Abstract
Cancer is expected to rank as the leading cause of death worldwide due to increasing morbidity and mortality. Long noncoding RNAs (lncRNAs) have been found to play pivotal roles in multiple biological processes, such as transcriptional interference, posttranscriptional regulation and epigenetic modification. Small nucleolar RNA host gene 15 (SNHG15), a snoRNA host gene which produces a short half-lived lncRNA, was reported to be upregulated in tumor cells and participate in the occurrence and development of multiple cancers. And more than half of the SNHG15 research in cancers has been published within the last 2 years. In this review, we summarized the current evidence concerning the biological functions and molecular mechanisms of SNHG15 in various cancers, including gastric, hepatocellular, pancreatic, colorectal, breast, and thyroid cancer, osteosarcoma, glioma, lung cancer, renal cell carcinoma, and epithelial ovarian cancer. SNHG15 plays critical roles in regulation of cell proliferation, migration and invasion of tumors via different potential mechanisms. Moreover, the abnormal expression of SNHG15 was associated with clinical features of patients with cancers. Consequently, SNHG15 could be considered as a promising biomarker for cancer diagnosis, prognosis or treatment.
Keywords: SNHG15, long noncoding RNA, cancer, biomarker
Introduction
As the incidence and mortality increase, cancer has become a major public health problem and ranked as the leading cause of death all over the world. It is estimated that 18,078,957 new cancer cases and 9,555,027 cancer deaths will happen in the world in 20181 and 1,735,350 new cancer cases and 609,640 cancer deaths will occur in the United States in 2018.2 Although great advancement has been achieved in the treatment of cancer, such as surgery, chemotherapy, radiotherapy, immunotherapy and targeted therapy, advanced-stage cancers still have a poor 5-year survival rate and high death rate.3 Therefore, there is a pressing need to identify potential diagnostic and prognostic biomarkers as well as investigate the molecular mechanisms in cancer, which may play an essential role in the clinical treatment of cancer patients.
According to eukaryote whole-genome sequencing, only 2% genes are related to protein coding, while the rest are noncoding RNAs (ncRNAs).4 Among these ncRNAs, lncRNAs are initially considered as transcriptional noise with >200 nucleotides and lack protein-coding potential.5–7 Recently, increasing evidence reveals that lncRNAs function as promising oncogenes, or tumor-suppressor genes,8,9 taking part in various biological processes, such as cell proliferation, migration, invasion, cell cycle arrest, apoptosis, metastasis, etc.10–13 For instance, MALAT1 in NSCLC14 and ESCC,15 ZFAS1 in breast cancer16 and etc. Till now, only a few of lncRNAs have been well studied, and the majority of the lncRNAs still remain unknown, which needs to be further investigated.
Small nucleolar RNA host gene 15 (SNHG15), located on chromosome 7p13, which was first characterized by Tani et al in the study of cellular stress responses as a short half-life lncRNA,17–19 is a novel lncRNA. Recently, it has been reported that SNHG15 is associated with various kinds of cancers. Chen et al first found that SNHG15 was overexpressed in gastric cancer (GC) in 201620 and confirmed to be associated with cancer progression. Soon afterward, overexpression of SNHG15 was proved to be linked with poor survival in many human malignancies, including breast cancer, colorectal cancer (CRC), GC, glioma, hepatocellular cancer, lung cancer, osteosarcoma (OS), ovarian cancer, pancreatic cancer (PC), renal cell carcinoma (RCC) and thyroid cancer.20–36 Overview of SNHG15 in human cancers may provide a new perspective to study the mechanisms of cancer development. In this review, we mainly summarize the recent progression of SNHG15 correlated with cancer clinicopathological features and regulation mechanisms in the development and progression of multiple cancers (Tables 1 and 2).
Table 1.
