Significance
Copy number variation describes the degree to which contiguous genomic regions differ in their number of copies among individuals. Copy number variable regions can drive ecological adaptation, particularly when they contain genes. Here, we compare differences in gene copy numbers among 17 polar bear and 9 brown bear individuals to evaluate the impact of copy number variation on polar bear evolution. Polar bears and brown bears are ideal species for such an analysis as they are closely related, yet ecologically distinct. Our analysis identified variation in copy number for genes linked to dietary and ecological requirements of the bear species. These results suggest that genic copy number variation has played an important role in polar bear adaptation to the Arctic.
Keywords: copy number variation, population genomics, adaptive evolution
Abstract
Polar bear (Ursus maritimus) and brown bear (Ursus arctos) are recently diverged species that inhabit vastly differing habitats. Thus, analysis of the polar bear and brown bear genomes represents a unique opportunity to investigate the evolutionary mechanisms and genetic underpinnings of rapid ecological adaptation in mammals. Copy number (CN) differences in genomic regions between closely related species can underlie adaptive phenotypes and this form of genetic variation has not been explored in the context of polar bear evolution. Here, we analyzed the CN profiles of 17 polar bears, 9 brown bears, and 2 black bears (Ursus americanus). We identified an average of 318 genes per individual that showed evidence of CN variation (CNV). Nearly 200 genes displayed species-specific CN differences between polar bear and brown bear species. Principal component analysis of gene CN provides strong evidence that CNV evolved rapidly in the polar bear lineage and mainly resulted in CN loss. Olfactory receptors composed 47% of CN differentiated genes, with the majority of these genes being at lower CN in the polar bear. Additionally, we found significantly fewer copies of several genes involved in fatty acid metabolism as well as AMY1B, the salivary amylase-encoding gene in the polar bear. These results suggest that natural selection shaped patterns of CNV in response to the transition from an omnivorous to primarily carnivorous diet during polar bear evolution. Our analyses of CNV shed light on the genomic underpinnings of ecological adaptation during polar bear evolution.
Brown bear (Ursus arctos) and polar bear (Ursus maritimus) diverged less than 500 kya (1–4). Despite this recent split, the polar bear has rapidly evolved unique morphological, physiological, and behavioral characteristics in response to the polar climate and ecology. The most obvious adaptation is the lack of fur pigmentation, which aids in camouflage (5). Additionally, polar bear claws are shorter, sharper, and more curved than those of the brown bear, adaptations that at once facilitate locomotion on icy surfaces and are better suited for grabbing and securely holding prey (5). Polar bears also have shortened tails and smaller ears to reduce heat loss, specialized front paws for swimming, and greater adipose deposits under the skin for thermal regulation (5–7). Polar bears and brown bears also have drastically different diets owing to their differing ecologies. The polar bear has a lipid-rich diet consisting almost exclusively of marine mammals, while the brown bear is an omnivore with the vast majority of its diet consisting of plant material (5, 8–10). The hypercarnivorous polar bear diet has resulted in several craniodental adaptations including a sharpening of the molars and a gap between canines and molars that allows for deeper canine penetration in prey (5, 9, 11).
Population genomic studies have identified genomic signatures of recent positive selection in polar bears suggesting genetic underpinnings to some adaptive phenotypes (3, 4). Miller et al. (4) sequenced the genomes of >20 polar and brown bear individuals, identifying hundreds of fixed missense substitutions in polar bears, as well as >1,000 genomic regions with highly divergent allele frequencies compared with brown bear populations. The functions of the genes identified in these analyses were involved in such processes as muscle formation, lactation, and fatty acid metabolism. Similar analyses performed by Liu et al. (3) on a different set of brown bear and polar bear individuals revealed functional associations of positively selected genes with adipose tissue development, fatty acid metabolism, heart function, and fur pigmentation. Together, these studies reveal a cohesive set of genes, pathways, and phenotypes that were likely shaped by natural selection during polar bear evolution.
These previous population genomic studies inferred recent positive selection solely from single-nucleotide polymorphisms (SNPs) and did not evaluate copy number variation (CNV) as an additional source of potential genomic divergence. CNV refers to differences in the number of repeats of a segment of DNA in the genome between individuals due to duplication, gain, or loss (12). CN variants have higher mutation rates than SNPs, and CNV loci together encompass an order of magnitude more nucleotides compared with SNPs (12, 13). CNV can influence phenotype, most commonly through changes in gene expression (14–16). For example, AMY1 encodes a salivary enzyme that catalyzes the initial step of starch digestion, and in humans the CN of AMY1 corresponds to increased transcript and protein expression and is greater in populations with starch-rich diets (17, 18). Numerous other examples of population-differentiated CN profiles that may be targets of natural selection have been identified in mammals (14, 19–23).
Here, we examine the extent of CNV in the polar bear and test the hypothesis that CN-variable genes reflect phenotypic adaptations. We generated whole-genome CN profiles for 17 polar bear and 9 brown bear individuals and identified genes with highly differentiated CN profiles between species that are indicative of recent positive selection. We observed a pervasive loss of genes involved in olfaction, as well as CN differences in genes underlying immune function, morphology, and diet. These results strongly suggest that gene CNV has contributed to polar bear adaptive evolution and illustrate the importance of including analysis of CN variants for comprehensive genomic scans of recent positive selection.
Results
Estimates of Genome-Wide CNV.
