerned or in the whole population; and Fw, the fixation index (Wright, 1951) defined as follows:
They were calculated using GENETIX v. 4.03 software (Belkhir et al., 1996–2004) based on the 43 598 diallelic selected markers.
The analysis consisting of identifying the diagnostic markers of the four basic taxa was mainly based on GST parameter estimations (Nei, 1973). GST is the coefficient of gene differentiation which measures differentiation among sub-populations. It is equivalent to Wright’s FST for two alleles and ranges from zero to one. The higher the value, the more differentiated the taxa. GST is defined as the ratio of inter-population diversity to total diversity:
where HeTot is the total genetic diversity of the whole population, Hs the average diversity within sub-populations and n is the number of sub-populations. In our study, we had four sub-populations comprising representative varieties of the four ancestral taxa.
H e is the expected proportion of heterozygous loci per individual under Hardy–Weinberg equilibrium (He = 1 − Σpi2, with pi the frequency of a given allele in the sub-population concerned or in the whole population HeTot).
The search for diagnostic SNPs for each taxon was based on GST parameter estimations for the taxa concerned considering two sub-populations: (1) the taxon concerned (Ti) and (2) a theoretical population of the three other basic taxa (T–i). Analyses were performed from the estimated allele frequency of each taxon considering the same population size for each taxon to estimate the frequency of the two sub-populations (Ti and T–i) and the frequency of the whole population (Tot):
Allele frequencies and GST estimation were computed in Excel from the genotyping matrix.
Analysis of population organization
We analysed the organization of genetic diversity of the 48 diploid varieties used in the study. A principal component analysis (PCA) was performed on them based on the 43 598 selected diallelic markers using the {ade4} (Chessel et al., 2004) R package.
Hierarchical ascending clustering was carried out for the representative accessions of the four ancestral taxa from the same matrix of diallelic markers. We produced a dissimilarity matrix by calculating the Euclidean distances between each pair of markers and hierarchical clustering using Ward’s method applied to the square of distances. Data were computed using the {stats} (R Core Team, 2017) R package, and the result was visualized using the {dendextend} (Galili, 2015) R package.
Identification of interspecific introgressions in representative varieties of the ancestral taxa and selection of DSNPs of the ancestral taxa
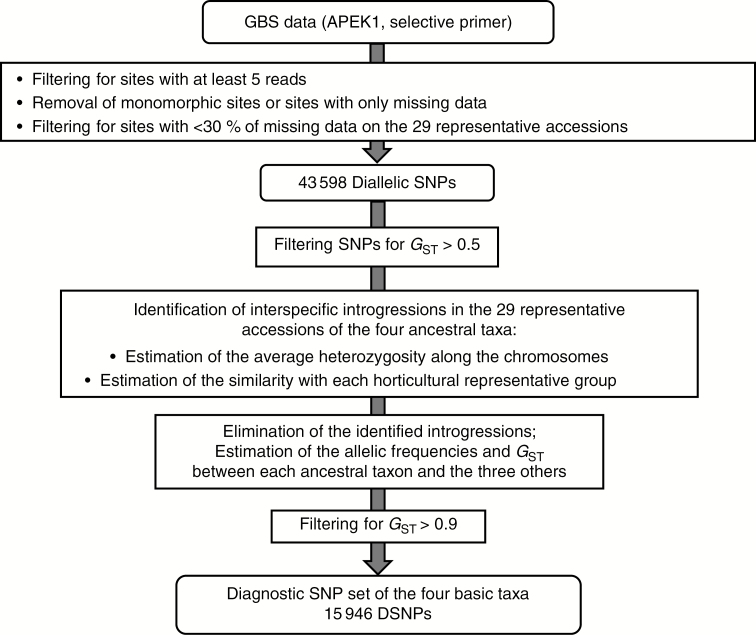
The identification of diagnostic markers of the four ancestral taxa from the GBS data is schematized in the workflow in Fig. 1. Some of the accessions cited above, mostly in the mandarin group, are already known to be non-pure (Curk et al., 2014; Wu et al., 2014, 2018). They were the result of a domestication process of the real ancestors which led to interspecific introgressions. Consequently, implementing a diagnostic marker set required the identification of interspecific introgressions among the varieties considered as representatives of the ancestral taxa, and of removing these regions of the variety under consideration from the analysis. This process provided a better estimation of the allelic frequencies of the ancestral taxa and hence of the GST parameter in the four basic taxa. The identification of the interspecific introgressed areas was based on the pattern of two parameters along the genome using consecutive non-sliding 20 SNP windows: (1) the average heterozygosity estimated from the matrix of SNP positions and (2) the similarity of the accession to the centroid of each of the four horticultural representative groups (the allelic frequencies of the centroid being the average frequency of the varieties of the considered group). It was expected that introgressed areas would display significant discontinuity of these patterns according to the level of differentiation between the two taxa involved. Indeed, heterozygous introgressions resulted in regions with an increase in heterozygosity and a decrease in the similarity, while homozygous introgressions resulted in a deep variation in the similarity patterns. To better visualize the pattern discontinuities, SNPs that were informative for the differentiation of one out of the four horticultural groups, representative of the ancestral taxon (GST >0.5), were filtered out. Once the interspecific introgressions were removed (considered as missing data), the allelic frequencies in the four ancestral taxa and the GST parameter between each ancestral taxon and the three others were estimated again. We then considered SNPs with a GST value (the taxon concerned relative to a sub-population of the three other ancestral taxa) >0.9 as diagnostic markers of a given taxon.
Fig. 1.
Workflow for the identification of diagnostic markers of the four ancestral taxa (C. maxima, C. reticulata, C. medica and C. micrantha) from GBS reads.
Analysis of the interspecific mosaic structure of complex genomes
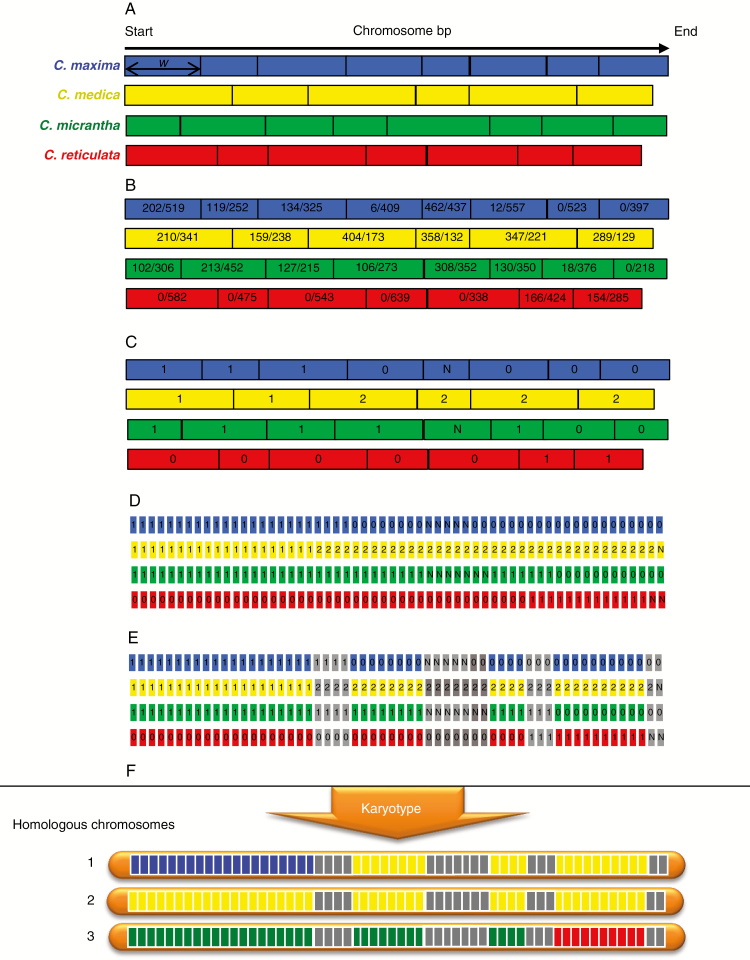
The objective was to develop a generic pipeline to decipher complex genomes resulting from reticulate evolution at diploid and polyploid levels, based on the availability of a set of diagnostic markers of the ancestral taxa involved (all along the genome) and GBS data of new populations obtained with the same experimental procedure as the reference DSNP set. According to our experimental data (see below) and reports in the literature (Bastien et al., 2018; McKinney et al., 2018), it is often difficult to estimate allelic doses at a single locus accurately in heterozygous polyploids from relative allele read frequencies resulting from GBS experiments. We developed an approach based on maximum likelihood analysis applied to multilocus numbers of reads of consecutive DSNPs of the same ancestor, that can be used for diploid, triploid and tetraploid plants. This approach is described below in the concrete case of citrus with four ancestral taxa, but the tool we developed can be used with models of any number of ancestral taxa. An illustration of the process for a triploid plant is provided in Fig. 2.
Fig. 2.
Example of local ancestor allele dose estimation for a triploid accession. (A) Definition of non-overlapping windows of ten DNSPs for each ancestral taxon: w, window of ten DSNPs. (B) Number of reads of the considered ancestor allele/number of reads of the alternative allele. (C) Estimation of allelic dosage of each ancestor per window of ten DSNPs [each pair of dose hypotheses are compared by maximum likelihood (LOD) test; if, for a pair including the more probable hypothesis, –3 < LOD < 3 → indeterminacy]. (D) Division of the chromosome into non-overlapping windows of 100 kb; the allelic dosage of this window is deduced from that of the ten DSNPs window that include the 100 kb window. (E) If the sum of allelic dosage of the four classes of DSNPs is different from the expected ploidy (here 3) → indeterminacy (grey). (F) Unphased karyotype automatic drawings. Blue, C. maxima; yellow, C. medica; green, C. micrantha; red, C. reticulata; grey, indeterminacy.
The first step aims to estimate the doses of the ancestral genome fragment along the genome. For each ancestral taxon, the citrus genome was segmented in windows of w consecutive DSNPs (Fig. 2A) and the doses of the ancestral taxon considered were estimated for each window by maximum likelihood analysis (Fig. 2C). The detail for the maximum likelihood analysis for diploid, triploid and tetraploid individuals is provided in Supplementary Data Text S1.
During the preceding step, the number and position of windows varied between the ancestral taxa according to the density and positions of the DSNPs. Therefore, the next step was to integrate the information obtained for the different ancestral taxa doses along the genome.
The genome was physically sub-divided into successive fragments of z kb (by default z = 100) (Fig. 2D). For each ancestor and for each genomic fragment, the corresponding window of w DSNPs was identified and the ancestral dose of this window was attributed to the genomic fragment. A non-phased representation of karyotypes with two, three and four chromosomes for diploid, triploid and tetraploid plants, respectively, was then generated from the ancestral doses of each genome fragment (Fig. 2F). For a given genome fragment, if the sum of the allelic doses of the different ancestors differed from the ploidy level of the plant concerned, the phylogenomic origin of the fragment was considered as undefined. Likewise, if one of the doses of the different ancestors was undefined, the phylogenomic origin of the fragment was considered as undefined (Fig. 2E). When phased haplotypes were known for the parental genomes, we proposed manually phased karyotypes for the concerned accession, assuming the lower number of recombination events as the best model.
The tool we developed (TraceAncestor) allows the user to define the number of DSNPs per window (by default: w = 10), the sequencing error rate (by default: e = 0.01) and the threshold for LOD values of the maximum likelihood test (by default: t = 3; the probability of the best hypothesis is >1000 times greater than the other one). There is no limit to the number of ancestral taxa considered (which is automatically defined by the reference matrix of DSNPs). This pipeline is available as a Galaxy workflow at http://galaxy.southgreen.fr/galaxy/ and for download at https://github.com/SouthGreenPlatform/galaxy-wrappers/tree/master/Galaxy_SouthGreen/traceancestor.