Functional characterization of SNHG15 in various cancers
| Cancer types | Expression | Functional roles | Related genes | Role | References |
|---|---|---|---|---|---|
| Breast cancer | Upregulated | Proliferation, migration, invasion, cell cycle arrest, apoptosis, EMT | MiR-211-3p, E-cadherin, vimentin, MMP2, MMP9, SNAI1 | Oncogenic | 21 |
| Colorectal cancer | Upregulated | Migration, EMT, tumor growth | Slug, E-cadherin | Oncogenic | 22,23 |
| Gastric cancer | Upregulated | Proliferation, migration, invasion, apoptosis, tumorigenesis | MMP2, MMP9 | Oncogenic | 20 |
| Glioma | Upregulated | Proliferation, migration, angiogenesis | MiR-153, VEGFA, Cdc42 | Oncogenic | 24 |
| Lung cancer | Upregulated | Proliferation, migration, invasion, cell cycle arrest, apoptosis, EMT | MiR-486, miRNA-211-3p, CDK14, BAX, Bcl-2, cleaved caspase-3, PARP, E-cadherin, N-cadherin, Vimentin, MMP-2, MMP-9 | Oncogenic | 26–28 |
| Osteosarcoma | Upregulated | Proliferation, migration, invasion, autophagy | MiR-141, Atg5, LC3-II, p62 | Oncogenic | 29 |
| Epithelial ovarian cancer | Upregulated | Proliferation, migration, invasion, drug resistance | — | Oncogenic | 30 |
| Pancreatic cancer | Upregulated | Proliferation, apoptosis, cell cycle arrest, tumorigenesis | EZH2, H3K27me3, P15, KLF2, CDK2, CDK4, cleaved caspase-3, cleaved caspase-9 | Oncogenic | 31,32 |
| Renal cell cancer | Upregulated | Proliferation, migration, invasion, cell cycle arrest, apoptosis, EMT | TNF-α, Snail1, Slug, ZEB1, E-cadherin, N-cadherin, Vimentin | Oncogenic | 33 |
| Thyroid cancer | Upregulated | Proliferation, migration, apoptosis, EMT | MiR-200a-3p, YAP1, MST1, LATS1, E-cadherin, N-cadherin, Vimentin, β-catenin | Oncogenic | 34 |
| Downregulated | Proliferation, migration, invasion | MiR-510-5p | Nononcogenic | 35,36 |
Abbreviations: EMT, Epithelial to mesenchymal transition; MMP, matrix metalloproteinase; SNAI1, snail family transcriptional repressor 1; VEGFA, vascular endothelial growth factor A; CDK, cyclin-dependent kinase; Bax, Bcl-2-associated X protein; Bcl-2, B-cell lymphoma 2; PARP, poly(ADP-ribose) polymerase; EZH2, enhancer of zeste homolog 2.
Table 2.
Clinical significance of SNHG15 in various cancers
| Cancer types | Clinicopathologic features | References |
|---|---|---|
| Breast cancer | Tumor size, lymph node metastasis, advanced TMN stage, poor survival | 21 |
| Colorectal cancer | Liver metastasis, lymph node metastasis, advanced TMN stage, poor overall survival | 22,23 |
| Gastric cancer | Invasion depth, advanced TNM stage, lymph node metastasis, poor overall survival, poor disease-free survival | 20 |
| Hepatocellular cancer | Histological grade, vein invasion, TNM stage, poor survival | 25 |
| Lung cancer | Tumor size, lymph node metastasis, advanced TMN stage, poor survival | 26–28 |
| Epithelial ovarian cancer | Cancer type, ascites, FIGO stage, poor overall survival, poor progression-free survival | 30 |
| Pancreatic cancer | Tumor size, lymph node metastasis, advanced TMN stage, tumor differentiation, poor overall survival | 31,32 |
| Renal cell cancer | Histological differentiation, advanced T stage, poor survival | 33 |
| Thyroid cancer | Tumor size, lymph node metastasis, advanced TMN stage, poor survival | 34 |
Abbreviations: TMN, tumor node metastasis; FIGO, Federation of Gynecology and Obstetrics.
SNHG15 in human cancers
Breast cancer
Breast cancer, one of the most frequent cancers in females, is the second mortality-related malignant cancers in women after lung cancer.37 Progressive therapeutics have revolutionized the treatment of breast cancer. However, distant metastasis still remains the major problem of deterioration in breast cancer patients.38
Kong et al21 found that SNHG15 was significantly increased in 58 breast cancer tissues and 5 cell lines, and upregulated SNHG15 expression was positively associated with larger tumor size, lymph node metastasis and advanced TMN stage. Besides, Kaplan–Meier analysis revealed that breast cancer patients with elevated SNHG15 expression had a worse survival. Then, knockdown of SNHG15 had a negative effect on cell proliferation, migration and invasion but a positive effect on cell cycle arrest and apoptosis of breast cancer cells. Further studies showed that cells with SNHG15-depleted had lower expression of MMP2, MMP9, SNAI1, vimentin but enhanced expression of E-cadherin, which indicated the inhibition of the cell migration and invasion, as well as epithelial to the mesenchymal transition (EMT) process. Furthermore, the mechanism investigation revealed that SNHG15 acts as a competing endogenous RNA (ceRNA) to sponge miR-211-3p, leading to the proliferation, migration and invasion of breast cancer cells.
In conclusion, SNHG15 may serve as a prognostic biomarker for breast cancer patients, and SNHG15 sponging miR-211-3p might play an essential role in the pathogenesis of breast cancer.
Colorectal cancer
CRC is the third most prevalent form of malignancies among males and females in the United States and ranked fourth in cancer patient death.39 Though incidence and mortality rates have been declining mainly due to early diagnoses by screening tests and improvements of standard treatment,40 the occurrence of relapse and metastasis still influence the prognosis of CRC patients.