We mapped Illumina whole-genome sequencing data from 17 polar bears, 9 brown bears, and 2 black bears (Ursus americanus) against both the reference polar bear genome (3) and, to account for reference genome bias, the reference black bear genome (24). We then estimated whole-genome CN profiles using Control-FREEC (25) and gene CN profiles using both Control-FREEC and a read-depth–based approach adapted from our previous work [hereafter referred to as background depth normalized (BDN)] (Materials and Methods) (26, 27). Mapping rates of all individuals to the polar bear and black bear reference genomes averaged 97.8% and 97.4%, respectively, with no individual sample falling below 90% against either reference genome (SI Appendix, Table S1). The frequencies and patterns of CNV were remarkably similar across bear species. Using whole-genome CN estimates from Control-FREEC, we found an average of 4,604 and 4,548 CNVs (gains and absences) accounting for 142 Mb and 144 Mb in the brown bear and polar bear, respectively. For clarity, we refer to CN of 0 as an “absence” rather than a deletion because the absence of a locus in an analyzed genome could be the result of a gain in the reference genome rather than a deletion in the analyzed genomes (28).
Ontology Enrichment Analysis for Genes Showing Intraspecific CN Variability.
We were primarily interested in identifying CN-variable genes because of their potential impact on phenotype. Thus, for each individual, we focused on CNVs that contained complete genes. We conservatively considered genes CNV if a gain or absence was independently predicted in each of the two CN estimation methods. We observed a mean of 266 and 373 duplicated genes and 13 and 21 absent genes in the polar bear and brown bear populations, respectively (SI Appendix, Fig. S1). The number of CNV genes per individual is consistent with results found in humans, mice, and horses (20, 26, 27). We identified a total of 1,168 and 36 genes that showed patterns of gain and absence, respectively, in at least one polar bear individual and 735 and 61 genes that showed patterns of gain and loss, respectively, in at least one brown bear individual (Dataset S1). Most notably, gene enrichment analysis of these gene sets revealed an overrepresentation of genes in the Gene Ontology (GO) categories associated with smell (SI Appendix, Tables S2–S5) (e.g., GO term sensory perception of smell: polar bear gene gain: P = 1.1e−3, brown bear gene gain: P = 4.1e−31, polar bear gene absence: P = 1e−4, and brown bear gene absence: P = 8e−11; SI Appendix, Tables S2–S5). We also identified several genes involved in vision (CACNA1F, CRX, GUCY2F, IRX5, NDP, OAT, OPN1LW, PAX6, PPEF1, RP2, RPGR, RS1, and VSX1), and hearing (POU3F4) that were CN-variable at the population level (Dataset S1), consistent with previous studies suggesting that CNV shapes the evolution of sensory perception in mammals (29, 30).
Gene CN Differentiation between Polar Bear and Brown Bear.
Positive selection can shape allele frequency differences between recently diverged populations and species that are subject to environmental differences (31). We used the VST measurement to identify highly divergent CN profiles between the polar bear and brown bear populations (32). VST is an estimate (analogous to FST) used for multiallelic genotype data such as CNVs and describes the proportion of population-level CNV due to differences between subpopulation. We first estimated genome-wide VST in 10-kb windows with a 2-kb step size across the reference polar bear genome (3). In total, we analyzed 1,143,840 windows, with an average VST value of 0.006 (SI Appendix, Fig. S2). We then independently calculated VST values across the 21,142 predicted polar bear protein-coding genes and observed an average VST of 0.018 using Control-FREEC (Fig. 1, Lower) and an average VST of 0.041 using the BDN approach (SI Appendix, Fig. S3). Similarly, we observed an average gene VST of 0.03 using Control-FREEC when black bear was used as the reference genome (SI Appendix, Fig. S4). These results indicate that the vast majority of the genome, including protein-coding genes, does not harbor differentiated CN profiles between polar bear and brown bear.
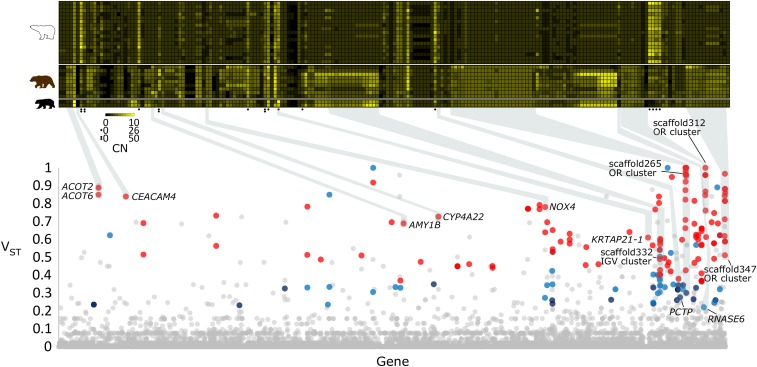
Fig. 1.
Genes with differentiated CN profiles between polar bear and brown bear. (Lower) A Manhattan plot of VST values (y axis) for each gene (n = 21,142) (x axis) in the reference polar bear genome. Genes are depicted by their VST value patterns across the Control-FREEC and BDN CN estimations: red = VST > 0.35 in both CN estimation methods, purple = VST > 0.22 and VST ≤ 0.35 in both CN estimation methods, blue = VST > 0.22 in one CN estimation method and VST > 0.35 in the other CN estimation, and gray = VST ≤ 0.22 in at one or both CN estimation methods. Genes of interest are labeled according to their respective human annotations, and are highlighted with gray boxes. (Upper) A heat map of diploid gene CN (columns) for each of the 197 differentiated genes in the 17 polar bear individuals, 9 brown bear individuals, and 2 black bear individual (rows). Genes with black circle notations represent high-CN genes and have different scales (see key). Black and yellow represent a CN of zero, and a high CN (8 or 50 depending on scale), respectively. Polar bear gene identifiers, along with either human or dog annotations when available, are noted for each gene. CNs are from Control-FREEC estimates. Individual samples identifiers and gene identifiers are provided in Dataset S1.