RESULTS
Genotype calling and varietal diversity
Figure 1 shows the workflow for the identification of diagnostic markers. The 53 varieties considered were part of two 55 plex libraries sequenced in two lanes of a Hiseq 2000 according to the Cornell GBS methodology (Elshire et al., 2011) using ApeKI as the restriction enzyme and a selective primer. A total of 344.8 million reads were obtained. The Tassel pipeline was used for genotype calling, and 314.2 million of these reads were validated (bar code, restriction site plus insert), and 290.7 million were mapped on the clementine reference genome (Wu et al., 2014). The average number of reads per variety was 2.2 million, ranging from 609 890 for ‘Meyer’ lemon to 5.68 million for ‘Shekwasha’ mandarin (Supplementary Data Fig. S1). A total of 2.045 million tags (unique sequence with at least five reads) were identified, of which half were only mapped once on the clementine reference genome. Genotype calling from the tags with a single hit map was undertaken considering a position with less than five reads as missing data. A total of 43 598 diallelic SNPs were selected, and filtered for sites with <30 % of missing data on the 29 representative accessions. The 35 and 84 % of the SNPs retained had, respectively, <5 % and <25 % of missing data (Supplementary Data Fig. S2A). At the individual level, 29.6 and 90.7 % of the varieties had, respectively, <5 % and <25 % of missing data (Supplementary Data Fig. S2B). ‘Meyer’ lemon had the highest rate of missing data: 35 %. The distribution of the read numbers per marker (Supplementary Data Fig. S3) appeared to be globally homogeneous among the nine chromosomes, with a mean value of 1024 reads. However, a decrease in the number of reads was observed in the middle of chromosomes 2, 4, 5, 8 and 9.
The distribution of the 43 598 mined polymorphisms on the nine chromosomes is reported in Table 1. The number of diallelic SNPs varied between 3611 SNPs on chromosome 8 and 7743 SNPs on chromosome 3. Little variation was observed among the expected heterozygosity values along the nine chromosomes, with an average of 0.309, or in the observed heterozygosity values which ranged between 0.197 (chromosome 2) and 0.227 (chromosome 6), with an average of 0.213. According to the Hardy–Weinberg equilibrium, the analysed population displayed a heterozygote deficiency with the Fw parameter equal to 0.282.
Table 1.
Polymorphisms mined from GBS data on 53 citrus varieties along the nine chromosome
| n | H o | H e | F w | |
|---|---|---|---|---|
| C1 | 4180 | 0.208 ± 0.115 | 0.314 ± 0.134 | 0.308 ± 0.279 |
| C2 | 5536 | 0.197 ± 0.106 | 0.312 ± 0.135 | 0.323 ± 0.278 |
| C3 | 7743 | 0.211 ± 0.122 | 0.308 ± 0.137 | 0.285 ± 0.280 |
| C4 | 4586 | 0.200 ± 0.109 | 0.307 ± 0.137 | 0.308 ± 0.265 |
| C5 | 5565 | 0.215 ± 0.135 | 0.311 ± .136 | 0.280 ± 0.317 |
| C6 | 3875 | 0.227 ± 0.130 | 0.309 ± 0.139 | 0.249 ± 0.264 |
| C7 | 3739 | 0.213 ± 0.117 | 0.306 ± 0.137 | 0.276 ± 0.261 |
| C8 | 3611 | 0.222 ± 0.127 | 0.309 ± 0.135 | 0.256 ± 0.282 |
| C9 | 4763 | 0.224 ± 0.133 | 0.309 ± 0.135 | 0.255 ± 0.295 |
| Total | 43 598 | 0.213 ± 0.01 | 0.309 ± 0.002 | 0.282 ± 0.025 |
n, number of polymorphisms; Ho, observed heterozygosity; He, expected heterozygosity; Fw, Wright fixation index; C1–C9. the nine chromosomes of the reference clementine genome (Wu et al., 2014).
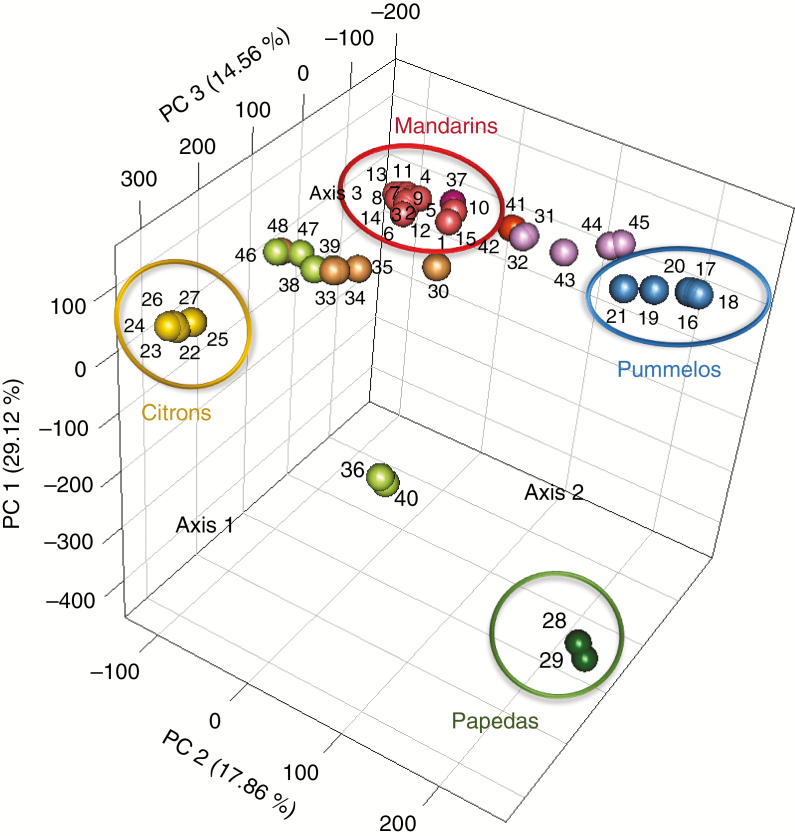
Based on the 43 598 diallelic SNPs, we performed a three-dimensional representation of the PCA to examine the genetic diversity of the 48 diploid citrus accessions (Fig. 3). The four main observed clusters corresponded to the four ancestral taxa (pummelos, mandarins, citrons and papedas). The first three axes represent 61.54 % of total diversity and clearly separate the four clusters of the ancestral taxa. Other clusters made of secondary species appeared between the ancestral clusters and revealed their genetic relationship. Lemons [‘Lisbon’ lemon (33), ‘Meyer’ lemon (34), ‘Eureka’ lemon (35), ‘Rough’ lemon (47) and ‘Volkamer’ lemon (48)], ‘Palestine’ sweet lime (38), ‘Marrakech’ limonette (39) and ‘Rangpur’ lime (46) were in an intermediate position between C. reticulata and C. medica clusters. Bergamot (30) was located close to the mandarin group but still in an intermediate position between the mandarin, pummelo and citron groups. Grapefruits [‘Duncan’ (43), ‘Marsh’ (44) and ‘Star Ruby’ (45)], sour oranges [‘Seville’ (31) and ‘Bouquetier de Nice’ (32)] and sweet oranges [‘Valencia late’ (41) and ‘Washington navel’ (42)], rather logically given their origin revealed by markers (Curk et al., 2015), previous GBS studies (Oueslati et al., 2017) and WGS analysis (Wu et al., 2014, 2018), were in an intermediate position between C. reticulata and C. maxima. ‘Nestour’ lime (36) and ‘Alemow’ (40) were located between C. medica and C. micrantha, in agreement with their origin proposed by Curk et al. (2016).
Fig. 3.
Principal component analysis (PCA) calculated from genotype data of the 48 diploid accessions corresponding to the 43 598 diallelic SNPs. 1, ‘Shekwasha’ mandarin; 2, ‘de Chios’ mandarin; 3, ‘Owari Satsuma’ mandarin; 4, ‘Nan feng mi chu’ mandarin; 5, ‘San Hu Hong Chu’ mandarin; 6, ‘Se Hui Gan’ mandarin; 7, ‘Szibat’ mandarin; 8, ‘Cleopatra’ mandarin; 9, ‘Dancy’ mandarin; 10, ‘Fuzhu’ mandarin; 11, ‘King’ mandarin; 12, ‘Ladu’ mandarin; 13, ‘Ponkan’ mandarin; 14, ‘Sunki’ mandarin; 15, ‘Willowleaf’ mandarin; 16, ‘Chandler’ pummelo; 17, ‘Timor’ pummelo; 18, ‘Deep red’ pummelo; 19, ‘Kao Pan’ pummelo; 20, ‘Pink’ pummelo; 21, ‘Tahitian’ pummelo; 22, ‘Corsican’ citron; 23, ‘Buddha’s Hand’ citron; 24, ‘Etrog’ citron; 25, ‘Humpang’ citron; 26, ‘Mac Veu de Montagne’ citron; 27, ‘Poncire commun’ citron; 28, ‘Small flower’ papeda 1; 29, ‘Small flower’ papeda 2; 30, Bergamot; 31, ‘Seville’ sour orange; 32, ‘Bouquetier de Nice’ sour orange; 33, ‘Lisbon’ lemon; 34, ‘Meyer’ lemon; 35, ‘Eureka’ lemon; 36, ‘Nestour’ lime; 37, ‘Nules’ clementine; 38, ‘Palestine’ sweet lime; 39, ‘Marrakech’ limonette; 40, ‘Alemow’; 41, ‘Valencia late’ sweet orange; 42, ‘Washington navel’ sweet orange; 43, ‘Duncan’ grapefruit; 44, ‘Marsh’ grapefruit; 45, ‘Star Ruby’ grapefruit; 46, ‘Rangpur’ lime; 47, ‘Rough’ lemon; 48, ‘Volkamer’ lemon.
Diversity among the four ancestral taxa and search for diagnostic markers
Genetic parameters
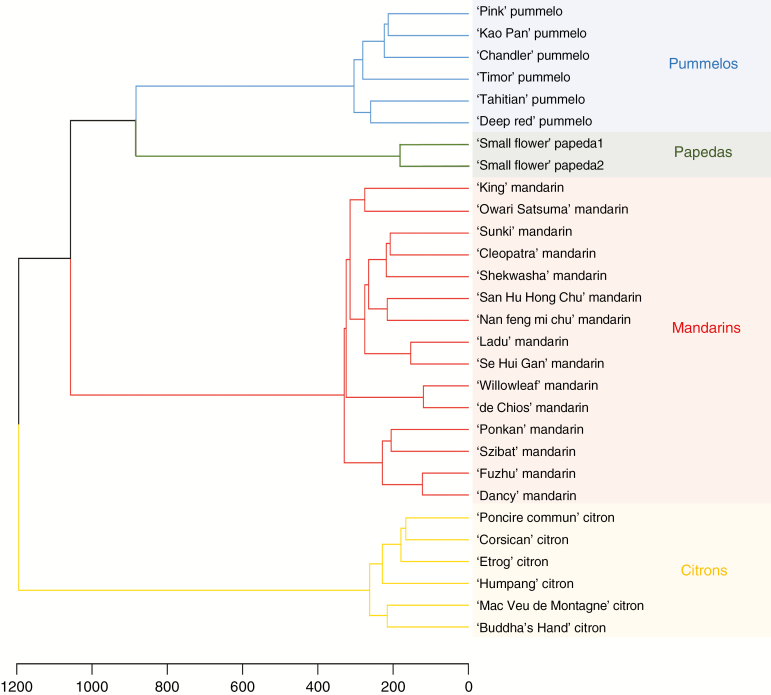
Analyses of the diversity among the 29 representative accessions (Table 2) revealed a marked difference in the number of polymorphic positions within each horticultural group: 18 567, 7325, 7156 and 2285 for mandarins, pummelos, citrons and papedas, respectively. The expected heterozygosity values (0.11, 0.07, 0.04 and 0.03 for mandarins, pummelos, citrons and papedas, respectively) ranked in the same order as the number of polymorphic loci. Thus, the mandarin set is the most polymorphic of the four representative sets. Conversely, papedas present the lowest intraspecific diversity, probably due to the fact that they are represented by only two accessions. The deficit of heterozygosity in citrons revealed by the positive Fw value can be explained by the cleistogamy of this group, while negative value observed in pummelos and mandarins could be related, respectively, to self-incompatibility and heterozygosity fixation by apomixes. The average values of the differentiation index (Fw = –0.12 and GST = 0.78) between the four representative sets revealed, as expected, marked genetic differentiation among the four populations. Hierarchical cluster analysis (Fig. 4), computed from the 43 598 diallelic SNPs, confirmed strong clustering of the four ancestral taxa and revealed greater differentiation between citrons and the other groups, and a closer relationship between pummelos and papedas.
Table 2.
Diversity of the 29 accessions representative of the four ancestral taxa
| n | H o | H e | F w | G ST | |
|---|---|---|---|---|---|
| Mandarins (Na = 15) | 18 567 | 0.121 ± 0.200 | 0.110 ± 0.162 | –0.107 | |
| Pummelos (Na = 6) | 7325 | 0.086 ± 0.212 | 0.070 ± 0.154 | –0.001 | |
| Citrons (Na = 6) | 7156 | 0.041 ± 0.163 | 0.044 ± 0.128 | 0.52 | |
| Papedas (Na = 2) | 2285 | 0.016 ± 0.068 | 0.028 ± 0.113 | –0.907 | |
| Total (Na = 29) | 35 333 | 0.066 ± 0.04 | 0.063 ± 0.031 | –0.1237 | 0.7831139 |
n, number of polymorphisms; Ho, observed heterozygosity; He, expected heterozygosity; Fw, Wright fixation index; GST, interpopulation differentiation parameter; Na, number of accessions per taxon.