Jiang et al22 reported that SNHG15 was noted to be upregulated in colon adenocarcinoma (COAD) samples according to TCGA datasets, and upregulated SNHG15 was also observed in higher grade colon cancer cells compared with lower grade cells. Analysis of clinical data from TCGA also demonstrated that upregulated SNHG15 expression was related to a worse overall survival in patients with COAD. In addition, overexpression of SNHG15 facilitated cell migration and accelerated tumor growth in vivo. Moreover, SNHG15 maintains the stability of Slug in colon cancer cells by inhibiting its ubiquitination and degradation via interaction with the zinc finger domain of Slug.
Huang et al23 similarly revealed that SNHG15 was overexpressed in 91 CRC tissues compared with those in paired normal tissues. Enhanced expression of SNHG15 was significantly linked with liver metastasis, lymph node metastasis, advanced TMN stage and poor overall survival.
Taken together, upregulated SNHG15 in CRC was an independent predictor of poor survival, and oncogenic lncRNA SNHG15 may be a therapeutic target in colon cancer patients.
Gastric cancer
According to the latest statistics, GC is the fourth leading cause of human malignant disease, and the second most common cancer death worldwide,1 with definitely high incidence in Eastern Asia.41,42 Although advanced diagnosed methods and treatment have been established, most patients are still diagnosed at an advanced stage, which results in dismal prognosis.43 Thus, it is of great significance to identify key biomarkers and illustrate the molecular mechanisms of GC progression.
Chen et al20 proved that SNHG15 was markedly overexpressed in 106 GC tissues and 5 cell lines compared with that in corresponding normal tissues and a normal gastric epithelial cell line, respectively. Upregulated SNHG15 was closely linked with invasion depth, advanced TNM stage and lymph node metastasis. Kaplan–Meier analysis showed that enhanced SNHG15 contributed to poor overall survival and poor disease-free survival of GC patients. Biologically, knocking down SNHG15 inhibited cell proliferation, migration and invasion but induced apoptosis in GC cell lines. Moreover, enhanced SNHG15 was verified to induce tumorigenesis of GC cells in vivo. More importantly, SNHG15 promoted cell proliferation and invasion in GC cell lines partly by increasing MMP2 and MMP9 protein expression.
These data suggested that SNHG15 displayed potential to work as a poor prognostic biomarker in GC patients.
Glioma
Glioma is believed to be the most common form of adult primary malignant brain tumor.44 Despite treated with surgery, chemotherapy and radiotherapy, glioma patients still have a high recurrent rate and the prognosis remains poor.45 Thus, it is urgent to elucidate the molecular mechanisms and seek effective biomarkers for the treatment of this disease.
Ma et al24 illustrated that SNHG15 expression levels were notably upregulated in glioma vascular endothelial cells compared with primary astrocyte cells. And silencing SNHG15 dramatically suppressed cell proliferation, migration and angiogenesis of glioma vascular endothelial cells. Moreover, miR-153 was low expressed in glioma vascular endothelial cells, while VEGFA and Cdc42 were upregulated. Additionally, patients with glioma of overexpressed SNHG15 tended to have poorer overall survival, while patients with overexpressed miR-153 tended to have better overall survival. Furthermore, SNHG15 was confirmed to bind with miR-153 and negatively regulating its function, and miR-153 was found to be directly targeted the 3ʹ-UTR of VEGFA and Cdc42 by bioinformatic databases and dual-luciferase reporter assay. Thus, knocking down SNHG15 downregulated VEGFA and Cdc42 expression via targeting miR-153, suppressing glioma vascular endothelial cell proliferation, migration and angiogenesis.
These findings indicated that SNHG15 and miR-153 could be novel potential therapeutic targets for antiangiogenesis therapy of glioma.
Hepatocellular cancer
Hepatocellular carcinoma (HCC) is considered to be the most prevalent primary malignancies of the liver and is the third leading cause of cancer-related deaths worldwide.46,47 Numerous patients are diagnosed at an advanced stage, which limits therapeutic options and affects prognosis.48 Therefore, identifying effective biomarkers for early diagnosis in HCC patients is urgently needed.
Zhang et al25 stated that the expression level of SNHG15 was significantly higher in 152 HCC tissues than in adjacent normal tissues. Overexpressed SNHG15 was correlated with histological grade, vein invasion and TNM stage. Kaplan–Meier analysis revealed that enhanced SNHG15 was associated with poor overall survival of patients.
Collectively, the above study showed that SNHG15 may act as an efficient biomarker for HCC patients’ prognosis and a therapeutic target for HCC.
Lung cancer
Lung cancer is the most aggressive malignant cancer among both men and women, leading to 11.6% new cases and 18.4% cancer deaths in 2018 worldwide.1 Non-small cell lung cancer (NSCLC) accounts for about 85% of lung cancers; though patients are treated with surgery, chemotherapy and radiotherapy, the overall survival rate of patients with late stage is still dissatisfying.49 Therefore, it is of great importance to identify novel biomarkers for lung cancer treatment.