To identify genes showing strong interspecific differences in CN, we performed permutation tests to classify genes as either “differentiated” (genes having a VST greater than the maximum 95% percentile permuted VST value) or “extremely differentiated” (greater than the maximum 99% percentile permuted VST value). Moreover, we only considered genes that met these criteria in both CN calling methods (SI Appendix, Fig. S3). This resulted in a set of 197 genes whose CN are differentiated between polar bear and brown bear (VST > 0.22; 0.93% of all genes), while 134 of these genes also qualified as being extremely differentiated (VST > 0.35; 0.63% of all genes) (Fig. 1 and Dataset S2). The number and percentages of differentiated genes are similar to those found between human populations (21). Interestingly, >80% of CN-differentiated genes had higher CNs in brown bears compared with polar bears. Furthermore, only 8.6% of CN-differentiated genes had a higher average CN in polar bear, as well as an average CN greater than 3 in polar bear.
Gene enrichment analysis of the differentiated and extremely differentiated gene sets again revealed a significant enrichment of genes in the GO Biological Process category detection of chemical stimulus involved in sensory perception of smell (differentiated P = 2.8e−39 and extremely differentiated P = 1.3e−21; SI Appendix, Tables S6 and S7). Overall gene enrichment results were highly similar when using the black bear as a reference genome (SI Appendix, Tables S8 and S9). Further, 92 of the 197 genes (47%) were annotated as olfactory receptors (ORs) via hmmscan (33), and 88% of these putative OR-encoding genes had lower CN in polar bears (SI Appendix, Fig. S5). Several of these differentiated OR genes are located in large genomic clusters. For example, the near entirety of the 72-kb scaffold265 and the 280-kb scaffold312 display differentiated CN profiles and contain 26 and 14 predicted OR-encoding genes, respectively (Fig. 1, SI Appendix, Fig. S5, and Datasets S1 and S2).
We identified other genes with differentiated CN profiles that may reflect ecological differences between brown bears and polar bears (Figs. 1 and 2). For instance, KRTAP21-1 is involved in hair shaft formation (34) (Fig. 1 and SI Appendix, Fig. S6) and found at significantly higher CN in polar bears. Additionally, several CN-differentiated genes are directly involved in metabolism. AMY1B, for example, encodes a salivary amylase involved in hydrolyzing starch, and fewer copies are present in polar bear (Figs. 1 and 2). Gene enrichment analysis of differentiated genes also showed an overrepresentation of genes involved in fatty acid metabolism (i.e., GO Biological Process: long-chain fatty acid metabolic process, P = 0.009; Reactome: fatty acid metabolism, P = 8.6e−4; GeneSetDB: arachidonic acid metabolism, P = 1.1e−6; and GO Biological Process: icosanoid metabolic process P = 0.038). These genes include ACOT2, ACOT6, CBR1, CYP4A22, CYP2A7, GSTM1, GSTM5, CBR3, GPX4, and PCTP (Fig. 1, SI Appendix, Fig. S2, and Dataset S2).
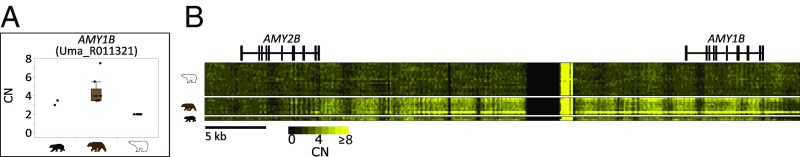
Fig. 2.
Polar bear possess fewer copies of AMY1B compared with brown bear. Box and whisker plot of gene CN for the salivary amylase-encoding gene, AMY1B (A). CNs are from Control-FREEC estimates. Each dot represents the diploid CN (y axis) for a given bear individual (x axis). The box plot displays the median value, first quartile, and third quartile. Whiskers are drawn to the furthest point with 1.5× the interquartile range (third quartile minus the first quartile) from the box. The black bear does not include a box and whisker plot because the analysis included only two individuals. (B) A heat map depicting the BDN CN estimates for nonoverlapping 100-bp windows in the AMY2B and AMY1B locus on scaffold70 (positions 1,331,300 to 1,380,700) of the reference polar bear genome.
Gene Loss in Polar Bears.
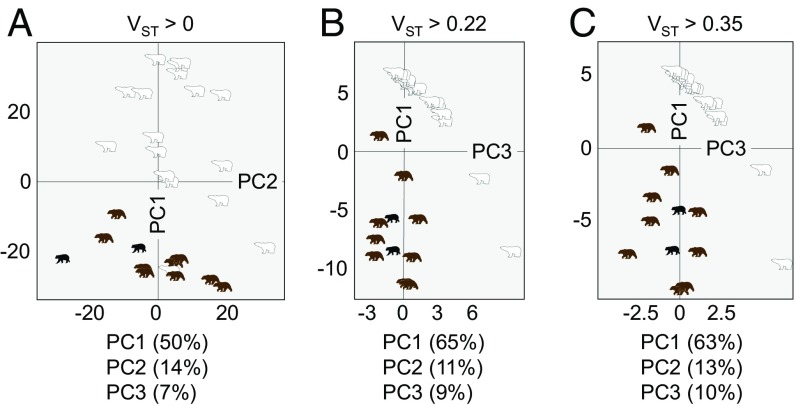
We observed that the majority of genes displaying differentiated CN have lower CN in polar bears. To evaluate whether this observation is more attributable to changes in the polar or brown bear lineage, we compared the gene CN profiles of brown bears and polar bears to that of the black bear, which diverged from brown/polar bears ∼3.5 to 5 Mya (2, 4). As a broad assessment of CN profiles between species, we first compared the CN distributions of differentiated genes between the polar bears, the brown bears, and the two black bears. As expected, this analysis revealed a statistically significant difference between polar bears and brown bears (Wilcoxon test; Control-FREEC P = 9e−180 and BDN P = 2e−173). Moreover, this analysis showed that although there is also a statistically significant difference between polar bears and black bears (Wilcoxon test; Control-FREEC P = 7e−75 and BDN P = 4e−75), there is no such difference between brown bears and black bears (Wilcoxon test; Control-FREEC P = 0.38 and BDN P = 0.60). Additionally, we performed principal component analysis (PCA) on the Control-FREEC–based CN estimates of (i) all genes, (ii) the differentiated gene set, and (iii) the extremely differentiated gene set including the black bears (Fig. 3). In each of the three PCAs, the majority of polar bear samples showed a clear separation from brown and black bears along the first principal component (PC1), with PC1 alone explaining at least 50% of the variance among individuals in each of the three analyses (Fig. 3 and SI Appendix, Fig. S7). Moreover, the black bear individuals consistently clustered with the brown bear individuals (Fig. 3 and SI Appendix, Fig. S7). We observed similar results when PCA was performed on the BDN estimates of gene CN (SI Appendix, Fig. S7). Together, these results show that brown and black bears share more similar CN profiles with one another than with the polar bear and suggest that CNV evolved rapidly in the polar bear lineage.