Fig. 4.
Hierarchical cluster analysis of the 29 representative accessions computed from the 43 598 diallelic SNPs.
Search for ancestral taxa diagnostic markers (DSNPs).
Removing the interspecific introgressed areas from the varieties representative of the four ancestral taxa was an important step to estimate effectively the allelic frequencies of the ancestral taxa and the differentiation parameter (GST) between the four ancestral taxa at each polymorphic position. The introgressions were identified through the analysis of the discontinuity in the pattern of two parameters along the genome: the heterozygosity and the similarity between the accession and the centroids of each horticultural group, representative of the ancestral taxa.
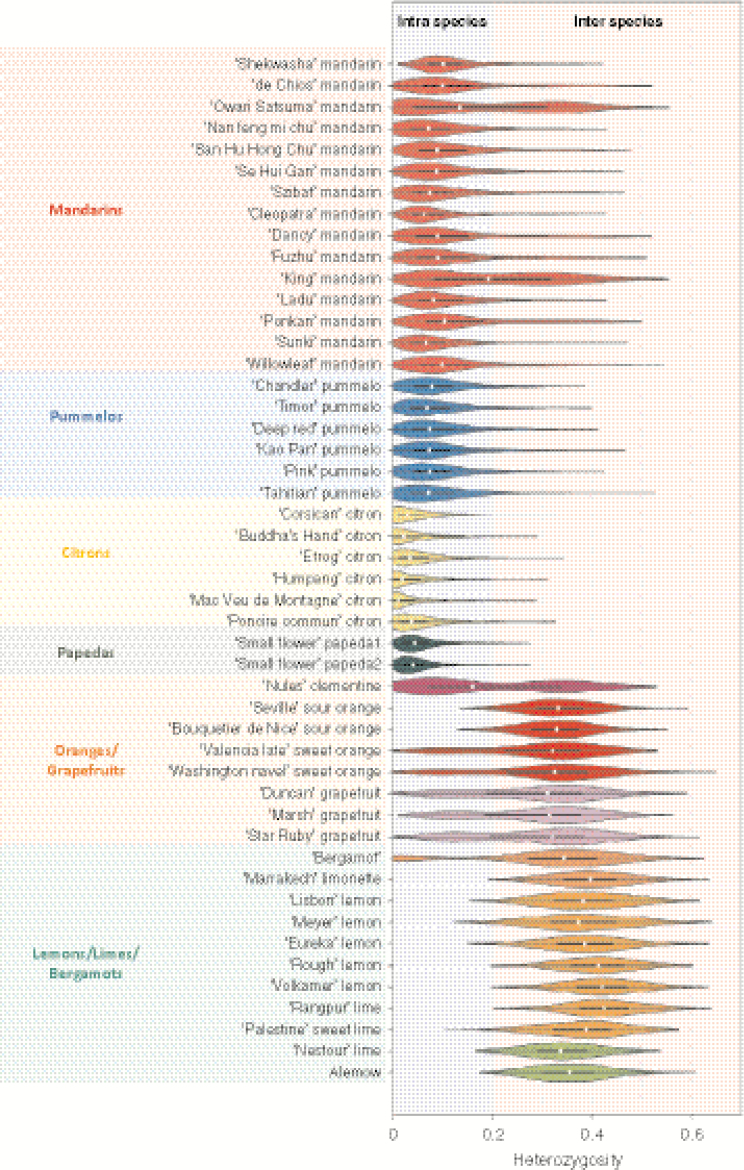
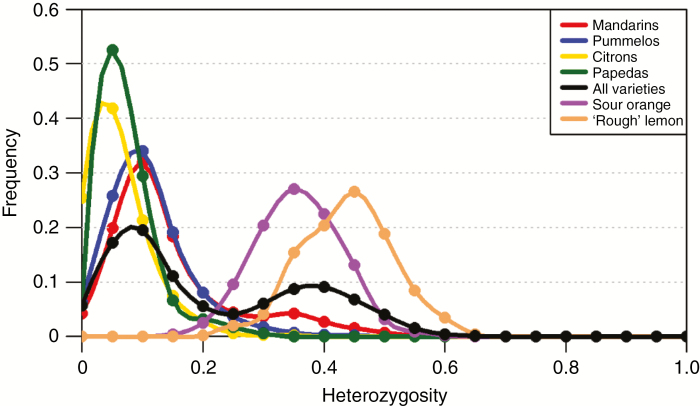
We examined the distribution of the observed heterozygosity of the diploid accessions with 100 polymorphic positions per window. Two main modes of distribution were observed among the varieties plotted individually (Fig. 5) or in sets (Fig. 6). These two modes correspond to intraspecific and interspecific heterozygosity, with values ranging between 0 and 0.2 and 0.2 and 0.7, respectively. Three distinct types of accessions were highlighted. The first type displayed a unimodal distribution with a high value (the average value of each accession was between 0.3 and 0.4) corresponding to interspecific heterozygosity. Accessions of this type probably result from direct two-way or three-way interspecific hybridization. Sour oranges, all lemons, ‘Marrakech’ limonette, ‘Rangpur’, ‘Palestine’ and ‘Nestour’ limes, as well as ‘Alemow’ displayed this pattern. A higher mid-value was observed for ‘Rough’ lemon than for sour orange [explained by the greater differentiation of C. reticulata (mandarins) with C. medica (citrons) than C. maxima (pummelos)]. Indeed, from WGS data, Wu et al. (2014, 2018) concluded that ‘Rough’ lemon and sour orange resulted from direct interspecific hybrids of C. reticulata with C. medica and C. maxima, respectively. The second type grouped the representative accessions of the basic taxa, except for the majority of mandarins. Pummelos, citrons and papedas displayed unimodal distribution, with average values of 0.09, 0.04 and 0.05, respectively. The representative mandarins belong to the third type of accessions with a bimodal distribution of heterozygosity, such as sweet orange, grapefruit, clementine and bergamot. The interspecific admixture among these accessions was highlighted. The same pattern of distribution of heterozygosity in sweet orange was reported in Wu et al. (2014, 2018) from WGS data and in Oueslati et al. (2017) from GBS data. More specifically, the set of mandarins showed a first peak around 0.1, close to the peak of the set of pummelos, and a second slight peak with a mode of approx. 0.3–0.35 (Fig. 6), as observed by Oueslati et al. (2017). At the individual level, the bimodal distribution in ‘Owari Satsuma’ mandarin and ‘King’ mandarin was particularly clear, a result consistent with those of Wu et al. (2018). As proposed by Wu et al. (2014) and adopted by Oueslati et al. (2017), when examining the representative accessions, we considered that regions with low heterozygosity represent diploid segments which combine two haplotypes from the same species, while regions with high heterozygosity were considered to be hybrid segments combining two haplotypes from two different species. Thus, regions with heterozygosity values >0.2 were assumed to be introgressed and were removed.
Fig. 5.
Violin plots of the heterozygosity distribution in the 48 diploid accessions computed from the average values in successive windows of 100 polymorphic positions along the genome. White dot, median; bar limits; upper and lower quartiles; whiskers, 1.5× interquartile range; light blue, intraspecies; light pink, interspecies.
Fig. 6.
Distribution of the heterozygosity in mandarins, pummelos, citrons, papedas, all the diploid varieties, the ‘Seville’ sour orange and the ‘Rough’ lemon computed from the average values in successive windows of 100 polymorphic positions along the genome.
The patterns of similarity between each accession and the centroid of the four horticultural groups were also examined. The regions with an increase in heterozygosity were associated with a decrease in similarity to their representative horticultural group and an increase in similarity to the horticultural group involved in the introgression. An example is given for chromosome 2 of the ‘King’ mandarin (Supplementary Data Fig. S4). A heterozygous introgression was clearly identified at the end of the chromosome. Heterozygosity increased with a decrease in similarity, starting at 25 Mb and continuing to the end of the chromosome. Similarity analysis was particularly useful to identify homozygous introgressions as described by Oueslati et al. (2017) for the ‘Ponkan’ variety. Indeed, respective similarity with the reference taxa and the introgressed taxa decreased and increased abruptly. The search for introgressions, based on the patterns of heterozygosity and similarities with centroids of the horticultural groups, was systematically performed on the nine chromosomes of the 29 representative accessions.
Allelic frequencies of the ancestral taxa and the differentiation parameter (GST) were then re-estimated considering the introgressed areas as missing data. All SNPs with GST >0.9 for one ancestral taxon compared with all others were considered as diagnostic of the taxon concerned. A total of 15 946 DSNPs of the four ancestral taxa distributed along the nine chromosomes (Table 3; Supplementary Data Table S2) were then identified. DSNPs of C. medica represented more than one-third (37.60%) of the total number of DSNPs. The low intraspecific heterozygosity of C. medica described above explains the higher number of diagnostic SNPs in this taxon (5997), and the same is true for the C. micrantha taxon whose DSNPs represent 27.41 % of the total. Citrus reticulata and C. maxima are represented by 21.9 and 13.09 %, respectively, of the total number of DSNPs. The distribution of the 15 946 DSNPS along the nine chromosomes closely resembled the distribution of the whole set of polymorphisms and is closely linked with the distribution of the gene sequences (Supplementary Data Fig. S5). The selected DSNPs were used to decipher the phylogenomic mosaic structures of the 53 varieties.
Table 3.
Distribution of the 15 946 diagnostic SNPs (DSNPs) per taxon and per chromosome along the nine chromosomes
| C. reticulata | C. maxima | C. medica | C. micrantha | Total | |
|---|---|---|---|---|---|
| C1 | 404 | 274 | 604 | 430 | 1712 |
| C2 | 429 | 257 | 826 | 555 | 2067 |
| C3 | 593 | 328 | 1089 | 817 | 2827 |
| C4 | 388 | 245 | 630 | 503 | 1766 |
| C5 | 423 | 264 | 719 | 490 | 1896 |
| C6 | 321 | 228 | 564 | 428 | 1541 |
| C7 | 318 | 179 | 494 | 343 | 1334 |
| C8 | 261 | 130 | 480 | 364 | 1235 |
| C9 | 354 | 182 | 591 | 441 | 1568 |
| Total | 3491 | 2087 | 5997 | 4371 | 15 946 |
| % | 21.9 | 13.09 | 37.6 | 27.41 | 100 |
C1–C9, the nine chromosomes of the reference clementine genome (Wu et al., 2014); %, percentage of DSNPs for the taxon.
Phylogenomic structure of modern varieties
Our main objective was to develop a pipeline for the analysis of GBS data which would make it possible to establish the phylogenomic karyotype in diploid, triploid and tetraploid germplasm. For polyploid germplasm, this requires the ability to estimate allelic doses for heterozygous genotypes. Looking at individual SNP loci for the DSNPs of C. medica in the triploid ‘Persian’ lime (Supplementary Data Fig. S6), the frequency of C. medica allele reads per locus did not display a clear bimodal distribution for heterozygous loci (Supplementary Data Fig. S6A) and, consequently, estimated allelic doses are subject to high uncertainty. When working with all reads of ten consecutive loci, the bimodal distribution of the C. medica allele frequency was much clearer (Supplementary Data Fig. S6B), enabling efficient estimation of the dose of C. medica (1/3 and 2/3) in the genomic fragment corresponding to the ten markers considered. For the analysis of diploid and triploid Citrus germplasm, we kept ten DSNPs per window as default to estimate the doses for each ancestral taxon.