Jin et al26 elucidated that SNHG15 was markedly increased in NSCLC tissue samples (p<0.01) and 4 NSCLC cell lines compared with those in corresponding normal tissue samples and a normal human bronchial epithelial cell line. Upregulated SNHG15 was associated with lymph node metastasis, advanced TMN stage and poor overall survival. Further experiments revealed that SNHG15 silencing inhibited cell proliferation but promoted apoptosis and induced cell cycle arrest at G0/G1 phase. Meanwhile, SNHG15 silencing also inhibited the tumor growth of NSCLC cells in vivo. Mechanically, miR-486 was revealed to target SNHG15 3′-UTR with molecular binding and 3′-UTR of CDK14 with complementary binding sites via bioinformatics prediction tools and luciferase reporter assay.
Dong et al27 also uncovered that the expression level of SNHG15 was significantly higher in 49 NSCLC tissues than in paired para-carcinoma tissue. Besides, upregulated SNHG15 was dramatically related to tumor size, lymph node metastasis, advanced TMN stage, poor disease-free survival and poor overall survival. Silencing SNHG15 inhibited proliferation, migration, invasion and EMT process but induced apoptosis.
Cui et al28 similarly revealed that SNHG15 was upregulated in 55 lung cancer tissues and 3 lung cancer cell lines, and patients with a high level of SNHG15 had worse clinical outcomes. Knocking down SNHG15 could depress the proliferation and migration and invasion of lung cancer cells and induced cell cycle arrest in G0/G1 phase. Furthermore, mechanical experiment indicated that SNHG15 promoted the occurrence of lung cancer through regulating miRNA-211-3p.
In conclusion, SNHG15 may serve as a novel pathogenesis and potential therapeutic target for lung cancer patients.
Osteosarcoma
OS is one of the most prevalent form of primary bone cancers worldwide that mostly affects children, adolescents and young adults.2 The treatment of OS has improved greatly during the last 40 years, including neoadjuvant chemotherapy followed by surgical treatment. However, large amounts of patients are insensitive to chemotherapy, and the recurrent rate is still very high.50 Therefore, identifying novel therapeutic biomarkers isurgently needed.
Liu et al29 corroborated that SNHG15 expression level was dramatically increased in 35 OS tissues and 5 OS cell lines compared with paired normal tissues and osteoblastic cell line. Meanwhile, expression of miR-141 was remarkably decreased in 35 OS tissues and SNHG15 was negatively linked with the expression of miR-141 in OS tissues. Biologically, SNHG15 knockdown notably attenuated cell proliferation, migration, invasion and autophagy. Mechanically, this study proved that SNHG15 promoted OS progression via directly targeting miR-141 and negatively regulating its expression.
As a conclusion, SNHG15 could potentially be a new prognostic biomarker and therapeutic target for OS patients.
Epithelial ovarian cancer
Epithelial ovarian cancer (EOC) is a common malignant tumor in female reproductive system, accounting for 5% malignancies in female patients.51 The molecular mechanisms underlying ovarian cancer are still unclear, and the 5-year survival rate of patients at advanced stage is only 40%.52 Thus, there is a critical need to find effective biomarker to help early diagnosis and treatment of EOC.
Qu et al30 observed that SNHG15 expression was remarkably higher in 182 EOC tissues and 6 EOC cell lines than in corresponding normal ovarian tissues and a normal cell line. Upregulated SNHG15 was notably correlated with cancer type, ascites and FIGO stage. Additionally, multivariate analysis revealed that enforced SNHG15 expression was an independent risk factor for poor overall survival and progression-free survival of EOC. Further studies verified that the abnormal expression of SNHG15 promoted proliferation, migration and invasion. Notably, SNHG15 expression was much higher in EOC cells that were cisplatin resistant than that in controls, indicating SNHG15 may contribute to the drug resistance of EOC cells.
In conclusion, SNHG15 exhibited a pivotal role in EOC progression, which may provide an opportunity for prognostic assessment and molecular target treatment in EOC patients.
Pancreatic cancer
PC remains one of the most challenging malignancies worldwide, and its incidence as well as death rate appears to be rising.53–55 Among all pancreatic carcinomas, pancreatic ductal adenocarcinoma (PDAC) represents around 90%. However, lack of diagnostic tests contributes to poor prognosis of PDAC.56 So, it is necessary to explore novel biomarkers, which will contribute a crucial role in the diagnosis and therapy of PC.
Ma et al31 confirmed that the expression level of SNHG15 was dramatically increased in PC tissues and 3 PC cell lines. Upregulated SNHG15 was remarkably correlated with tumor size, lymph node metastasis and advanced TNM stage in PC patients. Functionally, SNHG15 knockdown attenuated cell proliferation, while induced apoptosis and alter cell cycle arrest at G0/G1 phase. Besides, silencing SNHG15 was observed to inhibit tumorigenesis in vivo. Mechanically, SNHG15 was observed to inhibit P15 and KLF2 expression to promote PC cell proliferation via EZH2-mediated H3K27me3.