Fig. 3.
Polar bear exhibits divergent CN profiles compared with brown bear and black bear. PCA of gene CN across all bear individuals for genes showing agreement between the two CN estimation methods. (A) All CN-variable genes (VST > 0; 4,698 genes), (B) differentiated genes (VST > 0.22; 197 genes), and (C) highly differentiated genes (VST > 0.35; 134 genes). The percentage of variance explained by PC1, PC2, and PC3 are provided below each plot.
Discussion
Here, we conducted a population-level study to characterize genome-wide patterns of CNV in the polar bear and brown bear. CNV can drive ecological adaptation over short evolutionary periods (35–37). Polar bears and brown bears are excellent models for exploring the impact of natural selection on CNV, because they inhabit vastly different habitats yet are so recently diverged that they remain capable of producing fertile hybrid offspring (38, 39). Our analysis suggests that CNV is common in the Ursus genus. On average, ∼140 Mb of the polar bear and brown bear genome are CN-variable, accounting for ∼6% of the reference polar bear genome assembly. These findings are consistent with results observed in other mammals (∼5% in humans, ∼6.9% in mice, and ∼4% in cows) (12, 40, 41). Because CNV appears so abundant and dynamic within mammalian genomes, we explored the hypothesis that natural selection has acted on this form of genetic variation to facilitate adaptive refinements during polar bear evolution.
We identified 197 genes with differentiated CN profiles between polar bears and brown bears. Just 19% of these genes showed higher CN in polar bears, several of which are involved in immune function. Genes with immune response functions are overrepresented in CN-variable segments of mammalian genomes (14, 29, 32, 42, 43), and divergence of immunity-related genes is a common outcome of speciation (44, 45). For instance, relative to brown bears, polar bears have elevated CN of genes involved in such immune processes as antigen recognition (e.g., IGLV4-60) (46), the triggering of bacterial phagocytosis (e.g., CEACAM4) (47), the production of cytokines in response to viral infection (e.g., IFNA21) (48), and antibacterial activity in the urinary tract (RNASE6) (49, 50) (Fig. 1 and Dataset S2). We additionally discovered several large gene clusters containing Ig V-set domains on scaffold332 and scaffold342 that are present at lower CN in polar bears (Fig. 1 and Dataset S2). The differences observed in immune gene CN between polar bear and brown bear may be indicative of the differential pathogen pressures experienced after divergence.
Genes involved in fur pigmentation (e.g., EDNRB, TRMP1, LYST, and AIM1) were previously identified as examples of recent positive selection in polar bear (3, 4). We also identified an interesting fur-related gene, KRTAP21-1 (Uma_R019359), which was highly differentiated between bear species. KRTAP21-1 is involved in the formation of hair shafts and is a keratin-associated protein (KRTAP), a large and diverse gene family in mammals (51, 52). The higher gene CN of KRTAP21-1 in polar bears (Fig. 1 and SI Appendix, Fig. S6) may be related to the morphologic differences observed between brown and polar bear fur, with polar bear having both a more dense undercoat and a distinctively honeycombed cross-sectional structure to the fur itself (53). Differences in both fur pigmentation and fur structural morphology are likely adaptations to the arctic environment.
Polar bears have adapted to a diet exceptionally high in fat (5, 10), which is demonstrated by their ability to digest fat more efficiently than protein (54). Accordingly, a number of genes involved in cardiovascular function and fatty acid metabolism display signatures of recent positive selection (3, 4). Our results reinforce this finding. We identified CN differences between polar bears and brown bears in NOX4 (Uma_R015975), a fat storage-related gene (55). NOX4 is a regulator of metabolic homeostasis and is found at lower CN among polar bears (mean CN = 1.88) compared with brown bears (mean CN = 3.89) (Fig. 1, SI Appendix, Fig. S6, and Dataset S2). Importantly, NOX4 plays an antiadipogenic role in body composition (55), thus suggesting that the reduced CN of NOX4 in polar bear may be directly related to the need of this species to generate greater fat stores. Gene enrichment analysis of CN-differentiated genes also revealed an enrichment of genes involved in the categories of arachidonic acid and eicosanoid metabolism (SI Appendix, Tables S6–S9). Eicosanoids derive from arachidonic acid and play important roles in inflammation, thermoregulation, and cardiovascular function (56). Interestingly, most of the differentiated CN-variable genes involved in fatty acid metabolism are present at lower CN in polar bears and are perhaps related to the unique polar bear metabolic rates (57–59).