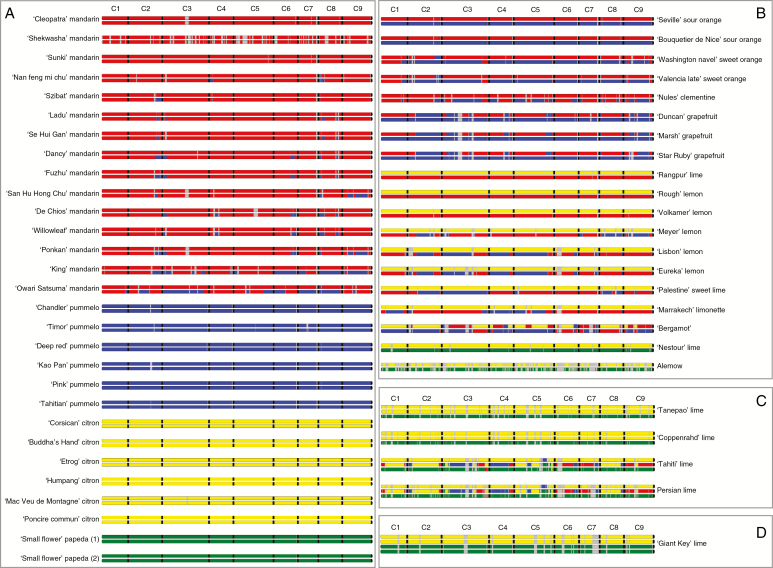
Using the TraceAncestor tool that we developed, we inferred the unphased phylogenomic karyotypes of the 53 accessions (Fig. 7). The average phylogenomic contributions of C. reticulata, C. maxima, C. medica and C. micrantha to the modern varieties are presented in Supplementary Data Text S2.
Fig. 7.
Unphased phylogenomic karyotypes of the 53 varieties of the study. (A) Karyotypes of the representative accessions of the four ancestral taxa. (B) Karyotypes of the secondary admixture species. (C) Karyotypes of the triploid hybrids. (D) Karyotype of the tetraploid hybrid lime. Red, C. reticulata; blue, C. maxima; yellow, C. medica; green, C. micrantha; grey, indeterminacy; black, separation between chromosomes.
Validation of the karyotypes inferred from GBS data
We compared karyotypes obtained from GBS data with those proposed by Wu et al. (2014, 2018) from WGS data for four citrons (‘Buddha’s Hand’, ‘Corsican’, ‘Humpang’ and ‘Mac Veu de Montagne’), C. micrantha, seven mandarins (‘Ponkan’, ‘Owari Satsuma’, ‘King’, ‘Dancy’, ‘Sunki’, ‘Cleopatra’ and ‘Willowleaf’), ‘Chandler’ pummelo, ‘Washington Navel’ sweet orange, ‘Seville’ sour orange, ‘Nules’ clementine, Marsh’ grapefruit, ‘Rough’ lemon, ‘Rangpur’ lime and ‘Eureka’ lemon. For example, Supplementary Data Fig. S7A shows the phylogenomic karyotypes of the ‘Washington Navel’ sweet orange and the ‘Owari Satsuma’ mandarin inferred from our GBS data and from WGS data (Wu et al., 2014). As concluded by Wu et al. (2018), the four citrons common to both studies and the two ‘small flower’ papeda were fully homozygous with C. medica and C. micrantha, respectively. Regarding ‘Chandler’ pummelo, only a small genomic area considered by Wu et al. (2014, 2018) to be introgressed in heterozygosity by C. reticulata on chromosome 2 (C2) coincided with an undetermined area in our karyotype generated from GBS data (Fig. 7A; Wu et al., 2018). For the rest of the genome, like Wu et al. (2014, 2018), we concluded homozygosity for C. maxima. For the representative mandarins, the karyotypes inferred from GBS data completely matched those inferred from WGS (Wu et al., 2014, 2018) except for two small genomic regions. A small C. reticulata homozygous fragment in the C6 of ‘Owari Satsuma’ mandarin and a small heterozygous introgression of C. maxima at the beginning of the C2 of ‘Willow leaf’ mandarin were not detected by the GBS analysis. Focusing on the areas determined in our GBS analysis, we detected no differences between our results for sweet orange, sour orange, clementine, grapefruit, lime and lemons common to both analyses (Fig. 7B, C) and those obtained by Wu et al. (2018). Moreover, we checked the repeatability of the analysis through three experimental replicates (three independent samples during preparation of the GBS library) of ‘Nules’ clementine. The determined areas of the three replicates displayed exactly the same pattern (Supplementary Data Fig. S7B). Overall, phylogenomic karyotypes were successfully inferred from GBS data but with more undetermined regions than those inferred from WGS data. Given these positive results, we considered that our GBS workflow was validated and the karyotypes inferred for all the remaining varieties as a good approximation of the phylogenomic structure.
New karyotypes of diploid varieties
The analysis of the additional varieties representative of the four ancestral taxa revealed introgressions of C. maxima fragments in all mandarins except ‘Shekwasha’ mandarin. It varied between 1.39 % for ‘Se Hui Gan’ mandarin to 4.41 % in ‘San Hu Hong Chu’ mandarin, with variable introgression positions in C2, C3, C4, C6, C8 and C9. ‘Shekwasha’ mandarin displayed a small introgression of C. micrantha in C3. In the case of pummelos, GBS data identified a small introgressed area by C. medica in the C7 of ‘Timor’ pummelo, while ‘Pink’, ‘Tahitian’, ‘Kao Pan’ and ‘Deep red’ pummelos appeared fully homozygous for C. maxima (Fig. 7A). In the same way, the two C. medica not analysed in the study of Wu et al. (2018) (‘Etrog’ and ‘Poncire commun’ citrons) appeared fully homozygous for C. medica.
For the secondary species, ‘Bouquetier de Nice’ sour orange displayed the same karyotype as ‘Seville’ sour orange with full C. reticulata/C. maxima heterozygosity. Examining the determined areas, ‘Valencia late’ sweet orange was found to be identical to ‘Washington navel’, displaying C. reticulata/C. maxima heterozygosity or C. reticulata homozygosity all along the genome except on two fragments on C2 and C8, which appeared in C. maxima homozygosity. In the same way, ‘Duncan’ and ‘Star Ruby’ grapefruits were found to be identical to ‘Marsh’ (Fig. 7B). ‘Volkamer’ lemon appeared to be fully heterozygous for C. reticulata/C. medica along the nine chromosomes, as previously observed for ‘Rangpur’ lime and ‘Rough’ lemon (Wu et al., 2018; this study). Karyotypes of ‘Palestine’ sweet lime, ‘Marrakech’ limonette, and ‘Meyer’ and ‘Lisbon’ lemons displayed interspecific heterozygous fragments of C. medica/C. reticulata and C. medica/C. maxima (Fig. 7B) as previously described for ‘Eureka’ lemon (Wu et al., 2018; our present results from GBS). Moreover ‘Lisbon’ and ‘Eureka’ lemons were strictly identical in their determined areas. Bergamot displayed a much more complex admixture of C. maxima, C. reticulata and C. medica genomes. Indeed, in addition to the C. medica/C. reticulata and C. medica/C. maxima heterozygosity regions, we found fragments in C. reticulata/C. maxima heterozygosity, C. reticulata homozygosity (C7) and C. maxima homozygosity (C3, C4, C6 and C7). Referring to the hypothesis that the bergamot comes from a hybridization between a sour orange and a lemon (Gallesio, 1811; Curk et al., 2016), we examined the ancestor allelic dosage of the 100 kb windows of this variety and its assumed parents. A total of 99.12 % of them completely fit with the hypothesis, each parental gamete bringing the ancestor allelic doses observed in the bergamot. The remaining 0.88 % corresponds to C. reticulata/C. maxima heterozygosity regions located in the C1 and C6 undetermined in lemon. Considering this origin hypothesis and the haplotype structure of the parental genomes, we have been able to draw the bergamot phased karyotype (Fig. 8; Supplementary Data Fig. S8). ‘Alemow’ and ‘Nestour’ lime displayed C. micrantha/C. medica heterozygosity for the nine chromosomes. It should be noted that ‘Alemow’ presented a relatively high proportion of undetermined areas (39.46 %), probably due to a low sequencing coverage (65 % of missing data at the SNP level).
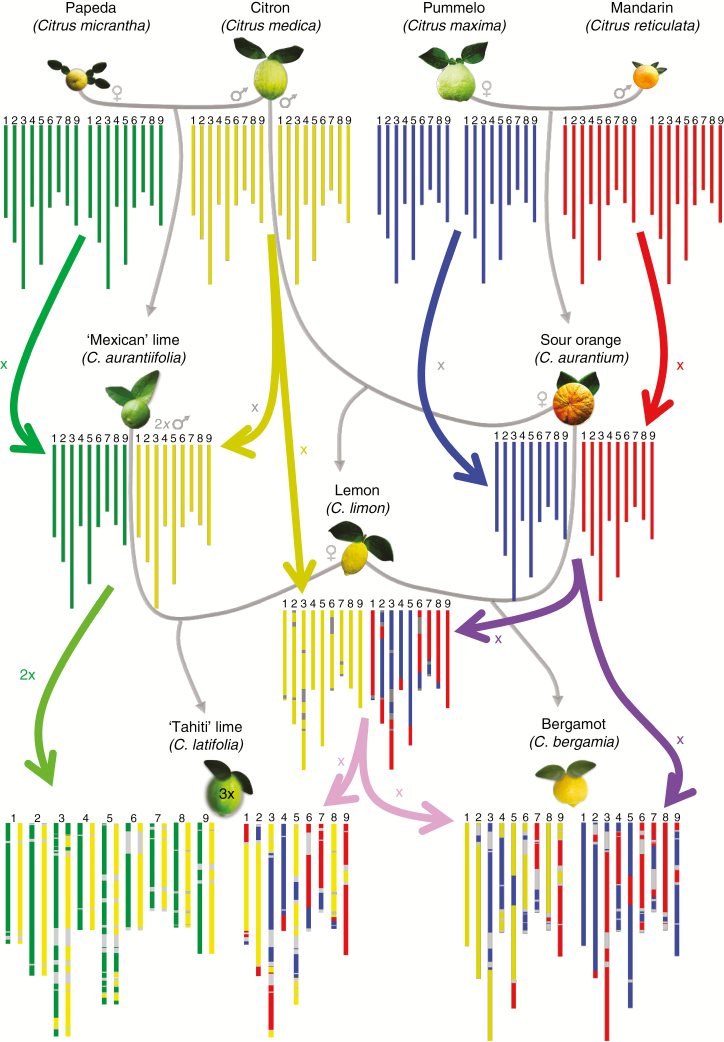
Fig. 8.
Phylogenetic origin and phased phylogenomic karyotypes of the sour orange (C. aurantium), the lemon (C. limon), the bergamot (C. bergamia) and the ‘Tahiti’ lime (C. latifolia). Red, C. reticulata; blue, C. maxima; yellow, C. medica; green, C. micrantha; grey, indeterminacy. The grey arrows indicate the cross between species, and the coloured arrows indicate whether the species contributes with x or 2x gametes.
Karyotypes of polyploid varieties
The phylogenomic structures of ‘Tanepao’, ‘Coppenrhad’, ‘Tahiti’ and ‘Persian’ triploid limes (Fig. 7C) and ‘Giant Key’ tetraploid lime were also inferred with the ‘TraceAncestor’ tool (Fig. 7D). ‘Tahiti’ and ‘Persian’ limes involving the contribution of the four basic taxa and, excluding undetermined areas, noticeably had the same phylogenomic karyotype. The quasi-systematic single dose of C. micrantha, the frequent double dose of C. medica and the occurrence of a double dose of C. micrantha (C3 and C5) and a triple dose of C. medica (C5) on small fragments, while C. reticulata and C. maxima were found only in single doses, fit the hypothesis that these limes derive from the union of a diploid ovule of ‘Mexican’ lime (C. aurantiifolia = C. micrantha × C. medica) and haploid pollen of lemon [C. limon = (C. maxima × C. reticulata) × C. medica] as proposed by Curk et al. (2016) and Rouiss et al. (2018). Therefore, following this hypothesis, we propose a phased karyotype identifying the haploid and diploid gametes from which this triploid lime originated (Fig. 8). For all the chromosomes, except C3 and C5, we observed a total restitution of the ‘Mexican’ lime-like parent by the diploid gamete. The representation of chromosomes 3 and 5 is just one of the different possibilities of C. medica and C. micrantha fragment phases in the diploid gamete. The interspecific recombination points in the diploid C. aurantiifolia and haploid C. limon gametes were clearly identified (Fig. 7C).
For determined areas, ‘Coppenrhad’ and ‘Tanepao’ limes displayed an identical pattern involving only C. medica and C. micrantha with single doses of C. micrantha and double doses of C. medica all along the nine chromosomes.
For the tetraploid ‘Giant key’ lime, the phylogenomic analysis with ten DSNPs per window produced many undetermined regions (60.58 %), due to a relatively low coverage (Supplementary Data Fig. S1) and the higher difficulty to distinguish 1/3, 2/2 and 3/1 doses for heterozygous genotypes. Therefore, we tested the inference with 20 and 30 DSNPs (Supplementary Data Fig. S9). The karyotype we obtained with 30 DSNP windows allowed a better estimation of the allelic doses and was able to reduce the undetermined regions to only 20 %. It showed full C. medica/C. micrantha heterozygosity along the genome.