Guo et al32 also detected that SNHG15 was overexpressed in PDAC tissues and in sera from PDAC patients compared with controls. Upregulated SNHG15 was positively associated with poor tumor differentiation, lymph node metastasis, advanced tumor stage and poorer overall survival in PDAC patients. Furthermore, Cox multivariate analyses proved that SNHG15 was an independent factor for PDAC patients’ prognosis.
These data indicated that SNHG15 could serve as a biomarker for detection and prognostic prediction for PC patients.
Renal cell cancer
RCC is believed to be the most predominant form of kidney malignancies, which represents 5% of all new cancer cases in 2018 in US.2 Though 5-year survival rates at diagnosis have improved, the overall prognosis for high-stage patients is still poor.57
Du et al33 identified that SNHG15 was remarkably upregulated in 96 RCC tissues and 5 cell lines compared with that in paired adjacent normal tissue samples and a proximal tubule epithelial cell line. Besides, upregulated SNHG15 was notably related to histological differentiation, advanced T stage and poorer survival. Knocking down SNHG15 significantly inhibited proliferation, migration, invasion and EMT process but increased apoptosis and induced cell cycle arrest at G0/G1 phase. Furthermore, this study indicated that SNHG15 may regulate RCC progression via regulating the NF-κB signaling pathway.
This study revealed that SNHG15 could be a new predictor of diagnosis and prognosis for RCC patients.
Thyroid cancer
Thyroid cancer is the most frequent endocrine malignant tumor and the ninth most common cancer for incidence worldwide. Though the mortality of thyroid cancer is low, the incidence of it is rising rapidly.1 Therefore, it is imperative to explore the molecular mechanisms to provide new insight into the treatment of thyroid cancer.
Wu et al34 found that SNHG15 served as an oncogene in papillary thyroid carcinoma (PTC). They indicated that the expression level of SNHG15 was higher in 92 human PTC tissues and 4 PTC cell lines than in controls. Overexpression of SNHG15 was closely associated with gender, larger tumor size, lymph node metastasis, advanced TNM stage and poorer overall survival. In addition, downregulation of SNHG15 attenuated cell proliferation, migration and EMT process but induced apoptosis in PTC cells. Mechanically, their study indicated that SNHG15 acts as a ceRNA to upregulate YAP1 signaling pathway via sponging miR-200a-3p.
However, Liu et al35 concluded that SNHG15 was downregulated in 50 thyroid cancer tissues and 4 cell lines and correlated with age, pathology classification, tumor size, lymph node metastasis, clinical stage, distant metastasis and disease‐free survival, indicating SNHG15 may act as an antitumor gene in thyroid cancer. This study also suggested that enhanced SNHG15 significantly suppressed proliferation, migration and invasion. Furthermore, they found that miR‐510‐5p promotes cell proliferation, migration and invasion via inhibiting SNHG15 in thyroid cancer.36
To conclude, the function of SNHG15 varies in different studies, which may be associated with the different subtypes of thyroid cancer. Further researches are needed to confirm these findings.
Conclusion and future perspectives
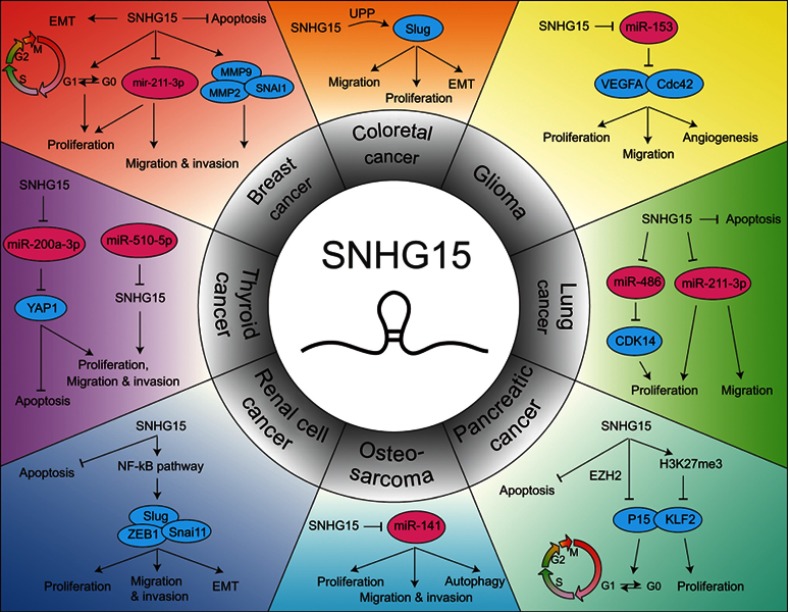
Dysregulated lncRNAs has been gradually identified as potential oncogenes or tumor-suppressor genes, as these RNAs play crucial regulatory roles in tumorigenesis and progression. As a novel lncRNA, SNHG15 is widely overexpressed in multiple cancers, such as breast cancer, CRC, GC, glioma, HCC, lung cancer, OS, EOC, PC, RCC and thyroid cancer (Figure 1). In contrast, SNHG15 is found downregulated in thyroid cancer by Liu et al. Upregulated SNHG15 was dramatically associated with multiple clinicopathological characteristics and prognosis, including TNM stage, lymph node metastasis and overall survival. In addition, SNHG15 is involved in cell proliferation, migration, invasion, apoptosis, angiogenesis, autophagy and EMT process. Mechanistically, SNHG15 could function as a ceRNA to directly interact with microRNAs (including miR-211-3p and miR-200a-3p), modifying the expression of the downstream target genes (such as SNAI1, MMP-2, MMP-9 and YAP1). SNHG15 also interacts with some proteins to modulate gene expression. Besides, SNHG15 could regulate some classical signaling pathways contributing to cancer progression. Collectively, SNHG15 may be considered as a promising biomarker for cancer diagnosis, prognostic or therapy.