Other aspects of our analysis furthered this association between diet, metabolism, and CNV. The most striking CN difference we observed was the widespread reduction of the OR gene CN in the polar bear lineage (Fig. 1 and Dataset S2). In vertebrates, OR genes encode G protein-coupled receptors and the OR gene family in mammals can comprise more than 1,000 genes in most species, including bears (60–63). Each OR is tuned to respond to only a specific set of odor molecules, with the diversity of a species’ OR repertoire reflecting the breadth of its olfactory-mediated chemoreceptive capacity (60, 63–65). ORs play essential roles in the detection of food, mates, and predators, and variations in the OR repertoire among species can reflect ecological differences (61, 66).
For polar bears, Liu et al. (3) previously identified two OR genes (OR5D13 and OR8B8) in their SNP-based positive selection screen, indicating that the OR family of genes have been a common target of selection during polar bear evolution. CNV among OR genes has been identified as a potential source of ecological adaptation in many species (20, 30, 44, 67–69). This is likely the case in polar bear evolution as well, where selective pressure to maintain a more diverse OR repertoire may be relaxed with the less complex chemical ecology of arctic environments (70). While several of the highly CN-differentiated OR genes have been associated with eating behavior, body fat (e.g., OR7G3, OR7E24, and OR7G1) (71), and reproduction (OR7A5) (72) in humans, most are directly related to olfaction (Dataset S2). Thus, polar bear olfaction appears to have evolved to become both more specific (i.e., a less diverse OR repertoire) and more acute [i.e., increased surface area of olfactory epithelium accommodated by enlarged olfactory turbinals (73)]. Both of these characteristics could be refinements toward the detection of mates and prey over greater distances.
Other highly differentiated genes were even more directly involved in sensory perception and dietary behavior. We also observed fewer copies of the salivary amylase (AMY1B) (Figs. 1 and 2). Higher amylase CN has been a common signature of selection in organisms with high-starch diets (18, 19, 74). Although the polar bear diet likely includes a small proportion of plant and fungal material, the vast majority of caloric intake comes from seal (10, 53, 75). Conversely, brown bears and black bears have a diverse omnivorous diet in which plant-based materials (i.e., grasses, herbs, fruits, roots, and corms) make up more than 70% of their diet (76). Accordingly, the limited plant carbohydrate content in the polar bear diet likely led to a loss of AMY1B CN. These results strongly suggest that reductions in CN of several genes central to dietary discernment and processing are consistent with a shift toward hypercarnivority in polar bears.
Our analyses reinforce the observation that CNV can contribute to rapid phenotypic diversity and ecological adaptation (19, 21, 30, 74, 77, 78). The strong selective pressure imposed by diet has shaped population-specific human genetic variants (18, 79–81), and similar evolutionary processes likely shaped the polar bear genome during the transition from an omnivorous diet to a mainly carnivorous diet. In agreement with previous studies, we posit that natural selection acting through structural variants can drive adaptive refinements over short evolutionary timescales (18, 19, 35, 74, 77, 78, 82–84).
Materials and Methods
Data Mining and Sequence Read Processing.
We used the high-quality draft polar bear genome for our primary mapping reference (3). We downloaded the genome sequence, protein sequences, and gene annotations from the GigaDB (http://gigadb.org/dataset/100008). We also used the black bear genome as a mapping reference (24) to cross-validate our results and limit bias that could be introduced through the usage of a single reference genome. The reference black bear genome and gtf file was downloaded from ftp://ftp.jax.org/maine_blackbear_project/. We downloaded whole-genome paired-end Illumina data for 17 polar bears, 9 brown bears, and 2 black bears with reported coverage values ≥10× from the NCBI Sequence Read Archive (polar bear BioSample accession nos.: SAMN02261811, SAMN02261819, SAMN02261821, SAMN02261826, SAMN02261840, SAMN02261845, SAMN02261851, SAMN02261853, SAMN02261854, SAMN02261856, SAMN02261858, SAMN02261865, SAMN02261868, SAMN02261870, SAMN02261871, SAMN02261878, and SAMN02261880; brown bear BioSample accession nos.: SAMN02256313, SAMN02256315, SAMN02256316, SAMN02256317, SAMN02256318, SAMN02256319, SAMN02256320, SAMN02256321, and SAMN02256322; black bear BioSample accession nos.: SAMN01057691 and SAMN10023688) (2, 3, 24). We performed quality trimming for each sample using Trim Galore (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Residual adapter sequences were removed from reads, and reads were trimmed such that they contained a minimum quality score of 30 at each nucleotide position. We discarded trimmed reads shorter than 50 nucleotides.
Estimating CNV.
Quality and adapter trimmed paired-end Illumina sequence reads were independently mapped against the reference polar bear and black bear genomes using the “sensitive” preset parameters in bowtie2 (85). SAM alignment files were converted into sorted BAM format using the view and sort functions in samtools (86). We used the read depth based approach implemented in Control-FREEC to estimate integer CNs for each 10-kb window with a 2-kb step size across the entire genome (25). CN variants were not predicted in reference scaffolds <10 kb. We used the following parameters in Control-FREEC: breakPointThreshold = 0.8, coefficientOfVariation = 0.062, minExpectedGC = 0.35, maxExpectedGC = 0.55, degree = 3, and telocentromeric = 0. From the Control-FREEC outputs and gene coordinate files, we used a custom Perl script to identify genes that were entirely overlapped by CN variants (script available at https://github.com/DaRinker/PolarBearCNV). Although infrequent, we observed some instances of genes with more than one CN estimate. These rare events were due to imperfect estimation of breakpoints given the resolution of our window size and sliding window, which led to overlapping boundaries between the end of one CNV and the beginning of the next CNV. In these instances, we used the average CN. Additionally, we employed an independent read depth-based approach (BDN) to estimate gene CN. This approach is similar to our previous work and the work of others (26, 27, 87, 88). For each sample, we extracted protein-coding gene coordinates from the polar bear reference gff file and calculated average coverage values for each gene using the samtools depth function. The median value of all gene average coverage values was used as a normalizing factor representing diploid CN of two. Gene CN was calculated as follows:
Identifying Genes Differentiated by CN between Polar Bear and Brown Bear.