DISCUSSION
The DSNP-based approach is powerful to decipher the admixture genomic structure in Citrus
Recent studies based on NGS (WGS and GBS) analysed the admixture of modern citrus varieties. They were based on the identification of diagnostic polymorphism (mainly SNPs) of the ancestral taxa considered. Wu et al. (2014) were the first to develop the DSNP approach to decipher the genomic structures of modern varieties originating from two ancestral taxa, C. reticulata and C. maxima, from WGS data. They used a small panel of mandarins (three varieties) and pummelos (two varieties), as representative of C. reticulata and C. maxima, to identify SNPs that distinguish these two ancestral taxa. The patterns of heterozygosity and similarity to the other mandarins and pummelos were used to identify introgressed areas in the different varieties in the two panels. The study revealed unexpected C. maxima introgressions in ‘Ponkan’ and ‘Willowleaf’ mandarins which were previously believed to be pure representatives of the C. reticulata taxon. The very large set of identified DSNPs was highly efficient to decipher the phylogenomic structures of clementine, sweet and sour oranges and ‘Afourer’ mandarin (W Murcott). Oueslati et al. (2017) showed that a similar approach can be used with GBS data using the ApeKI restriction enzyme. They expanded the phylogenomic analysis to 55 citrus varieties composed of representatives of C. maxima and C. reticulata taxa and hybrids assumed to derive from the admixture of these two taxa (mandarins, tangors, tangelos, orangelos and grapefruits). From a larger panel of representative mandarins (11 varieties) and pummelos (six varieties), these authors identified a set of 11 133 diagnostic polymorphisms, mostly SNPs (89 %), with a very similar pattern of distribution along the genome to those of gene sequences. This allowed them to infer the phylogenomic karyotypes of all the accessions by analysing the relative proportion of diagnostic markers homozygous for C. reticulata or C. maxima, or heterozygous in successive windows of 20 diagnostic markers.
Curk et al. (2015) were the first to publish sets of DSNPs for the four Citrus ancestral taxa. They identified 273 DSNPs from 454 amplicon sequencing data of 57 gene fragments dispersed on the nine chromosomes. They then developed allele competitive PCR markers (using KASPar technology) for 105 of these DSNPs and successfully analysed the interspecific origin of >200 Citrus accessions (Curk et al., 2015, 2016) and revealed systematic introgression of C. maxima in edible mandarins. However, the low number of DSNPs used in these studies did not make it possible to infer the phylogenomic karyotypes of the analysed varieties.
Wu et al. (2018) mined DSNPs which differentiate three of the four basic taxa (C. maxima, C. medica and C. reticulata) using only two pure Chinese mandarins, two citrons and three pummelos. They identified a total of 588 583 DSNPs (169 963 for C. reticulata, 116 803 for C. maxima and 301 817 for C. medica) and used them to decipher the phylogenomic karyotype of 47 Citrus varieties.
Whether the studies dealt with WGS (Wu et al., 2014, 2018), GBS (Oueslati et al., 2017) or DSNP markers (Curk et al., 2015, 2016), the analyses have always identified C. maxima introgressions in most cultivated mandarins. If the corresponding sequences are taken into account when estimating the allelic frequencies of the ancestral taxa, this introduces a bias in the estimation of the diversity parameters (allelic frequencies of the ancestral taxa and GST) and hence in the detection of diagnostic polymorphisms of the four ancestral taxa. This is why Wu et al. (2018) drastically limited their representative panel to only two pure genetically close mandarins. However, such a small panel could result in considering specific SNPs of the considered accessions as diagnostic of C. reticulata, whereas in fact polymorphisms existed within the species. Therefore, for our analysis, we preferred to keep the panel of representatives of the ancestral taxa as large as possible and used 15 mandarins, six pummelos, six citrons and two ‘small flowered’ papeda as representative of C. reticulata, C. maxima, C. medica and C. micrantha, respectively. Therefore, like Oueslati et al. (2017), we first identified introgression areas along the genome of the 29 representative accessions of the basic taxa according to the pattern along the genome of heterozygosity and to similarity with centroids of mandarins, pummelos, citrons and papedas. After removing the identified introgression regions, we computed the differentiation parameters again and filtered for polymorphic positions with GST >0.9. We selected 15 946 DSNPs and developed a pipeline to infer the phylogenomic structures of the 53 citrus accessions. Taking into account the difficulty involved in correctly estimating the allelic doses in triploid and tetraploid accessions at individual SNP loci (McKinney et al., 2018; Bastien et al., 2018; our data) and according to our choice of using the same analytical approach for diploids, triploids and tetraploids, we based our pipeline on the relative number of reads of each ancestor in windows of ten DSNPs of the considered taxon (while Wu et al., 2014, 2018 and Oueslati et al., 2017 performed their analysis in diploid accessions from genotyping data at individual loci) and on maximum likelihood analysis.
For diploid accessions common to both studies, our GBS data produced highly similar results to those obtained from WGS data (Wu et al., 2014, 2018), apart from the undetermined genomic areas, which were more frequent for GBS data, due to a lower density of DSNPs.
Therefore, GBS combined with our analytic pipeline proves to be a powerful approach to correctly analyse the phylogenomic admixture of diploid, triploid and tetraploid citrus varieties along the genome at significantly lower cost than the WGS approach. The panel of DSNPs can be used as reference for further GBS analyses using the same protocol (ApeKI; selection base A) to decipher the phylogenomic karyotypes in large citrus populations (germplasm or recombining populations). It opens the way for genetic association studies, quantitaive trait locus (QTL) analyses and genomic selection based on phylogenomics.
We developed a generic pipeline to decipher admixture in diploid, triploid and tetraploid genomes from an unlimited number of ancestors, allowing the user to define the number of DSNPs per window for the analysis of the dose contributed by each ancestor, the error rate considered for homozygous genotypes, the threshold for the LOD test of the maximum likelihood and the size of the window used to integrate information on the doses from the different ancestors to generate the phylogenomic karyotypes. This pipeline is available at http://galaxy.southgreen.fr/galaxy/ and should be useful for any species whose reproductive behaviour (vegetative propagation, preferential chromosome pairing associated with preferential disomic segregation) limited the number of interspecific recombinations after reticulation events and resulted in interspecific mosaics of large genomic fragments. It can also be used for the first generations of interspecific breeding schemes to identify interspecific recombination points. The selection of the ApeKI enzyme results in a marker density closely linked with gene sequence density and, consequently, in high coverage of the high recombining areas of the genome and low coverage of centromeric and paracentromeric areas with very low recombination rates (Aleza et al., 2015). This is a major advantage to trace interspecific recombination from GBS data efficiently. The main limitation of the approach is that it is based on the assumption of conserved physical genomic structure among the considered ancestors. In citrus, the overall high level of syntheny and conserved collinearity of markers observed for the genetic maps of clementine, sweet orange and pummelo (Ollitrault et al., 2012a), sour orange, pummelo, Poncirus trifoliata and ‘Fortune’ mandarin (Bernet et al., 2010), and sweet orange and Poncirus (Chen et al., 2008) justifies the use of the clementine reference genome as the genomic template to establish the phylogenomic karyotypes from either WGS data (Wu et al., 2014, 2018) or GBS data (Oueslati et al., 2017; this study). For plants with known large structural variations, a specific approach will be needed to describe the phylogenomic structures correctly in the genomic areas concerned.
The phylogenetic structures of 48 diploid varieties were deciphered; 16 for the first time
The representative accessions of the four basic taxa
We analysed 15 mandarins assumed to be good representatives of C. reticulata species. Twelve of them displayed C. maxima introgressions and one, ‘Shekwasha’ mandarin, has a small introgression of C. micrantha. No C. maxima introgressions were detected in ‘Shekwasha’, ‘Cleopatra’ and ‘Sunki’ mandarins. Limited introgressions were identified in ‘Szibat’ mandarin (1.49 %), ‘Ladu’ mandarin (1.72 %), ‘Nan Feng Mi Chu’ mandarin (1.74 %) and ‘Se Hui Gan’ mandarin (1.39 %). ‘Satsuma’ and ‘King’ mandarins were distinguished from all the other introgressed mandarins by their higher rate of C. maxima introgressions (22.6 and 19.5 %, respectively). Our results for newly studied varieties confirm that most edible mandarins are introgressed by C. maxima fragments as previously detected from WGS (Wu et al., 2014, 2018), 454 amplicon sequencing data (Curk et al., 2015) and GBS data (Oueslati et al., 2017). Wu et al. (2018) showed that some Chinese mandarins were not introgressed, and proposed three types of mandarins. The first type corresponds to unintrogressed genomes; type II includes mandarins with limited early introgression of the same two C. maxima haplotypes; and type III comprises mandarins derived from type II after more recent additional C. maxima introgression, probably resulting from hybridization with sweet orange. Based on our GBS analysis, ‘Szibat’, ‘Ladu’, ‘Nan Feng Mi Chu’ and ‘Se Hui Gan’ mandarins should be included in type II mandarins.
Despite the small C. reticulata introgressions in two pummelos (Wu et al., 2014, 2018; Oueslati et al., 2017) and the C. medica introgression in ‘Timor’ pummelo, our analysis confirms that modern pummelos can be considered as good representatives of the C. maxima species, as previously argued by several authors (Wu et al., 2014, 2018; Curk et al., 2015; Oueslati et al., 2017).
In our study, neither citrons nor ‘small flowered’ papedas displayed introgression areas. These results are in agreement with the conclusions drawn by Curk et al. (2015) and Wu et al. (2018). The analysed citrons and papedas therefore appear to be good representatives of the C. medica and C. micrantha species, respectively. Our results reveal the high level of homozygosity of citron accessions, including genomic areas with no revealed heterozygosity. Molecular marker studies (Barkley et al., 2006; Garcia-Lor et al., 2012; Luro et al., 2012; Curk et al., 2016) and WGS data (Wu et al., 2018) previously provided evidence for the low polymorphism of citrons and their high level of homozygosity. This can be linked with the cleistogamy of citron resulting in inbreeding and complete homozygosity of certain genome areas.
Secondary diploid species
The phylogenomic structures of accessions resulting from interspecific C. reticulata/C. maxima admixture are in full agreement with previous results and with hypotheses on their origins (Nicolosi et al., 2000; Ollitrault et al., 2012b; Curk et al., 2014; Wu et al., 2014, 2018; Oueslati et al., 2017). Thus, grapefruits, which are hybrids between C. maxima and sweet orange, display genome fragments in C. reticulata/C. maxima heterozygosity and C. maxima homozygosity. We found identical GBS-derived phylogenomic karyotypes for the three grapefruits analysed (‘Marsh’, ‘Duncan’ and ‘Star Ruby’) and that of ‘Marsh’ inferred from WGS (Wu et al., 2014, 2018). This confirms that these different cultivars derived from a single hybrid ancestor with no further sexual recombination. Citrus maxima and C. reticulata contributed equally to sour orange structure, and our results reveal an identical phylogenomic karyotype for ‘Bouquetier de Nice’ and ‘Seville’ sour oranges. The two sweet orange cultivars analysed displayed the same karyotypes with C. reticulata homozygosity fragments as well as C. maxima homozygosity and C. reticulata/C. maxima heterozygosity, in complete agreement with the study of ‘Washington Navel’ sweet orange by Wu et al. (2014). These results are evidence for the absence of sexual recombination during the diversification of these sweet oranges, whose polymorphisms are hypothesized to result from sporadic mutations, inheritable epigenetic changes and movements of transposable elements, as demonstrated for the anthocyanin content of blood oranges (Butelli et al., 2012).