Figure 1.
The molecular mechanisms of SNHG15 in different human cancers.
Abbreviations: SNHG15, small nucleolar RNA host gene 15; MALAT1, metastasis associated in lung adenocarcinoma transcript 1; ZFAS1, ZNFX1 antisense RNA 1; ceRNA, competing endogenous RNA; CRC, colorectal cancer; COAD, colon adenocarcinoma; GC, gastric cancer; HCC, hepatocellular carcinoma; NSCLC, non-small cell lung cancer; OS, osteosarcoma; EOC, epithelial ovarian cancer; PC, pancreatic cancer; PDAC, pancreatic ductal adenocarcinoma; PTC, papillary thyroid carcinoma; RCC, renal cell carcinoma; EMT, Epithelial to mesenchymal transition; MMP, matrix metalloproteinase; SNAI1, snail family transcriptional repressor 1; VEGFA, vascular endothelial growth factor A; CDK, cyclin-dependent kinase; Bax, Bcl-2-associated X protein; Bcl-2, B-cell lymphoma 2; PARP, poly(ADP-ribose) polymerase; EZH2, enhancer of zeste homolog 2.
However, the molecular mechanism of SNHG15 is still unknown in several cancers, such as hepatocellular cancer and EOC. Furthermore, upstream and downstream regulation mechanisms of SNHG15 remain relatively unknown. Thus, illumination of definitive molecular mechanisms of SNHG15 will help to better understand its role on cancer progression and to provide a new direction for clinical diagnosis and treatment of tumors.
Acknowledgments
This study was funded by the Medical and Health Research Project of Zhejiang Province (2018KY113).
Author contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 3.Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–289. doi: 10.3322/caac.21349 [DOI] [PubMed] [Google Scholar]
- 4.Dunham I, Kundaje A, Aldred SF, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. doi: 10.1038/nrg.2015.10 [DOI] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521 [DOI] [PubMed] [Google Scholar]
- 7.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 8.Dong D, Mu Z, Zhao C, Sun M. ZFAS1: a novel tumor-related long non-coding RNA. Cancer Cell Int. 2018;18:125. doi: 10.1186/s12935-018-0623-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghafouri-Fard S, Taheri M. Nuclear Enriched Abundant Transcript 1 (NEAT1): a long non-coding RNA with diverse functions in tumorigenesis. Biomed Pharmacother. 2018;111:51–59. doi: 10.1016/j.biopha.2018.12.070 [DOI] [PubMed] [Google Scholar]
- 10.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. doi: 10.1038/ng.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297–1325. doi: 10.1152/physrev.00041.2015 [DOI] [PubMed] [Google Scholar]
- 13.Gutschner T, Richtig G, Haemmerle M, Pichler M. From biomarkers to therapeutic targets-the promises and perils of long non-coding RNAs in cancer. Cancer Metastasis Rev. 2018;37(1):83–105. doi: 10.1007/s10555-017-9718-5 [DOI] [PubMed] [Google Scholar]
- 14.Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. doi: 10.1038/sj.onc.1206928 [DOI] [PubMed] [Google Scholar]
- 15.Hu L, Wu Y, Tan D, et al. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015;34:7. doi: 10.1186/s13046-015-0123-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Askarian-Amiri ME, Crawford J, French JD, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. Rna. 2011;17(5):878–891. doi: 10.1261/rna.2528811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tani H, Torimura M. Identification of short-lived long non-coding RNAs as surrogate indicators for chemical stress response. Biochem Biophys Res Commun. 2013;439(4):547–551. doi: 10.1016/j.bbrc.2013.09.006 [DOI] [PubMed] [Google Scholar]
- 18.Tani H, Torimura M. Development of cytotoxicity-sensitive human cells using overexpression of long non-coding RNAs. J Biosci Bioeng. 2015;119(5):604–608. doi: 10.1016/j.jbiosc.2014.10.012 [DOI] [PubMed] [Google Scholar]
- 19.Tani H, Okuda S, Nakamura K, Aoki M and Umemura T. Short-lived long non-coding RNAs as surrogate indicators for chemical exposure and LINC00152 and MALAT1 modulate their neighboring genes. PLoS One. 2017;12(7):e0181628. doi: 10.1371/journal.pone.0181628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen SX, Yin JF, Lin BC, et al. Upregulated expression of long noncoding RNA SNHG15 promotes cell proliferation and invasion through regulates MMP2/MMP9 in patients with GC. Tumour Biol. 2016;37(5):6801–6812. doi: 10.1007/s13277-015-4404-0 [DOI] [PubMed] [Google Scholar]