We calculated VST to identify divergent CNV profiles between the polar bear and brown bear populations (32). VST is a measurement specific for multiallelic genotype data such as microsatellites and CN variants and is analogous to FST. Both VST and FST consider how genetic variation can partition groups (populations or closely related species) (32) and range from 0 (no differentiation between groups) to 1 (complete differentiation between groups). We calculated VST as follows:
where Vtotal is total variance; Vpolar bear and Vbrown bear is the CN variance for the polar bear and brown bear populations, respectively; Npolar bear and Nbrown bear is the sample size for the polar bear and brown bear populations, respectively; and Ntotal is the total sample size. VST was calculated for sliding windows of 10-kb genomic bins, with a 2-kb step size across the reference polar bear genome (SI Appendix, Fig. S2). We also independently calculated VST across all genes using gene CN estimates obtained from Control-FREEC and our BDN method (SI Appendix, Fig. S4).
Permutation Testing for VST Differentiation.
To determine which genes displayed the greatest degree of observed interspecific CN variation that was likely not due to sampling bias, we performed permutation tests on the CN counts. Here, we randomly permuted all brown and polar bear individuals and calculated a new VST for every gene. This process was repeated 1,000 times creating a distribution of VST values for each gene (R script available at https://github.com/DaRinker/PolarBearCNV). We then selected those genes whose observed VST fell above the 95th and 99th percentile of the permuted VST distribution. These genes displayed strong intraspecific CN homogeneity, while also showing high degrees of interspecific differentiation (SI Appendix, Fig. S6). Finally, we took the maximum permuted VST observed in the 95th and 99th percentiles of all genes to establish a genome-wide standard cutoff (maximum 95th percentile: VST > 0.22; maximum 99th percentile: VST > 0.35) for all subsequent analysis. Gene VST values were considered significant when observed VST values were above the maximum 95% confidence interval cutoff in both gene CN estimate methods.
Protein Domain Classification and Gene Enrichment.
We used hmmscan to predict protein domains (33) and ShinyGO v0.50 to perform functional enrichment on the set of CN-variable genes with homology to the human genome (i.e., those genes with Ensembl gene identifiers) (89).
PCA of Gene CN.
We performed PCA on gene CN to evaluate whether overall CNV was sufficient to separate the polar bear, brown bear, and black bear species. The PCs were computed using the prcomp function in R using a data matrix containing the CNs of genes in all of the bear samples analyzed (90). CN counts were first scaled and centered before PC computation. Independent analyses were performed on the full set of genes with CN information (21,142 genes), as well as on subsets of genes thresholded at a minimal VST of 0.22 (197 genes) or 0.35 (134 genes) (Fig. 3 and SI Appendix, Table S8).
Supplementary Material
Acknowledgments
We thank Shiping Liu and Zijun Xiong for information regarding the polar bear reference genome, Elizabeth Weston for helpful comments on this manuscript, and two anonymous reviewers for constructive suggestions on an earlier version of this manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901093116/-/DCSupplemental.
References
- 1.Hailer F., et al. , Nuclear genomic sequences reveal that polar bears are an old and distinct bear lineage. Science 336, 344–347 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Kumar V., et al. , The evolutionary history of bears is characterized by gene flow across species. Sci. Rep. 7, 46487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu S., et al. , Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell 157, 785–794 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller W., et al. , Polar and brown bear genomes reveal ancient admixture and demographic footprints of past climate change. Proc. Natl. Acad. Sci. U.S.A. 109, E2382–E2390 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amstrup S. C., DeMaster D. P., “Polar bear, Ursus maritimus” in Wild Mammals of North America: Biology, Management, and Conservation, Feldhamer G. A., Thompson B. C., Chapman J. A., Eds. (Johns Hopkins University Press, Baltimore, 2003), ed. 2, pp. 587–610. [Google Scholar]
- 6.Lister A. M., Behavioural leads in evolution: Evidence from the fossil record. Biol. J. Linn. Soc. Lond. 112, 315–331 (2014). [Google Scholar]
- 7.Pond C. M., Mattacks C. A., Colby R. H., Ramsay M. A., The anatomy, chemical-composition, and metabolism of adipose-tissue in wild polar bears (Ursus-Maritimus). Can. J. Zool. 70, 326–341 (1992). [Google Scholar]
- 8.Naves J., Fernandez-Gil A., Rodriguez C., Delibes M., Brown bear food habits at the border of its range: A long-term study. J. Mammal. 87, 899–908 (2006). [Google Scholar]
- 9.Slater G. J., Figueirido B., Louis L., Yang P., Van Valkenburgh B., Biomechanical consequences of rapid evolution in the polar bear lineage. PLoS One 5, e13870 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derocher A. E., Wiig O., Andersen M., Diet composition of polar bears in Svalbard and the western Barents Sea. Polar Biol. 25, 448–452 (2002). [Google Scholar]
- 11.Figueirido B., Palmqvist P., Perez-Claros J. A., Ecomorphological correlates of craniodental variation in bears and paleobiological implications for extinct taxa: An approach based on geometric morphometrics. J. Zool. (Lond.) 277, 70–80 (2009). [Google Scholar]