The karyotype analysis of acidic citrus (limes and lemons) of Wu et al. (2018) was limited to ‘Rangpur’ and ‘Mexican’ limes and ‘Eureka’ and ‘Rough’ lemons. We expanded the analysis to ‘Alemow’, ‘Nestour’ lime, ‘Lisbon’, ‘Meyer’ and ‘Volkamer’ lemons, ‘Marrakech’ limonette and ‘Palestine’ sweet lime. ‘Rangpur’ lime, ‘Rough’ and ‘Volkamer’ lemons displayed the same pattern, with equal contributions of C. reticulata and C. medica along the nine chromosomes. These results support the hypothesis that they both derive from direct C. reticulata × C. medica hybridization as proposed by Curk et al. (2016) and Wu et al. (2018) for ‘Rangpur’ lime and ‘Rough’ lemon. In both previous studies, the contribution of C. medica as male parent was proved by chloroplast phylogeny. Our results also agree with the cytogenetic studies of Carvalho et al. (2005) in which ‘Alemow’ and ‘Nestour’ lime displayed the same pattern with C. micrantha/C. medica heterozygosity over all nine chromosomes, and confirm the hypothesis proposed by Curk et al. (2016), i.e. that these two acidic citrus resulted from direct hybridization between C. micrantha and C. medica. Using simple sequence repeat (SSR) markers in addition to DSNPs and cytoplasmic markers, Curk et al. (2016) also demonstrated that these two varieties resulted from independent reticulation events and that citron was the male parent. The phylogenomic karyotypes we obtained for ‘Eureka’ and ‘Lisbon’ lemons were identical and in full agreement with that proposed for ‘Eureka’ lemon by Wu et al. (2018). Probably, ‘Meyer’ lemon, ‘Palestine’ sweet lime and ‘Marrakech’ limonette involve the same three species C. maxima, C. reticulata and C. medica. Considering that C. medica is present as a single dose all over their genomes, we propose that they all result from hybridization between C. maxima/C. reticulata admixed genotypes and a C. medica. According to previous maternal phylogeny studies (Nicolosi et al., 2000; Luro et al., 2012; Carbonell-Caballero et al., 2015; Curk et al., 2016), C. medica is assumed to be the male parent in all cases. Previous molecular marker analyses of ‘Lisbon’ and ‘Eureka’ type yellow lemons (Nicolosi et al., 2000; Curk et al., 2016) suggested that they resulted from a single direct hybridization event between sour orange and citron. The same conclusion was drawn recently for ‘Eureka’ lemon based on WGS data (Wu et al., 2018). According to a maternal phylogenomic study (Curk et al., 2016) and nuclear data (Curk et al., 2016; this study), the ‘Marrakech’ limonette is hypothesized to have the same phylogenetic origin but to derive from an independent interspecific hybridization event. Maternal phylogeny studies revealed that ‘Meyer’ lemon and ‘Palestine’ sweet lime have the same cytoplasmic profile as sweet oranges and pummelos (Curk et al., 2016). However, their exact maternal parent remains to be determined.
The phylogenomic structure of bergamot also displays the admixture of the same three ancestral taxa, but the karyotype appears to be much more complex than that of the lemons, sweet lime and limonette discussed above. Many researchers have attempted to identify the origin of bergamot. In 1811, Gallesio proposed that it derives from a sour orange × lemon parentage. Several other origins have also been proposed, as reviewed and tested by Curk et al. (2016) in a nuclear and cytoplasmic marker study. Their results supported that proposed by Gallesio (1811). Our comparison of the karyotypes of bergamot and the karyotypes of sour orange and yellow lemons (‘Eureka’ and ‘Lisbon’) totally fits with the hypothesis that bergamot results from hybridization between a sour orange and a yellow lemon. It was therefore possible to draw a phased karyotype of the bergamot distinguishing the gamete originating from lemon and that originating from sour orange.
Considering their modern distribution, it is probable that bergamot and ‘Marrakech’ limonette resulted from hybridization that occurred in the Mediterranean Basin, where the presence of citrons dates from the second century BC and the introduction of sour orange dates to the Arab era in the seventh century (Webber et al., 1967; Swingle and Reece, 1967; Nicolosi et al., 2005). This confirms the importance of this region as a secondary area of citrus diversification.
The phylogenomic karyotype of triploid and tetraploid limes were deciphered
Leaving aside the undetermined regions, our phylogenomic inference resulted in identical structures for ‘Tahiti’ and ‘Persian’ limes, with a contribution of the four ancestral taxa. As reported in Curk et al. (2016), our results also revealed single or double doses of C. medica and C. micrantha, while C. maxima and C. reticulata contributed no dose or a single dose along the genome. Curk et al. (2016) proposed that this type of lime resulted from the fusion of a haploid lemon ovule and a diploid pollen of a diploid ‘Mexican’-like lime. Our analysis of the four ancestor doses all along the genome perfectly fits this hypothesis at the nuclear level. The diploid gamete of ‘Mexican’ lime type restituted 84.65 % of the parental interspecific heterozygosity and displayed only 2.47 and 0.80 % of C. micrantha and C. medica homozygote fragments, respectively. This high level of heterozygosity restitution and the heterozygosity for the centromeric areas of the nine chromosomes preclude the hypothesis of an unreduced gamete from a diploid ‘Mexican’ lime resulting from second division restitution (SDR) of the meiosis. They suggest that the diploid gamete comes from a doubled diploid parent with a preferential disomic segregation, or from first division restitution (FDR) of a diploid parent. Indeed, SDR 2n gametes contain sister chromatids and are homozygous from the centromere until the first crossing-over, while, under FDR, 2n gametes contain two non-sister chromatids allowing the entire conservation of parental heterozygosity from the centromere until the first crossing-over (Park et al., 2007; Ollitrault et al., 2008; Peloquin et al., 2008; Cuenca et al., 2011; Storme and Geelen, 2013); as a consequence, FDR gametes transmit 70–80 % of the parental heterozygosity, whereas this is only about 30–40 % for SDR (Barone et al., 1995; Douches and Mass, 1998; Dewitte et al., 2012; Aleza et al., 2016).
Molecular marker inheritance proved that doubled diploid ‘Mexican’ lime had preferential disomic inheritance with 90.2 % of heterozygosity restitution on average (Rouiss et al., 2018). Therefore, the phylogenomic karyotype of ‘Tahiti’ lime fits well with the interploid (diploid citron × tetraploid lime) origin hypothesis proposed by Rouiss et al. (2018). However, the unreduced FDR gamete hypothesis cannot be totally ruled out. Indeed, the FDR mechanism has been recently described at the origin of 2n pollen in citrus (Rouiss et al., 2017), and it can also result in a very high level of heterozygosity restitution.
‘Tanepao’ and ‘Coppenrhad’ limes presented identical patterns with single doses of C. micrantha and double doses of C. medica all along the nine chromosomes. Rouiss et al. (2018) observed that the preferential disomic inheritance of the doubled diploid ‘Mexican’ lime resulted in the production of 7 % of gametes with full interspecific heterozygosity. Therefore, an interploid backcross hybridization of a doubled diploid ‘Mexican’ lime type with a diploid citron may be at the origin of these limes, as proposed by Curk et al. (2016) and Rouiss et al. (2018). However, FDR coupled with asynapsis of ‘Mexican’ lime, which is dependent on low temperatures (Iwamasa and Iwasaki, 1963), could also produce fully heterozygous diploid gametes from a diploid ‘Mexican’ lime parent. Therefore, fertilization of an FDR ovule of ‘Mexican’ lime type by a haploid pollen of citron cannot be eliminated.
The tetraploid ‘Giant key’ lime displayed a full heterozygous pattern with a double dose of C. medica and a double dose of C. micrantha along its genome. In a molecular marker study, Curk et al. (2016) obtained identical patterns for ‘Giant key’ and ‘Mexican’ limes. They suggested that ‘Giant key’ lime emerged from the natural duplication of chromosomes of a ‘Mexican’ lime type which derives from a C. micrantha × C. medica natural hybridization. Our results agree with these conclusions. To limit the undetermined area for ‘Giant Key’ lime, we had to perform the likelihood analysis in windows of 30 DSNPs. This was required by the low coverage of this accession and also because more reads are necessary to conclude significantly between hypotheses of a 1/3, 2/2 and 3/1 ratio for heterozygous loci in tetraploids than a single homozygous/heterozygous distinction in diploid or 1/2 vs. 2/1 discrimination in triploids.
Conclusion
Genotyping by sequencing, using the ApeKI restriction enzyme, to focus on gene areas, and a selective base (A), to improve the depth of the analysis, was successfully applied to diploid, triploid and tetraploid citrus. The analysis of 29 representative accessions of the four citrus ancestral taxa allowed us to identify 15 946 DSNPs among 43 598 mined SNPs. The generic pipeline developed to infer phylogenomic karyotypes is based on the relative number of reads of ancestral and alternative alleles at DSNP loci. For each ancestral taxon, maximum likelihood tests were performed to infer doses of ancestral taxa in successive windows of ten DSNPs of the taxa considered. This approach provided results which closely resembled previously published results from WGS data. It revealed the phylogenomic structure for new diploid species and cultivars including direct interspecific hybrids such as C. limonia, ‘Volkamer’ lemon (C. reticulata × C. medica), C. macrophylla ‘Alemow’ and C. excelsa ‘Nestour’ lime (C. micrantha × C. medica), but also more complex structures involving three ancestors such as C. limetta ‘Marrakech’ limonette [(C. maxima × C. reticulata) × C. medica; sour orange × citron], C. limettiodes Tan. ‘Palestine’ Sweet lime and C. meyeri [(C. maxima × C. reticulata) × C. medica] or C. bergamia bergamot [(C. maxima × C. reticulata) × C. medica) × (C. maxima × C. reticulata); lemon × sour orange]. The phylogenomic karyotypes of triploid limes were also revealed, confirming the highly complex structure of ‘Tahiti’ and ‘Persian’ limes involving the four ancestral taxa [(C. maxima × C. reticulata) × C. medica) × (C. micrantha × C. medica); lemon haploid ovule × ‘Mexican’ lime-like diploid pollen], and are in agreement with the probable origin of ‘Tanepao’ and ‘Coppenrhad’ limes from the interploidy backcross [(C. micrantha × C. medica) × C. medica; ‘Mexican’ lime-like diploid ovule × citron haploid pollen]. The GBS approach and analytical pipeline combined with the reference DSNP matrix will be useful for any study of germplasm and hybrids resulting from breeding within the Citrus genus. The workflow implemented for mosaic genome analysis is available online and can also be used for other species with unlimited numbers of identified ancestral taxa, for diploid, triploid and tetraploid accessions. Considering the density of DSNPs along the genome revealed by GBS, it will probably be particularly useful for any species whose reproductive behaviour has limited the number of interspecific recombinations after reticulation events and resulted in interspecific mosaics of large genomic fragments. It can also be used to localize interspecific recombining points in the first generations of interspecific breeding schemes.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Text S1: maximum likelihood test for diploid, triploid and tetraploid individuals. Text S2: average phylogenomic contribution of the four ancestral taxa to the modern varieties. Figure S1: number of reads per accession. Figure S2: distribution of missing data among markers and individuals. Figure S3: distribution of the number of reads per marker along the nine chromosomes. Figure S4: identification of the interspecific introgressions in representative accessions of the ancestral taxa: example of the chromosome 2 of ‘King’ mandarin. Figure S5: distribution along the nine chromosomes of DSNPs, the whole set of diallelic SNPs and gene sequences. Figure S6: estimation of C. medica allele doses in triploid ‘Persian’ lime. Figure S7: validation of the GBS approach. Figure S8: phased karyotype of the bergamot based on the gametes of the lemon and the sour orange. Figure S9: the inferred karyotypes of the ‘Giant key’ lime. Table S1: plant material. Table S2: list of the 15 946 diagnostic markers, specifying their genomic position, their reference and alternative alleles, the ancestral taxon they are diagnostic for and the gene name where they are located if available.
FUNDING
This work was supported by the FEDER Guadeloupe ‘CAVALBIO’ project, by the FEDER Corsica ‘InnovAgrumes’ project and the Agropolis Fondation ‘GenomeHarvest project’ (ID 1504-006) through the ‘Investissements d’avenir’ programme (Labex Agro:ANR-10-LABX-0001-01).