- 21.Kong Q, Qiu M. Long noncoding RNA SNHG15 promotes human breast cancer proliferation, migration and invasion by sponging miR-211-3p. Biochem Biophys Res Commun. 2018;495(2):1594–1600. doi: 10.1016/j.bbrc.2017.12.013 [DOI] [PubMed] [Google Scholar]
- 22.Jiang H, Li T, Qu Y, et al. Long non-coding RNA SNHG15 interacts with and stabilizes transcription factor Slug and promotes colon cancer progression. Cancer Lett. 2018;425:78–87. doi: 10.1016/j.canlet.2018.03.038 [DOI] [PubMed] [Google Scholar]
- 23.Huang L, Lin H, Kang L, et al. Aberrant expression of long noncoding RNA SNHG15 correlates with liver metastasis and poor survival in colorectal cancer. J Cell Physiol. 2019;234(5):7032–7039. doi: 10.1002/jcp.27456 [DOI] [PubMed] [Google Scholar]
- 24.Ma Y, Xue Y, Liu X, et al. SNHG15 affects the growth of glioma microvascular endothelial cells by negatively regulating miR-153. Oncol Rep. 2017;38(5):3265–3277. doi: 10.3892/or.2017.5985 [DOI] [PubMed] [Google Scholar]
- 25.Zhang JH, Wei HW, Yang HG. Long noncoding RNA SNHG15, a potential prognostic biomarker for hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2016;20(9):1720–1724. [PubMed] [Google Scholar]
- 26.Jin B, Jin H, Wu HB, Xu JJ, Li B. Long non-coding RNA SNHG15 promotes CDK14 expression via miR-486 to accelerate non-small cell lung cancer cells progression and metastasis. J Cell Physiol. 2018;233(9):7164–7172. doi: 10.1002/jcp.26543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong YZ, Meng XM, Li GS. Long non-coding RNA SNHG15 indicates poor prognosis of non-small cell lung cancer and promotes cell proliferation and invasion. Eur Rev Med Pharmacol Sci. 2018;22(9):2671–2679. doi: 10.26355/eurrev_201805_14963 [DOI] [PubMed] [Google Scholar]
- 28.Cui HX, Zhang MY, Liu K, Liu J, Zhang ZL, Fu L. LncRNA SNHG15 promotes proliferation and migration of lung cancer via targeting microRNA-211-3p. Eur Rev Med Pharmacol Sci. 2018;22(20):6838–6844. doi: 10.26355/eurrev_201810_16152 [DOI] [PubMed] [Google Scholar]
- 29.Liu K, Hou Y, Liu Y, Zheng J. LncRNA SNHG15 contributes to proliferation, invasion and autophagy in osteosarcoma cells by sponging miR-141. J Biomed Sci. 2017;24(1):46. doi: 10.1186/s12929-017-0353-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qu C, Dai C, Guo Y, Qin R, Liu J. Long noncoding RNA SNHG15 serves as an oncogene and predicts poor prognosis in epithelial ovarian cancer. Onco Targets Ther. 2019;12:101–111. doi: 10.2147/OTT.S182657 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Ma Z, Huang H, Wang J, et al. Long non-coding RNA SNHG15 inhibits P15 and KLF2 expression to promote pancreatic cancer proliferation through EZH2-mediated H3K27me3. Oncotarget. 2017;8(48):84153–84167. doi: 10.18632/oncotarget.20359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo XB, Yin HS, Wang JY. Evaluating the diagnostic and prognostic value of long non-coding RNA SNHG15 in pancreatic ductal adenocarcinoma. Eur Rev Med Pharmacol Sci. 2018;22(18):5892–5898. doi: 10.26355/eurrev_201809_15917 [DOI] [PubMed] [Google Scholar]
- 33.Du Y, Kong C, Zhu Y, et al. Knockdown of SNHG15 suppresses renal cell carcinoma proliferation and EMT by regulating the NF-kappaB signaling pathway. Int J Oncol. 2018;53(1):384–394. doi: 10.3892/ijo.2018.4395 [DOI] [PubMed] [Google Scholar]
- 34.Wu DM, Wang S, Wen X, et al. LncRNA SNHG15 acts as a ceRNA to regulate YAP1-Hippo signaling pathway by sponging miR-200a-3p in papillary thyroid carcinoma. Cell Death Dis. 2018;9(10):947. doi: 10.1038/s41419-018-1111-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Li J, Li F, Li M, Shao Y, Wu L. SNHG15 functions as a tumor suppressor in thyroid cancer. J Cell Biochem. 2019;120(4):6120–6126. doi: 10.1002/jcb.27899 [DOI] [PubMed] [Google Scholar]