- 12.Zarrei M., MacDonald J. R., Merico D., Scherer S. W., A copy number variation map of the human genome. Nat. Rev. Genet. 16, 172–183 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Zhang F., Gu W., Hurles M. E., Lupski J. R., Copy number variation in human health, disease, and evolution. Annu. Rev. Genomics Hum. Genet. 10, 451–481 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handsaker R. E., et al. , Large multiallelic copy number variations in humans. Nat. Genet. 47, 296–303 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hastings P. J., Lupski J. R., Rosenberg S. M., Ira G., Mechanisms of change in gene copy number. Nat. Rev. Genet. 10, 551–564 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henrichsen C. N., Chaignat E., Reymond A., Copy number variants, diseases and gene expression. Hum. Mol. Genet. 18, R1–R8 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Carpenter D., Mitchell L. M., Armour J. A., Copy number variation of human AMY1 is a minor contributor to variation in salivary amylase expression and activity. Hum. Genomics 11, 2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perry G. H., et al. , Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 39, 1256–1260 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Axelsson E., et al. , The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature 495, 360–364 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Pezer Ž., Harr B., Teschke M., Babiker H., Tautz D., Divergence patterns of genic copy number variation in natural populations of the house mouse (Mus musculus domesticus) reveal three conserved genes with major population-specific expansions. Genome Res. 25, 1114–1124 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sudmant P. H., et al. , Global diversity, population stratification, and selection of human copy-number variation. Science 349, aab3761 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright D., et al. , Copy number variation in intron 1 of SOX5 causes the Pea-comb phenotype in chickens. PLoS Genet. 5, e1000512 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu L., et al. , Population-genetic properties of differentiated copy number variations in cattle. Sci. Rep. 6, 23161 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srivastava A., et al. , Genome assembly and gene expression in the American black bear provides new insights into the renal response to hibernation. DNA Res. 26, 37–44 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boeva V., et al. , Control-FREEC: A tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 28, 423–425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibbons J. G., Branco A. T., Godinho S. A., Yu S., Lemos B., Concerted copy number variation balances ribosomal DNA dosage in human and mouse genomes. Proc. Natl. Acad. Sci. U.S.A. 112, 2485–2490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibbons J. G., Branco A. T., Yu S., Lemos B., Ribosomal DNA copy number is coupled with gene expression variation and mitochondrial abundance in humans. Nat. Commun. 5, 4850 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Zhao S., Gibbons J. G., A population genomic characterization of copy number variation in the opportunistic fungal pathogen Aspergillus fumigatus. PLoS One 13, e0201611 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doan R., et al. , Identification of copy number variants in horses. Genome Res. 22, 899–907 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paudel Y., et al. , Copy number variation in the speciation of pigs: A possible prominent role for olfactory receptors. BMC Genomics 16, 330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabeti P. C., et al. , Positive natural selection in the human lineage. Science 312, 1614–1620 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Redon R., et al. , Global variation in copy number in the human genome. Nature 444, 444–454 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potter S. C., et al. , HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogers M. A., Langbein L., Praetzel-Wunder S., Winter H., Schweizer J., Human hair keratin-associated proteins (KAPs). Int. Rev. Cytol. 251, 209–263 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Iskow R. C., Gokcumen O., Lee C., Exploring the role of copy number variants in human adaptation. Trends Genet. 28, 245–257 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steenwyk J. L., Rokas A., Copy number variation in fungi and its implications for wine yeast genetic diversity and adaptation. Front. Microbiol. 9, 288 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang Y. C., Amon A., Gene copy-number alterations: A cost-benefit analysis. Cell 152, 394–405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abbott R., et al. , Hybridization and speciation. J. Evol. Biol. 26, 229–246 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Mallet J., Hybridization, ecological races and the nature of species: Empirical evidence for the ease of speciation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 2971–2986 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Locke M. E. O., et al. , Genomic copy number variation in Mus musculus. BMC Genomics 16, 497 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mielczarek M., et al. , Analysis of copy number variations in Holstein-Friesian cow genomes based on whole-genome sequence data. J. Dairy Sci. 100, 5515–5525 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Cooper G. M., Nickerson D. A., Eichler E. E., Mutational and selective effects on copy-number variants in the human genome. Nat. Genet. 39(suppl. 7), S22–S29 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Sudmant P. H., et al. ; Great Ape Genome Project Evolution and diversity of copy number variation in the great ape lineage. Genome Res. 23, 1373–1382 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malmstrøm M., et al. , Evolution of the immune system influences speciation rates in teleost fishes. Nat. Genet. 48, 1204–1210 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Nielsen R., et al. , A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 3, e170 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schroeder H. W. Jr, Cavacini L., Structure and function of immunoglobulins. J. Allergy Clin. Immunol. 125(suppl. 2), S41–S52 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Delgado Tascón J., et al. , The granulocyte orphan receptor CEACAM4 is able to trigger phagocytosis of bacteria. J. Leukoc. Biol. 97, 521–531 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manry J., et al. , Evolutionary genetic dissection of human interferons. J. Exp. Med. 208, 2747–2759 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Becknell B., et al. , Ribonucleases 6 and 7 have antimicrobial function in the human and murine urinary tract. Kidney Int. 87, 151–161 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lang D., Lim B. K., Gao Y., Wang X.. Adaptive evolutionary expansion of the Pancreatic ribonuclease 6 (RNase6) in Rodentia. Integr. Zool. 10.1111/1749-4877.12382 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Khan I., et al. , Mammalian keratin associated proteins (KRTAPs) subgenomes: Disentangling hair diversity and adaptation to terrestrial and aquatic environments. BMC Genomics 15, 779 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun X., et al. , Comparative genomics analyses of alpha-keratins reveal insights into evolutionary adaptation of marine mammals. Front. Zool. 14, 41 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Derocher A. E., Polar Bears: A Complete Guide to Their Biology and Behavior (The Johns Hopkins University Press, 2012). [Google Scholar]