LITERATURE CITED
- Abbott R, Albach D, Ansell S, et al. 2013. Hybridization and speciation. Journal of Evolutionary Biology 26: 229–246. [DOI] [PubMed] [Google Scholar]
- Abbott R, Hegarty M, Hiscock S, Brennan A. 2010. Homoploid hybrid speciation in action. Taxon 59: 1375–1386. [Google Scholar]
- Aleza P, Cuenca J, Hernández M, Juárez J, Navarro L, Ollitrault P. 2015. Genetic mapping of centromeres in the nine Citrus clementina chromosomes using half-tetrad analysis and recombination patterns in unreduced and haploid gametes. BMC Plant Biology 15: 80. doi: 10.1186/s12870-015-0464-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleza P, Cuenca J, Juarez J, Navarro L, Ollitrault P. 2016. Inheritance in doubled-diploid clementine and comparative study with SDR unreduced gametes of diploid clementine. Plant Cell Reports 35:1573–1586. [DOI] [PubMed] [Google Scholar]
- Almeida Balsalobre TW, Pereira G da S, Alves Margarido GR, et al. 2017. GBS-based single dosage markers for linkage and QTL mapping allow gene mining for yield-related traits in sugarcane. BMC Genomics 18: 72. doi: 10.1186/s12864-016-3383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold ML. 1997. Natural hybridization and evolution. Oxford: Oxford University Press. [Google Scholar]
- Arnold ML. 2006. Evolution through genetic exchange. Oxford: Oxford University Press. [Google Scholar]
- Arnold ML, Fogarty ND. 2009. Reticulate evolution and marine organisms: the final frontier? International Journal of Molecular Sciences 10: 3836–3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf BH, Jensen J, Asp T, Janss LL. 2014. Association studies using family pools of outcrossing crops based on allele-frequency estimates from DNA sequencing. Theoretical & Applied Genetics 127: 1331–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baack EJ, Rieseberg LH. 2007. A genomic view of introgression and hybrid speciation. Current Opinion in Genetics & Development 17: 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NA, Etter PD, Atwood TS, et al. 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3: e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkley NA, Roose ML, Krueger RR, Federici CT. 2006. Assessing genetic diversity and population structure in a citrus germplasm collection utilizing simple sequence repeat markers (SSRs). Theoretical & Applied Genetics 112: 1519–1531. [DOI] [PubMed] [Google Scholar]
- Barone A, Gebhardt C, Frusciante L. 1995. Heterozygosity in 2n gametes of potato evaluated by RFLP markers. Theoretical & Applied Genetics 91: 98–104. [DOI] [PubMed] [Google Scholar]
- Barton NH. 2001. The role of hybridization in evolution. Molecular Ecology 10: 551–568. [DOI] [PubMed] [Google Scholar]
- Barton NH, Hewitt GM. 1985. Analysis of hybrid zones. Annual Review of Ecology and Systematics 16: 113–148. [Google Scholar]
- Bastien M, Boudhrioua C, Fortin G, Belzile F. 2018. Exploring the potential and limitations of genotyping-by-sequencing for SNP discovery and genotyping in tetraploid potato. Genome 61: 449–456. [DOI] [PubMed] [Google Scholar]
- Baxter SW, Davey JW, Johnston JS, et al. 2011. Linkage mapping and comparative genomics using next-generation RAD sequencing of a non-model organism. PLoS One 6: e19315. doi: 10.1371/journal.pone.0019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beiko RG, Hamilton N. 2006. Phylogenetic identification of lateral genetic transfer events. BMC Evolutionary Biology 6: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. 1996–2004. GENETIX 4.05, logiciel sous Windows™ pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000. https://kimura.univ-montp2.fr/genetix/ [Google Scholar]
- Bernet GP, Fernandez-Ribacoba J, Carbonell EA, Asins MJ. 2010. Comparative genome-wide segregation analysis and map construction using a reciprocal cross design to facilitate citrus germplasm utilization. Molecular Breeding 25: 659–673. [Google Scholar]
- Butelli E, Licciardello C, Zhang Y, et al. 2012. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. The Plant Cell 24: 1242–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne S, Czaban A, Studer B, Panitz F, Bendixen C, Asp T. 2013. Genome wide allele frequency fingerprints (GWAFFs) of populations via genotyping by sequencing. PLoS One 8: e57438. doi: 10.1371/journal.pone.0057438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell-Caballero J, Alonso R, Ibañez V, Terol J, Talon M, Dopazo J. 2015. A phylogenetic analysis of 34 chloroplast genomes elucidates the relationships between wild and domestic species within the genus Citrus. Molecular Biology and Evolution 32: 2015–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho R, Soares WS, Brasileiro-Vidal AC, Guerra M. 2005. The relationships among lemons, limes and citron: a chromosomal comparison. Cytogenetic and Genome Research 109: 276–282. [DOI] [PubMed] [Google Scholar]
- Chen C, Lyon M, O’Malley D, et al. 2008. Origin and frequency of 2n gametes in Citrus sinensis × Poncirus trifoliata and their reciprocal crosses. Plant Science 174: 1–8. [Google Scholar]
- Chessel D, Dufour AB, Thioulouse J. 2004. The ade4 package-I-One-table methods. R News 4: 5–10. [Google Scholar]
- Clevenger JP, Ozias-Akins P. 2015. SWEEP: a tool for filtering high-quality SNPs in polyploid crops. G3: Genes Genomes Genetics 5: 1797–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois B, Audebert A, Dardou A, et al. 2013. Genome-wide association mapping of root traits in a japonica rice panel. PLoS One 8: e78037. doi: 10.1371/journal.pone.0078037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossa J, Beyene Y, Kassa S, et al. 2013. Genomic prediction in maize breeding populations with genotyping-by-sequencing. G3: Genes Genomes Genetics 3: 1903–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenca J, Froelicher Y, Aleza P, Juárez J, Navarro L, Ollitrault P. 2011. Multilocus half-tetrad analysis and centromere mapping in citrus: evidence of SDR mechanism for 2n megagametophyte production and partial chiasma interference in mandarin cv ‘Fortune.’ Heredity 107: 462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curk F, Ancillo G, Garcia-Lor A, et al. 2014. Next generation haplotyping to decipher nuclear genomic interspecific admixture in Citrus species: analysis of chromosome 2. BMC Genetics 15: 152. doi: 10.1186/s12863-014-0152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curk F, Ancillo G, Ollitrault F, et al. 2015. Nuclear species-diagnostic SNP markers mined from 454 amplicon sequencing reveal admixture genomic structure of modern citrus varieties. PLoS One 10: e0125628. doi: 10.1371/journal.pone.0125628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curk F, Ollitrault F, Garcia-Lor A, Luro F, Navarro L, Ollitrault P. 2016. Phylogenetic origin of limes and lemons revealed by cytoplasmic and nuclear markers. Annals of Botany 117: 565–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtolo M, Cristofani-Yaly M, Gazaffi R, Takita MA, Figueira A, Machado MA. 2017. QTL mapping for fruit quality in Citrus using DArTseq markers. BMC Genomics 18: 289. doi: 10.1186/s12864-017-3629-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, et al. 2011. The variant call format and VCFtools. Bioinformatics (Oxford, England) 27: 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey JW, Blaxter ML. 2011. RADSeq: next-generation population genetics. Briefings in Functional Genomics 10: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte A, Van Laere K, Van Huylenbroeck J. 2012. Use of 2n gametes in plant breeding. In: Abdurakhmonov I, ed. Plant breeding. InTech Open Access Publisher: 59–86. [Google Scholar]
- Doolittle WF. 1999. Phylogenetic classification and the universal tree. Science 284: 2124–2128. [DOI] [PubMed] [Google Scholar]
- Douches D, Maas DL. 1998. Comparison of FDR- and SDR-derived tetraploid progeny from 2x × 4x crosses using haploids of Solanum tuberosum L. that produce mixed modes of 2n eggs. Theoretical & Applied Genetics 97:1307–1313. [Google Scholar]
- Dowling TE, Secor CL. 1997. The role of hybridization and introgression in the diversification of animals. Annual Review of Ecology and Systematics 28: 593–619. [Google Scholar]
- Duan N, Bai Y, Sun H, et al. 2017. Genome re-sequencing reveals the history of apple and supports a two-stage model for fruit enlargement. Nature Communications 8: 249. doi: 10.1038/s41467-017-00336-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugrand-Judek A, Olry A, Hehn A, et al. 2015. The distribution of coumarins and furanocoumarins in Citrus species closely matches Citrus phylogeny and reflects the organization of biosynthetic pathways. PLoS One 10: e0142757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer RJ, Savolainen V, Schneider H. 2012. Apomixis and reticulate evolution in the Asplenium monanthes fern complex. Annals of Botany 110: 1515–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, et al. 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6: e19379. doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero M, Eaton DAR, Hahn M, Hipp AL. 2014. Genotyping-by-sequencing as a tool to infer phylogeny and ancestral hybridization: a case study in Carex (Cyperaceae). Molecular Phylogenetics and Evolution 79: 359–367. [DOI] [PubMed] [Google Scholar]
- Fanciullino AL, Dhuique-Mayer C, Luro F, Casanova J, Morillon R, Ollitrault P. 2006. Carotenoid diversity in cultivated citrus is highly influenced by genetic factors. Journal of Agricultural and Food Chemistry 54: 4397–4406. [DOI] [PubMed] [Google Scholar]
- Federici CT, Fang DQ, Scora RW, Roose ML. 1998. Phylogenetic relationships within the genus Citrus (Rutaceae) and related genera as revealed by RFLP and RAPD analysis. Theoretical & Applied Genetics 96: 812–822. [Google Scholar]
- Galili T. 2015. dendextend: an R package for visualizing, adjusting, and comparing trees of hierarchical clustering. Bioinformatics 31: 3718–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallesio G. 1811. Traité du citrus. Paris: Louis Fantin edn. chez Louis Fantin Libraire. [Google Scholar]
- Garcia-Lor A, Luro F, Navarro L, Ollitrault P. 2012. Comparative use of InDel and SSR markers in deciphering the interspecific structure of cultivated citrus genetic diversity: a perspective for genetic association studies. Molecular Genetics and Genomics 287: 77–94. [DOI] [PubMed] [Google Scholar]
- Garcia-Lor A, Curk F, Snoussi-Trifa H, et al. 2013. A nuclear phylogenetic analysis: SNPs, indels and SSRs deliver new insights into the relationships in the ‘true citrus fruit trees’ group (Citrinae, Rutaceae) and the origin of cultivated species. Annals of Botany 111: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaubitz JC, Casstevens TM, Lu F, et al. 2014. TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One 9: e90346. doi: 10.1371/journal.pone.0090346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant V. 1981. Plant speciation, 2nd edn. New York: Columbia University Press. [Google Scholar]
- Hamon P, Grover CE, Davis AP, et al. 2017. Genotyping-by-sequencing provides the first well-resolved phylogeny for coffee (Coffea) and insights into the evolution of caffeine content in its species: GBS coffee phylogeny and the evolution of caffeine content. Molecular Phylogenetics and Evolution 109: 351–361. [DOI] [PubMed] [Google Scholar]
- Heslot N, Rutkoski J, Poland J, Jannink J-L, Sorrells ME. 2013. Impact of marker ascertainment bias on genomic selection accuracy and estimates of genetic diversity. PLoS One 8: e74612. doi: 10.1371/journal.pone.0074612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Feng Q, Qian Q, et al. 2009. High-throughput genotyping by whole-genome resequencing. Genome Research 19: 1068–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamasa M, Iwasaki T. 1963. On the sterility phenomenon caused by low temperatures in the Mexican lime (Citrus aurantifolia Swing.). Bulletin of the Horticultural Research Station of Japan, Series B 2: 25–45. [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Xie R, Lu Z, Zhou Z. 2010. The origin of cultivated citrus as inferred from internal transcribed spacer and chloroplast DNA sequence and amplified fragment length polymorphism fingerprints. Journal of the American Society for Horticultural Science 135: 341–350. [Google Scholar]
- Linder CR, Rieseberg LH. 2004. Reconstructing patterns of reticulate evolution in plants. American Journal of Botany 91: 1700–1708. [PMC free article] [PubMed] [Google Scholar]
- Liu H, Bayer M, Druka A, et al. 2014. An evaluation of genotyping by sequencing (GBS) to map the Breviaristatum-e (ari-e) locus in cultivated barley. BMC Genomics 15: 104. doi: 10.1186/1471-2164-15-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luro F, Venturini N, Costantino G, Paolini J, Ollitrault P, Costa J. 2012. Genetic and chemical diversity of citron (Citrus medica L.) based on nuclear and cytoplasmic markers and leaf essential oil composition. Phytochemistry 77:186–196. [DOI] [PubMed] [Google Scholar]