- 36.Liu Y, Li J, Li M, Li F, Shao Y, Wu L. microRNA-510-5p promotes thyroid cancer cell proliferation, migration, and invasion through suppressing SNHG15. J Cell Biochem. 2019. doi: 10.1002/jcb.28454 [DOI] [PubMed] [Google Scholar]
- 37.DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67(6):439–448. doi: 10.3322/caac.21412 [DOI] [PubMed] [Google Scholar]
- 38.Kwan TT, Bardia A, Spring LM, et al. A digital RNA signature of circulating tumor cells predicting early therapeutic response in localized and metastatic breast cancer. Cancer Discov. 2018;8(10):1286–1299. doi: 10.1158/2159-8290.CD-18-0432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. doi: 10.3322/caac.21395 [DOI] [PubMed] [Google Scholar]
- 40.Volk RJ, Leal VB, Jacobs LE, et al. From guideline to practice: new shared decision-making tools for colorectal cancer screening from the American Cancer Society. CA Cancer J Clin. 2018;68(4):246–249. doi: 10.3322/caac.21459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 42.Ma W, Xu Z, Wang Y, et al. A positive feedback loop of SLP2 activates MAPK signaling pathway to promote gastric cancer progression. Theranostics. 2018;8(20):5744–5757. doi: 10.7150/thno.28898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gambardella V, Cervantes A. Precision medicine in the adjuvant treatment of gastric cancer. Lancet Oncol. 2018;19(5):583–584. doi: 10.1016/S1470-2045(18)30131-1 [DOI] [PubMed] [Google Scholar]
- 44.Ostrom QT, Gittleman H, Fulop J, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008-2012. Neuro Oncol. 2015;17(Suppl 4):iv1–iv62. doi: 10.1093/neuonc/nov189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stupp R. Drug development for glioma: are we repeating the same mistakes? Lancet Oncol. 2019;20(1):10–12. doi: 10.1016/S1470-2045(18)30827-1 [DOI] [PubMed] [Google Scholar]
- 46.Dawkins J, Webster RM. The hepatocellular carcinoma market. Nat Rev Drug Discov. 2019;18(1):13–14. doi: 10.1038/nrd.2018.146 [DOI] [PubMed] [Google Scholar]
- 47.Ringelhan M, Pfister D, O’Connor T, Pikarsky E, Heikenwalder M. The immunology of hepatocellular carcinoma. Nat Immunol. 2018;19(3):222–232. doi: 10.1038/s41590-018-0044-z [DOI] [PubMed] [Google Scholar]
- 48.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–1314. doi: 10.1016/S0140-6736(18)30010-2 [DOI] [PubMed] [Google Scholar]
- 49.Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1 [DOI] [PubMed] [Google Scholar]
- 50.Isakoff MS, Bielack SS, Meltzer P, Gorlick R. Osteosarcoma: current treatment and a collaborative pathway to success. J Clin Oncol. 2015;33(27):3029–3035. doi: 10.1200/JCO.2014.59.4895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Torre LA, Trabert B, DeSantis CE, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68(4):284–296. doi: 10.3322/caac.21456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su Z, Graybill WS, Zhu Y. Detection and monitoring of ovarian cancer. Clin Chim Acta. 2013;415:341–345. doi: 10.1016/j.cca.2012.10.058 [DOI] [PubMed] [Google Scholar]
- 53.Wolfgang CL, Herman JM, Laheru DA, et al. Recent progress in pancreatic cancer. CA Cancer J Clin. 2013;63(5):318–348. doi: 10.3322/caac.21190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kleeff J, Michl P. Targeted therapy of pancreatic cancer: biomarkers are needed. Lancet Oncol. 2017;18(4):421–422. doi: 10.1016/S1470-2045(17)30087-6 [DOI] [PubMed] [Google Scholar]
- 55.Zhu L, Staley C, Kooby D, El-Rays B, Mao H, Yang L. Current status of biomarker and targeted nanoparticle development: the precision oncology approach for pancreatic cancer therapy. Cancer Lett. 2017;388:139–148. doi: 10.1016/j.canlet.2016.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ray K. Pancreatic cancer: biomarkers for the early detection of PDAC. Nat Rev Gastroenterol Hepatol. 2017;14(9):504–505. doi: 10.1038/nrgastro.2017.111 [DOI] [PubMed] [Google Scholar]
- 57.Barata PC, Rini BI. Treatment of renal cell carcinoma: current status and future directions. CA Cancer J Clin. 2017;67(6):507–524. doi: 10.3322/caac.21411 [DOI] [PubMed] [Google Scholar]