- 54.Best R. C., Digestibility of ringed seals by the polar bear. Can. J. Zool. 63, 1033–1036 (1985). [Google Scholar]
- 55.Li Y., et al. , Deficiency in the NADPH oxidase 4 predisposes towards diet-induced obesity. Int. J. Obes. 36, 1503–1513 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Giroud S., et al. , Seasonal changes in eicosanoid metabolism in the brown bear. Sci. Nat., 105, 58 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nelson R. A., et al. , Behavior, biochemistry, and hibernation in black, grizzly, and polar bears. Bears Biol. Manage. 5, 284–290 (1983). [Google Scholar]
- 58.Pagano A. M., et al. , High-energy, high-fat lifestyle challenges an Arctic apex predator, the polar bear. Science 359, 568–572 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Welch A. J., et al. , Polar bears exhibit genome-wide signatures of bioenergetic adaptation to life in the arctic environment. Genome Biol. Evol. 6, 433–450 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hughes G. M., et al. , The birth and death of olfactory receptor gene families in mammalian niche adaptation. Mol. Biol. Evol. 35, 1390–1406 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mombaerts P., Genes and ligands for odorant, vomeronasal and taste receptors. Nat. Rev. Neurosci. 5, 263–278 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Niimura Y., Nei M., Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc. Natl. Acad. Sci. U.S.A. 102, 6039–6044 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Young J. M., Trask B. J., The sense of smell: Genomics of vertebrate odorant receptors. Hum. Mol. Genet. 11, 1153–1160 (2002). [DOI] [PubMed] [Google Scholar]
- 64.Buck L., Axel R., A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell 65, 175–187 (1991). [DOI] [PubMed] [Google Scholar]
- 65.Firestein S., How the olfactory system makes sense of scents. Nature 413, 211–218 (2001). [DOI] [PubMed] [Google Scholar]
- 66.Ache B. W., Young J. M., Olfaction: Diverse species, conserved principles. Neuron 48, 417–430 (2005). [DOI] [PubMed] [Google Scholar]
- 67.Freitag J., Ludwig G., Andreini I., Rössler P., Breer H., Olfactory receptors in aquatic and terrestrial vertebrates. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 183, 635–650 (1998). [DOI] [PubMed] [Google Scholar]
- 68.Gilad Y., Przeworski M., Lancet D., Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates. PLoS Biol. 2, E5 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McGowen M. R., Clark C., Gatesy J., The vestigial olfactory receptor subgenome of odontocete whales: Phylogenetic congruence between gene-tree reconciliation and supermatrix methods. Syst. Biol. 57, 574–590 (2008). [DOI] [PubMed] [Google Scholar]
- 70.Rosenzweig M. L., Species Diversity in Space and Time (Cambridge University Press, 1995). [Google Scholar]
- 71.Choquette A. C., et al. , Association between olfactory receptor genes, eating behavior traits and adiposity: Results from the Quebec Family Study. Physiol. Behav. 105, 772–776 (2012). [DOI] [PubMed] [Google Scholar]
- 72.Flegel C., et al. , Characterization of the olfactory receptors expressed in human Spermatozoa. Front. Mol. Biosci. 2, 73 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Green P. A., et al. , Respiratory and olfactory turbinal size in canid and arctoid carnivorans. J. Anat. 221, 609–621 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wingen L. U., et al. , Molecular genetic basis of pod corn (Tunicate maize). Proc. Natl. Acad. Sci. U.S.A. 109, 7115–7120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Iversen M., et al. , The diet of polar bears (Ursus maritimus) from Svalbard, Norway, inferred from scat analysis. Polar Biol. 36, 561–571 (2013). [Google Scholar]
- 76.McLellan B. N., Implications of a high-energy and low-protein diet on the body composition, fitness, and competitive abilities of black (Ursus americanus) and grizzly (Ursus arctos) bears. Can. J. Zool. 89, 546–558 (2011). [Google Scholar]
- 77.Cook D. E., et al. , Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338, 1206–1209 (2012). [DOI] [PubMed] [Google Scholar]
- 78.Gibbons J. G., et al. , The evolutionary imprint of domestication on genome variation and function of the filamentous fungus Aspergillus oryzae. Curr. Biol. 22, 1403–1409 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fumagalli M., et al. , Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science 349, 1343–1347 (2015). [DOI] [PubMed] [Google Scholar]
- 80.Hallmark B., et al. , Genomic evidence of local adaptation to climate and diet in indigenous Siberians. Mol. Biol. Evol. 36, 315–327 (2019). [DOI] [PubMed] [Google Scholar]
- 81.Tishkoff S. A., et al. , Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 39, 31–40 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nair S., et al. , Adaptive copy number evolution in malaria parasites. PLoS Genet. 4, e1000243 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Radke D. W., Lee C., Adaptive potential of genomic structural variation in human and mammalian evolution. Brief. Funct. Genomics 14, 358–368 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schrider D. R., Hahn M. W., Gene copy-number polymorphism in nature. P. Roy. Soc. B Biol. Sci. 277, 3213–3221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li H., et al. ; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Forche A., et al. , Rapid phenotypic and genotypic diversification after exposure to the oral host niche in Candida albicans. Genetics 209, 725–741 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Greenblum S., Carr R., Borenstein E., Extensive strain-level copy-number variation across human gut microbiome species. Cell 160, 583–594 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ge S., Jung D., ShinyGO: A graphical enrichment tool for animals and plants. bioRxiv:10.1101/315150 (4 May 2018).
- 90.Team R. C., R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna, 2018).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.