- Luro F, Bloquel E, Tomu B, et al. 2018. The INRA-CIRAD citrus germplasm collection of San Giuliano, Corsica. In: Zech-Matterne V, Fiorentino G, eds. AGRUMED: archaeology and history of citrus fruit in the Mediterranean: acclimatization, diversifications, uses. Naples: Publications du Centre Jean Bérard, 243–261. [Google Scholar]
- Ma X-F, Jensen E, Alexandrov N, et al. 2012. High resolution genetic mapping by genome sequencing reveals genome duplication and tetraploid genetic structure of the diploid Miscanthus sinensis. PLoS One 7: e33821. doi: 10.1371/journal.pone.0033821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarenkov V, Legendre P. 2004. From a phylogenetic tree to a reticulated network. Journal of Computational Biology 11: 195–212. [DOI] [PubMed] [Google Scholar]
- Mallet J. 2005. Hybridization as an invasion of the genome. Trends in Ecology & Evolution 20: 229–237. [DOI] [PubMed] [Google Scholar]
- Martin G, Baurens F-C, Droc G, et al. 2016. Improvement of the banana ‘Musa acuminata’ reference sequence using NGS data and semi-automated bioinformatics methods. BMC Genomics 17: 243. doi: 10.1186/s12864-016-2579-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M, Schuenemann VJ, Davidovich U, et al. 2016. Genomic analysis of 6,000-year-old cultivated grain illuminates the domestication history of barley. Nature Genetics 48: 1089–1093. [DOI] [PubMed] [Google Scholar]
- McKinney GJ, Waples RK, Pascal CE, Seeb LW, Seeb JE. 2018. Resolving allele dosage in duplicated loci using genotyping-by-sequencing data: a path forward for population genetic analysis. Molecular Ecology Resources 18: 570–579. [DOI] [PubMed] [Google Scholar]
- Meyer RS, Choi JY, Sanches M, et al. 2016. Domestication history and geographical adaptation inferred from a SNP map of African rice. Nature Genetics 48: 1083–1088. [DOI] [PubMed] [Google Scholar]
- Miller MR, Dunham JP, Amores A, Cresko WA, Johnson EA. 2007. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Research 17: 240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. 1973. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences, USA 70: 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolosi E, Deng ZN, Gentile A, La Malfa S, Continella G, Tribulato E. 2000. Citrus phylogeny and genetic origin of important species as investigated by molecular markers. Theoretical & Applied Genetics 100: 1155–1166. [Google Scholar]
- Nicolosi E, Malfa SL, El-Otmani M, Negbi M, Goldschmidt EE. 2005. The search for the authentic citron (Citrus medica L.): historic and genetic analysis. HortScience 40: 1963–1968. [Google Scholar]
- Ollitrault P, Jacquemond C, Dubois C, Luro F. 2003. Citrus. In: Hamon P, Seguin M, Perrier X, Glaszmann J-C, eds. Genetic diversity of cultivated tropical plants. Montpellier: Cirad, 193–217. [Google Scholar]
- Ollitrault P, Dambier D, Luro F, Froelicher Y. 2008. Ploidy manipulation for breeding seedless triploid citrus. Plant Breeding Reviews 30: 323–352. [Google Scholar]
- Ollitrault P, Terol J, Chen C, et al. 2012. a A reference genetic map of C. clementina hort. ex Tan.; citrus evolution inferences from comparative mapping. BMC Genomics 13: 593. doi: 10.1186/1471-2164-13-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollitrault P, Terol J, Garcia-Lor A, et al. 2012. b SNP mining in C. clementina BAC end sequences; transferability in the Citrus genus (Rutaceae), phylogenetic inferences and perspectives for genetic mapping. BMC Genomics 13: 13. doi: 10.1186/1471-2164-13-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP, Whitton J. 2000. Polyploid incidence and evolution. Annual Review of Genetics 34: 401–437. [DOI] [PubMed] [Google Scholar]
- Oueslati A, Salhi-Hannachi A, Luro F, Vignes H, Mournet P, Ollitrault P. 2017. Genotyping by sequencing reveals the interspecific C. maxima/C. reticulata admixture along the genomes of modern citrus varieties of mandarins, tangors, tangelos, orangelos and grapefruits. PloS One 12: e0185618. doi: 10.1371/journal.pone.0185618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamilo P, Nei M. 1988. Relationships between gene trees and species trees. Molecular Biology and Evolution 5: 568–583. [DOI] [PubMed] [Google Scholar]
- Pankin A, Altmüller J, Becker C, von Korff M. 2018. Targeted resequencing reveals genomic signatures of barley domestication. New Phytologist 218: 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T-H, Kim J-B, Hutten RCB, van Eck HJ, Jacobsen E, Visser RGF. 2007. Genetic positioning of centromeres using half-tetrad analysis in a 4x–2x cross population of potato. Genetics 176: 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peloquin SJ, Boiteux LS, Simon PW, Jansky SH. 2008. A chromosome-specific estimate of transmission of heterozygosity by 2n gametes in potato. Journal of Heredity 99: 177–181. [DOI] [PubMed] [Google Scholar]
- Penjor T, Mimura T, Matsumoto R, Yamamoto M, Nagano Y. 2014. Characterization of limes (Citrus aurantifolia) grown in Bhutan and Indonesia using high-throughput sequencing. Scientific Reports 4: 4853. doi: 10.1038/srep04853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penjor T, Mimura T, Kotoda N, et al. 2016. RAD-Seq analysis of typical and minor Citrus accessions, including Bhutanese varieties. Breeding Science 66: 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrier X, Bakry F, Carreel F, et al. 2009. Combining biological approaches to shed light on the evolution of edible bananas. Ethnobotany Research and Applications 7: 199–216. [Google Scholar]
- Perrier X, Langhe ED, Donohue M, et al. 2011. Multidisciplinary perspectives on banana (Musa spp.) domestication. Proceedings of the National Academy of Sciences, USA 108: 11311–11318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. 2012. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One 7: e37135. doi: 10.1371/journal.pone.0037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland JA, Brown PJ, Sorrells ME, Jannink J-L. 2012. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One 7: e32253. doi: 10.1371/journal.pone.0032253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. 2001. Intraspecific gene genealogies: trees grafting into networks. Trends in Ecology & Evolution 16: 37–45. [DOI] [PubMed] [Google Scholar]
- Ramadugu C, Pfeil BE, Keremane ML, Lee RF, Maureira-Butler IJ, Roose ML. 2013. A six nuclear gene phylogeny of Citrus (Rutaceae) taking into account hybridization and lineage sorting. PLoS One 8: e68410. doi: 10.1371/journal.pone.0068410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Madrigal J, Smith BD, Moreno-Mayar JV, et al. 2016. Genome sequence of a 5,310-year-old maize cob provides insights into the early stages of maize domestication. Current Biology 26: 3195–3201. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2017. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, URL https://www.R-project.org/. [Google Scholar]
- Rieseberg LH. 1997. Hybrid origins of plant species. Annual Review of Ecology and Systematics 28: 359–389. [Google Scholar]
- Rieseberg L, Soltis D. 1991. Phylogenetic consequences of cytoplasmic gene flow in plants. Evolutionary Trends in Plants 5: 65–84. [Google Scholar]
- Rocher S, Jean M, Castonguay Y, Belzile F. 2015. Validation of genotyping-by-sequencing analysis in populations of tetraploid alfalfa by 454 sequencing. PLoS One 10: e0131918. doi: 10.1371/journal.pone.0131918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouiss H, Cuenca J, Navarro L, Ollitrault P, Aleza P. 2017. Tetraploid citrus progenies arising from FDR and SDR unreduced pollen in 4x × 2x hybridizations. Tree Genetics and Genomes 13: 10. https://doi.org/10.1007/s11295-016-1094-8. [Google Scholar]
- Rouiss H, Bakry F, Froelicher Y, Navarro L, Aleza P, Ollitrault P. 2018. Origin of C. latifolia and C. aurantiifolia triploid limes: the preferential disomic inheritance of doubled-diploid ‘Mexican’ lime is consistent with an interploid hybridization hypothesis. Annals of Botany 121: 571–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scora RW. 1975. On the history and origin of citrus. Bulletin of the Torrey Botanical Club 102: 369–375. [Google Scholar]
- Sonah H, Bastien M, Iquira E, et al. 2013. An improved genotyping by sequencing (GBS) approach offering increased versatility and efficiency of SNP discovery and genotyping. PLoS One 8: e54603. doi: 10.1371/journal.pone.0054603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindel J, Wright M, Chen C, et al. 2013. Bridging the genotyping gap: using genotyping by sequencing (GBS) to add high-density SNP markers and new value to traditional bi-parental mapping and breeding populations. Theoretical & Applied Genetics 126: 2699–2716. [DOI] [PubMed] [Google Scholar]
- Stebbins G. 1950. Variation and evolution in plants. New York: Columbia University Press. [Google Scholar]
- Stebbins GL. 1959. The role of hybridization in evolution. Proceedings of the American Philosophical Society 103: 231–251. [Google Scholar]
- Stetter MG, Schmid KJ. 2017. Analysis of phylogenetic relationships and genome size evolution of the Amaranthus genus using GBS indicates the ancestors of an ancient crop. Molecular Phylogenetics and Evolution 109: 80–92. [DOI] [PubMed] [Google Scholar]
- Storme ND, Geelen D. 2013. Sexual polyploidization in plants – cytological mechanisms and molecular regulation. New Phytologist 198: 670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sverrisdóttir E, Byrne S, Sundmark EHR, et al. 2017. Genomic prediction of starch content and chipping quality in tetraploid potato using genotyping-by-sequencing. Theoretical & Applied Genetics 130: 2091–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swingle W, Reece P. 1967. The botany of citrus and its wild relatives. In: Reuther W, Webber HJ, Batchelor LD, eds. The citrus industry. The botany of Citrus and its wild relatives, Vol. 1 Berkeley, CA: University of California, 190–430. [Google Scholar]
- Tanaka T. 1954. Species problem in citrus: a critical study of wild and cultivated units of citrus, based upon field studies in their native homes (Revisio Aurantiacearum IX). Tokyo, Japan: Japanese Society for Promotion of Science, 141–152. [Google Scholar]
- Truong HT, Ramos AM, Yalcin F, et al. 2012. Sequence-based genotyping for marker discovery and co-dominant scoring in germplasm and populations. PLoS One 7: e37565. doi: 10.1371/journal.pone.0037565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uitdewilligen JGAML, Wolters A-MA, D’hoop BB, Borm TJA, Visser RGF, van Eck HJ. 2013. A next-generation sequencing method for genotyping-by-sequencing of highly heterozygous autotetraploid potato. PLoS One 8: e62355. doi: 10.1371/journal.pone.0062355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S, Gupta S, Bandhiwal N, Kumar T, Bharadwaj C, Bhatia S. 2015. High-density linkage map construction and mapping of seed trait QTLs in chickpea (Cicer arietinum L.) using Genotyping-by-Sequencing (GBS). Scientific Reports 5: 17512. doi: 10.1038/srep17512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li L, Zhang G. 2016. A high-density SNP genetic linkage map and QTL analysis of growth-related traits in a hybrid family of oysters (Crassostrea gigas × Crassostrea angulata) using genotyping-by-sequencing. G3: Genes, Genomes, Genetics 6: 1417–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JA, Bhangoo J, Fernandez-Fernandez F, et al. 2013. Saturated linkage map construction in Rubus idaeus using genotyping by sequencing and genome-independent imputation. BMC Genomics 14: 2. doi: 10.1186/1471-2164-14-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber HJ, Reuther W, Lawton H. 1967. History and development of the citrus industry. In: Reuther W, Webber HJ, Batchelor LD, eds. The citrus industry. The botany of Citrus and its wild relatives, Vol. 1 Berkeley, CA: University of California, 1–39. [Google Scholar]
- Wester PJ. 1915. The Philippine agricultural review, citrus fruits in the philippines. Quarterly Publication, VIII.1. [Google Scholar]
- Wright S. 1951. The genetical structure of populations. Annals of Eugenics 15: 323–354 [DOI] [PubMed] [Google Scholar]
- Wu GA, Prochnik S, Jenkins J, et al. 2014. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nature Biotechnology 32: 656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GA, Terol J, Ibanez V, et al. 2018. Genomics of the origin and evolution of Citrus. Nature 554: 311–316. [DOI] [PubMed] [Google Scholar]
- Xu Q, Chen L-L, Ruan X, et al. 2013. The draft genome of sweet orange (Citrus sinensis). Nature Genetics 45: 59–68. [DOI] [PubMed] [Google Scholar]
- Yakimowski SB, Rieseberg LH. 2014. The role of homoploid hybridization in evolution: a century of studies synthesizing genetics and ecology. American Journal of Botany 101: 1247–1258. [DOI] [PubMed] [Google Scholar]
- Yang X, Song J, You Q, Paudel DR, Zhang J, Wang J. 2017. Mining sequence variations in representative polyploid sugarcane germplasm accessions. BMC Genomics 18: 594. doi: 10.1186/s12864-017-3980-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Massonnet M, Sanjak JS, Cantu D, Gaut BS. 2017. Evolutionary genomics of grape (Vitis vinifera ssp. vinifera) domestication. Proceedings of the National Academy of Sciences, USA 114: 11715–11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.